Abstract
Aichi virus 1 (AiV-1), belonging to the genus Kobuvirus in the family Picornaviridae, has been proposed as a causative agent of human gastroenteritis potentially transmitted by fecal-oral routes through contaminated food or water. AiV-1 is globally distributed and has been detected in various types of environmental samples, such as sewage, river water, groundwater, and shellfish. Recent environmental studies revealed that this virus could be detected in higher frequency and greater abundance than other human enteric viruses. These findings suggest that AiV-1 could potentially be an appropriate indicator of viral contamination in the environment because of its high prevalence in water environments as well as structural and genetic similarity with some of the other important enteric viruses. Further studies on the occurrence and fate of AiV-1 in environments, even in combination with clinical studies of many regions, are needed for a better understanding of their epidemiology, temporal and geographical distribution, environmental stability, and potential health risks to humans.
Keywords: Aichivirus, kobuvirus, qPCR, occurrence, wastewater treatment, surface waters
1. Introduction
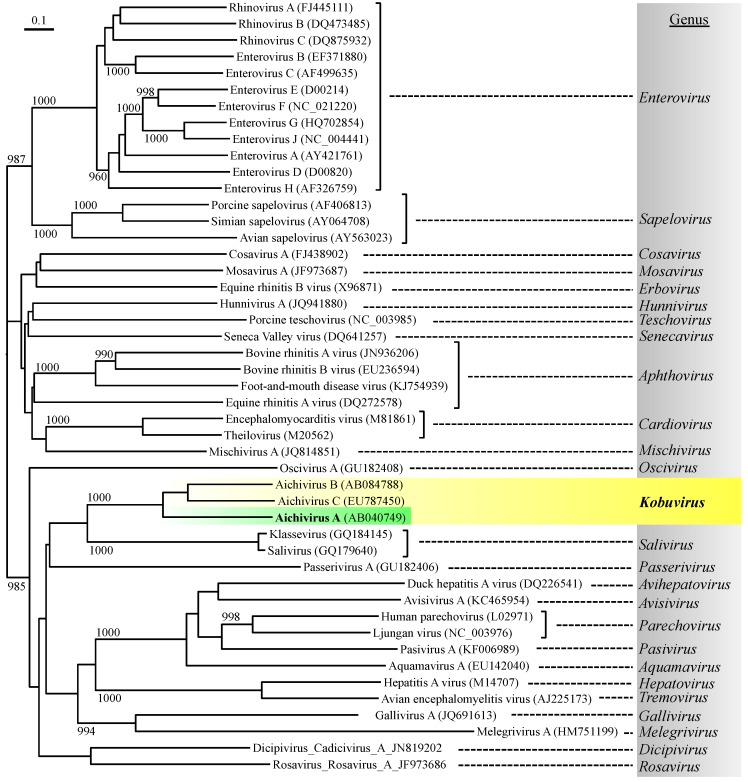
The family Picornaviridae currently consists of 26 recognized genera (International Committee on Taxonomy of Viruses, ICTV; http://www.ictvonline.org/) and contains important human enteric viruses that can cause waterborne infections to humans, such as enteroviruses and hepatitis A virus (Figure 1) [1]. Aichi virus 1 (AiV-1), a human enteric virus belonging to the genus Kobuvirus, is also a member of the family Picornaviridae [2]. The genus Kobuvirus, which is a newly recognized genus, consists of three species: Aichivirus A, Aichivirus B, and Aichivirus C, which were recently renamed and formerly called Aichi virus/Aichivirus, bovine kobuvirus, and porcine kobuviruses, respectively [3]. The species Aichivirus A consists of three genetically distinct members with different host species, namely, AiV-1 (Aichivirus in humans) [4], canine kobuvirus 1 [5], and murine kobuvirus 1 [6].
Figure 1.
Phylogenetic relationship of Aichi virus 1 with representative species from each picornavirus genus, based on amino acid similarity of the P1 (viral structural proteins) region using the neighbor-joining method. The numbers on each branch indicate the bootstrap values obtained from a bootstrap analysis with 1000 replicates, and the scale represents amino acid substitutions per site.
