Dear Editor,
It is now believed that the human genome contains a significant portion of regulatory bases, including a great number of transcription factor (TF)-binding cis-elements1, whose functions are largely unexplored due to the lack of convenient experimental methods to model element-specific loss-of-function. Based on recent technological breakthroughs adapting the bacterial type II CRISPR/Cas9 system for RNA-guided gene editing and transcriptional programming2, we tested the application of the non-cleavage dCas9/sgRNA as a sequence-specific cis-element-targeting platform in human cells. Our study established a convenient, efficient and flexible approach that may pave the way for functional annotation of TF-binding cis-elements in higher organisms.
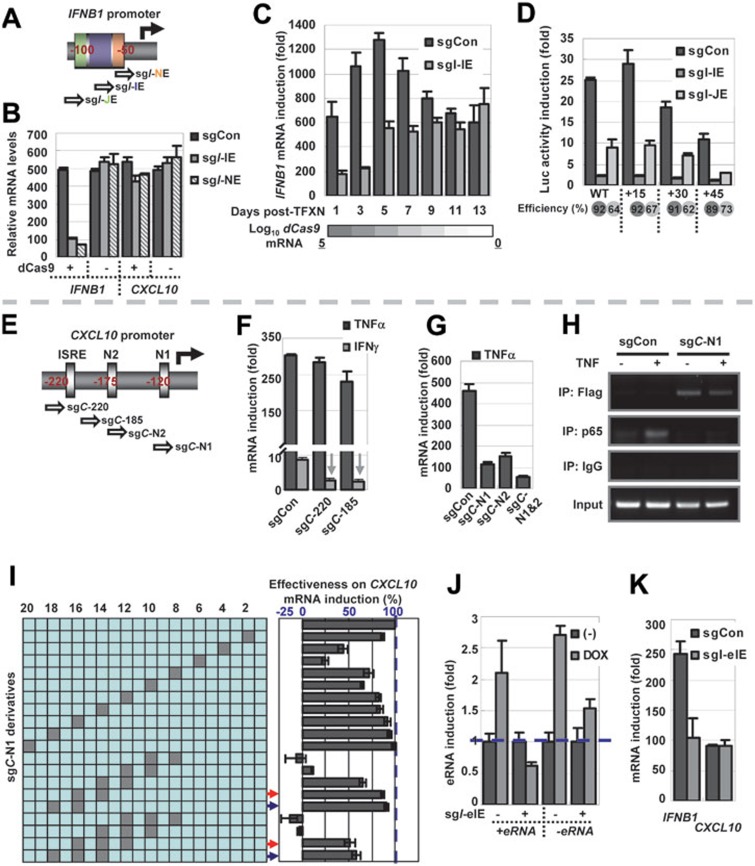
The sequence-specific DNA-binding property of dCas9/sgRNA was previously harnessed to map transcriptional cis-regulatory elements in E. coli and yeast3,4. Although a similar approach in human cells targeting cis-elements in the SV40 viral promoter of an SV-40-GFP reporter was inefficient4, no vigorous tests against endogenous cis-elements have been carried out. For proof-of-principle, we first focused on several cis-elements in proximal promoters, which mediate strong transcriptional outputs. Transcriptional activation of IFNB1 (gene for one of the anti-viral type I IFNs) was used as an initial test model due to the substantial understandings of the underlying mechanisms5. Signaling pathways triggered by virus-derived nucleic acid patterns commonly result in concurrent activation of TFs c-Jun/ATF2, IRF3/7 and NF-κB, which are in turn recruited to a tandem of closely situated cis-regulatory elements in the proximal promoter of IFNB1 to cooperatively activate its transcription (Supplementary information, Figure S1A). To mimic the latter process, we introduced a tet-on expression construct for a viral RNA-responsive signaling adaptor, i.e., MAVS5, into human embryonic kidney 293T cells. Upon deoxycyclin (DOX) administration, IFNB1 expression was robustly induced in these cells (Supplementary information, Figure S1B and S1C). We next designed sgRNAs largely covering each (or in one case two) of the TF-binding sites within the IFNB1 promoter (Figure 1A and Supplementary information, Figure S1A). Remarkably, these sgRNAs all effectively inhibited the DOX-induced activities of co-transfected IFNB1 promoter reporter (Supplementary information, Figure S1D, upper panel). Similar results were obtained when the activity of an integrated IFNB1 promoter reporter (Supplementary information, Figure S1E) or the endogenous IFNB1 mRNA level was analyzed (Figure 1B). The inhibition of IFNB1 mRNA induction by the sgRNAs was dCas9-dependent (Figure 1B) and accompanied by the inhibition of MX2, a classic example of IFNβ-stimulated genes (ISGs; Supplementary information, Figure S1F). Consistently, the culture supernatant from the cis-element (IFNB1)-targeted cells was markedly less active in increasing the protein levels of another ISG, i.e., STAT1, in naïve cells (Supplementary information, Figure S1G). Furthermore, the inhibitory effects by the transfected dCas9/sgRNA were dose- and time-dependent (Supplementary information, Figure S1H and Figure 1C), clearly demonstrating a tunable feature. In contrast to the drastic decrease of IFNB1 expression, the mRNA level of a chemokine gene CXCL10 was only slightly reduced (Figure 1B and Supplementary information, Figure S1H). Such a minor effect on a non-target gene (CXCL10) may possibly be attributed to autocrine regulation by type I IFN (Supplementary information, Figure S1I).
