Rahimpour et al. use MR1 tetramers to characterize the heterogeneous population of mouse MAIT cells and find a close resemblance to their human counterparts. These findings will provide the foundation for further investigation of MAIT cells in health and disease.
Abstract
Studies on the biology of mucosal-associated invariant T cells (MAIT cells) in mice have been hampered by a lack of specific reagents. Using MR1-antigen (Ag) tetramers that specifically bind to the MR1-restricted MAIT T cell receptors (TCRs), we demonstrate that MAIT cells are detectable in a broad range of tissues in C57BL/6 and BALB/c mice. These cells include CD4−CD8−, CD4−CD8+, and CD4+CD8− subsets, and their frequency varies in a tissue- and strain-specific manner. Mouse MAIT cells have a CD44hiCD62Llo memory phenotype and produce high levels of IL-17A, whereas other cytokines, including IFN-γ, IL-4, IL-10, IL-13, and GM-CSF, are produced at low to moderate levels. Consistent with high IL-17A production, most MAIT cells express high levels of retinoic acid–related orphan receptor γt (RORγt), whereas RORγtlo MAIT cells predominantly express T-bet and produce IFN-γ. Most MAIT cells express the promyelocytic leukemia zinc finger (PLZF) transcription factor, and their development is largely PLZF dependent. These observations contrast with previous reports that MAIT cells from Vα19 TCR transgenic mice are PLZF− and express a naive CD44lo phenotype. Accordingly, MAIT cells from normal mice more closely resemble human MAIT cells than previously appreciated, and this provides the foundation for further investigations of these cells in health and disease.
Mucosal-associated invariant T cells (MAIT cells) are T lymphocytes that express a semi-invariant TCR consisting of an “invariant” TCR-α chain comprised of Vα19 joined to Jα33 in mice or Vα7.2 joined to Jα33 or Jα12 or Jα20 in humans (Reantragoon et al., 2013; Lepore et al., 2014). These cells express a range of TCR-β chains, although they are biased toward Vβ6 and Vβ8 in mice and Vβ2 and Vβ13 in humans (Le Bourhis et al., 2013; Birkinshaw et al., 2014; Ussher et al., 2014). These TCRs imbue MAIT cells with the ability to detect microbially derived antigens (Ags) presented by the monomorphic Ag-presenting molecule MHC class I–related protein-1 (MR1) in mammals (Gold et al., 2010, 2014; Le Bourhis et al., 2010; Reantragoon et al., 2012).
Recent studies have demonstrated that MAIT cells recognize riboflavin (vitamin B2) metabolites as a class of MR1-restricted Ags (Kjer-Nielsen et al., 2012; Patel et al., 2013; Corbett et al., 2014; Eckle et al., 2014; McWilliam et al., 2015). Riboflavin is produced by many strains of bacteria and yeast, and the ability to synthesize riboflavin correlates closely with the ability of microbes to induce MAIT cell activation, suggesting that these metabolites are the major class of Ag for MAIT cells (Gold et al., 2010; Le Bourhis et al., 2010; Kjer-Nielsen et al., 2012; Corbett et al., 2014). A recent study demonstrated that MR1 presents a nonenzymatically generated complex derived from the riboflavin biosynthetic precursor 5-amino-6-d-ribitylaminouracil (5-A-RU), and methylglyoxal or glyoxal, to generate the MAIT cell Ags 5-(2-oxopropylideneamino)-6-d-ribityl-aminouracil (5-OP-RU) and 5-(2-oxoethylideneamino)-6-d-ribityl-aminouracil (5-OE-RU), respectively (Corbett et al., 2014; Rossjohn et al., 2015). Furthermore, tetramerized human and mouse MR1 molecules, expressed in soluble form and refolded with the 5-OP-RU Ag, are capable of detecting all MAIT cells in both species (Reantragoon et al., 2013; Corbett et al., 2014).
Before the generation of these MR1 tetramers, the study of human MAIT cells has progressed through the use of a surrogate staining approach where MAIT cells are typically identified as Vα7.2+CD161+ cells (Martin et al., 2009), and indeed, this population was largely coincident with the MR1-Ag tetramer+ population in humans (Reantragoon et al., 2013). Studies of mouse MAIT cells have previously been more difficult, in part because of the lack of a Vα19-specific antibody and also because of the relative scarcity of these cells (Tilloy et al., 1999; Treiner et al., 2003). The advent of Vα19 TCR transgenic mice has been a valuable addition to the field, facilitating their investigation using mouse models of T cell development (Kawachi et al., 2006; Martin et al., 2009; Seach et al., 2013), infection (Le Bourhis et al., 2010), and other noninfectious diseases (Croxford et al., 2006; Miyazaki et al., 2011). Studies of MAIT cells in non-Vα19 TCR transgenic mice have generally relied on polymerase chain reaction to identify the characteristic Vα19Jα33 invariant TCR-α chain, sometimes in conjunction with a surrogate αβTCR+CD4−CD8− phenotype (Tilloy et al., 1999; Meierovics et al., 2013). Initial experiments using mouse MR1-Ag tetramer demonstrated that MAIT cells could be clearly detected in Vα19 TCR transgenic mice, but they have yet to be investigated in non-TCR transgenic mice (Reantragoon et al., 2013). Interestingly, some MR1-Ag tetramer+ cells were detected even in Vα19 TCR Tg mice that lacked MR1, suggesting that some of these cells were not MR1-restricted MAIT cells (Reantragoon et al., 2013). Furthermore, an unexpected observation from an earlier study showed that although human MAIT cells were shown to express the promyelocytic leukemia zinc finger (PLZF) transcription factor, MAIT cells from Vα19 TCR transgenic mice lacked this factor (Martin et al., 2009). This is in contrast to NKT cells, which express PLZF in both species (Kovalovsky et al., 2008; Savage et al., 2008). Similarly, MAIT cells in Vα19 TCR transgenic mice exhibited a naive CD44-low phenotype, whereas human MAIT cells have a memory phenotype (Martin et al., 2009). Thus, despite the high level of TCR-α chain sequence conservation and recognition of common Ags, these studies have suggested a fundamental difference in the biology of MAIT cells between species. However, given that at least some of the MR1 tetramer+ cells that develop in the TCR transgenic mice are MR1 independent (Reantragoon et al., 2013), it remains to be determined whether these cells represent those that develop naturally in non-TCR transgenic mice. Thus, the frequency, diversity, subsets, and functional characteristics of MAIT cells in normal, nontransgenic mice remain unclear.
In this study, we have undertaken an MR1 tetramer–based investigation of mouse MAIT cells from normal mice, including analysis of subsets defined by CD4 and CD8, cytokine production, and transcription factor expression. These studies have revealed previously unrecognized characteristics of mouse MAIT cells, including a constitutive memory phenotype; the existence of a major population of IL-17–producing, retinoic acid–related orphan receptor γt (RORγt)+ and a minor population of IFN-γ–producing, T-bet+ MAIT cells; and high levels of PLZF expression and dependence on this transcription factor for MAIT cell development.
RESULTS
Identification of MAIT cells in WT mice
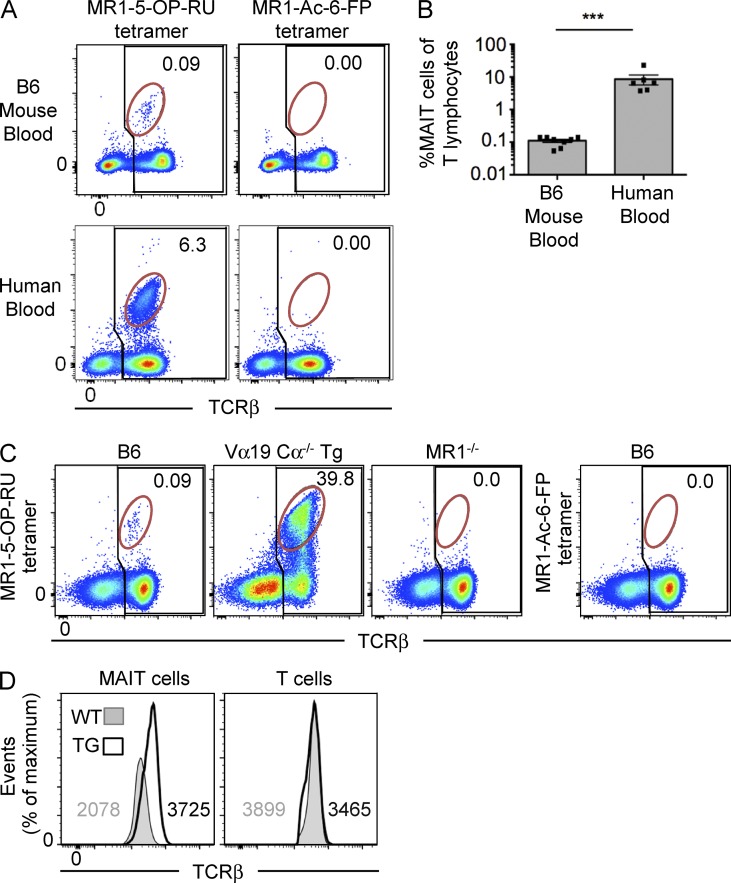
Using 5-OP-RU–loaded MR1 tetramers, MAIT cells can be readily identified in human peripheral blood and in Vα19 TCR transgenic mice (Reantragoon et al., 2013; Corbett et al., 2014). To directly compare human and mouse MAIT cells, we labeled peripheral blood lymphocytes from both species with either MR1–5-OP-RU (Corbett et al., 2014) or control nonantigenic MR1–acetyl-6-formylpterin (6-FP [Ac-6-FP]) tetramers (Eckle et al., 2014). Human MR1 tetramer was used for the human cells, and mouse MR1 tetramer was used for the mouse cells. These data showed that MAIT cells were much less abundant in B6 mouse blood, equating to ∼0.1% of T cells, compared with human blood where they comprised ∼6% of T cells, which falls within the usual range of human blood MAIT cells (Fig. 1 A; Le Bourhis et al., 2010; Reantragoon et al., 2013). To further investigate the specificity of the mouse MR1–5-OP-RU tetramer, we compared splenocytes from WT B6, Vα19 TCR transgenic.Cα−/− (Kawachi et al., 2006), and MR1−/− mice (Kawachi et al., 2006). In B6 mice, there was a clear population of TCRint tetramer+ cells, of similar frequency to those in peripheral blood, that was not detected with the control MR1–Ac-6-FP tetramer (Fig. 1 C). As expected, the MR1–5-OP-RU tetramer also stained an abundant population of cells in the TCR transgenic mice, yet no population was detected in the MR1−/− (Fig. 1 C), thus confirming the specificity of mouse MR1–5-OP-RU tetramer for MAIT cells. Staining of MR1−/− mouse cells with MR1–Ac-6-FP tetramer also failed to show a population of tetramer+ cells, as expected (not depicted). To formally establish that the population of MR1–5-OP-RU tetramer+ TCRint cells were indeed MAIT cells, single cells were sorted and TCRs sequenced after PCR amplification (Table 1). As expected, of the 21 cells where matched TCR-α and TCR-β chains were successfully identified, all expressed the MAIT cell invariant TRAV1-TRAJ33 (Vα19-Jα33) TCR-α chain, paired with a range of TCR-β chains that were highly enriched for TRBV13-3 and TRBV13-2 (Vβ8.1 or Vβ8.2), consistent with the previously documented MAIT TCR-β bias (Tilloy et al., 1999; Reantragoon et al., 2013; Lepore et al., 2014). In contrast to human MAIT cells, which can also use Jα12 and Jα20 instead of Jα33 (Reantragoon et al., 2013; Gold et al., 2014; Lepore et al., 2014), no alternate Jα gene usage was observed within the 21 productive TCR-α sequences derived from these experiments, suggesting that mouse MAIT cells have a strong dependence on this TCR element. Similar to NKT cells, MAIT cells expressed lower levels of TCR compared with conventional T cells, and furthermore, their TCR levels were also almost twofold lower in normal mice compared with Vα19 TCR transgenic Cα−/− mice (Fig. 1 D), which may reflect transgenic overexpression of the TCR in these mice. Accordingly, MR1–5-OP-RU tetramers can be used to specifically detect MAIT cells in WT mice.