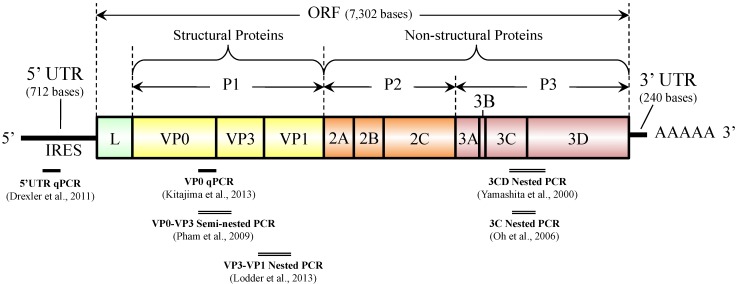
AiV-1 possesses a single-stranded, positive-sense RNA genome approximately 8.3 kb in length, which is comprised of a 5' untranslated region (UTR) with an internal ribosomal entry site (IRES), a large open reading frame (ORF) of approximately 7.3 kb encoding a single polyprotein (putative protein precursor), and a 3' UTR region [2,7,8,9]. As shown in Figure 2, a non-structural leader (L) protein is encoded at the N-terminus of the polyprotein, followed by viral structural proteins P1 (VP0, VP3, and VP1) and non-structural proteins P2 (2A, 2B, and 2C) and P3 (3A, 3B, 3C, and 3D) [2,7,8,9].
Figure 2.
Genome organization of Aichi virus 1 and target locations of diagnostic PCR assays.
The prototype AiV-1 strain A846/88 was first isolated in 1989 in Aichi prefecture, Japan, from a fecal sample of a patient with oyster-associated acute gastroenteritis [4]. AiV-1 has been divided into two genetically distinct genotypes (A and B) on the basis of nucleotide sequences of the 3C and 3D (3CD) junction region [9,10]. An AiV-1 strain classified into a distinct genetic cluster was identified in France and proposed to represent third genotype of AiV-1, genotype C [11].
AiV-1 has been detected in human fecal samples in Asian [12,13,14,15], European [11,16,17,18,19,20], South American [18], and African continents [21,22], suggesting their worldwide distribution. In addition, seroprevalence studies performed in Japan [23], Germany [18], France [24], Spain [25], and Tunisia [26] demonstrated a high prevalence of AiV-1 antibodies in adults (80%–99%), indicating widespread human exposure. In contrast, a number of clinical investigations demonstrated a low incidence of AiV-1 infection in patients with either sporadic or epidemic gastroenteritis [11,12,13,14,15,16,17,18,19,20,21,22]. It has been reported that clinical signs and symptoms of AiV-1 infection typically include diarrhea, abdominal pain, nausea, vomiting, and fever [4,27]. However, recent clinical studies have shown that AiV-1 is usually present together with other enteric viruses in fecal samples of gastroenteritis patients [11,20]. These findings suggest: (1) AiV-1 might be circulating without causing any symptoms; (2) AiV-1 could be responsible for a portion of subclinical gastroenteritis infections requiring no medical attention; and/or (3) AiV-1 may contribute to mixed viral infections leading to enteric disease.
Recently, it has been revealed that AiV-1 is frequently detected in various types of environmental samples, such as sewage, river water, groundwater, and shellfish, suggesting that this virus is an emerging viral pathogen associated with environmental contamination and potentially with water and food borne infections.
