Summary
Objective
To save time, healthcare providers frequently use abbreviations while authoring clinical documents. Nevertheless, abbreviations that authors deem unambiguous often confuse other readers, including clinicians, patients, and natural language processing (NLP) systems. Most current clinical NLP systems “post-process” notes long after clinicians enter them into electronic health record systems (EHRs). Such post-processing cannot guarantee 100% accuracy in abbreviation identification and disambiguation, since multiple alternative interpretations exist.
Methods
Authors describe a prototype system for real-time Clinical Abbreviation Recognition and Disambiguation (rCARD) – i.e., a system that interacts with authors during note generation to verify correct abbreviation senses. The rCARD system design anticipates future integration with web-based clinical documentation systems to improve quality of healthcare records. When clinicians enter documents, rCARD will automatically recognize each abbreviation. For abbreviations with multiple possible senses, rCARD will show a ranked list of possible meanings with the best predicted sense at the top. The prototype application embodies three word sense disambiguation (WSD) methods to predict the correct senses of abbreviations. We then conducted three experments to evaluate rCARD, including 1) a performance evaluation of different WSD methods; 2) a time evaluation of real-time WSD methods; and 3) a user study of typing clinical sentences with abbreviations using rCARD.
Results
Using 4,721 sentences containing 25 commonly observed, highly ambiguous clinical abbreviations, our evaluation showed that the best profile-based method implemented in rCARD achieved a reasonable WSD accuracy of 88.8% (comparable to SVM – 89.5%) and the cost of time for the different WSD methods are also acceptable (ranging from 0.630 to 1.649 milliseconds within the same network). The preliminary user study also showed that the extra time costs by rCARD were about 5% of total document entry time and users did not feel a significant delay when using rCARD for clinical document entry.
Conclusion
The study indicates that it is feasible to integrate a real-time, NLP-enabled abbreviation recognition and disambiguation module with clinical documentation systems.
Keywords: Clinical abbreviation, machine learning, clinical documentation system
Introduction
Effective communication within and across healthcare settings is a key prerequisite for high-quality healthcare delivery. Clinical notes in patients’ health records comprise a common, important form of inter-provider communication. Accurate and unambiguous clinical documentation is therefore critical in clinical information exchange. Nevertheless, pervasive use of abbreviations in clinical notes often hampers their interpretation. Acronyms and shortened words or phrases frequently convey important information, such as names of diseases and medications (e.g., “CAD – Coronary Artery Disease”, and “SL NTG – sublingual nitroglycerin”) [1–3]. A study of directly entered admission notes at New York Presbyterian Hospital (NYPH) showed that abbreviations constituted 17.1% of total word tokens in those documents [3]. Unfortunately, many abbreviations are ambiguous – one abbreviation can have multiple senses (e.g., “AA” can mean African American, abdominal aorta, Alcoholics Anonymous, etc.).
Clinicians use abbreviations to save time and space during documentation. Abbreviations that seem unambiguous to authors often confuse other healthcare providers [4]. Studies have shown that clinicians have difficulty recognizing the sense of abbreviations in clinical notes [5]. An Australian pediatric referral center record audit determined that use of abbreviations in hospital progress notes hindered effective communication. Manzar et al discovered that unacceptable abbreviations in daily progress reports occurred in a special care neonatal unit [6]. Sheppard and colleagues [7] assessed the ability of physicians from different sub-domains of medicine to interpret abbreviations in pediatric notes. Their study identified 2,286 and 3,668 occurrences of abbreviations within 25 pediatric signout sheets and 168 sets of medical notes, and found that pediatricians could identify the correct abbreviation senses 56–94% of the time, but clinicians from other disciplines did poorly (31–63%). Misunderstanding of abbreviations in clinical documents can lead to serious medical errors. The Institute for Safe Medication Practices reviewed abbreviations and dose expressions in medication orders, and showed that they led to misunderstandings by nurses, and to dangerous medication errors [8].
