Barnes et al. show that a bioactive lipid, lysophosphatidylserine, negatively influences T reg cell accumulation and activity through one of its receptors, GPR174. The authors speculate that GPR174 antagonism may be therapeutic for autoimmune diseases.
Abstract
Regulatory T cell (T reg cell) numbers and activities are tightly calibrated to maintain immune homeostasis, but the mechanisms involved are incompletely defined. Here, we report that the lysophosphatidylserine (LysoPS) receptor GPR174 is abundantly expressed in developing and mature T reg cells. In mice that lacked this X-linked gene, T reg cell generation in the thymus was intrinsically favored, and a higher fraction of peripheral T reg cells expressed CD103. LysoPS could act in vitro via GPR174 to suppress T cell proliferation and T reg cell generation. In vivo, LysoPS was detected in lymphoid organ and spinal cord tissues and was abundant in the colon. Gpr174−/Y mice were less susceptible to experimental autoimmune encephalomyelitis than wild-type mice, and GPR174 deficiency in T reg cells contributed to this phenotype. This study provides evidence that a bioactive lipid, LysoPS, negatively influences T reg cell accumulation and activity through GPR174. As such, GPR174 antagonists might have therapeutic potential for promoting immune regulation in the context of autoimmune disease.
Regulatory T cells (T reg cells) that express the transcription factor Foxp3 are tasked with the job of controlling aberrant immune responses. Accordingly, T reg cell abundance and activity are precisely calibrated, and even subtle changes in T reg cell homeostasis can potentiate or ameliorate immunopathology (Josefowicz et al., 2012). Many molecular signals that drive the development and maintenance of these cells have been deciphered, including TCR engagement, co-stimulation, and γ-chain cytokine signaling, most importantly by IL-2 (Josefowicz et al., 2012). Recently, retinoic acid, short-chain fatty acids, and sphingosine-1-phosphate, all small molecules that can be recognized by G protein–coupled receptors (GPCRs) or nuclear receptors, have been shown to modulate T reg cell development and activity (Liu et al., 2009; Hall et al., 2011; Smith et al., 2013). Thus, a paradigm is emerging whereby T reg cell populations are “tuned” by small molecules, such as metabolites, hormones, and bioactive lipids (Thorburn et al., 2014). The receptors for these molecules represent attractive therapeutic targets for modulating immunopathologies and immune responses.
GPR174 is one of four GPCRs known to be activated by the bioactive lipid lysophosphatidylserine (LysoPS; Inoue et al., 2012). Phospholipase A1 and A2 enzymes can catalyze the generation of LysoPS by hydrolyzing phosphatidylserine (PS) at the sn-1 or sn-2 position, respectively, resulting in the removal of an acyl chain (Grzelczyk and Gendaszewska-Darmach, 2013). The enzyme ABHD16a also exhibits PS lipase activity, and Abhd16a deficiency results in reduced LysoPS levels in vivo (Kamat et al., 2015). LysoPS species vary by acyl chain length and saturation, among which the 16:0, 18:0, and 18:1 isoforms are the most abundant in brain, heart, kidney, and lung tissues (Blankman et al., 2013). PS-PLA1, ABHD6, and ABHD12 can catalyze the degradation of LysoPS, and genetic deficiencies in the latter two enzymes have been linked to metabolic syndrome and inflammatory neurodegenerative disease, respectively (Sato et al., 1997; Blankman et al., 2013; Thomas et al., 2013). Roles for LysoPS in suppressing T cell proliferation in vitro (Bellini and Bruni, 1993) and activating mast cells (Martin and Lagunoff, 1979) have been described, but the mechanisms whereby it mediates these effects and its importance in vivo remain unclear.
The first LysoPS receptor to be deorphanized was GPR34, an X-linked GPCR that is most abundantly expressed in microglia, capable of coupling to Gαi-containing heterotrimers, and protective in the central nervous system (CNS) against Cryptococcus neoformans infection–induced pathology (Liebscher et al., 2011; Kitamura et al., 2012). Subsequently, three other GPCRs, GPR174, P2RY10, and P2RY10-L, were identified as selective and high-affinity LysoPS receptors using an in vitro screening approach (Inoue et al., 2012). These three receptors are all closely linked on the X chromosome, abundantly expressed by many immune cell types, and capable of signaling via Gα12/Gα13-containing heterotrimeric G proteins; GPR174 has also been suggested to have Gαs affinity (Sugita et al., 2013). Functions for these three receptors in the immune system have not yet been described.
Herein, we report that LysoPS is abundant in the thymus, peripheral lymphoid tissues, CNS, and colon, and that T reg cell homeostasis is altered in mice that lack the LysoPS receptor GPR174. In the thymus, T reg cells from Gpr174−/Y mice accumulated, and in the periphery, they showed increased CD103 expression; both phenotypes occurred in a cell-intrinsic manner. Furthermore, in the experimental autoimmune encephalomyelitis (EAE) model of CNS autoimmunity, GPR174-deficient T reg cells could limit immunopathology.
RESULTS AND DISCUSSION
Enriched GPR174 and LysoPS receptor expression in T reg cells
Our initial interest in GPR174 stemmed from an effort to identify GPCRs involved in regulating lymphocyte transit through lymphoid organs (Pham et al., 2008). Quantitative PCR analysis of the mRNA expression levels of 353 nonodorant GPCRs (Regard et al., 2008) in naive T and B cells identified Gpr174 (previously known as Fksg79) as one of the most abundantly expressed transcripts (not depicted). To characterize the role of this X-linked gene in the immune system, we replaced the single coding exon with a cassette encoding an “in-frame” dTomato allele (Fig. 1 A). Analysis of dTomato expression in Gpr174−/Y male mice (Fig. 1, B–D) confirmed high levels of GPR174 expression in naive T and B cells (Fig. 1, B and C), and dTomato expression patterns were similar to Gpr174 mRNA expression levels (Fig. 1, C and E). Naive T and B cell numbers and lymphoid tissue organization were normal in Gpr174−/Y mice (not depicted). In LN transit assays (Pham et al., 2008), no differences in trafficking between wild-type and Gpr174−/Y T or B cells were detected (not depicted). Further characterization of dTomato expression showed abundant GPR174 expression in CD25+ CD4 single-positive (SP) thymocytes and CD25+CD4+ T cells, populations that are highly enriched in T reg cells, compared with naive CD4+ T cells (Fig. 1, B and C). These Gpr174 gene expression patterns were confirmed by quantitative RT-PCR analysis of sorted cell populations from wild-type mice (Fig. 1 E). We observed that elevated Gpr174 expression correlated with Foxp3 expression in both immature T reg (Foxp3+CD25−) and mature T reg (Foxp3+CD25+) CD4 SP thymocytes, whereas levels were not elevated in Foxp3−CD25+ thymic T reg cell precursors (Fig. 1 E). In contrast, when we differentiated naive CD4+ T cells in Th1 or Th17 polarizing conditions, reduced GPR174-dTomato expression was observed compared with cells cultured in Th0 conditions (Fig. 1 D).
Figure 1.
Elevated expression of transcripts encoding GPR174 and other LysoPS receptors in T reg cells. (A) Diagram of the construct used to target the Gpr174 locus by homologous recombination. For more details, see Materials and methods. (B and C) Measurement of dTomato-GPR174 reporter allele expression in thymocytes (left) or splenocytes (right) by flow cytometry from 8-wk-old male Gpr174−/Y mice. (B) Thymocyte populations are as follows: gray shaded, CD4+CD8+ DP; blue dashed, CD8+ SP; purple dotted, CD25− CD4 SP; and red, CD25+ CD4 SP T reg cells. Splenocyte populations are as follows: gray shaded, background (splenocytes from wild-type mice); blue dashed, naive CD8+ T cells; purple dotted, CD25− naive CD4+ T cells; and red, CD25+CD4+ T reg cells. (C) The mean fluorescence intensity (MFI) of dTomato-GPR174 is shown for the indicated cell populations. B cells were identified as B220+IgDhigh splenocytes. Each dot represents a measurement from a separate mouse; n = 4. (D) Expression of dTomato-GPR174 was measured in naive CD4+ T cells cultured under Th0, Th1, or Th17 polarizing conditions for 5 d; representative flow cytometry data are shown. (E) The mRNA expression levels of the LysoPS receptors Gpr174, Gpr34, P2ry10, and P2ry10-L were measured by RT-PCR in the indicated sorted thymocyte and splenocyte populations from 8-wk-old wild-type mice. Populations were gated as described in Materials and methods. Cells were sorted in triplicate, and each dot represents the relative expression in a separate sorted cell population from a distinct mouse; n = 3; error bars show SD. All data in B–E are representative of at least three independent assays.