2. Detection Methods
AiV-1 was first identified as a cytopathic virus in BS-C-1 cells [4]. Since then, various methods have been used to identify AiV-1 in clinical stool specimens, such as virus isolation using BS-C-1 or Vero cells [4,14], the enzyme-linked immunosorbent assay (ELISA) [23], and conventional reverse transcription (RT)-polymerase chain reaction (PCR) [10,13,28]. Isolation of AiV-1 by cell culture is time-consuming (about 4 to 6 weeks) [23], and both virus isolation and ELISA are less sensitive than conventional RT-PCR [10]. The conventional RT-PCR assay is a widely used method for AiV-1 identification because of its high sensitivity and applicability for further genetic analysis to determine genotypes [10]. In both clinical and environmental studies, genotype of AiV-1 has been determined based on nucleotide sequence analysis of the PCR products of 3CD junction or the capsid region [9,11,12,13,15,16,18,19,22,29,30,31,32]. The 3CD junction region is conserved and suitable for the detection of a wider range of AiV-1 genotypes, but the sequence data of this region does not seem to provide sufficient sequence diversity for subtyping [33]. Meanwhile, the capsid region (VP1), coding for structural proteins that express the antigenicity of the virus, is more genetically diverse and particularly suitable for distinguishing subtypes of AiV-1 [33]. However, no phylogenetic evidence of recombination has been obtained between AiV-1 genotypes A and B in any part of the genome [33], which suggests that 3CD- and capsid-based genotyping can result in the same genotype. Nested PCR assays targeting the 3C, VP1, and VP3 regions have also been developed [18,32] (see Figure 2) and used for the detection of Aichivirus A in water [32]. Pham et al. reported a multiplex semi-nested PCR assay using genotype-specific primers that target VP0-VP3 region for differentiation of genotypes A and B [28].
Real-time quantitative PCR (qPCR) allows rapid and quantitative detection of viral genomes. It even has many other advantages over conventional PCR, including lower risk of contamination, better specificity, and capability of multiplex reaction using multiple probes labeled with different reporter dyes. RT-qPCR assays for AiV-1 targeting the highly conserved 5' untranslated region (5'-UTR), a common target region for broadly reactive assay for picornaviruses, have been developed and used for determination of viral RNA load in fecal samples [30,34]. More recently, Kitajima et al. developed an RT-qPCR system targeting VP0 region (see Figure 2), which is able to quantify AiV-1 and differentiate between genotypes A and B [35]. This system consists of two assays, an AiV-universal assay utilizing a universal primer pair, and a universal probe and a duplex genotype-specific assay utilizing the same primer pair and two genotype-specific probes. A side-by-side comparison of cycle threshold (CT) values between one of the 5'-UTR qPCR assays [30] and the VP0 qPCR system [35] in detecting cDNA of genotype A and B strains showed that CT values obtained from the two qPCR systems were comparable for both genotype A and B strains [35]. The design of the VP0 qPCR system has an advantage over the 5'-UTR qPCR, in that a single pair of primers is used not only to simplify the assay but also to enable simultaneous detection of two genotypes with a similar PCR efficiency [35]. The VP0 RT-qPCR system will be an efficient tool for routine diagnosis of AiV-1 in clinical stool specimens as well as in environmental samples, because of its ability for both quantification and genotyping of AiV-1 [35]. Molecular characterization of AiV-1 in environmental samples requires cloning of the PCR products, since usually multiple AiV-1 strains exist in an environmental sample [31]; however, these procedures are labor intensive, time consuming, and requiring expensive reagents and equipment. The VP0 RT-qPCR system, which allows rapid, sensitive, and specific detection and can be used for quantitative analysis as well as for differentiation of genotypes using multiplex qPCR format, would contribute to facilitate further investigation on the prevalence and genotype distribution of AiV-1 in human populations as well as in the environment.
Environmental detection of enteric viruses with PCR can be affected by inhibitors in environmental samples. In order to assess the RT-qPCR efficiency for the detection of AiV-1 in environmental samples, a primer sharing control (PSC) RNA for AiV-1 was developed [36]. PSC RNA is an artificial RNA that has same sequence as the target viral gene fraction except for the TaqMan probe hybridization site [37]. Therefore, both target viral RNA and PSC RNA are expected to be reverse transcribed and amplified with the same efficiency in the same reaction tube as the target gene even in cases where RT-qPCR inhibition occurs due to the presence of inhibitors present in environmental samples [37]. The AiV-PSC-RNA developed by Hata et al. is compatible with the VP0 RT-qPCR system [35]; the PSC has 108 nucleotides of a sequence identical to the partial sequence (nucleotide location 1882–1989) of the prototype AiV-1 strain A846/88 (GenBank accession number AB040749) except for the sequence AiV-specific probe hybridizes, which was replaced with a foreign genome sequence (i.e., complementary sequence of the murine norovirus-specific TaqMan probe MK-MNV-TP) [36].