Ambiguous abbreviations also bring challenges for natural language processing (NLP) systems for clinical text. Researchers have developed various word sense disambiguation (WSD) methods for abbreviations in clinical texts [9–13]. Supervised machine learning based methods (e.g., the support vector machines – SVMs) usually achieve better performance [12, 13]; but they require annotated samples for each ambiguous abbreviation, which makes it less practical. Pakhomov [14] proposed a method to generate simulated training data by replacing the expansions by abbreviations and showed reasonable performance on a set of six abbreviations. More recently, several studies showed that applying the vector space model to abbreviation context vectors can efficiently resolve ambiguity, especially when the training and test data are derived from difference sources [10, 11].
However, all current abbreviation identification systems perform their analyses in a “post-processing” manner, long after clinicians enter notes into electronic health record systems (EHRs). Since no existing NLP system has 100% accuracy [15], one cannot simply utilize NLP-identified abbreviation expansions post-facto to replace the abbreviations in their original texts. Doing so requires an additional step of clinician verification. The current study investigates the feasibility of a potentially more viable solution – integration of NLP-based abbreviation identification within the clinical documentation system that clinicians use to generate notes. In this model, the system will recognize and expand abbreviations as clinicians enter notes, immediately determining the intended sense for any ambiguous abbreviation. Different from the “post-processing” manner WSD methods, only the left-side context information is available in the real-time setting. Our ultimate goal is to elucidate the ability of advanced NLP technologies to improve the quality of health records during document generation.
NLP systems can be operated in clinical domains in real time settings to improve the patient recruitment efficiency [16], obtain quality feedback [17], reduce documentation errors [18] and enhance decision support systems [19]. Yet most such studies applied existing clinical NLP systems in a post-processing fashion. To the best of our knowledge, this is one of the first studies investigating the integration of sophisticated NLP methods (e.g., WSD algorithms) within the clinical documentation systems to resolve ambiguous abbreviations as clinicians enter text. The current preliminary study evaluated three aspects of real time clinical abbreviation disambiguation: 1) whether accuracy of different rapid-response WSD methods could be satisfactory; 2) whether system response time supports feasibility of implementing such systems in real time, and 3) whether users feel comfortable about this new documentation approach.
Methods
System overview
In this study, we developed the real-time Clinical Abbreviation Recognition and Disambiguation (rCARD) system, as a prototype clinical documentation tool that incorporates the ability to automatically recognize and expand abbreviations to their corresponding long forms in real-time, utilizing user feedback. ► Figure 1 shows a screen shot of the envisioned rCARD interface. When a user enters clinical text in the designated area, the system will automatically recognize each abbreviation (e.g., “PT” in the figure), and then display a ranked list of possible senses of the abbreviation, with the best-predicted sense at the top. The user would simply press the “space” key to accept the system-predicted sense (at the top) or select another sense using “up/down arrow” keys. The rCARD tool is Web-based using two major components: 1) the client-end interface and 2) the server-end WSD service. ► Figure 2 shows an overview of the components and data flow of the rCARD system. We describe each aspect in more details below.
Fig. 1.
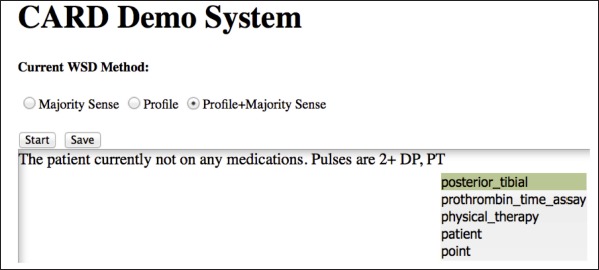
A screen shot of the rCARD interface.
Fig. 2.
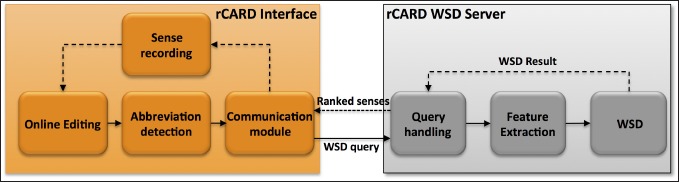
An overview of the rCARD system.
