Dear Editor,
Telomeres, the natural ends of linear eukaryotic chromosomes, are essential for cell viability and genome integrity [1]. In mammalian cells, telomere repeat factors 1 and 2 (TRF1 and TRF2) are the first two identified mammalian telomere-binding proteins that play essential roles in telomere homeostasis and maintenance2,3. Both TRF1 and TRF2 contain a central TRF homology (TRFH) domain and a C-terminal DNA-binding Myb domain (Supplementary information, Figure S1A)4. TRF1 and TRF2 only contain one Myb domain and they achieve high-affinity DNA association by homodimerization via their TRFH domains5. Myb domain-containing telomere-binding proteins have also been found in other organisms6,7. Fission yeast Schizosaccharomyces pombe (S. pombe) Taz1 associates with telomeric dsDNAs and plays an important role in telomere length regulation and protection [6,8,9,10,11,12]. In addition, similar to TRF1 and TRF2, Taz1 also negatively regulates telomere length via a “protein-counting” mechanism by recruiting Rap1 and Rif19,12. Therefore, Taz1 has been considered as the functional ortholog of mammalian TRF proteins.
In addition to the C-terminal DNA-binding Myb domain, sequence analysis also predicted a putative TRFH domain (residues 117-391) in Taz1 (Figure 1A), which can be aligned with the TRFH domains of TRF1 and TRF2 with low sequence similarity5,13. This observation has led to the proposal that Taz1 might also be a structural ortholog of mammalian TRF proteins. Limited proteolysis and MALDI mass spectrometry were employed to identify a protease-resistant core of Taz1 containing residues 127-388 (Supplementary information, Figure S1B). We crystalized Taz1127-388 and determined its structure at a resolution of 2.3 Å (Supplementary information, Table S1A). The calculated electron density map allowed unambiguous tracing of most part of Taz1127-388 except for a 27-residue disordered C-terminal tail (residues 362-388). The structure shows that Taz1127-361 adopts a compact globular fold with 14 α-helices tightly packed around a central hydrophobic core (Figure 1B).
Figure 1.
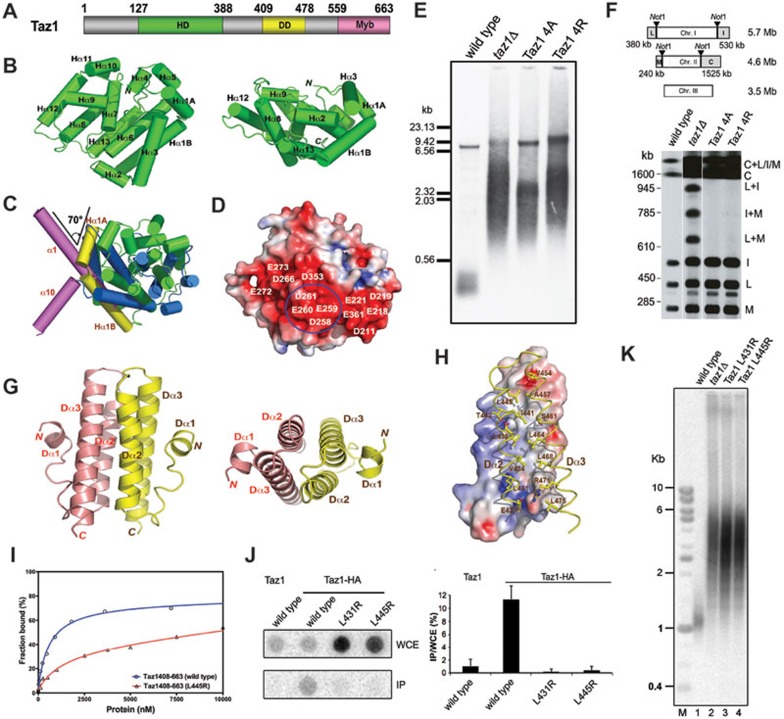
Structural and functional analyses of fission yeast telomere-binding protein Taz1. (A) Domain organization of the S. pombe Taz1. Numerals indicate residue numbers at the boundaries of subdivisions. The helical domain (HD) is colored in green, the dimerization domain (DD) in yellow, and the C-terminal Myb domain in pink. (B) Ribbon diagram of Taz1HD in two orthogonal views. Taz1HD is colored in green. The secondary structure elements are labeled. (C) Superposition of Taz1HD structure with TRFH domain structure indicates that Taz1HD is not structurally closely related with TRFH. Helices are shown as colored cylinders. Helices Hα1A and Hα1B in Taz1HD are colored in yellow and the rest 12 helices in green; helices α1 and α10 in the TRFH domain are in magenta and the rest eight helices in blue. (D) Taz1HD is shown in surface representation and colored according to its electrostatic potential (positive potential, blue; negative potential, red). (E) Southern blot analysis of the telomere DNA length of wild-type, taz1Δ, taz1-4A mutant, and taz1-4R mutant strains. (F) Telomere end protection in G1-arrested cells of wild-type, taz1Δ, taz1-4A mutant, and taz1-4R mutant strains. (G) Ribbon diagram of the Taz1DD dimer in two orthogonal views. The