3. Occurrence in the Environment
Table 1 summarizes recent reports on the occurrence of AiV-1 in the environment. AiV-1 excreted with human feces may contaminate surface waters directly or after discharge of treated or untreated sewage [31,32,36,38]. Fecal-oral AiV-1 transmission through contaminated food or water is indicated by AiV-1 detection in fecal samples of infected individuals as well as in raw and treated sewage [31,32,39], surface water [31,32,36,38], and shellfish [17,39,40]. Kitajima et al. and Lodder et al. reported the detection of both genotypes A and B in environmental samples in Japan and in the Netherlands, respectively [23,31,32], while the other environmental studies detected either genotype A or B. Genotype C has not been detected from environmental samples.
Table 1.
Detection of Aichi virus 1 (AiV-1) in water and shellfish.
| Sample | Detection Method | Positive Rate | Genotype | Concentration (copies/L) | Country | Reference | |
|---|---|---|---|---|---|---|---|
| Water | Raw sewage | Nested PCR-cloning-sequencing | 100% (12/12) | A + B | ND | Japan | [31] |
| PCR-direct sequencing | 8% (10/125) | A | ND | Tunisia | [39] | ||
| qPCR | 100% (12/12) | A | 1.4 × 105 to 2.2 × 107 | Japan | [35] | ||
| Nested PCR-cloning-sequencing | 100% (16/16) | A + B | ND | Netherlands | [32] | ||
| PCR-direct sequencing | 12.5% (6/48) | B | ND | Italy | [43] | ||
| qPCR | 100% (24/24) | AiV-1 | 1.2 × 104 to 4.0 × 106 | US | [44] | ||
| Viral metagenomics | 50% (2/4) | AiV-1 | ND | Spain, US | [45] | ||
| Viral metagenomics | 75% (3/4) | AiV-1 | ND | Nepal, Thailand, US | [46] | ||
| PCR-cloning-sequencing | 66.2% (137/207) | A | ND | Japan | [42] | ||
| Treated sewage | Nested PCR-cloning-sequencing | 92% (11/12) | A | ND | Japan | [31] | |
| PCR-direct sequencing | 4% (4/125) | A | ND | Tunisia | [39] | ||
| qPCR | 92% (11/12) | A | Up to 1.8 × 104 | Japan | [35] | ||
| qPCR | 100% (24/24) | AiV-1 | 2.0 × 103 to 4.0 × 105 | US | [44] | ||
| qPCR | 61% (61/100) | AiV-1 | Up to 103 | France | [47] | ||
| Reclaimed water | Viral metagenomics | 50% (1/2) | AiV-1 | ND | US | [48] | |
| River water | PCR-direct sequencing | 45% (5/11) | B | ND | Venezuela | [38] | |
| Nested PCR-cloning-sequencing | 60% (36/60) | A + B | ND | Japan | [31] | ||
| Nested PCR-cloning-sequencing | 85% (12/14) | A + B | ND | Netherlands | [32] | ||
| qPCR | 100% (29/29) | AiV-1 | 8.6 × 102 to 2.0 × 104 | Japan | [36] | ||
| qPCR | 11% (20/175) | ND | Up to 102 | France | [47] | ||
| Biosolids | Viral metagenomics | 100% (1/1) | ND | ND | US | [49] | |
| Viral metagenomics | 25% (3/12) | ND | ND | US | [50] | ||
| Shellfish | Clam | PCR-direct sequencing | 33% (19/57) | A | ND | Japan | [40] |
| Oyster | Nested PCR-direct sequencing | 8% (5/62) | A | ND | France | [17] | |
| Mussel, clam, cockle | Nested PCR-hybridization | 0% (0/41) | ND | ND | Spain | [51] | |
| Shellfish | PCR-direct sequencing | 6.6% (4/60) | A | ND | Tunisia | [39] | |
| Oyster, clam, cockle | qPCR | 0% (0/77) | ND | ND | Morocco | [52] | |
ND, not determined.