To characterize how T cells might sense LysoPS, we measured mRNA expression levels of the four known LysoPS receptors. Similar to Gpr174, its homologue P2ry10 also showed elevated mRNA levels in Foxp3+ T reg cells and thymocytes; however, unlike Gpr174, expression in double-positive (DP) thymocytes was also elevated (Fig. 1 E). Transcripts encoding P2ry10l were less abundant and not enriched in T reg cells or thymocytes. Gpr34 transcripts were detectable in Foxp3+ T reg cells, although their levels appeared to be at least an order of magnitude lower than those encoding Gpr174 or P2ry10 (Fig. 1 E).
Role of GPR174 in T reg cell accumulation and homeostasis
Because T reg cells expressed high amounts of LysoPS receptors, we characterized the effects of GPR174 deficiency on these cells. Initially, we took advantage of the linkage of Gpr174 to the X chromosome, for which one copy is subject to random inactivation in female cells. Therefore, approximately half of the hematopoietic cells in female Gpr174+/− mice express the Gpr174−-dTomato allele. Intriguingly, we consistently detected an increased frequency of dTomato+ cells among CD25+ compared with CD25− CD4 SP thymocytes (Fig. 2 A), suggesting that the fitness of T reg cells might be greater when GPR174 is absent. To quantify T reg cell abundance in Gpr174−/Y mice, we measured Foxp3 expression in the thymus, secondary lymphoid organs, and colon lamina propria. The frequency of Gpr174−/Y Foxp3+ CD4 SP cells was significantly increased in the thymus, the site where most T reg cells develop, and in the colon lamina propria, where both thymus-derived and peripherally induced T reg cells can accumulate in microbiota-colonized mice (Fig. 2 B; Bollrath and Powrie, 2013). In contrast, T reg cell frequencies were unaffected in the spleen and LNs (Fig. 2 B), where extrinsic factors that include γc-cytokines govern the niche size. Therefore, GPR174 deficiency appeared to favor T reg cell accumulation in specific tissues.
Figure 2.
GPR174 intrinsically constrains T reg cell accumulation and CD103 expression. (A) Flow cytometry analysis of the percentage of dTomato+ cells in CD25− and CD25+ CD4 SP T reg thymocytes in Gpr174+/− female mice. Lines link measurements of populations from the same mouse; n = 8. (B and C) Flow cytometry analysis of the frequency of Foxp3+ CD4 SP or CD4+ T cells in 8-wk-old wild-type and Gpr174−/Y littermate male mice (B) and of the percentage of Foxp3+ CD4 SP or CD4+ T cells that express CD103 (surface) or Helios (intracellular; C); n = 7 or 9. Horizontal lines indicate the mean. (D) Flow cytometry analysis of mice reconstituted with mixed wild-type and Gpr174−/Y bone marrow. Lethally irradiated Ly5-2 mice (CD45.1+) were reconstituted with a mixture of bone marrow from wild-type Ly5-1/-2 F1 (CD45.1+CD45.2+) and Gpr174−/Y (Ly5-1, CD45.2+) mice. Radioresistant cells (Ly5-2, CD45.1+) were excluded, and the contribution of Gpr174−/Y-derived cells (Ly5-1) to the indicated cell subsets is shown; n = 4; error bars show SD. (E) The contribution of Gpr174−/Y-derived cells to the CD103+ T reg cell population is shown for mice reconstituted as in D, or with mixed bone marrow from wild-type Ly5-1/-2 F1 (CD45.1+CD45.2+) and wild-type mice (Ly5-1, CD45.2+) mice; n = 4. All data are representative of at least three independent experiments and were evaluated using paired (A, E) or unpaired (B–D) Student’s t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
T reg cells can be subdivided into different subsets that have been associated with specialized regulatory functions (Campbell and Koch, 2011). To assess whether GPR174 deficiency favored the accumulation of a T reg cell subset, we analyzed a panel of cell surface molecules associated with T reg cell differentiation. No changes in T reg cell expression of CCR7, CD25, CD44, CD62L, CXCR3, CXCR6, or E- or P-selectin were detected in the thymus, spleen, or LN of Gpr174−/Y mice (not depicted). However, more Gpr174−/Y T reg cells expressed CD103, a marker associated in some contexts with augmented in vitro and in vivo suppressive activity and trafficking to nonlymphoid tissues (McHugh et al., 2002; Huehn et al., 2004; Suffia et al., 2005; Campbell and Koch, 2011; Geuking et al., 2011). This effect was not observed in Gpr174−/Y thymic T reg cells but could be observed in peripheral lymphoid tissues, suggesting that GPR174 constrains the expression of CD103 by T reg cells after they egress from the thymus (Fig. 2 C). The levels and frequencies of Helios expression, which have been associated with thymus-derived T reg cells, were similar in T reg cells from wild-type and Gpr174−/Y mice (Fig. 2 C), suggesting that GPR174 deficiency did not bias thymus-derived versus peripherally induced T reg cell accumulation under homeostatic conditions.
The T reg cell–constraining influence of GPR174 was cell intrinsic, as Gpr174−/Y Foxp3+ CD4 SP thymocyte frequencies were significantly higher than those of Gpr174−/Y DP and CD4 SP thymocytes in mixed bone marrow chimeric mice (Fig. 2 D). The contribution of Gpr174−/Y cells to the naive T cell compartment matched the extent of reconstitution of other lineages, such as granulocytes that lack GPR174 expression (not depicted), consistent with GPR174 having a selective role in T reg cells. Under these competitive conditions, Gpr174−/Y-derived T reg cells maintained their overrepresentation in peripheral lymphoid organs (Fig. 2 D). These data suggest that after wild-type or Gpr174−/Y T reg cell generation occurs in the thymus, the cells are equally competent to be maintained in the periphery. Moreover, the overall peripheral niche size available to T reg cells is not altered by GPR174 deficiency. The frequency of Gpr174−/Y-derived T reg cells expressing CD103 continued to be increased in mixed chimeras, indicating a cell-intrinsic bias for the accumulation of cells expressing this adhesion molecule (Fig. 2 E). No differences in Foxp3 or CD103 expression were detected in control wild-type (congenic) mixed bone marrow chimeras (Fig. 2 E and not depicted). The gene expression profile of Gpr34 led us to consider whether this LysoPS receptor also modulated T reg cell development; however, we found equivalent frequencies of Gpr34−/Y- and wild-type–derived Foxp3+ and Foxp3+CD103+ cells in the thymus and periphery of mixed bone marrow chimeras (not depicted).