Figure 1.
Identification of MAIT cells using MR1 tetramer in mice. (A) Detection of MAIT cells reactive to MR1–5-OP-RU tetramer in human and mouse blood. Flow cytometry analysis of human and mouse blood showing reactivity to MR1–5-OP-RU tetramer (left) or MR1–Ac-6-FP tetramer (negative control; right). Numbers indicate the percentage of MAIT cells (red gate) of total αβ T cells (black gate). Data are representative of four separate experiments with a combined total of n = 6 human blood samples and 9 mouse blood samples. (B) Scatter plot depicts MAIT cells as a proportion of T lymphocytes in mouse and human blood, gated as shown in A. Bars depict mean ± SEM of n = 6 human blood samples and 9 mouse blood samples derived from four separate experiments. ***, P < 0.001 using a Mann-Whitney rank sum U test. (C) B6, Vα19 Cα−/− transgenic (Tg) MR1+, or B6-MR1−/− spleen cells were stained with MR1–5-OP-RU tetramer (left three plots) or MR1–Ac-6-FP tetramer (far right plot). Numbers indicate the percentage of MAIT cells (red gate) of total αβ T cells (black gate). Data are representative of two independent experiments with a combined total of four mice. (D) Intensity of TCR-β staining on WT and transgenic MAIT cells is depicted as histograms, representative of four independent experiments with a combined total of six mice. Numbers depict mean fluorescence intensity.
Table 1.
TCR sequences of MR1–5-OP-RU tetramer+ TCR intermediate cells
| Seq | TRAV | TRAJ | CDR3α | TRBV | TRBJ | CDR3β |
| 1 | TRAV1 | TRAJ33 | CAVRDSNYQLIW | TRBV13-2 | TRBJ2-7 | CASGDGDWGREQYF |
| 2 | TRAV1 | TRAJ33 | CAVRDSNYQLIW | TRBV13-3 | TRBJ2-3 | CASSRXGGXSAETLYF |
| 3 | TRAV1 | TRAJ33 | CAVRDSNYQLIW | TRBV13-3 | TRBJ2-5 | CASSDGTXQDTQYF |
| 4 | TRAV1 | TRAJ33 | CAVRDSNYQLIW | TRBV13-3 | TRBJ1-3 | CASSGTSSGNTLYF |
| 5 | TRAV1 | TRAJ33 | CAVRDSNYQLIW | TRBV13-2 | TRBJ2-3 | CASGGGDSAETLYF |
| 6 | TRAV1 | TRAJ33 | CAVRDSNYQLIW | TRBV13-3 | TRBJ2-3 | CASSDTGGAQETLYF |
| 7 | TRAV1 | TRAJ33 | CAVRDSNYQLIW | TRBV1 | TRBJ2-4 | CTCSGDSQNTLYF |
| 8 | TRAV1 | TRAJ33 | CAVRDSNYQLIW | TRBV13-2 | TRBJ2-7 | CASGVGDYEQYF |
| 9 | TRAV1 | TRAJ33 | CAVRDSNYQLIW | TRBV13-2 | TRBJ2-7 | CASGDGGAPSYEQYF |
| 10 | TRAV1 | TRAJ33 | CAVRDSNYQLIW | TRBV13-2 | TRBJ2-3 | CASGDDWGGRAETLYF |
| 11 | TRAV1 | TRAJ33 | CAVRDSNYQLIW | TRBV13-2 | TRBJ2-5 | CASGLGGRQDTQYF |
| 12 | TRAV1 | TRAJ33 | CAVRDSNYQLIW | TRBV13-3 | TRBJ2-7 | CASSAGTSSYEQYF |
| 13 | TRAV1 | TRAJ33 | CAVMDSNYQLIW | TRBV13-2 | TRBJ2-3 | CASGGDGDSAETLYF |
| 14 | TRAV1 | TRAJ33 | CAVRDSNYQLIW | TRBV13-3 | TRBJ2-1 | CASSKGDYAEQFF |
| 15 | TRAV1 | TRAJ33 | CAVRDSNYQLIW | TRBV13-2 | TRBJ2-3 | CASGEETGGAETLYF |
| 16 | TRAV1 | TRAJ33 | CAVRDSNYQLIW | TRBV13-3 | TRBJ1-2 | CASSGTTNSDYTF |
| 17 | a) TRAV1 b) TRAV6-4a | TRAJ33 | CAVRDSNYQLIW | TRBV13-2 | TRBJ2-5 | CASGLGGRQDTQYF |
| 18 | a) TRAV1 b) TRAV6-5a | TRAJ33 | CAVRDSNYQLIW | TRBV13-2 | TRBJ2-3 | CASGDADRGQGAETLYF |
| 19 | a) TRAV1 b) TRAV3-3a | TRAJ33 | CAVRDSNYQLIW | TRBV13-3 | TRBJ2-4 | CASSDAASQNTLYF |
| 20 | a) TRAV1 b) TRAV9Na | TRAJ33 | CAVRDSNYQLIW | TRBV13-2 | TRBJ2-1 | CASGDGDSYAEQFF |
| 21 | a) TRAV1 b) TRAV12a | TRAJ33 | CAVRDSNYQLIW | TRBV13-2 | TRBV2-7 | CXSGDGGAPSYEQYF |
MR1–5-OP-RU tetramer+ TCR-β intermediate cells from B6 mouse spleen, gated in a similar manner to that shown in Fig. 1 C, were sorted as single cells, and TCR-α and -β chains were sequenced. Data are derived from two independent experiments with spleens from two mice per experiment. X, sequence unclear.
Mixed unproductive sequences.
Tissue distribution of mouse MAIT cells
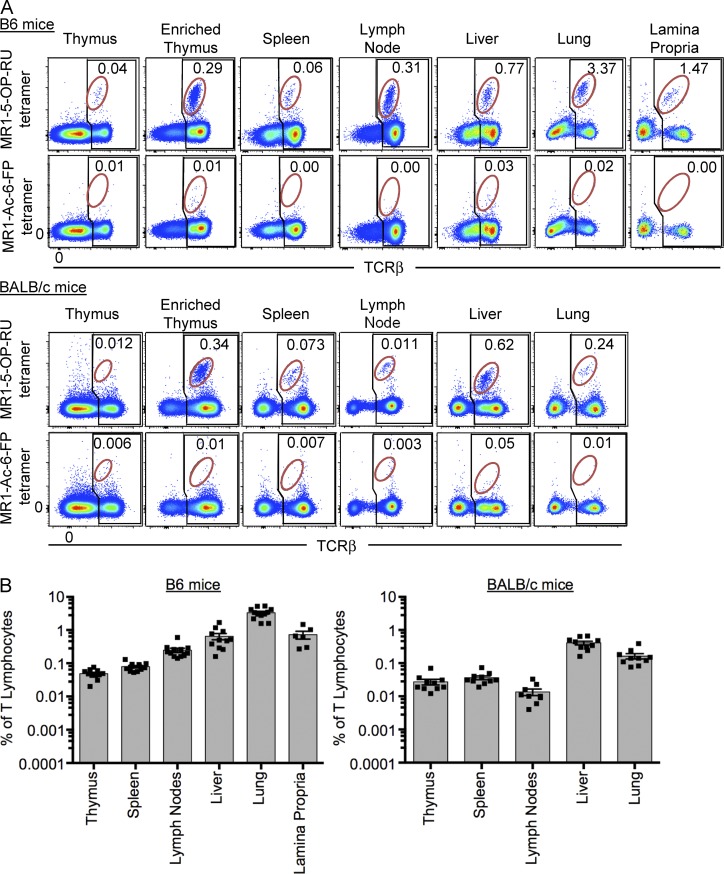
Having identified MAIT cells in blood from WT mice, we next examined these cells in other tissues of two commonly used mouse strains (B6 and BALB/c), including thymus, spleen, liver, lymph node (inguinal), lung, and lamina propria (Fig. 2, A and B). Negative controls included MR1–Ac-6-FP tetramer (Fig. 2 A) and MR1–5-OP-RU tetramer staining of tissues from MR1−/− mice (similar to Fig. 1 C and not depicted). The data indicate the percentage of MAIT cells of total αβTCR+ cells. MAIT cells were difficult to detect in whole mouse thymus (<0.1%), but after complement-mediated depletion of immature thymocytes using anti-CD24, a clear population of MAIT cells was observed, representing ∼0.3% of mature (CD24 depleted) αβTCR+ thymocytes. MAIT cells were identified in all other tissues, although their frequency varied considerably. In B6 mice, the highest percentage was detected in lung (mean 3.3%), followed by lamina propria (mean 0.7%), liver (mean 0.6%), peripheral lymph nodes (mean 0.2%), spleen (mean 0.08%), and thymus (mean 0.05%; Fig. 2 B). In BALB/c mice, MAIT cells were generally less frequent as a percentage of total αβTCR+ cells, although a similar distribution was observed. In this strain, the liver housed the highest frequency of MAIT cells (mean 0.4%), followed by lung (mean 0.2%), spleen (mean 0.04%), thymus (mean 0.02%), and lymph nodes (mean 0.01%; Fig. 2, A and B). BALB/c lamina propria was not tested. In BALB/c thymus, MAIT cells were only clearly detectable after depletion of immature thymocytes with anti-CD24 antibody (Fig. 2 A). We were unable to detect a clear population of MAIT cells within the intraepithelial compartment of the gastrointestinal tract (not depicted). Thus, in mice, MAIT cells appear to be distributed in the same tissues as conventional T cells, although their relative frequency appears to be differentially regulated compared with control conventional T cells.
