Abstract
Introduction
Diverse and multi-factorial processes contribute to the progression of cardiovascular disease. These processes affect cells involved in the development of this disease in varying ways, ultimately leading to atherothrombosis. The goal of our study was to compare the differential effects of specific stimuli – two bacterial infections and a Western diet – on platelet responses in ApoE-/- mice, specifically examining inflammatory function and gene expression. Results from murine studies were verified using platelets from participants of the Framingham Heart Study (FHS; n = 1819 participants).
Methods
Blood and spleen samples were collected at weeks 1 and 9 from ApoE-/- mice infected with Porphyromonas gingivalis or Chlamydia pneumoniae and from mice fed a Western diet for 9 weeks. Transcripts based on data from a Western diet in ApoE-/- mice were measured in platelet samples from FHS using high throughput qRT-PCR.
Results
At week 1, both bacterial infections increased circulating platelet-neutrophil aggregates. At week 9, these cells individually localized to the spleen, while Western diet resulted in increased platelet-neutrophil aggregates in the spleen only. Microarray analysis of platelet RNA from infected or Western diet-fed mice at week 1 and 9 showed differential profiles. Genes, such as Serpina1a, Ttr, Fgg, Rpl21, and Alb, were uniquely affected by infection and diet. Results were reinforced in platelets obtained from participants of the FHS.
Conclusion
Using both human studies and animal models, results demonstrate that variable sources of inflammatory stimuli have the ability to influence the platelet phenotype in distinct ways, indicative of the diverse function of platelets in thrombosis, hemostasis, and immunity.
Introduction
Cardiovascular disease (CVD) is a chronic inflammatory process with multiple etiologies, including genetic and environmental factors. Bacterial infections and obesity are two known pro-inflammatory contributors to the development of CVD. The effects of these inflammatory stimuli on vascular cells have been extensively studied, with the exception for the second most abundant cell in circulation, the platelet. Particularly, in light of work which shows the relationship between platelet transcripts and CVD in FHS [1,2] and their new role in inflammation [3,4,5], questions still remain as to how platelets contribute to the development of CVD. The goals of our study were to characterize the differential effects of two common bacterial infections and a Western diet on platelet inflammatory function and gene transcripts in ApoE-/- mice and in human samples. These results will help to better understand the connections between inflammatory stimuli, cardiovascular disease, and platelets.
One bacterium associated with CVD is the gram-negative oral pathogen Porphyromonas gingivalis (P. gingivalis), which induces low-grade chronic inflammation associated with oral bone loss, clinically known as periodontal disease. This bacterium expresses several outer membrane proteins which are recognized by the innate immune system through Toll-like receptor 2 (TLR2). In ApoE-/- mice, P. gingivalis accelerates atherosclerotic plaque size [6,7,8], with accumulation of lipids, T cells, and macrophages compared to uninfected mice [7]. Plaque collagen and elastin content is also altered with infection, suggesting that P. gingivalis infection affects the stability of atherosclerotic plaques [7]. In humans, P. gingivalis is detected in atherosclerotic plaques [9,10] and is associated with the development of carotid artery atherosclerosis [11].
Another bacterium associated with CVD is the obligate intracellular gram-negative bacterium, Chlamydia pneumoniae (C. pneumoniae). It is associated with atypical pneumonia, as well as pharyngitis, bronchitis, and sinusitis in humans. This bacterium is also recognized in part by TLR2 on innate immune cells. It has also been demonstrated to increase atherosclerotic plaque size, lipid content, and dendritic cells in ApoE-/- mice compared to uninfected mice [12]. In humans, C. pneumoniae antibody seropositivity is associated with the progression of coronary artery calcification [13] and risk of coronary heart disease and myocardial infarction[14]. C. pneumoniae has also been identified through PCR methodology in atherosclerotic plaques in humans [15,16].
Obesity, having a body mass index (BMI) ≥30 kg/m2, is another key risk factor associated with CVD. In the US, 35% of adults are considered obese [17]. For individuals ≥20 years of age surveyed between 2009 and 2010, 35.8% were obese and 72.5% had a health score of 0–1, classifying them in poor cardiovascular health [17]. Consistent with the importance of weight and diet, in animal models of atherosclerosis, feeding animals a high fat diet increases atherosclerotic plaque size, number, and severity [18,19,20].
We have previously demonstrated that in human and mouse platelets, P. gingivalis is recognized by TLR2 and induces aggregation [21] and heterotypic aggregate formation [3,4]. C. pneumoniae also binds to platelets to induce aggregation and P-selectin expression [22]. A Western diet in C57BL/6J mice increases thrombin-induced platelet adhesion [3]. The goal of this study was to characterize the differential effects of these two common bacterial infections and a Western diet on platelet inflammatory function and gene transcripts at different timepoints in a well-established atherosclerosis murine model prior to the formation of plaques. These results were also correlated with recent observations from aortic tissue following exposure to the same inflammatory stimuli. [23]. Our results suggest that both inflammatory stimuli and the kinetics of the response differentially influence platelet transcripts and inflammatory function. Furthermore, our results were corroborated in human studies and provide insight into how platelets alter the development of CVD.
Materials and Methods
Animals
Mice were cared for at the Boston University School of Medicine (BUSM) Vivarium. All work was approved by BUSM Institutional Animal Care and Use Committee. Mice were housed in a 12 hour light cycle at constant temperature and humidity with free access to food and water. During procedures, mice were monitored and removed if exhibiting signs of distress. P. gingivalis strain 381 and C. pneumoniae strain AO3 were grown and purified as previously described [24,25]. One set of ApoE-/- male mice (Jackson Laboratories) were treated with 4% sulfamethoxazole in their drinking water for 10 days to clear normal oral microflora [23]. These mice were then challenged 5 times per week for 3 weeks on the buccal surface of their maxilla with 1x109 CFU P. gingivalis in 2% carboxymethyl cellulose. An additional set of ApoE-/- male mice were challenged intranasally once per week for three weeks with 2x106 IFU C. pneumoniae suspended in sucrose-phosphate-glutamate buffer while under light anesthesia of ketamine and xylazine [23]. Mice were sacrificed by CO2 asphyxiation followed by cervical dislocation at an early timepoint 1 day after the last P. gingivalis challenge (~9 weeks of age at start) and 4 days after the last C. pneumoniae exposure (~9 weeks of age at start), with n = 3 for each group [23]. Another group was sacrificed 9 weeks later (~6 weeks of age at start for both groups), with n = 3 for each group, prior to the formation of overt aortic plaque formation as seen at later timepoints with these stimuli [7,12,23,26]. A final group of ApoE-/- male mice (~16 weeks of age at start) were fed a Western diet (17.3% protein, 48.5% carbohydrates, 1.2% fat, and 0.2% cholesterol; Teklad Harlan) for 9 weeks and then sacrificed with n = 6 for the Western diet [23]. Analysis was performed on samples collected from mice at each timepoint.
Whole Blood Cell and Platelet Counts
Blood collected from each mouse was analyzed for white blood cell (WBC) and platelet content using a COULTER AC.T Series Analyzer (Becton Dickinson) [3].
Heterotypic Aggregate Formation
A sample of whole blood from each mouse was dual stained with anti-mouse CD41 FITC-conjugated and anti-mouse Ly6G PE-Cy7-conjugated antibodies and appropriate isotype controls (eBioscience; Cat# 11–0411 and 25–5931, respectively). The percent of platelet-positive neutrophils was determined using a FACSCalibur Flow Cytometer with Cell Quest Software (BD Bioscience) [3,4].
Spleen Sectioning and Staining
Frozen spleen sections were stained with anti-mouse CD41 (Abcam; Cat# ab63983) and anti-mouse Ly6G and Ly6C (BD Bioscience; Cat# 553123) antibodies and appropriate isotype controls, followed by anti-rabbit IgG Alexa Fluor 405-conjugated and anti-rat Texas Red-X secondary antibodies (Life Technologies; Cat# A-31556 and T-6392, respectively). Samples were visualized using Leica TCS SP5 II Confocal Microscope with Leica Application Suite Advanced Fluorescence Version 2.6.0.7266 software then analyzed with ImageJ (v1.46r) and Adobe Photoshop CS3 (v13.0.1).
Platelet RNA Isolation
Platelets were isolated from blood samples using a series of centrifugation steps. Briefly, samples were centrifuged at 450xg for 7 minutes. The top layer consisting of platelet rich plasma (PRP) was removed and diluted 2.33X with platelet wash buffer (10 mM sodium citrate, 150 mM sodium chloride, 1 mM EDTA, 1% dextrose, pH 7.4) and 1:10000 prostaglandin E1 (Calbiochem). Samples were centrifuged at 300xg for 4 minutes to remove any contaminating WBC. The supernatant was further diluted 3X with platelet wash buffer and PGE1, then centrifuged at 3500xg for 10 minutes [3]. The resulting platelet pellet was lysed and RNA was isolated using the miRNeasy Kit (Qiagen).