The rCARD Interface
The rCARD prototype client-side interface provides basic functions typically found in online clinical documentation systems: clinicians can type, delete, copy and paste clinical texts. In addition to text editing functions, the rCARD prototype includes additional modules that: 1) detect abbreviations (using a predefined list of abbreviations) typed into the rCARD interface and record the abbreviation context (i.e., words before the abbreviation); 2) send and receive WSD requests (containing an abbreviation and its context) and responses (suggested senses) from the WSD server – using JQuery1; and 3) record the users’ responses (i.e., selections of the “correct” sense from the ranked list) – embedding the response into the clinical document using predefined XML tags. To facilitate selection of senses, the rCARD prototype allows users to simply press the “space” key to accept the top-ranked sense or to use “up/down arrow” keys to select among the other senses.
The rCARD WSD server
Once an abbreviation is recognized at the client-end, the abbreviation and its context are sent to the WSD server to perform disambiguation. All client queries are first handled by a “query handling” module, then processed by a feature extraction module to extract relevant features, and finally by a WSD algorithm to generate a ranked list of senses with the predicted one at the top. In this study, we investigated the accuracy and running time of three WSD algorithms.
WSD Features
Previous clinical abbreviation disambiguation studies used context at both the left and right side of the target abbreviation as features. However, in our real-time disambiguation system, only the left side of context is available for feature generation. Therefore, an interesting open question evaluated in this study was whether the left-sided context features can support abbreviation disambiguation. In this study, we investigated four different types of features. We describe the extracted features using the following sentence as an example : “… 81 year old woman with hx of lung ca, now with likely recurrence.”
Word features – stemmed words within a window size of 5 of the target abbreviation. The Snowball Stemmer from python NLTK (Natural Language Toolkit) package was used in this study: [woman, with, hx, of, lung, now, with, like, recurr];
Word feature with direction – The relative direction (left side or right side) of stemmed words in feature set 1 towards the target abbreviation : [L-woman, L-with, L-hx, L-of, L-lung, R-now, R-with, R-like, R-recurr];
Position feature – The distance between the feature word and the target abbreviation : [L5-woman, L4-with, L3-hx, L2-of, L1-lung, R1-now, R2-with, R3-like, R4-recurr];
Word formation features from the abbreviation itself – include: a) special characters such as “-” and “.”; b) features derived from the different combination of numbers and letters; c) features derived from the number of uppercase letters. We also extracted same types of features from both sides in order to compare its performance with left-side features only.
WSD Methods
Different WSD methods have been developed and applied to clinical abbreviations [9–11, 13]. Although supervised machine learning methods often show good performance, they require annotated data sets from the same corpus for every ambiguous abbreviation, which is not practical. Therefore, in this study, we implemented three simple but scalable WSD approaches, including a majority-sense based method and two profile-based methods that rely on the vector space model and have shown good performance as well as high transportability in a previous study [10]. We describe these three WSD methods as following:
The Majority-sense based method, which always takes the majority sense of an ambiguous abbreviation as the correct sense. This method is often used to serve as a baseline for WSD studies. We based majority senses of abbreviations on previously analyzed manually annotated clinical note data sets.
The Profile-based method, as described in our previous study [10]. For each sense of an ambiguous abbreviation, the algorithm searches for the sense string (long form of an abbreviation) in a corpus to automatically create the pseudo training samples, from which a profile vector is created for each sense. During disambiguation, a context vector of the testing sample is created and compared with each sense profile vector to calculate the cosine-similarities. The sense corresponding to the highest similarity score is selected as the correct sense. All the features described above were used to build the profiles. In this study, we used the manually annotated corpus to build profiles instead of searching the senses.
The Profile + Sense frequency method, which combines the profile-based method with the sense frequency distribution, as described in [10]. The overall similarity score of a candidate sense was considered to be the sum of the normalized similarity score from the profile-based classifier and the relative sense frequency from the estimated sense distribution. The sense with the highest overall similarity was selected as the correct sense.
The study used different strategies to rank the sense list for each WSD method. For majority sense-based method, the ranking was based on the sense frequency. For profile and profile + sense frequency methods, the ranking was based on the similarity scores between the sense profile and context vectors.