Figure 1.
A dCas9/sgRNA platform for efficient and flexible targeting of TF-responsive cis-elements. (A) A schematic display of the proximal promoter of IFNB1. The position of the nucleotides in relation to the TSS is presented in red. The sgRNAs (named according to their overlaps with IRF-responsive element, NFκB-responsive element and Jun/ATF2-responsive element as sgI-IE, sgI-NE and sgI-JE, respectively) are presented as small block arrows pointing towards the PAM. (B) MAVS tet-on cells were transfected with the indicated sgRNAs with or without dCas9 and subjected to drug selection. Twenty-four hours later, cells were treated with DOX for another 24 h. The cDNAs were analyzed in quadruplicates via qPCR. (C) Cells were transfected with sgCon or sgI-IE in combination with dCas9, and were drug-selected for the initial 48 h. At the indicated time points following the completion of drug selection, cells were treated with DOX for 24 h before harvest. The folds of IFNB1 mRNA induction are presented in bar graphs. The relative levels of dCas9 mRNA (log scale) were also determined and presented in a heat-map below (created by IrfanView). A 105-fold decrease of dCas9 mRNA level can be observed over two weeks. (D) A 15-, 30- or 45-bp spacer sequence was placed upstream of the TATA-box in the IFNB1-Luc reporter. MAVS tet-on cells were co-transfected with the indicated dCas9/sgRNA and each modified reporter in triplicates. The DOX-inducible reporter activity (24 h) was determined. The inhibitory efficiency (%) by either sgRNA on each reporter was calculated (numbers in shaded circles). (E) A schematic display of the endogenous CXCL10 proximal promoter is presented as in A. The two NFκB-responsive elements are marked as N1 and N2, respectively. The sgRNAs were named either according to the overlapped cis-elements (i.e., sgC-N1 and sgC-N2), or to their target positions (220 bp (sgC-220) and 185 bp (sgC-185) upstream of TSS). (F, G) Cells were transfected with the indicated dCas9/sgRNA. Following 36 h of drug selection, cells were treated with TNFα or IFNγ for 12 h. The levels of CXCL10 mRNA were measured. In F, notable inhibitory effects were marked with grey arrows. (H) ChIP was performed using antibodies against Flag (dCas9) or p65 (NFκB) in dCas9/sgRNA-transfected cells treated with or without TNFα for 3 h. Primers corresponding to the proximal promoter of CXCL10 were used for PCR analysis. (I) Single-, double- or triple-nucleotide mutations were introduced into various positions in sgC-N1. Experiments were performed similar to G. The inhibitory efficiency by the WT sgC-N1 (inhibition of TNFα-induced expression of CXCL10 mRNA) was set as 100%. Error bars represent normalized STDEV from results of quadruplicated qPCR reactions. (J, K) MAVS tet-on cells were transfected with sgRNA targeting the IFNB1 enhancer IRF3 element (sgI-eIE) together with dCas9, drug-selected for the initial 48 h and then treated with DOX for 24 h. The levels of indicated eRNAs (J) and mRNAs (K) were analyzed.
In comparison to previously described sgRNAs that enabled dCas9 to trigger strong interference with transcriptional initiation/elongation (targeting the −50 to +300 bp region relative to the transcription start site (TSS))3,6, some of the sgRNAs described thus far in our study, e.g., sgI-IE and sgI-JE, were designed against the IFNB1 promoter sequences upstream of the −50-bp position. Nevertheless, to confirm that sgRNAs could guide inhibition of IFNB1 transcription independently of general interference, we inserted a 15-, 30- or 45-bp spacer sequence to the 5′ side of the TATA-box in the IFNB1 luciferase reporter to move the clustered virus-responsive cis-elements further upstream (Figure 1D). The results demonstrated that the inclusion of none of the spacers affected the inhibitory efficiency of either sgI-IE or sgI-JE, strongly supporting that dCas9/sgRNA-mediated cis-element-specific blockage underlies the suppression of IFNB1 transcription.