RNA Microarray and Analysis
RNA from each mouse in each condition was pooled to generate at least 50 ng of RNA per sample. Samples were reverse transcribed using Ovation Pico WTA System V2 (Nugen) and purified using Agencourt RNA Clean XP Purification Beads. After SPIA amplification and purification, samples were labeled using the Encore Biotin Module (Nugen) and hybridized to the GenChip Mouse Gene 1.0 ST Array (Affymetrix), stained, and scanned using an Affymetrix GeneArray Scanner 3000 7G Plus. All procedures were performed by the Microarray Resource Facility at Boston University. Affymetrix CEL files were normalized to produce gene-level expression values using the Robust Multiarray Average (RMA) [27] in the Affymetrix Bioconductor (v1.28.1) [28] and an Entrez Gene-specific probe set (v14.0.0) mapping from the Molecular Behavioral Neuroscience Institute at the University of Michigan (http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF) [29]. Array quality was assessed by computing Relative Log Expression (RLE) and Normalized Unscaled Standard Error (NUSE) using the AffyPLM Bioconductor (v1.26.1) [30]. All microarray analyses were performed using the R environment (v2.15.1) for statistical computing. Gene Set Enrichment Analysis (GSEA; v2.0.13) [31] was used to identify biological terms, pathways, and processes. By using the normalized enrichment score (NES) of ≥2/-2, the false discovery rate (FDR q) of <0.05, and the nominal p-value (Nom p) of <0.05, we identified gene sets that were significantly enriched for and would have been affected the most by treatment. The Entrez Gene identifiers of the mouse homologs of the genes (as determined using HomoloGene v65 [32]) were ranked according to the log2-transformed fold change computed for each comparison; in cases where a given mouse gene mapped to more than one human gene, one homolog was randomly chosen. These ranked lists were then used to perform pre-ranked GSEA analyses (default parameters with random seed 1234) using Entrez Gene versions of the Biocarta, KEGG, Reactome, and Gene Ontology (GO) gene sets obtained from the Molecular Signature Database (MsigDB; v3.0) [33]. Post analysis information on transcript alternative names and functions were determined using genecard.org and ncbi.nlm.nih.gov. Heatmaps were created using MeV v4.9.0 [34].
qRT-PCR Verification
Isolated RNA was converted to cDNA using High-Capacity cDNA Reverse Transcription Kit (Life Technologies), followed by pre-amplification using TaqMan PreAmp Master Mix (Life Technologies). qRT-PCR was performed on the Applied Biosystems 7900 HT Fast Real-Time PCR System with SDS 2.2.2 Software (Life Technologies) using primers and probes for the following genes (Life Technologies): microfibrillar-associated protein 1a (MFAP1A), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β-actin (ACTB), and β2-microglobulin (B2M). Analysis included correcting by the mean of the three housekeeping genes (ΔCt)–GAPDH, ACTB, and B2M – followed by calculating the difference between treatment and Untreated Controls (ΔΔCt). Fold change (graphed) was then determined by calculating 2-ΔΔCt.
Framingham Heart Study
As previously described [1,2,3], platelet RNA was isolated from 1819 participants of the FHS Offspring Cohort Examination 8 using miRNeasy kit (Qiagen) and converted to cDNA. Pre-amplification was performed using TaqMan PreAmp Master Mix (Life Technologies). Quantitative RT-PCR was performed using the BioMark System (Fluidigm) with primers and probes from Life Technologies. Results were compared to data collected from the medical history and physical examination of each participant. Individuals were categorized as having hypertension if they had systolic blood pressure ≥140 mm Hg, diastolic blood pressure ≥90 mm Hg, or were receiving treatment for hypertension. They were categorized as having diabetes if their fasting blood glucose was ≥126 mg/dL, their non-fasting blood glucose was ≥200 mg/dL, or were being treated with insulin or hypoglycemic drugs. Finally, individuals were categorized as smokers if they had smoked 1 cigarette a day in the year prior to examination. All of this work with the FHS was approved by the University of Massachusetts Medical School and Boston University Medical Center Institutional Review Boards and all participants gave written informed consent [1,2,3]. Interleukin 6 (IL6) and C-reactive protein (CRP) levels were measured as previously described [2].
Statistics
Data from ApoE-/- mouse studies were reported as the mean ± the standard deviation. In some cases, the percent compared to Untreated Controls, which was set to 100%, was reported. All data were analyzed by ANOVA followed by Tukey’s Multiple Comparison Test using GraphPad Prism 5 software. Data were considered normally distributed, with a two-sided p<0.05 being significant.
For the FHS, descriptive statistics were reported as the mean ± standard deviation for continuous variable or the count (percent) for binary variables. Quantitative RT-PCR cycle threshold (Ct) values were corrected (ΔCT) by the geometric mean of three housekeeping genes–GAPDH, ACTB, B2M. Genes not expressed and failing to report a Ct value within a set period were assigned the maximum Ct value allowed in our procedure (28 cycles). Multivariable linear regression models for gene expression (ΔCt) were fitted adjusting for the following confounders–BMI, smoking status, total cholesterol, HDL cholesterol, triglycerides, systolic blood pressure, diastolic blood pressure, glucose levels, diabetes, coronary heart disease [angina pectoris, coronary insufficiency, myocardial infarction adjudicated by a review panel], lipid-lowering therapy, hormone replacement therapy, antihypertensive therapy, and regular aspirin therapy (at least 3 times a week). All statistical analysis was performed using Stata 12.0 software [1,2,3]. Statistical significance was assessed using two-sided p-value, with those <0.05 being considered statistically significant.
Results
Differential Platelet Gene Expression in ApoE-/- Mice Infected with Bacteria or Fed a Western Diet
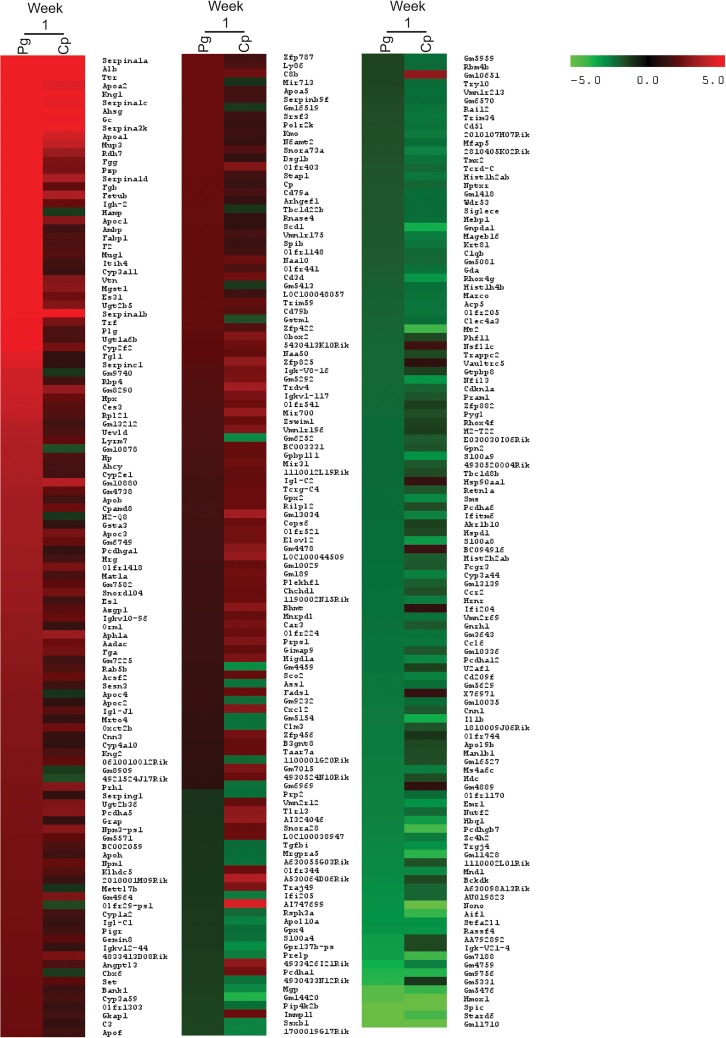
ApoE-/- mice (n = 3) were infected orally with P. gingivalis or intranasally with C. pneumoniae. One day after the last oral infection with P. gingivalis and 4 days after the last intranasal infection with C. pneumoniae (week 1), platelet RNA was isolated and pooled to generate a sufficient amount of RNA per condition for microarray analysis. Due to the small sample size, only genes up- or downregulated 2-fold or greater and pathways with a normalized enrichment score (NES) ≥ ~2/-2, nominal p-value (Nom p) <0.05, and a false discovery rate (FDR q) <0.05 were considered. Additionally, this data was intended to generate hypotheses on how platelets and platelet transcripts are affected by different inflammatory stimuli over time. During Week 1, we observed increased expression of 129 genes and decreased expression in 59 genes following P. gingivalis infection compared to Untreated Controls (Fig 1). The top five upregulated genes with known function were SERPINA1A (27.09-fold), ALB (17.83-fold), TTR (15.84-fold), APOA2 (15.08-fold), and KNG1 (13.01-fold). The top 5 downregulated genes were STARD6 (-5.49-fold), SPIC (-5.42-fold), HMOX1 (-4.60-fold), IGK-V21-4 (-3.08-fold), and RASSF4 (-2.89-fold). Using Gene Set Enrichment Analysis (GSEA), we identified gene sets with increased expression following P. gingivalis infection with an NES ≥2, a Nom p <0.05, and an FDR q <0.05. These gene sets included coagulation, lipids, signaling pathways, RNA/gene expression, and inflammation (S1 Table). Gene sets, which exhibited decreased expression, included those involved in enzyme activity, differentiation, metabolism, extracellular matrix related, signaling, and cytoskeleton pathways; however, these gene sets were not significant (NES ˃-2, FDR q ˃0.05; S2 Table).