Figure 2.
Tissue distribution of MAIT cells. (A) Flow cytometry analysis of naive mouse thymus, enriched thymus, spleen, lymph nodes (inguinal), liver, lung, and lamina propria, showing reactivity to MR1–5-OP-RU tetramer or MR1–Ac-6-FP tetramer. Plots depict lymphocytes with B220+ B cells excluded by electronic gating. B6 mice are depicted in the top group and BALB/c in the bottom group. Numbers indicate the percentage of MAIT cells (red gate) of total αβ T cells (black gate). (B) Scatter plots depict MAIT cells as a proportion of T lymphocytes in all tissues tested, gated as shown in A. Each symbol represents an individual mouse. Bars depict mean ± SEM, and data are derived from a minimum of three independent experiments for each strain, with a combined total of 6–12 mice per tissue.
Cell surface phenotype heterogeneity of mouse MAIT cells
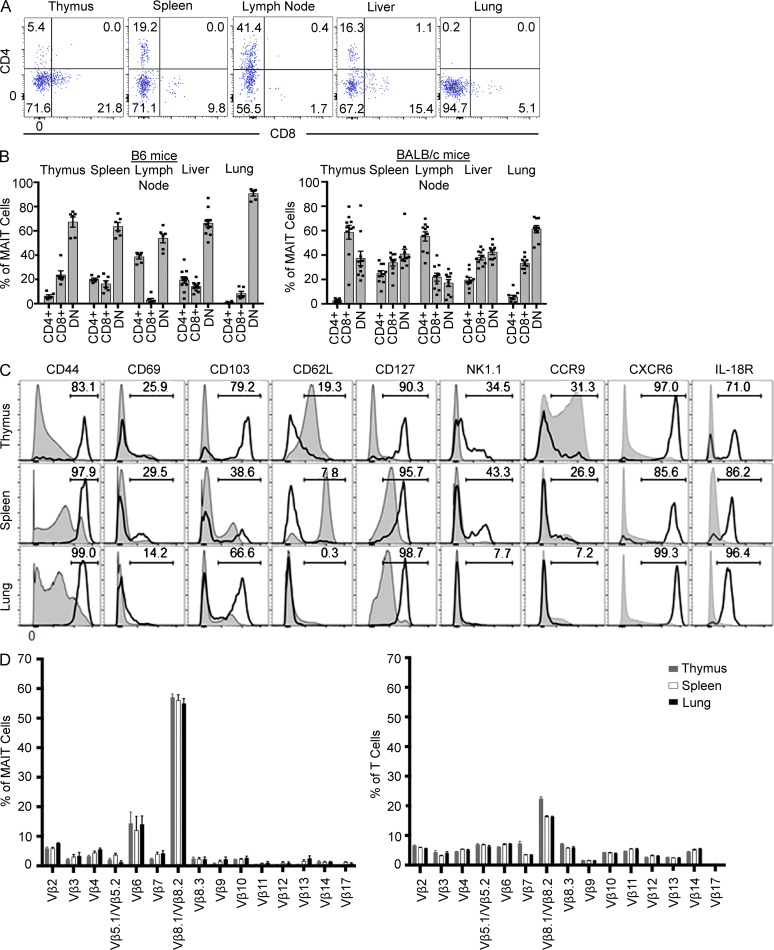
To investigate MAIT cell subsets and examine the activation and differentiation state of mouse MAIT cells, we colabeled MR1–5-OP-RU tetramer+ MAIT cells from B6 and BALB/c strains with antibodies specific for CD4 and CD8α (Fig. 3, A and B). In all tissues tested from B6 mice, the majority of MAIT cells were CD4−CD8−, although CD4+ and CD8+ subsets were also detected in lower proportions and with varied frequencies in a tissue-specific manner. For example, very few CD8+ MAIT cells (<5%) were detected in lymph nodes, whereas CD4+ MAIT cells were abundant (∼40%) in this tissue (Fig. 3, A and B), whereas CD4+ MAIT cells were virtually undetectable in lung. Furthermore, the expression level of CD4 and CD8 on MAIT cells were typically lower than on conventional T cells (Fig. 3 A and not depicted). CD8α+ MAIT cells in mice included a mix of both CD8α+β− and CD8α+β+ cells at roughly equal ratios, similar to human CD8+ MAIT cells (not depicted; Martin et al., 2009; Walker et al., 2012; Reantragoon et al., 2013). BALB/c MAIT cells also included the same three subsets based on CD4 and CD8 expression, although in this strain there were more CD8+ MAIT cells in thymus and a similar ratio of double-negative and CD8+ MAIT cells in spleen, lymph node, and liver (Fig. 3 B). Interestingly, similar to B6 mice, CD4+ MAIT cells were abundant (55%) in lymph nodes from BALB/c mice, suggesting that CD4+ MAIT cells preferentially reside in these organs.
Figure 3.
Phenotypic diversity within mouse MAIT cells. (A) Flow cytometry analysis of CD4 and CD8 coreceptor expression on MAIT cells from thymus, spleen, lymph nodes (inguinal), liver, and lung from B6 mice. (B) CD4/CD8 coreceptor–defined subsets of MAIT cells as a proportion of total MAIT cells in each tissue, gated as shown in A. Each symbol represents an individual mouse. Bar graphs depict mean ± SEM. Data are derived from a minimum of three independent experiments for each strain, with a combined total of 6–10 mice per tissue. (C) Phenotypic analysis of MAIT cells (thick black lines) from thymus, spleen, and lung compared with other (MR1–5-OP-RU tetramer− TCR-β+) T cells (shaded gray) depicted as histograms. Histograms for each marker, apart from CD218, show data concatenated from three separate mouse tissue samples from within one experiment and are representative of three similar experiments. Histograms for CD218 are representative of two experiments with four mice per experiment. (D) CD3+ and 5-OP-RU–reactive cells were stained for a panel of Vβ antibodies, and positive cells were plotted as a proportion of total MAIT cells. Bar graphs depict mean ± SEM from three independent experiments, each involving a pool of 10 mice.
We also examined other cell surface markers on MAIT cells from B6 mice, including activation and differentiation markers and chemokine and cytokine receptors: CD44, CD69, CD103, CD62L, CD127 (IL-7Rα), CD161 (NK1.1), CCR9, CXCR6, and CD218 (IL-18Rα; Fig. 3 C). MAIT cells were uniformly high for CD44 and low for CD62L expression, reflecting a memory phenotype, which contrasts with earlier studies of Vα19 TCR transgenic.Cα−/− MAIT cells, which were described as naive CD44lo cells (Kawachi et al., 2006; Martin et al., 2009). In contrast to NKT cells, which express CD69 (Godfrey et al., 2010), most MAIT cells were CD69lo, suggesting that they are not constitutively activated in mice. NK1.1 (CD161) was expressed by a subset (∼30–50%) of MAIT cells in thymus and spleen, whereas lung MAIT cells were negative for this marker. This contrasts with human MAIT cells, which are almost entirely CD161hi (Martin et al., 2009; Reantragoon et al., 2013). CD103, a mucosal homing receptor, showed bimodal expression on MAIT cells in each tissue tested, consistent with their ability to migrate to mucosal tissues. CD127 and CD218 were both expressed by the majority of MAIT cells, in line with the expression of these markers on human MAIT cells (Tang et al., 2013; Leeansyah et al., 2014). Of the chemokine receptors tested, MAIT cells were uniformly high for CXCR6 (similar to NKT cells [Geissmann et al., 2005]) and a subset expressed CCR9 (Fig. 3 C), but they were not observed to express other chemokine receptors including CCR4, CCR7, CXCR3, CX3CR1, and CXCR4 (not depicted). As CCR9 and CXCR6 are associated with homing to nonlymphoid tissue and mucosal tissue, this is consistent with the observed distribution of MAIT cells in mice.
Next we examined TCR Vβ usage by MAIT cells from B6 mice in different tissue compartments to determine whether the Vβ8 bias was consistent in all tissues. This was achieved by costaining with MR1-Ag tetramer and anti-Vβ–specific antibodies (Fig. 3 D). These data revealed that, consistent with the TCR sequencing data (Table 1) and as previously published (Tilloy et al., 1999; Kawachi et al., 2006; Reantragoon et al., 2013), a major population (∼55%) of MAIT cells use Vβ8.1/Vβ8.2 genes and a moderate bias toward the use of Vβ6 (∼15%), albeit less than was expected based on the original study of mouse MAIT cells (Tilloy et al., 1999). This may be simply caused by differences resulting from the use of T cell hybridomas for Vβ analysis in the earlier study (Tilloy et al., 1999), compared with freshly isolated MAIT cells here. Furthermore, we detected expression of a range of Vβ genes, including Vβ2, 3, 4, 5, 6, 7, 8.3, 9, 10, and 14, at low but detectable frequency. Moreover, similar Vβ distributions were observed for MAIT cells isolated from thymus, spleen, and lung, suggesting that recruitment to different tissues is not influenced by TCR-β chain repertoire.