Experiments and Evaluation
Data Set
The study used clinical documents from Vanderbilt Medical Center’s Synthetic Derivative (SD) database [20], which contains de-identified copies of the electronic health record documents at Vanderbilt University Hospital. The study was approved by the Vanderbilt Institutional Review Board. In a previous study [21], we defined a list of 25 frequently occuring ambiguous clinical abbreviations. This study used the same list of 25 clinical abbreviations. For each of the 25 clinical abbreviations, we randomly collected up to 200 instances/sentences from the Vanderbilt admission notes in the years of 2007–2009, resulting in 4,721 sentences in total. Three domain experts (authors JCD, STR, RAM) manually annotated the senses for all the abbreviation instances in this data set. These manually annotated sentences were then used for training and testing in this study.
Experiments
To evaluate rCARD, we conducted three experiments to assess different aspects of the system:
-
1) Performance evaluation of different WSD methods
For each abbreviation, we divided the instances (sentences) into 10 folds. We then used 9 folds of data to build models and tested it on the remaining one-fold data. We repeated it 10 times and finally reported results as an average of the 10 repeats. Building the profile based models was straightforward as there were no model parameters. To assess the upbound performance of the WSD task, we also evaluated the performance of an SVM-based WSD system. The implementation of the libsvm package2 was used in this study. For SVMs, we trained a specific SVMs model for each of the 25 abbreviations. The penalty parameter (c) and the termination parameter (e) were optimized according to the 10-fold cross validation for each abbreviation. We report the best average precision among all the abbreviations using the optimized parameters. Furthermore, we also evaluated the performance of the traditional WSD setting where both the left and right side context features are available, in addition to the real-time disambiguation setting where only left side context features are available. The primary measurement is the accuracy of the predicted sense by the different WSD methods.
-
2) Time evaluation of different real-time WSD methods:
To measure the cost of time for the different WSD methods, we conducted simulated experiments. Instead of manually typing sentences into the rCARD interface, we developed a pseudo client, which could send queries to the rCARD WSD server automatically. We then tracked the cost of time between sending a WSD request and receiving the ranked sense list for each abbreviation instance. As network communication time would be a major contributing factor of time in the client/server model, we evaluated the cost of time under three different network settings: 1) the client and server were on the same computer; 2) the client and server were on the same local network (e.g., same campus network), which is a typical setting for rCARD’s potential working environment; and 3) the client and server were on different local networks (e.g., access from outside of the campus).
-
3) A preliminary user study of rCARD:
We further conducted a user study to examine the feasibility of rCARD, in terms of timing and user experience. One hundred sentences were randomly selected for the twenty-five abbreviations (four sentences for each abbreviation). Two users (including a physician and a nurse) manually typed these sentences using the rCARD interface and resolved the ambiguous abbreviations using the disambiguation module. We developed recording functions in rCARD to count the overall typing time as well as the time spent on the disambiguation. In addition, we asked the two users to fill in an evaluation form to assess the rCARD system on five different aspects including the ease of use, ease of understanding the results, system results quality, disambiguation response time, and the overall burden, on a scale of 1–5.
Results
Three domain experts manually annotated all the randomly selected instances and constructed a sense inventory containing 103 unique senses for the 25 ambiguous clinical abbreviations. ► Table 1 shows the annotated senses and sense frequencies for the 25 ambiguous abbreviations. The number of possible senses for each abbreviation varied greatly (column “SEN”, ranges from 2 to 8). Column “OCC” denotes the number of occurrences for each abbreviation in the corpus (admission notes from the years 2007 - 2009).
Table 1.
