Abstract
Plants are an attractive host system for pharmaceutical protein production. Many therapeutic proteins have been produced and scaled up in plants at a low cost compared to the conventional microbial and animal-based systems. The main technical challenge during this process is to produce sufficient levels of recombinant proteins in plants. Low yield is generally caused by proteolytic degradation during expression and downstream processing of recombinant proteins. The yield of human therapeutic interleukin (IL)-10 produced in transgenic tobacco leaves was found to be below the critical level, and may be due to degradation by tobacco proteases. Here, we identified a total of 60 putative cysteine protease genes (CysP) in tobacco. Based on their predicted expression in leaf tissue, 10 candidate CysPs (CysP1-CysP10) were selected for further characterization. The effect of CysP gene silencing on IL-10 accumulation was examined in tobacco. It was found that the recombinant protein yield in tobacco could be increased by silencing CysP6. Transient expression of CysP6 silencing construct also showed an increase in IL-10 accumulation in comparison to the control. Moreover, CysP6 localizes to the endoplasmic reticulum (ER), suggesting that ER may be the site of IL-10 degradation. Overall results suggest that CysP6 is important in determining the yield of recombinant IL-10 in tobacco leaves.
Introduction
Plants are attractive biofactories for recombinant protein production. They offer several advantages such as reduction in cost of production and increased scalability over microbial and mammalian cell culture systems. This low cost of large scale protein production is due to significant reduction in capital investment and processing cost. Unlike cell culture based systems, plants do not require fermenters or highly skilled personnel to operate them [1]. Plants are also capable of carrying out complex post translational modifications (PTM), a process necessary for the biological activity of many eukaryotic proteins. Additionally, plant systems include reduced risk of contamination from animal-borne pathogens, and provide a platform to manufacture valuable pharmaceutical proteins that cannot be produced using fermenter-based systems [2,3]
Over the past two decades, a wide range of food and non-food crops such as tobacco, alfalfa, cereals (rice and maize), legumes (soybean), and fruits and vegetables (carrot, tomato, potato) have been used for recombinant protein production [1]. Being a non-food crop, tobacco (Nicotiana tabacum) offers a competitive edge over many other plant hosts in terms of biosafety. Tobacco provides a high yield of protein per green leaf biomass that can be harvested 3–4 times in a year, and has a prolific seed production ability [4]. Furthermore, molecular and genetic tools for transfer and expression of foreign genes in tobacco are well established. The first pharmaceutical protein produced in planta, human growth hormone (hGH), was expressed in the tobacco leaves [5]. Since then, many recombinant proteins such as monoclonal antibodies, recombinant subunit vaccines and cytokines have been successfully produced in tobacco [3,6–8].
Though plant bioreactors have emerged as an economical alternative, low accumulation of proteins is the major concern posing limitations for its use as a commercial protein production platform [9]. One of the major reasons for low yields of recombinant proteins is their degradation by plant proteases [10,11]. Protein degradation by plant proteases occurs both intracellularly and extracellularly. Inside the cell, degradations can occur after synthesis, assembly and in some cases during PTM in the endoplasmic reticulum (ER) and golgi apparatus [12]. Outside the cell, proteins are degraded by extracellular proteases in the apoplastic space or in the culture medium when they are synthesized as secreted proteins [13]. Degraded protein fragments have been observed in many plant hosts expressing recombinant proteins, such as tobacco [14–17], Arabidopsis [14], corn [18,19], potato [20] and alfalfa [21]. Among several different classes of plant proteases, cysteine proteases (CysPs) were identified as the enzymes responsible for the degradation of recombinant sea anemone protein equistatin when expressed in potato tubers [20], and for the degradation of the human cytokine, hGM-CSF, produced in rice cell suspension cultures [22].
CysPs are ubiquitous proteins found in organisms ranging from bacteria, fungi, viruses to plants and animals. In plants, CysPs play key roles in environmental stress response, nutrient remobilization and cellular housekeeping, and account for nearly 30% of the proteolytic activity in mature organs [23]. In Arabidopsis, CysPs are known to carry out multifarious roles in different tissues during development [24–26]. However, not much is known regarding the role of CysPs in tobacco. Few tobacco CysPs have been characterized which are implicated in stress response [27], protein degradation during programmed cell death [28], pollen grain development [29], and amino acid remobilization in senescing leaves [23].
Interleukin (IL)-10 is an immune-regulatory cytokine produced by immune cells that may turn into a new therapeutic target for the treatment of chronic inflammatory diseases [30]. IL-10 has been used as a model recombinant protein to study the factors that control the synthesis and accumulation of recombinant proteins in plants [31]. Despite several efforts towards increasing the production of IL-10 in tobacco [32–34], its accumulation during initial stable expression in whole plants was reported to be less than 1% of the total soluble protein, a level which is not sufficient for the viable commercial production of proteins. Here using a functional genomics approach, we identified a cysteine protease, CysP6, which affects IL-10 accumulation level in tobacco leaves. Furthermore, CysP6 is localized to the ER, indicating that the ER is the potential site of IL-10 proteolysis.
Materials and Methods
Plant materials and growth conditions
Seeds of tobacco (N. tabacum) cv. 81V9 and the transgenic line G7 homozygous for the IL-10 transgene (hereby called as IL-10 control) were grown in a green house at 24°C and 16 h daylight, with 60–70% relative humidity. The IL-10 control tobacco line overexpresses IL-10 that is designed to accumulate in the ER [32]. N. benthamiana plants were grown in a growth chamber under similar conditions.
In silico analysis
To identify CysP genes present in tobacco, a keyword search using “cysteine protease” was used against the DFCI tobacco gene index database (http://compbio.dfci.harvard.edu/tgi/). The database contains complete or partial CysP sequences obtained from sequencing of tobacco cDNA libraries. Candidate genes were selected for the study according to their expression in the leaves and their complete sequence availability in the database. The amino acid sequences of the full length CysPs were predicted using ExPASy software (http://web.expasy.org/translate/) and were aligned using ClustalW [35]. The domain organizations of putative CysPs were predicted using Pfam 27.0 (http://pfam.xfam.org/) and the subcellular localizations were predicted using PSORT (http://psort.hgc.jp/form.html).
Plasmid construction
To produce RNAi expression clones targeting candidate CysP genes, the gene fragments were PCR amplified using gene-specific primers (S1 Table) and cloned into pDONR/Zeo vector using the BP clonase reaction mix (Invitrogen, USA), sequence confirmed, and recombined into the RNAi destination vectors pB7GWIWG2(II),0 or pK7GWIWG2D(II),0 using the LR clonase reaction mix (Invitrogen, USA). The orientation of inserted CysP fragments in the destination vectors was confirmed using the gene-specific forward primers (CysPF) and the vector-specific chloramphenicol resistance (Cmr) primers (S1 Table). The recombinant destination vectors were transformed into A. tumefaciens GV3101 and used for plant transformations.
To study the subcellular localization, full-length CysP6 was amplified using CysP6-OE-F/R1 primers (S1 Table). The PCR product was recombined into the Gateway entry vector pDONR/Zeo, followed by recombination into the destination vector pEarlyGate101 to create a pEG101-CysP6-YFP.
Tobacco transformation
For stable and transient expression of CysP silencing constructs, A. tumefaciens strain GV3101 harbouring the constructs were grown in infiltration culture media (LB medium with 10 mM morpholino-ethanesulfonic acid [MES] pH 5.6, 100 μM acetosyringone), supplemented with rifampicin (10 μg/mL), gentamycin (50 μg/mL), and kanamycin (50 μg/mL for pK7GWIWG2D(II),0-CysP plasmids) or spectinomycin (50 μg/mL for pB7GWIWG2(II),0-CysP plasmids). The cultures were grown to an optical density (OD600) of 0.5–0.8 and centrifuged at 3000 g for 30 min. The pellets were resuspended in Gamborg’s solution (3.2 g/L Gamborg’s B5 and vitamins, 20 g/L sucrose, 10 mM MES pH, 5.6, 200 μM acetosyringone) to a final OD600 of 1, and incubated at room temperature for 2 h with gentle agitation to activate the virulence gene required for transformation.
