Abstract
MicroRNA (miRNA) play a major role in the post-transcriptional regulation of gene expression. Mammalian miRNA biogenesis begins with co-transcriptional cleavage of RNA polymerase II (Pol II) transcripts by the Microprocessor complex. While most miRNA are located within introns of protein coding genes, a substantial minority of miRNA originate from long non coding (lnc) RNA where transcript processing is largely uncharacterized. We show, by detailed characterization of liver-specific lnc-pri-miR-122 and genome-wide analysis in human cell lines, that most lnc-pri-miRNA do not use the canonical cleavage and polyadenylation (CPA) pathway, but instead use Microprocessor cleavage to terminate transcription. This Microprocessor inactivation leads to extensive transcriptional readthrough of lnc-pri-miRNA and transcriptional interference with downstream genes. Consequently we define a novel RNase III-mediated, polyadenylation-independent mechanism of Pol II transcription termination in mammalian cells.
MicroRNA (miRNA) are ~22 nucleotide (nt) RNA that play a major role in the regulation of gene expression in most eukaryotic species. In humans, miRNA are thought to post-transcriptionally repress at least 60% of mRNA by binding to targets in the 3′ untranslated region (UTR)1. miRNA show both tissue- and developmental stage-specific expression patterns. Tight regulation of biogenesis of individual miRNA occurs both at the level of transcription and at downstream processing steps, and is essential for normal development and physiology2.
miRNA transcription is generally mediated by RNA polymerase II (Pol II), which synthesizes primary (pri-) miRNA transcripts that can extend to several kilobases in length and are typically capped and polyadenylated3,4. The mature miRNA is located within a hairpin structure that is recognized by the Microprocessor complex, comprising the dsRNA binding protein DGCR8 and the RNase III endonuclease Drosha5. Microprocessor cleavage releases a ~70nt hairpin precursor (pre-) miRNA, which is exported and processed by Dicer to generate a mature miRNA5. In a few cases, pre-miRNA are generated independent of Microprocessor, either from small debranched introns (mirtrons)6, or from unusually short Pol II transcripts7.
Similar to RNA processing events required for mRNA generation, excision of pre-miRNA is co-transcriptional8,9. Most miRNA derive from introns of protein coding genes, where co-transcriptional Microprocessor cleavage does not inhibit splicing, allowing co-expression of miRNA and mRNA from the same host transcript10,11. In contrast, Drosha processing of a miRNA located in a protein coding gene exon can inhibit production of the spliced host mRNA12. Importantly, 17.5% of miRNA are located in long non coding (lnc)RNA (Supplementary Fig. 1), which we define as lnc-pri-miRNA. Processing of these transcripts has not been characterized.
Transcriptional termination of Pol II transcribed genes is tightly coupled to 3′ end processing13. mRNA 3′ end formation occurs co-transcriptionally by a Pol II-associated cleavage and polyadenylation (CPA) mechanism. This involves recognition of the polyadenylation site (PAS), including a canonical AAUAAA sequence and additional, more degenerate, sequence elements. RNA cleavage occurs 10-30 nucleotides downstream of the AAUAAA sequence, followed by the addition of a polyA (pA) tail to the resulting 3′ end. Multiple protein complexes are required for this process, with endonucleolytic cleavage at the PAS mediated by cleavage and pA-specific factor, CPSF-7314. CPA creates an entry site for the 5′-3′ exonuclease Xrn2 (Rat1 in yeast) which acts as a ‘torpedo’, degrading the nascent transcript and contributing to the displacement of Pol II15,16, while the CPA factor Pcf11 also contributes to transcription termination by associating with the Pol II CTD and dismantling the elongation complex17,18. In budding yeast, the RNase III protein Rnt1 can mediate polyadenylation-independent 3′ end formation on pre-mRNA as well as Pol I transcripts19-22, but a similar mammalian pathway has not been identified.
In this study, we aimed to characterize the processing of lnc-pri-miRNA transcripts. Focusing on the liver-specific lncRNA transcript that produces miR-122, which is important for cholesterol metabolism and hepatitis C virus (HCV) replication23-26, we found that Microprocessor cleavage at the pre-miR-122 hairpin mediates transcription termination. By genome-wide nascent RNA-sequencing, we show that this mechanism is also used by most lnc-pri-miRNA, but not protein coding pri-miRNA. We identified a biological role for Microprocessor-mediated termination in preventing transcriptional interference with downstream genes.
Results
Lnc-pri-miR-122 transcripts are capped but not polyadenylated
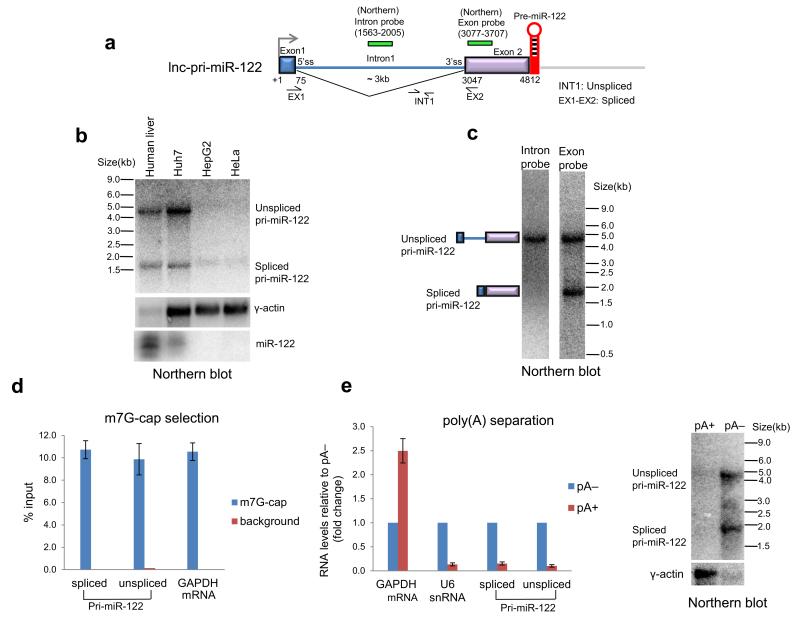
First, we characterized the processing of lnc-pri-miR-122 (Fig. 1a). By northern analysis, we identified mature miR-122 and two lnc-pri-miR-122 transcripts of ~4.8 and ~1.9 kilobase (kb) in total RNA from human liver and the human hepatocellular carcinoma cell line Huh7, but not HeLa or HepG2 cells (Fig. 1b). Intron and exon specific probes showed that the larger lnc-pri-miR-122 transcript was unspliced, while the smaller transcript corresponded to inefficiently spliced RNA that lacks an internal 3 kb intron (Fig. 1c). The transcript size indicated that the 3′ end lay close to the pre-miR-122 hairpin, ~2.5kb upstream of a previously identified polyadenylated 3′ end27 (Fig. 1a). We did not detect any longer lnc-pri-miR-122 transcripts. Immunoprecipitation with an antibody directed against the m7G cap demonstrated that lnc-pri-miR-122 was capped, similar to GAPDH mRNA and in agreement with existing CAGE data28 (Fig. 1d). However, quantitative RT-PCR (RT-qPCR) and northern analysis of pA-selected RNA indicated that lnc-pri-miR-122 was non-polyadenylated, in contrast to GAPDH mRNA, but similar to U6 snRNA (Fig. 1e).
Figure 1. Lnc-pri-miR-122 transcripts are capped but not polyadenylated.
a. Gene map showing exon-intron structure and 3′ positioned pre-miR-122 hairpin. Position of primers and Northern probes are indicated. b. Northern blot showing spliced and unspliced lnc-pri-miR-122 from human liver and three cell lines as indicated. Levels of γ-actin mRNA and mature miR-122 were also measured. Note reduced levels of liver RNA were employed. c. Northern blot detecting unspliced and spliced lnc-pri-miR-122 in human liver RNA using intron and exon-specific probes. d. Levels of m7G-cap immunoselected RNA measured by RT-qPCR as indicated. e. Proportion of pA+ RNA relative to pA− for transcripts indicated measured by RT-qPCR, and for lnc-pri-miR-122 and γ-actin measured by northern blotting. Error bars represent s.d. of an average (n=3 independent experiments).
