Abstract
Chromatin Assembly Factor 1 (CAF-1) is a histone chaperone that assembles acetylated histones H3/H4 onto newly synthesized DNA, allowing the de novo assembly of nucleosomes during replication. CAF-1 is an evolutionary conserved heterotrimeric protein complex. In Arabidopsis, the three CAF-1 subunits are encoded by FAS1, FAS2 and MSI1. Atfas1-4 mutants have reduced fertility due to a decrease in the number of cells that enter meiosis. Interestingly, the number of DNA double-strand breaks (DSBs), measured by scoring the presence of γH2AX, AtRAD51 and AtDMC1 foci, is higher than in wild-type (WT) plants, and meiotic recombination genes such AtCOM1/SAE2, AtBRCA1, AtRAD51 and AtDMC1 are overexpressed. An increase in DSBs in this mutant does not have a significant effect in the mean chiasma frequency at metaphase I, nor a different number of AtMLH1 nor AtMUS81 foci per cell compared to WT at pachytene. Nevertheless, this mutant does show a higher gene conversion (GC) frequency. To examine how an increase in DSBs influences meiotic recombination and synaptonemal complex (SC) formation, we analyzed double mutants defective for AtFAS1 and different homologous recombination (HR) proteins. Most showed significant increases in both the mean number of synapsis initiation points (SIPs) and the total length of AtZYP1 stretches in comparison with the corresponding single mutants. These experiments also provide new insight into the relationships between the recombinases in Arabidopsis, suggesting a prominent role for AtDMC1 versus AtRAD51 in establishing interhomolog interactions. In Arabidopsis an increase in the number of DSBs does not translate to an increase in the number of crossovers (COs) but instead in a higher GC frequency. We discuss different mechanisms to explain these results including the possible existence of CO homeostasis in plants.
Author Summary
Meiosis is a special cell division common in all sexually reproducing organisms. It consists of two successive rounds of chromosome segregation, preceded by a single DNA replication event. Homologous recombination is a key process that occurs during the first meiotic division. It guarantees the association of the homologous chromosomes by chiasmata, the cytological manifestations of reciprocal interchanges (crossovers, COs). The formation of COs during meiosis is fine-tuned by several mechanisms. One of them, reported in some model organisms, is CO homeostasis, which ensures a consistent number of COs despite variability in early recombination events. Here we described the analysis of Atfas1-4, a mutant defective for the histone chaperone CAF-1 (Chromatin Assembly Factor 1), which has an increase in double-strand breaks (DSBs), without a corresponding increase in COs. Interestingly, Atfas1-4 has a higher gene conversion (GC) frequency. These results demonstrate that Arabidopsis meiocytes are able to maintain WT levels of COs even when DSBs numbers are increased. Furthermore, we provide evidence for a prominent role for AtDMC1 in establishing interhomolog interactions in Arabidopsis.
Introduction
Histone chaperones are a family of proteins that facilitate appropriate interactions between histones and DNA by regulating the assembly and disassembly of chromatin in response to cellular requirements [1–3]. CAF-1 is a heterotrimeric histone chaperone complex that mediates nucleosome assembly on newly replicated DNA in fungi, animals and plants [4].
The CAF-1 complex is composed of: FASCIATA 1 (AtFAS1), AtFAS2, and the MULTICOPY SUPPRESOR OF IRA1 (AtMSI1) [5]. The large subunit AtFAS1 binds acetylated histones H3/H4 and interacts with Proliferating Cell Antigen (PCNA) [6, 7]. The AtFAS2 subunit enables protein-protein interactions within CAF-1 and with Anti-Silencing Function 1 (ASF1), another major evolutionarily conserved H3/H4 histone chaperone, which also participates in replication-independent nucleosome assembly [8–11]. MSI1, the small subunit of CAF-1, forms part of other complexes involved in chromatin dynamics [12]. Mutants deficient in AtFAS1 and AtFAS2 show a fasciated phenotype characterized by shoot apical meristem defects including altered phyllotaxy, stem broadening and bifurcation, and alterations in floral organ numbers [13]. In addition, CAF-1 components also control genome replication at multiple developmental steps [14, 15], and maintain the correct pattern of heterochromatin silencing [16]. For instance, loss of AtFAS1 leads to a switch to the endocycle program [17]. Genomic DNA from these mutants is hypersensitive to DNA digestion, suggesting a less compact chromatin conformation [16, 18]. Moreover, Atfas1 and Atfas2 are hypersentive to genotoxic agents and DSB-inducing treatments [4, 18, 19]. Depletion of either subunit increased the frequency of somatic homologous recombination (HR) ~40-fold, as well as increased T-DNA integration [19]. These findings suggest that the loss of CAF-1 activity produces defects in chromatin assembly that lead to genomic instability. Atfas1-4 appears to be the strongest allele since it exhibits a severe developmental phenotype and ~96-fold higher intrachromosomal HR [5, 18]. Since several HR genes display normal expression in the Atfas1-4 mutant, it has been suggested that chromatin conformation is a key factor limiting HR in plants [18]. By contrast, single mutants in the ASF1 subunits AtASF1a or AtASF1b exhibit no apparent somatic abnormalities. However, double mutants show reduced growth and defects in organ development, and overexpression of genes involved in S-phase checkpoints and DNA repair by HR [11].
Additionally, CAF-1 deficiency leads to cell cycle arrest during post-meiotic pollen development resulting in formation of only one sperm cell [20]. Here, we use the Atfas1-4 mutant to investigate the influence of an increase in DSB frequency on meiotic phenotypes in Arabidopsis. The characterization of double mutants defective for both AtFAS1 and either AtRAD51 or AtDMC1 allowed us to infer new insights about the interplay between the action of recombinases and the regulation of synapsis. Furthermore, the analysis of Atfas1-4, with a normal chiasma frequency, and a significant increase in the frequency of gene conversion (GC) events, suggests the possible existence of a homeostatic control of crossovers (COs) in Arabidopsis.
Results
Fertility phenotype of Atfas1-4 plants
Atfas1-4 plants have significantly fewer ovules per gynecium (31.58 ± 2.76 vs. 50.75 ± 2.04; W = -72.02; P < 10−4), and pollen grains per anther (14.17 ± 1.69 vs. 43.3 ± 1.95; W = 12.5; P = 3.9 x 10−3) compared to wild-type (WT) plants (S1 Fig). Likewise, the mean silique length (cm) and number of seeds are also significantly diminished in Atfas1-4 compared to WT (0.71 ± 0.02 vs 1.30 ± 0.02; t = 18.804; P < 10−4; and 11.58 ± 0.4 vs. 48.8 ± 1.59; t = 22.046; P < 10−4, respectively). Consistent with previous phenotypic analysis of Atfas1 plants, we observed that the flowers had only five anthers instead of the six consistently observed in WT (see Figure S1A in [4]). These results indicate that Atfas1-4 plants have reduced fertility.
Meiosis in pollen mother cells (PMCs)
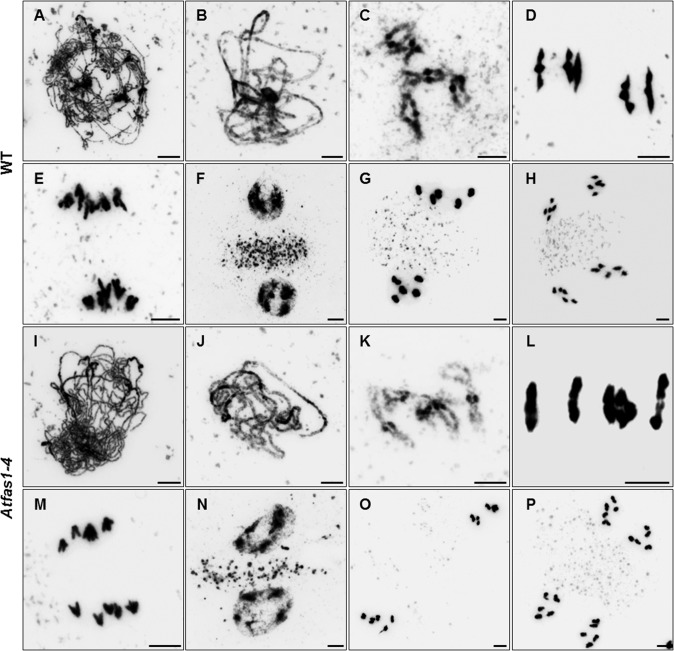
To assess the ability of Atfas1-4 plants to progress through meiosis we compared 4’6-diamidino-2-phenylindole (DAPI) stained chromosome spreads of PMCs from WT and mutant plants. The cytological analysis revealed that in Atfas1-4 chromosome pairing, synapsis and recombination between homologs occurred normally, as did the first and second divisions and tetrad formation (n > 400; Fig 1). Normal meioses were also observed in Atfas2-1, and in a line in which AtASF1a and AtASF1-b were inactivated by RNAi (S2 Fig). Therefore, the fertility decrease exhibited by these mutants is not due to a defect in meiosis.
Fig 1. Atfas1-4 does not show cytological meiotic alterations.
(A-H) Chromosome spread preparations from WT and (I-P) Atfas1-4 PMCs. (A, I) Leptotene. (B, J) Pachytene. (C, K) Diplotene. (D, L) Metaphase I. Five ring bivalents in WT and four ring bivalents in Atfas1-4. (E, M) Anaphase I. (F, N) Prophase II. (G, O) Metaphase II. (H, P) Anaphase II. Bars = 5 µm.
Since Atfas1-4 displays a more “open chromatin” configuration in somatic cells [19], we looked for changes in the chromatin of meiotic cells, particularly during pachytene, a stage when bivalents are individualized and decondensed. We hybridized chromosome spreads with the centromeric probe pAL1 (180 bp) and observed fading of the chromomeric pattern. The centromeric fluorescence in situ hybridization (FISH) signals are weaker, less defined, and more extended in Atfas1-4 than in WT (1.26 ± 0.59 µm vs. 0.854 ± 0.059 µm; t = -2.126; P = 0.040; n = 30; S3 Fig). These observations suggest a gross change in chromosome condensation states during pachytene.
Expression analysis of meiotic genes involved in HR
Kirik and colleagues [18] reported that Atfas1-4 displays one of the strongest intra-chromosomal HR phenotype of all chromatin mutants analyzed in plants to date. Therefore, we tested whether meiotic recombination is also enhanced. As a first approximation, we used real-time polymerase chain reaction (qPCR) to quantify the transcripts of several meiotic recombination genes in flower bud samples. After analyzing 15 measurements for each target gene from three experimental and five biological replicates, we detected a statistically significant overexpression of the meiosis-specific gene AtDMC1 (3.842 ± 0.794), and also of DNA repair genes, including: AtCOM1 (3.460 ± 0.304), AtBRCA1 (3.379 ± 0.025), AtRAD51 (5.043 ± 0.656), AtSMC6A (1.719 ± 0.114), and AtSMC6B (1.664 ± 0.075) (numbers represent fold variation over WT and after normalization; Figs 2A and S4). The expression levels of three other genes: AtMND1 (1.687 ± 0.206 vs. 1 ± 0.340), AtBLAP75 (1.665 ± 0.138 vs.1 ± 0.404) and AtTOP3α (1.736 ± 0.141 vs. 1 ± 0.235) were suggestive of increases, but not statistically significant (see Materials and Methods for more details).
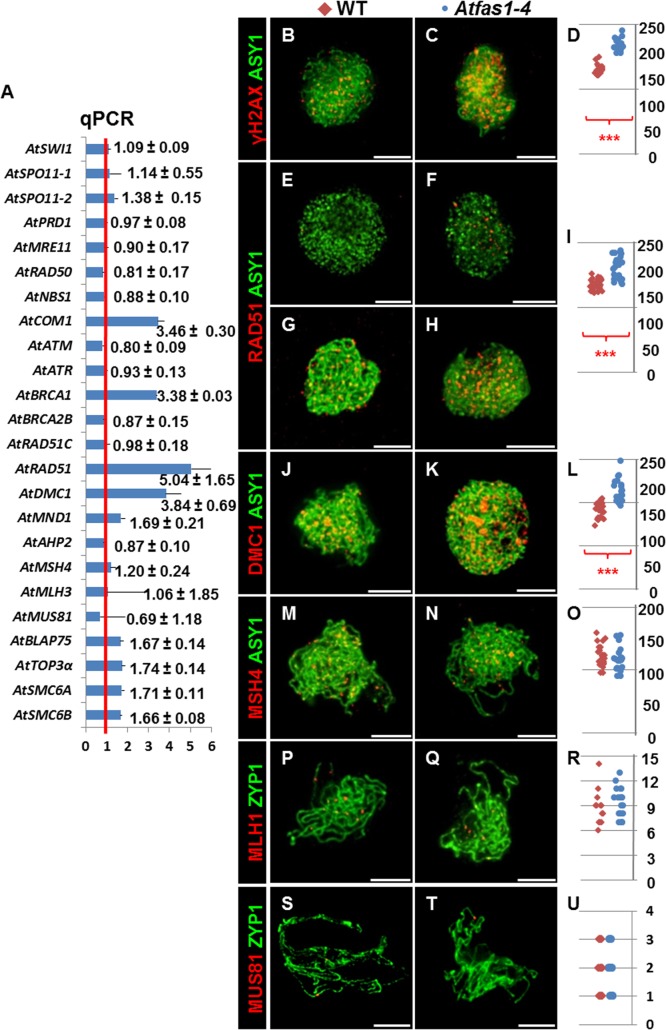
Fig 2. Gene expression and protein quantification for several genes involved in HR in Atfas1-4.