PLZF expression by mouse MAIT cells
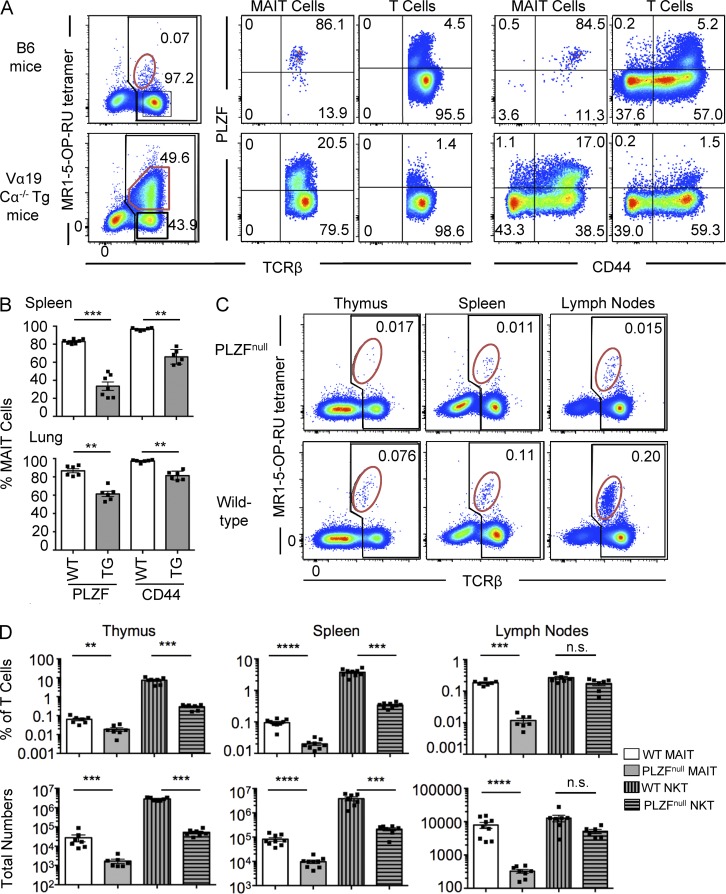
The transcription factor PLZF is considered to be a master regulator of innate-like T cells, including NKT cells (Kovalovsky et al., 2008; Savage et al., 2008) and γδ T cells (Kreslavsky et al., 2009). We examined PLZF expression in mouse MAIT cells from spleens of B6 WT and Vα19 TCR transgenic.Cα−/− mice (Fig. 4 A). Whereas the WT MAIT cells were almost all PLZFhi, most of the TCR transgenic MAIT cells from spleen were PLZFlo, which is reminiscent of a previous study suggesting that Vα19 TCR transgenic.Cα−/− MAIT cells lacked this transcription factor (Martin et al., 2009). Interestingly, the percentage of Vα19 TCR transgenic MAIT cells that were PLZFhi was higher in lung (mean 61%) compared with spleen (mean 33%; Fig. 4 B). Nearly all (>95%) of the PLZFhi MAIT cells also expressed high levels of the memory T cell marker CD44, regardless of whether they were from WT or Vα19 TCR transgenic mice, whereas many but not all of the PLZFlo MAIT cells that were abundant in the TCR transgenic mice also lacked CD44 expression (Fig. 4, A and B).
Figure 4.
MAIT cells express PLZF and are PLZF dependent. (A) MAIT cells from WT B6 mice (top row) and Vα19 TCR Tg Cα−/− mice (bottom row). Plots in first column depict lymphocytes with B220+ B cells excluded by electronic gating. Numbers indicate the percentage of MAIT cells (red gate) of total αβ T cells (black gate). MAIT cells and non-MAIT “T cells,” gated as shown in the first column, were examined for PLZF versus TCR-β (second and third columns) and PLZF versus CD44 (fourth and fifth columns) expression. Data are representative of four separate experiments with a combined total of six mice per group. (B) Scatter plots depict the percentage of PLZF+ and CD44+ MAIT cells from spleen and lung of WT and Vα19 TCR transgenic Cα−/− mice. Each symbol represents an individual mouse. Bars depict mean ± SEM from a total of six separate mice. (C) Presence of MAIT cells in thymus, spleen, and lymph nodes of B6 WT and PLZFnull mice. Plots depict lymphocytes with B220+ B cells excluded by electronic gating. Top row shows PLZFnull mice, and bottom row shows WT mice. Numbers indicate the percentage of MAIT cells (red gate) of total αβ T cells (black gate). Data are representative of three independent experiments with a combined total of six to seven mice per group. (D) Percentage (top row) and total numbers (bottom row) of MAIT and NKT cells (CD1d–α-GalCer tetramer+ T cells) of total lymphocytes in thymus, spleen, and lymph nodes. Each symbol represents a different mouse. Bars depict mean ± SEM. For C and D, data are representative of three independent experiments with a combined total of six to seven mice per group. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 using a Mann–Whitney rank sum U test.
Given the high expression of PLZF by mouse MAIT cells, this raised the question of whether the development of these cells was dependent on this transcription factor, which is known to control the development of NKT cells (Kovalovsky et al., 2008; Savage et al., 2008). To directly test this, we examined MAIT cells in mice with a T to A transversion in exon 2 of the Plzf gene that changes a tyrosine to a premature stop codon (Y70Stop; not depicted; PLZFnull mice). MAIT cells were almost completely absent in thymus, spleen, and lymph nodes of these mice, although a residual population (<10% of the number in WT B6 mice) remained (Fig. 4, C and D). Single cell TCR sequencing of these residual cells revealed that they expressed the invariant TRAV1-TRAJ33 (Vα19-Jα33) TCR-α chain and were enriched for TRBV13-3 and TRBV13-2 (Vβ8.1 or Vβ8.2; Table 2), supporting the concept that they were indeed MAIT cells. Interestingly, these residual MAIT cells were enriched for the CD4+ MAIT cell subset (not depicted), suggesting that at least some CD4+ MAIT cells may be developmentally distinct to CD4− MAIT cells. Accordingly, these experiments indicated that, like NKT cells, MAIT cells express high levels of the transcription factor PLZF and, furthermore, that the development of these cells is highly, although not absolutely, dependent on PLZF expression.
Table 2.
TCR sequences of MR1–5-OP-RU tetramer+ cells in PLZF KO mice
| Seq | TRAV | TRAJ | CDR3α | TRBV | TRBJ | CDR3β |
| 1 | TRAV1 | TRAJ33 | CAVRDSNYQLIW | TRBV13-2 | TRBJ2-1 | CASGDAEDYAEQFF |
| 2 | TRAV1 | TRAJ33 | CAVRDSNYQLIW | TRBV13-3 | TRBJ2-4 | CASSDRGXGSQNTLYF |
| 3 | TRAV1 | TRAJ33 | CAVRDSNYQLIW | TRBV13-3 | TRBJ2-7 | CASGDAEDYAEQFF |
| 4 | TRAV1 | TRAJ33 | CAVRDSNYQLIW | TRBV19 | TRBJ2-7 | CASSIGDTYEQYF |
| 5 | TRAV1 | TRAJ33 | CAVRDSNYQLIW | TRBV19 | TRBJ2 | CASSPTGGEGYAEQFF |
| 6 | TRAV1 | TRAJ33 | CAVRDSNYQLIW | TRBV13-3 | TRBJ1-2 | CASGDGDSYAEQFF |
| 7 | a) TRAV1 b) TRAV6a | TRAJ33 | CAVRDSNYQLIW | TRBV13-3 | TRBJ2-2 | CASSEGRDTGQLYF |
| 8 | TRAV1 | TRAJ33 | CAVRDSNYQLIW | TRBV14 | TRBJ2-7 | CASSFDIYEQYF |
| 9 | TRAV1 | TRAJ33 | CAVRDSNYQLIW | TRBV13-3 | TRBJ2-1 | CASSGTVYAEQFF |
| 10 | TRAV1 | TRAJ33 | CAVRDSNYQLIW | TRBV13-2 | TRBJ2-1 | CASGGTYYAEQFF |
| 11 | TRAV1 | TRAJ33 | CAVRDSNYQLIW | TRBV13-3 | TRBJ2-7 | CASTGTGISYEQYF |
| 12 | TRAV1 | TRAJ33 | CAVRDSNYQLIW | ND | ||
| 13 | TRAV1 | TRAJ33 | CAVRDSNYQLIW | TRBV13-3 | TRBJ2-4 | CASXXXGGASQNTLYF |
| 14 | a) TRAV1 b) TRAV5Da | TRAJ33 | CAVRDSNYQLIW | TRBV13-3 | TRBJ2-5 | CASSDRGQDTQYF |
| 15 | a) TRAV1 b) TRAV10a | TRAJ33 | CAVRDSNYQLIW | TRBV13-2 | TRBJ2-1 | CASGDGGAPSYEQYF |
| 16 | a) TRAV1 b) TRAV10a | TRAJ33 | CAVRDSNYQLIW | TRBV1 | TRBJ2-4 | CTCSAVWFSQNTLYF |
MR1–5-OP-RU tetramer+ TCR intermediate cells from PLZFnull mouse spleen, gated in a similar manner to that shown in Fig. 4 C, were sorted as single cells, and TCR-α and -β chains were sequenced. Cells were derived from spleens from two mice per group. X, sequence unclear.
Mixed unproductive sequences.