Microprocessor cleavage generates lnc-pri-miR-122 3′ end
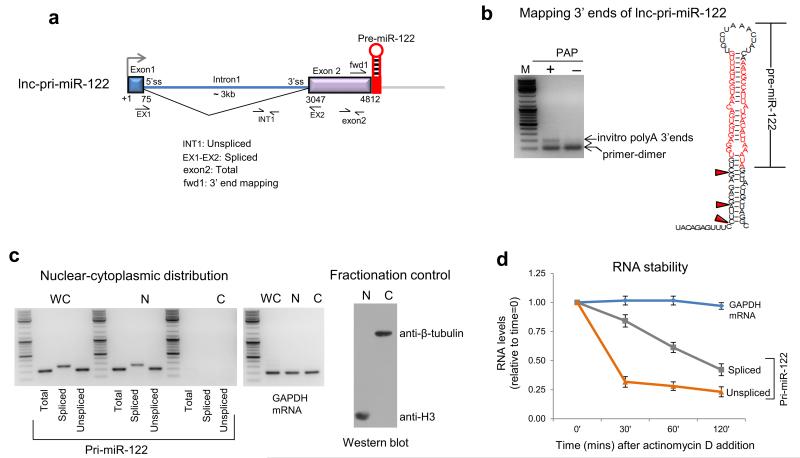
To map lnc-pri-miR-122 3′ ends at nucleotide resolution, we developed a pA tail-independent 3′RACE technique. We mapped lnc-pri-miR-122 3′ ends to just upstream of the site of Drosha cleavage on the 5′ arm of the pre-miR-122 hairpin. We observed some heterogeneity of 3′ end location, presumably due to exonucleolytic trimming following Drosha cleavage (Fig. 2b, Supplementary Fig. 2). Both unspliced and spliced lnc-pri-miR-122 were retained in the nucleus and rapidly degraded (Fig. 2c,d), as expected for non-polyadenylated transcripts.
Figure 2. 3′ end mapping, subcellular distribution and rapid turnover of lnc-pri-miR-122.
a. Gene map of lnc-pri-miR-122 showing position of primers for RT-qPCR and 3′ end mapping. b. PolyA polymerase (PAP)-dependent 3′ end mapping using 3′ RACE. Position of 3′ RACE PCR product shown by gel fractionation and location of mapped 3′ end cleavage products are shown by red arrow heads on the pre-miR-122 hairpin structure (see Supplementary Fig. 2). c. Pri-miR-122 and GAPDH mRNA distribution were determined between cell fractions (WC denotes whole cell, N nuclear and C cytoplasmic) by RT-PCR using indicated primers. Western blot shows purity of nuclear and cytoplasmic fractions by use of cytoplasmic and nuclear specific protein antibodies. d. RNA stability following actinomycin D inhibition of transcription at various time points measured by RT-qPCR of lnc-pri-miR-122 transcripts versus GAPDH mRNA. RNA levels are expressed relative to the levels at time=0, which were set to 1. Huh7 cells were used in all experiments. Error bars represent s.d. of an average (n=3 independent experiments).
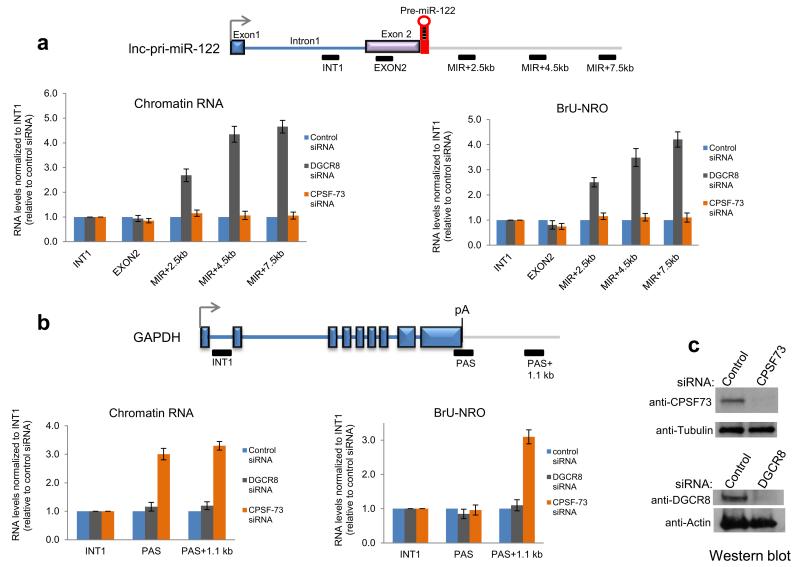
These results suggested that the lnc-pri-miR-122 3′ end is generated by Drosha cleavage and not by CPA. To characterize the role of the Microprocessor in lnc-pri-miR-122 3′ end formation, we compared the effects of siRNA-mediated knockdown in Huh7 cells of DGCR8 versus the CPA endonuclease CPSF-73. Depletion of both proteins was effective (Fig. 3c). RT-qPCR analysis of RNA isolated from nuclear chromatin, which is enriched in nascent transcripts29, was used to compare profiles across lnc-pri-miRNA-122 and the protein coding gene GAPDH. The level of each RT-qPCR product was normalized to the intron 1 product as a measure of basal nascent transcription, and is shown relative to control siRNA-treated cells. These nascent transcript profiles showed a clear increase in level downstream of the pre-miR-122 sequence in DGCR8 but not CPSF-73 depleted cells (Fig. 3a). We found that Drosha depletion elicited readthrough transcription similar to DGCR8 depletion, while Dicer depletion had no effect (Supplementary Figure 3a). This argues against indirect effects of miRNA depletion on termination and strongly suggests a direct role for the Microprocessor in lnc-pri-miR-122 transcription termination. In contrast, GAPDH nascent transcripts showed no effect of DGCR8 depletion, but as expected showed increased level downstream of the PAS indicating a strong termination defect following CPSF-73 depletion (Fig. 3b). Nuclear run on (NRO) analysis confirmed that DGCR8 knockdown caused a termination defect for lnc-pri-miR-122 but not GAPDH, while CPSF-73 knockdown inhibited termination in GAPDH but not lnc-pri-miR-122 (Fig. 3a,b). Pol II chromatin-immunoprecipitation (ChIP) also confirmed transcriptional readthrough on lnc-pri-miR-122 following DGCR8 depletion (Supplementary Fig. 3b). In sum, we establish that the Microprocessor dictates lnc-pri-miR-122 3′ end formation and transcriptional termination, in contrast to GAPDH which relies on the orthodox CPA complex.
Figure 3. Microprocessor defines transcriptional termination of lnc-pri-miR-122.
a, b. Mapping nascent transcription across lnc-pri-miR-122 versus GAPDH in Huh7 cells. Gene maps show positions of amplicons used in analysis of chromatin RNA levels (left) and bromo UTP-labeled nuclear run on RNA (right). c. Western blot showing effective depletion of DGCR8 and CPSF-73 by siRNA transfection in Huh7 cells. Error bars represent s.d. of an average (n=3 independent experiments).
CPA does not occur on lnc-pri-miR-122 transcripts
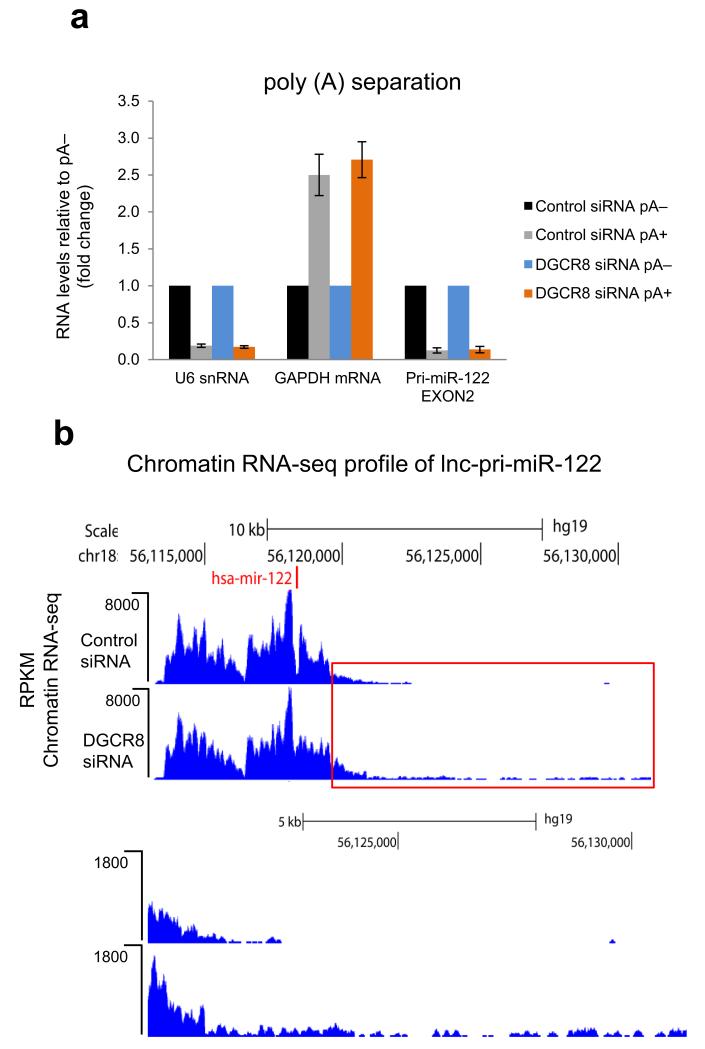
The transcriptional readthrough we observed following DGCR8 knockdown implied that lnc-pri-miR-122 does not switch to the efficient CPA mechanism of transcriptional termination when the Microprocessor mechanism is inhibited. To address this question directly, we pA-selected RNA from Huh7 cells with or without DGCR8 knockdown. Similarly to U6 snRNA, lnc-pri-miR-122 transcripts remained pA− even when their 3′ end formation was compromised by DGCR8 depletion. In contrast, GAPDH mRNA was strongly pA+ under both conditions (Fig. 4a). By next generation sequencing analysis of chromatin-associated RNA from Huh7 cells (chromatin RNA-seq), we found that DGCR8 depletion leads to extensive transcriptional readthrough for over 5kb downstream of the pre-miR-122 hairpin in lnc-pri-miR-122 (Fig. 4b), despite the presence of several consensus PAS.