25 abbreviations and their sense frequencies.
| ABB | SEN | Annotated senses and their frequencies | OCC |
|---|---|---|---|
| HD | 8 | “high dose”-56%, “hemodialysis”-39.5%, “hydrocodone”-1.5%, “hospital day”-1%, “heart disease”-0.5%, “hemodynamic”-0.5%, “hemodynamically”-0.5%, “Hodgkin’s Disease”-0.5% | 6,672 |
| BM | 7 | “bowel movement”-64%, “bone marrow”-27.5%, “bimanual”-5.5%, “bare metal”-1%, “breast milk”-1%, “black male”-0.5%, “Bilateral Myringotomy”-0.5% | 2,133 |
| HS | 7 | “at bedtime”-73.5%, “high school”-12%, “history”-5%, “heart sounds”-5%, “has”-2.5%, “heel-to-shin”-1.5%, “Henoch Schonlein”-0.5% | 1,523 |
| AD | 7 | “ad”-91%, “antidiarrheal”-3%, “and”-2%, “advertisement”-1.5%, “anterior descending”- 1.5%, “as”-0.5%, “alzheimer’s disease”-0.5% | 565 |
| CA | 6 | “calcium” – 64%, “cancer”-18.5%, “carbohydrate antigen”-9.5%, “carcinoma”-4%, “child’s age”-3.5%, “California”-0.5%, | 6,921 |
| SS | 6 | “Hemoglobin SS”-59%, “single strength”-24%, “sliding scale”-14%, “social security”-1.5%, “serosanguinous”-1%, “sickle cell disease”-0.5% | 1,634 |
| PT | 5 | “patient”-89.5%, “posterior tibial”-4.5%, “physical therapy”-3.5%, “point”-1.5%, “prothrombin time assay”-1% | 114,726 |
| MED | 5 | “medical”-48%, “medication”-43%, “medicine”-6%, “road name”-2%, “medial”-1% | 13,155 |
| PE | 5 | “physical exam”-32.5%, “pulmonary embolism”-30.5%, “pressure-equalization”-26.5%, “physical exercise”-8%, “pseudoephredrine”-2.5%, | 4,566 |
| LS | 5 | “lumbosacral”-86%, “left side”-9%, “lung sounds”-2%, “Lichen sclerosus at atrophicus”-1.5%, “lumbar spine”-1.5% | 62 |
| LAD | 4 | “Anterior descending branch of left coronary artery”-75%, “lymphadenopathy”-24%, “left atrial dilation”-0.5%, “left axis deviation”-0.5% | 9,169 |
| CC | 4 | “cubic centimeter”-62%, “chief complaint”-21.5%, “with meals”-12.5%, “calcaneus cuboid”-4% | 7,098 |
| GTT | 4 | “drop”-71.5%, “drip”-23%, “glucose tolerance test”-4.5%, “gamma glutamyl transpeptidase”-1% | 5,469 |
| RA | 3 | “room air”-74.5%, “rheumatoid arthritis”-18.5%, “right atrium”-7% | 8,374 |
| ICD | 3 | “implantable cardioverter defibrillator”-97.5%, “international classification of disease code”-2%, “inherited or intrinsic cardiac disease”-0.5% | 5,065 |
| SI | 3 | “sacroiliac”-88.5%, “suicidal ideation”-10%, “stress incontinence”-1.5%, | 1,899 |
| AG | 3 | “antigen”-68%, “anion gap”-30%, “age”-2% | 779 |
| LN | 3 | “lymph node”-94%, “line”-3%, “lane”-3% | 311 |
| MG | 2 | “milligram”-98.5%, “magnesium”-1.5% | 491,419 |
| CM | 2 | “centimeter”-99%, “costal margin”-1% | 17,551 |
| DM | 2 | “diabetes mellitus”-85%, “dextromethorphan”-15% | 9,763 |
| LE | 2 | “lower extremity”-89%, “leukocyte esterase”-11% | 6,542 |
| MI | 2 | “myocardial infarction”-99.5%, “Michigan”-0.5% | 2,948 |
| TIA | 2 | “transient ischemic attack”-99.5%, “[patient name]”-0.5% | 1,172 |
| SLE | 2 | “systemic lupus erythematosus”-96%, “Ophthalmic”-4% | 713 |
ABB: abbreviation; SEN: number of annotated senses; OCC: number occurrences from three year’s notes
► Table 2 shows the accuracy of three implemented WSD methods, as well as the SVM-based WSD method. The baseline method (majority sense) achieved an average accuracy of 72.57%. The upbound performance by SVMs was 89.25%. The “profile + sense frequency” method achieved an average accuracy of 88.80%, which is slightly lower than SVMs. An interesting finding was that the performance of using the left side context features did not drop much compared to the performance where both context sides were used as features (e.g., 89.25% vs. 90.76% for SVMs and 88.80% vs. 90.06% for “profile + sense frequency”). In the profile-based approach, using the left side context features actually increased the performance.