Fig 1. Genes upregulated and downregulated at week 1 in ApoE-/- mice with bacterial infection.
Heatmap shows the transcripts identified through microarray as upregulated or downregulated 2-fold or more with either P. gingivalis (Pg) or C. pneumoniae (Cp) infection compared to Untreated Control in the ApoE-/- mice. Each condition represents RNA from 3 mice pooled.
In ApoE-/- mice infected with C. pneumoniae at week 1, there were 87 platelet transcripts increased and 70 transcripts decreased 2-fold or more compared to Untreated Control (Fig 1). The top 5 genes that exhibited an increase in expression were SERPINA1A (13.17-fold), ALB (7.75-fold), SERPINA3K (6.47-fold), TTR (6.20-fold), and KNG1 (5.14-fold). The top 5 genes that exhibited a decrease in expression were NONO (-10.89-fold), SPIC (-6.92-fold), HMOX1 (-5.15-fold), PCDHGB7 (-4.09-fold), and STARD6 (-4.09-fold). GSEA identified the following positively enriched gene sets: coagulation, lipids, metabolism, signaling pathways, cytoskeleton, and RNA/gene expression (NES ≥2, Nom p <0.05, FDR q <0.05; S3 Table). As seen with P. gingivalis, negatively enriched genes, including those involved in enzyme activity, signaling, and extracellular matrix, were affected but not significantly (NES >-2, FDR q >0.05; S4 Table).
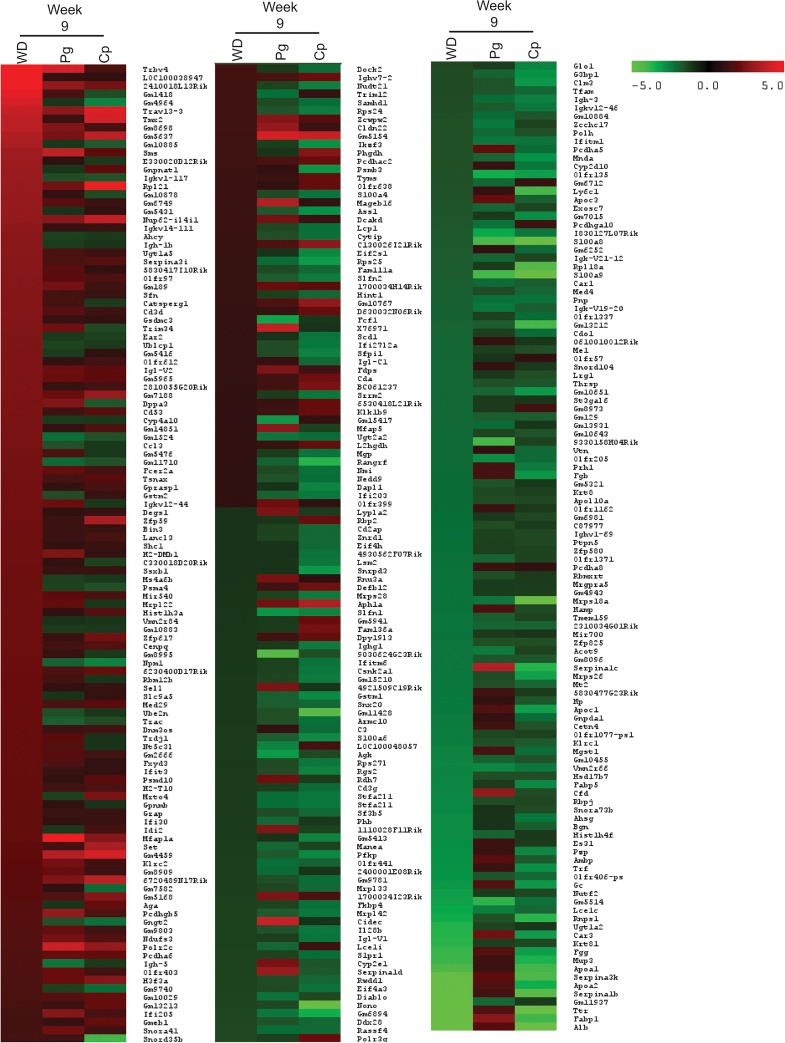
Nine weeks following the last infection with P. gingivalis, C. pneumoniae, or 9 weeks of feeding a Western diet, ApoE-/- mice were sacrificed in order to understand the changes in platelet transcripts before the formation of overt plaques. Following P. gingivalis infection, only 41 genes were increased and 32 were decreased (Fig 2). The top 5 genes that exhibited increased expression were MFAP1A (5.31-fold), TRBV4 (4.21-fold), POLR2C (3.99-fold), CIDEC (3.97-fold), and SMS (3.75-fold). Top 5 genes that exhibited decreased expression were S100A8 (-4.15-fold), S100A9 (-4.08-fold), OLFR135 (-3.45-fold), FCF1 (-3.26-fold), and AGK (-3.01-fold). GSEA identified numerous gene sets positively enriched, including coagulation and signaling pathways; however, these gene sets were not significantly altered by P. gingivalis infection (NES >-2, FDR q >0.05; S5 Table). It was the negatively enriched gene sets that were more significantly affected, including RNA/gene expression (NES ≤-2, Nom p <0.05, FDR q <0.05; S6 Table).
Fig 2. Genes upregulated and downregulated at week 9 in ApoE-/- mice with bacterial infection or a Western diet.
Heatmap shows the transcripts identified through microarray as upregulated or downregulated 2-fold or more by infection with P. gingivalis (Pg), C. pneumoniae (Cp), or fed a Western diet (WD) compared to Untreated Control in the ApoE-/- Mice. Each condition represents RNA from 3 mice pooled.
Nine weeks following the last infections with C.pneumoniae, there was differential effects on gene expression profiles as compared to that observed early after infection. Twenty-seven genes were increased and 109 decreased at week 9 compared to Untreated Control (Fig 2). The top 5 genes with increased expression at week 9 were RPL21 (5.52-fold), TRAV13-3 (4.34-fold), TMX2 (4.16-fold), NUP62-IL4I1 (3.81-fold), and APH1A (3.48-fold). The top 5 genes with decreased expression at week 9 were ALB (-16.46-fold), S100A9 (-9.38-fold), S100A8 (-9.38-fold), NONO (-6.79-fold), and TTR (-5.46-fold). Similarly to P. gingivalis at week 9, GSEA identified numerous gene sets positively enriched but that were not significantly affected by C. pneumoniae (S7 Table). Gene sets negatively enriched at week 9 were significant, including coagulation, RNA/gene expression, and inflammation (NES <-2, Nom p <0.01, FDR q <0.05; S8 Table).
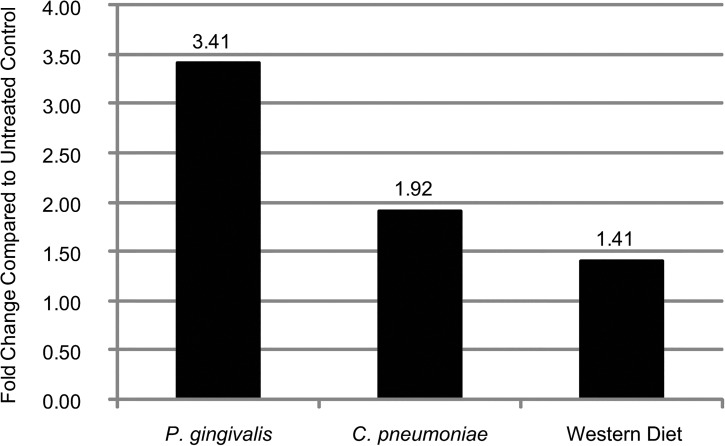
In mice fed a Western diet for 9 weeks, there were 67 genes increased and 67 genes decreased (Fig 2). The top 5 genes that exhibited increased expression as a result of a Western diet were TRBV4 (7.14-fold), TRAV13-3 (3.90-fold), TMX2 (3.74-fold), SMS (3.14-fold), and GNPNAT1 (3.05-fold). The top 5 genes with decreased expression with a Western diet were ALB (-18.34-fold), FABP1 (-6.89-fold), TTR (-5.51-fold), SERPINA1B (-4.95-fold), and APOA2 (-4.95-fold). Numerous gene sets were positively enriched, including those associated with DNA/proliferation, inflammation, RNA/gene expression, and signaling (NES >2, Nom p <0.05, FDR q <0.05; S9 Table). Only a few gene sets were negatively enriched, including those involved in coagulation and lipid (NES <-2, Nom p <0.01, FDR q <0.05; S10 Table). As a way to verify these results, qRT-PCR was run on week 9 samples for MFAP1A. As seen in Fig 3, there is an increase in its expression with P. gingivalis infection that is greater than the increase with C. pneumoniae infection or a Western diet, similar to the findings in the microarray.