Figure 4. Microprocessor depletion leads to generation of pA− transcriptional readthrough products on lnc-pri-miR-122.
a. RT-qPCR analysis of lnc-pri-miR-122 versus U6 snRNA or GAPDH mRNA controls using pA+ or pA− fractionated RNA from siRNA-treated Huh7 cells. pA+ RNA levels are expressed relative to pA− which were set to 1. Error bars represent s.d. of an average (n=3 independent experiments). b. Chromatin RNA-seq analysis of lnc-pri-miR-122 in control or DGCR8 siRNA-treated Huh7 cells. Red boxed 3′ region is shown below at higher magnification. Position of miRNA (miR-122) is shown by red vertical line.
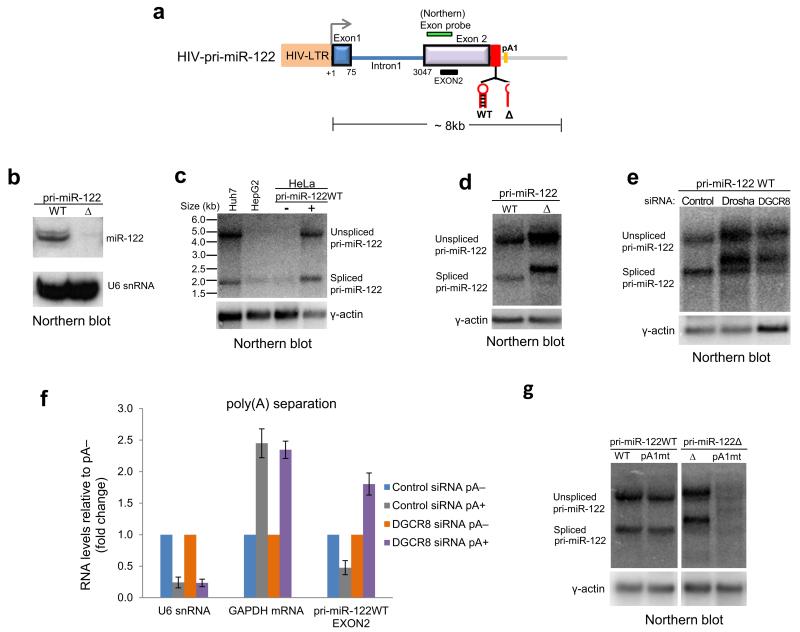
These results indicated that Pol II transcribing lnc-pri-miR-122 fails to recognize PAS even when Microprocessor cleavage is inhibited. This surprising finding suggested that Pol II might be recruited to the endogenous lnc-pri-miR-122 promoter in a CPA refractory form. To investigate this further, we cloned lnc-pri-miR-122 under the control of the human immunodeficiency virus (HIV) long terminal repeat (LTR) promoter with or without pre-miR-122 hairpin deletion (Fig. 5a). The resulting wild type (WT) and deleted (Δ) plasmids were transfected into HeLa cells, which do not express endogenous lnc-pri-miR-122 (Fig. 1b), together with a plasmid encoding the Tat transcriptional activator. We confirmed that mature miR-122 was expressed from the WT but not Δ plasmid (Fig. 5b). Unspliced and spliced lnc-pri-miR-122 transcripts generated from the WT plasmid were the same size as the endogenous transcripts in Huh7 cells (Fig. 5c), indicating that the site of 3′ end formation is independent of promoter or cell type. Deletion of the pre-miR-122 hairpin or depletion of Drosha or DGCR8 led to production of unspliced and spliced lnc-pri-miR-122 transcripts that migrated at a higher molecular weight than wildtype transcripts (Fig. 5d,e). pA fractionation indicated that lnc-pri-miR-122 RNA generated from the WT plasmid was pA−, similar to U6 snRNA, but became pA+ when DGCR8 was depleted, similar to GAPDH mRNA (Fig. 5f). The 3′ end of Δ RNA was mapped by 3′RACE to a PAS 91nt downstream of the pre-miR-122 hairpin (pA1, Fig 5a). Mutagenesis indicated that this PAS was necessary for 3′ end generation in Δ, but not WT, lnc-pri-miR-122 transcripts (Fig. 5g). This confirmed that in a plasmid context, lnc-pri-miR-122 3′ ends were generated by the Microprocessor, but in the absence of this processing, CPA occurred at a site downstream. Importantly, this was in contrast to the endogenous transcripts, where transcriptional readthrough occurred when Microprocessor cleavage was inhibited, and the PAS was not used (Fig. 3,4).
Figure 5. Ectopically expressed lnc-pri-miR-122 switches to CPA when Microprocessor activity is inhibited.
a. Schematic of lnc-pri-miR-122 expression construct driven under HIV-LTR promoter, with locations of northern probe, PCR amplicon and PAS (pA1). WT and Δ plasmids were generated with and without the pre-miR-122 hairpin. b. Northern blot showing that mature miR-122 is expressed from the WT, but not Δ, plasmid following transfection into HeLa cells. U6 snRNA is loading control. c. Northern blot showing spliced and unspliced lnc-pri-miR-122 transcripts generated from the WT plasmid transfected into HeLa cells are the same size as endogenous transcripts in Huh7 cells. d. Northern blot showing spliced and unspliced lnc-pri-miR-122 transcripts generated from the Δ plasmid are larger than the WT transcripts upon transfection into HeLa cells. e. Northern blot showing transcripts generated from the WT plasmid increase in size following Drosha or DGCR8 depletion in HeLa cells. f. RT-qPCR analysis of pA+ or pA− fractionated RNA extracted from siRNA-treated HeLa cells transfected with the WT plasmid. pA+ RNA levels of U6 snRNA, GAPDH mRNA and lnc-pri-miR-122 are expressed relative to pA− which were set to 1. Error bars represent s.d. of an average (n=3 independent experiments). g. Northern blot showing that mutation of PAS (pA1) does not affect migration of WT lnc-pri-miR-122, but leads to loss of unspliced and spliced transcripts generated from the Δ plasmid upon transfection into HeLa cells.