Table 2.
Average accuracy of different WSD methods, across 4,721 sentences of 25 abbreviations, using left context only or both side context.
| Left features only (%) | Left and right features (%) | |
|---|---|---|
| Majority sense | 72.57 | 72.57 |
| Profile | 76.16 | 74.52 |
| Profile + Sense frequency | 88.80 | 90.06 |
| SVMs | 89.25 | 90.76 |
All 4,721 abbreviation instances were used to evaluate the time of the rCARD system using the pseudo client. ► Table 3 shows the average cost of time (in millisecond) for recognizing and disambiguating one clinical abbreviation. The majority sense was the most efficient method, with 0.630 milliseconds in the same network setting. Profile and Profile + Sense Frequency methods took more time, with 1.589 and 1.649 milliseconds respectively. When accessing from outside of a network, the cost of time was much larger, ranging from 8.527–11.830 milliseconds for each abbreviation.
Table 3.
Cost of WSD time (in millisecond) per abbreviation at different network settings.
| Same Machine | Same Network | Across Network | |
|---|---|---|---|
| Majority sense | 0.260 | 0.630 | 8.527 |
| Profile | 1.070 | 1.589 | 9.987 |
| Profile + Sense frequency | 1.091 | 1.649 | 11.830 |
► Table 4 shows the costs of time in the user study. The typing speeds varied between the two users, which were calculated as 50.4 tokens/minute for user 1 and 32.7 tokens/minute for user 2, correspondingly. However, the percentages of time for abbreviation disambiguation were similar between the two users (5.1% for user 1 and 5.7% for user 2). ► Table 5 shows the evaluation scores from the survey. The survey results show that both users thought that the extra burden caused by the real-time WSD module was acceptable. Most of the time, users only needed to press the “space” key to accept the default sense as the rCARD showed a decent performance.
Table 4.
Costs of time in the user study.
| #Tokens | Total time | Disambiguation time | Disambiguation time in percentage | |
|---|---|---|---|---|
| User 1 | 2,455 | 2,922 seconds | 148 seconds | 5.1% |
| User 2 | 2,455 | 4,500 minutes | 259 seconds | 5.7% |
Table 5.
Evaluation scores from the user survey.
| Easy of use | Easy of understanding | System results quality | Response time | Overall burden | |
|---|---|---|---|---|---|
| User 1 | 3 | 1 | 2 | 2 | 3 |
| User 2 | 1 | 2 | 2 | 1 | 2 |
The survey used score scale of 1–5 (1 is the highest score and 5 is the lowest score)
Discussion
This paper presents a preliminary study relevant to developing a real-time clinical abbreviation recognition and disambiguation system to be embedded in clinical documentation systems. The system’s ultimate objective is to automatically predict the most likely sense of an ambiguous abbreviation and display it to clinicians for validation as they enter a note. Using a previously annotated test data set of 25 common clinical abbreviations, we show that the simple “profile+sense frequency” WSD method achieved reasonable performance (an accuracy of 88.8%, slightly lower than SVM’s performance of 89.25%), when only left-side context information was used. We further evaluated the additional time required for running the rCARD prototype and demonstrated that it was potentially feasible to integrate it with clinical documentation systems in a manner that might not impede the efficiency of note generation. This system is one of the first to attempt real-time disambiguation for clinical abbreviations, and the results using our prototype are preliminary. Further work must occur to realize the goal of improving the health record quality at the time of input by using advanced NLP methods in real-time.