Fig 3. Verification of microarray analysis through qRT-PCR.
Gene expression for MFAP1A at Week 9 identified through the microarray were verified using qRT-PCR. Each condition represents RNA from 3 mice pooled.
Differential Physiological Effects of Bacterial Infections or a Western Diet in ApoE-/- Mice
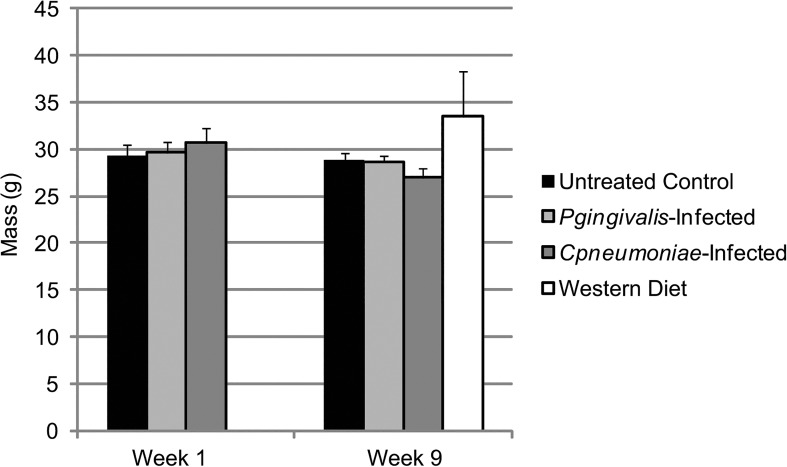
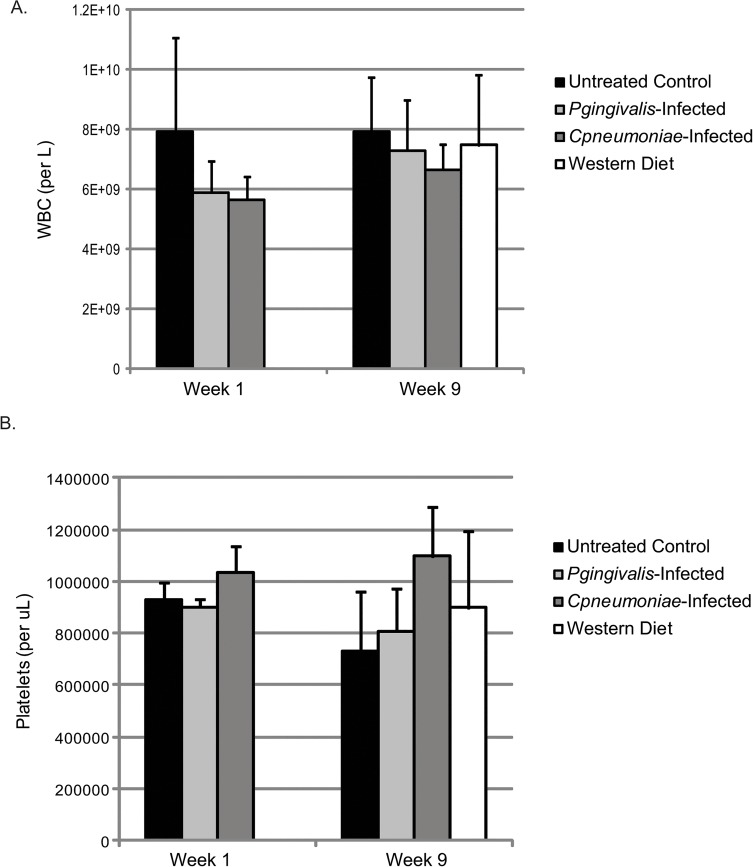
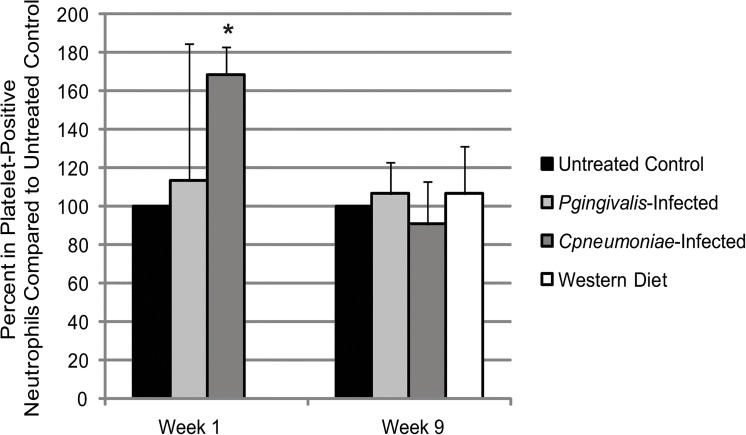
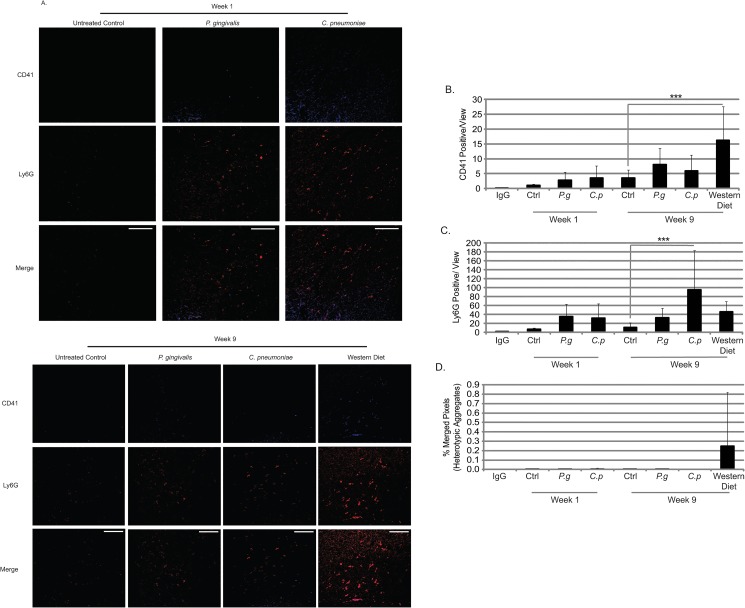
There were no changes in body mass (Fig 4), or increases in white blood cell (WBC) counts (Fig 5A) at week 1 in ApoE-/- mice infected with either P. gingivalis or C. pneumoniae. Only C. pneumoniae altered platelet counts (Fig 5B) compared to Untreated Control, although not significantly, suggesting this bacterial infection altered megakaryocyte maturation and platelet production and/or platelet clearance. Circulating heterotypic aggregates (platelet-neutrophil aggregates) were significantly increased in C. pneumoniae infected mice at week 1 (Fig 6); however, there were inconsistencies with the P. gingivalis-infected ApoE-/- mice, but the data suggest that there was an increase in aggregate formation as well. Analysis of spleen sections from P. gingivalis or C. pneumoniae infected mice revealed increased CD41 levels, a platelet marker, and Ly6G levels, a neutrophil marker (Fig 7A–7C; IgG controls in S1 Fig). However, there were no heterotypic aggregates as indicated by the lack of overlap between the two signals when merged (Fig 7A and 7D).
Fig 4. Change in ApoE-/- mouse body mass with infection and diet.
Mass was measured on Week 1, which was 24 h after the last P. gingivalis challenge and 4 days after C. pneumoniae exposure (n = 3 for each group), and on Week 9 (n = 3 for Untreated Control, P. gingivalis-Infected, and C. pneumoniae-Infected; n = 6 for Western Diet). Data is normally distributed and analyzed using an ANOVA.
Fig 5. WBC and platelet count changes with infection and diet in the ApoE-/- mice.
Whole blood cell counts were assessed to determine the number of WBC and platelets. Measurements were performed on Week 1, which was 24 h after the last P. gingivalis challenge and 4 days after exposure to C. pneumoniae (n = 3 for each group), and on Week 9 (n = 3 for Untreated Control, P. gingivalis-Infected, and C. pneumoniae-Infected; n = 6 for Western Diet). Data is normally distributed and analyzed using an ANOVA.