Microprocessor terminates transcription of most lnc-pri-miRNA
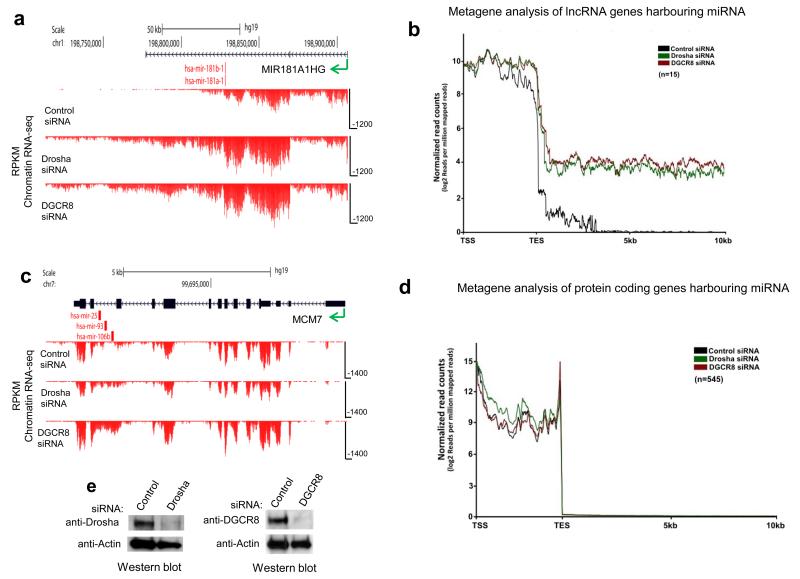
Having demonstrated that chromatin RNA-seq is an effective method of identifying defects in transcriptional termination following Microprocessor depletion (Fig. 4b), we extended this analysis to HeLa cells on a genome-wide scale. HeLa cells were chosen because they express a relatively high number of miRNA. We found that either Drosha or DGCR8 depletion, by siRNA treatment (Fig. 6e), resulted in transcriptional readthrough in most of the expressed lnc-pri-miRNA (Supplementary Table 1). MIR181A1HG and MIR17HG are shown as specific examples (Fig. 6a, Supplementary Fig. 4a), while metagene analysis showed a general termination defect with transcription extending more than 10 kb downstream of lnc-pri-miRNA following Microprocessor depletion (Fig. 6b). A few lnc-pri-miRNA did not shown transcriptional readthrough following Microprocessor depletion (shaded area, Supplementary Table 1); MIRLET7BHG is shown as an example (Supplementary Fig. 4b). Importantly, Dicer knockdown did not affect lnc-pri-miRNA transcriptional termination, confirming that the effects of Microprocessor depletion are direct and not due to loss of mature miRNA (Supplementary Fig. 5).
Figure 6. Microprocessor dependent chromatin RNA-seq profiles across pri-miRNA from HeLa cells.
a. Reads (RPKM) across (MIR181A1HG) showing readthrough profiles following Microprocessor depletion. b. Metagene analysis of all expressed lnc-pri-miRNA. c. Reads across a protein coding gene (MCM7) harboring an intronic miRNA cluster showing intron accumulation following Microprocessor depletion d. Metagene analysis of all expressed protein coding genes harboring miRNAs. Direction of transcription indicated by green arrow and positions of miRNA by red vertical lines in a and c. Metagene profiles show transcription unit (between transcription start site, TSS and transcription end site, TES) followed by 10 kb of 3′ flanking region in b and d (Mann–Whitney U-test, P < 0.0001, two-tailed, n = 1400, for both cases). e. Western blot showing effective depletion of Drosha and DGCR8 by siRNA transfection in HeLa cells.
In marked contrast, protein coding genes that harbor intronic pre-miRNA showed no termination defect following Microprocessor depletion. Rather, intronic sequence containing pre-miRNA was stabilized (higher reads) as shown for MCM7 (Fig. 6c), which has three miRNA (miR-25, 93 and 106b) in its penultimate intron. Presumably this selective intron stabilization was caused by the loss of Drosha-mediated co-transcriptional cleavage8. Metagene analysis revealed that Microprocessor activity generally had no effect on transcriptional termination for these protein coding miRNA genes (Fig. 6d). This implies a functional difference between the processing of miRNA in introns of protein coding transcripts, which use CPA-mediated termination irrespective of internal Microprocessor cleavage, in contrast to lnc-pri-miRNA transcripts, which rely on the Microprocessor for efficient termination. Chromatin RNA-seq analysis in Huh7 cells also identified transcriptional readthrough in lnc-pri-miRNA, but not protein coding pri-miRNA, following DGCR8 depletion. Although the pattern of pri-miRNA expression differs between Huh7 and HeLa, many of the same lnc-pri-miRNA were affected in both cell lines (Supplementary Table 2).
We also investigated the effect of Microprocessor depletion on levels of transcription at gene 5′ ends (TSS). While no change in nascent transcript level was observed for lnc-pri-miRNA genes, protein coding genes hosting pre-miRNA showed transcript reduction following DGCR8 and especially Drosha knockdown (Supplementary Fig. 6a,b). This suggests that the Microprocessor may have a positive influence on gene transcription, as previously noted30. It also suggests that transcriptional initiation differs at lnc-pri-miRNA and protein coding pri-miRNA gene promoters. As a control for the quality of our chromatin RNA-seq data we showed that duplicate libraries display high correlation (Supplementary Fig. 7).
Microprocessor prevents transcriptional interference
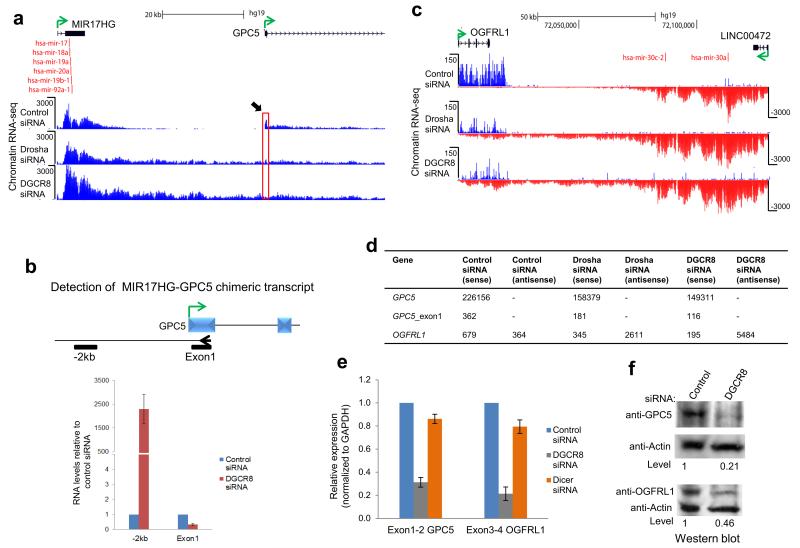
We identified specific examples of lnc-pri-miRNA genes in which the transcriptional readthrough induced by Microprocessor depletion extended into a downstream protein coding gene, either in convergent or tandem orientation. We reasoned that such readthrough transcription might downregulate the invaded gene by a transcriptional interference mechanism31. For the tandem MIR17HG-GPC5 locus, Microprocessor depletion caused the MIR17HG transcript to extend over 20 kb, reading into GPC5 (Fig. 7a, Supplementary Fig. 8). Chimeric transcripts were readily detected, as was a substantial reduction in GPC5 exon 1 RNA levels (Fig. 7b, d, Supplementary Fig. 8). Both GPC5 mRNA and protein levels were more than 70% reduced (Fig. 7e,f), indicating a clear transcriptional interference effect caused by loss of Microprocessor-mediated termination. For the convergent OGFRL1-LINC00472 locus, loss of Microprocessor caused LINC00472 transcripts to read through into OGFRL1, again causing transcriptional down-regulation (Fig. 7c,d). OGFRL1 mRNA levels dropped 80% while protein levels were 50% lower (Fig. 7e,f). Possibly this protein has higher stability than GPC5. These data imply that convergent transcription can also induce gene inactivation, possibly by Pol II collision effects32. Notably Dicer depletion had no effect on GPC5 or OGFRL1 mRNA level (Fig. 7e), indicating that the effects of DGCR8 knockdown are due to transcriptional interference and not miRNA-mediated mRNA destabilization.
Figure 7. Microprocessor dependent termination prevents transcriptional interference.
a. Chromatin RNA-seq profiles across MIR17HG-GPC5 tandem gene locus showing readthrough transcription following Microprocessor depletion. Black arrow and red box highlights reduction in GPC5 exon 1 peak following Microprocessor depletion. b. RT-qPCR analysis of chimeric transcripts versus GPC5 exon 1. Gene specific RT primer (arrowhead) in exon 1 and PCR amplicons are indicated by black bars. c. Chromatin RNA-seq profiles across convergent OGFRL1-LINC00472 gene locus. d. Read quantification for protein coding genes subject to transcriptional interference following Microprocessor depletion. e. mRNA levels of GPC5 and OGFRL1 were determined by RT-qPCR using exon specific primers. RNA levels are expressed relative to control siRNA which were set to 1. All values are normalized to GAPDH mRNA. f. Protein levels of GPC5 and OGFRL1 are reduced by transcriptional interference. Direction of transcription is indicated by green arrow and miRNA by red vertical lines. All experiments used HeLa cells. Error bars represent s.d. of an average (n=3 independent experiments).