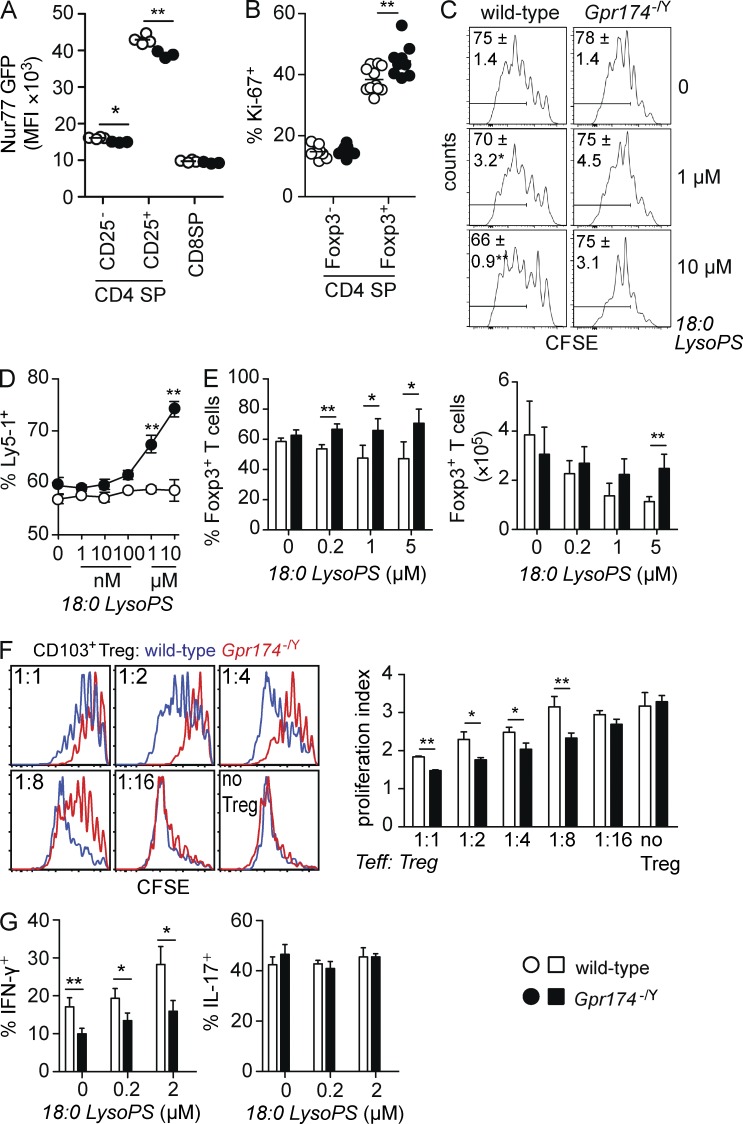
To better characterize the development of thymic Gpr174−/Y T reg cells, we crossed Gpr174−/Y mice to animals expressing a Nur77-GFP transgene (Moran et al., 2011). Levels of Nur77 increase in positively selected thymic T reg cells and CD25+Foxp3− T reg cell precursors in response to recent MHC class II–dependent signaling. Importantly, Nur77 levels are reduced in conditions that favor enhanced T reg cell development, such as in response to exogenous TNFR family–mediated co-stimulation (Mahmud et al., 2014). CD25+ CD4 SP thymocytes in Gpr174−/Y mice expressed lower levels of Nur77-GFP, consistent with a model in which GPR174 deficiency results in the exposure of developing thymocytes to T reg cell–favoring signals (Fig. 3 A). Additionally, we noted that CD25− CD4 SP thymocytes showed a very slight reduction in Nur77-GFP levels (Fig. 3 A). Because TNF receptor family members contribute to thymic T reg cell development and reduced Nur77-GFP abundance (Mahmud et al., 2014), we measured levels of glucocorticoid-induced TNFR family–related gene (GITR), OX40, and TNFR2 in the Foxp3+ and Foxp3− CD4 SP subsets and detected no differences between wild-type and Gpr174−/Y thymocytes (not depicted). In line with the accumulation of Gpr174−/Y thymic T reg cells, more Foxp3+, but not Foxp3− CD4 SP thymocytes expressed Ki-67, a marker of cells that have recently divided, compared with cells in wild-type littermates (Fig. 3 B). This effect was most obvious in the thymus of mice younger than 8 wk old (not depicted). Together, these data suggest that GPR174 signaling might intrinsically constrain thymic T reg cell proliferation.
Figure 3.
CD4+ T cells show diminished Nur77 levels and enhanced proliferation in Gpr174−/Y mice. (A) Flow cytometry analysis of the mean fluorescence intensity (MFI) of Nur77-GFP expression in thymocytes from 6-wk-old wild-type and Gpr174−/Y Nur77-GFP+ littermate male mice. Each dot represents an individual mouse; n = 4. (B) Intracellular levels of Ki-67 expression in the indicated thymocyte populations from 6-wk-old wild-type and Gpr174−/Y littermate male mice were determined using flow cytometry. The percentage of Ki-67+ cells among Foxp3+ T reg and Foxp3− CD4 SP thymocytes is shown; n = 9 or 10. (A and B) Horizontal lines indicate the mean. (C) The effects of LysoPS on T cell proliferation under neutral conditions. CFSE-labeled LN cells from wild-type or Gpr174−/Y mice were cultured in round-bottom plates with 0.25 µg/ml soluble anti-CD3 and the indicated amounts of LysoPS. Cell proliferation was assessed 3 d later based on CFSE dilution that was measured by flow cytometry; live CD4+TCR-β+ T cells are shown. Gates indicate the percentage of cells that divided at least three times, and means ± SD are shown; n = 4. (D and E) Direct effects of GPR174 and its ligand LysoPS on proliferation and Foxp3 induction. In flat 96-well plates coated with anti-CD3 and anti-CD28 mAbs (both 2 µg/ml), a mixture of either wild-type (Ly5-1+) or Gpr174−/Y (Ly5-1+) with congenic wild-type (Ly5-2+) naive CD4+ T cells (D) or either wild-type or Gpr174−/Y naive CD4+ T cells (E) were added. Cells were cultured in the presence of 200 U/ml IL-2 (D) or 1 ng/ml TGF-β (E) and the indicated concentrations of 18:0 LysoPS for 4 d. Expression of Ly5-1 and Ly5-2 (D) or Foxp3 (E) was measured by flow cytometry; n = 4; error bars indicate SD. Counts were quantified based on the number of events acquired on a flow cytometer run for 60 s per sample (E). Cells used in E were isolated from mixed bone marrow chimeric mice to minimize extrinsic effects on naive T cells. (F) In an in vitro T reg cell suppressor assay, CD4+CD25+CD45RBhighCD103− T reg cells were sorted from spleens of wild-type and Gpr174−/Y littermate mice. T reg cells were cultured at the indicated ratios with 105 CFSE-labeled naive CD45.1+CD4+ T cells and 2 × 104 CD11c-enriched DCs in the presence of 0.5 µg/ml soluble anti-CD3ε mAb for 3 d. The proliferation index of CFSE-labeled cells was determined in triplicate cultures, and representative CFSE flow cytometry plots are shown. (G) A total of 2 × 105 naive CD4+ T cells from CD45.2+ wild-type or Gpr174−/Y littermate mice were cultured along with an equivalent number of cells from CD45.1+ wild-type mice under Th1 or Th17 polarizing conditions for 5 d. After restimulation with PMA and ionomycin, the percentage of CD45.2+ cells that secreted IFN-γ or IL-17 was determined by intracellular cytokine staining and flow cytometry analysis. (F and G) Error bars show SD. Data were evaluated by unpaired Student’s t test: *, P < 0.05; **, P < 0.01. All data are representative of three or more independent experiments.
LysoPS suppresses CD4+ T cell proliferation and T reg cell differentiation via GPR174
LysoPS was previously reported to suppress lymphocyte proliferation in human PBMC cultures (Bellini and Bruni, 1993). To test whether LysoPS could suppress mouse T cell proliferation, we cultured total LN cells with mitogenic soluble anti-CD3 mAb in the presence of 1 or 10 µM LysoPS. The proliferation of wild-type CD4+ T cells could be suppressed by LysoPS, and this effect was GPR174 dependent (Fig. 3 C). Next, we sorted naive CD4+ T cells and measured the proliferation of co-cultured wild-type and Gpr174−/Y cells after stimulation with plate-bound anti-CD3 and anti-CD28 mAbs in the presence of exogenous IL-2. In this setting, LysoPS effectively suppressed the proliferation of wild-type, but not Gpr174−/Y cells, suggesting that LysoPS can directly inhibit T cell proliferation in vitro (Fig. 3 D). However, the in vivo proliferation of Gpr174−/Y OT-II T cells 3 d after intraperitoneal immunization with OVA in alum adjuvant occurred comparably with that of wild-type OT-II T cells (not depicted), indicating that this repressive pathway might only act in certain contexts. To test whether LysoPS could affect the induction of Foxp3-expressing cells in vitro, naive T cells were cultured under TGF-β–mediated T reg cell–skewing conditions. The addition of LysoPS reduced the number and frequency of Foxp3+ cells in wild-type T cell cultures, whereas these effects were not observed in Gpr174−/Y T cell cultures (Fig. 3 E). These influences of LysoPS signaling via GPR174 might contribute to the T reg cell accumulation observed in Gpr174−/Y mice.
Enhanced T reg cell activity and reduced Th1 responses of GPR174−/Y T cells
In addition to affecting T reg cell development and turnover, we considered that GPR174 might alter T reg cell functionality. In vitro CD4+ T cell suppression assays can reveal some aspects of T reg cell function (Thornton and Shevach, 1998). In this assay, GPR174-deficient T reg cells could more potently suppress the anti-CD3 and DC-induced proliferation of naive CD4+ responder T cells compared with wild-type control T reg cells (Fig. 3 F). When we separated T reg cells into CD103+ and CD103− subsets, we found that the CD103− subset of Gpr174−/Y T reg cells were better suppressors than their wild-type counterparts (Fig. 3 F). Furthermore, CD103+ T reg cells were more potent suppressors than CD103− T reg cells for both wild-type and Gpr174−/Y cells (not depicted), consistent with some studies for wild-type cells (Lehmann et al., 2002; McHugh et al., 2002). These data suggest that Gpr174−/Y mice might have enhanced immune regulation compared with wild-type mice as the result of both increased CD103− T reg cell activity and increased CD103+ T reg cell numbers.