3.1. Sewage
The presence of viruses in sewage reflects the actual prevalence of the viruses in a community [41]. Kitajima et al. and Yamashita et al. reported that AiV-1 RNA was detected in 100% (12/12) and 66.2% (137/207), respectively, of raw sewage samples collected in Japan [31,42]. In the Netherlands, AiV-1 RNA was detected in 100% (16/16) of raw sewage samples by nested RT-PCR assays [32]. Sdiri-Lourizi et al. and Di Martino et al. also identified AiV-1 RNA in 15 of 250 (6%) raw sewage samples in Tunisia and 6 of 48 (12.5%) raw sewage samples in Italy, respectively [39,43]; the positive rate was far lower than that reported by Kitajima et al. [31] and Lodder et al. [32]. Sdiri-Lourizi et al. [39] and Di Martino et al. [43] probably underestimated the positive rate because they did not perform the second PCR that substantially increases detection sensitivity.
Kitajima et al. quantitatively detected AiV-1 RNA in raw sewage in Japan by RT-qPCR and reported its concentration ranging from 1.4 × 105 to 2.2 × 107 copies/L, which was the first report quantifying AiV-1 RNA in water [35]. Subsequently, the concentration of AiV-1 RNA in sewage in Arizona, USA was investigated using the VP0 RT-qPCR system [44]. In this study, AiV-1 was found in greater abundance without clear seasonality, and showed a lower reduction during wastewater treatment (0.94 ± 0.33 and 0.99 ± 0.12 log10 reduction by the whole wastewater treatment processes using activated sludge and trickling filter, respectively) than other human enteric viruses, such as noroviruses, sapoviruses, enteroviruses [44]. The results demonstrated the difficulty of achieving substantial physical removal of AiV-1 by the conventional wastewater treatment. Viruses discharged from wastewater treatment plants (WWTPs) can pose a risk of infection via various routes such as recreational water contact or virus accumulation in shellfish. WWTPs are one of the most effective modes of controlling circulation of viruses among human populations and aquatic environments; the more efficient removal of viruses from WWTP effluents before it is discharged into aquatic environments will directly contribute to reduction of viruses in the environment. Since recent studies demonstrated that AiV-1 showed relatively high abundance in both raw and treated wastewater [35,44], it could potentially be used as a conservative indicator of wastewater treatment system performance with respect to virus occurrence and removal.
3.2. River Water
Viral contamination of river water is of etiological significance, because river water is an important source of drinking water production for many regions of the world and often used for recreational purposes, such as swimming and bathing. AiV-1 was detected in 5 out of 11 (45%) sewage-polluted river water samples in Venezuela using the 3CD nested RT-PCR with a modified nested primer set (AiF2-AiR2b), which was the first study reporting the identification of AiV-1 in environmental water [38]. AiV-1 genotype identified in this study by direct sequencing of RT-PCR products was only genotype B. The detection and molecular characterization of AiV-1 in river water was subsequently reported from Japan, which identified AiV-1 in 36 out of 60 (60%) river water samples [31]. In this study, a total of 135 AiV-1 strains were identified from the river water by nucleotide sequencing of cloned nested RT-PCR products, of which only three clones obtained from one river sample were classified into genotype B and the rest of the strains were genotype A [31]. Interestingly, this study demonstrated more frequent detection of AiV-1 than human noroviruses and sapoviruses in the same set of river water samples [31].