Most lnc-pri-miRNA remain pA− after Microprocessor depletion
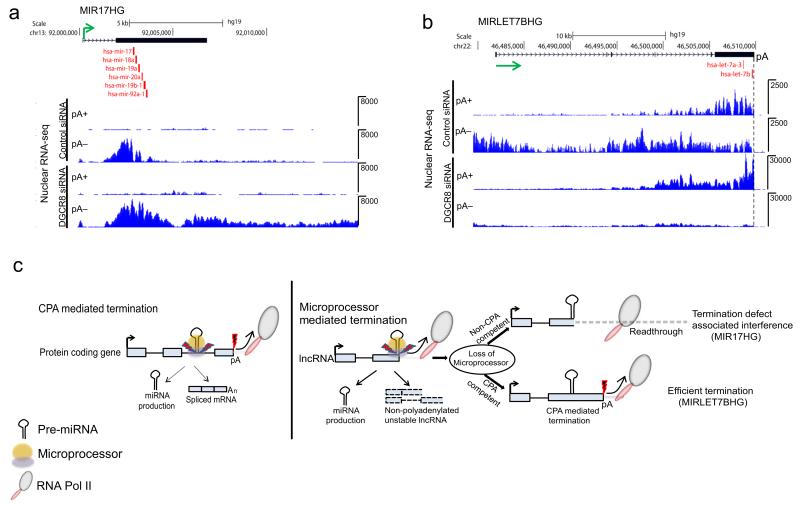
Similar to endogenous lnc-pri-miR-122, Microprocessor-terminated lnc-pri-miRNA appears to be insensitive to the presence of cryptic PAS, invariably present within their gene and 3′ flanking regions. To further investigate the use of PAS in pri-miRNA, we performed nuclear pA+ and pA− RNA-seq in HeLa cells with or without DGCR8 knockdown. We found that the majority of lnc-pri-miRNA existed as predominantly pA− transcripts, and those that showed extensive readthrough upon loss of Microprocessor remained pA− (Supplementary Table 3). MIR17HG is shown as a specific example (Fig. 8a). It is remarkable that for these Pol II transcripts PAS remain opaque to RNA processing by the CPA complex. However, a few lnc-pri-miRNA utilize PAS to some extent, especially following Microprocessor inactivation. Thus MIRLET7BHG transcripts switched from mainly pA− to pA+ following Microprocessor depletion (Fig. 8b, Supplementary Table 3), and efficient termination occurred at a canonical PAS positioned immediately downstream of pre-miR-let7b. This is similar to the switch to CPA at a downstream PAS that we observed in ectopically expressed lnc-pri-miR-122 (Fig. 5), indicating that this distinction is biologically relevant.
Figure 8. Lnc-pri-miRNA may be CPA-incompetent or competent following Microprocessor depletion.
RNA-seq of pA fractionated nuclear RNA from siRNA-treated HeLa cells. a. Nuclear RNA-seq profile of MIR17HG pA+ and pA− RNA following DGCR8 depletion. b. Nuclear RNA-seq profile of MIRLET7BHG pA+ and pA− RNA following DGCR8 depletion. Position of the PAS (pA) is marked by a dashed vertical line. Direction of transcription indicated by green arrow and positions of miRNA by red vertical lines in a and b. c. Model showing CPA in protein coding pri-miRNA allows generation of spliced mRNA and miRNA, while Microprocessor-driven termination in lnc-pri-miRNA generates miRNA and a pA− host transcript that is rapidly degraded. Lnc-pri-miRNA may be CPA-incompetent, leading to transcriptional readthrough and interference when Microprocessor cleavage is inhibited, or CPA-competent allowing effective transcription termination even in the absence of Microprocessor. Red thunderbolt with black fill depicts CPA-mediated cleavage. Blue thunderbolt with red fill depicts Microprocessor-mediated cleavage.
Discussion
We have identified a CPSF-73 independent, Microprocessor-driven transcription termination mechanism for pri-miR-122 lncRNA. This results in the production of unstable nuclear unspliced and spliced lnc-pri-miR-122 transcripts with 3′ ends defined by Drosha cleavage (Fig. 1,2). By genome-wide analysis, we found that transcriptional termination by the Microprocessor is a feature shared with most other lnc-pri-miRNA genes in both HeLa and Huh7 cells (Fig. 6, Supplementary Table 1,2).
Previous evidence indicated that pri-miRNA are typical capped, polyadenylated Pol II transcripts. This is clearly established for protein coding genes containing intronic miRNA8, but is also true of the few lnc-pri-miRNA for which the 3′ end has been characterized, such as pri-miR-214,33 and C. elegans let-734. Our genome-wide analysis confirmed that CPA does occur in a minority of lnc-pri-miRNA (Supplementary Table 3), but showed that the Microprocessor-mediated termination mechanism predominates (Supplementary Table 1,2). A previous study showed that Drosha processing of pre-miRNA hairpins can attenuate downstream transcription by providing an entry site for Xrn2. However, these experiments were carried out using plasmid constructs that lacked a PAS, and did not provide evidence that Microprocessor cleavage could actually replace CPA as a mode of transcriptional termination9. A role for Drosha cleavage at the HIV LTR in preventing productive transcription elongation in the absence of Tat also indicates that the Microprocessor can disrupt the transcription machinery35, but does not connect miRNA processing with transcriptional termination. In contrast, we have demonstrated that Microprocessor cleavage mediates transcriptional termination on endogenous pri-miRNA transcripts, and moreover that this is limited to lnc-pri-miRNA. This departs from the clear current consensus that pri-miRNA are typical capped and polyadenylated transcripts (see recent review5), a view derived from analysis of protein coding pri-miRNA and confirmed for protein coding genes by our genome-wide analysis.
The Microprocessor termination pathway adds to a short list of non-canonical mechanisms of Pol II transcriptional termination. Termination of histone mRNA does not involve polyadenylation, similar to pri-miR-122, but requires cleavage by CPSF-73, in common with CPA36. In yeast, the Nrd1-Nab3-Sen1 pathway terminates Pol II transcription of small nuclear (sn)RNA, small nucleolar (sno)RNA and cryptic unstable transcripts (CUTs)37-39, while in mammals the Integrator complex mediates transcriptional termination and subsequent processing of snRNA40,41. Importantly, both the Nrd1 and Integrator pathways are only used for termination on short transcripts. In contrast, the Microprocessor mechanism described here provides an alternative to CPA in terminating transcription several kilobases downstream of initiation.
The closest parallel to this Microprocessor-dependent termination pathway is in budding yeast, where Rnt1 cleavage leads to pA-independent termination of Pol II transcription when CPA fails, preventing readthrough transcription and subsequent transcriptional interference20,22. Rnt1-terminated transcripts are rapidly degraded, similar to lnc-pri-miR-122. Transcriptional termination following either Rnt1 cleavage or CPA occurs as a result of Rat1 or Xrn2 degradation of the nascent transcript downstream of the cleavage site15,16,20. However, we observed no effect of Xrn2 depletion on lnc-pri-miR-122 transcriptional termination (data not shown), in contrast to the role for Xrn2 in RNA degradation following Drosha cleavage of an intergenic clustered pri-miRNA9. The mechanism of Pol II termination following Microprocessor cleavage of lnc-pri-miR-122 may involve other nucleases or termination factors, such as Pcf1118,42 or the mammalian ortholog of Sen1, Senataxin43. Of note, we observed variable levels of chromatin RNA-seq signal downstream of the pre-miRNA hairpin following DGCR8 knockdown among different Microprocessor-terminated lnc-pri-miRNA. For example, the amplitude of readthrough transcription is higher in MIR181A1HG and MIR17HG than lnc-pri-miR-122 (Fig. 4b,6, Supplementary Fig. 4, Supplementary Table 1,2). The decrease in lnc-pri-miR-122 readthrough transcription may result from low-level Microprocessor-driven termination mediated by residual DGCR8 following siRNA transfection, with gene-specific differences possibly due to some pre-miRNA hairpins competing more effectively for the remaining Microprocessor. Alternatively, it is possible that other, non-CPA, mechanisms can displace transcribing Pol II from these genes.