Fig 6. Heterotypic aggregate formation in ApoE-/- mice with infection and diet.
Whole blood samples taken on Week 1 (24 h after the last P. gingivalis challenge and 4 days after exposure to C. pneumoniae; n = 3 for each group) and on Week 9 (n = 3 for each group) and dual stained for platelet marker (CD41) and neutrophil marker (Ly6G). The percent of platelet-positive neutrophils were determined through flow cytometry and normalized to Untreated Control at each timepoint. Data is normally distributed and analyzed using an ANOVA. *p<0.05 compared to Untreated Control at Week 1.
Fig 7. Platelet and neutrophil content in spleens in ApoE-/- mice with infection and diet.
(A) Spleen sections from ApoE-/- mice at week 1 and week 9 were stained for CD41 (platelet marker; Alexa Fluor 405) and Ly6G (neutrophils; Texas Red) and visualized by confocal microscopy at 63X. The amount of CD41 staining (area; B), Ly6G staining (area; C), and the overlap of each signal (% pixels with overlapping signal); (D) were quantified. Data is normally distributed and analyzed using an ANOVA. n = 3 for each condition; ***p<0.001 compared to Untreated Control. Scale Bar in merged images = 50.0 μm.
At week 9, P. gingivalis or C. pneumoniae infections did not alter ApoE-/- mouse body mass (Fig 4). However, in mice fed a Western diet, there was an increase in body mass by 5.2g. There was no effect on WBC counts at week 9 with either bacteria or a Western diet (Fig 5A). Platelet counts were still elevated following C. pneumoniae infection (Fig 5B). The increased circulating heterotypic aggregates seen with bacterial infections at week 1 were gone by week 9 (Fig 6). There were a greater number of platelets in the spleen sections from P. gingivalis or C. pneumoniae infected mice at week 9 as compared to week 1 (Fig 7A and 7B). P. gingivalis infection maintained the same amount of neutrophils in the spleen from week 1 to 9 (Fig 7A and 7C). For C. pneumoniae, there was a significantly greater number of neutrophils compared to Untreated Control at week 9 than that observed at week 1 (Fig 7A and 7C). A Western diet at week 9 did not increase circulating heterotypic aggregate formation. There was a significant increase in platelets in the spleens of mice fed a Western diet (Fig 7A and 7B) and a non-significant increase in neutrophils (Fig 7A and 7C). Unlike that observed following bacterial infections, there were heterotypic aggregates present in the spleens of mice fed a Western diet (Fig 7A and 7D).
Changes in Platelet Transcripts Due to Diet in Participants of the Framingham Heart Study
To verify the relevance of the in vivo mouse observations, select platelet transcripts altered by a Western diet in the ApoE-/- mice were measured in participants of the FHS (n = 1819; Table 1). These genes were either increased or decreased in the ApoE-/- mice fed a Western diet, including albumin (ALB), α-1-microglobulin (AMBP), chemokine ligand 3 (CCL3), CD3 antigen (CD3D), CD53 antigen (CD53), fatty acid binding protein 1 (FABP1), fatty acid binding protein 5 (FABP5), fibrinogen γ chain (FGG), mitochondrial ribosomal protein S26 (MRPS26), nuclear receptor subfamily 2 (NR2F6), ribosomal protein L21 (RPL21), α-1-antitrypsin (SERPINA1), and vitronectin (VTN). These genes encompass various functional groups identified in the microarray, including coagulation, gene expression, inflammation, and lipids. These transcripts were compared to variables measured in the FHS that were associated with increased obesity [1,35] and a Western diet [26,36] in prior publications, including cholesterol, BMI, lipids, and triglyceride levels. A complete list of genes and their relationship with all studied clinical variables is found in S11 Table.
Table 1. Framingham heart study offspring cohort examination 8 characteristics.
| Variables | Mean±Standard Deviation/Number (%) |
|---|---|
| Sample Size | 1819 |
| Women | 993 (51) |
| Age (y) | 67±9 |
| BMI (kg/m2) | 28.3±5.3 |
| Systolic Blood Pressure (mm Hg) | 129±17 |
| Diastolic Blood Pressure (mm Hg) | 73±10 |
| Lipid Treatment | 798 (44) |
| Total Cholesterol/HDL Ratio (mg/100 mL) | 3.5±1.1 |
| Triglycerides (mg/100 mL) | 116±67 |
| Glucose (mg/dL) | 107±25 |
| Antihypertensive Treatment | 919 (51) |
| Aspirin (3 times/week) | 823 (45) |
| Current Hormone Replacement Therapy | 104 (6) |
| Diabetes mellitus | 255 (14) |
| Prevalent Coronary Heart Disease | 199 (11) |
| Smoker | 155 (8.5) |
Of the genes decreased in ApoE-/- mice fed a Western diet, 4 out of the 7 (ALB, AMBP, FGG, and VTN) were also significantly decreased with a higher mean BMI or triglycerides in the FHS (Table 2 and S11 Table). FABP1 was not associated with cholesterol, BMI, or triglycerides, while FABP5 and SERPINA1 were increased with a higher mean in cholesterol or BMI. As for the genes that increased with a Western diet in the ApoE-/- mice, 4 of the 5 (CD3D, CD53, NR2F6, and RPL21) were also increased significantly with a higher mean BMI, triglycerides, and/or cholesterol (Table 2 and S11 Table). CD53 and NR2F6 were increased significantly with an increased mean total cholesterol:HDL ratio and BMI but decreased significantly with increased mean triglyceride levels. One transcript, CCL3, although increased in the mice, was not increased in individuals in the FHS with these variables but was associated with other factors, such as diabetes (S11 Table).
Table 2. The relation between gene expression, clinical variables, and inflammatory markers in the FHS.
| FHS Fold Change (95% Conference Interval) | |||||||
|---|---|---|---|---|---|---|---|
| Gene | Gene Function | Mouse Microarray Data (Fold Change Compared to UC) | Total Cholesterol: HDL 1 | Triglycerides 1 | BMI 2 | CRP 2 | IL6 2 |
| ALB | Fatty Acid Transport | 0.055 | — | 0.90 (0.81, 0.99)* | — | — | — |
| FABP1 | Fatty Acid Transport/ Metabolism | 0.15 | — | — | — | 0.82 (0.74, 0.91)*** | 0.71 (0.60, 0.83)*** |
| SERPINA1 | Inflammation | 0.20 | — | — | 1.13 (1.04, 1.23)** | 1.29 (1.19, 1.40)*** | 1.50 (1.32, 1.72)*** |
| FGG | Coagulation | 0.26 | — | — | 0.87 (0.78, 0.97)* | 0.83 (0.74, 0.92)*** | 0.68 (0.58, 0.80)*** |
| AMBP | Coagulation | 0.35 | — | — | 0.87 (0.78, 0.98)* | 0.80 (0.72, 0.89)*** | 0.68 (0.57, 0.81)*** |
| FABP5 | Fatty Acid Transport/ Metabolism | 0.39 | 1.15 (1.04, 1.26)** | — | — | 1.10 (1.02, 1.18)* | 1.12 (1.00, 1.26)* |
| MRPS26 | Gene Expression- Mitoribosomes | 0.43 | — | — | — | 1.14 (1.05, 1.22)*** | 1.22 (1.09, 1.38)*** |
| VTN | Coagulation | 0.49 | — | 0.87 (0.79, 0.96)** | — | — | 0.85 (0.73, 0.99)* |
| CCL3 | Inflammation | 2.40 | — | — | — | 1.14 (1.05, 1.23)** | 1.27 (1.13, 1.44)*** |
| CD53 | Inflammation | 2.48 | 1.13 (1.02, 1.24)* | 0.92 (0.85, 1.00)* | 1.08 (1.00, 1.17)* | 1.14 (1.05, 1.23)*** | 1.24 (1.10, 1.40)*** |
| NR2F6 | Gene Expression–Inflammation | 2.55 | 1.15 (1.04, 1.27)** | 0.92 (0.85, 1.00)* | 1.11 (1.03, 1.20)** | 1.17 (1.09, 1.26)*** | 1.26 (1.12, 1.42)*** |
| CD3D | Inflammation | 2.63 | — | — | 1.10 (1.01, 1.19)* | 1.14 (1.05, 1.24)** | 1.21 (1.07, 1.38)** |
| RPL21 | Translation | 3.00 | — | — | 1.11 (1.03, 1.19)** | 1.12 (1.04, 1.20)** | 1.21 (1.08, 1.35)** |
1Fold Change for FHS: Total Cholesterol:HDL, BMI, Triglycerides – regression coefficient transformed (2-β) to express fold change in gene expression associated with 1 unit change in clinical covariate, with the exception in BMI (5 points) and triglycerides (50 points). *p<0.05, **p<0.01, ***p<0.001.