(A) Expression analysis of genes encoding meiotic proteins in WT and Atfas1-4 bud samples. Values are the average of assays carried out in triplicate using five different cDNA preparations. The red line is the reference for the fold change respect to the WT after normalization to 18S rRNA. The means corresponding to changes in gene expression and their standard errors are indicated. (B-Q) Dual immunolocalization of AtASY1 and AtZYP1 with HR proteins in WT and Atfas1-4 prophase I nuclei. (B, C) AtASY1 (green) and γH2AX (red) on WT and Atfas1-4 at leptotene. (E, G) AtASY1 (green) and AtRAD51 (red) on WT and (F, H) on Atfas1-4 at G2 or leptotene, respectively. (J, K) AtASY1 (green) and AtDMC1 (red) on WT and Atfas1-4 at leptotene. (M, N) AtASY1 (green) and AtMSH4 (red) on WT and Atfas1-4 at late zygotene. (P, Q) AtZYP1 (green) and AtMLH1 (red) on WT and Atfas1-4 at pachytene. (S, T) AtZYP1 (green) and AtMUS81 (red) on WT and Atfas1-4 at pachytene. Bars = 5 µm. (D, I, L, O, R, U) Total foci per nucleus in WT (red) and Atfas1-4 (blue) PMCs are indicated. Each dot is the count from a single nucleus. P values are from two-sided Wilcoxon Mann-Whitney tests (***P < 0.001).
Immunodetection of meiotic proteins in PMCs
To estimate the number of DSBs, we counted foci corresponding to phosphorylated histone H2AX (γH2AX), a sensitive marker that can be used to examine the DNA damage produced and the subsequent repair of the DNA lesion, and the recombinases AtRAD51 and AtDMC1 which are involved in DNA strand invasion during the beginning of the recombination process (S4 Fig). We also carried out immunolocalization of AtMSH4, which promotes the formation of Type I COs (CO1) that are sensitive to CO interference at zygotene, and AtMLH1 which marks CO1s at pachytene. To establish the chronology of prophase I we used AtASY1, a structural protein related to the axial/lateral element (AE/LE) of the synaptonemal complex (SC), and AtZYP1, which forms part of the central element (CE) of the SC, as cytological markers. We used Atspo11-1-5 as a negative control, as DSB formation is blocked in this mutant (S5 Fig).
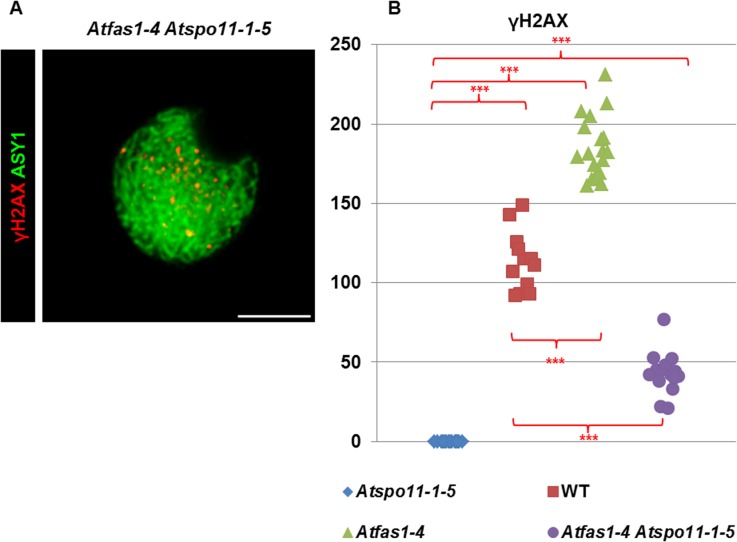
In both Atfas1-4 and WT, AtASY1 adopted a linear configuration, and appeared in intimate association with the AEs of the SC at leptotene (Fig 2B–2N). However, the signal strength of γH2AX foci was more intense in the mutant (Fig 2B and 2C). As expected, no γH2AX signal was observed in Atspo11-1-5 (n = 15; S5 Fig). Quantification of γH2AX foci suggests that there are significantly more DSBs in Atfas1-4 compared to WT: 57.70% (W = 112.0; P = 1.42 x 10−5; Fig 2D and S1 Table).
AtRAD51 and AtDMC1 were detected in both Atfas1-4 and WT at early prophase (Fig 2E–2K). Again as expected, AtRAD51 and AtDMC1 foci were absent in Atspo11-1-5 (S5 Fig). Furthermore, in Atfas1-4 abundant AtRAD51 signals appeared at G2, before leptotene (Fig 2F) whereas they were scarce in WT (Fig 2E). We also quantified the number of foci corresponding to both recombinases. In Atfas1-4 the number of foci of AtRAD51 and AtDMC1 was significantly higher than in WT, 41.13% (W = 442.5; P = 3.98 x 10−9; Fig 2I), and 33.56% (W = 180.5; P = 2.24 x 10−6; Fig 2L), respectively (S1 Table).
We also carried out simultaneous detection of AtMSH4 and AtASY1, which showed diffuse and discontinuous signals in chromosome regions which had started to synapse (Fig 2M and 2N). AtMLH1 and AtMUS81 localizations were performed with AtZYP1, which appears as a linear signal at pachytene (Fig 2P, 2Q, 2S and 2T). No significant differences for AtMSH4 (W = -34.5; P = 0.31; Fig 2O), AtMLH1 (W = 13.5; P = 0.66; Fig 2R), or AtMUS81 were observed, (W = -9.0; P = 0.71; Fig 2U) (S1 Table).
Chiasma analysis
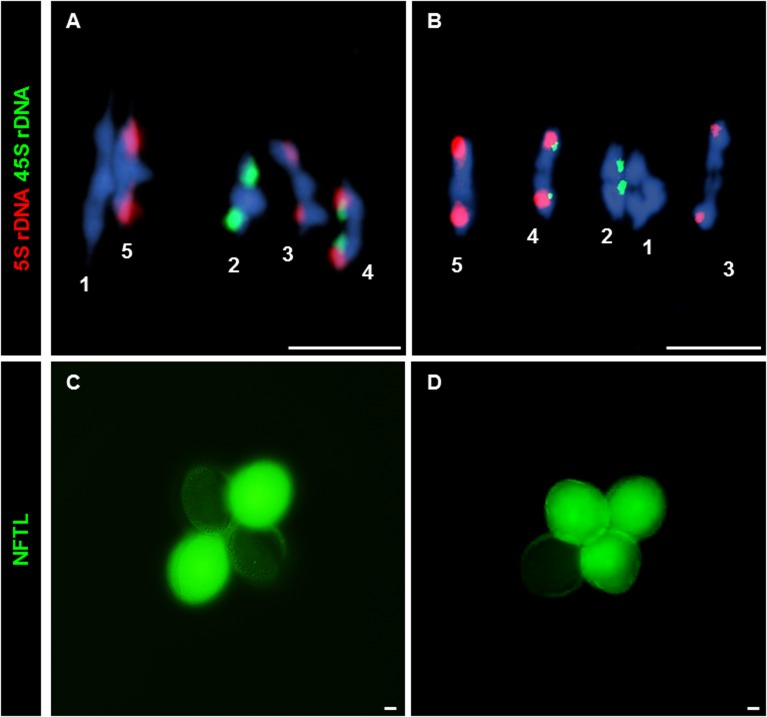
We used FISH with probes for 45S and 5S rDNA to distinguish the chromosomes of Arabidopsis, and counted chiasmata to estimate the mean total COs per cell [21–23] (Fig 3A and 3B). Data were collected from three plants per genotype. Since there were not significant differences in the mean number of chiasmata (the cytological expression of COs) per cell between plants, data were grouped. The mean chiasma frequencies per cell, per bivalent and per bivalent arm are shown in Table 1. There were no significant differences between WT and Atfas1-4 for any of these parameters. Furthermore, we did not detect changes in the distribution of COs over the chromosomes, which are located in either distal or subdistal regions. These results are consistent with the previous AtMLH1 and AtMUS81 immunolocalization data (Fig 2P–2U).
Fig 3. CO and NCO measurements: Chiasma and GC scoring by FISH and NFTLs.
(A, B) FISH using 5S (red) and 45S (green) rDNA probes for chromosome identification and chiasma scoring. (A) WT metaphase I. (B) Atfas1-4 metaphase I. Four ring bivalents (with at least one chiasma in both arms) and one rod bivalent (chromosome 3) are observed. (C, D) Two different examples for the GC test loci using NFTL. (C) Tetrad without GC (2:2). (D) Tetrad with GC (3:1). Bars = 5 µm.
Table 1. Mean chiasma frequencies per cell, per bivalent, and per bivalent arm.
| Bivalent | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | Total | n c | ||||||
| S a | L b | S | L | S | L | S | L | S | L | |||
| 0.99 | 1.54 | 0.61 | 1.14 | 0.9 | 1.26 | 0.48 | 1.01 | 0.97 | 1.3 | |||
| WT | 2.52 | 1.75 | 2.16 | 1.49 | 2.28 | 10.2 ± 0.4 | 69 | |||||
| (0.25) | (0.17) | (0.21) | (0.15) | (0.22) | ||||||||
| 0.87 | 1.50 | 0.77 | 1.17 | 0.90 | 1.27 | 0.77 | 1.10 | 0.93 | 1.33 | |||
| Atfas1-4 | 2.37 | 1.93 | 2.17 | 1.87 | 2.27 | 10.6 ± 0.28 | 30 | |||||
| (0.22) | (0.18) | (0.20) | (0.18) | (0.21) | ||||||||
a Short arm
b Long arm
c Number of cells analyzed
Tetrad analysis of meiotic GC events
Because we observed results consistent with increased levels of DSBs but did not detect an increase in COs we tested whether GC frequencies change in Atfas1-4 relative to WT. GCs can accompany either COs or non-crossovers (NCOs), but in the absence of an increase in COs an increase in GCs can be interpreted as an increase in NCO repair of DSBs. To detect GCs we used the quartet1 non-fluorescent tagged lines (NFTLs) in a system described by Sun and colleagues [24]. Pollen tetrads from plants heterozygous for fluorescent and non-fluorescent alleles at a transgene locus will segregate fluorescence in a 2:2 ratio (Fig 3C). If GC occurs at the test locus, a non-Mendelian 3:1 ratio is observed (Fig 3D). The chromosomal localization of the different NFTL alleles is shown in S6 Fig. We analyzed five plants obtained from different crosses (S2 Table). We found no difference between plants so we grouped the data (Table 2).
Table 2. GC frequencies in Atfas1-4.
Comparison of Atfas1-4 data with previously published WT data revealed a 9-fold increase in GC frequency at the chromosome 1 test locus in Atfas1-4 (χ2 = 49.43; P < 10−4). A test locus on chromosome 2 also showed a 4.5-fold increase (χ2 = 15.34; P = 10−4). A third test locus on chromosome 4 showed a slight (1.53-fold) but non-significant increase (χ2 = 1.04; P = 0.308). This result may be explained by the fact that the chromosome 4 locus has the highest GC frequency in WT. It should also be noted that GC frequencies in WT are not uniform between loci [24] (χ2 = 51.98; P < 10−4), but that such differences were not observed in Atfas1-4 (χ2 = 0.21; P = 0.902).
Meiotic analysis in PMCs of different double mutants
To determine if the additional DSBs observed in Atfas1-4, as measured by counting γH2AX foci, are dependent on AtSPO11-1 we obtained an Atfas1-4 Atspo11-1-5 double mutant. We compared the phenotype of this double mutant to WT and to other Atfas1-4 double mutants in genes involved in downstream steps in the meiotic recombination process, including Atdmc1-2 (knockout allele, KO), Atrad51-2 (knockdown allele, KD) and Atrad51-3 (KO).