Although Microprocessor-driven termination is a common feature of lnc-pri-miRNA, it is not universal. This raises the question of whether specific genetic features are necessary for Microprocessor-mediated termination to occur. We find that this mechanism can be used by lncRNA irrespective of whether the pre-miRNA is located in an exon or intron (Supplementary Table 4c), at a range of distances from the TSS (Supplementary Table 4a,b). It is possible that the efficiency of termination is affected by the Microprocessor cleavage event itself. For example, protein cofactors are known to assist in Microprocessor release of specific pre-miRNA5, while sequence features surrounding the pre-miRNA hairpin can influence the efficiency of processing44. Many Microprocessor-terminated lnc-pri-miRNA contain clustered pre-miRNA hairpins, which raises the interesting question of whether Microprocessor cleavage at a specific pre-miRNA drives transcription termination. As some transcriptional readthrough occurs following Microprocessor cleavage, similar to the continued Pol II transcription following cleavage at a PAS in CPA-dependent termination, it is not possible to precisely define which cleavage event drives termination based on our chromatin RNA-seq data. A recent study defined a ‘Microprocessing index’ (MPI) that shows variable cleavage efficiency for different pre-miRNA45. Of the 13 HeLa lnc-pri-miRNA detected in this study, 11 contain a pre-miRNA hairpin with MPI <-1.0 indicating efficient co-transcriptional processing, and 7 of these contain a hairpin with MPI <-3.0, indicating highly efficient processing45. Although the pool of lnc-pri-miRNA that we detect in HeLa is too small to draw statistically robust conclusions, this raises the possibility that rapid Microprocessor cleavage may be important for transcriptional termination.
We found that Microprocessor-driven transcriptional termination is used by most lnc-pri-miRNA (73%), but not protein coding pri-miRNA (Fig. 6, Supplementary Table 1,2), demonstrating a fundamental difference in RNA processing between lncRNA and protein coding transcripts. The Microprocessor mechanism has parallels to another CPA-independent mechanism of transcriptional termination used by the lncRNA MALAT1, where 3′ end formation and concomitant release of a small RNA is mediated by the tRNA biogenesis endonucleases RNase P and RNase Z46. However, lncRNA derived from bi-directional firing at protein coding gene promoters, known as upstream antisense (ua)RNA transcripts, terminate transcription by CPA and tend to use promoter proximal PAS47,48. The difference between pA− lnc-pri-miRNA transcripts as described in this study and CPA-competent lncRNA derived from antisense promoter activity may relate to promoter specificity. Pol II elongation complexes set up on a protein coding gene promoter may be CPA-responsive irrespective of promoter directionality. In contrast, lnc-pri-miRNA promoters may form a different type of Pol II elongation complex that is CPA non-responsive but Microprocessor-active. A role for the promoter is supported by our observation that most lnc-pri-miRNA, including lnc-pri-miR-122, do not use CPA even when the Microprocessor is depleted, instead showing extensive transcriptional readthrough and remaining pA− (Fig. 8a, Supplementary Table 3). In contrast, ectopically expressed lnc-pri-miR-122 uses Microprocessor cleavage to mediate transcriptional termination but switches to CPA when this mechanism is inhibited (Fig. 5). Therefore, the PAS downstream of pre-miR-122 can function, but not in the context of the endogenous gene. The biological relevance of this distinction is clear from our observation of a switch to CPA in MIRLET7BHG transcripts following Microprocessor depletion (Fig. 8b).
LncRNA genes are generally thought to be similar to mRNA genes, with similar chromatin profiles, transcriptional regulation and splice signals. However, lncRNA genes tend to have fewer and longer exons than mRNA genes, and there is a trend for less efficient splicing of lncRNA than of mRNA49,50. We find that CPA is inefficient on lnc-pri-miRNA (Supplementary Table 3). As splicing and CPA function to generate a stable cytoplasmic coding transcript, there would be little need for these processes to occur on most lncRNA. Microprocessor-driven transcriptional termination occurs on lncRNA genes with either intronic or exonic miRNA (Supplementary Table 4c), suggesting that splicing in lncRNA transcripts is not functionally important. Possibly spliced transcripts generated from lncRNA genes such as pri-miR-122 are simply a by-product of transcription; as the splicing machinery is recruited co-transcriptionally, some default recognition and processing of splice sites occurs. LncRNA remain relatively uncharacterized, and it is possible that multiple classes of lncRNA may exist that use different mechanisms of RNA processing.
The use of Microprocessor-driven 3′ end formation by a subset of pri-miRNA genes raises the question of why these genes use this mechanism. A major consequence is the generation of pA− lnc-pri-miR-122 transcripts that are rapidly degraded (Fig. 1,2). This suggests that this mode of termination might have evolved to limit the production of spliced cytoplasmic pri-miRNA transcripts. The concurrent generation of a spliced mRNA that occurs from intronic coding miRNA genes may not be desirable for all miRNA genes. miR-122 expression is very high in liver cells, with an average of 66,000 copies per cell23, and the accumulation of a similar number of copies of the spliced host transcript might be problematic. The miRNA genes we identify as using the Microprocessor-mediated transcriptional termination pathway include others that can be highly expressed and are biologically important, such as the miR-17~92a (MIR17HG) cluster which is highly expressed in embryonic cells and cancer51.
Our genome-wide analysis identified a few examples of Microprocessor depletion leading to transcriptional interference with downstream genes in either tandem or convergent orientation (Fig. 7, Supplementary Fig. 8). For two such genes, we observed a strong reduction in mRNA and protein levels following DGCR8 knockdown, confirming that gene expression is affected. It is unclear how widespread this transcriptional interference is. It is likely that it will be influenced by the relative transcriptional activity of the upstream and downstream genes, the distance between them and their chromatin context, and that tissue-specific examples will exist. As Drosha expression changes in different tissues and during differentiation52, transcriptional interference may also differ in these situations.
In conclusion, we have identified a novel RNase III-mediated transcriptional termination pathway in mammalian cells. This Microprocessor mechanism provides an alternative to CPA, generating unspliced and spliced transcripts of multiple kb, and is specific to lncRNA. We propose a model in which Microprocessor-driven transcriptional termination of lnc-pri-miRNA prevents readthrough and interference with downstream genes. At the same time, a miRNA is produced, while the host transcript is not polyadenylated and so rapidly turned over in the nucleus (Fig. 8c). This may be valuable for highly expressed miRNA such as miR-122, allowing the cell to achieve high levels of the miRNA without concomitant generation of high levels of an unwanted host transcript.
Online Methods
PCR primers and siRNA sequences
Antibodies
anti-Glypican5 (ab124886; Abcam), anti-OGFRL1 (SC-137654; Santa Cruz), anti-Actin (A2103; Sigma), anti-Drosha (ab12286; Abcam), anti-DGCR8 (10996-1-AP; Proteintech), anti-CPSF73 (A301-090A; Bethyl), anti-α-Tubulin (T5168; Sigma), anti-H3 (ab1791; Abcam), anti-β-tubulin (ab6046; Abcam), anti-Dicer (13D6; Abcam). Validation of primary antibodies is provided on manufacturers’ websites.
Plasmid Constructs
Construction of βwt (formerly labeled HIVβ) has been described previously53. The pri-miR-122WT construct was made by insertion of a genomic PCR fragment generated using the primers Pri122qF/Pri122qR on Huh7 genomic DNA. The resulting ~8kb PCR fragment was ligated into a cloning vector prepared by long range PCR amplification of βwt with primers Open_BetaqR/B10 qF using Prime STAR HS DNA polymerase (Takara). The Quick change II XL site directed mutagenesis kit (Stratagene) was used to generate the various mutants using the following primer sets: pri-miR-122Δ (DELTA mirqF/DELTA mirqR); pA1mt (pA1MTqF/pA1MTqR).
Cell culture and transfection of siRNA and plasmids
HeLa, Huh7 and HepG2 cells were maintained in DMEM supplemented with 10% fetal bovine serum. For Huh7 and HepG2 culture, 1% non-essential amino acids (Invitrogen) were also included in the culture media. RNAi was performed using lipofectamine RNAiMax (Invitrogen), with siRNA delivered at 30nM final concentration. A second siRNA treatment was performed at 48h, and cells harvested at 72h after the first hit. Lipofectamine 2000 (Invitrogen) was used to deliver 0.1μg pri-miR-122 plasmid and 0.025μg pTAT per well of a 6 well plate.
RNA isolation and northern blot
RNA was isolated using TRIzol reagent (Ambion) according to the manufacturer’s instructions. Total RNA from human liver was purchased from Agilent Technologies (Cat. No. 540017). Northern blotting was carried out using standard procedures on equal molar quantities of RNA. Membranes were probed with a random-primed 32P-labelled DNA fragment corresponding to nt 3077-3707 (exon probe) and nt 1563-2005 (intron probe) of pri-miR-122 in Ultrahyb (Ambion). A fragment corresponding to nt 685-1171 of γ-actin was used as a loading control. For miRNA northern blots, the small RNA fraction was isolated by dissolving total RNA in 300μL of TE buffer with addition of equal amounts of PEG solution (20% PEG 8000, 2M NaCl). Samples were mixed and incubated for at least 30 min on ice, followed by centrifugation at 14,000g for 15min and isopropanol precipitation of the supernatant. Small RNA were run on a 18% polyacrylamide (19:1) urea gel and analyzed as described before54 using a 32P-end-labeled oligonucleotide complementary to miR-122.
