Ungerbäck et al. show that transcription factors Ebf1 and Pax5 act in a coordinated, dose-dependent manner to preserve B-lineage cell fate. Combined heterozygous loss of both transcription factors results in increased T cell lineage skewing in B cell progenitors.
Abstract
To investigate how transcription factor levels impact B-lymphocyte development, we generated mice carrying transheterozygous mutations in the Pax5 and Ebf1 genes. Whereas combined reduction of Pax5 and Ebf1 had minimal impact on the development of the earliest CD19+ progenitors, these cells displayed an increased T cell potential in vivo and in vitro. The alteration in lineage fate depended on a Notch1-mediated conversion process, whereas no signs of de-differentiation could be detected. The differences in functional response to Notch signaling in Wt and Pax5+/−Ebf1+/− pro–B cells were reflected in the transcriptional response. Both genotypes responded by the generation of intracellular Notch1 and activation of a set of target genes, but only the Pax5+/−Ebf1+/− pro–B cells down-regulated genes central for the preservation of stable B cell identity. This report stresses the importance of the levels of transcription factor expression during lymphocyte development, and suggests that Pax5 and Ebf1 collaborate to modulate the transcriptional response to Notch signaling. This provides an insight on how transcription factors like Ebf1 and Pax5 preserve cellular identity during differentiation.
B-lymphocyte development is regulated by the orchestrated action of transcription factors coordinating the activation and silencing of genes crucial for normal differentiation. Two central proteins in this process are Ebf1 and Pax5, both critically important for normal B-lymphocyte development (Urbánek et al., 1994; Lin and Grosschedl, 1995). Even though both these transcription factors are crucial for the development of CD19-expressing B cell progenitors, high-resolution analysis of early B cell differentiation has revealed that Ebf1 and Pax5 are expressed and act in a sequential manner during the differentiation process (Nutt et al., 1997, 1998; Mansson et al., 2010; Zandi et al., 2012). In the absence of Ebf1, lymphoid progenitor cells fail to initiate transcription of B-lineage genes (Lin and Grosschedl, 1995; Zandi et al., 2008), revealing that Ebf1 is crucial for B-lineage specification, including initiation of Pax5 expression. In the absence of Pax5, a B-lineage–specific transcriptional program is initiated (Nutt et al., 1997; Zandi et al., 2012); however, Pax5-deficient cells are not stably committed and external signals such as cytokine stimulation or Notch signaling is sufficient to drive these cells into alternative cell fates in vitro and in vivo (Nutt et al., 1999; Rolink et al., 1999; Heavey et al., 2003; Höflinger et al., 2004; Cobaleda et al., 2007; Zandi et al., 2012). Using conditional targeting of the Pax5 or Ebf1 genes, it has been reported that inactivation of either of these proteins in CD19+ cells results in disruptions in the genetic program and loss of B cell identity, allowing the cells to adopt alternative cell fates (Cobaleda et al., 2007; Nechanitzky et al., 2013). Analysis of progenitor compartments and developmental processes has provided evidence that this involves dedifferentiation of the CD19+ cells into immature multipotent progenitors in the BM, allowing the generation of multiple hematopoietic lineages (Cobaleda et al., 2007; Nechanitzky et al., 2013). Even though Ebf1 and Pax5 act in a hierarchical manner, they share several target genes (Lin et al., 2010; Treiber et al., 2010; Revilla-I-Domingo et al., 2012; Vilagos et al., 2012) and activate as well as repress transcription in a coordinated manner. Furthermore, the collaboration between these two proteins has been suggested to create a positive feedback loop where Pax5 regulates expression of Ebf1 and Ebf1 interact with enhancer elements in the Pax5 gene (O’Riordan and Grosschedl, 1999; Roessler et al., 2007; Pongubala et al., 2008; Decker et al., 2009). Even though the importance of this autoregulatory loop is somewhat disputed because loss of Ebf1 does not have any major impact on Pax5 expression (Nechanitzky et al., 2013), ectopic expression of Ebf1, in Pax5-deficient cells displaying reduced Ebf1 levels, results in lineage restriction (Pongubala et al., 2008). Thus, Pax5 and Ebf1 participate in a complex interplay in the specification and commitment of lymphoid progenitors in the B-lineage pathway.
Although the complete absence of either Ebf1 or Pax5 results in total disruption of B cell development, a reduction of the functional dose of any of these factors as a consequence of a mutation of only one allele of the coding genes results in more subtle phenotypes (Urbánek et al., 1994; Lin and Grosschedl, 1995; O’Riordan and Grosschedl, 1999; Lukin et al., 2011; Åhsberg et al., 2013). Whereas heterozygous loss of Pax5 has a minimal impact on B cell development (Urbánek et al., 1994), loss of one allele of Ebf1 results in a significant reduction of the pre–B cell compartment (O’Riordan and Grosschedl, 1999; Lukin et al., 2011; Åhsberg et al., 2013). The Ebf1+/− phenotype is enhanced by combined heterozygous deletions of either E2a (O’Riordan and Grosschedl, 1999) or Runx1 (Lukin et al., 2010), highlighting the importance of transcription factor dose in normal B cell development. The identification of heterozygous mutations in the PAX5 and EBF1 genes in human B-lineage acute lymphoblastic leukemia (B-ALL; Mullighan et al., 2007) suggests that transcription factor dose is of crucial importance in the prevention against B-lineage malignancies as well. This idea was supported by analysis of a mouse model where the expression of a constitutively active Stat5 was combined with heterozygous mutations in either the Pax5 or Ebf1 genes (Heltemes-Harris et al., 2011). These mice developed B cell leukemia, revealing that mutations in either of these transcription factors can synergize with a proliferation signal such as that provided by activated Stat5 in the generation of malignant disease. Hence, transcription factor dose creates a link between development and disease.
While the importance of Ebf1 and Pax5 in lineage restriction has been elegantly demonstrated, the crucial function of either of these proteins for B cell identity and survival complicates the analysis of the collaboration between these factors in normal B cell development. Hence, to investigate how Ebf1 and Pax5 collaborate in the induction and regulation of B-lineage development, we crossed Ebf1+/− mice to Pax5+/− mice to generate animals carrying transheterozygous mutations in these genes. The combined heterozygous deletion of Pax5 and Ebf1 does not impair the earliest stages of B cell development or the establishment of the transcriptional program defining the identity of the early B-lineage cells. However, the Pax5+/−Ebf1+/− Lin−B220+CD19+CD43highIgM− (pro–B cells) display a dramatically increased ability to generate T-lineage cells both in vivo and in vitro. In vitro differentiation experiments suggest that this is critically dependent on a Notch signal and a result of lineage conversion rather than dedifferentiation. Hence, our data suggest that Pax5 and Ebf1 collaborate to reduce the impact of active Notch signaling, thereby maintaining B-lineage identity in early progenitor cells.
RESULTS
Combined heterozygous deletion of Pax5 and Ebf1 results in a reduced pre–B cell compartment and increased plasticity of B cell progenitors
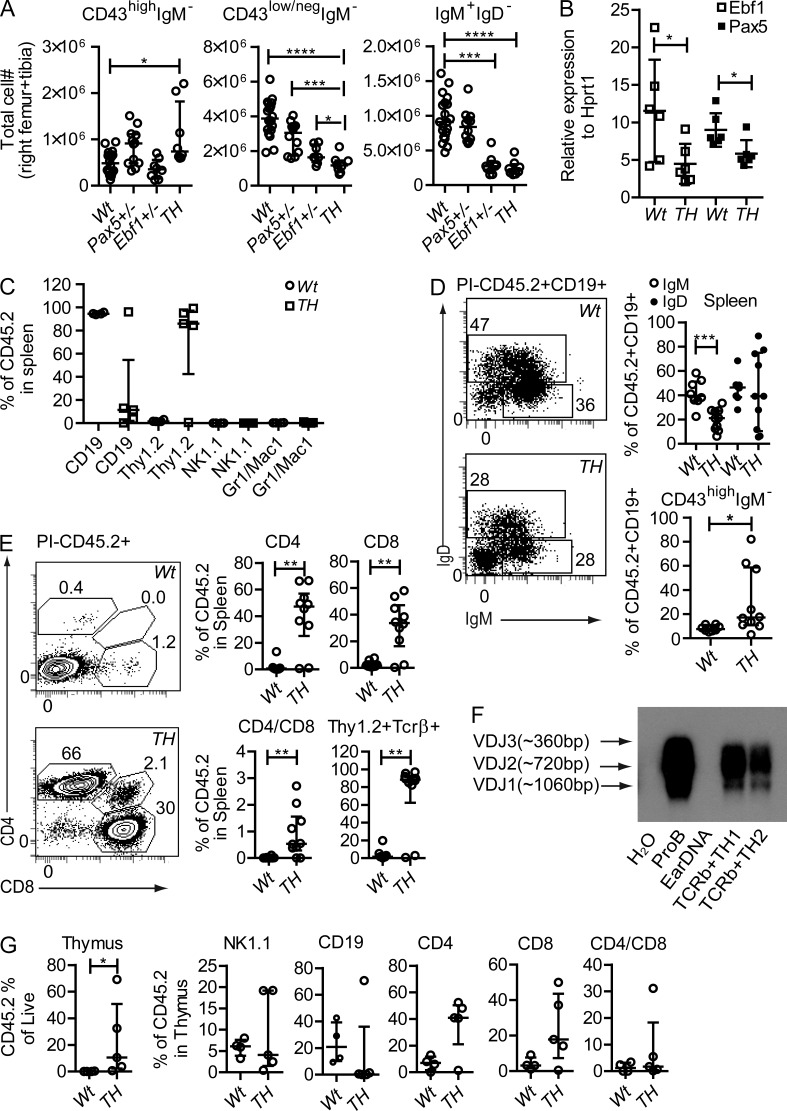
To investigate the functional consequence of combined reductions of Ebf1 and Pax5 dose, we generated mice transheterozygous (TH) for mutations in the Ebf1 and Pax5 genes. These animals did not display any decrease in the total number of the pro–B cells as compared with that of the Wt or the single mutant mice (Fig. 1 A). In contrast, the Lin-B220+CD19+CD43low/negIgM− (pre–B cell) compartment was reduced in TH mice as compared with what was observed in any of the single mutant mice. The number of Lin−B220+CD19+CD43low/negIgM+IgD− cells was reduced to levels comparable to single Ebf1+/− mice in the Pax5+/−Ebf1+/− animals (Fig. 1 A). The formation of early progenitor cells in the Pax5+/−Ebf1+/− mice suggested that the combined dose reduction in Ebf1 and Pax5 would not result in a complete disruption of the autoregulatory loop between Ebf1 and Pax5 and collapse of the genetic program. Consistent with this notion, Q-PCR analysis of primary pro–B cells from Wt and Pax5+/−Ebf1+/− mice (Fig. 1 B) suggested a twofold down-regulation of Ebf1 and Pax5 transcripts, respectively in pro–B cells from the TH mice, as would be expected from loss of one functional allele.
Figure 1.