Atfas1-4 Atspo11-1-5
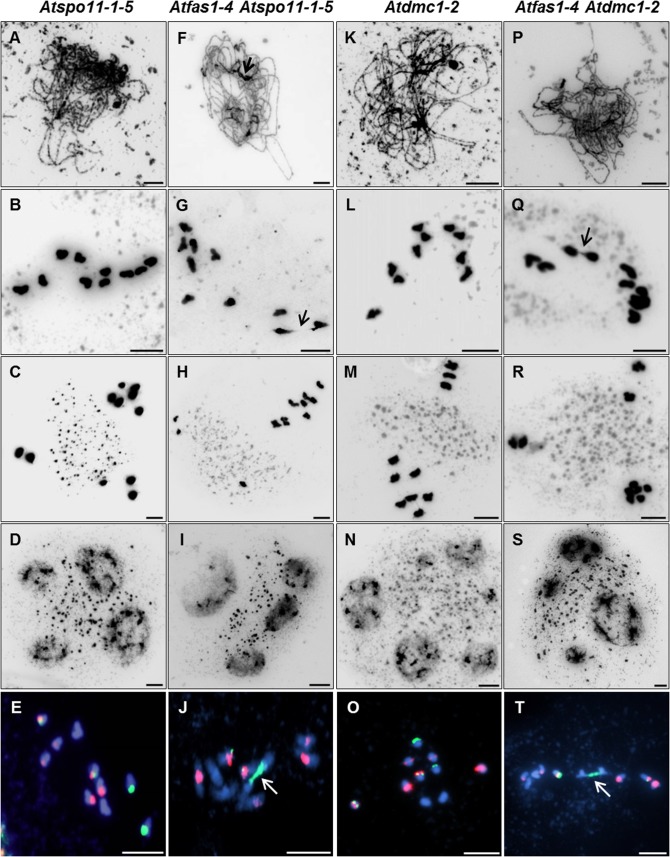
DSBs fail to form in Atspo11-1-5 and as a result, synapsis does not occur (Fig 4A) and ten univalents are observed at diplotene-metaphase I (Fig 4B) leading to unbalanced chromosome segregation after meiosis II (Fig 4C and 4D). However, in the double mutant Atfas1-4 Atspo11-1-5 we observed very small pairing stretches (Fig 4F, arrow), later confirmed by immunodetection of AtZYP1 (see below), and some chromosome associations at metaphase I (Fig 4G, arrow). Unbalanced segregation at meiosis II and tetrads with unequal sized nuclei were also observed (Fig 4H and 4I). Mean chiasma frequency per cell in the double mutant was 0.11 ± 0.05 (n = 45) and in 11% of the cells at least one chiasma was detected (Fig 4J). These results suggest that some DSBs are being produced in Atfas1-4 Atspo11-1-5. We further investigated the level of these residual DSBs by immunolocalization of γH2AX (Fig 5A). We observed 42.81 ± 3.18 γH2AX foci (n = 16) in the double mutant, whereas no foci could be detected in the single Atspo11-1-5 mutant (n = 20; S5 Fig). Fig 5B also illustrates the overall comparison between all the backgrounds analyzed. The number of DSBs was 64% less in the double Atfas1-4 Atspo11-1-5 mutant than in WT. Thus, it is not possible to restore the meiotic phenotype of Atspo11-1-5 by deleting AtFAS1. It is also noteworthy that the number of AtSPO11-independent DSBs detected in Atfas1-4 Atspo11-1-5 (43) is lower than the increase in DSBs observed in the single Atfas1-4 mutant (180.05 ± 5.7) with respect to WT (114.17 ± 5.29) (S1 Table). This suggests that the additional DSBs produced in Atfas1-4 may be both AtSPO11-dependent and independent.
Fig 4. Atfas1-4 Atspo11-1-5 and Atfas1-4 Atdmc1-2 display CO on some occasions.
Meiotic spreads of PMCs stained with DAPI. (A-D) Atspo11-1-5. (F-I) Atfas1-4 Atspo11-1-5. (K-L) Atdmc1-2. (P-S) Atfas1-4 Atdmc1-2. (E, J, O, T) FISH metaphases I using 5S (red) and 45S (green) rDNA probes for chromosome identification in these four backgrounds. Bivalents are observed in the two double mutants (arrows). Bars = 5 µm.
Fig 5. DSB formation in Atfas1-4, Atspo11-1-5, and in the double mutant Atfas1-4 Atspo11-1-5.
(A) Dual immunolocalization of AtASY1 (green) and γH2AX (red) in Atfas1-4 Atspo11-1-5. Bar = 5 µm. (B) Quantification of γH2AX foci in the different backgrounds. Each dot is the count from a single nucleus. P values are from two-sided Wilcoxon Mann-Whitney tests (***P < 0.001).
Atfas1-4 Atdmc1-2
Atdmc1-2 plants fail to synapse at pachytene and have ten univalents at diplotene-zygotene (Fig 4K and 4L). This leads to abnormal segregation at the second division and polyad formation (Fig 4M and 4N). We did not observe any pairing stretches in Atfas1-4 Atdmc1-2 after DAPI staining (Fig 4P). However, we detected some SC stretches after AtZYP1 immunodetection (see below). We found chiasmatic associations at metaphase I in 14% of the cells analyzed (Fig 4Q). The mean chiasma frequency per cell was 0.13 ± 0.07 (n = 36). The majority of chiasmata occurred between 45S rDNA loci (Fig 4T). Second division was similar to that of Atdmc1-2 (Fig 4R and 4S). These results show that all Atfas1-4 Atdmc1-2 DSBs are repaired by an AtDMC1 independent mechanism that is capable of forming some COs.
Atfas1-4 Atrad51-2 and Atfas1-4 Atrad51-3
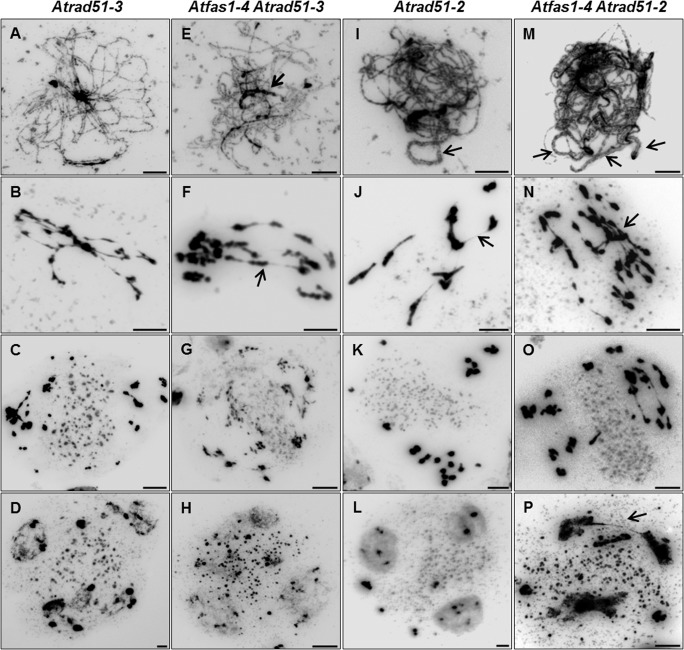
Pairing defects and asynapsis were observed at post-leptotene in Atrad51-3 (Fig 6A), and chromosomes were entangled and interconnected at metaphase I (Fig 6B). After the second division we observed chromosome/chromatid fragments (Fig 6C) and polyads containing micronuclei (Fig 6D). The meiotic phenotype of Atfas1-4 Atrad51-3 was slightly different: pairing stretches at prophase I (Fig 6E, arrow) and some conspicuous chromosome associations at metaphase I (Fig 6F, arrow). The number of chromosome/chromatid fragments and polyads observed at second division appeared to be higher than in Atrad51-3 (Fig 6G and 6H).
Fig 6. Atfas1-4 Atrad51 genetic analysis. (A-D) Atrad51-3 KO mutant plants.
Homologous chromosomes do not pair, and severe chromatin fragmentation is observed. (E-H) Atfas1-4 Atrad51-3. Certain regions appear to be paired (arrow) at post-leptotene and some associated chromosomes are bioriented at metaphase I (arrow). (I-L) Atrad51-2 KD mutant plants display some short SC stretches (arrow) and chromosome associations at metaphase I (arrow). (M-P) The double mutant Atfas1-4 Atrad51-2 also shows short SC stretches (arrows) at post-leptotene and bioriented associated chromosomes at metaphase I (arrow). Bars = 5 µm.
Atrad51-2 meiosis was previously described by Pradillo and colleagues [25]: i) few pairing stretches at post-leptotene (Fig 6I); ii) univalents, well-defined homologous and non-homologous bivalents, multivalents, and small chromosome/chromatid fragments at metaphase I (Fig 6J); iii) chromosomes, chromosome fragments and chromatids lagged in the organelle zone at metaphase II (Fig 6K); and iv) polyads with micronuclei (Fig 6L). Atfas1-4 Atrad51-2 meiocytes presented partial post-leptotene pairing (Fig 6M). At metaphase I the number of fragments and associations with bioriented chromosomes were higher than in Atrad51-2 (Fig 6N), and the elevated level of fragmentation persisted into the second division (Fig 6O). The number of nuclei in polyads was also higher than in the single mutant (Fig 6P).
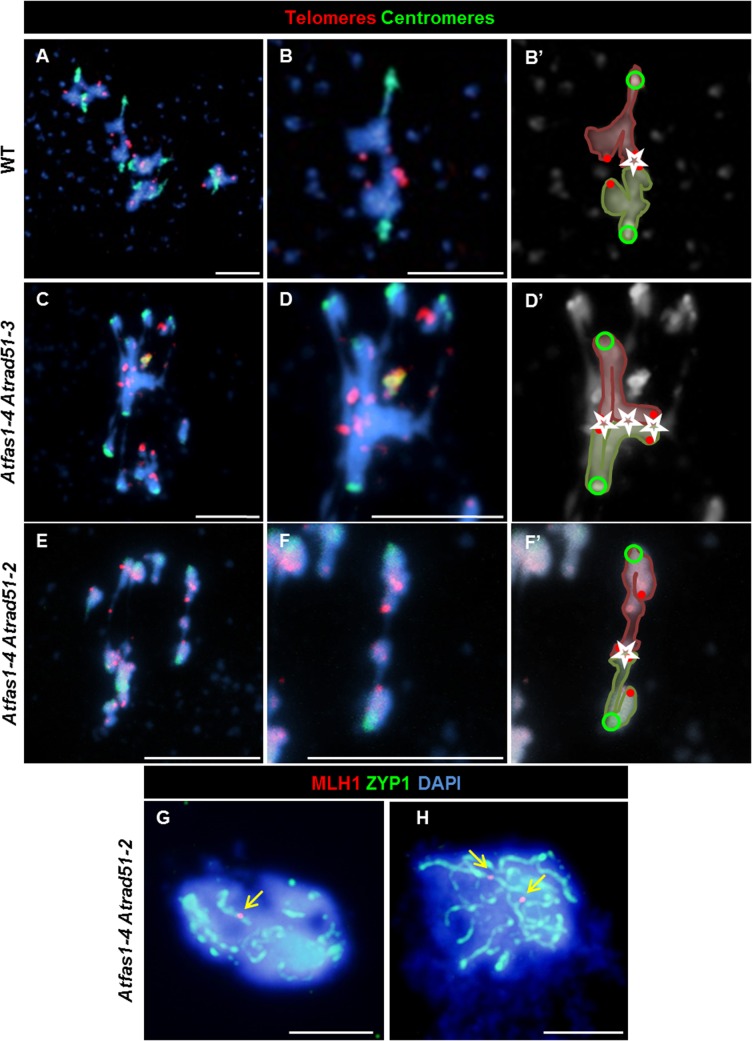
The centromeric tension and orientation of chromosomes towards opposite poles observed in Atfas1-4 Atrad51-2 suggests the existence of chiasmatic associations at metaphase I (Fig 6N). To address this question, we carried out FISH analysis with telomere and centromere probes to confirm the nature of these associations. The presence of an unlabeled DAPI stained strip running longitudinally through the center of the terminal associations and the telomeric signals located in the outer part of chromosome junctions constitute cytological evidence of subterminal chiasmata (Fig 7A–7F) (see for discussion [25, 26]). Visualization of AtMLH1 foci provided additional evidence that these associations are chiasmata (Fig 7G and 7H). On these grounds, the mean chiasma frequency per cell in Atfas1-4 Atrad51-2 plants was 2.08 ± 0.19 (n = 30), which is significantly higher than the result obtained for Atrad51-2 1.55 ± 0.09 (n = 30; t = 2.54; P = 0.01). Likewise, both values are significantly higher than the mean observed in Atfas1-4 Atrad51-3, 0.91 ± 0.15 (n = 27) (t = -5.18; P < 10−4, and t = -3.78; P = 4.16 x 10−3, respectively).