Although GPR174 expression was lower on effector Th1 and Th17 cells than on nonpolarized T cells (Fig. 1 D), we were interested to determine whether LysoPS could also act via GPR174 to alter the differentiation of effector CD4+ T cell subsets. Surprisingly, Gpr174−/Y naive T cells were less prone to differentiate into Th1 cells that could secrete IFN-γ (Fig. 3 G). This phenotype was observed even in the absence of exogenous LysoPS. The addition of 200 nM or 2 µM LysoPS increased the frequency of Th1 cells, but unlike its activities in T reg cell–skewing cultures, it did not reveal GPR174-specific effects as the frequency of Th1 cells increased similarly in both wild-type and Gpr174−/Y cell cultures (Fig. 3 G). In contrast, neither GPR174 deficiency nor exogenous LysoPS affected in vitro Th17 cell differentiation (Fig. 3 G). The reduced Th1 cell differentiation propensity of Gpr174−/Y T cells could be a consequence of LysoPS influences during T cell development, as suggested by the CD25− CD4 SP thymocyte Nur77-GFP expression profile (Fig. 3 A); alternatively, GPR174 might have a LysoPS-independent influence on T cells in the early phase of Th1 cultures.
LysoPS abundance and lipase expression in lymphoid organs
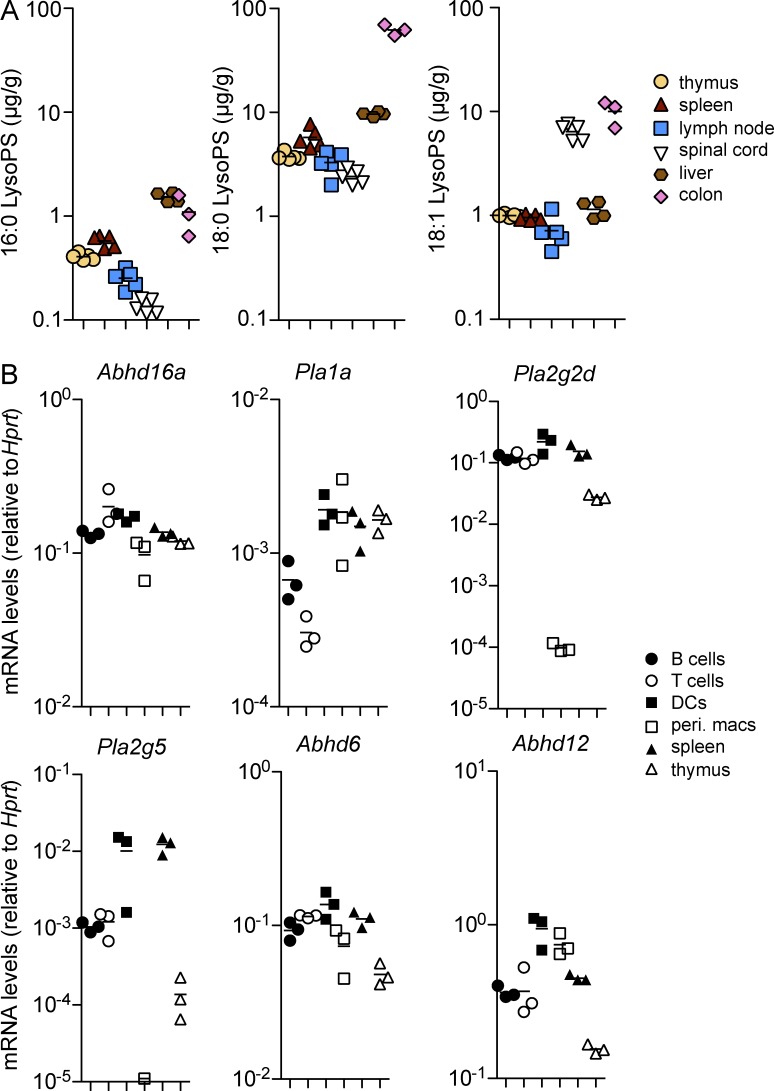
Given the effects of LysoPS on T cells, we next investigated whether LysoPS was present in lymphoid organs under homeostatic conditions. Previously, LysoPS concentrations were reported for selected tissues, including the liver (Blankman et al., 2013), in which 18:0, 18:1, and 16:0 LysoPS were among the most common isoforms. We developed a liquid chromatography followed by tandem mass spectrometry (LC-MS/MS) procedure for measuring LysoPS and detected concentrations of the 16:0, 18:0, and 18:1 isoforms in the liver that were similar to those measured by Blankman et al. (2013; Fig. 4 A). The thymus, spleen, and LNs each contained abundant LysoPS, with the 18:0 isoform being the most common, followed by 18:1 and 16:0 (Fig. 4 A). Although this analysis did not permit the extra- and intracellular pools of LysoPS to be distinguished, it is notable that a concentration of 1 µg/g corresponds to ∼2 µM LysoPS. Plasma contained much lower concentrations of all three LysoPS isoforms that were below the limit of detection for our assay (not depicted). We also analyzed LysoPS concentrations in two common sites of immunopathology. Spinal cord tissues exhibited slightly lower 18:0 and 16:0 LysoPS concentrations compared with lymphoid tissues but contained almost an order of magnitude more 18:1 LysoPS (Fig. 4 A). Colon tissues contained an order of magnitude more 18:0 LysoPS compared with any other tissue measured, in addition to high levels of 18:1 and 16:0 LysoPS (Fig. 4 A).
Figure 4.
LysoPS is abundant in lymphoid tissues and can be generated by various immune cell types. (A) Concentrations of 16:0, 18:0, and 18:1 LysoPS were measured under homeostatic conditions in the indicated tissues using LC-MS/MS. (B) Transcript levels of LysoPS synthetic and metabolic enzymes were measured by quantitative RT-PCR in the indicated cell populations and tissues. In both panels, each dot represents a separate mouse; n = 3–5. (A and B) Horizontal lines indicate the mean.
The network of enzymes that regulates the generation and degradation of LysoPS is likely to be complex. To assess the abundance of known enzymes with effects on PS→LysoPS reactions or the degradation of LysoPS in immune tissues, we performed quantitative PCR analyses. ABHD16a, an enzyme with established activity in generating LysoPS in vivo (Kamat et al., 2015), was broadly expressed in DCs, macrophages, lymphocytes, and lymphoid organs (Fig. 4 B). Transcripts for Pla1a mRNA, which encode an enzyme that can both generate and degrade LysoPS (Sato et al., 1997), were most abundant in myeloid cells. Levels of Pla2g2d and Pla2g5 mRNA transcripts were high in lymphocytes and DCs but low in peritoneal macrophages. Pla2g5 mRNA transcript levels were markedly lower in the thymus than in the spleen. Transcripts encoding ABHD6 and ABHD12, enzymes that have been shown in vitro and in vivo to degrade LysoPS (Blankman et al., 2013; Thomas et al., 2013), could also be detected in lymphoid cells and spleen but were notably underexpressed in the thymus (Fig. 4 B). The expression in DCs of several LysoPS synthetic enzymes (Abhd16a, Pla2g2d, and Pla2g5) raises the possibility that LysoPS was acting in a paracrine fashion in the in vitro suppressor experiments (Fig. 3 F). Although we were not able to measure changes in LysoPS in the culture supernatants with our current LC-MS/MS procedure (not depicted), it is notable that activated myeloid cells can secrete LysoPS (Kamat et al., 2015), which might allow for localized changes in ligand abundance. Future studies of mice lacking individual phospholipase enzymes will be needed to elucidate the key enzymes responsible for LysoPS production and metabolism in lymphoid cells and tissues.