Combined transheterozygous loss of Pax5 and Ebf1 results in a partial block at the pro–B cell stage and lineage plasticity in CD19+ cells. (A) Graphs displaying the absolute numbers of 7AAD− pro–B cells (Lin−B220+CD19+CD43highIgM−), pre–B cells (Lin−B220+CD19+CD43low/negIgM−), and IgM+ B cells (Lin−B220+CD19+CD43low/negIgM+) in bone marrow from Wt (n = 15), Pax5+/− (n = 10), Ebf1+/− (n = 7), and Pax5+/−Ebf1+/− (TH) (n = 6) mice. (B) Q-PCR analysis of Ebf1 and Pax5 expression in pro–B cells from WT and Pax5+/−Ebf1+/− (TH) mice. Each square indicates a biological replicate analyzed by triplicate Q-PCR reactions. Mean of all mice is presented as a horizontal line. Statistical analysis was performed using unpaired Student’s t test. (C) Graphs illustrating the percent of donor (CD45.2+) of CD19+Thy1.2−, CD19−Thy1.2+, NK1.1+, and Gr1/Mac1+ cells in spleen of Rag1−/− CD45.1+ recipient mice 6–9 wk after transplantation with 2 × 106 sorted bone marrow CD19+IgM− cells from Wt (n = 8) or Pax5+/−Ebf1+/− (TH; n = 9) mice. Mice were from two independent transplantation experiments (4 Wt and 4 TH in experiment one and 4 Wt and 5 TH transplantations in experiment two). (D) Graphs and representative FACS plot illustrating the percent of IgM+, IgM+IgD+, and CD43highIgM− pro–B cells out of CD45.2+CD19+ from Rag1−/− mice as described in A. (E) Diagrams and representative FACS plot illustrating the percent of donor (CD45.2+) of CD4+CD8−, CD4−CD8+, CD4+CD8+, and Thy1.2+Tcrβ+ cells in the spleen of Rag1−/− mice. (F) Southern blot of generated PCR products analyzing immunoglobulin heavy chain VDJ recombination using genomic DNA from Wt pro–B, mouse ear, or sorted TCRβ+ cells from TH transplanted Rag1−/− mice. (G) Diagrams illustrating total CD45.2+ out of live cells in thymus, also the percent of donor (CD45.2+) of CD19+, NK1.1+, CD4+CD8−, CD4−CD8+, and CD4+CD8+ in the thymus of Rag1−/− mice as described in A. In A, C, E, and G, each open circle/square represents one mouse (Wt, n = 4; TH, n = 5) and the data are presented as median (horizontal line) ± interquartile range. Statistical analysis was performed using Mann-Whitney U test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
To investigate the functional potential of Pax5+/−Ebf1+/− B cell progenitors in vivo, we transplanted CD19+IgM− cells from Wt and Pax5+/−Ebf1+/− CD45.2 mice into Rag1−/− CD45.1 animals. Six to nine weeks after transplantation, we analyzed the cellular composition of CD45.2+ cells in the spleen of the transplanted mice. While over 93% of the cells in the mice transplanted with Wt progenitors expressed CD19, only one of the analyzed mice transplanted with Pax5+/−Ebf1+/− B cell progenitors presented with large numbers of CD19+ cells (Fig. 1 C). The majority of the mice transplanted with Pax5+/−Ebf1+/− progenitors presented a large fraction of Thy1.2+ cells, representing a most limited population in mice transplanted with Wt cells (Fig. 1 C). Donor cells expressing NK1.1 or Gr1/Mac1 were the most limited or undetectable (Fig. 1 C). Analysis of the CD19+ compartment revealed that the major part of the CD19+ cells generated from both Wt and Pax5+/−Ebf1+/− B cell progenitors represented IgM+ or IgM+IgD+ B cells (Fig. 1 D). However, in the three Pax5+/−Ebf1+/− progenitor transplanted mice displaying the largest fraction of CD19+ cells, a major part of the cells displayed the phenotype of pro–B cells with lack of IgM expression and high expression of CD43 (Fig. 1 D). These data reveal that Pax5+/−Ebf1+/− B cell progenitors are deficient in their ability to generate mature B-lineage cells, and the presence of large amounts of pro–B cells in the spleen of three of the transplanted mice indicate that these cells may be prone to malignant transformation. Whereas the majority of the mice transplanted with Wt cells presented a limited frequency of Thy1.2 high cells, this marker was highly expressed on the majority of the cells generated from Pax5+/−Ebf1+/− B cell progenitors. High Thy1.2 expression is normally restricted to T-lineage cells and to investigate the identity of these cells in more detail, we stained spleen cells with antibodies detecting CD4, CD8, and TCRβ. Virtually all the Thy1.2+ cells expressed TCRβ, and we could observe CD4 and CD8 single-positive as well as double-positive donor cells in the spleen (Fig. 1 E), strongly supporting the notion that the B cell progenitors from Pax5+/−Ebf1+/− mice had preferentially generated T-lineage cells after transplantation in vivo.
To investigate if we could detect evidence of immunoglobulin heavy chain VDJ recombination in the generated T-lineage cells, we sorted CD3+TCRβ+ cells from mice transplanted with TH cells, extracted DNA, and investigated the presence of VDJ recombination by PCR using one primer in J3 in combination with three degenerated primers directed toward the V-gene families J558, Q52, and 7183 (Schlissel et al., 1991). This indicated that the T cell generated from transplanted Pax5+/−Ebf1+/− cells carried detectable polyclonal immunoglobulin VDJ recombination events, supporting the notion that the cells were generated from B-lineage progenitors (Fig. 1 F). Analysis of the thymus of transplanted mice revealed that Pax5+/−Ebf1+/− B cell progenitors repopulated the thymus more efficiently than Wt cells (Fig. 1 G). Although the cells from Wt progenitors displayed a substantial amount of CD19+ cells in the thymus of the recipients, with the exception of one mouse with peripheral pro–B cell expansion, Pax5+/−Ebf1+/− B cell progenitors generated Thy1.2+ cells being either CD4 or CD8 single positive. A small fraction of double-positive cells could also be detected in the thymus of transplanted mice. In contrast to what was observed in the spleen, we could also detect a small fraction of NK1.1+ cells generated from both Wt and Pax5+/−Ebf1+/− B cell progenitors. Even though we did detect T-lineage cells in mice transplanted with Wt CD19+IgM− cells, the frequency was significantly lower than what we observed in mice transplanted with Pax5+/−Ebf1+/− progenitors, revealing that combined reduction of Pax5 and Ebf1 dose results in abnormal lineage plasticity in vivo after transplantation to lymphocyte-deficient mice.
CD19+ pro–B cells from Pax5+/−Ebf1+/− mice display increased lineage plasticity as compared with single heterozygote cells in the presence of a strong Notch signal
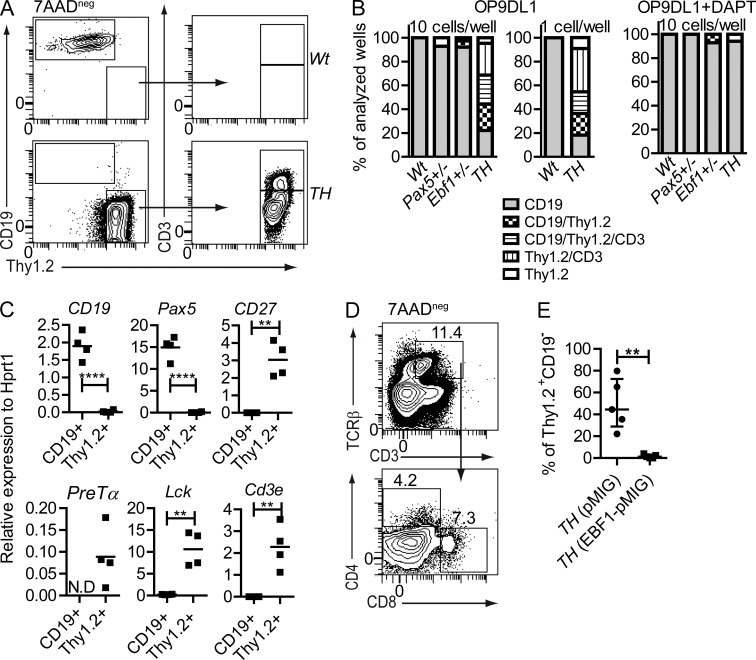
The finding that CD19+ progenitors from Pax5+/−Ebf1+/− mice generated T-lineage cells in vivo suggest that even though these cells express CD19, they are not stably committed to B-lineage cell fate. To investigate the lineage fidelity of B cell progenitors generated with reduced levels of Pax5 and Ebf1 in more detail, we sorted pro–B cells and seeded them on OP9-DL1 stroma cells, generating conditions permissive for T-lineage development of noncommitted progenitors (Schmitt and Zuñiga-Pflucker, 2002). Seeding 10 Wt pro–B cells per well, we detected growth in 32% of the wells. The overall cloning frequency of Pax5+/− cells was 7%, a decrease as compared with Wt cells (P < 0.0001), whereas ∼24% of the wells seeded with Ebf1+/− cells generated colonies, suggesting that the cloning frequency was reduced by loss of one allele of Pax5. Pax5+/−Ebf1+/− cells generated colonies in almost 50% of the wells. Upon analysis of the cellular content of the cultures, 100% of the colonies generated from Wt progenitors contained only CD19+ cells, whereas wells seeded with Pax5+/− or Ebf1+/− cells generated a low frequency of colonies with CD19− cells expressing Thy1.2 (Fig. 2, A and B). Analysis of the content in wells seeded with pro–B cells from Pax5+/−Ebf1+/− mice revealed that the majority of these cultures were composed of a mixture of cells, including CD19−Thy1.2+ and CD3+ cells (Fig. 2, A and B), supporting an increased T cell potential in Pax5+/−Ebf1+/− as compared with Wt (P < 0.0001) or single heterozygous mice. To verify plasticity at the single-cell level, we seeded single Wt or Pax5+/−Ebf1+/− pro–B cells on OP9-DL1 stromal cells. Analysis of the cellular content of the single cell cultures revealed that although all the Wt cells generated CD19+ cells, a majority of the Pax5+/−Ebf1+/− cells generated mixed colonies (Fig. 2 B), revealing plasticity at the single-cell level. Inclusion of the γ-secretase inhibitor DAPT in the cell cultures inhibited the formation of CD19−Thy1.2+ cells, confirming a need of Notch signaling to initiate the phenotypic change (Fig. 2 B). We could also verify that the plasticity was critically dependent on the Ebf1 dose in the pro–B cells because retroviral transduction of Pax5+/−Ebf1+/− pro–B cells with an Ebf1 encoding retrovirus blocked the formation of CD19−Thy1.2+ cells (Fig. 2 E). Even though high expression of Thy1.2 and CD3 normally serves as reliable markers for cells that have entered the T-lineage pathway, we extended our analysis by performing a Q-PCR analysis of colonies containing either CD19+ or Thy1.2+ cells generated from Wt pro–B cells or from Pax5+/−Ebf1+/− pro–B cells, respectively. Although the expression of B-lineage genes such as CD19 and Pax5 was decreased as compared with Wt cells cultured under the same conditions (Fig. 2 C), analysis of the levels of T cell–associated transcripts including CD3e, Pre-Tα, Lck, and CD27 suggested that these genes were all expressed in the generated Thy1.2+ cells, verifying that the cells had initiated development toward T-lineage. Culturing Wt or Pax5+/−Ebf1+/− pro–B cells on OP9-DL1 cells for 21 d, followed by replacement of IL-7 with IL-2 for 10 additional days, resulted in cell death among the Wt cells, whereas the Pax5+/−Ebf1+/− cells generated offspring expressing TCRβ on the surface (Fig. 2 D).