two monomers are colored in salmon and yellow, respectively. (H) The dimer interface. One Taz1DD molecule is in surface representation and colored according to its electrostatic potential. The other molecule is in ribbon representation and colored in yellow. Residues important for dimerization are shown as stick models. (I) Equilibrium-binding curves for the binding of wild-type (blue) and L445R mutant (red) Taz1408-663 to telomeric single-stranded DNA. The solid lines represent theoretical-binding curves fit to the data for wild-type and L445R mutant Taz1408-663. (J) Levels of telomeric DNA enrichment in Taz1-HA immunoprecipitates derived from asynchronously growing cells analyzed by ChIP followed by dot blot analysis with the telomere-specific probe. Dot blot results are quantified as the ratio of signal intensity in immunoprecipitated fragments (IP) to that in the whole-cell extract (WCE). (K) Southern blot analysis of the telomere DNA length of wild-type, taz1Δ, taz1-L431R, and taz1-L445R strains.
Structural comparison shows that there are significant structural differences between Taz1127-388 and the TRFH domains of TRF1 and TRF2. First, only six helices in Taz1127-388 can be roughly matched to the TRFH domains (Figure 1C and Supplementary information, Figure S1C). Structural-based sequence analysis only shows < 6% identity between Taz1127-388 and TRFH with many gaps in the alignment (Supplementary information, Figure S1C). Second, unlike the TRFH domain that forms a stable homodimmer, Taz1127-388 adopts a monomeric conformation in the crystal. In the TRFH domains, the N-terminus of helix α1 extends outside of the core and interacts with helix α10 from the other molecule in the dimer (Supplementary information, Figure S1D)5. In contrast, the α1 helix of Taz1127-388 is severely bent (∼ 70°) (Figure 1C). Consequently, the α1 helix in Taz1127-388 is now composed of two helices α1A and α1B, with a one-residue intervening linker (Figure 1C). The bent α1A helix and the N-terminal tail of Taz1127-388 fit into a hydrophobic groove formed by helices α3, α4, and α5 (Supplementary information, Figure S1E). In addition, the C-terminal 27-residue tail of Taz1127-388 (residues 362-388) that corresponds to helix α10 in TRF1 and TRF2 is disordered in the structure. Thus, Taz1127-388 lacks the dimeric interface mediated by helices α1 and α10 in the structures of the TRFH domains of TRF1 and TRF25. Consistent with this observation, purified Taz1127-388 is also a monomer in solution as revealed by gel filtration chromatographic analysis (Supplementary information, Figure S1F). Another important difference between Taz1127-388 and TRFH comes from the concave side of Taz1127-388 and TRFH domains. A 15-residue-loop L34 together with two helices α2 and α3 in TRFH constitute a peptide-binding pocket that recruits telomere-associated factors to the chromosome ends14. However, Taz1127-388 lacks a long loop equivalent to L34 in the TRFH domains, suggesting that it is unlikely that Taz1127-388 possesses a TRFH-like peptide-binding pocket. Nevertheless, it is possible that other region of Taz1 might still possess a peptide-binding pocket. Taken together, we conclude that Taz1127-388 and the mammalian TRFH domains are not structurally closely related as previously thought. Hereafter, we will refer to Taz1127-388 as the helical domain (HD) of Taz1 (Taz1HD; Figure 1A).