Most mouse MAIT cells produce IL-17A and express RORγt
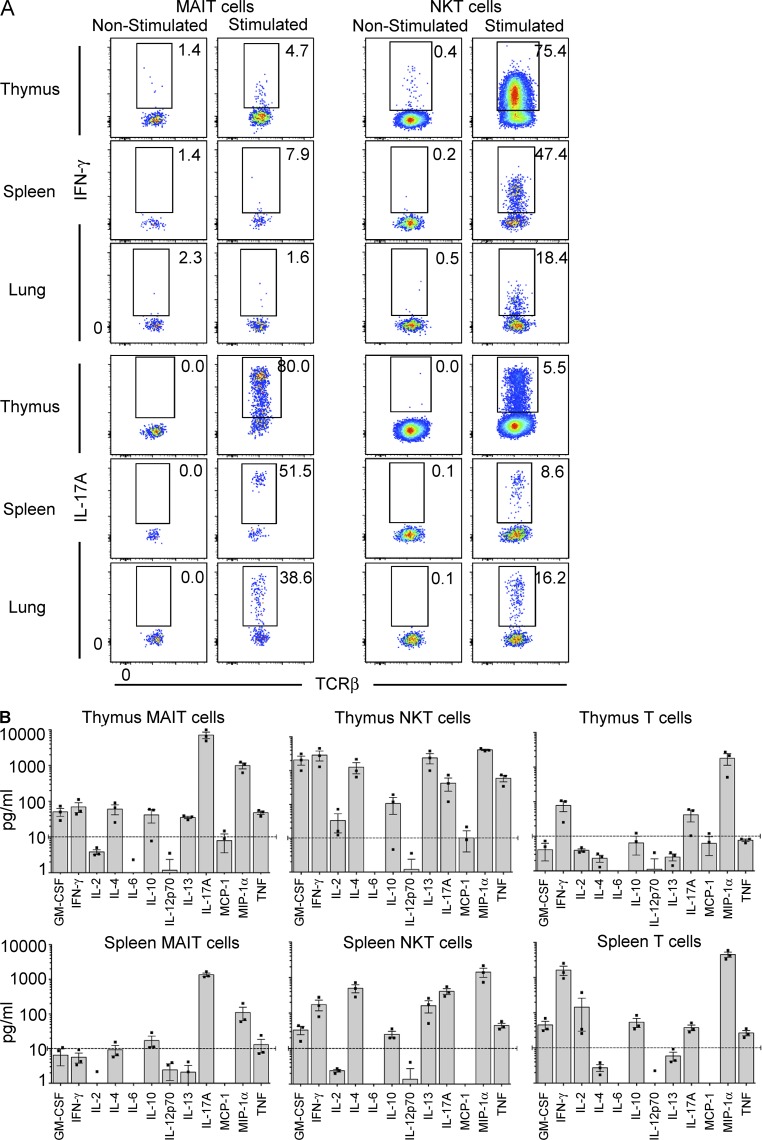
Previous studies of Vα19 TCR transgenic mice have suggested that, similar to NKT cells, mouse MAIT cells can produce high levels of a range of cytokines including IL-4, IL-5, IL-10, IFN-γ, and TNF (Kawachi et al., 2006). Here, we stimulated MAIT cells from thymus, lung, and spleen of WT mice using PMA and ionomycin for 4 h and measured acute cytokine production by intracellular cytokine staining (Fig. 5 A and not depicted). These data suggested that very few MAIT cells were able to produce IFN-γ (Fig. 5 A) and IL-4 (not depicted). In contrast, the majority of NKT cells in the same experiments were clearly capable of producing both IFN-γ (Fig. 5 A) and IL-4 (not depicted). However, the majority of MAIT cells from the thymus, spleen, and lung were able to produce very high levels of the proinflammatory cytokine IL-17A, whereas expression of this cytokine was limited to a small subset of NKT cells from the same organs (Fig. 5 A). We also examined production of these cytokines by MAIT cell subsets defined by CD4/CD8 expression but did not observe any clear differences (not depicted). To investigate cytokine production in response to TCR ligation, we next isolated MAIT cells from thymus, spleen, and lung and stimulated them in vitro with plate-bound CD3 and CD28 antibodies, measured cytokine secretion into the culture supernatant after 24 h (Fig. 5 B and not depicted), and compared them with NKT cells and conventional T cells. This confirmed that thymic MAIT cells are capable of producing a broad range of cytokines including IL-4, IL-10, IL-13, GM-CSF, IFN-γ, and TNF, although generally at low levels (<100 pg/ml). In line with the intracellular cytokine staining data, IL-17A was produced at extremely high levels by thymic MAIT cells, at least 10-fold higher than that of NKT cells and conventional T cells in the same experiments (Fig. 5 B). Generally lower levels of cytokines were detected from spleen (Fig. 5 B) and lung MAIT cells (not depicted) although IL-17A was the main cytokine produced by these cells in these assays.
Figure 5.
Cytokine production by MAIT cells. (A) CD24-depleted mouse thymocytes were stimulated for 4 h in PMA/ionomycin, after which time the cells were labeled with MR1-Ag tetramer and then fixed and permeabilized before staining with anti–IFN-γ or IL-17A. Data are representative of three experiments with a combined total of six mice per group. (B) MAIT cells, NKT cells (CD1d–α-GalCer tetramer+ T cells), and conventional MR1 tetramer− TCR-β+ T cells were sorted from WT spleen and thymus and then added at 10,000 cells/well and stimulated by plate-bound CD3 and CD28. Supernatants were harvested at 24 h, and cytokines were analyzed by CBA. Graphs depict the mean concentration of cytokines ± SEM from three independent experiments for thymus and spleen, each using MAIT cells pooled from 10 mice. Dashed lines are the cutoff at 10 pg/ml for clearly detectable cytokine production.
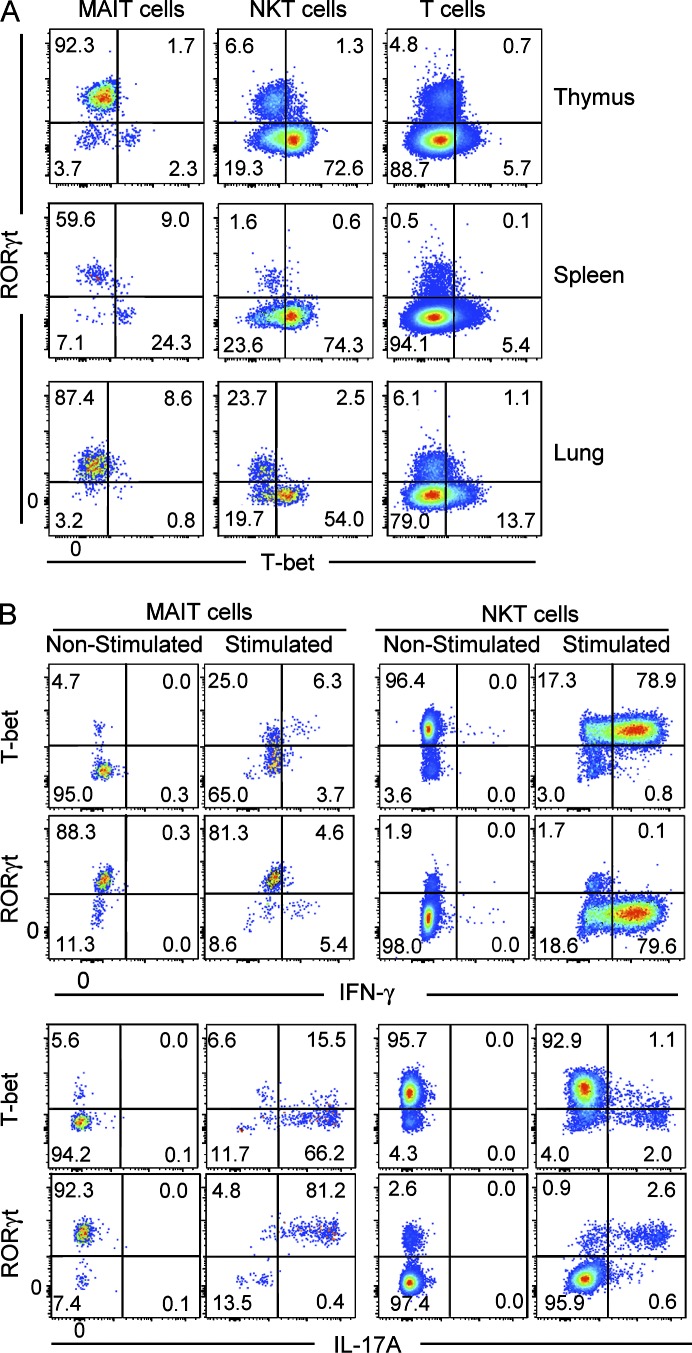
Given that MAIT cells express high levels of IL-17A and low levels of IFN-γ, we next examined the transcription factors RORγt and T-bet, which in other cell types including NKT cells are associated with production of these cytokines, respectively (Lee et al., 2013). These results showed that most MAIT cells expressed RORγt, whereas only a small subset (∼5–30%) expressed T-bet, and these transcription factors appeared to be mutually exclusive (Fig. 6 A). In contrast, most NKT cells expressed T-bet and only a small subset expressed RORγt, whereas conventional T cells mostly lacked these transcription factors (Fig. 6 A). To directly test whether these transcription factors are associated with the cytokine profile of mouse MAIT cells, we investigated cytokine production by RORγt+ and T-bet+ MAIT cells isolated from thymus. These data indicated that the rare IFN-γ+ MAIT cells were T-bet+RORγtlo, whereas IL-17A was exclusively produced by the RORγthi MAIT cells. Although some of the IL-17A+ MAIT cells also appeared to express low levels of T-bet (Fig. 6 B), we noted that T-bet expression moderately increased on MAIT cells upon stimulation (Fig. 6 B). Thus, it is likely that the IL-17A+RORγthi MAIT cells were derived from the predominant T-bet− MAIT cell population, whereas IFN-γ was produced by the T-bet+RORγtlo MAIT cells (Fig. 6 B). Collectively, these results suggested the existence of two functionally distinct populations of MAIT cells, governed by differential transcription factor expression in mice.
Figure 6.
Most MAIT cells are RORγthi and produce IL-17A. (A) MAIT cells, NKT cells (CD1d–α-GalCer tetramer+), and conventional TCR-β+ T cells were labeled with antibodies specific for RORγt and T-bet. (B) Samples from thymus, spleen, and lung were stimulated for 4 h in vitro with PMA/ionomycin, and expression of RORγt and T-bet and production of IFN-γ and IL-17A by MAIT and NKT cells were analyzed by flow cytometry. Data are representative of four experiments, with a combined total of eight mice.
Mouse MAIT cells respond to Ag-mediated stimulation
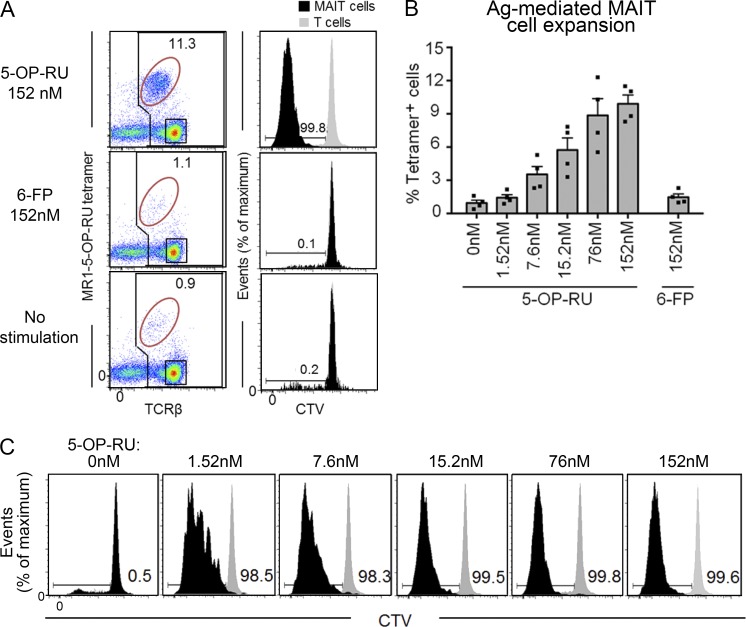
To determine whether WT mouse MAIT cells can respond directly to challenge with the 5-OP-RU Ag, B6 mouse splenocytes were cultured in the presence of graded doses of this Ag for 3 d, and MAIT cell proliferation was measured by dilution of the cell trace violet (CTV) fluorescent dye (Fig. 7, A and B). These data show that mouse MAIT cells responded in a dose-dependent manner (Fig. 7, B and C) to the 5-OP-RU Ag, whereas they did not respond to the control MR1-binding nonagonist 6-FP (Fig. 7, A and B). Cytokines, including IFN-γ, were also detected in these cultures (not depicted), although the amounts were lower and more variable than in cultures of purified MAIT cells (Fig. 5), likely because of the low frequency of MAIT cells in the bulk culture. When MAIT cells were purified by flow cytometric sorting of MR1–5-OP-RU tetramer+TCR-β+ cells and stimulated with 5-OP-RU Ag in the presence of MR1+ Ag-presenting cells, we detected clear production of IL-17A in response to Ag, albeit at lower levels than that seen after CD3 and CD28 stimulation (not depicted). Accordingly, these data demonstrate that WT mouse MAIT cells can recognize and respond specifically to the agonist ligand, 5-OP-RU in a dose-dependent manner.