Figure 2.
Combined heterozygous loss of Pax5 and Ebf1 results in Notch-dependent lineage plasticity in pro–B cells at the single cell level. (A) Representative FACS plots of sorted Wt and Pax5+/−Ebf1+/− pro–B cells after 14 d of T cell–inducing co-culture on OP9-DL1 stroma cells. (B) Cellular composition of clones from 10 Wt, Pax5+/−, Ebf1+/−, or Pax5+/Ebf1+/− (TH) or single Wt or Pax5+/−Ebf1+/− pro–B cells (Lin−B220+CD19+CD43highIgM−) incubated for 14 d on OP9-DL1 cells to stimulate T cell development, with or without the γ-secretase inhibitor DAPT. Total number of wells analyzed from co-culture of OP9-DL1 and 10 pro–B cells are Wt (51), Pax5+/− (14), Ebf1+/− (38), and Pax5+/−Ebf1+/− (86) and co-culture of OP9-DL1 and one pro–B cell are Wt (12) and Pax5+/−Ebf1+/− (15) collected from 2–3 independent experiments. Similarly, the number of wells analyzed after co-culture of OP9-DL1 and 10 pro–B cells in the presence of DAPT are Wt (21), Pax5+/− (2), Ebf1+/− (14), Pax5+/−Ebf1+/− (17). CD19 cells were scored as CD19+Thy1−CD3−, Thy1 cells as CD19−Thy1+CD3−, and CD3 as CD19−Thy1+CD3+. (C) The graphs display gene expression analyzed by Q-PCR in cultures derived from Wt (4 wells) and Pax5+/−Ebf1+/− (4 wells) after 14 d of T cell–inducing co-culture on OP9-DL1 stroma cells. Each square represents one well, analyzed in triplicate Q-PCR reactions. Mean of all wells is presented as a horizontal line. Statistical analysis was performed using unpaired Student’s t test. N.D = no samples in this group showed detectable expression after 45 cycles of PCR. (D) Representative FACS plot of Pax5+/−Ebf1+/−(TH) CD43highIgM− cells cultivated for 31 d on OP9-DL1 supplied with IL-7, kit ligand, and Flt3 ligand day 0–21. On days 22–31, IL-7 was substituted with IL-2 to further stimulate T cell development. The data are representative of three cultures from two experiments. (E) Dot plots representing the cellular content of cell cultures after transduction with either a pMIG (GFP) control or Ebf1 encoding retrovirus (Ebf1-pMIG) in Pax5+/Ebf1+/− (TH) pro–B cells after 14 d of co-culture with OP9-DL1. The data are collected from pro–B sorted from five different animals. Statistical analysis was performed using unpaired Student’s t test. **, P < 0.01; ****, P < 0.0001.
Calculating the efficiency of the seeded pro–B cells to generate CD3+ offspring in vitro revealed that although none of the Wt cells generated CD3+ cells, 1 out of 1,819 of the Ebf1+/− and 1 out of 2,223 Pax5+/− cells displayed this ability. Among the seeded Pax5+/−Ebf1+/− cells, 1 out of 37 seeded cells was able to generate CD3+ progeny. Hence, even though the overall cloning frequency differed (1/25 for Wt, 1/39 for Ebf1+/−, 1/166 for Pax+/−, and 1/19 for Pax5+/−Ebf1+/− pro–B cells), the combined reduction of Ebf1 and Pax5 dose results in a synergistic increase in T cell potential in CD19+ B cell progenitors. This argues against the idea that the change in lineage would be the result of selection of a rare population of plastic cells enriched in the Pax5+/−Ebf1+/− pro–B cell population.
Combined reduction of Ebf1 and Pax5 dose impacts the expression of shared target genes but does not cause a collapse of the B-lineage program
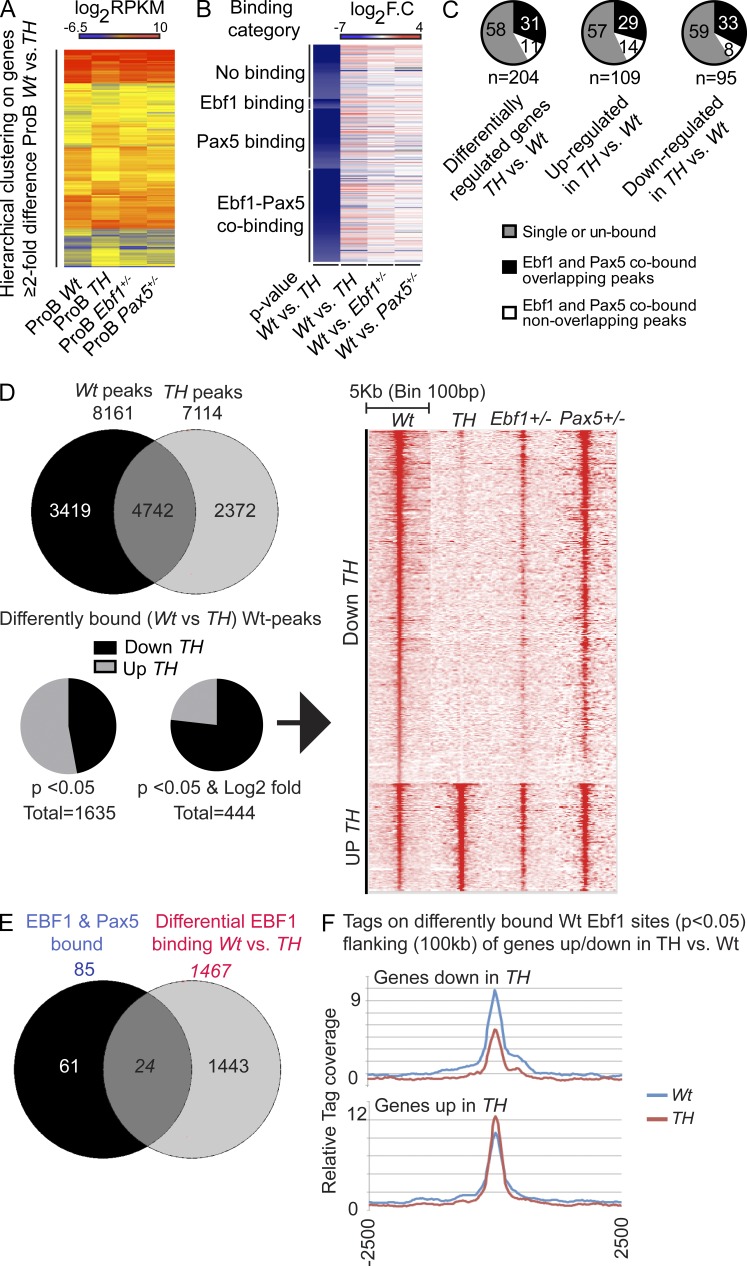
Lineage plasticity in Ebf1−/− or Pax5−/− B cell progenitors has been linked to rather drastic changes in gene expression patterns, including increased expression of T-lineage associated genes such as Notch1 (Souabni et al., 2002; Nechanitzky et al., 2013). To investigate how the combined dose reduction of Pax5 and Ebf1 would impact the transcriptional program in the pro–B cell compartment, we sorted pro–B cells from Wt, Ebf1+/−, Pax5+/−, and Pax5+/−Ebf1+/− animals and performed RNA sequencing analysis. Analysis of the data suggested that even though the transcriptome of Wt and Pax5+/−Ebf1+/− pro–B cells differed in expression at 204 genes (Fig. 3 A), the levels of classical B- or T-lineage genes, including Ebf1 and Pax5 target genes such as CD79α, CD79β, Notch1, or Gata3, were not significantly altered in mice carrying combined heterozygous deletion of Ebf1 and Pax5. K.E.G.G pathway analysis of the differentially expressed genes highlighted a significant difference (Benjamini-Hochberg corrected, P < 0.05) in the categories Cytokine–cytokine receptor interactions, Acute Myeloid Leukemia, Nod-like receptor signaling pathway, Leukocyte transendothelial migration, and NK cell–mediated cytotoxicity. Even though genes involved in cellular signaling were differentially expressed, we did not detect any obvious changes that could be linked to the cellular plasticity observed in Pax5+/−Ebf1+/− pro–B cells. Analysis of the gene expression data suggested that a major part of the genes differentially expressed in the Pax5+/−Ebf1+/− cells was dependent on the combined loss of Ebf1 and Pax5, as the RNA levels were not significantly altered in the single heterozygous cells. To investigate if the differentially expressed genes were direct targets for Ebf1, Pax5, or both, we investigated the binding of these transcription factors in the proximity of the differentially expressed genes using existing Rag2−/− pro–B cell ChIP-sequencing data (Revilla-I-Domingo et al., 2012; Vilagos et al., 2012). This indicated that 85 (42%) of the differentially expressed genes (≥2-fold) contained either overlapping or nonoverlapping binding sites for both Ebf1 and Pax5 (Fig. 3, B and C), whereas an additional 32% contained binding sites for either Ebf1 or Pax5 (Fig. 3 B). Hence, the majority of the differentially regulated genes are direct targets for Ebf1, Pax5, or both. 29% of the up-regulated and 33% of the down-regulated genes contained overlapping peaks (Fig. 3 C), supporting the notion that Pax5 and Ebf1 share a substantial number of regulatory elements.
Figure 3.