Stable transformation of tobacco leaves was performed using a leaf disc transformation method. Briefly, approximately 5 mm2 tobacco leaf explants were submerged in the recombinant A. tumefaciens suspension cultures, blotted on a filter paper and placed on plates containing MS medium supplemented with naphthalene acetic acid (1 mg/L) and benzyl aminopurine (98 μg/L) (MST-agar). After 2–3 days of co-culture, the leaf explants were transferred to a new MST-agar plate containing timentin (500 mg/L) and BASTA (4 mg/L). The explants were then transferred to a new MST/BASTA plate after 3 days and sub-cultured in a new plate every week until calli were produced from the transformed plant cells. After the calli differentiated into independent shoots, the shoots were cut and transferred to a rooting MST-agar medium. CysP silenced shoots were transferred to soil after sufficient roots were produced. Individual lines were grown in different pots and transferred to the greenhouse (T0). T1 plants were also generated for a selected number of T0 lines. Both T0 and T1 plants were grown together with IL-10 control plants and used for the evaluation of IL-10 levels.
For transient silencing of CysP6, A. tumefaciences GV3101 harbouring the silencing construct was infiltrated into 7-week old tobacco plants. Fully expanded 4th, 5th and 6th leaves from the top were selected for infiltration, and the tissues were collected four days post-infiltration for total RNA and protein extraction.
RNA isolation, RT-PCR and qPCR
Total RNA was extracted from tobacco leaves using an RNeasy Plant Mini kit (Qiagen Inc., USA). An on-column DNase1 digestion was performed to eliminate genomic DNA contamination (Promega, USA). Total RNA (1 μg) was used for cDNA synthesis using a QuantiTect Reverse transcription Kit (Qiagen Inc., USA). A quantitative RT-PCR (qRT-PCR) was performed to check the expression of CysP6 using SsoFast EvaGreen Supermix (Bio-Rad Laboratories, Inc., USA) and a CFX96 real-time PCR detection system (Bio-Rad Laboratories, Inc., USA). CysP6 was amplified using the primers, 5’- CACCATATCCCTACTTCTCCTCCTC- 3’ and, 5’- CCATGTTCGACTAGCCATGACTC-3’. The amplicon was cloned into pGEM-T Easy vector (Promega, USA) and sequence verified. Tobacco actin (Gene bank ID: AB158612.1) was used as a reference gene and was amplified using the primers 5’-GGTTGGTATGGGTCAAAAGGATCA-3’ and 5’-GGAGCAACACGCAACTCATTG-3’.
RT-PCR was performed to check the expression of CysP6 in tobacco after transient expression of CysP6 silencing construct. Gene specific primers (S1 Table) were used for 36 cycles of PCR and the amplified product was visualized on a 1% agarose gel.
Protein extraction and ELISA
Protein extraction was performed in 1X phosphate buffer saline pH 7.4 containing 0.1% Tween-20, 1mM EDTA, 100 mM sodium ascorbate, 2% PVPP, 1 mM PMSF and 100 μg leupeptin. Total soluble protein in each sample was measured using the Bradford dye reagent (Bio-Rad Laboratories Inc., USA). IL-10 levels were checked using anti-human IL-10 monoclonal antibodies in a double sandwich ELISA following manufacturer’s instructions (BD Biosciences, Canada). Purified human IL-10 was used as standards in the ELISA (BD Biosciences, Canada).
Subcellular localization
For subcellular localization, 4–6 week-old N. benthamiana leaves were infiltrated with the Agrobacterium culture containing pEG101-CysP6-YFP construct. Transient expression of CysP6-YFP was visualized using a Leica TCS SP2 inverted microscope. A cyan fluorescent protein (CFP) targeted to retain the ER was used as a control to verify the probable localization of CysP6-YFP in the ER. The control construct was obtained from Arabidopsis Biological Resource Center (ABRC, clone name: ER-CK) [36]. For co-localization, Agrobacterium cultures containing pEG101-CysP6-YFP and pEG101-ERCFP constructs were mixed in a ratio of 1:1 and co-infiltrated into tobacco leaves followed by confocal microscopy. A 63X water immersion lens was used at an excitation wavelength of 514 nm for YFP and 434 nm for CFP. The emission wavelengths for YFP and CFP were between 530–560 nm and 460–490 nm, respectively.
Results
Tobacco genome contains at least 60 putative CysPs
An in silico analysis was performed using the DFCI tobacco gene index (TGI) database that contains 324,058 tobacco expressed sequence tags (ESTs). A total of 55 putative CysP genes were identified in the database using the key word ‘cysteine protease’. Out of 55 putative CysP sequences, 32 were tentative contig sequences which were created by assembling ESTs into virtual transcripts. The remaining 23 were singleton sequences, each of which represented a unique EST in the database. These putative CysPs were derived from tobacco cDNA libraries constructed from a variety of tissues including leaf, flower, root, whole seedling or cultured tobacco cell suspension cv Bright Yellow-2. Table 1 provides a list of putative CysP genes as represented by their tentative contig or singleton sequence identification numbers, their tissue specific expression and predicted protein sizes for those where complete coding regions were available. Of the 55 putative CysPs, 10 were selected as candidates for further study based on their complete sequence availability and expression in leaf tissue. Leaf specific CysPs were of main interest, as the principal objective of this research was to identify potential CysPs involved in determining IL-10 yield in the leaf tissue. The candidate CysPs were named as CysP1 to CysP10.
Table 1. List of putative CysPs, tissue expression and predicted protein size of candidate CysPs selected for silencing in tobacco.
| TC sequence | Most expressed in | Candidate CysP | Predicted Protein size (AA) | Singleton Sequence | Most expressed in | Protein size (AA) |
|---|---|---|---|---|---|---|
| TC122594 | R,L | CysP1 (NTCP23) | 360 | BP132783 | BY-2 | _ |
| TC123875 | R | _ | BP529990 | BY-2 | _ | |
| TC124565 | L,F,R | CysP2 | 240 | CN498801 | L | _ |
| TC124614 | R,L | _ | DV159633 | Seed | _ | |
| TC124631 | L,F,R | _ | DW001815 | L,R | _ | |
| TC126531 | L,F,R | CysP3 | _ | AM837057 | L | _ |
| TC128311 | L,F,R | _ | AM829003 | L | _ | |
| TC129167 | L,F,R | _ | AM831937 | L | _ | |
| TC129618 | L,F,R, BY-2 | CysP5 | 360 | AM791536 | L | _ |
| TC130400 | R | _ | AM787277 | L | _ | |
| TC130996 | L,F,R | _ | AM846161 | W | _ | |
| TC132948 | L,F,R | CysP6 (NTCP6) | 466 | AM834885 | W | _ |
| TC133645 | L,F,R | CysP4 | 349 | AM836936 | W | _ |
| TC133786 | BY-2 | _ | AM801664 | L | _ | |
| TC135985 | L,R | _ | AM787303 | W | _ | |
| TC139681 | L,F,R | _ | AM823454 | W | _ | |
| TC140530 | L,R,W | _ | AM801795 | W | _ | |
| TC141516 | L,F,R | CysP7 | 355 | AM844688 | W | _ |
| TC143494 | L,F,R | CysP8 | 434 | FG634183 | L | _ |
| TC143787 | L,F,R | CysP9 | 301 | FG644916 | L | _ |
| TC144748 | L, R,W | _ | FG167001 | R | _ | |
| TC145665 | L,R | _ | FG172560 | R | _ | |
| TC151953 | W | _ | FG190220 | L,F,R | _ | |
| TC152484 | L,F,R,W | _ | ||||
| TC154564 | L,R | _ | ||||
| TC155175 | L,F,R,W | _ | ||||
| TC162422 | L | _ | ||||
| TC163159 | L,W | _ | ||||
| TC164523 | L | _ | ||||
| TC164665 | L,F,R | _ | ||||
| TC166013 | Proembryo | _ | ||||
| TC166795 | L,F,R | CysP10 | 361 |
TC, tentative contig sequence; AA, amino acid; L, leaf; F, flower; R, root; W, whole plant, BY-2, Nicotiana tabacum cv. Bright Yellow cell line;-, partial CysP sequences
In addition to the 55 putative CysPs identified through a database search, five other CysP sequences were found through a literature search [29,37,38]. Their accession numbers and the paralog tentative contig sequences are shown in Table 2. Among them, NtCP1 was expressed only in senescing leaves while NtCP2, CyP7 and CyP8 in mature leaves. NtCP56 was strongly expressed in anthers. The untranslated region of NtCP56 and CysP10 are not conserved suggesting these two genes are not alleles. A very high degree of amino acid identity was found between NtCP56 and CysP10 (99.17%), which indicates that they might share functional similarity or belong to the members of same gene family in tobacco.