Figure 7.
MAIT cells proliferate upon Ag stimulation in vitro. Splenocytes were labeled with CTV and cultured in the presence of 5-OP-RU, 6-FP, or no Ag. After 72 h, MAIT cell numbers and CTV dilution were measured by flow cytometry. (A) Left column shows representative flow cytometry profiles showing percentage of MAIT cells (MR1–5-OP-RU-tetramer+) of total T cells. Plots depict lymphocytes with B220+ B cells excluded by electronic gating. Right column shows MAIT cell and T cell proliferation based on decreased CTV dye dilution. Black histograms, MAIT cells; gray histograms, MR1 tetramer− T cells. Data are representative of a minimum of two separate experiments with a combined total of four mice. (B) Bar graph depicts percentage of expanded MAIT cells, gated as shown in A (mean ± SEM) from two independent experiments and a combined total of four mice. (C) Representative flow cytometry profiles showing CTV dilution of MAIT cells at each Ag dose. Data are representative of a minimum of two separate experiments with a combined total of four mice. Black histograms, MAIT cells; gray histograms, MR1 tetramer− T cells.
DISCUSSION
MR1-restricted, riboflavin metabolite–reactive, MAIT cells are a unique T cell lineage that is present in mice and humans and other mammals (Le Bourhis et al., 2013; Birkinshaw et al., 2014; Ussher et al., 2014). Despite their discovery 15 years ago (Tilloy et al., 1999), the biology of these cells remains poorly defined, in part because of difficulties in identifying them in mice, thereby limiting the study of MAIT cell function in mouse models of development and disease. Some studies have used Vα19 TCR transgenic Cα−/− mice (Okamoto et al., 2005; Kawachi et al., 2006; Martin et al., 2009), although how these compare with MAIT cells in WT mice was unclear. Our experiments on MAIT cells, using MR1–5-OP-RU tetramer in WT mice including two commonly used strains (B6 and BALB/c), revealed that these cells are relatively rare compared with MAIT cells in humans, although they clearly are present in all tissues where T cells are detected.
In this study, we have demonstrated the existence of multiple subsets of mouse MAIT cells defined by CD4/CD8 expression, differential expression of the transcription factors RORγt and T-bet, and an associated differential production of the cytokines IL-17A and IFN-γ, respectively. Although our studies of cytokine production were partly in agreement with a previous study with rapid production of a range of cytokines (Kawachi et al., 2006), we observed that the most prominent cytokine produced by these cells was IL-17A, a proinflammatory cytokine that has previously been identified as a product of human MAIT cells (Dusseaux et al., 2011; Walker et al., 2012; Tang et al., 2013). Furthermore, IL-17A–producing MAIT cells are prominent in psoriatic skin lesions in humans (Teunissen et al., 2014), suggesting an important role for these cells in this disease. Indeed, mouse MAIT cells produced far higher levels of IL-17A than NKT cells and conventional T cells in parallel cultures, and the majority of MAIT cells produced this factor. Consistent with the high IL-17A production, most mouse MAIT cells expressed high levels of the transcription factor RORγt, and it was these MAIT cells that produced IL-17A, whereas a smaller subset of the mouse MAIT cells that were RORγtlo instead expressed T-bet. The latter population was responsible for the production of IFN-γ and notably did not coexpress IL-17A. Given that these two populations were both detected in thymus, these data are reminiscent of a recent study that shows multiple developmentally distinct subsets of NKT cells that are thought to develop as separate lineages during intrathymic development (Lee et al., 2013). Accordingly, these data highlight the existence of at least two functionally distinct subpopulations of mouse MAIT cells: a major RORγt+T-betlo IL-17A–producing subset and a minor RORγtloT-bet+ IFN-γ–producing subset. It will be important to examine these subsets separately in future studies of MAIT cell biology.
We have detected some important differences between MAIT cells in WT mice and those that develop in Vα19 TCR transgenic.Cα−/− mice. In the TCR transgenic mice, MAIT cells were previously reported as having a naive CD44lo phenotype and lacking the transcription factor PLZF (Martin et al., 2009). Here, using MR1–5-OP-RU tetramer we also observed that many, though not all, MAIT cells in TCR transgenic mice were CD44loPLZFlo. In contrast, nearly all of the MAIT cells detected in WT mice exhibited a memory CD44hi, CD62Llo phenotype and were PLZFhi. This indicates that MAIT cells in WT mice are more comparable with human MAIT cells than was previously realized because human MAIT cells also exhibit a memory phenotype and are PLZF+ (Martin et al., 2009). It is not clear why MAIT cells in WT mice are so different from the Vα19 TCR transgenic.Cα−/− mice, but it may be that aberrant timing or expression levels of the TCR transgene, or the absence of T cells expressing other TCR-α chains in the Cα−/− background, disrupts the normal MAIT cell developmental pathway. Regardless, these findings suggest that some aspects of MAIT cell development and function in a TCR transgenic background may not reflect MAIT cell biology in normal mice and humans.
There are many similarities between MAIT cells in WT mice and humans. As previously established, they express orthologous TRAV1-TRAJ33 TCR-α chains (Tilloy et al., 1999), and as confirmed here, they recognize the same riboflavin metabolite–based Ag (5-OP-RU) presented by MR1, and they include CD4+, CD4−CD8−, and CD8+ subsets. Furthermore, both mouse and human MAIT cells express PLZF, they produce high levels of IL-17A, and they share cytokine receptors, including CD127 (IL7Rα) and CD218 (IL18Rα), and chemokine receptors (CCR9 and CXCR6; Dusseaux et al., 2011). A striking difference between MAIT cells in mice and humans is their frequency within the T cell population. Although in humans MAIT cells are highly abundant, representing up to 10% of T cells in blood (Le Bourhis et al., 2010) and up to 45% of T cells in liver (Dusseaux et al., 2011), in mice MAIT cells are far less abundant in all tissues tested, including liver, rarely representing more than 1% of T lymphocytes. Given the known role for microbial stimulation in MAIT cell expansion (Treiner et al., 2003), and the fact that MAIT cells are less abundant in human thymus and cord blood (Martin et al., 2009), it is tempting to speculate that one reason for the differences in MAIT cell numbers between laboratory mice and humans is that humans are far more likely to suffer from microbial infections than laboratory mice from clean animal facilities.
The expression of high levels of PLZF by the vast majority of MAIT cells was consistent with the concept that these cells undergo a similar developmental programming to other “innate-like” T cells, including NKT cells, γδ T cells, and other, as-yet-undefined, innate-like T cells, that also express this transcription factor (Kovalovsky et al., 2008; Savage et al., 2008; Kreslavsky et al., 2009; Prince et al., 2014). This is not unexpected because positive selection of MAIT cells requires an interaction with MR1 expressed by CD4+CD8+ double-positive thymocytes (Seach et al., 2013), and this is a SAP-dependent process (Martin et al., 2009). These factors appear to initiate the alternate developmental pathway that results in the potent cytokine-producing, CD44hi memory phenotype (Godfrey et al., 2010), which is shared by MAIT and NKT cells. Indeed, our observation that MAIT cells are almost entirely depleted in PLZF mutant mice highlights the important role that PLZF plays in the generation of these cells. Interestingly, the fact that the few remaining PLZF-independent MAIT cells still expressed the invariant TRAV1-TRAJ33 TCR-α chain, yet were enriched for CD4+CD8−, suggested that this subset of MAIT cells either represents a developmentally distinct sublineage or a precursor that requires PLZF for further maturation. Very little is known about CD4+ MAIT cells, but their overrepresentation in lymph nodes and in PLZF mutant mice suggests that they may play a distinct role in the immune system, and future studies should compare the development and function of these cells with double-negative and CD8+ MAIT cells.
Collectively, this study provides a comprehensive analysis of MAIT cells in WT mice using MR1-Ag tetramers. We have revealed previously unrecognized subsets of MAIT cells in mice, including a major population of RORγt+ IL-17A–producing MAIT cells and a minor population of T-bet+ IFN-γ–producing MAIT cells. Furthermore, we have shown that mouse MAIT cells have a memory phenotype, they express the transcription factor PLZF, and have a strong dependence on this factor for their normal development. This study therefore demonstrates that mouse MAIT cells are far more aligned with human MAIT cells than previously recognized and thus provides an important foundation for future studies of MAIT cell development and function in mouse models of health and disease.
MATERIALS AND METHODS
Mice.
C57BL/6 (B6), MR1−/−, and BALB/c mice were bred in house at the Department of Microbiology and Immunology Animal House, University of Melbourne. Vα19 TCR transgenic Cα−/− and MR1−/− mice were backcrossed at least 10 times to the B6 background and were a gift from S. Gilfillan (Washington University in St. Louis School of Medicine, St. Louis, MO). The Plzf−/− mice were identified in an N-ethyl-N-nitrosourea (ENU) random mutagenesis screen because of their skeletal abnormality. The strain was initially generated on a mixed C57BL/6 and CBA.H background and backcrossed for 12 generations to C57BL/6. The phenotype was linked to an interval between SNP accession numbers rs3667699 and rs13480241 on chromosome 9. PLZF was selected as a candidate gene in the interval and the causal mutation was found to be a T to A transversion in exon 2 that changes a tyrosine to a premature stop codon (Y70Stop). The Plzf−/− mice are freely available through the Australian Phenome Bank. All procedures on mice were approved by the University of Melbourne Animal Ethics Committee or the Australian National University animal experimentation ethics committee, respectively.
Cell suspensions.