We implemented the prototype rCARD system using traditional Client/Server architecture, which can easily incorporate more sophisticated WSD methods in the future. Our approach might cost much time than a client only model, due to network communication delays. Our results in ► Table 3 verified that network communication time could degrade performance dramatically when accessing a busy or degraded network. However, the cost of time within the same campus network was very efficient (0.630–1.649 milliseconds per abbreviation). Based on the study by [22], the average typing speed for a clinician was 30 words/minute and the maximum typing speed of clinicians was about 80 words/minute (750 milliseconds per word) [23]. So we expect that the additional 2 milliseconds delay on disambiguating an abbreviation probably will not make any noticeable changes to a physicians’ regular typing speed. The AJAX (Asynchronous JavaScript and XML) technique is a feasible solution to manipulate the client interface dynamically and to reduce the response time during real-time WSD. Using the implementation of JQuery, we were able to extract features from the client, access or modify the html elements dynamically.
To identify abbreviations while typing a note, the right-side features are not available for disambiguation. An interesting finding from this study is that using left-side features did not drop the WSD performance much (reduced from 90.76% to 89.25% in the SVMs and from 90% to 88.80% in the “profile + sense frequency” method). In the profile only method, using left-side features actually increased the WSD performance by 1.6%. This result indicates that left-side features are more important in WSD tasks. A previous WSD study also reported that a larger left-side window and a smaller right-side window achieved better performance [13]. The supervised machine learning method (SVMs) achieved the best accuracy under both of the settings (89.25% for left-side feature only and 90.76% for all features). However, we found that the SVMs are very sensitive to the model parameters. For example, the precision of SVMs are 79.12% for left-side features and 78.54 for all features using the default parameters in libsvm (c=1 and e=0.001). Thus, it’s very important to optimize the parameters using cross validation.
The user study shows that the costs of time by the real-time WSD module (about 5% of the overall typing time) are acceptable. In reality, as not every sentence in a clinical document contains ambiguous abbreviations, the real cost could be lower than that in the user study.
This study has several limitations. We evaluated the rCARD system using only 25 highly ambiguous abbreviations, which may not be good representative examples of all clinical abbreviations. Moreover, the evaluation was conducted at the sentence level in a simulated fashion, using the one-server and one-client setting. Further evaluation will be needed when many clients are opened at the same time to enter clinical documents. Furthermore, to apply this system to real clinical settings, we need a more comprehensive list of common abbreviations and their possible senses.
Conclusions
In this study, we implemented rCARD, a system that automatically disambiguates clinical abbreviations and interacts with authors during note generation to verify correct abbreviation senses. Our evaluation shows that it has a reasonable WSD performance and a very efficient response time, which suggests that it is feasible to integrate existing WSD methods with clinical documentation systems in a real-time manner that might not impede efficiency of note generation.
Acknowledgments
This study is supported in part by grants from NLM R01LM010681–05, NIGMS 1R01GM102282, and NCI R01CA141307.
Footnotes
Clinical Relevance Statement
NLP systems operated in clinical domains in real time settings could improve the patient recruitment efficiency, obtain quality feedback, reduce documentation errors and enhance decision support systems. Our research shows that it is feasible to integrate existing WSD methods with clinical documentation systems.
Conflict of Interest
The authors declare that they have no competing interests.
Human Subjects Protections
The study was approved by the Vanderbilt Institutional Review Board.