Table 2. Comparison of published tobacco CysP sequences with tentative contig sequences from tobacco EST database.
| CysP genes | Accession # | Closest tentative contig a | Amino Acid identity (%) | Reference |
|---|---|---|---|---|
| NtCP1 | AY881011 | TC133645 (CysP4) | 94.84 | [37] |
| NtCP2 | AY881010.1 | TC124565 (CysP2) | 96.6 | [37] |
| CyP7 | Z13959.1 | TC131927 | 97.8 | [38] |
| CyP8 | Z13964.1 | TC131927 | 97.5 | [38] |
| NtCP56 | EU429306.1 | TC166795 (CysP10) | 99.17 | [29] |
aClosed tentative contig in bold letters (candidate CysPs in parenthesis) share a high degree of amino acid identity with the respective published CysP sequences
Catalytic triad and non-contiguous ERFNIN motif are conserved in candidate CysPs
To determine the domain composition of the candidate CysP proteins in [32] tobacco, we performed the domain analysis using pfam database (http://pfam.xfam.org/). The analysis revealed that CysP1-CysP8 and CysP10 each contain a common papain domain, shown to be responsible for the endo-peptidase activity of CysPs. Proteins containing such domains are classified under the papain or peptidase_C1 family and are synthesized as precursor CysPs [39,40]. Precursor CysPs have an additional propeptide sequence preceding their papain domain, called I29 or the cathepsin propeptide inhibitor domain (Fig 1). CysP6 and CysP8 have an additional granulin-like repeat that is known to play a role in targeting and activity regulation of certain CysPs [41]. CysP9 contains a single large ovarian tumor (OTU) domain suggesting it may belong to the otubain or peptidase_C65, and possibly possess functions similar to deubiquitinating enzymes of the peptidase_C65 family. Detailed information regarding putative domain structure of all candidate CysPs is shown in Table 3.
Fig 1. Schematic representation of different domains in papain and otubain family CysPs.
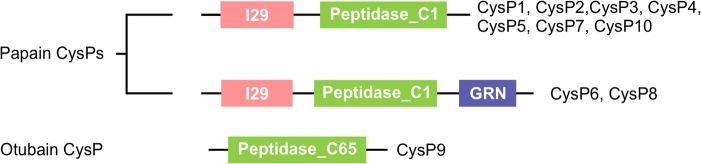
Candidate CysPs having specific domain types are indicated. The diagram is not drawn to scale. I29, Cathepsin propeptide inhibitor domain; Peptidase_C1, Papain family domain; GRN, Granulin like repeats; Peptidase_C65, Otubain family domain.
Table 3. Detail domain information of candidate tobacco CysPs.
| Candidate CysPs | Cathepsin propeptide inhibitor domain | Papain family CysP domain | Granulin like repeats | Otubain domain |
|---|---|---|---|---|
| CysP1(NTCP23) | 61–117 | 143–358 | X | X |
| CysP2 | Cleaved | 5–221 | X | X |
| CysP3 | 53–110 | 141–246* | X | X |
| CysP4 | 42–99 | 131–348 | X | X |
| CysP5 | 61–117 | 143–358 | X | X |
| CysP6(NTCP6) | 50–107 | 138–353 | 387–435 | X |
| CysP7 | 49–105 | 137–352 | X | X |
| CysP8 | 1–41 | 75–292 | 339–387 | X |
| CysP9 | X | X | X | 45–300 |
| CysP10 | 38–93 | 126–342 | X | X |
Numbers in the domains represent amino acids.
*complete coding region sequence was not available.
X, absent in respective CysPs.
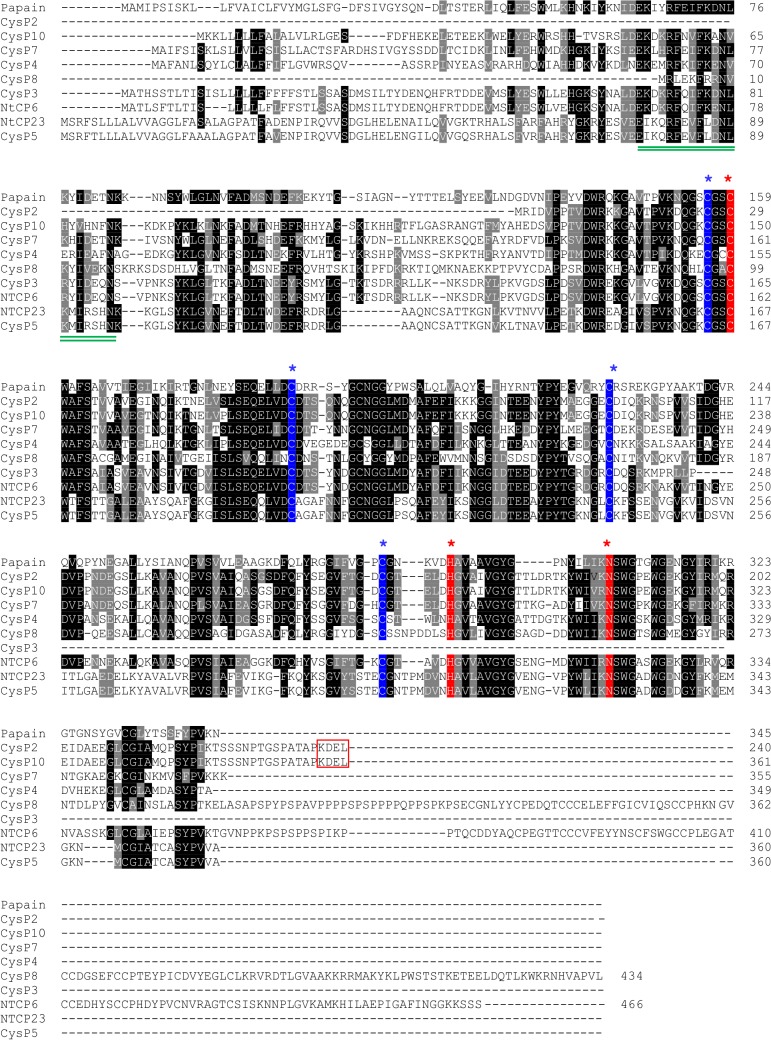
To identify catalytic residues in the CysPs, the deduced amino acid sequences of papain family CysPs identified in this study were aligned, along with papain and a previously characterized tobacco CysP, NTCP23 (Fig 2). A catalytic triad comprised of Cys25, His159 and Asn175 (papain numbering) was found in the papain domain of all aligned CysPs. Enzymatic activity of a CysP is dependent on Cys25 and His159 residues, which exist as ion-pairs in a pH interval of 3.5–8 [42]. Besides the catalytic Cys residues, four other Cys residues were also identified to be conserved in the papain domain of all aligned CysPs. These residues are likely involved in disulfide bridge formation leading to proper folding and activity of the enzymes. The alignment shown in Fig 2 excludes CysP3 as it lacked several N-terminal amino acid residues, and critical residues Asn and His in the catalytic triad. A highly conserved block of amino acids interspersed with variable residues, EX3 RX3 FX2 NX3 I/VX3 N (ERFNIN motif) is present in the propeptide region of all papain family CysPs [43].