Combined dose reduction of Ebf1 and Pax5 results in alterations in gene expression patterns and Ebf1 binding but does not result in collapse of the B-lineage transcriptional program. (A) Heatmap of 204 Wt versus TH (Ebf1+/−Pax5+/−) differentially expressed genes in primary ex vivo sorted pro–B cells. Log2 RPKM values of Wt (n = 4), TH (n = 4), Ebf1+/− (n = 2), and Pax5+/− (n = 2), respectively, are displayed. Differentially expressed genes have been hierarchically clustered with the Cluster 3.0 software (Euclidean distance with average linkage). (B) Heatmap of 204 Wt versus TH differentially expressed genes in primary sorted probe cells. Log2 fold-changes between Wt (n = 4) and TH (n = 4), Wt and Ebf1+/− (n = 2), Wt and Pax5+/− (n = 2), respectively, are displayed. Differentially expressed genes have been clustered on Ebf1+/− and Pax5+/− (Revilla-I-Domingo et al., 2012; Vilagos et al., 2012) binding categories and sorted after increasing P values (top to bottom) as defined by Student’s t test (Benjamini-Hoeschberg corrected) for TH versus Wt differential gene expression. (C) Pie-charts displaying overlapping and nonoverlapping Ebf1+/− and Pax5+/− peaks (Revilla-I-Domingo et al., 2012; Vilagos et al., 2012) in the 204 TH versus Wt differentially regulated genes, as well as the 109 up-regulated and 95 down-regulated genes. Numbers in the pie-charts are percent of total peaks in each group. (D) Venn diagram representing the total number of differentially and shared Ebf1 bound sites determined by ChIP-seq analysis of Wt and TH pro–B cells. The bottom pie-charts represent the number of peaks detected in Wt cells displaying statistically significant changed binding in TH cells and the number of sites with P < 0.05 and fourfold of differentially bound Ebf1 peaks (444 peaks in total). Chromosomes X, Y, M, and Random were filtered out from the Wt and TH peak lists before analysis. The heatmap shows Ebf1 binding on the 444 peaks differentially bound in TH centered on the Wt Ebf1 peaks with a window of 5-kb and a bin size of 100 bp. Data were collected from two independent experiments from each genotype. (E) A list of genes (1467) closest to the 1635 differentially bound (P < 0.05) Ebf1 peaks was compared with the list of genes that are differentially expressed between Wt and TH and co-bound by both Pax5 and Ebf1 as defined by ChIP seq from (Revilla-I-Domingo et al., 2012; Vilagos et al., 2012). (F) Tags from Ebf1 ChIP-seq in Wt and TH samples were plotted centered on peaks defined by the P < 0.05 differentially bound peak list, found within 100 kb of genes that were up or down-regulated in TH versus Wt as indicated.
To investigate how a reduction in Ebf1 dose impacts DNA binding, we performed Ebf1-ChIP-seq analysis on Wt, Ebf1+/−, Pax5+/−, as well as Ebf1+/−Pax5+/− pro–B cells. This allowed for the identification of 8161 peaks in the Wt samples, whereas 7114 peaks were detected in the TH cells (Fig. 3 D). 4742 of the peaks were detected as bound by Ebf1 in both Wt and Ebf1+/−Pax5+/− pro–B cells, whereas 3419 and 2373 unique peaks were identified in the Wt and TH, respectively. Motif enrichment analysis suggested that the transcription factor dose had a limited impact on the selection of either Ebf1 or associated binding sites (Table S1). Sites conforming to the Ebf1 consensus site were detected in 64% (P = 10−4878) of the Wt peaks and in 73% (P = 10−5013) of the peaks generated from TH cells. In addition to the Ebf1 consensus site, we detected enrichment in binding sites for Ets and Runx proteins as well as an E-box motif, independently of genotype (Table S1). To investigate how Ebf1 binding to regulatory regions was modulated by a reduction in transcription factor dose, we compared Ebf1 binding in Wt and TH cells on sites occupied by Ebf1 in Wt cells. This revealed that a total of 1,635 sites were differentially bound (P < 0.05), with approximately as many displaying reduced as increased binding in the TH cells. Increasing the stringency in the analysis, demanding a fourfold change in binding, identified 444 peaks. Investigating Ebf1 binding in a 5-kb window centered on peaks defined as Wt peaks with differentially binding revealed that a majority (341) of these 444 differentially bound peaks had a decrease in Ebf1 binding in TH cells as compared with Wt cells (Fig. 3 D). 104 of the genes displayed increased binding of Ebf1 in the TH as compared with the Wt cells. To link changes in gene expression to Ebf1 binding, we identified the genes closest to the 1,653 sites differentially bound by Ebf1 in Wt and TH cells. This resulted in the identification of 1,467 genes (Fig. 3 E), out of which 24 were among the 85 genes identified as differentially expressed direct targets for both Ebf1 and Pax5 in Fig. 3 B. Ebf1 binding was studied in a 5-kb window around differentially bound Ebf1 peaks between Wt and TH (P < 0.05) found within a 100-kb window of up- or down-regulated genes. The result suggests that genes down-regulated in the Ebf1+/−Pax5+/− pro–B cells displayed reduced Ebf1 binding on these sites, whereas up-regulated genes instead displayed an increased Ebf1 binding (Fig. 3 E), suggesting a correlation with changes in Ebf1 binding and gene expression. Hence, although the consensus-binding sites remain the same, combined heterozygote deletion of the Ebf1 and Pax5 genes impacts the binding patterns of Ebf1 to target genes in the pro–B cells.
The generation of T-lineage cells from Pax5+/−Ebf1+/− pro–B cells is a result of lineage conversion
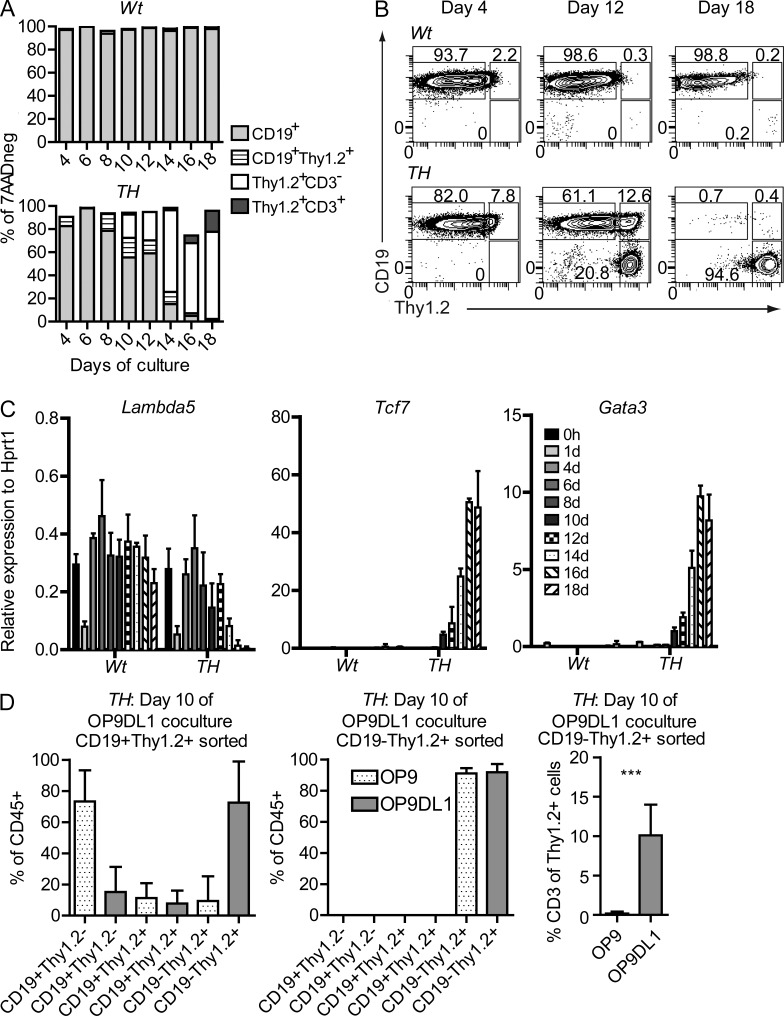
Although the generation of T-lineage cells from Pax5 and Ebf1 deficient B or pro–B cells has been reported to be a result of dedifferentiation (Cobaleda et al., 2007; Nechanitzky et al., 2013), the generation of macrophages from B-lineage cells has been suggested to be a result of direct lineage conversion (Xie et al., 2004). To understand the basic mechanism for the generation of T-lineage cells from Pax5+/−Ebf1+/− pro–B cells, we analyzed the donor derived progenitor compartments in transplanted Rag1−/− mice. However, we did not detect CD45.2+ common lymphoid progenitors (CLPs) or lymphoid-primed multipotent progenitors (LMPPs) in either BM or spleen. Hence, we were unable to detect signs of dedifferentiation of pro–B cells into classical hematopoietic progenitor compartments, arguing against the formation of T-lineage cells involving genuine dedifferentiation into conventional hematopoietic progenitors. To investigate the process in more detail, we sorted Wt and Pax5+/−Ebf1+/− pro–B cells, seeded them on OP9-DL1 cells, and followed the formation of T-lineage cells over time (Fig. 4, A and B). After 4 d of culture, the major part of the cells expressed CD19 but no, or low, levels of Thy1.2. After 8 d of incubation, we noted an increase in Thy1.2 expression generating CD19+Thy1.2+ cells, and after 10 d of incubation, a subpopulation of the cells lost expression of CD19. This subpopulation increased with time and, after 14 d of incubation, this was the dominant population in the cultures. At 16 d after incubation, we observed an increasing population of CD3+ cells. Initiation of normal T cell development has been shown to involve Gata3 and TCF7 (Weber et al., 2011; Banerjee et al., 2013); however, not until 10 d after the initiation of the experiments, at a time when we started to observe CD19− cells (Fig. 4, A and B), did we detect increased expression of Gata3 and Tcf7 in the Pax5+/−Ebf1+/− cells by Q-PCR (Fig. 4 C). This indicates that the expression of these transcription factors is not significantly preceding the development of CD19− cells in this process suggesting that the T cells are generated via a Thy1.2+CD19+ double positive state, indicative of lineage conversion rather than dedifferentiation. To establish links between the functional lineage conversion process and the defined populations, we sorted CD19+Thy1High, as well as CD19−Thy1High cells from a 10-d OP9-DL1 culture of Pax5+/−Ebf1+/− pro–B cells. The cells were then reseeded on either OP9 or OP9-DL1 cells and analyzed for the formation of B- or T-lineage cells after an additional 10 d in culture (Fig. 4 D). This revealed that CD19+Thy1High cells were able to generate CD19+ cells when reseeded on OP9 cells, whereas, if seeded on OP9-DL1 cells, the absolute majority of the CD19+Thy1High cells lost the expression of CD19. CD19−Thy1High cells were unable to generate CD19+ cells even when cultured in the absence of Delta ligand; however, only OP9-DL1 cells supported the formation of CD3-expressing cells (Fig. 4 D). These data support the idea that the conversion process involves the formation of a bivalent state capable of adopting either B-or T-lineage cell fate.
Figure 4.
Pax5+/−Ebf1+/− pro–B cells undergo lineage conversion into T cells via a bivalent state. (A) Bars describe the expression change of surface markers CD19, Thy1.2, and CD3 d 4–18 after that freshly sorted Lin−B220+CD19+CD43highIgM− Wt of Pax5+/−Ebf1+/−were placed on OP9-DL1 (day 0) stroma cells under conditions permissive for T cell development. 1,000,000 cells were sorted from 2 Pax5+/−Ebf1+/− (TH) and 2 Wt animals, which were further divided into 2 replicates per mouse before plating onto OP9-DL1. (B) Representative FACS plots of CD19 and Thy1.2 surface expression at days 4, 12, and 18 d after seeding onto OP9-DL1 as explained in A. (C) Changes of relative gene expression of Lambda5, Tcf7, and Gata3 0–18 d after Lin−B220+CD19+CD43highIgM− Wt or Pax5+/−Ebf1+/− (TH) were placed on OP9-DL1. Data are shown as median expression levels relative to Hprt1 from four samples analyzed in triplicate Q-PCR reactions (two of each genotype in duplicate). Error bars describe interquartile range. (D) Diagrams describing the percentage CD19+Thy1.2−, CD19+Thy1.2+, or CD19−Thy1.2+ of total CD45+ cells in OP9 (dotted bars) or OP9-DL1 (gray bars) co-cultures from sorted CD19+Thy1.2+ or CD19−Thy1.2+ cells generated by preincubation of primary sorted Pax5+/−Ebf1+/− (TH) pro–B cells on OP9-DL1 for 10 d. The experiment is based on independently sorted cells from two mice and analysis of a total of six co-cultures for each condition. ***, P < 0.001.