Thymus, spleen, and lymph node cell suspensions were prepared by gently grinding each organ through a 40-µM nylon cell strainer into ice-cold FACS buffer (PBS with 2% FBS). Spleen suspensions were treated with red blood cell lysis buffer and further washed with FACS buffer. Mature thymocytes were isolated by complement-mediated depletion of CD24+ cells, and hepatic leukocytes were isolated as previously described (Pellicci et al., 2002). After mice were killed, lung tissues were perfused before removal and digested enzymatically in collagenase type III (Worthington Biochemical Corporation; 3 mg/ml in RPMI-1640 supplemented with 2% FCS) for 60 min at 37°C. Small intestines were dissected, Peyer’s patches removed, and intestines cut longitudinally into 1-cm-long pieces. To remove intraepithelial lymphocytes, gut tissues were incubated with 15.4 mg/ml dithioerythritol in 10% of 1× HBSS/Hepes bicarbonate buffer for 30 min at 37°C on a shaker (200 rpm). Gut tissues were then treated with 0.5 mg/ml collagenase D (Roche), 3 mg/ml dispase II (Roche), and 0.5 mg/ml DNase I (Roche) for 30 min in 37°C on a shaker (200 rpm). Lymphocytes were subsequently purified using 44/67% Percoll gradient (800 g at 20°C for 20 min; Weigmann et al., 2007). Healthy human PBMCs were obtained from the Australian Red Cross, ethics approval 13-04VIC-07 (Australian Red Cross) and 1035100.1 (University of Melbourne). PBMCs were isolated using a histopaque-1077 (Sigma-Aldrich) density gradient centrifugation before staining for flow cytometric analysis.
Tetramers.
Mouse and human MR1–5-OP-RU tetramers were generated by refolding MR1 monomers with 5-A-RU and methylglyoxal, as previously described (Corbett et al., 2014). Mouse and human MR1–Ac-6-FP tetramers were also generated as previously described (Eckle et al., 2014). C-terminal cysteine-tagged MR1–5-OP-RU was generated as described previously (Corbett et al., 2014) and then reduced with 5 mM DTT for 20 min before buffer exchange into PBS using a PD-10 column (GE Healthcare) and biotinylated with Maleimide-PEG2 biotin (Thermo Fisher Scientific) with a 30:1 molar ratio of biotin/protein at 4°C for 16 h in the dark. Biotinylated MR1 was subjected to S200 10/300 GL (GE Healthcare) chromatography to remove excess biotin. Biotinylated MR1–5-OP-RU monomers were tetramerized with streptavidin conjugated to either PE (SA-PE) or Brilliant Violet 421 (SA-BV; BD). Soluble mouse CD1d/β2m was prepared as previously described (Pellicci et al., 2009). The α-GalCer analogue PBS-44 with a C24:1 acyl chain, a gift of P. Savage (Brigham Young University, Provo, UT), was incubated with purified CD1d-biotin at a 6:1 (lipid/protein) molar ratio at 37°C overnight. SA-PE (BD) or SA-BV (BioLegend) was added sequentially (1/8 the required volume), including a series of 10-min incubations at 4°C.
Single cell TCR sequence determination.
cDNA from sorted MR1–5-OP-RU tetramer+ TCR-β+ intermediate cells was generated by the addition of 2 µl per well of buffer containing SuperScript VILO (Invitrogen) and 0.1% Triton X-100 (Sigma-Aldrich) and incubated according to the manufacturer’s instructions. Transcripts encoding Vα and Vβ were amplified by two rounds of nested PCR as previously described (Dash et al., 2011). PCR products were separated on a 1.5% agarose gel and sequenced (Applied Genetics Diagnostics, University of Melbourne).
CTV labeling and proliferation assay.
Splenocytes from WT C57BL/6 mice were labeled with CTV and incubated for 10 min at 37°C. Labeled cells were cultured for 3 d in the presence of graded doses of 5-OP-RU Ag or Ac-6-FP or no stimulation. For proliferative analysis, cells were harvested after 72 h and stained with anti–TCR-β and MR1–5-OP-RU tetramer. Generational divisions of proliferating MAIT cells were determined with FlowJo software (Tree Star).
Cell surface staining.
In vitro stimulated cells were stained with MR1-Ag tetramer, or α-GalCer (PBS-44)–loaded CD1d tetramer conjugated to PE, 7-aminoactinomycin D (Sigma-Aldrich), anti–CD45R-allophycocyanin (APC)-Cy7 (BD) and anti–TCR-β–APC (BD). For Vβ analysis, anti–TCR-β was excluded from the antibody cocktail, instead, and a secondary stain was performed using anti–Vβ-FITC (BD). Proliferative responses were assessed using LSR II or LSR Fortessa flow cytometers (BD) and FlowJo software. Tetramer staining experiments were performed using an LSR Fortessa (BD) equipped with a yellow laser.
Intracellular cytokine staining.
Lymphocytes were cultured in cell culture medium containing 10 ng/ml PMA (Sigma-Aldrich), 1 µg/ml ionomycin (Sigma-Aldrich), and 2 μM monensin (GolgiStop; BD) for 4 h. Cells were then washed and labeled with cell surface antibodies before fixation and permeabilization using the Cytofix/Cytoperm staining kit (BD) as per the manufacturer’s instructions. Cells were subsequently washed and stained for intracellular anti–IFN-γ and anti–IL-17A (BD) for flow cytometric analysis.
Intracellular transcription factor staining.
To detect intracellular transcription factors, anti–mouse Foxp3 staining kits (eBioscience) were used after appropriate surface staining. Staining was performed according to the manufacturer’s instructions. In brief, cells were resuspended in fixation/permeabilization buffer and incubated at 4°C, in the dark for 1 h. Cells were then washed with Perm/Wash buffer (eBioscience) before being labeled with anti-PLZF (eBioscience), anti–T-Bet (eBioscience), and anti-RORγt (eBioscience). This approach was also used for intracellular transcription factor and cytokine costaining.
Cytometric bead array (CBA).
CD24 plus complement-depleted mature thymocytes and B cell–depleted splenocytes from WT C57BL/6 mice were labeled with MR1–5-OP-RU tetramer and anti–TCR-β, and MR1–5-OP-RU tetramer+ TCR-β+ cells were sorted using a flow cytometer (FACSAria; BD). Sorted cells were then cultured in the presence of plate-bound anti-CD3 (5 µg/ml) and anti-CD28 (10 µg/ml). Culture supernatants were collected after 24 h and analyzed with CBA flex set for mice (BD).
Acknowledgments
The authors thank Lars Kjer-Nielsen and Mehmet Yabas for helpful advice and technical assistance, Tina Luke, flow cytometry facility staff, and Michael Thomson (St. Vincent’s Institute Flow Cytometry Facility) for flow cytometry assistance, David Taylor and animal house staff for animal husbandry assistance, and Paul Savage for providing α-GalCer (PBS-44).
This work was supported by a project grant and program grants from the National Health and Medical Research Council of Australia (NHMRC; 108394, 1013667, and 1016629) and the Australian Research Council (ARC; CE140100011 and LE110100106). A. Enders is supported by an NHMRC Career Development Fellowship (1035858); D.G. Pellicci is supported by an NHMRC Early Career Fellowship (1054431); A.P. Uldrich is supported by an ARC Future Fellowship (FT140100278); D.I. Godfrey and D.P. Fairlie are supported by NHMRC Senior Principal Research Fellowships (1020770 and 1027369); J. Rossjohn is supported by an NHMRC Australia Fellowship (AF50).
The authors declare no competing financial interests.
Author contributions: A. Rahimpour, H.F. Koay, and R. Clanchy performed experiments and prepared figures. A. Enders, S.B.G. Eckle, B. Meehan, Z. Chen, B. Whittle, L. Liu, D.P. Fairlie, and C.C. Goodnow provided key reagents and tissue samples. J. McCluskey, J. Rossjohn, A.P. Uldrich, D.G. Pellicci, and D.I. Godfrey planned experiments and worked together with data interpretation and manuscript preparation. D.I. Godfrey wrote the paper and led the investigation.