Reverse transcription and real-time qPCR analysis
Total RNA was treated with DNase I (Roche) and reverse-transcribed using SuperScript Reverse Transcriptase III (Invitrogen) and random primers (Invitrogen). Real-time quantitative PCR (qPCR) was performed with 2x Sensimix SYBR mastermix (Bioline) and analyzed on a Corbett Research Rotor-Gene GG-3000 machine.
Nuclear-cytoplasmic fractionation
The procedure for isolating nuclear and cytoplasmic RNA has been described elsewhere55.
m7G cap selection
Capped nuclear RNA was immunoprecipitated using a mouse monoclonal antibody against the 5′-terminal m7G cap (Cat.No.201001; SYSY Synaptic Systems) according to the manufacturer’s protocol. RNA was analyzed by qPCR as described above and compared to 10% input.
PolyA+ and polyA− RNA separation
DNase I-treated nuclear RNA was incubated with oligodT magnetic beads (Dynabeads mRNA purification kit, Invitrogen) to isolate either polyA+ RNA, which was bound to beads, or polyA− RNA, which was present in the flowthrough after incubation. OligodT magnetic bead selection was performed twice to ensure pure polyA+ or polyA− populations. The polyA− RNA population was further processed with the Ribo-Zero Magnetic Kit (Human/Mouse/Rat, Epicentre) to deplete most of the abundant ribosomal RNA.
In vitro polyA tailing and 3′ RACE
To detect the 3′ end of pri-miR-122, in vitro polyA tailing of nuclear RNA from Huh7 was carried out first using the polyA tailing kit (AM1350; Ambion) according to the manufacturer’s protocol. Purified RNA was reverse-transcribed using an oligodT24V anchor using SuperScript III (Invitrogen). Compatible linker and sense primer (fwd1) were used for PCR. PCR fragments were gel purified and cloned into TA cloning vector (StrataClone PCR cloning kit) for sequence verification.
RNA stability
Transcript half-life was estimated after actinomycin-D (5μg/ml) treatment of Huh7 cells. RNA was extracted from cells at different time points after addition of actinomycin-D using TRIzol. Transcript levels at the indicated time points were analyzed by RT-qPCR. The relative levels of expression of each transcript at different time points were plotted relative to the levels at time=0, which were set to 1.
Western blot
Cells extracts were prepared in 15mM HEPES (pH 7.5), 0.25M NaCl, 0.5% NP-40, 10% glycerol, 1x protease inhibitor (Roche) and 1mM phenyl methyl sulfonyl fluoride. Proteins were separated by 4-12% tris-glycine SDS–PAGE and transferred to nitrocellulose (0.45 μM, Amersham Biosciences), and protein detection was carried out with standard western blotting techniques. Secondary antibodies were anti-mouse (Sigma) and anti-rabbit (Sigma). Signals were detected with an ECL kit (GE Healthcare) and quantified using ImageJ software.
Br-UTP nuclear run-on analysis
The Br-UTP NRO was carried out largely as described30 followed by RT-qPCR analysis as described above. The primers used are listed in Supplementary Table 6.
Chromatin RNA isolation
The procedures for separating nuclear RNA into chromatin-associated and released fractions have been described before56. Chromatin-associated RNA from Huh7 cells was analyzed using RT-qPCR as detailed above. The primers used are listed in Supplementary Table 6.
ChIP analysis
ChIP analysis was carried out as previously described before15. 5μg antibody was used per ChIP. The following antibodies were used for ChIP: rabbit total RNAPII (N-20; sc-899; Santa Cruz Biotechnology). Immunoprecipitated DNAs were used as templates for qPCR.
RNA-sequencing
Chromatin-associated RNA was isolated as described above with the omission of tRNA in solution preparation. Nuclear polyA− or polyA+ RNA were prepared as described above. RNA-seq was performed by the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics, University of Oxford. RNA Samples were ribodepleted using Ribo-Zero rRNA removal kit (Human/Mouse/Rat, EpiCentre RZH110424). Libraries were prepared using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina, v1.0 (cat # E7420) using manufacturer’s guidelines with an exception of using our own 8bp tags for indexing according to57. Libraries were sequenced on an Illumina HiSeq-2000 using 100bp and 50bp paired end reads, v3 chemistry.
Bioinformatic analysis
Datasets
All human miRNAs and their genomic coordinates were obtained from miRBase release 2058. Annotation for protein coding genes (GRCh37) was obtained from Ensembl, release 7459. Annotation for long noncoding RNA (lncRNA) was obtained from GENCODE v7 catalog of human long noncoding RNA49.
We separated the miRNA into two groups: lnc-pri-miRNA (if the miRNA overlapped with an lncRNA) and protein coding pri-miRNA (if the miRNA overlapped with an Ensembl protein coding gene), based on the genomic coordinates and taking the strand orientation into account. miRNA harboring genes having length ≥200 bp were considered for this study. This resulted in 112 lnc-pri-miRNA and 967 protein coding pri-miRNA. An additional 18 lnc-pri-miRNA were added to this list which mapped to genes having ‘lincRNA’ as Ensembl gene biotype and gene length ≥200 bp.
Genomic distribution of miRNA as belonging to protein coding genes and long non-coding genes was based on Ensembl gene biotype annotation (Supplementary Fig.1).
Mapping of sequencing reads
Paired-end reads for each sample were mapped to the human genome reference assembly GRh37/hg19 (build 37.2, Feb 2009) using the Bowtie2 alignment software60. Prior to alignment, the first 12 nucleotides were trimmed from all the reads owing to the low quality of the bases. Uniquely mapped reads with no more than two mismatches were retained for further analysis. For nuclear polyA+ data, we filtered out reads that had 8 or more genomically encoded A–stretch at their 3′ ends. A statistical summary of read alignments can be found in Supplementary Table 5 for HeLa and Huh7 chromatin RNA-seq and HeLa nuclear polyA+ and polyA− RNA-seq.
Calculation of metagene profiles
We used the Ensembl gene annotation to define transcription start and end sites. In-house Perl and Python scripts were used to compute metagene profiles. To get a list of miRNA expressed in HeLa cells, miRNA FPKM was calculated using Cufflinks on small RNA-seq data for HeLa cells downloaded from the ENCODE Experiment Matrix available as ENCODE Project at UCSC61. This resulted in 15 lncRNA expressing 34 miRNA in HeLa cells. For the protein coding dataset, miRNA harboring genes with length ≥2 kb and the gene body RPKM ≥ 1 were considered. 545 genes were identified in this category.
Metagene profile (Figure 6) for Control siRNA, Drosha siRNA and DGCR8 siRNA in HeLa cells was calculated for a region from the start of the miRNA host gene to 1 kb upstream of the start of the next downstream gene. For this, read counts were normalized to total sequencing depth. The region extending from TSS to TES was scaled to 4 kb and the region from downstream of TES to 1 kb upstream of the start of the next gene was scaled to 10 kb. Normalized read counts were plotted for each 10 bp bin.
To investigate the profile surrounding TSS upon Microprocessor depletion, we plotted normalized read count across a region of 1kb upstream and downstream of annotated TSS for Control siRNA, Drosha siRNA and DGCR8 siRNA in HeLa cells.
For supplementary Table 1 and 2, normalized read count (RPKM) was calculated over a region from the TES (3′ end) of the lnc-pri-miRNA to 1 kb upstream of the TSS of the next downstream gene for miRNA harboring lncRNA genes that are expressed (RPKM≥1) in HeLa and Huh7 cell lines.
Classification of polyA+, polyA− and bimorphic transcripts
Classification of transcripts into polyA+, polyA− and bimorphic was done as described62. Briefly, all expressed transcripts were classified as polyA+, polyA− and bimorphic predominant transcripts based on their relative abundance, calculated using BPKM (bases per kilobase of gene model per million mapped bases) in the polyA+ and polyA− sample for each condition (Control siRNA and DGCR8 siRNA). See Supplementary Table 3.
Modified images
Original images of gels, autoradiographs and blots used in this study can be found in Supplementary Data Set 1.