Combined dose reduction of Ebf1 and Pax5 alters the cellular response to Notch signaling
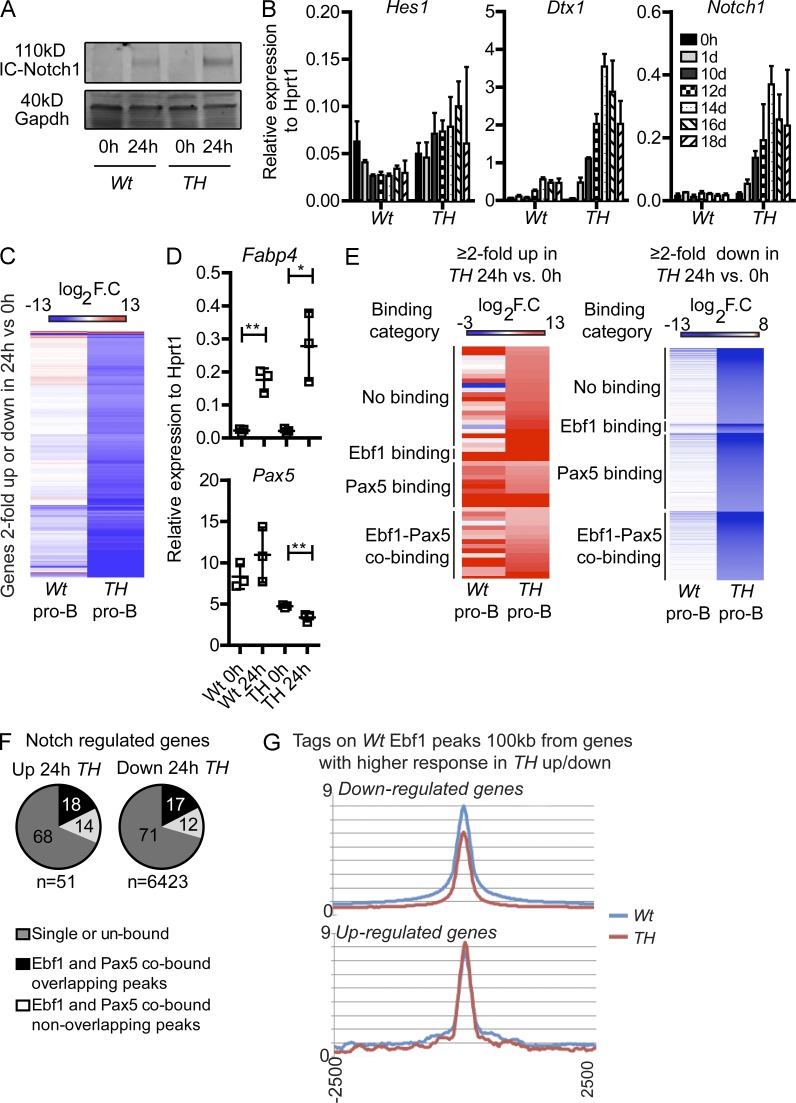
The change of lineage fate in Pax5−/− or Ebf1−/− mice has been attributed to increased expression of Notch1, presumably making the cells more sensitive to Notch ligand in the environment (Souabni et al., 2002; Nechanitzky et al., 2013). However, in contrast to what has been reported from these models of differentiation, we did not detect any significant increase in Notch1 in Pax5+/−Ebf1+/− pro–B cells. Hence, another mechanism of action should be operating in this model system. To investigate the direct cellular response to Notch ligand, we incubated Wt and Pax5+/−Ebf1+/− pro–B cells on OP9-DL1 cells for 24 h and analyzed the formation of intracellular Notch (ICN) in the two cell types (Fig. 5 A). This suggested that both the Wt and the Pax5+/−Ebf1+/− pro–B cells responded with the formation of ICN; however, Q-PCR analysis suggested that the Notch target genes Deltex1 and Notch1 were not induced in the Wt cells as observed in the Pax5+/−Ebf1+/− pro–B cells (Fig. 5 B). To investigate the response to Notch signals in more detail, we analyzed the induction of genes in response to Notch signal by RNA sequencing analysis in Wt and Pax5+/−Ebf1+/− pro–B cells 24 h after seeding onto OP9-DL1 cells (Fig. 5 C). This revealed that whereas both Wt and Pax5+/−Ebf1+/− pro–B cells responded to the Notch signal by up-regulation of known Notch target genes, such as Fosl1, Fabp4, Dll1, and Heyl, only the Pax5+/−Ebf1+/− cells up-regulated the expression of Dtx1 (Fig. 5 B). To verify the RNA seq data, we performed Q-PCR analysis examining the expression of Fabp4 (Fig. 5 D). This verified the RNA seq data, suggesting that both Wt and TH cells responded by induction of Fabp4 transcription (Fig. 5 D), whereas Deltex1 was selectively activated in the Pax5+/−Ebf1+/− cells (Fig. 5 B). This supports the idea that both Wt and Pax5+/−Ebf1+/− cells respond to Notch signals but that the response differs in regard to the activation of target genes. The induction of several genes, including Fabp4, Fosl1, and Dll1 were as strong or stronger than what we observed for Pax5+/−Ebf1+/− cells, arguing against substantial difference in signal strength as an explanation for the difference in Notch response. Our analysis, suggested that Wt cells in total up-regulated 180 genes more than twofold whereas Pax5+/−Ebf1+/− cells up-regulated 51 genes (Fig. 5 C). Extracting information about genes more than twofold down-regulated by Notch signaling in the Wt and Pax5+/−Ebf1+/− cells suggested that although 6,423 genes were down-regulated by Notch signaling in the TH cells, only 245 genes were down-regulated in the Wt cells. K.E.G.G pathway analysis suggested that 46 of the genes down-regulated in the Pax5+/−Ebf1+/− cells belonged to the category B cell receptor-signaling pathway (P = 7.0 × 10−7). Analysis of the data revealed that the down-regulated genes included CD19, CD79α, CD79β, as well as Ebf1 and Pax5 themselves, whereas none of these genes were affected in the Wt cells. This idea was verified by Q-PCR analysis revealing significant down-regulation of Pax5 after Notch activation selectively in the TH cells (Fig. 5 D). Hence, Pax5+/−Ebf1+/− cells respond to Notch signaling by a general down-regulation of the transcriptional program defining the early B-lineage cells in a manner not observed in Wt cells. To investigate if the modulated genes are direct targets of Ebf1 and/or Pax5, we analyzed this in relation to the Rag2−/− pro–B GEO ChIP seq datasets (Revilla-I-Domingo et al., 2012; Vilagos et al., 2012), which revealed that a majority (4,351 out of 6,474) of these differentially regulated genes were directly bound by either one or both of these transcription factors (Fig. 5, E and F). 18% of the genes up-regulated and 17% of those down-regulated displayed overlapping peaks (Fig. 5 F). Performing KEGG pathway analysis of up-regulated genes with combined Ebf1/Pax5 binding revealed that the only significantly ordered group of genes was in the Notch signaling pathway. Performing the same type of analysis on down-regulated genes revealed the B cell receptor signaling pathway as the most significantly affected pathway. To investigate if the differential response to Notch signaling was reflected in differential Ebf1 binding before Notch activation, we investigated the binding of Ebf1 (defined by the total filtered Wt list) in a 100-kb window, to genes with a stronger response (twofold) to Notch stimuli in TH that were either up- or down-regulated in the TH cells as compared with nonstimulated cells. This revealed that whereas Ebf1 binding to genes up-regulated by Notch signaling was not decreased, the binding to genes down-regulated displayed reduced Ebf1 binding, even before Notch activation (Fig. 5 G).
Figure 5.
The cellular response to Notch signaling is altered in pro–B cells carrying combined dose reduction of Ebf1 and Pax5. (A) The levels of intracellular Notch1 determined with Western blot 0 and 24 h after exposure to OP9-DL1 cells. The experiment was performed in triplicate and GAPDH was used as a loading control. (B) Q-PCR detecting changes of relative gene expression of Deltex1, Notch1, and Hes1 0–18 d after CD43highIgM− Pax5+/−Ebf1+/− (TH) or CD43highIgM− Wt were placed on OP9-DL1. Data are shown as median expression levels relative to Hprt1 from four samples analyzed in triplicate Q-PCR reactions (two of each genotype in duplicate). Error bars describe interquartile range. (C) Heatmap of log2 fold change gene expression values in Wt (n = 4) and TH (n = 4) pro–B cells displaying 6,474 genes with expression over 0.5 RPKM linear scale and a 2-fold change in expression in TH cells after 24 h of exposure to OP9-DL1. Hierarchical clustering was performed with the Cluster 3.0 software (Euclidean distance with average linkage). (D) Q-PCR detecting changes of relative gene expression of Fabp4 and Pax5 24 h after that CD19+CD43highIgM− WT or Pax5+/−Ebf1+/− (TH) cells expanded in OP9 cultures were reseeded on OP9-DL1 cells. Data are shown as median expression levels relative to Hprt1 from three independent samples analyzed in triplicate Q-PCR reactions. Error bars describe interquartile range. (E) Heatmaps of Wt and TH samples on genes with differential expression in TH (24 vs. 0 h on OP9-DL1) were grouped according to Ebf1 and Pax5 (Revilla-I-Domingo et al., 2012; Vilagos et al., 2012) binding categories and sorted based on increasing fold change in TH. (left) The genes ≥2-fold in TH 24 versus 0 h; (right) genes ≥2-fold in TH 24 versus 0 h. (F) Overlapping and nonoverlapping Ebf1+/− and Pax5+/− peaks (Revilla-I-Domingo et al., 2012; Vilagos et al., 2012) in the 6,474 TH 24 versus 0 h on OP9-DL1 differentially regulated genes. Numbers in the pie graphs show percent of total peaks. (G) Tags from Ebf1 ChIP-seq in Wt and TH samples were plotted on peak center ± 2,500 bp. Peaks used were derived from the total Wt sample peak list (Chr, X, Y, M, and random are filtered out) and found within 100 kb of genes that had a higher (≥2-fold) response to OP9-DL1 stimuli in TH compared with Wt. *, P < 0.05; **, P < 0.01.