3.3. Groundwater
One of our previous studies assessed the occurrence and removal of viruses at full-scale managed aquifer recharge (MAR) systems in three different regions of USA [53]. The concentration of AiV-1 in groundwater was up to 1.52 × 104 copies/L. We also found that pepper mild mottle virus (PMMoV), a plant virus, was more abundant in groundwater than AiV-1 or any other enteric viruses tested. Since this is the only study investigating the occurrence of AiV-1 in groundwater so far, the behavior of AiV-1 in soil aquifer has not been well understood and warrants further investigation.
3.4. Shellfish
AiV-1 RNA was detected in 19 out of 57 (33%) packages of Japanese clams by RT-PCR, and only genotype A strains with over 95% nucleotide homology were identified by direct sequencing of the RT-PCR products [40]. Le Guyader et al. reported that AiV-1 genotype A was detected in 5 out of 62 (8%) oyster samples in France [17]. Sdiri-Loulizi et al. also reported that AiV-1 genotype A was detected in 4 out of 60 (6.6%) shellfish in Tunisia [39]. In contrast, Vilariño et al. and Bonabbes et al. reported that AiV-1 was not detected in shellfish from Spain and Morocco, respectively [51,52].
3.5. Persistence
AiV-1 RNA has been detected by PCR-based methods, but no previous study confirmed whether the viral RNA was derived from infectious particles. Nevertheless, these results indicate a potential risk of viral infection when treated wastewater is discharged into recreational areas or used to produce reclaimed water. Therefore, our future efforts should focus on developing methodologies for eliminating AiV-1 from these aquatic environments. Fortunately, AiV-1 can be easily propagated and assayed with routine cell culture system using Vero cells [23], which allows us to evaluate the effectiveness of disinfectants such as chlorine, UV, ozone in inactivating AiV-1.
It was suggested that AiV-1 could be readily inactivated within a day by dairy manure-based composting conditions mandated by the U.S. Environmental Protection Agency for Class A biosolids [54]. A few previous studies reported that AiV-1 was completely resistant to hydrostatic pressure treatments of up to 800 MPa, exhibiting generally higher resistance than other enteric viruses and surrogate viruses, such as coxsackieviruses, human parechovirus, feline calicivirus, murine norovirus, porcine enteric calicivirus, and Tulane virus [55,56,57].
AiV-1 was reported to be stable under a wide range of pH (2 to 10) and resistant (<0.5 log10 inactivation) against alcohol treatments with up to 90% isopropanol or ethanol for up to 5 min; in contrast, heat treatment at 56°C for 20 min readily inactivated AiV-1 (>4 log10 inactivation) [57]. AiV-1 on stainless steel disks seems to be insensitive to chlorine treatment (1.3 log10 inactivation by 1000 ppm for 5 min) [57].
4. Conclusion
AiV-1 has been proposed as a causative agent of human gastroenteritis potentially transmitted by fecal-oral routes through contaminated food or water; however, the epidemiology and etiological role of AiV-1 is, to a large extent, unknown. Recent environmental studies revealed that this virus could be detected in higher frequency and greater abundance than other human enteric viruses [31,36,44]. These findings suggest that AiV-1 could potentially be an appropriate indicator of viral contamination in the environment because of their high prevalence in water environments as well as structural and genetic similarity with some of other important enteric viruses. AiV-1 is a small round virus possessing a single-stranded RNA genome and is a member of the family Picornaviridae that includes enteroviruses and hepatitis A virus; these two viruses are listed in the latest U.S. Environmental Protection Agency’s Contaminant Candidate List (CCL3), which identifies emerging contaminants of aquatic environments that may pose a public health risk [58]. Further studies on the occurrence, behavior, and genotype distribution of AiV-1 in the environment, even in combination with clinical studies, of many regions are needed for a better understanding of its epidemiology, temporal and geographical distribution, environmental stability, and potential health risks to humans.
Acknowledgments
The authors thank Eiji Haramoto at Yamanashi University for his assistance in generating the phylogenetic dendrogram of picornaviruses.