Fig 7. Atrad51 mutants display truly COs.
(A-F) FISH of metaphase I spreads of WT, Atfas1-4 Atrad51-3 and Atfas1-4 Atrad51-2 PMCs using telomeric (red) and centromeric (green) probes. (A) WT metaphase I. (B, B’) Magnification of one bivalent from (A). The external localization of the telomeres (red dots) shows a chiasmatic association (star) between two bioriented homologous chromosomes (centromeres are represented as green circles). Representative images of metaphase I cells from (C) Atfas1-4 Atrad51-3 and (E) Atfas1-4 Atrad51-2. (D, D’, F, F’) Bivalent magnification and diagrammatic chiasma interpretation. (G, H) CO confirmation (arrow) in Atfas1-4 Atrad51-2 detected by immunolocalization of AtMLH1 (red) and AtZYP1 (green) and counterstained with DAPI (blue). Bars = 5 µm.
SC formation
To determine if the small pairing patches detected by DAPI staining were truly SC stretches we used immunodetection of AtZYP1. We estimated two different parameters (Tables 3 and 4 and S3 and S4): i) we quantified the number of synaptic initiation points (SIPs) per cell in PMCs that had AtZYP1 signal covering at most 10% of the total of the chromosome axis; ii) we measured the total SC length in PMCs that had AtZYP1 signal covering all the chromosome axes. The 10% criterion was not applied to Atspo11-1-5, Atdmc1-2, Atrad51-3, Atfas1-4 Atspo11-1-5 and Atfas1-4 Atdmc1-2 because synapsis rarely goes beyond this percentage.
Table 3. Mean numbers of SIPs per cell in the mutants analyzed.
| Mean number of SIPs | Range | n | |
|---|---|---|---|
| Col | 8.89 ± 1.46 | 3–18 | 12 |
| Atfas1-4 | 9.54 ± 1.12 | 4–16 | 11 |
| Atspo11-1-5 | 1.65 ± 0.29 | 0–4 | 27 |
| Atfas1-4 Atspo11-1-5 | 13.54 ± 1.82 | 3–24 | 15 |
| Atdmc1-2 | 5.23 ± 0.44 | 0–14 | 56 |
| Atfas1-4 Atdmc1-2 | 12.50 ± 1.54 | 4–23 | 15 |
| Atrad51-3 | 8.07 ± 0.55 | 0–18 | 57 |
| Atfas1-4 Atrad51-3 | 13.89 ± 1.15 | 1–26 | 26 |
| Atrad51-2 | 9.57 ± 1.12 | 0–28 | 30 |
| Atfas1-4 Atrad51-2 | 18.07 ± 1.16 | 4–28 | 30 |
Table 4. Comparison of SC lengths among different mutants.
| SC mean length (µm) | Range | Maximun percentage of synapsis observed | n | |
|---|---|---|---|---|
| Col | 165.40 ± 4.79 | 140.35–188.77 | 100 | 20 |
| Atfas1-4 | 160.67 ± 8.32 | 143.60–183.50 | 100 | 12 |
| Atspo11-1-5 | 1.15 ± 0.28 | 0–4.63 | 2.89 | 27 |
| Atfas1-4 Atspo11-1-5 | 6.98 ± 0.55 | 1.83–10.82 | 6.54 | 24 |
| Atdmc1-2 | 2.66 ± 0.28 | 0–10.77 | 6.50 | 56 |
| Atfas1-4 Atdmc1-2 | 5.89 ± 2.29 | 0.37–29.80 | 18.03 | 15 |
| Atrad51-3 | 5.35 ± 0.60 | 0–25.62 | 15.50 | 57 |
| Atfas1-4 Atrad51-3 | 14.64 ± 1.77 | 2.59–35.34 | 21.39 | 28 |
| Atrad51-2 | 9.57 ± 1.74 | 0–38.35 | 23.21 | 30 |
| Atfas1-4 Atrad51-2 | 40.67 ± 4.97 | 2.36–114.16 | 68.98 | 30 |
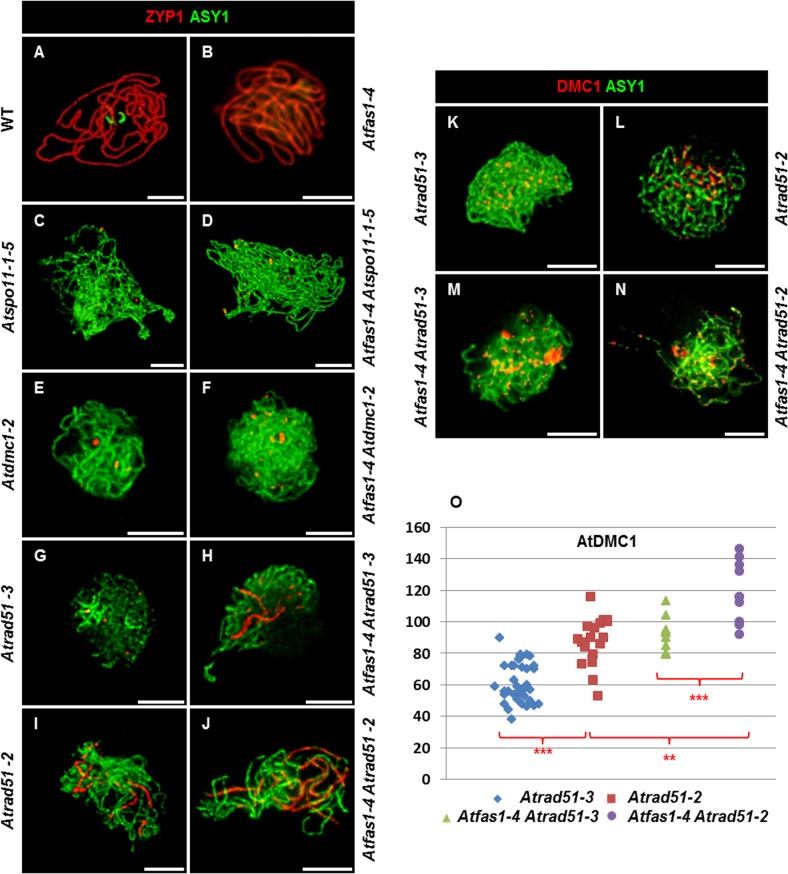
Both WT (Fig 8A) and Atfas1-4 (Fig 8B) achieved full synapsis at pachytene. The results obtained for the different double mutants can be summarized as follows:
Fig 8. AtRAD51 and AtDMC1 could play different roles in the synaptic process.
(A-J) Dual immunolocalization of AtASY1 (green) and AtZYP1 (red) on (A) WT, (B) Atfas1-4, (C) Atspo11-1-5, (D) Atfas1-4 Atspo11-1-5, (E) Atdmc1-2, (F) Atfas1-4 Atdmc1-2, (G) Atrad51-3, (H) Atfas1-4 Atrad51-3, (I) Atrad51-2 and on (J) Atfas1-4 Atrad51-2. Dual immunolocalization of AtASY1 (green) and AtDMC1 (red) on (K) Atrad51-3, (L) Atrad51-2, (M) Atfas1-4 Atrad51-3, and on (N) Atfas1-4 Atrad51-2. Bars = 5 µm. (O) Dispersion diagram for the total foci per nucleus in Atrad51-3 (blue), Atrad51-2 (red), Atfas1-4 Atrad51-3 (green), and Atfas1-4 Atrad51-2 (purple) PMCs are indicated. Each dot is the count from a single nucleus. P values are from two-sided Wilcoxon Mann-Whitney tests (**P < 0.01; ***P < 0.001).
Atfas1-4 Atspo11-1-5
Atspo11-1-5 had the lowest number of SIPs and the shortest SC length (Fig 8C). However, in Atfas1-4 Atspo11-1-5 the number of SIPs was restored to that observed in WT (8.89 ± 1.46; Fig 8D). This is reflected in the significant increase in synapsis extension observed in the double mutant (2.89% in Atspo11-1-5 vs. 6.54% in Atfas1-4 Atspo11-1-5). Hence, AtSPO11 independent DSBs are apparently being processed by the meiotic machinery, as shown by the residual COs detected.
Atfas1-4 Atdmc1-2
In Atdmc1-2 the number of SIPs was very low and SC formation very limited (Fig 8E). In Atfas1-4 Atdmc1-2 the number of SIPs was restored to normal levels, but not SC extension (Fig 8F). It is noteworthy that both the mean and the maximum SIP values in these mutants were lower than those defective for AtRAD51.
Atfas1-4 Atrad51-3
No differences in the number of SIPs were found between Atrad51-3 (Fig 8G) and WT, although, at most 15.5% of the complement was synapsed in this mutant. The mean length of AtZYP1 signals was similar between Atrad51-3 and Atfas1-4 Atdmc1-2. In contrast, both mutants have significantly shorter SCs than Atfas1-4 Atrad51-3 (Fig 8H).
Atfas1-4 Atrad51-2
The mean number of SIPs in Atrad51-2 (Fig 8I) was similar to WT, but some PMCs showed more than 20 SIPs. The maximum SC length observed increased up to 23.2%. Consequently, the proportion of the chromosome complement that achieved synapsis in Atfas1-4 Atrad51-2 (Fig 8J) was longer than in Atfas1-4 Atrad51-3 (Fig 8H) and increased up to 68.98%.
Immunolocalization of AtDMC1
The dynamics of synapsis described above suggests that the differences between mutants might be due to variation in the number of DSBs formed and/or the balance between RAD51 and DMC1 during early prophase I. To test this, we measured the number of AtDMC1 foci in mutants defective for AtFAS1 and AtRAD51.
There was a significant increase in the number of leptotene AtDMC1 foci in Atfas1-4 Atrad51-3 (Fig 8M) and Atfas1-4 Atrad51-2 (Fig 8N) when compared to the respective single mutants (Fig 8K and 8L and S1 Table). Also, the mean number of AtDMC1 foci in PMCs of Atrad51-3 was significant lower than in Atrad51-2 (W = 183.5; P = 3.30 x 10−6), and between Atfas1-4 Atrad51-3 and Atfas1-4 Atrad51-2 (W = -40.0; P = 2.81 x 10−3), as well as between Atrad51-3 and Atfas1-4 Atrad51-2 (W = 190.0; P = 1.47 x 10−7) (Fig 8O). However, there were no differences between Atrad51-2 and Atfas1-4 Atrad51-3 (W = 13.5; P = 0.567). Among all of these genotypes, Atfas1-4 Atrad51-2 showed the highest number of AtDMC1 foci but it was always lower than those in WT and Atfas1-4 (W = 51.5; P = 0.046 and W = 89.0; P = 2.17 x 10−5, respectively).
Discussion
Meiotic DSB frequency is increased in Atfas1-4
Reciprocal recombination events (COs) between homologous chromosomes are necessary in many organisms to ensure correct chromosome segregation at first meiotic division. This is accomplished using highly regulated pathways initiated by programmed DSBs catalyzed by SPO11 (S4 Fig, [27]). Typically, an excess of DSBs are generated, and only a small fraction mature as COs (cytologically manifested as chiasmata), the rest being repaired by other pathways. There are differences in the DSBs/CO ratio among species [28]. Neither physical genome size nor chromosome number can explain these differences. Budding yeast has about two times more DSBs than COs [29], while Arabidopsis has approximately 15 times more DSBs than COs [30, 31]. In Arabidopsis the number of DSBs can be estimated using recombination protein foci (e.g. γH2AX, AtRAD51 and AtDMC1) as a proxy. As a result, different studies, using different antibodies for immunolocalization have estimated different numbers [see 30, 32–36]. Nevertheless, using the same technique it is possible to compare WT with different mutant backgrounds accurately.