GPR174 deficiency in T reg cells protects from neuroinflammation
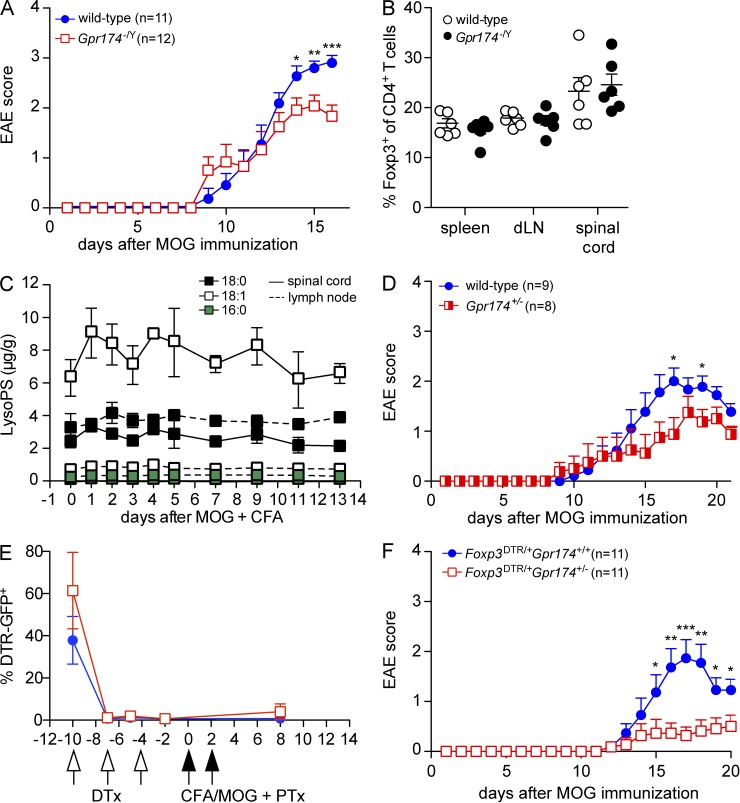
Finally, given the constraining influence of GPR174 on T reg cell development and functionality, we examined whether GPR174 deficiency might protect mice from autoimmune pathology. To induce EAE, we administered myelin oligodendrocyte glycoprotein (MOG) peptide in CFA along with pertussis toxin. In this disease model, Th1 and Th17 cells drive CNS inflammation, whereas T reg cells can limit disease severity and promote the remission of symptoms (Stromnes and Goverman, 2006). Although the day of disease onset was unaffected, Gpr174−/Y male mice showed significantly reduced peak disease severity compared with wild-type littermates (Fig. 5 A). At day 12, a time point when Gpr174−/Y and wild-type mice showed a similar disease score, the frequency of Foxp3+ T reg cells was not significantly affected by GPR174 deficiency in the secondary lymphoid organs or spinal cord (Fig. 5 B); however, the frequency of CD103+ cells among T reg cells remained elevated in the draining LN at days 8–12 (not depicted). Moreover, levels of 16:0, 18:0, and 18:1 LysoPS did not appear to markedly change in the draining LN or spinal cord throughout the experiment (Fig. 5 C). These whole tissue analyses do not exclude the possibility that there are changes in other LysoPS isoforms (Blankman et al., 2013; Kamat et al., 2015) or local changes in interstitial LysoPS concentrations that affect cell behavior.
Figure 5.
GPR174 deficiency in T reg cells limits the severity of EAE. (A) EAE was induced in 10-wk-old male wild-type or Gpr174−/Y littermate mice by injecting a MOG35–55 + CFA emulsion (s.c.) followed by pertussis toxin (i.v.). Mice were scored daily for disease symptoms; mean disease scores are shown and error bars indicate SEM. (B) The frequency of Foxp3+ T reg cells among total CD4+ T cells was determined in the indicated tissues 12 d after EAE induction in 10-wk-old male wild-type or Gpr174−/Y littermate mice by flow cytometry. (C) EAE was induced in a cohort of wild-type mice, and levels of LysoPS were measured by LC-MS/MS in the inguinal LN that drained the MOG + CFA emulsion (dotted lines) and in the spinal cord (solid lines) throughout the course of disease; n = 3–5. (B and C) Error bars show SD. (D) EAE was induced in 10-wk-old female Gpr174+/− or wild-type littermate mice as in A; error bars show SEM. (E and F) In Gpr174+/− or wild-type female mice heterozygous for an X-linked Foxp3-DTR allele, diphtheria toxin (DTx) was injected on days −10, −7, and −4. EAE was induced as in A on day 0. The ablation efficiency was assessed by analyzing the frequency of DTR-GFP+ cells among blood CD4+CD25+ T reg cells at the indicated time points (E); error bars show SD. Disease was scored daily after MOG immunization (F); n = 11; error bars show SEM. Data are representative of two (E and F) or three or more (A–D) independent experiments. Differences between groups were compared using unpaired Student’s t test for each time point: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To test whether intrinsic effects on T reg cells contributed to the reduced disease severity, we examined female Gpr174+/− mice because they have a mixture of wild-type and GPR174-deficient T reg and naive T cells as a consequence of X chromosome inactivation (Fig. 2 A). We predicted that if GPR174-deficient T reg cells in these mice had more potent activity, they might act in trans to suppress EAE. Indeed, disease was modestly, but significantly, reduced in Gpr174+/− female mice compared with wild-type littermates (Fig. 5 D). The frequency of inactivation of the wild-type versus the GPR174-deficient X chromosome showed a considerable range in Gpr174+/− females (Fig. 2 A). To make the T reg cell compartment in Gpr174+/− females nearly completely GPR174 deficient, we crossed Gpr174+/− females to Foxp3DTR/Y males to yield offspring with a Gpr174− allele on one X chromosome and a Foxp3DTR allele on the other. Administering diphtheria toxin to these mice selectively ablated the Gpr174+Foxp3DTR T reg cells, in accord with past work (Fig. 5 E; Pierson et al., 2013), and allowed GPR174-deficient T reg cells to fill the niche (not depicted). At day 10 after diphtheria toxin treatment, we induced EAE as before and found that T reg cell–specific GPR174 deficiency significantly protected mice from EAE (Fig. 5 F). Therefore, we identify T reg cell expression of GPR174 as a nonredundant factor that can contribute to immunopathology.
Concluding remarks
Our in vitro and in vivo findings show that GPR174 acts in Foxp3+ T cells to constrain their development and immune regulatory function. Thus, under inflammatory conditions where LysoPS production can increase (Kamat et al., 2015), GPR174 could contribute to down-modulating T reg cell activity to favor effector responses. Although this may be beneficial during responses to infections, it might allow more severe tissue damage during autoimmune responses. The complete mechanism whereby GPR174 signaling restrains T reg cell activities, and whether this involves Gα12, Gα13, or Gαs, is not yet clear and will require further investigation. Our in vitro experiments also suggest that there could be conditions in vivo whereby LysoPS exerts a direct T cell proliferation–inhibitory or Th1-skewing effect via GPR174 independently of its actions on T reg cells. Finally, although our findings are consistent with GPR174 functioning as a LysoPS receptor, we do not exclude the possibility that other physiologically relevant ligands exist for this receptor. Overall, our findings inform recent genome-wide association studies that linked human SNPs in or near the GPR174 locus to the autoimmune conditions Graves’ disease and rheumatoid arthritis (Chu et al., 2013; Zhao et al., 2013; Okada et al., 2014). Given its function in T reg cells, studies of GPR174 antagonists to treat such conditions might be warranted.
MATERIALS AND METHODS
Mice.
To generate Gpr174−/Y mice, the Gpr174 locus was targeted using a standard homologous recombination approach in 129-background embryonic stem cells, as shown in Fig. 1 A. The entire Gpr174 open reading frame is depicted by a black box, and gray boxes represent untranslated regions. Dashed arrows indicate the homologous regions of the arms of the targeting construct. EcoRI sites that were used for the Southern blot confirmation of homologous recombination and the size of the predicted DNA fragments are indicated. In the Gpr174 open reading frame, we inserted a dTomato allele to track fluorescence as a surrogate for Gpr174 expression levels. Gpr174−/Y mice were backcrossed to the C57BL/6J background (000664; The Jackson Laboratory) for 12 generations. Nur77-GFP (018974; The Jackson Laboratory) and Foxp3–diphtheria toxin receptor (DTR; 016958; The Jackson Laboratory) mice were generated by the Hogquist (University of Minnesota, Minneapolis, MN) and Rudensky (Memorial Sloan-Kettering Cancer Center, New York, NY) laboratories, respectively. Gpr34−/Y mice (Liebscher et al., 2011) were provided by T. Schoeberg and A. Schulz (Institut für Biochemie, Universität Leipzig, Leipzig, Germany).
To produce bone marrow chimeric mice, B6-LY5.2/Cr mice (01B96; The National Cancer Institute) were lethally irradiated with a split dose of 1,100 rads γ-radiation and then were i.v. injected with a mixture of bone marrow cells from F1 C57BL/6J × C57BL/6-BoyJ (002014; The Jackson Laboratory) mice and either Gpr174−/Y or wild-type littermate mice. After allowing 10 wk for reconstitution, the contribution of Ly5-1+ cells to the indicated reconstituted cell populations was determined. Radioresistant CD45.1+ and dead cells were excluded. In mixed chimeras, the degree of reconstitution by the two types of donor bone marrow cells varied with each mixture, possibly as a result of cell counting variability or differences in the proportions of mature and hematopoietic stem cells in the donor bone marrow.