2 Fold Change for FHS: CRP and IL6 – regression coefficient transformed (2-β) to express fold change in gene expression associated with loge-CRP or loge-IL6 serum levels adjusted for multiple covariates. *p<0.05, **p<0.01, ***p<0.001.
Abbreviations: BMI – Body mass index, CRP – C-reactive protein, FHS – Framingham Heart Study, HDL – High density lipoprotein, IL6 – Interleukin 6, Total Cholesterol:HDL – Ratio between total and HDL cholesterol, UC – Untreated control.
Transcripts altered by a Western diet that were examined in FHS were also compared to circulating levels of CRP and IL6 (Table 2), inflammatory markers shown to be increased in individuals with CVD [37,38], obesity [35,39], and a Western diet [36,40,41]. As previously shown, these cytokines also associated significantly with inflammatory-related transcripts from platelets in FHS [2]. All but one transcript was significantly associated with higher mean levels of CRP and/or IL6. AMBP, FABP1, FGG, and VTN levels decreased with a higher mean level of CRP and/or IL6, which were all decreased in ApoE-/- mice fed a Western diet and some in the FHS. CCL3, CD3D, CD53, FABP5, MRPS26, NR2F6, RPL21, and SERPINA1 were increased significantly with a higher mean CRP and/or IL6, which the majority were also increased in either FHS or in ApoE-/- mice fed a Western diet. Only expression of ALB was not significantly associated with either of these cytokines.
Discussion
The goal of the study was to characterize the effects of inflammatory stimuli–bacterial infections and a Western diet–on platelet transcripts and inflammatory function over time in a murine model of CVD. Similar effects on platelets were observed following infection with P. gingivalis and C. pneumoniae early after infection as determined by both transcriptional profiles and heterotypic aggregate formation. However, at a later timepoint (week 9), their effects on transcription diverged, with C. pneumoniae infection affecting platelets more similarly to a Western diet. The effects of a Western diet on platelet transcripts were confirmed in platelet samples from FHS.
Platelet transcripts and inflammatory function were measured at week 1 following either P. gingivalis or C. pneumoniae infection to establish the early responses to each bacterium in ApoE-/- mice. Platelet transcripts were affected similarly by each bacterial infection, which suggests that each stimuli was activating through common pathways, most likely the TLR2-NFКB pathway, in the platelet precursor cell, the megakaryocyte, which we previously published on [42]. It is hypothesized that bacteria are recognized by megakaryocytes through their innate immune receptors. This hypothesis has been explored using Pam3CSK, a TLR2 synthetic agonist, in human and mouse megakaryocytes, showing upregulation of inflammatory and coagulation related transcripts [42]. Genes associated with RNA/gene expression and molecular transport, were also increased in platelets following infection with P. gingivalis and C. penumoniae. It is hypothesized the increase in gene expression correlates to an increase in mRNA translation within the platelets, as has been seen with platelets in response to cytokines [2,43]. Additionally, the increased number of transcripts in both of these groups could be transferred to other vascular cells, such as endothelial cells or monocytes [44], to increase translation in these cells in response to infection. We hypothesized transcriptional changes would be reflective with functional modifications in the platelets. As seen at week 1, there was an increase in circulating heterotypic aggregates and positive enrichment of inflammatory gene sets (i.e., complement cascade). Not only is platelet production affected but so is the platelet response. As previously shown, platelets respond to Pam3CSK4, P. gingivalis [3,4], and C. pneumoniae (data not shown) through TLR2 and IL1R1. This results in platelets binding to neutrophils to form heterotypic aggregates, as well as increased P-selectin surface expression, and adhesion to collagen, increasing the risk of thrombosis as seen in human studies [22,45].
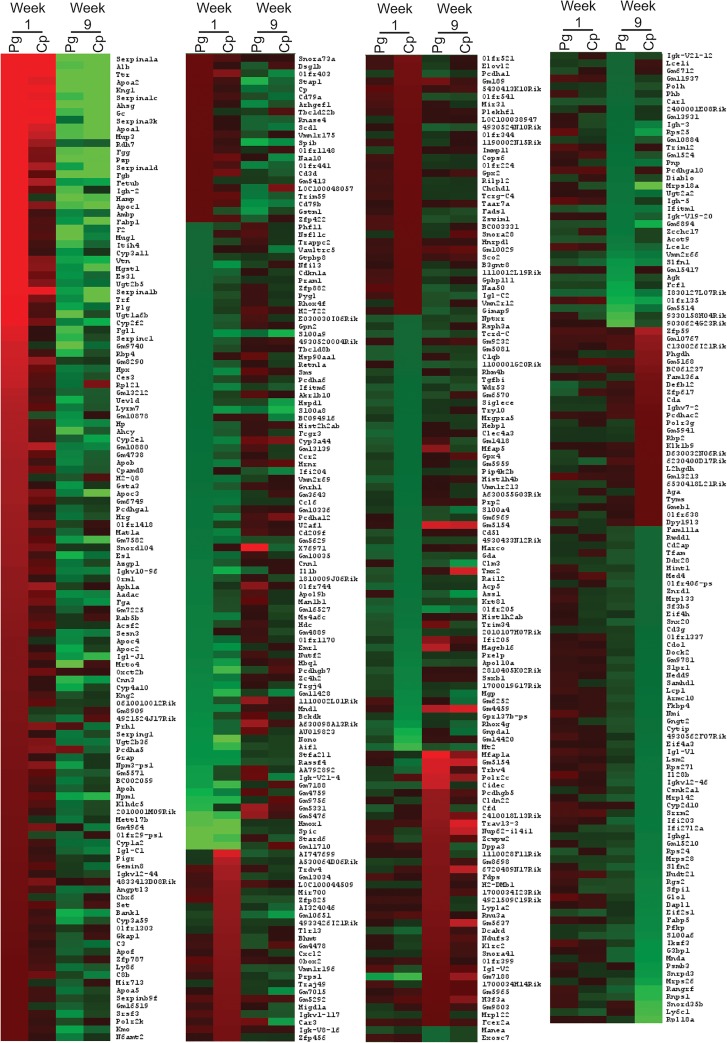
At week 9, following P. gingivalis and C. pneumoniae infections, before overt plaque formation had yet to occur in the aorta, alterations in both platelet transcripts and inflammatory function are still occurring. Although not directly measured in our study, previous work by our group and others have shown that each bacterium is still present in the systems of these mice. An increase in circulating P. gingivalis-specific IgG has been detected through 13 weeks post infection [46] and by 24 weeks, this bacterium is identified in aorta, heart, and liver tissue [47]. As for C. pneumoniae, there is a measurable serum IgG level up to 16 weeks, with bacteria detected in the lungs, aorta, heart, and spleen [48]. Overall, both bacteria upregulated fewer genes at the later timepoint compared that observed at week 1 (Fig 8). Infection with C. pneumoniae resulted in more downregulation in gene expression, which was further supported by GSEA, showing significant negative enrichment in gene sets. Unlike with C. pneumoniae, P. gingivalis infection alterations in gene transcript changes were diminished at week 9. It is hypothesized that over the course of the infection, megakaryocytes are slowly returning to baseline in terms of the types of RNA produced, with fewer genes and gene groups being altered. Further, changes in transcripts are also reflective of platelet function. Platelet-neutrophil aggregates are no longer in circulation at week 9. The cells were individually present in the spleens obtained from these mice. It is possible that we no longer see these cells as aggregates even in the spleens because the interaction between the platelets and neutrophils is transient, a brief interaction that results in activation of either/both cells and transfer of information on the current environment. With more platelets in the spleen, it is expected that there would be a decrease in circulating platelet levels. Instead, the platelet concentrations stay steady if not increase. This data suggest megakaryocytes are still responding to the infection, directly or indirectly.
Fig 8. Comparison of genes affected by both bacterial infections at week 1 and week 9.
Heatmap shows the transcripts identified through microarray as upregulated or downregulated 2-fold or more with either P. gingivalis (Pg) or C. pneumoniae (Cp) infection compared to Untreated Control in the ApoE-/- mice at week 1 and 9. Each condition represents RNA from 3 mice pooled.
With a Western diet, it was expected that lipid associated genes would be upregulated in platelets, but instead, as seen in FHS [1,2,3], there was an increase in inflammatory related transcripts, as well as signaling and proliferation. As previously seen with the FHS [1], the changes associated with a Western diet on platelets can either be due to the diet, change in body weight, or the proinflammatory setting. Further work is needed to understand this relationship. Interestingly, many genes and/or gene function groups altered by a Western diet were also altered following a C. pneumoniae infection. Genes downregulated following a Western diet or C. pneumoniae infection included TTR, GC, ALB, APOA2, and FABP1. Functionally, the Western diet did not produce circulating heterotypic aggregates but did promote the formation of aggregates to the vessel wall, as seen in the spleens. It is hypothesized that a Western diet, promotes platelets adhesion to the blood vessels, which promotes adhesion of immune cells as well, leading to the formation of atherosclerosis [49].