The number of γH2AX, AtRAD51 and AtDMC1 foci in Atfas1-4 was significantly higher than in WT (Fig 2 and S1 Table). The most increase of DSBs might be a consequence of breaks produced during pre-meiotic DNA replication as occurs in the somatic line [17]. In fact, AtRAD51 foci appeared in Atfas1-4 earlier than in WT (Fig 2E and 2F), in a stage (pre-meiotic G2) when DSBs catalyzed by AtSPO11 have not yet been produced. These DSBs are likely repaired by HR with the sister chromatid serving as template, since AtDMC1 is unavailable at G2. Alternatively, the increase in DSBs may be related to a more open chromatin configuration exhibited by the mutant that may facilitate access by topoisomerases (S3 Fig; [18]). The residual DSBs detected by immunolocalization of γH2AX (Fig 5A) in Atfas1-4 Atspo11-1-5 and its comparison with those observed in the single mutant Atfas1-4 (Fig 2C) demonstrate that the increase in DSBs produced in Atfas1-4 most likely involves both mechanisms. Furthermore, the analysis of the double Atfas1-4 Atspo11-1-5 mutant revealed that a portion of these AtSPO11 independent DSBs may be processed as genuine programmed DSBs, similar to those produced by cisplatin [30], since, in contrast to Atspo11-1-5, chiasmata and SC were observed. Another conclusion that comes from this result is that a minimum number of DSBs is required to guarantee full synapsis and the obligatory CO. According to γH2AX immunolocalization, the number of DSBs in Atfas1-4 Atspo11-1-5 (42.81 ± 3.1) represents less than the 40% of the total DSBs produced in WT (114.17 ± 5.29). However, this number is enough to restore a normal SIPs phenotype (Fig 8D and Table 3). These findings could indicate that SC nucleation and elongation are dependent on the number of DSBs, which in Atfas1-4 Atspo11-1-5 are insufficient to achieve both full synapsis and the obligate CO [37, 38]. In this context, we cannot exclude the possibility that the excess of DSBs in Atfas1-4 derives from a delayed prophase I.
“Extra” DSBs of Atfas1-4 are repaired by HR
At least four genes involved in HR are overexpressed in this mutant: AtBRCA1, AtCOM1, AtRAD51 and AtDMC1 (Fig 2A). AtBRCA1 loads recombinases on to DNA [39, 40]. The expression of this gene depends on DNA damage, for instance Lafarge and Montané [41] reported that a 100 Gy dose produces about 92-fold overexpression of this gene. In mammals BRCA1 interacts with MRN (MRE11-RAD50-NBS1), CtIP (COM1-SAE2), and Retinoblastoma, to activate G2 cell cycle check-point by means of ATM phosphorylation [42, 43].
In Atcom1-1 there is an accumulation of AtSPO11 at prophase of meiosis I without formation of AtRAD51 foci, despite the presence of unrepaired DSBs. AtCOM1 acts downstream of AtSPO11-1 and upstream of AtBRCA1 and AtDMC1. Thus, it is necessary for regular turnover of AtSPO11-1 and processing of meiotic DSBs [44]. Absence of AtCOM1 causes chromosome fragmentation like other mutants defective in creation of 3’ ssDNA tails via resection [45, 46]. Furthermore, DNA fragmentation in the mutant is suppressed by eliminating AtSPO11-1 [44]. The overexpression of AtCOM1, observed in Atfas1-4, might be facilitating the correct processing (resection) of the extra DSBs.
Once 3’ ssDNA tails are generated at the DSB site, RAD51 and DMC1 catalyze a strand exchange reaction with an intact DNA duplex to produce joint molecule intermediates (see S4 Fig; [47]). These recombinases bind the ssDNA tails to produce presynaptic filaments. It has been proposed that DMC1 is essential to establish preferred interactions between homologs, whereas the catalytic activity of RAD51 alone results in sister chromatid exchanges [28, 30, 35, 48–50]. However, the exact action of both recombinases is not fully understood (see [27] for review in Arabidopsis). AtDMC1, as well as AtCOM1 and AtBRCA1, are about 3-fold overexpressed in Atfas1-4. However, AtRAD51 is about 5-fold overexpressed (Fig 2A), an increase that could be related to the extra AtSPO11-independent DSB repair, in concordance with the early meiotic appearance of AtRAD51 in Atfas1-4 compared to WT (Fig 2E and 2F). Increases of AtRAD51 and AtDMC1 mRNA were accompanied with those of their respective protein products (Fig 2I and 2L). The abundance of these proteins may help prevent chromosome fragmentation despite an increase in DSBs. The overexpression of AtSMC6 in the mutant could be also related to the repair of non-programmed DSBs [51–53]. Accordingly, it is tempting to speculate about the existence of an increase in inter-sister recombination. On the other hand, although the most likely explanation for the overexpression of HR genes is the requirement for repairing the “extra” DSBs, another possibility is an extended prophase I in Atfas1-4. Since only some genes are overexpressed, a global effect on transcription because of changes in chromatin structure does not seem to be probable.
Increase of GC events in Atfas1-4
COs can be produced by at least two pathways in Arabidopsis. Type I COs are subject to positive interference and are dependent on the ZMM proteins [54–56]. Type II COs are insensitive to interference, are randomly distributed and are dependent on MUS81 and MMS4 proteins for their formation [57, 58]. In Atfas1-4, genes involved in either Type I COs (AtMLH3) or Type II COs (AtMUS81) were not overexpressed (Fig 2A). Likewise, the number of both AtMLH1 (Type I) and AtMUS81 foci (Type II) was similar to WT (Fig 2R and 2U). These results are consistent with the absence of differences in chiasma frequency between the mutant and WT (Table 1). According to the model proposed by Berchowitz and Copenhaver [59], if an increase in DSBs is not translated to a CO increase it implies that there is an increase either in the number of recombination intermediates in the Synthesis Dependent Strand Annealing (SDSA) pathway or in the frequency of double Holliday Junctions (dHJs) resolved as NCOs (S4 Fig). Although during meiosis there is a bias against inter-sister events, an alternative explanation is an increase in inter-sister repair events [60].
AtMSH4 and AtMSH5 form a complex that loads and stabilizes dHJs [61–63]. Since the number of AtMSH4 foci was similar in Atfas1-4 and WT (Fig 2O), we conclude that an overall change in the number AtMSH4 foci did not take place. Cole et al. [64] showed that mice that over-express SPO11β (2X higher expression) have twice the level of γH2AX, whereas they have only 7% more MSH4. In Atfas1-4 an increase of 58% in DSBs does not change either the number of AtMSH4 foci (Table 1) or the number of SIPs (Tables 3 and S3). However, since our observations correspond to mid-late zygotene we cannot exclude a possible increase of AtMSH4 foci in early stages. Additionally, it is interesting to note that mRNA levels of AtBLAP75 and AtTOP3α have suggestive (but not statistically significant) increases in expression levels, and that both are involved in dHJ dissolution leading to NCOs [65, 66].
Sun and colleagues [24] reported differences in the GC frequency at several loci in WT, for instance GC events were most frequent in the short arm of chromosome 4 followed by those produced in chromosomes 2 and 1 (4 > 2 >1). The situation in Atfas1-4 is exactly the opposite since the magnitude of GC events was 1 > 2 > 4 (Table 2). It seems that in Atfas1-4 those regions that are less prone to GC in WT are more prone to processing excess DSBs with a mechanism that also yields GCs. Local specific changes in chromatin conformation could also affect these GC frequencies.
Martini and colleagues [67] reported in yeast that a decrease in the number of DSBs observed in different spo11 alleles did not affect the overall CO frequency at the expense of a decrease in the number of NCO events. This phenomenon was called “CO homeostasis”. On the other hand, in mice overexpressing SPO11 the mean number of RAD51 and DMC1 was higher than in WT, whereas MLH1 foci number was similar [65]. Our results are in some way similar to these because the increase in the number of DSBs observed in Atfas1-4 had no influence on the frequency of COs (scored as either chiasmata or AtMLH1/AtMUS81 foci), or on their distribution (Figs 2P–2U and 3). However, the excess of DSBs produces an increase in the frequency of GC events. We cannot exclude that the increase in GC frequency could be due to mismatch repair defects or alterations in inter-homolog/inter-sister interactions. Nevertheless, the existence of CO homeostasis, as a consequence of positive CO interference, is a likely explanation. Joshi et al. [68] have recently demonstrated that recombination outcome is dependent on global DSB levels. Thus, early and low-abundant DSBs lack homolog bias. This could be the case for pre-meiotic G2 DSBs observed in Atfas1-4. Following the model proposed by Joshi et al. [68], the gradual implementation of interhomolog bias would produce an increase in interhomolog interactions throughout prophase I in Atfas1-4, with an increase in NCOs vs. CO compensating for increased homolog bias.
The interplay between AtRAD51 and AtDMC1 in Atfas1-4
The double mutants Atfas1-4 Atrad51-2, Atfas1-4 Atrad51-3, and Atfas1-4 Atdmc1-2 enabled functional analysis of the interaction between AtRAD51 and AtDMC1. An increase in DSBs in absence of AtFAS1 is accompanied by an excess of AtDMC1 in Atfas1-4 Atrad51, and AtRAD51 in Atfas1-4 Atdmc1-2. Bishop and colleagues [69] and Tsubouchi and Roeder [70] demonstrated in yeast that high levels of inter-homolog recombination could be achieved in the absence of Dmc1 upon either overexpression of Rad51 or by stimulating its partner Rad54. Recently, Cloud and colleagues [48] reported that the activity of Dmc1, but not that of Rad51, is necessary for meiotic recombination in yeast. Thus, Rad51 may be an accessory factor which contributes to homolog bias independently of strand exchange activity. However, it may also be relevant when Dmc1 fails. On the other hand, Hong and colleagues [71] and Lao and colleagues [72] have provided evidence for the inhibitory role of Dmc1 in Rad51 activity, and that the default option for recombination is homolog bias, independent of whether strand exchange is promoted by either DMC1 or RAD51. Observations of da Ines and colleagues [50] in Arabidopsis support an accessory role of AtRAD51. However, Pradillo and colleagues [25] observed homologous and non-homologous recombination in a KD Atrad51 mutant, which also had less chromosome fragmentation than the KO mutant. These results suggest that some DSBs can be processed by interhomolog recombination in the mutant, although a critical level of this protein is required to ensure the fidelity of HR during inter-chromosomal exchanges. On the other hand, Kurzbauer and colleagues [35] have reported that Atrad51 is defective in AtDMC1 loading. Likewise, Uanschou and colleagues [73] have pointed out that AtDMC1 might have a role as an inhibitor of AtRAD51 [27].
Atfas1-4 Atdmc1-2 displayed a similar meiotic phenotype compared to the single mutant Atdcm1-2 (Fig 4, [74]). However, a few bivalents were observed at metaphase I (Fig 4T). Thus, AtRAD51 is capable of repairing the excess DSBs produced in Atfas1-4, and may also permit the formation of some COs independently of AtDMC1. Nevertheless, the presence of other regulatory factors involved in the decision dictating sister chromatid vs. inter-homolog exchange cannot be ruled out.
On the other hand, decreased (Atrad51-2) or elimination (Atrad51-3) of AtRAD51 leads to chromosome fragmentation (Fig 6), the effects being more drastic in Atfas1-4 Atrad51-3 than in Atfas1-4 Atrad51-2. These results constitute additional evidence of excess DSBs in Atfas1-4. In these double mutants, the number of AtDMC1 foci was higher than in Atrad51 but lower than WT because, despite AtDMC1 overexpression, AtRAD51 is necessary to correct AtDMC1 loading on chromosome axes [32, 35]. The presence of a certain amount of AtRAD51 in Atfas1-4 Atrad51-2 promoted COs dependent on AtMLH1. Moreover, the fact that chiasma frequency was higher in Atfas1-4 Atrad51 than in Atfas1-4 Atdmc1-2 revealed the important role of AtDMC1 in homologous interchanges. This suggests that AtDMC1 is more efficient than AtRAD51 in promoting homologous interchanges [75].
AtRAD51, AtDMC1 and SC formation
The relationship between synapsis initiation at recombination initiation sites and its ultimate progression varies depending on species, sex, and other factors [37, 61]. In Arabidopsis, there are differences in chiasma frequency and SC length between PMCs and megaspore mother cells (MMCs) of the same plant, and between different accessions [23, 76–78]. SC length is also determined by genome size, and in Arabidopsis this relation, measured as cM/Mb, is the highest described in any plant species to date [23]. However, the chromatin decondensation exhibited by Atfas1-4 did not imply any changes in either the initial SC nucleation or total SC length (Tables 3 and 4).