DISCUSSION
The data presented in this report support the idea that Ebf1 and Pax5 act in a coordinated, dose-dependent manner to preserve B-lineage cell fate. This is well in line with the findings that Ebf1 and Pax5 act in an autoregulatory loop and that complete loss of function of either of these proteins results in increased plasticity of B cell progenitors. Just as for normal T cell development (Radtke et al., 1999; Wilson et al., 2001), Notch signaling had an essential role in the lineage conversion process in vitro because the phenotypic changes were only observed on OP9-DL1 cells and prevented by the inclusion of DAPT (Fig. 2 B). It is reasonable to presume that activation of Notch signaling would be a requirement in vivo where the cells could be exposed to Notch ligands in the thymus (Felli et al., 1999; Schmitt et al., 2004) but also in other locations such as the spleen (Caton et al., 2007). Exposure to Notch ligands in peripheral organs might also explain the presence of CD4CD8 double positive cells in the spleens of Rag mice transplanted with Ebf1+/−Pax5+/− pro–B cells (Fig. 1 E). Even though we do detect some nonconventional populations of both B and T cells in the periphery of the transplanted mice, the majority of the TCRβ-expressing cells are either CD4 or CD8 single-positive, resembling major T cell populations in normal mice. In contrast, the majority of the TCRβ+ cells generated in vitro (Fig. 2 D) did not express either CD4 or CD8. Even though CD3+TCR+CD4−CD8− immunoregulatory T cells has been identified in the periphery of normal mice and humans (Hillhouse and Lesage, 2013), the generation of both CD4 and CD8 single-positive T cells in vivo argues against this indicating that the Ebf1+/−Pax5+/− pro–B cells would be primed toward development into any specific T cell subtype.
It has been shown that the generation of T-lineage cells from Pax5−/− (Cobaleda et al., 2007) or Ebf1−/− (Nechanitzky et al., 2013) B cell progenitors involves a component of dedifferentiation. This is somewhat in contrast to our data suggesting that the generation of T-lineage cells from Ebf1+/−Pax5+/− CD19+ B cell progenitors instead involves the formation of a bivalent state displaying features of both B and T-lineage cells and with ability to adopt either of the two cell fates. This does in many regards resemble the lineage conversion process reported to generate macrophages from B cells in response to expression of Cebpα (Xie et al., 2004; Di Tullio et al., 2011). However, it should be noted that the dedifferentiation results reported in Pax5- and Ebf1-deficient mice involves the generation of rather atypical progenitor cells in some aspects resembling early pro–B cells (Cobaleda et al., 2007; Nechanitzky et al., 2013) and that these cells may in some regards correspond to the CD19+ pro–B cells we detect several weeks after transplantation of Pax5+/−Ebf1+/− cells (Fig. 1 D). We could, however, not detect the development of myeloid cells in any of our mice, in contrast to what has been reported for Pax5−/− B cells (Cobaleda et al., 2007). A potential discrepancy could be a result of that as opposed to what has been reported for Pax5−/− or Ebf1−/− cells (Cobaleda et al., 2007; Nechanitzky et al., 2013) the progenitor compartments in Pax5+/−Ebf1+/− mice express a full B-lineage program and possess normal survival capability, eliminating the need of the transgenic expression of Bcl2 used in previous studies (Cobaleda et al., 2007; Nechanitzky et al., 2013). Even though differences in survival and potential competition between different populations in the cell cultures make it difficult to exactly estimate the frequency of Pax5+/−Ebf1+/− CD19+ progenitor cells that can undergo the conversion into T-linage cells, our data argue against the idea that this would be the result of selection of a small population of plastic cells. The in vivo experiments generated T cells with polyclonal VDJ rearrangements and 80% of the single cells able to generate colonies generated T-lineage cells in vitro. The calculated T cell cloning frequency of 1/37 was comparable to that of CD19+ cells in the same experiment, hence, even though we do not want to conclude that all cells possess the ability to change lineage, a significant portion of the CD19+ progenitors clearly display a plasticity toward T-lineage development.
Whereas the complete loss of function of either Ebf1 or Pax5 has been shown to result in increased expression of genes associated with other cell fates, including Notch1 (Souabni et al., 2002; Nechanitzky et al., 2013), the combined heterozygote loss of Ebf1 and Pax5 does not appear to cause any major disruptions in the genetic program of the pro–B cells (Fig. 3 A). Even though we did detect significant differential expression of a set of genes, several of which appear as direct targets for Ebf1 and Pax5, we were unable to find significant changes in genes that, to our knowledge, directly repress functional Notch signaling. Even though the complexity of this signaling pathway calls for caution in the interpretation of negative data, we believe that our short term gene expression analysis provide support for the idea that both Wt and Ebf1+/−Pax5+/− cells respond to Delta1 (Dll1) stimulation. Wt and Ebf1+/−Pax5+/− cells responded by increased expression of a set of genes suggested to be Notch targets, including Fabp4 (Harjes et al., 2014) and Heyl (Maier and Gessler, 2000), and also of the Notch ligand Dll1. In contrast, only the Ebf1+/−Pax5+/− cells responded by induction of the Notch target Deltex1 (Deftos et al., 2000). This was rather unexpected considering that Deltex1 is a powerful inhibitor of Notch signaling, driving progenitor cells into a B cell fate (Izon et al., 2002) and capable of preventing the induction of T cell fate in Pax5−/− cells (Zandi et al., 2012). However, because this gene has regions of overlapping binding of Ebf1 and Pax5, it is likely directly under the control of these transcription factors in normal cells. Binding of Ebf1 and or Pax5 could be detected in 27 of the 50 genes up-regulated by the Notch signal in the Ebf1+/−Pax5+/− cells; however, because 14 of these 27 also responded in the Wt cells, binding of Ebf1 or Pax5 is not fully correlated to selective activation in the mutant cells. Even though there were differences in the spectra of activated genes, the most dramatic discrepancy between Wt and Ebf1+/−Pax5+/− cells was the down-regulation of a large number of genes, including a large part of the B cell–specific program, in the TH cells in response to the Notch signal. This included down-regulation of Pax5 and Ebf1 as well as of a large set of target genes, indicating that the Notch signal directly targets the Ebf1–Pax5 network. Notch1 has been shown to target Ebf1 posttranscriptionally (Smith et al., 2005), possibly adding to any direct effects on Ebf1 or Pax5 transcription. This, together with the finding that Gata3 and Tcf7 are significantly up-regulated first after 10–12 d of OP9-DL1 stimulation (Fig. 4 C), indicates that the initial response of the CD19+CD43+IgM− TH cells to the Notch signal is to repress the B cell program, whereas the T cell identity genes in general requires more time and are up-regulated at a later phase of the conversion process.
In conclusion, we believe that our data support the idea that Ebf1 and Pax5 collaborate to preserve lineage identity and homeostasis in early B cell development. Even though this is well in line with previous work in this area, the fact that we are able to look at lineage conversion in cells with a well preserved transcriptional program allows us to obtain an increased understanding of a lineage specification process. This insight strongly suggests that Ebf1 and Pax5 counteract an activated Notch signal by repressing the functional response initiated by the ICN protein, thereby preserving the lineage identity of B cell progenitors.
MATERIALS AND METHODS
Animal models.
Pax5+/− (Urbánek et al., 1994) or Ebf1+/− (Lin and Grosschedl, 1995) were all on C57BL/6 (CD45.2) background (Backcrossed for >10 generations) whereas Rag1−/− animals were on C57BL/6 (CD45.1). Animal procedures were performed with consent from the local ethics committee at Linköping University (Linköping, Sweden).
Transplantation procedures.
Adoptive transfers were performed by tail vein injection. Sublethally irradiated (4.5 Gy) Rag1−/− CD45.1 animals were transplanted with ∼2 million Lin−B220+CD19+IgM− Pax5+/−Ebf1+/− or Wt cells as controls.
FACS staining and sorting of hematopoietic cells.
For analysis and cell sorting of B lineage cells, CD16/CD32 (FC)-blocked (93; eBioscience) cells were stained with antibodies against lineage markers CD11b/Mac1 (M1/70), Gr1 (RB6-8C5), TER119 (Ter119), CD3 (17A2; BD), CD11c (N418), and NK1.1 (PK136), followed by additional staining with CD19 (ID3), CD45R/B220 (RA3-6B2), CD43 (S7), IgM (RMM-1), and IgD (11–26; eBioscience). Analysis and cell sorting was performed on a FACSAria (BD) using propidium iodide (PI; Invitrogen) as a viability marker. Samples in Fig. 1 A were analyzed with a FACSCantoII (BD) and dead cells were excluded with 7-aminoactinomycin D (7-AAD; Sigma-Aldrich). Lymph node cells were FC-blocked and stained with CD11b/Mac1 (M1/70), Gr1 (RB6-8C5), CD3 (17A2), CD19 (ID3), CD45R/B220 (RA3-6B2), CD43 (S7), IgM (RMM-1), and propidium iodide and analyzed and sorted on FACSAria. Samples from transplanted mice were analyzed on a FACSAria with antibodies for CD45.1 (A20), CD45.2 (104), NK1.1 (PK136), CD3 (17A2), CD19 (ID3), CD45R/B220 (RA3-6B2), CD43 (S7), and IgM (RMM-1). All antibodies were purchased from BioLegend unless stated otherwise. Gates were set according to fluorescence minus one (FMO) controls. The cellular composition in the transplanted Rag1−/− mice was analyzed using frozen single-cell suspensions from thymus, BM, and spleen.
RNA-sequencing and data analysis.
Total RNA was isolated from four Wt CD19+CD43highIgM− and four Pax5+/−Ebf1+/− pro–B samples using RNAeasy MicroKit (QIAGEN) according to the manufacturer’s recommendations. RNA was sent to UCLA Clinical Microarray Core for library preparation and subjected to 50 cycles of HiSeq 2000 SBS sequencing generating 20–30 million reads/sample. RNA sequencing of OP9-DL1 stimulated cells was performed using 50 cycles of Next Seq 500 sequencing generating 20–30 million reads from four independent experiments. Data analysis was performed with Arraystar (DNASTAR). For analysis of RNA-seq experiments the reads were aligned to mouse reference genome (mm9, NCBI 37) and RPKM normalized. Statistical analysis in Arraystar was performed with a Student’s t test with correction for Multiple Testing (Benjamini Hochberg). P < 0.05 was considered to be statistically significant.
In vitro evaluation of lineage potentials.
For evaluation of B and T cell potential, cells were deposited (using FACSAria) directly into 96-well plates containing preplated (2,000 cells/well) stroma cells. T cell cultures (on OP9-DL1 stroma layers) were supplemented with 10 ng/ml KIT ligand, 10 ng/ml Fms-like tyrosine kinase 3 ligand (FLT3L), and 10 ng/ml IL-7. All cytokines were acquired from PeproTech. Cultures were substituted with fresh cytokines after 7 d. OptiMEM supplemented with 10% heat-inactivated fetal calf serum, 25 mM Hepes, 50 µg/ml Gentamicin, and 50 µM β-mercaptoethanol was used for maintaining the OP9/OP9-DL1 stroma cell-lines (Schmitt and Zuñiga-Pflucker, 2002), as well as for the cocultures. Cocultures were evaluated at day 14 by FACS staining with CD19 (ID3), CD90.2/Thy1.2 (53–2.1), CD3 (17A2) and 7-AAD using a FACSCantoII (BD).
Quantitative RT-PCR.