Fig 2. Multiple sequence alignment of Peptidase_C1 (Papain) family candidate CysPs.
Predicted amino acid sequences of a papain, NTCP23 (CysP1), CysP2, CysP4, CysP5, NTCP6 (CysP6), CysP7, CysP8 and CysP10 were aligned using CLUSTALW and shaded using BOXSHADE 3.21. Identical and conserved amino acids are shaded by dark and light grey, respectively. (*) indicates catalytic residues cysteine (C), histidine (H) and asparagine (N). (*) indicates the disulfide bridge forming cysteine residues. Red box indicates endoplasmic reticulum retention signal KDEL in CysP2 and CysP10. The conserved ERFNIN motif in the propeptide region is double-underlined. C-terminal amino acids of CysP6 and CysP8 beyond the positions 410 and 362, respectively, are not shown in the alignment.
RNAi silencing of candidate CysP genes and generation of stable CysP silenced tobacco lines
To identify CysPs that influence IL-10 accumulation in tobacco, RNAi silencing of selected CysPs was performed. Unique regions were identified for each of the CysP sequences to facilitate targeted gene silencing. RNAi vectors with the specific CysP inserts were screened using two different primer combinations by PCR, which produced amplicons of different sizes. The first amplicon contained the larger intron (In-1) and the second amplicon contained the smaller intron (In-2) region (Fig 3A). A low alkaloid tobacco G7 line constitutively overexpressing IL-10 [32] was used for transforming the silencing constructs. Fig 3B outlines the process of generating CysP silenced tobacco plants and the approximate time required to proceed from one stage to the other. In approximately 16 weeks from initial transformation, fully grown plants were obtained. A total of 82 independent transgenic lines were generated, and utilized for the evaluation of CysP transcript and IL-10 levels.
Fig 3. RNAi silencing in tobacco.
RNAi constructs were generated for all candidate CysPs and the presence of CysP double inserts in the silencing vectors were confirmed using primer combinations as shown by the arrowheads (A). Stable CysP silenced tobacco lines were generated using the RNAi constructs (B). G7 tobacco lines overexpressing IL-10 protein were grown in magenta boxes. Leaf discs were cut and co-cultured with the Agrobacterium cultures containing the RNAi construct. Calli were regenerated from transformed cells and subcultured in differentiation medium containing BASTA. Independent CysP silenced transgenic lines were obtained from the individual callus and transferred to the rooting media before transferring them to the greenhouse.
In addition to the targeted silencing of specific candidate CysPs, a conserved region of CysP1 to CysP5 genes was also chosen for silencing. Two silenced lines were generated for CysPAll-Si. Since no RNAi line could be generated for CysP5, further work on this gene was not carried out.
Silencing of CysP6 increases IL-10 accumulation
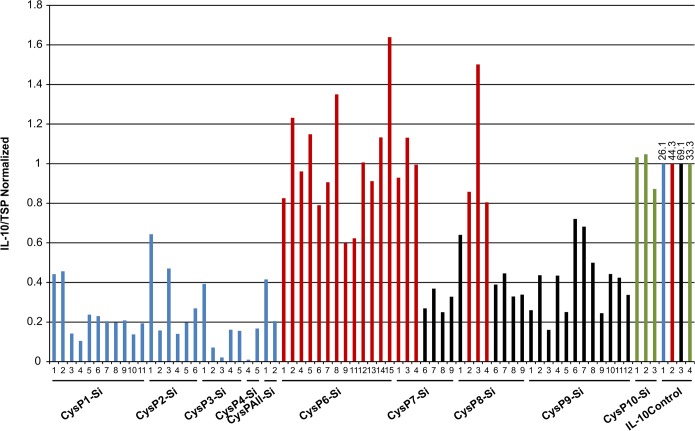
To determine the effect of CysP silencing in the accumulation of recombinant IL-10, we measured IL-10 levels in multiple independent transgenic silencing lines for CysP1-CysP4, CysP6-CysP10 and CysPAll. Except for several CysP6Si lines and CysP8Si-3, there was no increase in IL-10 accumulation compared to the IL-10 control plants in any other lines (Fig 4). Since these CysP silenced lines were generated over different periods of time, IL-10 control plants were also grown at the same time for an appropriate comparison of IL-10 levels. It was found that the accumulation of IL-10 varied among the controls, even though the plants were grown under similar conditions. Thus, IL-10 accumulations for different T0 lines are shown in normalized fold levels.
Fig 4. IL-10 accumulation in candidate CysP silenced T0 tobacco lines.
IL-10 accumulation remains lower in comparison to the controls in T0 generation of CysP1-CysP10 (except CysP6) silenced lines. Y-axis represents the normalized value of IL-10/TSP for different independent transgenic lines grown at different time period. Blue, red, black and green bars are normalized against their respective color coded IL-10 level in IL-10 control plant. Numbers above the bar represent actual IL-10 level in ng/mg of total soluble protein in control plants. Si, silenced.
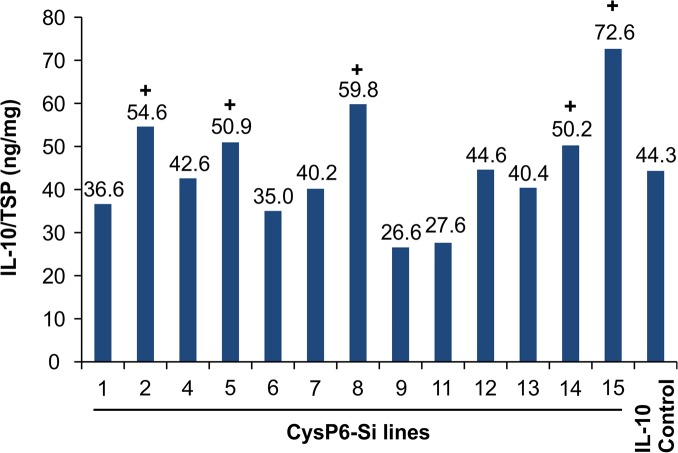
A closer look at CysP6Si lines revealed that IL-10 levels were higher in 5 out of 13 CysP6 silenced lines compared to the control (Fig 5). IL-10 level was increased by 1.6- fold in CysP6Si-15 as compared to the control. Three lines, CysP6Si-4, CysP6Si-7 and CysP6Si-13, displayed similar level of IL-10 accumulation in comparison to the control. The results indicated that silencing of CysP6 positively affects IL-10 accumulation in T0 transgenic tobacco lines.
Fig 5. IL-10 accumulation in CysP6 silenced T0 tobacco lines.
Measurement of IL-10 level in independent T0 CysP6 silenced lines by dsELISA. Data label above the bar represents IL-10 level of individual CysP6 silenced lines expressed as ng/mg of total soluble protein. + indicates the CysP6 silenced lines with higher level of IL-10 accumulation in comparison to the IL-10 control. TSP, total soluble protein; Si, silenced
Transient silencing of CysP6 increases IL-10 accumulation
To further examine the effect of CysP6 silencing on IL-10 accumulation levels, transient silencing experiments were performed in tobacco plants that were 1.5 ft tall. A. tumefaciens containing CysP6-Si construct was infiltrated into tobacco leaf epidermal cells and samples were collected for evaluating CysP6 transcript and IL-10 protein accumulation. Our RT-PCR analysis using gene specific primers did not detect any CysP6 transcript in all 7 biological replicates whereas it was present in vector-only plants (Fig 6A). This suggests that CysP6 was efficiently silenced by transient expression of the silencing construct. Furthermore, IL-10 levels were significantly higher in all 7 biological replicates which were infiltrated with the CysP6 silencing construct, than tissues infiltrated with the vector-only constructs (Fig 6B). These results demonstrated that transient silencing CysP6 results in the increase in IL-10 accumulation in G7 tobacco lines.