In yeast and mouse, MSH4 localizes over SIPs and it has been proposed that this protein cooperates with RAD51 and DMC1 in SC nucleation [79, 80]. In Atfas1-4 as well as in WT plants, the number of AtMSH4 and AtMSH5 foci estimated by immunolocalization greatly exceed that of SIPs and COs (Table 3; [61–63]). This fact maybe explained by the necessity of establishing efficient homologous interactions at prophase I, not necessarily essential to achieve full synapsis [61–63, 81]. The increase of DSBs in Atfas1-4, either dependent or independent of AtSPO11, did not produce changes in the frequency of SIPs, SC length or the number of late AtMSH4 foci. As we discussed above, less than the 40% of the total DSBs produced in WT are enough to restore a normal number of SIPs in Atfas1-4 Atspo11-1-5 (Fig 8D and Table 3). However, while there were not differences in the number of SIPs between Atrad51-3 and WT, Atdmc1-2 displayed lower values (Fig 8 and Tables 3 and S3). Thus, both recombinases are probably able to catalyze HR and to be involved in the SC initiation as occurs in yeast [73, 82, 83], although AtDMC1 seems to be the most efficient. The normal number of SIPs was restored in Atfas1-4 Atdmc1-2, but in Atfas1-4 Atrad51-3 the mean number of SIPs greatly exceeded normal levels [23, 76–78].
In any of the mutants analyzed, AtZYP1 localized along the entire length of AEs, indicating that both recombinases are indispensable for achieving full synapsis. However, the presence of only one of them generated differences in synapsis progression rates. Thus, there were no differences in SC length between Atdmc1-2 and Atfas1-4 Atdmc1-2, but they existed between Atrad51-3 and Atfas1-4 Atrad51-3. A possible interpretation of these results is that although AtRAD51 overexpression does not produce changes in SC lengths, AtDMC1 does (Fig 8 and Tables 4 and S4). AtDMC1 activity may also be favored by a certain amount of AtRAD51 because the number of SIPs and SC length are higher in Atfas1-4 Atrad51-2 than in Atfas1-4 Atrad51-3, perhaps due to AtRAD51 contribution to AtDMC1 loading on chromosome axes (Fig 8; [32, 35]). Finally, it is noteworthy that in all mutants analyzed there was a positive relationship between the number of AtDMC1 foci and the development of the synaptic process. That is, the higher number of AtDMC1 signals the higher the number of SIPs and the longer the SC length (Fig 8, S1 and S3 and S4 Tables). Another issue to consider for the interpretation of this result is the balance between both recombinases, which influences correct SC assembly [30, 35]. In Atfas1-4 Atrad51-2, AtDMC1 is overexpressed and there is presumably more AtRAD51 than in the single mutant Atrad51-2. These observations could reflect the interplay between control of chromosome axes and HR, a system in which AtASY1, AtASY3 and AtDMC1 would have similar roles to Hop1, Red1 and Dmc1 in yeast [30, 33, 84].
Concluding remarks
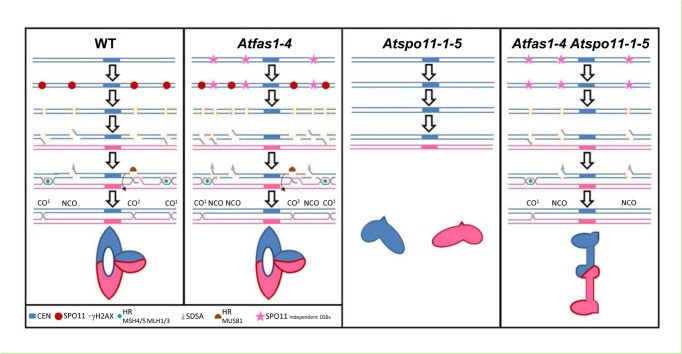
Summing up, our results show that the number of COs can be constrained in plant species even when the number of DSBs increases during meiosis. Fig 9 shows different models for this regulation in the different backgrounds analyzed. In WT meiosis, DSBs catalyzed by AtSPO11 can be processed by different pathways; mainly by HR producing mostly COs and possibly some NCOs, or by SDSA producing NCOs. In absence of AtSPO11-catalyzed DSBs, there is no HR, no COs are formed and the homologous chromosomes are not linked by chiasmata at metaphase I. In a double mutant Atspo11-1-5 Atfas1-4, AtSPO11-independent DSBs can be processed as NCOs and COs producing some bivalents at metaphase I. Thus, some AtSPO11-independent DSBs can be processed by HR to form COs and produce chiasmata between homologous chromosomes. In the single Atfas1-4 mutant, where AtSPO11-dependent DSBs and extra AtSPO11-independent DSBs are formed, the number of COs at metaphase I is the same that in the WT. The extra DSBs appear to be processed to NCOs in this mutant in order to keep the same CO frequency compared to the WT, producing an increase in GC frequency. Taken together, these results highlight the complex regulation of CO formation in Arabidopsis meiosis.
Fig 9. Meiotic DNA DSB outcomes in Arabidopsis.
The model shows the different meiotic fates of DSBs (AtSPO11-dependent and independent) in WT, Atfas1-4, Atspo11-1-5; and in the double mutant Atfas1-4 Atspo11-1-5. (CEN: centromere; HR: Homologous Recombination; SDSA: Synthesis Dependent Strand Annealing).
WT CO model
The model shows the meiotic processing of AtSPO11-dependent DSBs using two homologous chromosomes (blue and pink). AtSPO11 (red circle) binds to the chromatin along the chromosomes and produces DSBs. The phosphorylation of H2AX (γH2AX, orange squares) occurs rapidly around the DSB chromatin region. The DSBs are processed by the MRN complex that resects the 5’ ends of the DSBs allowing the 3’ ends to be exposed as single strand DNA (ssDNA) tails which could follow different pathways: HR (green circle, CO1), SDSA (grey triangle) or AtMUS81 interference-independent CO pathway (CO2). During HR the 3’ SSD tails will associate to AtRAD51 and AtDMC1. These recombinases are involved in the ssDNA invasion to search for the homologous sequences to process the DSBs with the homologous chromosome. If they are successful, they will form different recombination intermediates using proteins like AtMSH4/5 and AtMLH1/3 that will end into a double Holliday Junction (dHJ) which could have different outputs: i) dHJ resolution, producing mostly CO1 and very little NCOs. These COs are subjected to interference. ii) dHJ dissolution which using proteins like BLAP75 and TOP3α would produce NCOs. iii) The ssDNA tail invasion might not follow the HR pathway and form a dHJ which would be processed by AtMUS81 to produce COs not subjected to interference (CO2). iv) If the ssDNA tail invasion is not stable and the 3’ end gets liberated from the initial intermediate with the homologous chromosome, SDSA could process this DSB and repaired it producing a NCO. A metaphase I ring bivalent configuration bearing one chiasma in one arm and two chiasmata in the other is showed.
Atfas1-4 CO model
The model shows the meiotic processing of both, the AtSPO11-dependent (red circles) and AtSPO11-independent (pink stars) DSBs present in this mutant. This mutant presents an increase in DSBs visualized by an increase of -γH2AX (orange squares). These extra DSBs could be processed by the different pathways showed in WT but nevertheless, the outcome of these extra DSBs is to produce NCOs. Thus, keeping the amount of COs to the same quantity of that present in the WT even when the number of DSBs is increased (CO homeostasis). A metaphase I ring bivalent configuration bearing three chiasmata is showed.
Atspo11-1-5
DSBs are not formed and no HR can be achieved. Two univalents are showed at metaphase I.
Atspo11-1-5 Atfas1-4
This model shows the meiotic processing of AtSPO11 independent (pink stars) DSBs. Lacking AtSPO11-1 means that not meiotic DSBs are produced and obviously not recombination between the homologous chromosomes can be achieved. Chromosomes at metaphase I cannot form bivalents and only univalents are observed. Nevertheless, in Atspo11-1-5 Atfas1-4 some bivalents can be formed. In this double mutant AtSPO11-dependent DSBs are not formed but, due to the lack of AtFAS1 protein, some DSBs are produced (probably during or just after the pre-meiotic S phase due to DNA replication errors). These AtSPO11 independent DSBs can be processed to NCOs and also as COs. A metaphase I rod bivalent bearing one distal chiasma is showed.
Materials and Methods
Plant materials
Plants were grown, material harvested and DNA extracted as described previously by Pradillo and colleagues [25]. The Arabidopsis thaliana Columbia (Col-0) accession was used as a control. Seeds of most mutants were provided by the Nottingham Arabidopsis Stock Center (Nottingham, UK). Dr. Crisanto Gutiérrez kindly donated Atfas1-4 (SAIL_662_D10; [4]), Atfas2-1 [85], and RNAi Atasf1a Atasf1b plants with decreased levels of both ASF1A and ASF1B [86]. For double mutant analysis, T-DNA alleles previously described by Pradillo and colleagues [25] were used: Atspo11-1-5 (SALK_009440), Atrad51-2, Atrad51-3 (SAIL_873_C08) and Atdmc1-2 (SAIL_170_F08). To score GC in Atfas1-4 we used three NFTLs: NFTL 567-GC1, NFTL 3411-GC1 and NFTL 424-GC1 [24]. Genotyping of double mutants was performed by PCR using a combination of three primers, one T-DNA specific primer and two specific primers for the corresponding lines (S5 Table).
Cytological analysis
Observations of gynoecia were carried out using a squash procedure with fixed buds. Sepals, petals, and stamens were dissected from flower buds in 100 µl of acetic carmine solution (Sigma) and the number of ovules was scored. Pollen tetrads were collected by dipping flowers in 10 µl of Pollen Growth Media [24]. Fixation, PMC slide preparation and FISH were carried out as described by Sánchez-Morán and colleagues [21]. The DNA probes used were: 45S rDNA (pTa71, [87]), 5S rDNA (pCT4.2, [88]), centromeres (pAL1, [89]), and telomeres (pLT11, [90]). Microscopy was carried out using an Olympus BX-60 microscope equipped with an Olympus DP71 digital camera controlled by analysis software (Soft Imaging System). Images were analyzed and processed with Adobe Photoshop CS4.
The spreading immunolocalization technique previously described by Armstrong and colleagues [91] was used to detect SC and HR related proteins. Prof. Chris Franklin kindly donated primary antibodies: anti-γH2AX (Ser 139, catalog no. 07–164 Upstate Biotechnology; rabbit, 1:250 dilution), anti-AtASY1 (rat; 1:1000 dilution), anti-AtZYP1 (rabbit; 1:500), anti-AtRAD51 (rabbit; 1:250), anti-AtDMC1 (rabbit; 1:250), anti-AtMSH4 (rabbit; 1:250), and anti-AtMLH1 (rabbit; 1:250) [30, 37, 61, 92–94]. Protein foci were manually counted using the Count tool in Adobe Photoshop CS4. SC length was measured using Image J software (http://rsbweb.nih.gov/ij/).
RNA extraction and quantitative PCR
Total RNA was isolated from young flower buds using an RNeasy kit (Qiagen). Quantitative PCR was performed with the Fast Start Taq Man Probe Master kit using UPL probes and specific primers designed by the UPL Assay Design Center (http://www.roche-applied-science.com/sis/rtpcr/upl/index.jsp?id=UP030000). See S6 Table for qPCR primers and UPL probe information. Relative quantification of mRNA was calculated using fold variation over a calibrator with the standard curves method [95] after normalization to 18S rRNA as an internal control (Hs99999901_s1; Applied Biosystems, http://www.appliedbiosystems.com). Three experimental replicates of five independent RNA extractions were carried out for each target gene.
Statistical methods
Statistical comparisons of the number of ovules and data on SC formation and HR were performed using a non-parametric Wilcoxon Mann-Whitney test with a confidence interval of 95%. Statistical analysis of chiasma counts was done as described in Sánchez-Morán and colleagues [22, 23] and López and colleagues [24]. We compared GC frequencies between WT and Atfas1-4 plants using chi-square tests. To evaluate the significance of relative gene expression differences between mutant and WT plants, 95% confidence intervals were defined for the average expression of each gene.
Supporting Information
Squash procedure to visualize and estimate the number of ovules per gynoecium (A) on WT and (B) on Atfas1-4.
(TIF)
Chromosome spread preparations from (A, C, E, G) Atfas1-2 and (B, D, F, H) AtASF1a/AtASF1b RNAi PMCs. (A, B) Pachytene. (C, D) Metaphase I: four ring bivalents and one rod bivalent on each. (E, F) Prophase II. (G, H) Tetrad. Bars = 5µm.
(TIF)
FISH using the centromeric probe pAL1 (180 bp) (A) on WT and (B) on Atfas1-4 at pachytene. In the mutant the centromeric FISH signal is weaker, less defined and more extended than in WT.