Q-PCR analysis of sorted cells was performed as previously described (Mansson et al., 2008). Assays-on-Demand probes (Applied Biosystems) used were: Hprt; Mm00446968_m1, Pax5; Mm00515420_m1, Cd79α/Mb-1;Mm00432423_m1, CD79β/B29;Mm00434143_m1, CD19 Mm00515420_m1, CD27 Mm01185210_m1, PreTa Mm00478361_m1, Lck Mm00802897_m1, CD3e Mm00599683_m1, TCF7 Mm493445_m1, Gata3 Mm00484683_m1, Notch1 Mm00435245_m1, Hes1 Mm00468601_m1, Deltex1 Mm00492294_m1. Assay on demands, Lambda5 (Igll1) (Mansson et al., 2010) and Ebf1, Forward primer, 5′-TCATGTTTGGGATCCAGGAAAG-3′; Reverse primer, 5′-GTTGGATTTCCGCAGGTTAGA-3′.
VDJ recombination analysis.
Live cells were sorted and DNA was extracted and subjected to PCR based VDJ analysis as in (Schlissel et al., 1991). The Schlissel et al. (1991)–generated PCR products were transferred to a nylon membrane and hybridized to a P32-labeled oligonucleotide acting as an internal J3 probe.
Chromatin immunoprecipitation.
Cultivated CD43+IgM− cells were spun down and fixed in 1 mg/ml DSG (Thermo Fisher Scientific) in PBS for 30 min at RT, followed by addition of up to 1% formaldehyde and an additional 10 min fixation at RT. The cross-link reaction was quenched with 0.125 M glycine, washed in PBS, and snap frozen or used immediately for ChIP.
Nuclei for ChIP were isolated by 10 min incubation in Nuclei Isolation buffer (50 mM Tris, pH 8.0, 60 mM KCl, and 0.5% NP-40) + protease inhibitor cocktail (PIC; Roche) on ice. Pelleted nuclei were dissolved in Lysis buffer (0.5% SDS, 10 mM EDTA, 0.5 mM EGTA, 50 mM Tris-HCl, pH 8.0) + PIC and sonicated on a Bioruptor (Diagenode) 18 cycles, max power for 30 s followed by 30 s of rest. Sonication was followed by pelleting of debris, and then the supernatant was transferred to new tube and chromatin was diluted 5X in Dilution Buffer (1% Triton, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl, pH 8.0 + PIC). 5 µg anti-Ebf1 (ABE1294), rabbit, poly, 1 mg/ml (lot Q2399134) was hybridized to 20 µl Dynabeads M-280 sheep anti–rabbit IgG (Life Technologies) in 100 µl PBS + PIC for 4 h, washed twice with PBS, resuspended in 20 µl PBS + PIC and added to the diluted chromatin. ChIP was performed over night at 4°C. and subsequently washed (1 time with 500 µl Low Salt Immune Complex Wash Buffer, 1 time with 200 µl High Salt Immune Complex Wash Buffer, 1 time with 200 µl LiCl Immune Complex Wash Buffer, and 2 times with 200 µl TE buffer) and eluted for 4 h at 65°C (20 mM Tris-HCl, pH 7.5, 5 mM EDTA, 50 mM NaCl, 1% SDS, 100 µg RNase A, and 50 µg proteinase K), treated, and cleaned up using Zymo ChIP DNA Clean & Concentrator before ChIP-seq libary preparation. 50-bp single-read sequencing was performed on an Illumina HiSeq 2000 at 23–39 million reads per sample.
Analysis of ChIP seq data.
Reads from high-throughput sequencing were aligned to a mouse reference genome (mm9, NCBI 37) using Bowtie (Langmead et al., 2009) with best match parameters (bowtie -m 1–sam–best–strata -v 2). Further analyses were made using the HOMER package (Heinz et al., 2010). Replicate Ebf1 ChIP-seq runs on each genotype (Wt, TH, Ebf1+/−, and Pax5+/−) and corresponding inputs were pooled into one dataset and analyzed as one combined sample per genotype.
Peaks were identified using findPeaks.pl with the –style factor parameter and normalized to sequencing inputs. Motif enrichment analysis was performed with the findMotifsGenome.pl command of the HOMER package using a 200-bp window.
Because cells in the different cell populations could be from male or female mice, for a more detailed comparison of Wt and TH samples, new peak-lists were constructed by filtering out peaks on Chr. X, Chr. Y, Chr. M, and Random Chr from original peak-files. Overlapping peaks between Wt and TH samples were identified using mergePeaks.pl (default parameters). Sequencing data are deposited at the Gene Expression Omnibus under accession no. GSE69227.
To find differentially bound peaks between Wt and TH, the modified Wt peakfile was annotated with the tag counts from all experiments using annotatePeaks.pl with –noadj. Next, the differentially bound peaks (P < 0.05 and P < 0.05 + fourfold F.C) were called with the getDiffExpression.pl with the –peak option. Tag density plots and heatmaps were created with annotatePeaks.pl (–hist or –hist & -ghist, respectively), normalizing data to 10 million mapped reads per experiment, and visualized using Excel and Java Treeview (Version 1.1.6r4; Saldanha, 2004).
To analyze peaks around a certain gene, a customized Perl script was used to identify peaks flanking each defined gene in a 100-kb window.
Venn diagrams were created using Venn Diagram (http://www.bioinformatics.lu/venn.php) with lists for closest gene from the HOMER-annotated peak file in comparison with gene lists from RNA-seq experiments.
In addition, Rag2−/− pro–B Ebf1 and Pax5 ChIP-seq data (Revilla-I-Domingo et al., 2012; Vilagos et al., 2012) were obtained from the Gene Expression Omnibus (under accession nos. GSM932921, GSM932922, GSM932923, GSM932924, GSM876622, GSM876623, GSM876635, GSM876636, GSM876637, GSM876638, GSM876639, and GSM876640) and pooled into one dataset per ChIP and further analyzed using the HOMER package (Heinz et al., 2010) as described above. Annotated peak-lists from these datasets were used to overlay closest genes to peaks with lists from RNA-sequencing datasets. Overlapping Ebf1 and Pax5 peaks were identified with mergePeaks.pl (default parameters).
Western blot analysis of intracellular Notch1.
The intracellular Notch 1 level was determined by Western blot analysis. Pro–B cells were cultured on a monolayer for OP9DL1 stroma cells for 24 h and the stimulation was terminated by placing the cells on ice. The pro–B cells were removed carefully without disturbing the OP9DL1 stroma cells, centrifuged, and lysed in RIPA buffer supplemented with Protease inhibitor (Roche; 11697498001) and PMSF (1 µM; Sigma-Aldrich; P7626). Protein concentration was determined by Bradford reagent (Sigma-Aldrich; B6916), 50 µg of protein from each sample was boiled for 5 min in Laemmli buffer, and the proteins were separated on SDS-PAGE (456–8081; Bio-Rad Laboratories). The proteins were electrophoretically transferred to PVDF membranes (Immobulin FL; Millipore). Membranes were blocked with 5% BSA and probed with antibodies against intracellular Notch1 (Val1744; Cell Signaling Technology; 2421; 1:250 dilution), and equal loading was determined by probing blots with antibodies against GAPDH (ab9483; Abcam). Proteins were detected by IR dyes (Odyssey) and the membrane were scanned using Odyssey CLx Infrared Imaging System.
Transduction of Pax5+/−Ebf1+/− pro–B cells by Ebf1 retrovirus.
Retrovirus transduction was performed as in (Zandi et al., 2012) using either a MIGR1-GFP control virus or a virus where the open reading frame of Ebf1 was cloned upsteam of the IRES and GFP reporter gene. The transduced cells were sorted for GFP expression and incubated on OP9DL1 stroma cells before FACS analysis.
Online supplemental material.
Table S1 shows an analysis of Ebf1 ChIP-seq data in Wt, TH (Ebf1+/−Pax5+/−), Ebf1+/−, and Pax5+/− pro–B cells. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20132100/DC1.
Supplementary Material
Acknowledgments
We wish to thank our colleges for help with transgenic mice and cell lines. We also wish to thank Liselotte Lenner and Linda Bergström for advice and assistance.
This work was supported by grants from the Swedish Cancer Society, the Swedish Research Council, including a Center grant to Hematolinne, Knut and Alice Wallenbers Stiftelse, The Swedish Childhood cancer foundation, and Linköping University.
The authors declare no competing financial interests.
Author contributions: J. Ungerbäck, J. Åhsberg, R. Somasundaram, T. Strid, and M. Sigvardsson conducted, designed, and interpreted the analysis of the mouse model. All authors participated in the writing of the manuscript.