Close examination of the structure revealed a large stripe of a negatively charged surface on Taz1HD (Figure 1D). Particularly, four acidic residues (258DEED261) in the loop between helices α8 and α9 are all solvent-exposed and make only two electrostatic contacts with the rest of the protein (Figure 1D and Supplementary information, Figure S1G). To investigate the possible role of these residues in telomere regulation, we substituted them with either four alanine or four arginine residues and examined telomere length in yeast cells. Both mutant proteins were expressed at near wild-type levels, suggesting that these acidic residues are not required for protein stability (data not shown). The 4R mutation resulted in a dramatic increase in telomere length and length heterogeneity as severely as in taz1Δ cells (Figure 1E). Intriguingly, the 4A mutant retained partial function in suppressing telomere elongation, although taz1-4A cells still exhibited extremely heterogeneous telomeres similar to taz1Δ and taz1-4R cells (Figure 1E). Next, the effects of mutations on the telomere association of Taz1-GFP were assessed using ChIP analysis. Both Taz1 mutants exhibited partial loss of telomere association in a manner that is consistent with the severity of the telomere length defects (Supplementary information, Figure S1H). These results support the notion that acidic residues 258DEED161 on Taz1 are necessary for both telomere recruitment of Taz1 and telomere length regulation. To analyze the roles of the acidic surface in telomere protection, we assessed telomere stability of the 4A and 4R mutants by pulsed field gel electrophoresis of NotI-digested chromosomal DNA followed by Southern blotting. Although taz1Δ cells exhibited clear chromosome fusion bands, the mutations did not result in telomere fusions (Figure 1F). We conclude that acidic residues 258DEED161 of Taz1 are not required for telomere protection and the small amount of telomere-bound mutant proteins are enough to protect telomeres from fusion.
Given that Taz1 binds to telomeric DNAs as a homodimer15, our data suggest that dimerization of Taz1 is likely mediated by another region outside of Taz1HD. Various fragments were evaluated for their ability to form a homodimer in solution by gel filtration chromatography and chemical cross-linking analyses (Supplementary information, Figure S1I). We found that residues 395-490 of Taz1 (Taz1DD, the dimerization domain (DD) of Taz1) constitute the minimal core for Taz1 dimerization (Figure 1A and Supplementary information, Figure S1I). To understand the mechanism of Taz1 dimerization, we determined the structure of Taz1DD at a resolution of 1.5 Å (Supplementary information, Table S1B). Two molecules of Taz1DD form a homodimer, resulting in the burial of a total of 2 515 Å2 of the solvent-accessible surface area. Each Taz1DD monomer consists of three helices (Figure 1G). Helices Dα2 and Dα3 pack closely against each other, forming an antiparallel coiled-coil structure that mediates the dimerization through a two-fold axis (Figure 1G). The dimer interface consists of seven layers of two-fold-symmetry-related interdigitating residues from helices Dα2 and Dα3 (Supplementary information, Figure S1J). Although the dimeric interface is predominantly hydrophobic, intermolecular electrostatic interactions provide additional specificity and stability to the structure (Figure 1H and Supplementary information, Figure S1J).
To investigate the importance of the dimeric interface observed in the Taz1DD crystal structure in Taz1 dimerization, the effects of mutations at the hydrophobic dimeric interface (L431R, V434W, L438W, and L445R) on the dimer formation of Taz1DD were tested in a yeast two-hybrid assay. Three mutants (L431R, V434W, and L445R) exhibited a complete loss of the dimeric interaction, whereas mutant L438W still maintained a detectable dimeric interaction (Supplementary information, Figure S1K). These results confirmed that the interface observed in the Taz1DD crystal structure is indeed responsible for Taz1 dimerization.
A common feature of the telomeric dsDNA-binding proteins is that they all bind to the DNA via multiple Myb domains. Since Taz1 exists as a homodimer15 (Figure 1G), and each monomer contains only one Myb domain, we hypothesized that Taz1 homodimerization is also essential for its telomere association. To test this hypothesis, the L445R mutation that disrupts the Taz1 dimerization in yeast two-hybrid assay was analyzed for its effect on the DNA binding ability of Taz1 by electrophoretic mobility shift assay (EMSA). While wild-type Taz1 bound to DNA with an equilibrium dissociation constant (Kd) of ∼600 nM (Figure 1I), the L445R mutation caused a 10-fold decrease in DNA binding with a Kd of ∼7 μM (Figure 1I), suggesting that Taz1 homodimerization is required for its efficient association with the telomeric DNA in vitro. To examine whether Taz1 dimerization is also crucial for its telomere association in vivo, the DNA-binding abilities of two dimerization-deficient mutants (L431R and L445R) were assessed using ChIP. HA-tagged wild-type Taz1 or its dimerization-deficient mutants were expressed in cells from taz1 endogenous locus. Telomere DNA was precipitated from cell extracts in an HA-tag-dependent manner followed by dot blot analysis using telomere-specific probe (Figure 1J). Mutant proteins were expressed at a comparable level as wild-type Taz1 (data not shown). Consistent with the in vitro data, both the L431R and L445R mutants of Taz1 exhibited a complete loss of telomere association (Figure 1J), indicating that the homodimerization of Taz1 is necessary for its telomere association in vivo. Thus, we reasoned that the dimerization-deficient mutants of Taz1 would have the same phenotype as that of taz1Δ. Indeed, yeast cells expressing the L431R and L445R mutants exhibited significant loss of function in telomere length regulation and showed long and highly heterogenous telomeres that were as severe as that in taz1Δ cells (Figure 1K). Notably, the telomere length analysis of these mutants is also consistent with the strong signal of the whole-cell extracts derived from these mutant cells in the dot blot assay, which indicates long telomere length (Figure 1J). Taken together, we conclude that Taz1 dimerization is critical for both telomere localization of Taz1 and telomere length regulation.