(TIF)
Single strands of DNA are shown as either blue (parent 1) or red (parent 2) rods. AtSPO11 helped by other proteins initiates programmed DSBs. H2AX phosphorylation occurs at the break zone and DSBs are resected 5’ to 3’ to produce single ssDNA tails by the MRN complex and COM1. One of these ends invades the homologous duplex DNA, giving raise a D loop intermediate mediated by AtRAD51 and AtDMC1 and other proteins. If the second end is captured and the broken DNA strands are ligated, a dHJ is formed. This intermediate is resolved as CO1, sensitive to interference, or NCO upon appropriate resolution of the two HJs. On the other hand, this dHJ can be dissolved as a NCO. Alternatively, the D loop can be processed to generate a CO2 (insensitive to interference). When the D-loop is dissociated before the second end capture SDSA pathway occurs, the invading strand dissociates after DNA synthesis. This strand then re-anneals to the original parent, resulting in repair of the DSB and a heteroduplex DNA. This pathway is always resolved as a NCO.
(TIF)
(A-C) Dual immunolocalization on Atspo11-1-5 nuclei. (A) AtASY1 (green) and γH2AX (red). (B) AtASY1 (green) and AtRAD51 (red). (C) AtASY1 (green) and AtDMC1 (red). Bars = 5µm.
(TIF)
(TIF)
(PDF)
All the analyzed plants were heterozygous for the fluorescent and non-fluorescent allele and homozygous for Atfas1-4.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
We are grateful to Crisanto Gutiérrez (CBGP, Madrid, Spain) for providing several mutant lines and Chris Franklin (University of Birmingham, UK) for providing antibodies used in this study. We also thank Javier Silva (CBGP, Madrid, Spain) for qPCR technical support. We are grateful to anonymous referees for their comments to improve the final version of the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
ESM´s research is funded by BBSRC. GPC lab is founded by the United States National Science Foundation (MCB-1121563). JLS and MP are funded by grants from Ministerio de Economía y Competitividad of Spain (AGL2012-38852) and the European Union FP7 (Meiosys-KBBE-2009-222883). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Eitoku M, Sato L, Senda T, Horikoshi M. Histone chaperones: 30 years from isolation to elucidation of the mechanisms of nucleosome assembly and disassembly. Cell Mol Life Sci. 2008;65: 414–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Das C, Tyler JK, Churchill ME. The histone shuffle: histone chaperones in an energetic dance. Trends Biochem Sci. 2010;35: 476–489. 10.1016/j.tibs.2010.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ransom M, Dennehey BK, Tyler JK. Chaperoning histones during DNA replication and repair. Cell. 2010;140: 183–195. 10.1016/j.cell.2010.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ramírez-Parra E, Gutierrez C. E2F regulates FASCIATA1, a chromatin assembly gene whose loss switches on the endocycle and activates gene expression by changing the epigenetic status. Plant Physiol. 2007;144: 105–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kaya H, Shibahara KI, Taoka KI, Iwabuchi M, Stillman B, Araki T. FASCIATA genes for chromatin assembly factor-1 in arabidopsis maintain the cellular organization of apical meristems. Cell. 2001;104: 131–142. [DOI] [PubMed] [Google Scholar]
- 6. Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96: 575–585. [DOI] [PubMed] [Google Scholar]
- 7. Polo SE, Almouzni G. Chromatin assembly: a basic recipe with various flavours. Curr Opin Genet Dev. 2006;16: 104–111. [DOI] [PubMed] [Google Scholar]
- 8. Le S, Davis C, Konopka JB, Sternglanz R. Two new S-phase-specific genes from Saccharomyces cerevisiae . Yeast. 1997;13: 1029–1042. [DOI] [PubMed] [Google Scholar]
- 9. Tyler JK, Collins KA, Prasad-Sinha J, Amiott E, Bulger M, Harte PJ, et al. Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol Cell Biol. 2001;21: 6574–6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mello JA, Silljé HHW, Roche DMJ, Kirschner DB, Nigg EA, Almouzni G. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Reports. 2002;3: 329–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhu Y, Weng M, Yang Y, Zhang C, Li Z, Shen WH, et al. Arabidopsis homologues of the histone chaperone ASF1 are crucial for chromatin replication and cell proliferation in plant development. Plant J. 2011;66: 443–455. 10.1111/j.1365-313X.2011.04504.x [DOI] [PubMed] [Google Scholar]
- 12. Hennig L, Bouveret R, Gruissem W. MSI1-like proteins: An escort service for chromatin assembly and remodelling complexes. Trends Cell Biol. 2005;15: 295–302. [DOI] [PubMed] [Google Scholar]
- 13. Worsdell WC. “FASCIATION”: its Meaning and Origin. New Phytol. 1905;4: 55–74. [Google Scholar]
- 14. Exner V, Taranto P, Schönrock N, Gruissem, Hennig L. Chromatin assembly factor CAF-1 is required for cellular differentiation during plant development. Development. 2006;133: 4163–4172. [DOI] [PubMed] [Google Scholar]
- 15. Sánchez MDLP, Caro E, Desvoyes B, Ramirez-Parra E, Gutierrez C. Chromatin dynamics during the plant cell cycle. Semin Cell Dev Biol. 2008;19: 537–546. 10.1016/j.semcdb.2008.07.014 [DOI] [PubMed] [Google Scholar]
- 16. Ono T, Kaya H, Takeda S, Abe M, Ogawa Y, Kato M, et al. Chromatin assembly factor 1 ensures the stable maintenance of silent chromatin states in Arabidopsis . Genes Cells. 2006;11: 153–162. [DOI] [PubMed] [Google Scholar]
- 17. Ramírez-Parra E, Gutierrez C. The many faces of chromatin assembly factor 1. Trends Plant Sci. 2007;12: 570–576. [DOI] [PubMed] [Google Scholar]
- 18. Kirik A, Pecinka A, Wendeler E, Reiss B. The chromatin assembly factor subunit FASCIATA1 is involved in homologous recombination in plants. Plant Cell. 2006;18: 2431–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Endo M, Ishikawa Y, Osakabe K, Nakayama S, Kaya H, Araki T, et al. Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. EMBO J. 2006;25: 5579–5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen Z, Tan JLH, Ingouff M, Sundaresan V, Berger F. Chromatin assembly factor 1 regulates the cell cycle but not cell fate during male gametogenesis in Arabidopsis thaliana . Development. 2008;135: 65–73. [DOI] [PubMed] [Google Scholar]
- 21. Sánchez-Morán E, Armstrong SJ, Santos JL, Franklin FC, Jones GH. Chiasma formation in Arabidopsis thaliana accession Wassileskija and in two meiotic mutants. Chromosome Res. 2001;9: 121–128. [DOI] [PubMed] [Google Scholar]
- 22. Sánchez-Morán E, Armstrong SJ, Santos JL, Franklin FCH, Jones GH. Variation in chiasma frequency among eight accessions of Arabidopsis thaliana . Genetics. 2002;162: 1415–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. López E, Pradillo M, Romero C, Santos JL, Cuñado N. Pairing and synapsis in wild type Arabidopsis thaliana . Chromosome Res. 2008;16: 701–708. 10.1007/s10577-008-1220-z [DOI] [PubMed] [Google Scholar]
- 24. Sun Y, Ambrose JH, Haughey BS, Webster TD, Pierrie SN, Muñoz DF, et al. Deep genome-wide measurement of meiotic gene conversion using tetrad analysis in Arabidopsis thaliana . PLoS Genet 2012;8: e1002968 10.1371/journal.pgen.1002968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pradillo M, López E, Linacero R, Romero C, Cuñado N, Sánchez-Morán E, et al. Together yes, but not coupled: new insights into the roles of RAD51 and DMC1 in plant meiotic recombination. Plant J. 2012;69: 921–933. 10.1111/j.1365-313X.2011.04845.x [DOI] [PubMed] [Google Scholar]
- 26. Jones GH. Giemsa C-banding of rye meiotic chromosomes and the nature of “terminal” chiasmata. Chromosoma 1978;66: 45–57. [Google Scholar]
- 27. Pradillo M, Varas J, Oliver C, Santos JL. On the role of AtDMC1, AtRAD51 and its paralogs during Arabidopsis meiosis. Front Plant Sci 2014;5: 23 10.3389/fpls.2014.00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Serrentino ME, Borde V. The spatial regulation of meiotic recombination hotspots: are all DSB hotspots crossover hotspots? Exp Cell Res. 2012;318: 1347–1352. 10.1016/j.yexcr.2012.03.025 [DOI] [PubMed] [Google Scholar]
- 29. Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 2008;454: 479–485. 10.1038/nature07135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sánchez-Morán E, Santos JL, Jones GH, Franklin FCH. ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis . Genes Dev. 2007;21: 2220–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baudat F, de Massy B. Regulating double strand break repair towards crossover and noncrossover during mammalian meiosis. Chromosome Res. 2007;15: 567–577. [DOI] [PubMed] [Google Scholar]
- 32. Vignard J, Siwiec T, Chelysheva L, Vrielynck N, Gonord F, Armstrong, et al. The interplay of RecA-related proteins and the MND1-HOP2 complex during meiosis in Arabidopsis thaliana . PLoS Genet 2007;3: e176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ferdous M, Higgins JD, Osman K, Lambing C, Roitinger E, Mechtler K, et al. Inter-homolog crossing-over and synapsis in Arabidopsis meiosis are dependent on the chromosome axis protein AtASY3. PLoS Genet 2012;8: e1002507 10.1371/journal.pgen.1002507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Knoll A, Higgins JD, Seeliger K, Reha SJ, Dangel NJ, Bauknecht M, et al. The Fanconi anemia ortholog FANCM ensures ordered homologous recombination in both somatic and meiotic cells in Arabidopsis . Plant Cell 2012;24: 1448–1464. 10.1105/tpc.112.096644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kurzbauer MT, Uanschou C, Chen D, Schlögelhofer P. The recombinases DMC1 and RAD51 are functionally and spatially separated during meiosis in Arabidopsis . Plant Cell. 2012;24: 2058–2070. 10.1105/tpc.112.098459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Crismani W, Girard C, Froger N, Pradillo M, Santos JL, Chelysheva L, et al. FANCM limits meiotic crossovers. Science 2012;336: 1588–1590. 10.1126/science.1220381 [DOI] [PubMed] [Google Scholar]
- 37. Higgins JD, Sánchez-Moran E, Armstrong SJ, Jones GH, Franklin FCH. The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 2005;19: 2488–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kauppi L, Barchi M, Lange J, Baudat F, Jasin M, Keeney S. Numerical constraints and feedback control of double-strand breaks in mouse meiosis. Genes Dev. 2013;27: 873–886. 10.1101/gad.213652.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Siaud N, Dray E, Gy I, Gérard E, Takvorian N, Doutriaux MP. Brca2 is involved in meiosis in Arabidopsis thaliana as suggested by its interaction with Dmc1. EMBO J. 2004;23: 1392–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Seeliger K, Dukowic-Schulze S, Wurz-Wildersinn R, Pacher M, Puchta H. BRCA2 is a mediator of RAD51- and DMC1-facilitated homologous recombination in Arabidopsis thaliana . New Phytol. 2012;193: 364–375. 10.1111/j.1469-8137.2011.03947.x [DOI] [PubMed] [Google Scholar]
- 41. Lafarge S, Montané MH. Characterization of Arabidopsis thaliana ortholog of the human breast cancer susceptibility gene 1: AtBRCA1, strongly induced by gamma rays. Nucleic Acids Res. 2003;31: 1148–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Foray N, Marot D, Gabriel A, Randrianarison V, Carr AM, Perricaudet M, et al. A subset of ATM- and ATR-dependent phosphorylation events requires the BRCA1 protein. EMBO J. 2003;22: 2860–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen Z, Higgins JD, Hui JTL, Li J, Franklin FCH, Berger F. Retinoblastoma protein is essential for early meiotic events in Arabidopsis . EMBO J. 2011;30: 744–755. 10.1038/emboj.2010.344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Uanschou C, Siwiec T, Pedrosa-Harand A, Kerzendorfer C, Sánchez-Morán E, Novatchkova M, et al. A novel plant gene essential for meiosis is related to the human CtIP and the yeast COM1/SAE2 gene. EMBO J. 2007;26: 5061–5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bleuyard JY, Gallego ME, White CI. Meiotic defects in the Arabidopsis rad50 mutant point to conservation of the MRX complex function in early stages of meiotic recombination. Chromosoma. 2004;113: 197–203. [DOI] [PubMed] [Google Scholar]