Footnotes
Abbreviations used:
- 5-OP-RU
- 5-(2-oxopropylideneamino)-6-d-ribityl-aminouracil
- 6-FP
- 6-formylpterin
- Ac-6-FP
- acetyl-6-FP
- Ag
- antigen
- CBA
- cytometric bead array
- CTV
- cell trace violet
- MAIT cell
- mucosal-associated invariant T cell
- PLZF
- promyelocytic leukemia zinc finger
- RORγt
- retinoic acid–related orphan receptor γt
References
- Birkinshaw R.W., Kjer-Nielsen L., Eckle S.B., McCluskey J., and Rossjohn J.. 2014. MAITs, MR1 and vitamin B metabolites. Curr. Opin. Immunol. 26:7–13. 10.1016/j.coi.2013.09.007 [DOI] [PubMed] [Google Scholar]
- Corbett A.J., Eckle S.B., Birkinshaw R.W., Liu L., Patel O., Mahony J., Chen Z., Reantragoon R., Meehan B., Cao H., et al. 2014. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. 509:361–365. 10.1038/nature13160 [DOI] [PubMed] [Google Scholar]
- Croxford J.L., Miyake S., Huang Y.Y., Shimamura M., and Yamamura T.. 2006. Invariant Vα19i T cells regulate autoimmune inflammation. Nat. Immunol. 7:987–994. 10.1038/ni1370 [DOI] [PubMed] [Google Scholar]
- Dash P., McClaren J.L., Oguin T.H. III, Rothwell W., Todd B., Morris M.Y., Becksfort J., Reynolds C., Brown S.A., Doherty P.C., and Thomas P.G.. 2011. Paired analysis of TCRα and TCRβ chains at the single-cell level in mice. J. Clin. Invest. 121:288–295. 10.1172/JCI44752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusseaux M., Martin E., Serriari N., Péguillet I., Premel V., Louis D., Milder M., Le Bourhis L., Soudais C., Treiner E., and Lantz O.. 2011. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17–secreting T cells. Blood. 117:1250–1259. 10.1182/blood-2010-08-303339 [DOI] [PubMed] [Google Scholar]
- Eckle S.B., Birkinshaw R.W., Kostenko L., Corbett A.J., McWilliam H.E., Reantragoon R., Chen Z., Gherardin N.A., Beddoe T., Liu L., et al. 2014. A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells. J. Exp. Med. 211:1585–1600. 10.1084/jem.20140484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann F., Cameron T.O., Sidobre S., Manlongat N., Kronenberg M., Briskin M.J., Dustin M.L., and Littman D.R.. 2005. Intravascular immune surveillance by CXCR6+ NKT cells patrolling liver sinusoids. PLoS Biol. 3:e113 10.1371/journal.pbio.0030113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey D.I., Stankovic S., and Baxter A.G.. 2010. Raising the NKT cell family. Nat. Immunol. 11:197–206. 10.1038/ni.1841 [DOI] [PubMed] [Google Scholar]
- Gold M.C., Cerri S., Smyk-Pearson S., Cansler M.E., Vogt T.M., Delepine J., Winata E., Swarbrick G.M., Chua W.J., Yu Y.Y., et al. 2010. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol. 8:e1000407 10.1371/journal.pbio.1000407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold M.C., McLaren J.E., Reistetter J.A., Smyk-Pearson S., Ladell K., Swarbrick G.M., Yu Y.Y., Hansen T.H., Lund O., Nielsen M., et al. 2014. MR1-restricted MAIT cells display ligand discrimination and pathogen selectivity through distinct T cell receptor usage. J. Exp. Med. 211:1601–1610. 10.1084/jem.20140507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawachi I., Maldonado J., Strader C., and Gilfillan S.. 2006. MR1-restricted Vα19i mucosal-associated invariant T cells are innate T cells in the gut lamina propria that provide a rapid and diverse cytokine response. J. Immunol. 176:1618–1627. 10.4049/jimmunol.176.3.1618 [DOI] [PubMed] [Google Scholar]
- Kjer-Nielsen L., Patel O., Corbett A.J., Le Nours J., Meehan B., Liu L., Bhati M., Chen Z., Kostenko L., Reantragoon R., et al. 2012. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 491:717–723. 10.1038/nature11605 [DOI] [PubMed] [Google Scholar]
- Kovalovsky D., Uche O.U., Eladad S., Hobbs R.M., Yi W., Alonzo E., Chua K., Eidson M., Kim H.J., Im J.S., et al. 2008. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat. Immunol. 9:1055–1064. 10.1038/ni.1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreslavsky T., Savage A.K., Hobbs R., Gounari F., Bronson R., Pereira P., Pandolfi P.P., Bendelac A., and von Boehmer H.. 2009. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of γδ T cells with restricted TCR diversity. Proc. Natl. Acad. Sci. USA. 106:12453–12458. 10.1073/pnas.0903895106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bourhis L., Martin E., Péguillet I., Guihot A., Froux N., Coré M., Lévy E., Dusseaux M., Meyssonnier V., Premel V., et al. 2010. Antimicrobial activity of mucosal-associated invariant T cells. Nat. Immunol. 11:701–708. (published erratum appears in Nat. Immunol. 2010. 11:969) 10.1038/ni.1890 [DOI] [PubMed] [Google Scholar]
- Le Bourhis L., Mburu Y.K., and Lantz O.. 2013. MAIT cells, surveyors of a new class of antigen: development and functions. Curr. Opin. Immunol. 25:174–180. 10.1016/j.coi.2013.01.005 [DOI] [PubMed] [Google Scholar]
- Lee Y.J., Holzapfel K.L., Zhu J., Jameson S.C., and Hogquist K.A.. 2013. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat. Immunol. 14:1146–1154. 10.1038/ni.2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeansyah E., Loh L., Nixon D.F., and Sandberg J.K.. 2014. Acquisition of innate-like microbial reactivity in mucosal tissues during human fetal MAIT-cell development. Nat. Commun. 5:3143 10.1038/ncomms4143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepore M., Kalinichenko A., Colone A., Paleja B., Singhal A., Tschumi A., Lee B., Poidinger M., Zolezzi F., Quagliata L., et al. 2014. Parallel T-cell cloning and deep sequencing of human MAIT cells reveal stable oligoclonal TCRβ repertoire. Nat. Commun. 5:3866 (published erratum appears in Nat. Commun. 2014. 5:4493) 10.1038/ncomms4866 [DOI] [PubMed] [Google Scholar]
- Martin E., Treiner E., Duban L., Guerri L., Laude H., Toly C., Premel V., Devys A., Moura I.C., Tilloy F., et al. 2009. Stepwise development of MAIT cells in mouse and human. PLoS Biol. 7:e54 10.1371/journal.pbio.1000054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliam H.E., Birkinshaw R.W., Villadangos J.A., McCluskey J., and Rossjohn J.. 2015. MR1 presentation of vitamin B-based metabolite ligands. Curr. Opin. Immunol. 34:28–34. 10.1016/j.coi.2014.12.004 [DOI] [PubMed] [Google Scholar]
- Meierovics A., Yankelevich W.J., and Cowley S.C.. 2013. MAIT cells are critical for optimal mucosal immune responses during in vivo pulmonary bacterial infection. Proc. Natl. Acad. Sci. USA. 110:E3119–E3128. 10.1073/pnas.1302799110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki Y., Miyake S., Chiba A., Lantz O., and Yamamura T.. 2011. Mucosal-associated invariant T cells regulate Th1 response in multiple sclerosis. Int. Immunol. 23:529–535. 10.1093/intimm/dxr047 [DOI] [PubMed] [Google Scholar]
- Okamoto N., Kanie O., Huang Y.Y., Fujii R., Watanabe H., and Shimamura M.. 2005. Synthetic α-mannosyl ceramide as a potent stimulant for an NKT cell repertoire bearing the invariant Vα19-Jα26 TCR α chain. Chem. Biol. 12:677–683. 10.1016/j.chembiol.2005.04.014 [DOI] [PubMed] [Google Scholar]
- Patel O., Kjer-Nielsen L., Le Nours J., Eckle S.B., Birkinshaw R., Beddoe T., Corbett A.J., Liu L., Miles J.J., Meehan B., et al. 2013. Recognition of vitamin B metabolites by mucosal-associated invariant T cells. Nat. Commun. 4:2142 10.1038/ncomms3142 [DOI] [PubMed] [Google Scholar]
- Pellicci D.G., Hammond K.J.L., Uldrich A.P., Baxter A.G., Smyth M.J., and Godfrey D.I.. 2002. A natural killer T (NKT) cell developmental pathway involving a thymus-dependent NK1.1−CD4+ CD1d-dependent precursor stage. J. Exp. Med. 195:835–844. 10.1084/jem.20011544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicci D.G., Patel O., Kjer-Nielsen L., Pang S.S., Sullivan L.C., Kyparissoudis K., Brooks A.G., Reid H.H., Gras S., Lucet I.S., et al. 2009. Differential recognition of CD1d-α-galactosyl ceramide by the Vβ8.2 and Vβ7 semi-invariant NKT T cell receptors. Immunity. 31:47–59. 10.1016/j.immuni.2009.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince A.L., Watkin L.B., Yin C.C., Selin L.K., Kang J., Schwartzberg P.L., and Berg L.J.. 2014. Innate PLZF+CD4+ αβ T cells develop and expand in the absence of Itk. J. Immunol. 193:673–687. 10.4049/jimmunol.1302058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reantragoon R., Kjer-Nielsen L., Patel O., Chen Z., Illing P.T., Bhati M., Kostenko L., Bharadwaj M., Meehan B., Hansen T.H., et al. 2012. Structural insight into MR1-mediated recognition of the mucosal associated invariant T cell receptor. J. Exp. Med. 209:761–774. 10.1084/jem.20112095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reantragoon R., Corbett A.J., Sakala I.G., Gherardin N.A., Furness J.B., Chen Z., Eckle S.B., Uldrich A.P., Birkinshaw R.W., Patel O., et al. 2013. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J. Exp. Med. 210:2305–2320. 10.1084/jem.20130958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossjohn J., Gras S., Miles J.J., Turner S.J., Godfrey D.I., and McCluskey J.. 2015. T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 33:169–200. 10.1146/annurev-immunol-032414-112334 [DOI] [PubMed] [Google Scholar]
- Savage A.K., Constantinides M.G., Han J., Picard D., Martin E., Li B., Lantz O., and Bendelac A.. 2008. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 29:391–403. 10.1016/j.immuni.2008.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seach N., Guerri L., Le Bourhis L., Mburu Y., Cui Y., Bessoles S., Soudais C., and Lantz O.. 2013. Double-positive thymocytes select mucosal-associated invariant T cells. J. Immunol. 191:6002–6009. 10.4049/jimmunol.1301212 [DOI] [PubMed] [Google Scholar]
- Tang X.Z., Jo J., Tan A.T., Sandalova E., Chia A., Tan K.C., Lee K.H., Gehring A.J., De Libero G., and Bertoletti A.. 2013. IL-7 licenses activation of human liver intrasinusoidal mucosal-associated invariant T cells. J. Immunol. 190:3142–3152. 10.4049/jimmunol.1203218 [DOI] [PubMed] [Google Scholar]
- Teunissen M.B., Yeremenko N.G., Baeten D.L., Chielie S., Spuls P.I., de Rie M.A., Lantz O., and Res P.C.. 2014. The IL-17A-producing CD8+ T-cell population in psoriatic lesional skin comprises mucosa-associated invariant T cells and conventional T cells. J. Invest. Dermatol. 134:2898–2907. 10.1038/jid.2014.261 [DOI] [PubMed] [Google Scholar]
- Tilloy F., Treiner E., Park S.H., Garcia C., Lemonnier F., de la Salle H., Bendelac A., Bonneville M., and Lantz O.. 1999. An invariant T cell receptor α chain defines a novel TAP-independent major histocompatibility complex class Ib–restricted α/β T cell subpopulation in mammals. J. Exp. Med. 189:1907–1921. 10.1084/jem.189.12.1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiner E., Duban L., Bahram S., Radosavljevic M., Wanner V., Tilloy F., Affaticati P., Gilfillan S., and Lantz O.. 2003. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. 422:164–169. 10.1038/nature01433 [DOI] [PubMed] [Google Scholar]
- Ussher J.E., Klenerman P., and Willberg C.B.. 2014. Mucosal-associated invariant T-cells: new players in anti-bacterial immunity. Front. Immunol. 5:450 10.3389/fimmu.2014.00450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L.J., Kang Y.H., Smith M.O., Tharmalingham H., Ramamurthy N., Fleming V.M., Sahgal N., Leslie A., Oo Y., Geremia A., et al. 2012. Human MAIT and CD8αα cells develop from a pool of type-17 precommitted CD8+ T cells. Blood. 119:422–433. 10.1182/blood-2011-05-353789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigmann B., Tubbe I., Seidel D., Nicolaev A., Becker C., and Neurath M.F.. 2007. Isolation and subsequent analysis of murine lamina propria mononuclear cells from colonic tissue. Nat. Protoc. 2:2307–2311. 10.1038/nprot.2007.315 [DOI] [PubMed] [Google Scholar]