Animals were housed in specific pathogen–free enclosures at the University of California, San Francisco (UCSF), Laboratory Animal Research Center, and all experiments complied with animal protocols that were approved by the UCSF Institutional Animal Care and Use Committee.
Cell sorting.
To obtain sorted thymocyte and splenocyte populations for measurements of LysoPS receptor mRNA transcript levels, cells from wild-type mice were sorted using a FACSAria III (BD). The sorted populations were as follows: DP, CD4+CD8+; CD8 SP, CD4−CD8+TCR-β+; CD4 SP, CD4+CD8−TCR-β+, and subdivided further based on Foxp3-GFP and CD25 expression; CD8+ naive T cell, CD8+CD44lowCD62LhighTCR-β+; CD4+ naive T cell, CD4+CD25−Foxp3-GFP−CD44lowCD62LhighTCR-β+; CD4+ Foxp3+ T reg cell, CD4+CD25+CD44lowCD62LhighFoxp3-GFP+TCR-β+; and B cell, B220+IgDhigh. DAPI was used to exclude dead cells, and cell purity was routinely at least 99%. Foxp3-GFP+ indicates GFP-positive cells sorted from unmanipulated Foxp3DTR/DTR mice that express GFP in all Foxp3+ cells.
To obtain naive CD4+ T cells for in vitro assays, splenocyte and LN cells were mashed through a 70-µm strainer to create a single-cell suspension. Cells were incubated with a lineage cocktail of biotin-conjugated mAbs (anti-B220, CD8α, DX5, GR1, I-A(b), and TER115; all BioLegend) in MACS buffer on ice for 20 min and then were incubated with streptavidin-conjugated microbeads (Miltenyi Biotech) in MACS buffer on ice for 15 min, with gentle agitation every 5 min. The MACS buffer contained PBS with 2% FBS, 1 µM EDTA, and 1× penicillin + streptomycin. CD4+ T cells were enriched by negative selection on LS columns (Miltenyi Biotech) under a magnetic field. Enriched cells were stained with CD4–Pacific blue, CD25-APC, CD44-FITC, and CD62L-PE in MACS buffer on ice for 20 min. Cells were sorted using a FACSAria III to obtain CD4+CD44lowCD62LhighCD25− naive T cells that were routinely at least 99% pure.
Flow cytometry.
For cell surface staining, empirically determined dilutions of primary mAbs were used to stain single cell suspensions on ice for 20 min in FACS buffer (PBS with 2% FBS, 0.1% NaN3, and 1 µM EDTA). All mAbs were purchased from BioLegend unless otherwise indicated. The following mAb clones were used: B220-PE-Cy7, CD4-PE-Cy7, CD8α-PerCP-Cy5.5, CD11b-PE-Cy7, CD11c-FITC, CD25–Alexa Fluor 647, CD44-FITC, CD45.1-FITC, CD45.2-BV605, CD62L-PE-Cy7, CD103-biotin (followed by streptavidin-BV711; BD), Foxp3-PE (eBioscience), Helios-APC, I-A(b)-PE, Ki-67–Alexa Fluor 647, and TCR-β–Pacific blue. Dead cells were excluded using Fixable Viability Dye eFluor780 (eBioscience). Intracellular staining to detect Foxp3 expression was performed according to the manufacturer’s instructions, using the manufacturer’s protocol for Foxp3 staining in 96-well plates (eBioscience). Cells were analyzed using an LSR-II flow cytometer (BD) equipped with 405-, 488-, 552-, and 640-nm lasers. Flow cytometry data were processed using FlowJo version 9.7.5 software (Tree Star).
Sample preparation for LC-MS/MS.
Tissue samples were homogenized using a Precellys 24 homogenizer with a Cryolys cooling unit (Bertin Technologies). Individual mouse tissues were placed in 2-ml sample vials with 10 vol water (10-fold dilution) and seven homogenization beads. Homogenization was conducted by three cycles for 20 s at 5,000 rpm (with 30-s breaks) at a temperature lower than 10°C.
After sample homogenization, 20 µl homogenized samples were each pipetted into 13 × 100 mm tubes. Next, 20 µl water and 20 µl internal standard solution (0.1 µg/ml) were added, and the solution was vortexed for 10 s. To precipitate proteins, 100 µl acetonitrile (ACN; C2H3N) was added, and the solution was centrifuged at 3,000 rpm for 10 min. The supernatant was injected into an LC-MS/MS system.
LC-MS/MS.
Samples (10-µl volume) were injected into a Shimadzu Prominence UFLC system equipped with a binary pump and a SIL-20AC autosampler. Separation was achieved on a Zic-HILIC column (4.6 × 50 mm, 3.5 µm; SaQuant). A gradient separation was used consisting of mobile phase A (ACN/H2O [95%/5%, vol/vol] containing 1 mM ammonium formate [NH4HCO2]) and mobile phase B (ACN/H2O [10%/90%, vol/vol] containing 1 mM NH4HCO2) delivered at a flow rate of 1 ml/min. Mobile phase B was used at 0% from 0 to 2.0 min, and then was increased linearly to 20% B within 3.4 min, followed by a quick ramp within 0.1 min to 100% B, which was maintained at 100% B for 1.5 min. After a quick ramp back to 0% mobile phase B, it was maintained at 0% for another 2.9 min until the end of the analysis.
Mass spectrometric detection was performed using an Applied Biosystems/MDS SCIEX. An API 5000 triple quadrupole mass spectrometer was operated in multiple reaction monitoring (MRM) mode via the negative electrospray ionization interface using the transitions (m/z) 496→153, 524→153, 522→153, and 508→153 for 16:0 LysoPS, 18:0 LysoPS, 18:1 LysoPS, and 17:1 LysoPS (as an internal standard), respectively. The ion source temperature was maintained at 400°C. The spraying needle voltage was set at −4.5 kV in positive mode. Curtain gas, collision gas (CAD), gas 1, and gas 2 were set at 20, 4, 50, and 50, respectively. The entrance potential (EP) was set at –10 V. The declustering potential and collision cell exit potential (CXE) were –150 V and –15 V, respectively. Collision energy (CE) was –35 eV for all compounds. Quantitation was performed based on the peak area ratio. Data acquisition and quantitative processing were accomplished using the Applied Biosystems Analyst version 1.5.1 software.
Measurements of gene expression.
Total RNA was isolated using an RNeasy kit and in-column DNA digestion according to the manufacturer’s instructions (QIAGEN). For measurements of GPCR gene expression levels, RNA was isolated from 106 sorted cells for each thymocyte or splenocyte population. For phospholipase enzyme measurements, RNA was isolated from 106 MACS-enriched B cells, T cells, and DCs, day 4 thioglycolate–elicited peritoneal macrophages, and total spleen and thymus tissues. Gene expression levels were measured by real-time PCR using SYBR green PCR mix (Roche) on an ABI Prism 7300 sequence detection system (Applied Biosystems). Values were normalized to those of a housekeeping gene, Hprt. The following primers were used: Abhd6, (F) 5′-AAGACCAGGTGCTTGATGTGTCC-3′ and (R) 5′-TCCATCACTACCGAATGGCCACAG-3′; Abhd12, (F) 5′-TCCAGTTTATCCCCTTTCAC-3′ and (R) 5′-CGGTTCCGACTTCCCTA-3′; Abhd16a, (F) 5′-ATGTTTGTGGACCGACGG-3′ and (R) 5′-CTTCTAGGGGTGTGGAGACA-3′; Gpr34, (F) 5′-TGTATTTCCTGATGTCCAGTAAT-3′ and (R) 5′-GCTTTCACTTCTGCTTGCTT-3′; Gpr174, (F) 5′-GCCTTATGGGTATTTTACGG-3′ and (R) 5′-AGTCAGCAATGGCTAGGTTT-3′; Hprt, (F) 5′-AGGTTGCAAGCTTGCTGGT-3′ and (R) 5′-TGAAGTACTCATTATAGTCAAGGGCA-3′; P2ry10, (F) 5′-AGTCTTCGTTATCTGCTTCACT-3′ and (R) 5′-GATGGAAATACAGGGTACTTTT-3′; P2ry10l, (F) 5′-TTTTCTGCTGCTTCCACTGA-3′ and (R) 5′-CCCATCTTGAGTGTTTATGTTCC-3′; Pla1a, (F) 5′-TGGAGTTTTATTTGAAGGAGA-3′ and (R) 5′-GTGGGTTAGGATGAGCCAT-3′; Pla2g2d, (F) 5′-GCTCTGGGCTGGAACTATGA-3′ and (R) 5′-CCTGGGTTGCAGTTATACCG-3′; and Pla2g5, (F) 5′-CTCACACTGGCTTGGTTCCT-3′ and (R) 5′-CATGGACTTGAGTTCTAGCAAGC-3′.