- 46. Puizina J, Siroky J, Mokros P, Schweizer D, Riha K. Mre11 deficiency in Arabidopsisis associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant Cell. 2004;16: 1968–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Neale MJ, Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442: 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cloud V, Chan YL, Grubb J, Budke B, Bishop DK. Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science. 2012;337: 1222–1225. 10.1126/science.1219379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nimonkar AV Dombrowski CC, Siino JS, Stasiak AZ, Stasiak A, Kowalczykowski SC. Saccharomyces cerevisiae Dmc1 and Rad51 proteins preferentially function with Tid1 and Rad54 proteins, respectively, to promote DNA strand invasion during genetic recombination. J Biol Chem. 2012;287 28727–28737. 10.1074/jbc.M112.373290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Da Ines O, Degroote F, Goubely C, Amiard S, Gallego ME, White CI. Meiotic recombination in Arabidopsis is catalysed by DMC1, with RAD51 playing a supporting role. PLoS Genet. 2013;9: e1003787 10.1371/journal.pgen.1003787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43: 525–558. 10.1146/annurev-genet-102108-134233 [DOI] [PubMed] [Google Scholar]
- 52. Schubert V, Weissleder A, Ali H, Fuchs J, Lermontova I, Meister A, et al. Cohesin gene defects may impair sister chromatid alignment and genome stability in Arabidopsis thaliana . Chromosoma. 2009;118: 591–605. 10.1007/s00412-009-0220-x [DOI] [PubMed] [Google Scholar]
- 53. Watanabe K, Pacher M, Dukowic S, Schubert V, Puchta H, Schubert I. The STRUCTURAL MAINTENANCE OF CHROMOSOMES 5/6 complex promotes sister chromatid alignment and homologous recombination after DNA damage in Arabidopsis thaliana . Plant Cell. 2009;21:2688–2699. 10.1105/tpc.108.060525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Copenhaver GP, Housworth E, Stahl FW. Crossover interference in Arabidopsis . Genetics. 2002;160: 1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Börner GV, Kleckner N, Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117: 29–45. [DOI] [PubMed] [Google Scholar]
- 56. Lam WS, Yang X, Makaroff CA. Characterization of Arabidopsis thaliana SMC1 and SMC3: evidence that AtSMC3 may function beyond chromosome cohesion. J Cell Sci. 2005;118: 3037–3048. [DOI] [PubMed] [Google Scholar]
- 57. de los Santos T, Hunter N, Lee C, Larkin B, Loidl J, Hollingsworth NM. The Mus81/Mms4 endonuclease acts independently of double Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics. 2003;164: 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Berchowitz LE, Francis KE, Bey AL, Copenhaver GP. The role of AtMUS81 in interference-insensitive crossovers in A. thaliana . PLoS Genet. 2007;3: e132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Berchowitz LE, Copenhaver GP. Genetic interference: don’t stand so close to me. Curr Genomics. 2010;11: 91–102. 10.2174/138920210790886835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pradillo M, Santos JL. The template choice decision in meiosis: is the sister important? Chromosoma. 2011;120: 447–454. 10.1007/s00412-011-0336-7 [DOI] [PubMed] [Google Scholar]
- 61. Higgins JD, Armstrong SJ, Franklin FCH, Jones GH. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis . Genes Dev. 2004;18: 2557–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Higgins JD, Buckling EF, Franklin FC, Jones GH. Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J. 2008;54: 152–162. 10.1111/j.1365-313X.2008.03403.x [DOI] [PubMed] [Google Scholar]
- 63. Higgins JD, Vignard J, Mercier R, Pugh AG, Franklin FCH, Jones GH. AtMSH5 partners AtMSH4 in the class I meiotic crossover pathway in Arabidopsis thaliana, but is not required for synapsis. Plant J. 2008;55: 28–39. 10.1111/j.1365-313X.2008.03470.x [DOI] [PubMed] [Google Scholar]
- 64. Cole F, Kauppi L, Lange J, Roig I, Wang R, Keeney S. Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat Cell Biol. 2012;14: 424–430. 10.1038/ncb2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chelysheva L,Vezon D, Belcram K, Gendrot G, Grelon M. The Arabidopsis BLAP75/Rmi1 homologue plays crucial roles in meiotic double-strand break repair. PLoS Genet. 2008;4: e1000309 10.1371/journal.pgen.1000309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Knoll A, Schröpfer S, Puchta H. The RTR complex as caretaker of genome stability and its unique meiotic function in plants. Front Plant Sci. 2014;5: 33 10.3389/fpls.2014.00033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Martini E, Diaz RL, Hunter N, Keeney S. Crossover homeostasis in yeast meiosis. Cell. 2006;126: 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Joshi N, Brown MS, Bishop DK, Börner GV. Gradual implementation of the meiotic recombination program via checkpoint pathways controlled by global DSB levels. Mol Cell. 2015;57:797–811. 10.1016/j.molcel.2014.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bishop DK, Nikolski Y, Oshiro J, Chon J, Shinohara M, Chen X. High copy number suppression of the meiotic arrest caused by a dmc1 mutation: REC114 imposes an early recombination block and RAD54 promotes a DMC1-independent DSB repair pathway. Genes Cells. 1999;4: 425–44. [DOI] [PubMed] [Google Scholar]
- 70. Tsubouchi H, Roeder GS. The importance of genetic recombination for fidelity of chromosome pairing in meiosis. Dev Cell. 2003;5: 915–925. [DOI] [PubMed] [Google Scholar]
- 71. Hong S, Sung Y, Yu M, Lee M, Kleckner N, Kim KP. The logic and mechanism of homologous recombination partner choice. Mol Cell. 2013;51: 440–53. 10.1016/j.molcel.2013.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lao JP, Cloud V, Huang CC, Grubb J, Thacker D, Lee CY, et al. Meiotic crossover control by concerted action of rad51-dmc1 in homolog template bias and robust homeostatic regulation. PLoS Genet. 2013;9: e1003978 10.1371/journal.pgen.1003978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Uanschou C, Ronceret A, Von Harder M, De Muyt A, Vezon D, Pereira L, et al. Sufficient amounts of functional HOP2/MND1 complex promote interhomolog DNA repair but are dispensable for intersister DNA repair during meiosis in Arabidopsis . Plant Cell. 2013;25: 4924–4940. 10.1105/tpc.113.118521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Couteau F, Belzile F, Horlow C, Grandjean O, Vezon D, Doutriaux MP. Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis . Plant Cell. 1999;11: 1623–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Liu Y, Gaines WA, Callender T, Busygina V, Oke A, Sung P, et al. Down-regulation of Rad51 activity during meiosis in yeast prevents competition with Dmc1 for repair of double-strand breaks. PLoS Genet. 2014;10: e1004005 10.1371/journal.pgen.1004005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Albini SM. A karyotype of the Arabidopsis thaliana genome derived from synaptonemal complex analysis at prophase I of meiosis. Plant J. 1994;5: 665–672. [Google Scholar]
- 77. Chelysheva L, Gendrot G, Vezon D, Doutriaux MP, Mercier R, Grelon M. Zip4/Spo22 is required for class I CO formation but not for synapsis completion in Arabidopsis thaliana . PLoS Genet. 2007;3: e83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Drouaud J, Mercier R, Chelysheva L, Bérard A, Falque M, Martin O, et al. Sex-specific crossover distributions and variations in interference level along Arabidopsis thaliana chromosome 4. PLoS Genet. 2007;3: e106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Novak JE, Ross-Macdonald PB, Roeder GS. The budding yeast Msh4 protein functions in chromosome synapsis and the regulation of crossover distribution. Genetics. 2001;158: 1013–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Neyton S, Lespinasse F, Moens PB, Paul R, Gaudray P, Paquis-Flucklinger V, et al. Association between MSH4 (MutS homologue 4) and the DNA strand-exchange RAD51 and DMC1 proteins during mammalian meiosis. Mol Hum Reprod. 2004;10: 917–924. [DOI] [PubMed] [Google Scholar]
- 81. Zickler D, Kleckner N. Integrating Structure and Function. Annu Rev Genet. 1999;33:603–754. [DOI] [PubMed] [Google Scholar]
- 82. Bishop DK. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell. 1994;79: 1081–1092. [DOI] [PubMed] [Google Scholar]
- 83. San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77: 229–257. 10.1146/annurev.biochem.77.061306.125255 [DOI] [PubMed] [Google Scholar]
- 84. Carballo J, Johnson AL, Sedgwick SG, Cha RS. Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell. 2008;132: 758–770. 10.1016/j.cell.2008.01.035 [DOI] [PubMed] [Google Scholar]
- 85. Leyser O, Furner IJ. Characterisation of three shoot apical meristem mutants of Arabidopsis thaliana . Development. 1992;116: 397–403 [Google Scholar]
- 86. Lario LD, Ramirez-Parra E, Gutierrez C, Spampinato CP, Casati P. ANTI-SILENCING FUNCTION1 proteins are involved in ultraviolet-induced DNA damage repair and are cell cycle regulated by E2F transcription factors in Arabidopsis . Plant Physiol. 2013;162: 1164–1177. 10.1104/pp.112.212837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Gerlach WL, Bedbrook JR. Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res. 1979;7: 1869–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Campell BR, Song Y, Posch TE, Cullis CA, Town CD. Sequence and organization of 5S ribosomal RNA-encoding genes of Arabidopsis thaliana . Gene. 1992;112: 225–228. [DOI] [PubMed] [Google Scholar]
- 89. Martínez-Zapater JM, Estelle MA, Somerville C. A highly repeated DNA sequence in Arabidopsis thaliana . Mol Gen Genet. 1986;204: 417–423. [Google Scholar]
- 90. Richards EJ, Ausubel FM. Isolation of a higher eukaryotic telomere from Arabidopsis thaliana . Cell. 1988;53: 127–136. [DOI] [PubMed] [Google Scholar]
- 91. Armstrong SJ, Sánchez-Morán E, Franklin FCH. Cytological analysis of Arabidopsis thaliana meiotic chromosomes. Methods Mol Biol. 2009;558: 131–145. 10.1007/978-1-60761-103-5_9 [DOI] [PubMed] [Google Scholar]
- 92. Armstrong SJ, Caryl AP, Jones GH, Franklin FCH. Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica . J Cell Sci. 2002;115: 3645–3655. [DOI] [PubMed] [Google Scholar]
- 93. Mercier R, Armstrong SJ, Horlow C, Jackson NP, Makaroff CA, Vezon D, et al. The meiotic protein SWI1 is required for axial element formation and recombination initiation in Arabidopsis . Development. 2003;130: 3309–3318. [DOI] [PubMed] [Google Scholar]
- 94. Jackson N, Sánchez-Moran E, Buckling E, Armstrong SJ, Jones GH, Franklin FC. Reduced meiotic crossovers and delayed prophase I progression in AtMLH3-deficient Arabidopsis . EMBO J. 2006;25: 1315–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Larionov A, Krause A, Miller W. A standard curve based method for relative real time PCR data processing. BMC Bioinformatics. 2005;21: 6–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Squash procedure to visualize and estimate the number of ovules per gynoecium (A) on WT and (B) on Atfas1-4.
(TIF)
Chromosome spread preparations from (A, C, E, G) Atfas1-2 and (B, D, F, H) AtASF1a/AtASF1b RNAi PMCs. (A, B) Pachytene. (C, D) Metaphase I: four ring bivalents and one rod bivalent on each. (E, F) Prophase II. (G, H) Tetrad. Bars = 5µm.
(TIF)
FISH using the centromeric probe pAL1 (180 bp) (A) on WT and (B) on Atfas1-4 at pachytene. In the mutant the centromeric FISH signal is weaker, less defined and more extended than in WT.
(TIF)
Single strands of DNA are shown as either blue (parent 1) or red (parent 2) rods. AtSPO11 helped by other proteins initiates programmed DSBs. H2AX phosphorylation occurs at the break zone and DSBs are resected 5’ to 3’ to produce single ssDNA tails by the MRN complex and COM1. One of these ends invades the homologous duplex DNA, giving raise a D loop intermediate mediated by AtRAD51 and AtDMC1 and other proteins. If the second end is captured and the broken DNA strands are ligated, a dHJ is formed. This intermediate is resolved as CO1, sensitive to interference, or NCO upon appropriate resolution of the two HJs. On the other hand, this dHJ can be dissolved as a NCO. Alternatively, the D loop can be processed to generate a CO2 (insensitive to interference). When the D-loop is dissociated before the second end capture SDSA pathway occurs, the invading strand dissociates after DNA synthesis. This strand then re-anneals to the original parent, resulting in repair of the DSB and a heteroduplex DNA. This pathway is always resolved as a NCO.
(TIF)
(A-C) Dual immunolocalization on Atspo11-1-5 nuclei. (A) AtASY1 (green) and γH2AX (red). (B) AtASY1 (green) and AtRAD51 (red). (C) AtASY1 (green) and AtDMC1 (red). Bars = 5µm.
(TIF)
(TIF)
(PDF)
All the analyzed plants were heterozygous for the fluorescent and non-fluorescent allele and homozygous for Atfas1-4.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.