Supplementary Material
Acknowledgements
We thank members of the NJP lab for advice and encouragement. This work was supported by a Programme grant from the Wellcome Trust and a European Research Council Advanced Award to NJP and a Biotechnology and Biological Sciences Research Council David Phillips Fellowship (BB/F02360X/1) to CLJ. High throughput sequencing was performed by the Genomics group at The Oxford Wellcome Centre for Human Genetics.
Footnotes
Accession codes
RNA-seq data have been deposited in the Gene Expression Omnibus database under accession number GSE58838.
References
- 1.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 3.Lee Y, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–66. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–24. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 6.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–6. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie M, et al. Mammalian 5′-Capped MicroRNA Precursors that Generate a Single MicroRNA. Cell. 2013;155:1568–80. doi: 10.1016/j.cell.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morlando M, et al. Primary microRNA transcripts are processed cotranscriptionally. Nat. Struct. Mol. Biol. 2008;15:902–9. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ballarino M, et al. Coupled RNA processing and transcription of intergenic primary microRNAs. Mol. Cell. Biol. 2009;29:5632–8. doi: 10.1128/MCB.00664-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–83. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–7. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sundaram GM, et al. ‘See-saw’ expression of microRNA-198 and FSTL1 from a single transcript in wound healing. Nature. 2013;495:103–6. doi: 10.1038/nature11890. [DOI] [PubMed] [Google Scholar]
- 13.Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–82. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mandel CR, et al. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature. 2006;444:953–6. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.West S, Gromak N, Proudfoot NJ. Human 5′ --> 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–5. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- 16.Kim M, et al. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–22. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z, Fu J, Gilmour DS. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′-end processing factor, Pcf11. Genes Dev. 2005;19:1572–80. doi: 10.1101/gad.1296305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z, Gilmour DS. Pcf11 is a termination factor in Drosophila that dismantles the elongation complex by bridging the CTD of RNA polymerase II to the nascent transcript. Mol. Cell. 2006;21:65–74. doi: 10.1016/j.molcel.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Kawauchi J, Mischo H, Braglia P, Rondon A, Proudfoot NJ. Budding yeast RNA polymerases I and II employ parallel mechanisms of transcriptional termination. Genes Dev. 2008;22:1082–92. doi: 10.1101/gad.463408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rondon AG, Mischo HE, Kawauchi J, Proudfoot NJ. Fail-safe transcriptional termination for protein-coding genes in S. cerevisiae. Mol. Cell. 2009;36:88–98. doi: 10.1016/j.molcel.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El Hage A, Koper M, Kufel J, Tollervey D. Efficient termination of transcription by RNA polymerase I requires the 5′ exonuclease Rat1 in yeast. Genes Dev. 2008;22:1069–81. doi: 10.1101/gad.463708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghazal G, et al. Yeast RNase III triggers polyadenylation-independent transcription termination. Mol. Cell. 2009;36:99–109. doi: 10.1016/j.molcel.2009.07.029. [DOI] [PubMed] [Google Scholar]
- 23.Chang J, Nicolas E, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biology. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 24.Esau C, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 25.Elmen J, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–9. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 26.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–81. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 27.Li ZY, et al. Positive regulation of hepatic miR-122 expression by HNF4alpha. J. Hepatol. 2011;55:602–11. doi: 10.1016/j.jhep.2010.12.023. [DOI] [PubMed] [Google Scholar]
- 28.Chien CH, et al. Identifying transcriptional start sites of human microRNAs based on high-throughput sequencing data. Nucleic Acids Res. 2011;39:9345–56. doi: 10.1093/nar/gkr604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang IX, et al. RNA-DNA Differences Are Generated in Human Cells within Seconds after RNA Exits Polymerase II. Cell Rep. 2014;6:906–15. doi: 10.1016/j.celrep.2014.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gromak N, et al. Drosha regulates gene expression independently of RNA cleavage function. Cell Rep. 2013;5:1–12. doi: 10.1016/j.celrep.2013.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greger IH, Aranda A, Proudfoot N. Balancing transcriptional interference and initiation on the GAL7 promoter of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2000;97:8415–20. doi: 10.1073/pnas.140217697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prescott EM, Proudfoot NJ. Transcriptional collision between convergent genes in budding yeast. Proc. Natl. Acad. Sci. USA. 2002;99:8796–801. doi: 10.1073/pnas.132270899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ribas J, et al. A novel source for miR-21 expression through the alternative polyadenylation of VMP1 gene transcripts. Nucleic Acids Res. 2012;40:6821–33. doi: 10.1093/nar/gks308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bracht J, Hunter S, Eachus R, Weeks P, Pasquinelli AE. Trans-splicing and polyadenylation of let-7 microRNA primary transcripts. RNA. 2004;10:1586–94. doi: 10.1261/rna.7122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagschal A, et al. Microprocessor, Setx, Xrn2, and Rrp6 co-operate to induce premature termination of transcription by RNAPII. Cell. 2012;150:1147–57. doi: 10.1016/j.cell.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolev NG, Yario TA, Benson E, Steitz JA. Conserved motifs in both CPSF73 and CPSF100 are required to assemble the active endonuclease for histone mRNA 3′-end maturation. EMBO Rep. 2008;9:1013–8. doi: 10.1038/embor.2008.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steinmetz EJ, Conrad NK, Brow DA, Corden JL. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature. 2001;413:327–31. doi: 10.1038/35095090. [DOI] [PubMed] [Google Scholar]
- 38.Arigo JT, Eyler DE, Carroll KL, Corden JL. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol. Cell. 2006;23:841–51. doi: 10.1016/j.molcel.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 39.Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat. Struct. Mol. Biol. 2008;15:795–804. doi: 10.1038/nsmb.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baillat D, et al. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–76. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 41.O’Reilly D, et al. Human snRNA genes use polyadenylation factors to promote efficient transcription termination. Nucleic Acids Res. 2014;42:264–75. doi: 10.1093/nar/gkt892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim M, et al. Distinct pathways for snoRNA and mRNA termination. Mol. Cell. 2006;24:723–34. doi: 10.1016/j.molcel.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 43.Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell. 2011;42:794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Auyeung VC, Ulitsky I, McGeary SE, Bartel DP. Beyond Secondary Structure: Primary-Sequence Determinants License pri-miRNA Hairpins for Processing. Cell. 2013;152:844–58. doi: 10.1016/j.cell.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Conrad T, Marsico A, Gehre M, Orom UA. Microprocessor Activity Controls Differential miRNA Biogenesis In Vivo. Cell Rep. 2014;9:542–54. doi: 10.1016/j.celrep.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 46.Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–32. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Almada AE, Wu X, Kriz AJ, Burge CB, Sharp PA. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013;499:360–3. doi: 10.1038/nature12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ntini E, et al. Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nat. Struct. Mol. Biol. 2013;20:923–8. doi: 10.1038/nsmb.2640. [DOI] [PubMed] [Google Scholar]
- 49.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tilgner H, et al. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res. 2012;22:1616–25. doi: 10.1101/gr.134445.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20:1603–14. doi: 10.1038/cdd.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sperber H, et al. miRNA sensitivity to Drosha levels correlates with pre-miRNA secondary structure. RNA. 2014;20:621–31. doi: 10.1261/rna.043943.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
References for online methods
- 53.Dye MJ, Proudfoot NJ. Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol. Cell. 1999;3:371–78. doi: 10.1016/s1097-2765(00)80464-5. [DOI] [PubMed] [Google Scholar]
- 54.Pall GS, Hamilton AJ. Improved northern blot method for enhanced detection of small RNA. Nat. Protoc. 2008;3:1077–84. doi: 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
- 55.Dye MJ, Gromak N, Proudfoot NJ. Exon tethering in transcription by RNA polymerase II. Mol. Cell. 2006;21:849–59. doi: 10.1016/j.molcel.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 56.West S, Proudfoot NJ, Dye MJ. Molecular dissection of mammalian RNA polymerase II transcriptional termination. Mol. Cell. 2008;29:600–10. doi: 10.1016/j.molcel.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lamble S, et al. Improved workflows for high throughput library preparation using the transposome-based nextera system. BMC Biotechnol. 2013;13:104. doi: 10.1186/1472-6750-13-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2014;39:D152–7. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Flicek P, et al. Ensembl 2014. Nucleic Acids Res. 2014;42:D749–55. doi: 10.1093/nar/gkt1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang L, Duff MO, Graveley BR, Carmichael GG, Chen LL. Genomewide characterization of non-polyadenylated RNAs. Genome Biol. 2011;12:R16. doi: 10.1186/gb-2011-12-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.