Fig 6. Silencing of CysP6 in tobacco using transient assay and stable transgenic lines.
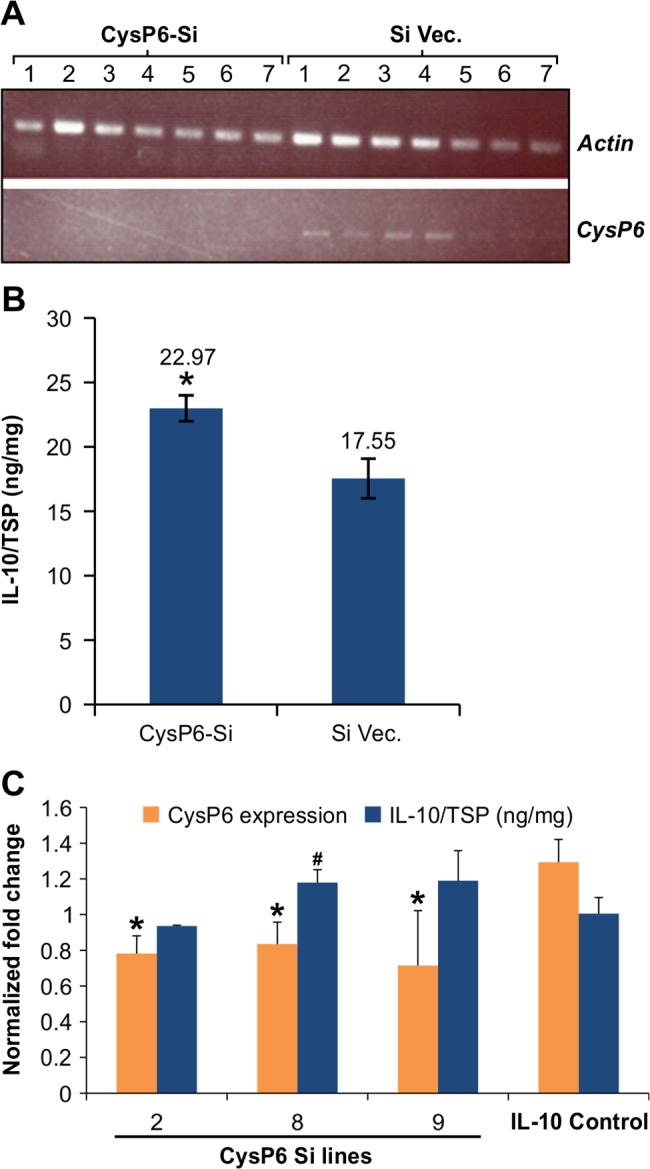
Leaves from seven weeks old G7 tobacco plants (4th, 5th and 6th from the top) were infiltrated with the CysP6 silencing (CysP6-Si) and empty vector constructs (Si vec.), and the tissues were used to determine the CysP6 transcript and IL-10 accumulation levels. CysP6 expressions were checked in CysP6-Si and Si Vec. infiltrated tissues, 4 days post infiltration. The numbers 1–7 indicate the biological replicates (B). The accumulation of IL-10 was measured in all the CysP6 silencing and vector only infiltrated tissues. The IL-10/TSP values are average of 7 biological replicates and asterisk (*) represents the significant difference in IL-10 level between CysP6-Si and vector only control using student t-test at 95% confidence level. Si, Silenced; Vec., Vector (C). Comparison of CysP6 expression and IL-10 accumulation in CysP6Si T1 lines. Blue bars represent IL-10/TSP normalized to IL-10 control and orange bars represent fold expression of CysP6 normalized to Actin. Three biological and three technical replicates were used. Error bars represent the standard errors of the biological and technical replicates (D). Asterisks (*) indicates the significant difference in CysP6 expression as compared to control, and hashtags (#) indicates the significant difference in IL-10 levels between CysP6-Si lines and IL-10 controls using Mann-Whitney U test (for qPCR data) and students t-test (for ELISA data) at P < 0.05.
Transcript abundance was also checked in selected stable CysP6 silenced T1 lines that showed higher IL-10 accumulation in T0 generation compared to control lines. As shown in Fig 6C, the lines 2, 8 and 9 showed reduced CysP6 expression in comparison to the IL-10 controls. CysP6 silencing significantly influenced IL-10 accumulation in the lines 8. However, IL-10 level was not altered significantly with the reduction of CysP6 transcript in the line 2 and 9. Despite this variability, we conclude that IL-10 accumulation was lower in the lines expressing higher CysP6 in both transient and stable lines.
CysP6 localizes to the ER
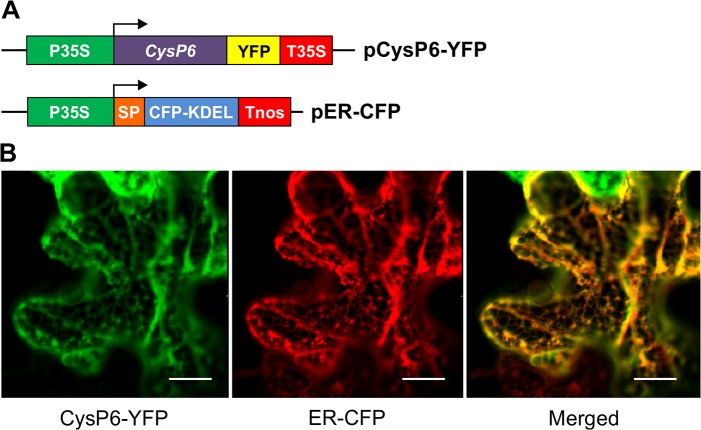
To evaluate the possibility that IL-10 is degraded by CysP6 in the secretory pathway, the subcellular localization of CysP6 was determined. A translational fusion of CysP6 and YFP was created (Fig 7A), and transiently co-expressed with ER-targeted CFP (control) in N. benthamiana leaf epidermal cells, followed by confocal microscopy. A typical ER-pattern showing net-like structures and co-localization of CysP6-YFP protein with ER-targeted CFP was observed, indicating that CysP6 localizes in the ER in tobacco leaf cells (Fig 7B).
Fig 7. CysP6 localizes to the ER.
Agrobacterium the carrying plasmids pCysP6-YFP and pER-CFP (ER control) were infiltrated in Nicotiana benthamiana (A). The localization was visualized after 48 hours using confocal microscopy (B). CysP6-YFP was visualized in the networks of the ER and co-localizes with the control, ER-CFP. Scale bars, 15 μm.
IL-10 accumulation increases with plant maturity in tobacco
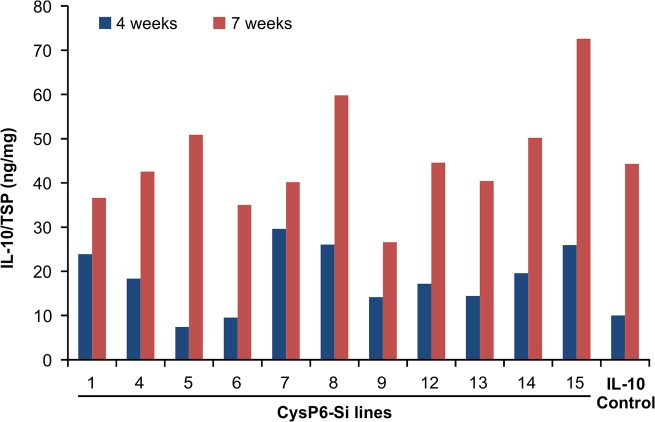
To evaluate if the age of transgenic tobacco plants affects the production of recombinant protein, IL-10 levels in CysP6 silenced lines and IL-10 controls were compared at two different time points. The plants were grown simultaneously in the greenhouse and IL-10 level was measured when they were either 4- or 7-weeks old (time after the transfer of plants from rooting media to soil). The plants were approximately 0.5 ft. tall at 4 weeks of age and approximately 1.5 ft. tall at 7 weeks of age. As shown in Fig 8, the levels of IL-10 accumulation in the leaf tissue increased as plants progressed to maturity in both CysP6 silenced and IL-10 control plants. Moreover, neither abnormal growth nor any morphological differences were observed for the CysP silenced lines compared to IL-10 controls.