The effects of a Western diet in the ApoE-/- mice were verified using platelet transcripts from the FHS. Thirteen genes were identified from the microarray data, including ALB, AMBP, CCL3, CD3D, CD53, FABP1, FABP5, FGG, MRPS26, NR2F6, RPL21, SERPINA1, and VTN. Twelve out of the 13 genes had altered gene expression that correlated to one or more clinical variables associated with adiposity, including BMI, triglycerides, and cholesterol. Additionally, inflammatory related cytokines, CRP and IL6, which are increased with CVD and associate significantly with inflammatory related transcripts from platelets in FHS [2], were associated with changes in 12 of the 13 identified transcripts, again, supporting the notion that platelets react with an inflammatory stimuli resembles their reaction to a Western diet.
In conjunction with the analysis of platelets in these ApoE-/- mice, aortic tissue, consisting of endothelial cells, immune cells, and smooth muscle cells, were also examined by microarray. At week 1, P. gingivalis infection had little effect on gene expression in the aortic tissue. C. pneumoniae infection upregulated genes associated with GPCR signaling, viral myocarditis, antigen processing/presentation, and membranes in the entire aortic tissue [23], which is in contrast to what is shown here in platelets. When comparing the platelet data to the results from the aortic tissue at week 9, we see there are no gene sets similarly affected with either bacterial infection [23]. In fact, two pathways positively enriched in aortic tissue, PPAR signaling and peptide chain formation, are negatively enriched in platelets. As for a Western diet, there were only 4 similarities found in the positively enriched gene sets, with consisted of B cell receptor signaling pathways, host interactions of HIV factors, cell cycle checkpoints, and M-G1 transition.
Strengths and Limitations
Our study has substantial advantages including a translational breadth with experimental and epidemiological data relating transcripts from mouse and human platelets and mouse platelet function, human lipid, adiposity, and inflammatory data from FHS. However, we note the following caveats in our study. Our bacterial and Western diet exposure data were conducted in 3 mice, so we may have been underpowered to detect modest effect sizes. We could not verify any confounding attributions due to antibiotic pre-treatment with the P. gingivalis group for all genes identified; however, we were able to show in a couple of genes they were not altered by antibiotics (sham treatment) compared to Untreated Control in C57BL/6J mice (S2 Fig). Additionally, there are discrepancies between human and mouse data due to species differences [50]. Our FHS data were observational and cross-sectional–we cannot rule out residual confounding or establish the temporality or causal nature of the associations we report. In addition, our cohort was constituted of largely middle-aged to older adults of European ancestry; we cannot determine the generalizability of our findings to younger individuals or other races/ethnicities. Finally, it should be noted that we did not correct for the number of statistical comparisons made when examining the relationships between each gene and clinical covariates in the FHS samples. To reduce the probability of type I error, we focused interpretation on only those covariates of most relevance given the mouse experiments examining a Western diet.
Conclusion
Overall, as has been seen with other immune cells, platelets respond differentially to distinct inflammatory etiologies. Platelets early response to an infection with an oral bacterium or a respiratory bacterium, both associated with CVD, were similar; however, over time, the effects on platelets by each bacterium diverge. Interestingly, C. pneumoniae alters platelet transcripts similarly to a Western diet at week 9. In conclusion, the inflammatory-induced transcriptional and functional changes measured in platelets appear to reflect systemic changes occurring with disease development, and provides insight into progression and regression.
Supporting Information
(EPS)
(EPS)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability
All files containing the full microarray data are available from the GEO database (assigned Series ID GSE68835).
Funding Statement
The work presented was supported by N01-HC 25195, RFA-HL-12-008 (JEF, EM), RFA-RM-12-013 (JEF), P-01-HL085381 (JEF), P01 AI078894 (LMB, CDK, EOW, XH, SM, RI, CAG, and JEF), 1RO1 HL64753, R01 HL076784, and 1 R01 AG028321 (EJB), from the National Institutes of Health (www.nih.gov). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Freedman JE, Larson MG, Tanriverdi K, O'Donnell CJ, Morin K, Hakanson AS, et al. Relation of platelet and leukocyte inflammatory transcripts to body mass index in the Framingham heart study. Circulation 2010; 122: 119–129. 10.1161/CIRCULATIONAHA.109.928192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McManus DD, Beaulieu LM, Mick E, Tanriverdi K, Larson MG, Keaney JF Jr, et al. Relationship among circulating inflammatory proteins, platelet gene expression, and cardiovascular risk. Arterioscler Thromb Vasc Biol 2013; 33: 2666–2673. 10.1161/ATVBAHA.112.301112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beaulieu LM, Lin E, Mick E, Koupenova M, Weinberg EO, Kramer CD, et al. Interleukin 1 receptor 1 and interleukin 1beta regulate megakaryocyte maturation, platelet activation, and transcript profile during inflammation in mice and humans. Arterioscler Thromb Vasc Biol 2014; 34: 552–564. 10.1161/ATVBAHA.113.302700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blair P, Rex S, Vitseva O, Beaulieu L, Tanriverdi K, Chakrabarti S, et al. Stimulation of Toll-like receptor 2 in human platelets induces a thromboinflammatory response through activation of phosphoinositide 3-kinase. Circ Res 2009; 104: 346–354. 10.1161/CIRCRESAHA.108.185785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Semple JW, Italiano JE Jr, Freedman J. Platelets and the immune continuum. Nat Rev Immunol 2011; 11: 264–274. 10.1038/nri2956 [DOI] [PubMed] [Google Scholar]
- 6. Gibson FC, Hong C, Chou HH, Yumoto H, Chen JQ, Lien E, et al. Innate immune recognition of invasive bacteria accelerates atherosclerosis in apolipoprotein E-deficient mice. Circulation 2004; 109: 2801–2806. [DOI] [PubMed] [Google Scholar]
- 7. Hayashi C, Viereck J, Hua N, Phinikaridou A, Madrigal AG, Gibson FC 3rd, et al. Porphyromonas gingivalis accelerates inflammatory atherosclerosis in the innominate artery of ApoE deficient mice. Atherosclerosis 2011; 215: 52–59. 10.1016/j.atherosclerosis.2010.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Velsko IM, Chukkapalli SS, Rivera MF, Lee JY, Chen H, Zheng D, et al. Active invasion of oral and aortic tissues by Porphyromonas gingivalis in mice causally links periodontitis and atherosclerosis. PLoS One 2014; 9: e97811 10.1371/journal.pone.0097811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Haraszthy VI, Zambon JJ, Trevisan M, Zeid M, Genco RJ. Identification of periodontal pathogens in atheromatous plaques. J Periodontol 2000; 71: 1554–1560. [DOI] [PubMed] [Google Scholar]
- 10. Nakano K, Inaba H, Nomura R, Nemoto H, Takeuchi H, Yoshioka H, et al. Distribution of Porphyromonas gingivalis fimA genotypes in cardiovascular specimens from Japanese patients. Oral Microbiol Immunol 2008; 23: 170–172. 10.1111/j.1399-302X.2007.00406.x [DOI] [PubMed] [Google Scholar]
- 11. Yakob M, Soder B, Meurman JH, Jogestrand T, Nowak J, Soder PO. Prevotella nigrescens and Porphyromonas gingivalis are associated with signs of carotid atherosclerosis in subjects with and without periodontitis. J Periodontal Res 2011; 46: 749–755. 10.1111/j.1600-0765.2011.01398.x [DOI] [PubMed] [Google Scholar]
- 12. Naiki Y, Sorrentino R, Wong MH, Michelsen KS, Shimada K, Chen S, et al. TLR/MyD88 and liver X receptor alpha signaling pathways reciprocally control Chlamydia pneumoniae-induced acceleration of atherosclerosis. J Immunol 2008; 181: 7176–7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Player MS, Mainous AG 3rd, Everett CJ, Diaz VA, Knoll ME, Wright RU. Chlamydia pneumoniae and progression of subclinical atherosclerosis. Eur J Prev Cardiol 2014; 21: 559–565. 10.1177/2047487312472078 [DOI] [PubMed] [Google Scholar]
- 14. Sakurai-Komada N, Iso H, Koike KA, Ikeda A, Umesawa M, Ikehara S, et al. Association between Chlamydophila pneumoniae infection and risk of coronary heart disease for Japanese: the JPHC study. Atherosclerosis 2014; 233: 338–342. 10.1016/j.atherosclerosis.2014.01.023 [DOI] [PubMed] [Google Scholar]