Ex vivo T cell differentiation and proliferation.
For T cell proliferation assays using total LN cells, peripheral LNs were mashed through 100-µm filters. Cells were resuspended in PBS + 0.1% BSA and labeled with 5 µM CFSE at 37°C in the dark for 10 min. Then, 4 × 105 cells were added to round-bottom 96-well plates along with 0.25 µg/ml anti-CD3ε mAb (LEAF grade; BioLegend) in RPMI 1640 media with 10% FBS, 1× penicillin + streptomycin, 2 mM l-glutamine, 10 mM Hepes, and 50 µg/ml 2-mercaptoethanol. To some cultures, 18:0 LysoPS (Avanti Polar Lipids) that was maintained as a 5 mM stock solution in cell culture grade water with 0.5% DMSO was added to wells at the indicated concentrations. Finally, 3 d later, cells were harvested and CFSE dilution of CD4+ T cells was quantified by flow cytometry.
For proliferation assays of sorted naive CD4+ T cells, 24-well plates were coated with 2 µg/ml anti-CD3 and 2 µg/ml anti-CD28 mAbs (both LEAF purified from BioLegend) in PBS overnight at 4°C. Then, a mixture of 2 × 105 sorted naive Ly5.1+ and Ly5.2+ CD44highCD62LlowCD25−CD4+ T cells were plated in RPMI 1640 media with 10% FBS, 1× penicillin + streptomycin, 2 mM l-glutamine, 10 mM Hepes, and 50 µg/ml 2-mercaptoethanol. Additionally, 200 U/ml recombinant human IL-2 (National Cancer Institute) was added to cultures. To some cultures, 18:0 LysoPS (Avanti Polar Lipids) that was maintained as a 5 mM stock solution in cell culture grade water with 0.5% DMSO was added to wells at the indicated concentrations. Then, 3 d later, cells were harvested and Ly5-1 and Ly5-2 expression was analyzed by flow cytometry.
For T reg cell differentiation assays, 24-well plates were coated with 2 µg/ml anti-CD3 and 2 µg/ml anti-CD28 mAbs (both LEAF purified from BioLegend) in PBS overnight at 4°C. Then, 2 × 105 sorted naive CD44highCD62LlowCD25−CD4+ T cells were plated in RPMI 1640 media with 10% FBS, 1× penicillin + streptomycin, 2 mM l-glutamine, 10 mM Hepes, and 50 µg/ml 2-mercaptoethanol. Recombinant human TGF-β1 (PeproTech) was added to T reg cell–skewing cultures at a concentration of 1 ng/ml. To some cultures, 18:0 LysoPS (Avanti Polar Lipids) that was maintained as a 5 mM stock solution in cell culture–grade water with 0.5% DMSO was added to wells at the indicated concentrations. Then, 4 d later, cells were harvested and Foxp3 expression was analyzed by flow cytometry.
For Th1 and Th17 cultures, cells were sorted and cultured on plates coated with 2 µg/ml anti-CD3 and 2 µg/ml anti-CD28 mAbs as above. For Th1 cultures, 10 ng/ml IL-12 (PeproTech) and 10 µg/ml anti–IL-4 mAb were added. For Th17 cultures, 5 ng/ml TGF-β1, 10 ng/ml IL-1β, 20 ng/ml IL-6, and 10 ng/ml IL-23 (PeproTech), along with anti–IFN-γ, anti–IL-2, anti–IL-4, and anti–IL-12 mAbs (all 10 µg/ml) were added.
For the in vitro T reg cell suppressor assays, populations of CD103+ or CD103− CD4+CD25+CD45RBlow T reg cells were sorted along with congenically distinct CD4+CD25−CD44lowCD62Lhigh naive T cells. After CFSE labeling, 105 naive T cells were cultured in round-bottom 96-well plates along with T reg cells at the indicated ratios in the presence of 2 × 104 DCs (positively selected from splenocytes using anti-CD11c microbeads; Miltenyi Biotec) and 0.5 µg/ml anti-CD3ε (LEAF grade; BioLegend). After 3.5 d of culture, effector T cell proliferation was assessed by CFSE dilution using flow cytometry and gating on live CD4+CD45.1+ T cells.
Induction of EAE.
CNS inflammation was induced using a well-established model of EAE (Stromnes and Goverman, 2006). In brief, groups of 10-wk-old age- and sex-matched mice were s.c. injected in the flank with 100 mg MOG35–55 peptide (Genemed) in a 1:1 (vol/vol) water and CFA (Chondrex) emulsion using a 26 1/2–G needle. The same day and 2 d later, 200 ng pertussis toxin (List Biological Labs) in 200 µl physiological saline was injected i.v. Mice were monitored daily for signs of disease and/or weight loss. Disease severity was scored as follows: 0, normal; 1, limp tail; 2, tail paralysis and hind-limb weakness; 3, hind-limb paralysis; 4, paralysis of all limbs; and 5, moribund. If a mouse partially met the criteria for a disease score, an intermediate value was assigned to that mouse; e.g., a mouse with tail paralysis and the complete paralysis of only one hind-limb was scored as 2.5.
For Foxp3DTR/+ mice, 8-wk-old females were injected i.p. on days −10, −7, and −4 with 500 ng diphtheria toxin (List Biological Labs). Then EAE was induced as described above.
Statistical analyses.
Data were analyzed using paired or unpaired Student’s t test as appropriate. Instances for which a statistically significant difference was detected are indicated by asterisks: *, P < 0.05; **, P < 0.01; ***, P < 0.001. Prism version 5 (GraphPad Software) was used for all statistical analyses and to generate plots. Each experiment was repeated at least three times, unless otherwise indicated in the figure legends.
Acknowledgments
We would like to thank members of the Cyster laboratory for helpful discussions, Andrea Reboldi and Hayakazu Sumida for critically reading this manuscript, and Scott Zamvil for advice regarding the EAE experiments.