Fig 8. IL-10 accumulation in tobacco plants of different stages.
IL-10 accumulation in 4 weeks and 7 weeks old CysP6 silenced tobacco lines. IL-10 accumulation increased consistently in CysP6 silenced lines and IL-10 controls with the age. The numbers indicate different independent transgenic lines carrying CysP6 silencing construct. TSP, total soluble protein; Si, silenced.
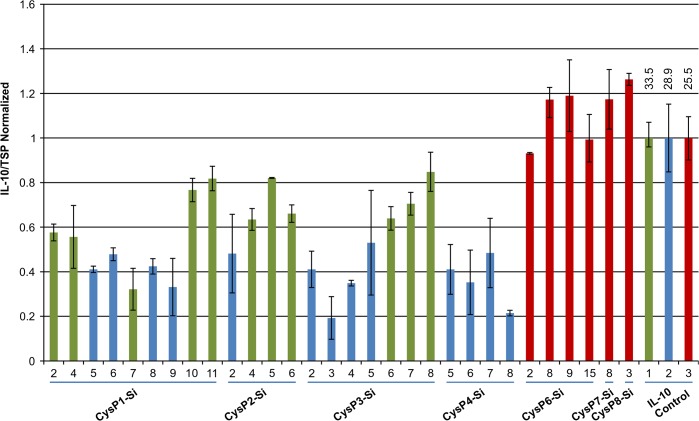
Comparison of IL-10 accumulation in CysP silenced T0 and T1 tobacco lines
To study the stability of IL-10 accumulation in CysP silenced tobacco plants, IL-10 levels were measured for a total of 30 different lines in both T0 and T1 generations. The accumulation of IL-10 was stable in most of the lines over successive generations of tobacco (Fig 9). CysP silenced T1 lines, CysP1-Si, CysP2-Si, CysP3-Si and CysP4-Si, all showed lower levels of IL-10 in comparison to their respective IL-10 control plants (represented by same color bars in Fig 9). Lower IL-10 accumulation was also seen for these lines in the T0 generation (Fig 4). Similarly, CysP6 silenced T1 lines showed nearly equal (lines 2 and 15) or higher (lines 8 and 9) levels of IL-10 in comparison to the respective control plant. Both CysP6-Si lines 2 and 15 accumulated higher IL-10 in T0 generation (Fig 9).This suggests that, the effect of CysP silencing on IL-10 accumulation in tobacco remains similar over generations.
Fig 9. IL-10 accumulation in CysP silenced T1 tobacco lines.
Normalized IL-10 levels of different CysP-Si T1 lines and controls. Green, blue and red bars are normalized to the IL-10 levels in their respective control plants (IL-10 control, on the right). Numbers above the bar represent the actual IL-10 levels in ng/mg of total soluble proteins. Error bars represent the mean of at least two biological replicates for all the lines except for CysP6-Si lines 8 and 15 (3 biological replicates).
Discussion
Tobacco CysPs belong to a large gene family with at least 60 members, and is comparable to that of other plants such as Arabidopsis (38 CysPs) and Populus (44 CysPs) [44]. Evolutionarily, tobacco (N. tabacum) originated from an interspecific cross of two wild forms, N. sylvestris and N. tomentosiformis. The interspecific cross between two diploid species with equal number of chromosomes (12 pairs each) resulted into a fertile amphiploid carrying a total of 24 pairs of chromosomes [45]. The larger number of CysPs in tobacco could have resulted from the preservation of CysPs from both of the parents. CysPs play critical roles in plant growth and development and during responses to different biotic and abiotic factors. Such roles for CysP have been reported in many plants including tobacco [23,38,46–49]. To identify the CysP genes that may be involved in IL-10 degradation, expressed tobacco CysPs were first identified using the TGI database. Since the tobacco genome sequence has not been published yet, the TGI database was the only major source of information regarding gene sequences and their tissue specific expression. A total of 60 putative CysPs were identified from the database and the literature search. The total number of CysPs may increase as more unique ESTs are sequenced or as the whole genome sequence of tobacco becomes available.
The papain family tobacco CysPs contained a long propeptide sequence (38–250 amino acids) and a papain domain (220–260 amino acids) (Table 3). Activation of these enzymes can occur by an intra- or inter-molecular proteolysis, where the propeptide sequence is cleaved off [39]. The propeptide sequence also showed a conserved ERFNIN motif which is shared by CysPs from diverse group of species [43]. Mammalian cathepsins with ERFNIN are known to carry out different intracellular and extracellular functions that require the right balance of enzyme activation. Intracellularly, they function in apoptosis and antigen processing, whereas extracellularly, they contribute directly to the degradation of foreign proteins and the extracellular matrix [50]. The signature ERFNIN motif is thought to be associated mostly with inhibiting protease activity before enzyme activation is required. CysP6 and CysP8 contain an extra GRN motif, the role of which in plants is not well understood, but is implicated in protein-protein interactions, targeting and/or activity regulation inside the cell [41].
Quantitative analysis of IL-10 levels in CysP6-silenced tobacco lines showed increased accumulation compared to the IL-10 control. Some CysP6-silenced lines showed no change in IL-10 accumulation while others accumulated lower levels in comparison to the IL-10 control plants. The accumulation of recombinant protein varied across different CysP6 silenced lines, both in T0 and T1 generation. The variation in the IL-10 levels could have resulted from genomic position effects of the inserted CysP or differential levels of CysP6 gene silencing. Such a position effect is a common character of RNAi-mediated silencing and has been described in several transgenic plants and animals [51]. Multiple genes were silenced in Arabidopsis using different RNAi constructs, which showed significant transcript variability between the independent RNAi lines of the same target gene [52]. In tobacco, the degree of gene silencing was analyzed in three independent lines carrying β-Glucuronidase (GUS) transgenes, and it was found that the amount of antisense GUS RNA correlated with the extent of post-transcriptional silencing in each line [53]. Differential silencing was also observed in the expression of rice CysPs, as small interfering (si) RNA was targeted to silence Rep1 and EP3A in transgenic rice cell lines expressing recombinant hGM-CSF [22].
Transient expression of the CysP6 RNAi construct resulted in near to invisible levels of CysP6 transcript and significantly increased the accumulation of IL-10 in tobacco. This also supports the fact that CysP6 is involved in regulating IL-10 accumulation in stable tobacco lines. With the post-genomic era and a flooding of gene sequences, several ways have been explored to assign putative gene functions. Because of its efficiency and specificity, RNAi is one of the widely used techniques in determining gene function. Most of the RNAi approaches utilize generation of stable transgenic lines and observation of RNAi phenotypes related to the particular genes. However, generation of stable lines usually requires a long time, which may range from 2–6 months depending on the type of plants used (4 months in tobacco, Fig 3B). Using Agrobacterium-mediated transient expression of the silencing construct, gene functions can be determined in a short period of time. As seen in this study, transient silencing of CysP6 could be achieved within a week, which is very short compared to the time required to generate stable transgenic tobacco lines. Transient silencing of indigenous genes via agro-infiltration had been reported so far in plant species such as Vitis vinifera [54], but not in tobacco. Particularly with the results seen here, transient RNAi can be encouraging for gene functional assays in tobacco.
Silencing of the candidate CysPs (CysP1-CysP5 and CysP7-CysP10) in tobacco did not increase IL-10 accumulation. Surprisingly, in all of these CysP silenced T0 independent transgenic lines, the level of IL-10 accumulation was lower than that of the IL-10 control plant. A detailed transcript analysis of all candidate CysPs may shed light on the extent these CysPs are silenced, and how the silencing relates to IL-10 accumulation seen in the individual silenced lines.