- 15. Jha HC, Srivastava P, Divya A, Prasad J, Mittal A. Prevalence of Chlamydophila pneumoniae is higher in aorta and coronary artery than in carotid artery of coronary artery disease patients. APMIS 2009; 117: 905–911. 10.1111/j.1600-0463.2009.02553.x [DOI] [PubMed] [Google Scholar]
- 16. Ciervo A, Mancini F, Sale P, Russo A, Cassone A. Real-time polymerase chain reaction and laser capture microdissection: an efficient combination tool for Chlamydophila pneumoniae DNA quantification and localization of infection in atherosclerotic lesions. Int J Immunopathol Pharmacol 2008; 21: 421–428. [DOI] [PubMed] [Google Scholar]
- 17. Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Blaha MJ, et al. Heart disease and stroke statistics—2014 update: a report from the American Heart Association. Circulation 2014; 129: e28–e292. 10.1161/01.cir.0000441139.02102.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nakashima Y, Plump AS, Raines EW, Breslow JL, Ross R. ApoE-deficient mice develop lesions of all phases of atherosclerosis throughout the arterial tree. Arterioscler Thromb 1994; 14: 133–140. [DOI] [PubMed] [Google Scholar]
- 19. Sullivan PM, Mezdour H, Aratani Y, Knouff C, Najib J, Reddick RL, et al. Targeted replacement of the mouse apolipoprotein E gene with the common human APOE3 allele enhances diet-induced hypercholesterolemia and atherosclerosis. J Biol Chem 1997; 272: 17972–17980. [DOI] [PubMed] [Google Scholar]
- 20. Eitzman DT, Westrick RJ, Xu Z, Tyson J, Ginsburg D. Hyperlipidemia promotes thrombosis after injury to atherosclerotic vessels in apolipoprotein E-deficient mice. Arterioscler Thromb Vasc Biol 2000; 20: 1831–1834. [DOI] [PubMed] [Google Scholar]
- 21. Hayashi C, Gudino CV, Gibson FC 3rd, Genco CA. Review: Pathogen-induced inflammation at sites distant from oral infection: bacterial persistence and induction of cell-specific innate immune inflammatory pathways. Mol Oral Microbiol 2010; 25: 305–316. 10.1111/j.2041-1014.2010.00582.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kalvegren H, Majeed M, Bengtsson T. Chlamydia pneumoniae binds to platelets and triggers P-selectin expression and aggregation: a causal role in cardiovascular disease? Arterioscler Thromb Vasc Biol 2003; 23: 1677–1683. [DOI] [PubMed] [Google Scholar]
- 23. Kramer CD, Weinberg EO, Gower AC, He X, Mekasha S, Slocum C, et al. Distinct gene signatures in aortic tissue from ApoE-/- mice exposed to pathogens or Western diet. BMC Genomics 2014; 15: 1176 10.1186/1471-2164-15-1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nassar H, Chou H-H, Khlgatian M, Gibson FC, Dyke TEV, Genco CA. Role for Fimbriae and Lysine-Specific Cysteine Proteinase Gingipain K in Expression of Interleukin-8 and Monocyte Chemoattractant Protein in Porphyromonas gingivalis-Infected Endothelial Cells. Infection and Immunity 2002; 70: 268–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. He X, Mekasha S, Mavrogiorgos N, Fitzgerald KA, Lien E, Ingalls RR. Inflammation and fibrosis during Chlamydia pneumoniae infection is regulated by IL-1 and the NLRP3/ASC inflammasome. J Immunol 2010; 184: 5743–5754. 10.4049/jimmunol.0903937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moustardas P, Kadoglou NP, Katsimpoulas M, Kapelouzou A, Kostomitsopoulos N, Karayannacos PE, et al. The complementary effects of atorvastatin and exercise treatment on the composition and stability of the atherosclerotic plaques in ApoE knockout mice. PLoS One 2014; 9: e108240 10.1371/journal.pone.0108240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003; 4: 249–264. [DOI] [PubMed] [Google Scholar]
- 28. Gautier L, Cope L, Bolstad BM, Irizarry RA. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004; 20: 307–315. [DOI] [PubMed] [Google Scholar]
- 29. Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res 2005; 33: e175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brettschneider J, Collin F, Bolstad BM, Speed TP. Quality Assessment for Short Oligonucleotide Microarray Data. Technometrics 2008; 50: 241–264. [Google Scholar]
- 31. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005; 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 2013; 41: D8–D20. 10.1093/nar/gks1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics 2007; 23: 3251–3253. [DOI] [PubMed] [Google Scholar]
- 34. Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 2003; 34: 374–378. [DOI] [PubMed] [Google Scholar]
- 35. Bertola A, Bonnafous S, Anty R, Patouraux S, Saint-Paul MC, Iannelli A, et al. Hepatic expression patterns of inflammatory and immune response genes associated with obesity and NASH in morbidly obese patients. PLoS One 2010; 5: e13577 10.1371/journal.pone.0013577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Scott NJ, Cameron VA, Raudsepp S, Lewis LK, Simpson ER, Richards AM, et al. Generation and characterization of a mouse model of the metabolic syndrome: apolipoprotein E and aromatase double knockout mice. Am J Physiol Endocrinol Metab 2012; 302: E576–584. 10.1152/ajpendo.00222.2011 [DOI] [PubMed] [Google Scholar]
- 37. Dhingra R, Gona P, Nam BH, D'Agostino RB Sr., Wilson PW, Benjamin EJ, et al. C-reactive protein, inflammatory conditions, and cardiovascular disease risk. Am J Med 2007; 120: 1054–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sarwar N, Butterworth AS, Freitag DF, Gregson J, Willeit P, Gorman DN, et al. Interleukin-6 receptor pathways in coronary heart disease: a collaborative meta-analysis of 82 studies. Lancet 2012; 379: 1205–1213. 10.1016/S0140-6736(11)61931-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Browning LM, Krebs JD, Magee EC, Fruhbeck G, Jebb SA. Circulating markers of inflammation and their link to indices of adiposity. Obes Facts 2008; 1: 259–265. 10.1159/000169832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cheng C, Tempel D, Den Dekker WK, Haasdijk R, Chrifi I, Bos FL, et al. Ets2 determines the inflammatory state of endothelial cells in advanced atherosclerotic lesions. Circ Res 2011; 109: 382–395. 10.1161/CIRCRESAHA.111.243444 [DOI] [PubMed] [Google Scholar]
- 41. Nie P, Li D, Hu L, Jin S, Yu Y, Cai Z, et al. Atorvastatin improves plaque stability in ApoE-knockout mice by regulating chemokines and chemokine receptors. PLoS One 2014; 9: e97009 10.1371/journal.pone.0097009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Beaulieu LM, Lin E, Morin KM, Tanriverdi K, Freedman JE. Regulatory effects of TLR2 on megakaryocytic cell function. Blood 2011; 117: 5963–5974. 10.1182/blood-2010-09-304949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lindemann S, Tolley ND, Dixon DA, McIntyre TM, Prescott SM, Zimmerman GA, et al. Activated platelets mediate inflammatory signaling by regulated interleukin 1beta synthesis. J Cell Biol 2001; 154: 485–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Risitano A, Beaulieu LM, Vitseva O, Freedman JE. Platelets and platelet-like particles mediate intercellular RNA transfer. Blood 2012; 119: 6288–6295. 10.1182/blood-2011-12-396440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ohki T, Itabashi Y, Kohno T, Yoshizawa A, Nishikubo S, Watanabe S, et al. Detection of periodontal bacteria in thrombi of patients with acute myocardial infarction by polymerase chain reaction. Am Heart J 2012; 163: 164–167. 10.1016/j.ahj.2011.10.012 [DOI] [PubMed] [Google Scholar]
- 46. Hayashi C, Papadopoulos G, Gudino CV, Weinberg EO, Barth KR, Madrigal AG, et al. Protective role for TLR4 signaling in atherosclerosis progression as revealed by infection with a common oral pathogen. J Immunol 2012; 189: 3681–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li L, Messas E, Batista EL Jr, Levine RA, Amar S. Porphyromonas gingivalis infection accelerates the progression of atherosclerosis in a heterozygous apolipoprotein E-deficient murine model. Circulation 2002; 105: 861–867. [DOI] [PubMed] [Google Scholar]
- 48. Moazed TC, Kuo C, Grayston JT, Campbell LA. Murine models of Chlamydia pneumoniae infection and atherosclerosis. J Infect Dis 1997; 175: 883–890. [DOI] [PubMed] [Google Scholar]
- 49.Karshovska E, Zhao Z, Blanchet X, Schmitt MM, Bidzhekov K, Soehnlein O, et al. Hyperreactivity of Junctional Adhesion Molecule A-Deficient Platelets Accelerates Atherosclerosis in Hyperlipidemic Mice. Circ Res 2014. [DOI] [PubMed]
- 50. Rowley JW, Oler AJ, Tolley ND, Hunter BN, Low EN, Nix DA, et al. Genome-wide RNA-seq analysis of human and mouse platelet transcriptomes. Blood 2011; 118: e101–111. 10.1182/blood-2011-03-339705 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(EPS)
(EPS)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All files containing the full microarray data are available from the GEO database (assigned Series ID GSE68835).