Since the clustered TF-binding sites within the proximal promoter of IFNB1 function cooperatively5, we seek other systems to examine the spatial rule, where the dCas9/sgRNA complex may selectively block a cis-element without affecting another one nearby. We constructed an artificial double-cis-element reporter with an upstream two-copy NFκB-responsive element (2×NκRE) and a downstream (following a 60-bp spacer) five-copy interferon stimulation-responsive element (5×ISRE7), which are respectively targeted by TNFα and IFNα (Supplementary information, Figure S2A). Two separate sgRNAs against the 2×NκRE were both effective in abrogating TNFα-induced reporter activity. In contrast, neither of these two sgRNAs affected IFNα-induced reporter activity (5×ISRE-mediated), clearly demonstrating an example of sub-promoter, cis-element-level functional selectivity of dCas9/sgRNA (Supplementary information, Figure S2B). Next, to probe the nucleotide-level resolution of dCas9/sgRNA-mediated cis-element targeting, we focused on the endogenous promoter of CXCL10 that harbors an array of single-copy cis-elements, including two proximal NκREs (denoted as N1 and N2) and an upstream ISRE, respectively activated by TNFα and IFNγ8. We designed several sgRNAs targeting these cis-regulatory elements (Figure 1E). Indeed, two sgRNAs closely flanking the CXCL10 ISRE (sgC-220 and sgC-185) substantially inhibited IFNγ-induced CXCL10 expression (Figure 1F). In contrast, neither of these two ISRE-oriented sgRNAs markedly affected TNFα-induced CXCL10 expression (Figure 1F). Since the base-paired target of sgC-185 is only 10 bp away from the N2 element that is TNFα-responsive (Figure 1E), the latter result suggests that the dCas9/sgRNA complex exhibits a rather localized, ∼40-nt cis-element-targeting resolution (20-nt paired bases plus 10 flanking nucleotides on either side) in live cells. Such results are in general agreement with the spatial details of CRISPR/Cas9 foot-printing data in vitro9.
Since TNFα acts via both of the NκREs within the CXCL10 promoter8, we designed two corresponding sgRNAs (sgC-N1 and sgC-N2, Figure 1E). These two sgRNAs were both effective in inhibiting TNFα-induced CXCL10 expression (Figure 1G). Additionally, ChIP experiments confirmed the binding of dCas9/sgC-N1 to the proximal promoter of CXCL10, accompanied by a concomitant loss of TNFα-dependent NFκB recruitment (Figure 1H), providing direct evidence that dCas9/sgRNA inhibits cis-element activities via a mechanism of exclusive binding. Furthermore, when sgC-N1 and sgC-N2 were applied together to simultaneously target both the N1 and N2 elements, an additive inhibitory effect on TNFα-induced CXCL10 expression was observed (Figure 1G), consistent with the notion that dCas9/sgRNA-mediated targeting of cis-elements is multiplexible.
To further assess the fidelity of dCas9/sgRNA-mediated cis-element inhibition, we made a series of single-, double- and triple-nucleotide mutations in sgC-N1 and analyzed their effects on endogenous CXCL10 mRNA induction upon TNFα treatment. For this particular target site, single-nucleotide mismatches at most of the positions (except for positions 4 and 6) were tolerated (Figure 1I). In contrast, double- or triple-nucleotide mismatches around the center of the target site abrogated the targeting activities (Figure 1I). Although sgC-N1 variants with double or triple mismatches near the 5′ end were still relatively effective, triple mismatches therein were apparently less tolerated than the double mismatches (see paired comparisons indicated by red and blue arrows, Figure 1I). The overall trend was reproduced using similar mutants of sgI-IE targeting the IFNB1 promoter reporter, although in the latter case, a generally more stringent base-pairing requirement was observed (Supplementary information, Figure S2D). Therefore, CRISPR/dCas9-mediated non-cleavage cis-element targeting is dependent on extensive base matches between the sgRNA and target DNA. Consistently, the cis-element-targeting efficiencies of sgC-N1 variants when coexpressed with dCas9 largely correlated with their indel-inducing activities when coexpressed with Cas9 (Supplementary information, Figure S2C).