Footnotes
Abbreviations used:
- B-ALL
- B-lineage acute lymphoid leukemia
- CLP
- common lymphoid progenitor
- Ebf1
- early B cell factor 1
- LMPP
- lymphoid primed multipotent progenitor
- OP9-DL1
- OP9 Delta ligand 1
- Pax5
- paired box protein 5TH, transheterozygous
References
- Åhsberg J., Ungerbäck J., Strid T., Welinder E., Stjernberg J., Larsson M., Qian H., and Sigvardsson M.. 2013. Early B-cell factor 1 regulates the expansion of B-cell progenitors in a dose-dependent manner. J. Biol. Chem. 288:33449–33461. 10.1074/jbc.M113.506261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Northrup D., Boukarabila H., Jacobsen S.E., and Allman D.. 2013. Transcriptional repression of Gata3 is essential for early B cell commitment. Immunity. 38:930–942. 10.1016/j.immuni.2013.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton M.L., Smith-Raska M.R., and Reizis B.. 2007. Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J. Exp. Med. 204:1653–1664. 10.1084/jem.20062648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobaleda C., Jochum W., and Busslinger M.. 2007. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature. 449:473–477. 10.1038/nature06159 [DOI] [PubMed] [Google Scholar]
- Decker T., Pasca di Magliano M., McManus S., Sun Q., Bonifer C., Tagoh H., and Busslinger M.. 2009. Stepwise activation of enhancer and promoter regions of the B cell commitment gene Pax5 in early lymphopoiesis. Immunity. 30:508–520. 10.1016/j.immuni.2009.01.012 [DOI] [PubMed] [Google Scholar]
- Deftos M.L., Huang E., Ojala E.W., Forbush K.A., and Bevan M.J.. 2000. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity. 13:73–84. 10.1016/S1074-7613(00)00009-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Tullio A., Vu Manh T.P., Schubert A., Castellano G., Månsson R., and Graf T.. 2011. CCAAT/enhancer binding protein alpha (C/EBP(alpha))-induced transdifferentiation of pre-B cells into macrophages involves no overt retrodifferentiation. Proc. Natl. Acad. Sci. USA. 108:17016–17021. 10.1073/pnas.1112169108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felli M.P., Maroder M., Mitsiadis T.A., Campese A.F., Bellavia D., Vacca A., Mann R.S., Frati L., Lendahl U., Gulino A., and Screpanti I.. 1999. Expression pattern of notch1, 2 and 3 and Jagged1 and 2 in lymphoid and stromal thymus components: distinct ligand-receptor interactions in intrathymic T cell development. Int. Immunol. 11:1017–1025. 10.1093/intimm/11.7.1017 [DOI] [PubMed] [Google Scholar]
- Harjes U., Bridges E., McIntyre A., Fielding B.A., and Harris A.L.. 2014. Fatty acid-binding protein 4, a point of convergence for angiogenic and metabolic signaling pathways in endothelial cells. J. Biol. Chem. 289:23168–23176. 10.1074/jbc.M114.576512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heavey B., Charalambous C., Cobaleda C., and Busslinger M.. 2003. Myeloid lineage switch of Pax5 mutant but not wild-type B cell progenitors by C/EBPalpha and GATA factors. EMBO J. 22:3887–3897. 10.1093/emboj/cdg380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., and Glass C.K.. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38:576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heltemes-Harris L.M., Willette M.J., Ramsey L.B., Qiu Y.H., Neeley E.S., Zhang N., Thomas D.A., Koeuth T., Baechler E.C., Kornblau S.M., and Farrar M.A.. 2011. Ebf1 or Pax5 haploinsufficiency synergizes with STAT5 activation to initiate acute lymphoblastic leukemia. J. Exp. Med. 208:1135–1149. 10.1084/jem.20101947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillhouse E.E., and Lesage S.. 2013. A comprehensive review of the phenotype and function of antigen-specific immunoregulatory double negative T cells. J. Autoimmun. 40:58–65. 10.1016/j.jaut.2012.07.010 [DOI] [PubMed] [Google Scholar]
- Höflinger S., Kesavan K., Fuxa M., Hutter C., Heavey B., Radtke F., and Busslinger M.. 2004. Analysis of Notch1 function by in vitro T cell differentiation of Pax5 mutant lymphoid progenitors. J. Immunol. 173:3935–3944. 10.4049/jimmunol.173.6.3935 [DOI] [PubMed] [Google Scholar]
- Izon D.J., Aster J.C., He Y., Weng A., Karnell F.G., Patriub V., Xu L., Bakkour S., Rodriguez C., Allman D., and Pear W.S.. 2002. Deltex1 redirects lymphoid progenitors to the B cell lineage by antagonizing Notch1. Immunity. 16:231–243. 10.1016/S1074-7613(02)00271-6 [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., and Salzberg S.L.. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., and Grosschedl R.. 1995. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 376:263–267. 10.1038/376263a0 [DOI] [PubMed] [Google Scholar]
- Lin Y.C., Jhunjhunwala S., Benner C., Heinz S., Welinder E., Mansson R., Sigvardsson M., Hagman J., Espinoza C.A., Dutkowski J., et al. 2010. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat. Immunol. 11:635–643. 10.1038/ni.1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukin K., Fields S., Lopez D., Cherrier M., Ternyak K., Ramírez J., Feeney A.J., and Hagman J.. 2010. Compound haploinsufficiencies of Ebf1 and Runx1 genes impede B cell lineage progression. Proc. Natl. Acad. Sci. USA. 107:7869–7874. 10.1073/pnas.1003525107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukin K., Fields S., Guerrettaz L., Straign D., Rodriguez V., Zandi S., Månsson R., Cambier J.C., Sigvardsson M., and Hagman J.. 2011. A dose-dependent role for EBF1 in repressing non-B-cell-specific genes. Eur. J. Immunol. 41:1787–1793. 10.1002/eji.201041137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier M.M., and Gessler M.. 2000. Comparative analysis of the human and mouse Hey1 promoter: Hey genes are new Notch target genes. Biochem. Biophys. Res. Commun. 275:652–660. 10.1006/bbrc.2000.3354 [DOI] [PubMed] [Google Scholar]
- Mansson R., Zandi S., Anderson K., Martensson I.L., Jacobsen S.E., Bryder D., and Sigvardsson M.. 2008. B-lineage commitment prior to surface expression of B220 and CD19 on hematopoietic progenitor cells. Blood. 112:1048–1055. 10.1182/blood-2007-11-125385 [DOI] [PubMed] [Google Scholar]
- Mansson R., Zandi S., Welinder E., Tsapogas P., Sakaguchi N., Bryder D., and Sigvardsson M.. 2010. Single-cell analysis of the common lymphoid progenitor compartment reveals functional and molecular heterogeneity. Blood. 115:2601–2609. 10.1182/blood-2009-08-236398 [DOI] [PubMed] [Google Scholar]
- Mullighan C.G., Goorha S., Radtke I., Miller C.B., Coustan-Smith E., Dalton J.D., Girtman K., Mathew S., Ma J., Pounds S.B., et al. 2007. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 446:758–764. 10.1038/nature05690 [DOI] [PubMed] [Google Scholar]
- Nechanitzky R., Akbas D., Scherer S., Györy I., Hoyler T., Ramamoorthy S., Diefenbach A., and Grosschedl R.. 2013. Transcription factor EBF1 is essential for the maintenance of B cell identity and prevention of alternative fates in committed cells. Nat. Immunol. 14:867–875. 10.1038/ni.2641 [DOI] [PubMed] [Google Scholar]
- Nutt S.L., Urbánek P., Rolink A., and Busslinger M.. 1997. Essential functions of Pax5 (BSAP) in pro-B cell development: difference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes Dev. 11:476–491. 10.1101/gad.11.4.476 [DOI] [PubMed] [Google Scholar]
- Nutt S.L., Morrison A.M., Dörfler P., Rolink A., and Busslinger M.. 1998. Identification of BSAP (Pax-5) target genes in early B-cell development by loss- and gain-of-function experiments. EMBO J. 17:2319–2333. 10.1093/emboj/17.8.2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt S.L., Heavey B., Rolink A.G., and Busslinger M.. 1999. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 401:556–562. 10.1038/44076 [DOI] [PubMed] [Google Scholar]
- O’Riordan M., and Grosschedl R.. 1999. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity. 11:21–31. 10.1016/S1074-7613(00)80078-3 [DOI] [PubMed] [Google Scholar]
- Pongubala J.M., Northrup D.L., Lancki D.W., Medina K.L., Treiber T., Bertolino E., Thomas M., Grosschedl R., Allman D., and Singh H.. 2008. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat. Immunol. 9:203–215. 10.1038/ni1555 [DOI] [PubMed] [Google Scholar]
- Radtke F., Wilson A., Stark G., Bauer M., van Meerwijk J., MacDonald H.R., and Aguet M.. 1999. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 10:547–558. 10.1016/S1074-7613(00)80054-0 [DOI] [PubMed] [Google Scholar]
- Revilla-I-Domingo R., Bilic I., Vilagos B., Tagoh H., Ebert A., Tamir I.M., Smeenk L., Trupke J., Sommer A., Jaritz M., and Busslinger M.. 2012. The B-cell identity factor Pax5 regulates distinct transcriptional programmes in early and late B lymphopoiesis. EMBO J. 31:3130–3146. 10.1038/emboj.2012.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler S., Györy I., Imhof S., Spivakov M., Williams R.R., Busslinger M., Fisher A.G., and Grosschedl R.. 2007. Distinct promoters mediate the regulation of Ebf1 gene expression by interleukin-7 and Pax5. Mol. Cell. Biol. 27:579–594. 10.1128/MCB.01192-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolink A.G., Nutt S.L., Melchers F., and Busslinger M.. 1999. Long-term in vivo reconstitution of T-cell development by Pax5-deficient B-cell progenitors. Nature. 401:603–606. 10.1038/44164 [DOI] [PubMed] [Google Scholar]
- Saldanha A.J. 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics. 20:3246–3248. 10.1093/bioinformatics/bth349 [DOI] [PubMed] [Google Scholar]
- Schlissel M.S., Corcoran L.M., and Baltimore D.. 1991. Virus-transformed pre-B cells show ordered activation but not inactivation of immunoglobulin gene rearrangement and transcription. J. Exp. Med. 173:711–720. 10.1084/jem.173.3.711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt T.M., and Zúñiga-Pflücker J.C.. 2002. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 17:749–756. 10.1016/S1074-7613(02)00474-0 [DOI] [PubMed] [Google Scholar]
- Schmitt T.M., Ciofani M., Petrie H.T., and Zúñiga-Pflücker J.C.. 2004. Maintenance of T cell specification and differentiation requires recurrent notch receptor-ligand interactions. J. Exp. Med. 200:469–479. 10.1084/jem.20040394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.M., Akerblad P., Kadesch T., Axelson H., and Sigvardsson M.. 2005. Inhibition of EBF function by active Notch signaling reveals a novel regulatory pathway in early B-cell development. Blood. 106:1995–2001. 10.1182/blood-2004-12-4744 [DOI] [PubMed] [Google Scholar]
- Souabni A., Cobaleda C., Schebesta M., and Busslinger M.. 2002. Pax5 promotes B lymphopoiesis and blocks T cell development by repressing Notch1. Immunity. 17:781–793. 10.1016/S1074-7613(02)00472-7 [DOI] [PubMed] [Google Scholar]
- Treiber T., Mandel E.M., Pott S., Györy I., Firner S., Liu E.T., and Grosschedl R.. 2010. Early B cell factor 1 regulates B cell gene networks by activation, repression, and transcription- independent poising of chromatin. Immunity. 32:714–725. 10.1016/j.immuni.2010.04.013 [DOI] [PubMed] [Google Scholar]
- Urbánek P., Wang Z.-Q., Fetka I., Wagner E.F., and Busslinger M.. 1994. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell. 79:901–912. 10.1016/0092-8674(94)90079-5 [DOI] [PubMed] [Google Scholar]
- Vilagos B., Hoffmann M., Souabni A., Sun Q., Werner B., Medvedovic J., Bilic I., Minnich M., Axelsson E., Jaritz M., and Busslinger M.. 2012. Essential role of EBF1 in the generation and function of distinct mature B cell types. J. Exp. Med. 209:775–792. 10.1084/jem.20112422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber B.N., Chi A.W., Chavez A., Yashiro-Ohtani Y., Yang Q., Shestova O., and Bhandoola A.. 2011. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 476:63–68. 10.1038/nature10279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A., MacDonald H.R., and Radtke F.. 2001. Notch 1-deficient common lymphoid precursors adopt a B cell fate in the thymus. J. Exp. Med. 194:1003–1012. 10.1084/jem.194.7.1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H., Ye M., Feng R., and Graf T.. 2004. Stepwise reprogramming of B cells into macrophages. Cell. 117:663–676. 10.1016/S0092-8674(04)00419-2 [DOI] [PubMed] [Google Scholar]
- Zandi S., Mansson R., Tsapogas P., Zetterblad J., Bryder D., and Sigvardsson M.. 2008. EBF1 is essential for B-lineage priming and establishment of a transcription factor network in common lymphoid progenitors. J. Immunol. 181:3364–3372. 10.4049/jimmunol.181.5.3364 [DOI] [PubMed] [Google Scholar]
- Zandi S., Ahsberg J., Tsapogas P., Stjernberg J., Qian H., and Sigvardsson M.. 2012. Single-cell analysis of early B-lymphocyte development suggests independent regulation of lineage specification and commitment in vivo. Proc. Natl. Acad. Sci. USA. 109:15871–15876. 10.1073/pnas.1210144109 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.