CysP6 fused to YFP showed that it is localized to the ER. It was interesting to see the localization pattern of CysP6, without the presence of the C-terminal ER retrieval signal sequence KDEL or HDEL (Fig 2). Several ER resident proteins that lack the retention signal are known to localize in the ER in a signal-independent fashion [55]. Arabidopsis RD21 is one of such papain protein without KDEL sequence, and is localized to the vacuole and ER in response to osmotic stress and wounding [55]. Some proteins retained in the ER also play a role in quality control through an interaction with ER chaperones, whose function is to retain and subsequently degrade assembly-defective proteins [56]. Localization in the ER could also suggest similar functions for CysP6 in tobacco.
Though recombinant proteins have been targeted to the ER for a higher yield, their production has been reported to be affected by plant proteases [57]. It was shown that IgG1 targeted to accumulate in the ER is possibly degraded by acidic proteases, most of which consisted of senescence associated CysPs [57]. Localization of CysP6 to the ER provides a good argument for its involvement in IL-10 degradation, since IL-10 accumulates in the ER as well. The degradation process might be occurring along the secretory route when IL-10 is being processed or accumulated. However, a detailed study is required regarding the fate of IL-10, to show that IL-10 degradation occurs within the ER thus affecting its overall yield in tobacco. This study identified and down-regulated several tobacco CysPs, and found CysP6 most likely to be IL-10- specific, is the initial step towards this goal.
Supporting Information
(DOCX)
Acknowledgments
We thank Alex Molnar for his help with the preparation of the figures.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Agriculture and Agri-Food Canada's Abase grant number J-000158.
References
- 1. Ma JK, Drake PM, Christou P (2003) The production of recombinant pharmaceutical proteins in plants. Nature reviews Genetics 4: 794–805. [DOI] [PubMed] [Google Scholar]
- 2. Sack M, Hofbauer A, Fischer R, Stoger E (2015) The increasing value of plant-made proteins. Current Opinion in Biotechnology 32: 163–170. 10.1016/j.copbio.2014.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stoger E, Fischer R, Moloney M, Ma JK-C (2014) Plant Molecular Pharming for the Treatment of Chronic and Infectious Diseases. Annual Review of Plant Biology 65: 743–768. 10.1146/annurev-arplant-050213-035850 [DOI] [PubMed] [Google Scholar]
- 4. Conley AJ, Zhu H, Le LC, Jevnikar AM, Lee BH, Brandle JE, et al. (2011) Recombinant protein production in a variety of Nicotiana hosts: a comparative analysis. Plant Biotechnology Journal 9: 434–444. 10.1111/j.1467-7652.2010.00563.x [DOI] [PubMed] [Google Scholar]
- 5. Barta A, Sommergruber K, Thompson D, Hartmuth K, Matzke MA, Matzke AJ, et al. (1986) The expression of a nopaline synthase—human growth hormone chimaeric gene in transformed tobacco and sunflower callus tissue. Plant Mol Biol 6: 347–357. 10.1007/BF00034942 [DOI] [PubMed] [Google Scholar]
- 6. Kolotilin I, Topp E, Cox E, Devriendt B, Conrad U, Joensuu J, et al. (2014) Plant-based solutions for veterinary immunotherapeutics and prophylactics. Vet Res 45: 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fischer R, Schillberg S, Buyel JF, Twyman RM (2013) Commercial aspects of pharmaceutical protein production in plants. Curr Pharm Des 19: 5471–5477. [DOI] [PubMed] [Google Scholar]
- 8. Fischer R, Schillberg S, Hellwig S, Twyman RM, Drossard J (2012) GMP issues for recombinant plant-derived pharmaceutical proteins. Biotechnology Advances 30: 434–439. 10.1016/j.biotechadv.2011.08.007 [DOI] [PubMed] [Google Scholar]
- 9. Fischer R, Stoger E, Schillberg S, Christou P, Twyman RM (2004) Plant-based production of biopharmaceuticals. Current Opinion in Plant Biology 7: 152–158. [DOI] [PubMed] [Google Scholar]
- 10. Faye L, Boulaflous A, Benchabane M, Gomord V, Michaud D (2005) Protein modifications in the plant secretory pathway: current status and practical implications in molecular pharming. Vaccine 23: 1770–1778. [DOI] [PubMed] [Google Scholar]
- 11. Doran PM (2006) Foreign protein degradation and instability in plants and plant tissue cultures. Trends in Biotechnology 24: 426–432. [DOI] [PubMed] [Google Scholar]
- 12. Sharp JM, Doran PM (2001) Characterization of monoclonal antibody fragments produced by plant cells. Biotechnology and Bioengineering 73: 338–346. [DOI] [PubMed] [Google Scholar]
- 13. Hehle VK, Paul MJ, Drake PM, Ma JK, van Dolleweerd CJ (2011) Antibody degradation in tobacco plants: a predominantly apoplastic process. BMC Biotechnol 11: 128 10.1186/1472-6750-11-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. De Neve M, De Loose M, Jacobs A, Van Houdt H, Kaluza B, Weidle U, et al. (1993) Assembly of an antibody and its derived antibody fragment in Nicotiana and Arabidopsis. Transgenic Res 2: 227–237. [DOI] [PubMed] [Google Scholar]
- 15. van Engelen FA, Schouten A, Molthoff JW, Roosien J, Salinas J, Dirkse WG, et al. (1994) Coordinate expression of antibody subunit genes yields high levels of functional antibodies in roots of transgenic tobacco. Plant Mol Biol 26: 1701–1710. [DOI] [PubMed] [Google Scholar]
- 16. Schiermeyer A, Schinkel H, Apel S, Fischer R, Schillberg S (2005) Production of Desmodus rotundus salivary plasminogen activator alpha1 (DSPAalpha1) in tobacco is hampered by proteolysis. Biotechnology and Bioengineering 89: 848–858. [DOI] [PubMed] [Google Scholar]
- 17. Stevens LH, Stoopen GM, Elbers IJ, Molthoff JW, Bakker HA, Lommen A, et al. (2000) Effect of climate conditions and plant developmental stage on the stability of antibodies expressed in transgenic tobacco. Plant Physiology 124: 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Russell DA (1999) Feasibility of antibody production in plants for human therapeutic use. Curr Top Microbiol Immunol 240: 119–138. [DOI] [PubMed] [Google Scholar]
- 19. Russell DA, Spatola LA, Dian T, Paradkar VM, Dufield DR, Carroll JA, et al. (2005) Host limits to accurate human growth hormone production in multiple plant systems. Biotechnology and Bioengineering 89: 775–782. [DOI] [PubMed] [Google Scholar]
- 20. Outchkourov NS, Rogelj B, Strukelj B, Jongsma MA (2003) Expression of sea anemone equistatin in potato. Effects of plant proteases on heterologous protein production. Plant Physiology 133: 379–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Khoudi H, Laberge S, Ferullo JM, Bazin R, Darveau A, Castonguay Y, et al. (1999) Production of a diagnostic monoclonal antibody in perennial alfalfa plants. Biotechnol Bioeng 64: 135–143. [DOI] [PubMed] [Google Scholar]
- 22. Kim NS, Kim TG, Kim OH, Ko EM, Jang YS, Jung ES, et al. (2008) Improvement of recombinant hGM-CSF production by suppression of cysteine proteinase gene expression using RNA interference in a transgenic rice culture. Plant Mol Biol 68: 263–275. 10.1007/s11103-008-9367-8 [DOI] [PubMed] [Google Scholar]
- 23. Ueda T, Seo S, Ohashi Y, Hashimoto J (2000) Circadian and senescence-enhanced expression of a tobacco cysteine protease gene. Plant molecular biology 44: 649–657. [DOI] [PubMed] [Google Scholar]