More than a decade of structural and functional studies have revealed that the OB folds and the Myb domains function as evolutionarily conserved protein motifs utilized by a set of telomere-binding proteins to bind to single-stranded or double-stranded telomeric DNAs. However, it is not known whether evolutionarily conserved protein-protein interaction motifs also exist among telomere proteins. The TRFH domain in mammalian TRF proteins was proposed to represent such motif, since a putative TRFH domain was identified in S. pombe Taz113. However, our current structural studies of Taz1 dispelled this notion, because this putative TRFH domain (Taz1HD) has a different 3D structure and does not contain a peptide-binding pocket found in mammalian TRFH-containing proteins. In addition, unlike TRF proteins, Taz1HD does not mediate the homodimerization of Taz1 as previously thought. Instead, Taz1 utilizes a separate 2-helix bundle to mediate the dimerization. Notably, similar to TRF1 and TRF2, dimerization plays an essential role in telomere localization of Taz1 and telomere length regulation. These results highlight the remarkable structural plasticity of functionally conserved telomere-binding proteins. Except for the DNA-binding activities, many important functions might be preserved through structurally distinct modules of telomere-binding proteins in different organisms.
Accession numbers
Atomic coordinates and structure factors of Taz1HD and Taz1DD have been deposited into the Protein Data Bank with accession codes 4ZMI and 4ZMK.
Acknowledgments
We thank Mr Ke Wan and Drs Yong Chen and Yuting Yang for constructive suggestions and help during this study. We thank Dr Julie Cooper for discussion and allowing some experiments to be performed in her lab. We are grateful to the National Centre for Protein Science Shanghai (Protein Expression and Purification system) for the instrument support and technical assistance. This work was supported by the Ministry of Science and Technology of China (2013CB910402), the National Natural Science Foundation of China (31330040), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB08010201) and Howard Hughes Medical Institute.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
(A) Organization of the human TRF1 and TRF2 polypeptide chains.
References
- Cech TR. Cell. 2004. pp. 273–279. [DOI] [PubMed]
- van Steensel B, de Lange T. Nature. 1997. pp. 740–743. [DOI] [PubMed]
- van Steensel B, Smogorzewska A, de Lange T. Cell. 1998. pp. 401–413. [DOI] [PubMed]
- Court R, Chapman L, Fairall L, Rhodes D. EMBO Rep. 2005. pp. 39–45. [DOI] [PMC free article] [PubMed]
- Fairall L, Chapman L, Moss H, et al. Mol Cell. 2001. pp. 351–361. [DOI] [PubMed]
- Cooper JP, Nimmo ER, Allshire RC, et al. Nature. 1997. pp. 744–747. [DOI] [PubMed]
- Li B, Espinal A, Cross GA. Mol Cell Biol. 2005. pp. 5011–5021. [DOI] [PMC free article] [PubMed]
- Cooper JP, Watanabe Y, Nurse P. Nature. 1998. pp. 828–831. [DOI] [PubMed]
- Kanoh J, Ishikawa F. Curr Biol. 2001. pp. 1624–1630. [DOI] [PubMed]
- Miller KM, Rog O, Cooper JP. Nature. 2006. pp. 824–828. [DOI] [PubMed]
- Tomita K, Matsuura A, Caspari T, et al. Mol Cell Biol. 2003. pp. 5186–5197. [DOI] [PMC free article] [PubMed]
- Miller KM, Ferreira MG, Cooper JP. EMBO J. 2005. pp. 3128–3135. [DOI] [PMC free article] [PubMed]
- Li B, Oestreich S, de Lange T. Cell. 2000. pp. 471–483. [DOI] [PubMed]
- Chen Y, Yang Y, van Overbeek M, et al. Science. 2008. pp. 1092–1096. [DOI] [PubMed]
- Spink KG, Evans RJ, Chambers A. Nucleic Acids Res. 2000. pp. 527–533. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Organization of the human TRF1 and TRF2 polypeptide chains.