J.G. Cyster is an Investigator of the Howard Hughes Medical Institute. M.J. Barnes was supported by National Institutes of Health (NIH) grant T32 AI 7334-23. This work was supported in part by NIH grant AI45073.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ACN
- acetonitrile
- CNS
- central nervous system
- DP
- double positive
- DTR
- diphtheria toxin receptor
- EAE
- experimental autoimmune encephalomyelitis
- GPCR
- G protein–coupled receptor
- LysoPS
- lysophosphatidylserine
- MOG
- myelin oligodendrocyte glycoprotein
- PS
- phosphatidylserine
- SP
- single positive
- T reg cell
- regulatory T cell
References
- Bellini F., and Bruni A.. 1993. Role of a serum phospholipase A1 in the phosphatidylserine-induced T cell inhibition. FEBS Lett. 316:1–4. 10.1016/0014-5793(93)81724-E [DOI] [PubMed] [Google Scholar]
- Blankman J.L., Long J.Z., Trauger S.A., Siuzdak G., and Cravatt B.F.. 2013. ABHD12 controls brain lysophosphatidylserine pathways that are deregulated in a murine model of the neurodegenerative disease PHARC. Proc. Natl. Acad. Sci. USA. 110:1500–1505. 10.1073/pnas.1217121110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollrath J., and Powrie F.M.. 2013. Controlling the frontier: regulatory T-cells and intestinal homeostasis. Semin. Immunol. 25:352–357. 10.1016/j.smim.2013.09.002 [DOI] [PubMed] [Google Scholar]
- Campbell D.J., and Koch M.A.. 2011. Phenotypical and functional specialization of FOXP3+ regulatory T cells. Nat. Rev. Immunol. 11:119–130. 10.1038/nri2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu X., Shen M., Xie F., Miao X.J., Shou W.H., Liu L., Yang P.P., Bai Y.N., Zhang K.Y., Yang L., et al. 2013. An X chromosome-wide association analysis identifies variants in GPR174 as a risk factor for Graves’ disease. J. Med. Genet. 50:479–485. 10.1136/jmedgenet-2013-101595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuking M.B., Cahenzli J., Lawson M.A., Ng D.C., Slack E., Hapfelmeier S., McCoy K.D., and Macpherson A.J.. 2011. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 34:794–806. 10.1016/j.immuni.2011.03.021 [DOI] [PubMed] [Google Scholar]
- Grzelczyk A., and Gendaszewska-Darmach E.. 2013. Novel bioactive glycerol-based lysophospholipids: new data — new insight into their function. Biochimie. 95:667–679. 10.1016/j.biochi.2012.10.009 [DOI] [PubMed] [Google Scholar]
- Hall J.A., Grainger J.R., Spencer S.P., and Belkaid Y.. 2011. The role of retinoic acid in tolerance and immunity. Immunity. 35:13–22. 10.1016/j.immuni.2011.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huehn J., Siegmund K., Lehmann J.C., Siewert C., Haubold U., Feuerer M., Debes G.F., Lauber J., Frey O., Przybylski G.K., et al. 2004. Developmental stage, phenotype, and migration distinguish naive- and effector/memory-like CD4+ regulatory T cells. J. Exp. Med. 199:303–313. 10.1084/jem.20031562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A., Ishiguro J., Kitamura H., Arima N., Okutani M., Shuto A., Higashiyama S., Ohwada T., Arai H., Makide K., and Aoki J.. 2012. TGFα shedding assay: an accurate and versatile method for detecting GPCR activation. Nat. Methods. 9:1021–1029. 10.1038/nmeth.2172 [DOI] [PubMed] [Google Scholar]
- Josefowicz S.Z., Lu L.F., and Rudensky A.Y.. 2012. Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 30:531–564. 10.1146/annurev.immunol.25.022106.141623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamat S.S., Camara K., Parsons W.H., Chen D.H., Dix M.M., Bird T.D., Howell A.R., and Cravatt B.F.. 2015. Immunomodulatory lysophosphatidylserines are regulated by ABHD16A and ABHD12 interplay. Nat. Chem. Biol. 11:164–171. 10.1038/nchembio.1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura H., Makide K., Shuto A., Ikubo M., Inoue A., Suzuki K., Sato Y., Nakamura S., Otani Y., Ohwada T., and Aoki J.. 2012. GPR34 is a receptor for lysophosphatidylserine with a fatty acid at the sn-2 position. J. Biochem. 151:511–518. 10.1093/jb/mvs011 [DOI] [PubMed] [Google Scholar]
- Lehmann J., Huehn J., de la Rosa M., Maszyna F., Kretschmer U., Krenn V., Brunner M., Scheffold A., and Hamann A.. 2002. Expression of the integrin αEβ7 identifies unique subsets of CD25+ as well as CD25− regulatory T cells. Proc. Natl. Acad. Sci. USA. 99:13031–13036. 10.1073/pnas.192162899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebscher I., Müller U., Teupser D., Engemaier E., Engel K.M., Ritscher L., Thor D., Sangkuhl K., Ricken A., Wurm A., et al. 2011. Altered immune response in mice deficient for the G protein-coupled receptor GPR34. J. Biol. Chem. 286:2101–2110. 10.1074/jbc.M110.196659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Burns S., Huang G., Boyd K., Proia R.L., Flavell R.A., and Chi H.. 2009. The receptor S1P1 overrides regulatory T cell–mediated immune suppression through Akt-mTOR. Nat. Immunol. 10:769–777. 10.1038/ni.1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmud S.A., Manlove L.S., Schmitz H.M., Xing Y., Wang Y., Owen D.L., Schenkel J.M., Boomer J.S., Green J.M., Yagita H., et al. 2014. Costimulation via the tumor-necrosis factor receptor superfamily couples TCR signal strength to the thymic differentiation of regulatory T cells. Nat. Immunol. 15:473–481. 10.1038/ni.2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T.W., and Lagunoff D.. 1979. Interactions of lysophospholipids and mast cells. Nature. 279:250–252. 10.1038/279250a0 [DOI] [PubMed] [Google Scholar]
- McHugh R.S., Whitters M.J., Piccirillo C.A., Young D.A., Shevach E.M., Collins M., and Byrne M.C.. 2002. CD4+CD25+ immunoregulatory T cells: gene expression analysis reveals a functional role for the glucocorticoid-induced TNF receptor. Immunity. 16:311–323. 10.1016/S1074-7613(02)00280-7 [DOI] [PubMed] [Google Scholar]
- Moran A.E., Holzapfel K.L., Xing Y., Cunningham N.R., Maltzman J.S., Punt J., and Hogquist K.A.. 2011. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J. Exp. Med. 208:1279–1289. 10.1084/jem.20110308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y., Wu D., Trynka G., Raj T., Terao C., Ikari K., Kochi Y., Ohmura K., Suzuki A., Yoshida S., et al. GARNET consortium. 2014. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 506:376–381. 10.1038/nature12873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham T.H., Okada T., Matloubian M., Lo C.G., and Cyster J.G.. 2008. S1P1 receptor signaling overrides retention mediated by Gαi-coupled receptors to promote T cell egress. Immunity. 28:122–133. 10.1016/j.immuni.2007.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson W., Cauwe B., Policheni A., Schlenner S.M., Franckaert D., Berges J., Humblet-Baron S., Schönefeldt S., Herold M.J., Hildeman D., et al. 2013. Antiapoptotic Mcl-1 is critical for the survival and niche-filling capacity of Foxp3+ regulatory T cells. Nat. Immunol. 14:959–965. 10.1038/ni.2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regard J.B., Sato I.T., and Coughlin S.R.. 2008. Anatomical profiling of G protein-coupled receptor expression. Cell. 135:561–571. 10.1016/j.cell.2008.08.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Aoki J., Nagai Y., Dohmae N., Takio K., Doi T., Arai H., and Inoue K.. 1997. Serine phospholipid-specific phospholipase A that is secreted from activated platelets. A new member of the lipase family. J. Biol. Chem. 272:2192–2198. 10.1074/jbc.272.4.2192 [DOI] [PubMed] [Google Scholar]
- Smith P.M., Howitt M.R., Panikov N., Michaud M., Gallini C.A., Bohlooly-Y M., Glickman J.N., and Garrett W.S.. 2013. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 341:569–573. 10.1126/science.1241165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromnes I.M., and Goverman J.M.. 2006. Active induction of experimental allergic encephalomyelitis. Nat. Protoc. 1:1810–1819. 10.1038/nprot.2006.285 [DOI] [PubMed] [Google Scholar]
- Suffia I., Reckling S.K., Salay G., and Belkaid Y.. 2005. A role for CD103 in the retention of CD4+CD25+ Treg and control of Leishmania major infection. J. Immunol. 174:5444–5455. 10.4049/jimmunol.174.9.5444 [DOI] [PubMed] [Google Scholar]
- Sugita K., Yamamura C., Tabata K., and Fujita N.. 2013. Expression of orphan G-protein coupled receptor GPR174 in CHO cells induced morphological changes and proliferation delay via increasing intracellular cAMP. Biochem. Biophys. Res. Commun. 430:190–195. 10.1016/j.bbrc.2012.11.046 [DOI] [PubMed] [Google Scholar]
- Thomas G., Betters J.L., Lord C.C., Brown A.L., Marshall S., Ferguson D., Sawyer J., Davis M.A., Melchior J.T., Blume L.C., et al. 2013. The serine hydrolase ABHD6 is a critical regulator of the metabolic syndrome. Cell Reports. 5:508–520. 10.1016/j.celrep.2013.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorburn A.N., Macia L., and Mackay C.R.. 2014. Diet, metabolites, and “western-lifestyle” inflammatory diseases. Immunity. 40:833–842. 10.1016/j.immuni.2014.05.014 [DOI] [PubMed] [Google Scholar]
- Thornton A.M., and Shevach E.M.. 1998. CD4+CD25+ immunoregulatory T cells suppress polyclonal T cell activation in vitro by inhibiting interleukin 2 production. J. Exp. Med. 188:287–296. 10.1084/jem.188.2.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S.X., Xue L.Q., Liu W., Gu Z.H., Pan C.M., Yang S.Y., Zhan M., Wang H.N., Liang J., Gao G.Q., et al. China Consortium for the Genetics of Autoimmune Thyroid Disease. 2013. Robust evidence for five new Graves’ disease risk loci from a staged genome-wide association analysis. Hum. Mol. Genet. 22:3347–3362. 10.1093/hmg/ddt183 [DOI] [PubMed] [Google Scholar]