Lastly, we tested the application of the dCas9/sgRNA system on a non-promoter, TF-responsive cis-element. In principle, such cis-elements are commonly situated within accessible chromatin domains1 permissive to dCas9/sgRNA binding10,11. It came to our attention that a very recent report identified a novel upstream (∼20 kb) enhancer region for human IFNB1, where a bi-directional eRNA transcription was also observed following viral stimulation12. Interestingly, the enhancer region contains an IRF3-binding site located between the bi-directional eRNAs, potentially serving as a critical element that drives eRNA transcription. To probe the physiological connections among the events of IRF3 binding, eRNA transcription and enhancer activation, we designed dCas9/sgRNA against this novel IFNB1 enhancer IRF3-responsive element (I-eIE) in the MAVS tet-on cells (Supplementary information, Figure S2E). We found that the I-eIE-targeting dCas9/sgRNA significantly inhibited DOX-induced expression of both eRNA strands (Figure 1J). Additionally, the same dCas9/sgRNA also partially reduced DOX-induced IFNB1, but not CXCL10 mRNA levels (Figure 1K), confirming the role of this enhancer-born IRF3 cis-element in downstream transcription of IFNB112. Therefore, our data suggest that the dCas9/sgRNA platform is also suitable for mapping non-promoter, TF-responsive cis-elements at their endogenous loci.
In summary, by using a simple non-cleavage dCas9/sgRNA platform with minimal manipulation and optimization, we have consistently (11 out of 11 sgRNAs tested) achieved effective cis-element targeting in several promoter/enhancer models in a human cell line. Such efficiencies are remarkable, considering that no additional repressor domains2 were fused to dCas9 and that the method was designed to directly limit the access of endogenous TFs to the targeted cis-elements. The latter attribute translates into a direct, cis-element-oriented, loss-of-function perturbation strategy that is indeed suitable for faithful functional annotation of novel genomic loci. Our data further demonstrate that in addition to high resolution and multiplexibility, dCas9/sgRNA-mediated cis-element mapping also features good functional fidelity, consistent with a recent report on CRISPRi analyses6. Moreover, dCas9/sgRNA-mediated cis-element targeting is intrinsically reversible and tunable, owing to the underlying non-cleavage, site-blockage mechanism. Therefore, the present study establishes a useful and flexible method that may be further exploited in relevant cell types and even in animal models to ultimately characterize the large-numbered, function- and disease-associated cis-regulatory elements revealed by accumulating human genomics studies1. Such a new approach shall make significant contributions to diverse areas of basic and applied research in biology and medicine.
Acknowledgments
We thank members of X Huang and J Liu labs for helpful discussions and Y Cao and Z Qin lab for generous equipment sharing (Nanjing University). We thank Dr Curt Horvath (Northwestern University) for the original ISRE reporter. We are grateful to Jingmei Chen and Yanlan Cao for excellent technical assistance. This work was supported by the National Basic Research Program of China (973 Program; 2011CB944104 to JL) and the National Natural Science Foundation of China (31271499 to JL and 31471400 to XH).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Supplementary figures S1 and S2 and methods
References
- Consortium EP, Bernstein BE, Birney E, et al. Nature 201248957–74.22955616
- Hsu PD, Lander ES, Zhang F. Cell. 2014. pp. 1262–1278. [DOI] [PMC free article] [PubMed]
- Qi LS, Larson MH, Gilbert LA, et al. Cell. 2013. pp. 1173–183. [DOI] [PMC free article] [PubMed]
- Gilbert LA, Larson MH, Morsut L, et al. Cell. 2013. pp. 442–451. [DOI] [PMC free article] [PubMed]
- Honda K, Takaoka A, Taniguchi T. Immunity. 2006. pp. 349–360. [DOI] [PubMed]
- Gilbert LA, Horlbeck MA, Adamson B, et al. Cell. 2014. pp. 1–15.
- Parisien JP, Lau JF, Rodriguez JJ, et al. J Virol. 2002. pp. 4190–4198. [DOI] [PMC free article] [PubMed]
- Majumder S, Zhou LZ, Chaturvedi P, et al. J Immunol. 1998. pp. 4736–4744. [PubMed]
- Jinek M, Jiang F, Taylor DW, et al. Science. 2014. p. 1247997. [DOI] [PMC free article] [PubMed]
- Kuscu C, Arslan S, Singh R, et al. Nat Biotechnol. 2014. pp. 677–683. [DOI] [PubMed]
- Wu X, Scott DA, Kriz AJ, et al. Nat Biotechnol. 2014. pp. 670–676. [DOI] [PMC free article] [PubMed]
- Banerjee AR, Kim YJ, Kim TH. Nucleic Acids Res. 2014. pp. 12537–12554. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures S1 and S2 and methods