- 24. Lee S, Jung KH, An G, Chung YY (2004) Isolation and characterization of a rice cysteine protease gene, OsCP1, using T-DNA gene-trap system. Plant molecular biology 54: 755–765. [DOI] [PubMed] [Google Scholar]
- 25. Gan S, Amasino RM (1995) Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270: 1986–1988. [DOI] [PubMed] [Google Scholar]
- 26. Gietl C, Schmid M (2001) Ricinosomes: an organelle for developmentally regulated programmed cell death in senescing plant tissues. Naturwissenschaften 88: 49–58. [DOI] [PubMed] [Google Scholar]
- 27. Linthorst HM, van der Does C, Brederode F, Bol J (1993) Circadian expression and induction by wounding of tobacco genes for cysteine proteinase. Plant Molecular Biology 21: 685–694. [DOI] [PubMed] [Google Scholar]
- 28. Zhao P, Zhou XM, Zhang LY, Wang W, Ma LG, Yang LB, et al. (2013) A bipartite molecular module controls cell death activation in the Basal cell lineage of plant embryos. PLoS Biol 11: e1001655 10.1371/journal.pbio.1001655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang XM, Wang Y, Lv XM, Li H, Sun P, Lu H, et al. (2009) NtCP56, a new cysteine protease in Nicotiana tabacum L., involved in pollen grain development. Journal of Experimental Botany 60: 1569–1577. 10.1093/jxb/erp022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Asadullah K, Sterry W, Volk HD (2003) Interleukin-10 Therapy—Review of a New Approach. Pharmacological Reviews 55: 241–269. [DOI] [PubMed] [Google Scholar]
- 31.Chen L, Dempsey BR, Gyenis L, Menassa R, Brandle JE, Dhaubhadel S (2013) Identification of the factors that control synthesis and accumulation of a therapeutic protein, human immune-regulatory interleukin-10, in Arabidopsis thaliana. Plant Biotechnology Journal. [DOI] [PubMed]
- 32. Menassa R, Nguyen V, Jevnikar A, Brandle J (2001) A self-contained system for the field production of plant recombinant interleukin-10. Molecular Breeding 8: 177–185. [Google Scholar]
- 33. Menassa R, Kennette W, Nguyen V, Rymerson R, Jevnikar A, Brandle J (2004) Subcellular targeting of human interleukin-10 in plants. Journal of Biotechnology 108: 179–183. [DOI] [PubMed] [Google Scholar]
- 34. Westerhof LB, Wilbers RHP, Roosien J, van de Velde J, Goverse A, Bakker J, et al. (2012) 3D Domain Swapping Causes Extensive Multimerisation of Human Interleukin-10 When Expressed In Planta. PLoS ONE 7: e46460 10.1371/journal.pone.0046460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, et al. (2013) Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res 41: W597–600. 10.1093/nar/gkt376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. The Plant Journal 51: 1126–1136. [DOI] [PubMed] [Google Scholar]
- 37. Beyene G, Foyer CH, Kunert KJ (2006) Two new cysteine proteinases with specific expression patterns in mature and senescent tobacco (Nicotiana tabacum L.) leaves. Journal of Experimental Botany 57: 1431–1443. [DOI] [PubMed] [Google Scholar]
- 38. Linthorst HJ, van der Does C, Brederode FT, Bol JF (1993) Circadian expression and induction by wounding of tobacco genes for cysteine proteinase. Plant Mol Biol 21: 685–694. [DOI] [PubMed] [Google Scholar]
- 39. Wiederanders B, Kaulmann G, Schilling K (2003) Functions of propeptide parts in cysteine proteases. Curr Protein Pept Sci 4: 309–326. [DOI] [PubMed] [Google Scholar]
- 40. Grudkowska M, Zagdanska B (2004) Multifunctional role of plant cysteine proteinases. Acta Biochim Pol 51: 609–624. [PubMed] [Google Scholar]
- 41. Tolkatchev D, Xu P, Ni F (2001) A peptide derived from the C-terminal part of a plant cysteine protease folds into a stack of two beta-hairpins, a scaffold present in the emerging family of granulin-like growth factors. J Pept Res 57: 227–233. [DOI] [PubMed] [Google Scholar]
- 42. Grzonka Z, Jankowska E, Kasprzykowski F, Kasprzykowska R, Lankiewicz L, Wiczk W, et al. (2001) Structural studies of cysteine proteases and their inhibitors. Acta Biochim Pol 48: 1–20. [PubMed] [Google Scholar]
- 43. Karrer KM, Peiffer SL, DiTomas ME (1993) Two distinct gene subfamilies within the family of cysteine protease genes. Proc Natl Acad Sci U S A 90: 3063–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Garcia-Lorenzo M, Sjodin A, Jansson S, Funk C (2006) Protease gene families in Populus and Arabidopsis. BMC Plant Biol 6: 30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Leitch IJ, Hanson L, Lim KY, Kovarik A, Chase MW, Clarkson JJ, et al. (2008) The ups and downs of genome size evolution in polyploid species of Nicotiana (Solanaceae). Ann Bot 101: 805–814. 10.1093/aob/mcm326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ho SL, Tong WF, Yu SM (2000) Multiple mode regulation of a cysteine proteinase gene expression in rice. Plant Physiol 122: 57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Koizumi M, Yamaguchi-Shinozaki K, Tsuji H, Shinozaki K (1993) Structure and expression of two genes that encode distinct drought-inducible cysteine proteinases in Arabidopsis thaliana. Gene 129: 175–182. [DOI] [PubMed] [Google Scholar]
- 48. Schaffer MA, Fischer RL (1988) Analysis of mRNAs that Accumulate in Response to Low Temperature Identifies a Thiol Protease Gene in Tomato. Plant Physiol 87: 431–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bernoux M, Timmers T, Jauneau A, Briere C, de Wit PJ, Marco Y, et al. (2008) RD19, an Arabidopsis cysteine protease required for RRS1-R-mediated resistance, is relocalized to the nucleus by the Ralstonia solanacearum PopP2 effector. Plant Cell 20: 2252–2264. 10.1105/tpc.108.058685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dickinson DP (2002) Cysteine peptidases of mammals: their biological roles and potential effects in the oral cavity and other tissues in health and disease. Crit Rev Oral Biol Med 13: 238–275. [DOI] [PubMed] [Google Scholar]
- 51. Matzke AJ, Matzke MA (1998) Position effects and epigenetic silencing of plant transgenes. Curr Opin Plant Biol 1: 142–148. [DOI] [PubMed] [Google Scholar]
- 52. Kerschen A, Napoli CA, Jorgensen RA, Muller AE (2004) Effectiveness of RNA interference in transgenic plants. FEBS Lett 566: 223–228. [DOI] [PubMed] [Google Scholar]
- 53. Hamilton AJ, Baulcombe DC (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286: 950–952. [DOI] [PubMed] [Google Scholar]
- 54. Bertazzon N, Raiola A, Castiglioni C, Gardiman M, Angelini E, Borgo M, et al. (2012) Transient silencing of the grapevine gene VvPGIP1 by agroinfiltration with a construct for RNA interference. Plant Cell Rep 31: 133–143. 10.1007/s00299-011-1147-2 [DOI] [PubMed] [Google Scholar]
- 55. Gu C, Shabab M, Strasser R, Wolters PJ, Shindo T, Niemer M, et al. (2012) Post-translational regulation and trafficking of the granulin-containing protease RD21 of Arabidopsis thaliana. PLoS One 7: e32422 10.1371/journal.pone.0032422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hammond C, Helenius A (1995) Quality control in the secretory pathway. Curr Opin Cell Biol 7: 523–529. [DOI] [PubMed] [Google Scholar]
- 57. Stevens LH, Stoopen GM, Elbers IJ, Molthoff JW, Bakker HA, Lommen A, et al. (2000) Effect of climate conditions and plant developmental stage on the stability of antibodies expressed in transgenic tobacco. Plant Physiol 124: 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.