References
- 1.Berman JJ. Pathology abbreviated: a long review of short terms. Arch Pathol Lab Med 2004; 128(3): 347-352. [DOI] [PubMed] [Google Scholar]
- 2.Stetson PD, Johnson SB, Scotch M, Hripcsak G. The sublanguage of cross-coverage. Proc AMIA Symp 2002: 742-746. [PMC free article] [PubMed] [Google Scholar]
- 3.Xu H, Stetson PD, Friedman C. A study of abbreviations in clinical notes. AMIA Annu Symp Proc 2007: 821-825. [PMC free article] [PubMed] [Google Scholar]
- 4.Dawson Kp Fau – Capaldi N, Capaldi N Fau – Haydon M, Haydon M Fau – Penna AC, Penna AC. The paediatric hospital medical record: a quality assessment. 19920731 DCOM- 19920731(0726–3139 (Print)). [PubMed] [Google Scholar]
- 5.Walsh KE, Gurwitz JH. Medical abbreviations: writing little and communicating less. Archives of disease in childhood 2008; 93(10): 816-817. [DOI] [PubMed] [Google Scholar]
- 6.Manzar S, Nair AK, Govind Pai M, Al-Khusaiby S. Use of abbreviations in daily progress notes. Arch Dis Child Fetal Neonatal Ed 2004; 89(4): F374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheppard JE, Weidner LC, Zakai S, Fountain-Polley S, Williams J. Ambiguous abbreviations: an audit of abbreviations in paediatric note keeping. Archives of disease in childhood 2008; 93(3): 204-206. [DOI] [PubMed] [Google Scholar]
- 8.ISMP Medication Safety Alert – May 2, 2001. The Institute for Safe Medication Practices; 2001. [Google Scholar]
- 9.Schuemie MJ, Kors JA, Mons B. Word sense disambiguation in the biomedical domain: an overview. J Comput Biol 2005; 12(5): 554-565. [DOI] [PubMed] [Google Scholar]
- 10.Xu H, Stetson PD, Friedman C. Combining corpus-derived sense profiles with estimated frequency information to disambiguate clinical abbreviations. AMIA Annu Symp Proc 2012; 2012: 1004-1013. [PMC free article] [PubMed] [Google Scholar]
- 11.Pakhomov S, Pedersen T, Chute CG. Abbreviation and acronym disambiguation in clinical discourse. AMIA Annu Symp Proc 2005: 589-593. [PMC free article] [PubMed] [Google Scholar]
- 12.Liu H, Teller V, Friedman C. A multi-aspect comparison study of supervised word sense disambiguation. J Am Med Inform Assoc 200; 11(4): 320-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moon S, Pakhomov S, Melton GB. Automated disambiguation of acronyms and abbreviations in clinical texts: window and training size considerations. AMIA Annu Symp Proc 2012; 2012: 1310-1319. [PMC free article] [PubMed] [Google Scholar]
- 14.Pakhomov S. Semi-supervised Maximum Entropy based approach to acronym and abbreviation normalization in medical texts. Proceedings of the 40th Annual Meeting on Association for Computational Linguistics. Philadelphia, Pennsylvania: Association for Computational Linguistics 2002: 160-167. [Google Scholar]
- 15.Wu Y, Denny JC, Rosenbloom ST, Miller RA, Giuse DA, Xu H. A comparative study of current Clinical Natural Language Processing systems on handling abbreviations in discharge summaries. AMIA Annu Symp Proc 2012; 2012: 997–1003. [PMC free article] [PubMed] [Google Scholar]
- 16.Weng C, Batres C, Borda T, et al. A real-time screening alert improves patient recruitment efficiency. AMIA Annu Symp Proc 2011; 2011: 1489-1498. [PMC free article] [PubMed] [Google Scholar]
- 17.Filip D, Gao X, Angulo-Rodriguez L, et al. Colometer: a real-time quality feedback system for screening colonoscopy. World journal of gastroenterology: WJG 2012; 18(32): 4270-4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sandberg WS, Sandberg EH, Seim AR, et al. Real-time checking of electronic anesthesia records for documentation errors and automatically text messaging clinicians improves quality of documentation. Anesthesia and analgesia 2008; 106(1):192–201, table of contents. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Fong S, Fiaidhi J, Mohammed S. Real-time clinical decision support system with data stream mining. Journal of biomedicine & biotechnology 2012; 2012: 580186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roden DM, Pulley JM, Basford MA, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther 2008; 84(3): 362-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu H, Wu Y, Elhadad N, Stetson PD, Friedman C. A new clustering method for detecting rare senses of abbreviations in clinical notes. J Biomed Inform 2012; 45(6): 1075-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dawes M, Chan D. Knowing we practise good medicine: implementing the electronic medical record in family practice. Canadian family physician Medecin de famille canadien 2010; 56(1):15–6, e1-e3. [PMC free article] [PubMed] [Google Scholar]
- 23.Westby GF. 2011. [cited 2015; Available from: http://drwes.blogspot.com/2011/01/results-are-in-age-vs-typing-speed-in.html