Author Contributions
M.K. and C.P.G. conceived the work and wrote the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Tapparel C., Siegrist F., Petty T.J., Kaiser L. Picornavirus and enterovirus diversity with associated human diseases. Infect. Genet. Evol. 2013;14:282–293. doi: 10.1016/j.meegid.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 2.Reuter G., Boros A., Pankovics P. Kobuviruses—A comprehensive review. Rev. Med. Virol. 2011;21:32–41. doi: 10.1002/rmv.677. [DOI] [PubMed] [Google Scholar]
- 3.Adams M.J., King A.M.Q., Carstens E.B. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2013) Arch. Virol. 2013;158:2023–2030. doi: 10.1007/s00705-013-1688-5. [DOI] [PubMed] [Google Scholar]
- 4.Yamashita T., Kobayashi S., Sakae K., Nakata S., Chiba S., Ishihara Y., Isomura S. Isolation of cytopathic small round viruses with BS-C-1 cells from patients with gastroenteritis. J. Infect. Dis. 1991;164:954–957. doi: 10.1093/infdis/164.5.954. [DOI] [PubMed] [Google Scholar]
- 5.Kapoor A., Simmonds P., Dubovi E.J., Qaisar N., Henriquez J.A., Medina J., Shields S., Lipkin W.I. Characterization of a canine homolog of human Aichivirus. J. Virol. 2011;85:11520–11525. doi: 10.1128/JVI.05317-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phan T.G., Kapusinszky B., Wang C., Rose R.K., Lipton H.L., Delwart E.L. The fecal viral flora of wild rodents. PLoS Pathog. 2011;7:e1002218. doi: 10.1371/journal.ppat.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamashita T., Sakae K., Tsuzuki H., Suzuki Y., Ishikawa N., Takeda N., Miyamura T., Yamazaki S. Complete nucleotide sequence and genetic organization of Aichi virus, a distinct member of the Picornaviridae associated with acute gastroenteritis in humans. J. Virol. 1998;72:8408–8412. doi: 10.1128/jvi.72.10.8408-8412.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sasaki J., Kusuhara Y., Maeno Y., Kobayashi N., Yamashita T., Sakae K. Construction of an infectious cDNA clone of Aichi virus (a new member of the family Picornaviridae) and mutational analysis of a stem-loop structure at the 5ʹ end of the genome. J. Virol. 2001;75:8021–8030. doi: 10.1128/JVI.75.17.8021-8030.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamashita T., Sakae K. Molecular biology and epidemiology of Aichi virus and other diarrhoeogenic enteroviruses. In: Desselberger U., Gray J., editors. Perspectives in Medical Virology. Elsevier; Amsterdam, The Netherlands: 2003. pp. 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamashita T., Sugiyama M., Tsuzuki H., Sakae K., Suzuki Y., Miyazaki Y. Application of a reverse transcription-PCR for identification and differentiation of Aichi virus, a new member of the Picornavirus family associated with gastroenteritis in humans. J. Clin. Microbiol. 2000;38:2955–2961. doi: 10.1128/jcm.38.8.2955-2961.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ambert-Balay K., Lorrot M., Bon F., Giraudon H., Kaplon J., Wolfer M., Lebon P., Gendrel D., Pothier P. Prevalence and genetic diversity of Aichi virus strains in stool samples from community and hospitalized patients. J. Clin. Microbiol. 2008;46:1252–1258. doi: 10.1128/JCM.02140-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pham N.T.K., Khamrin P., Nguyen T.A., Kanti D.S., Phan T.G., Okitsu S., Ushijima H. Isolation and molecular characterization of Aichi viruses from fecal specimens collected in Japan, Bangladesh, Thailand, and Vietnam. J. Clin. Microbiol. 2007;45:2287–2288. doi: 10.1128/JCM.00525-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pham N.T.K., Trinh Q.D., Khamrin P., Nguyen T.A., Dey S.K., Phan T.G., Hoang L.P., Maneekarn N., Okitsu S., Mizuguchi M., et al. Sequence analysis of the capsid gene of Aichi viruses detected from Japan, Bangladesh, Thailand, and Vietnam. J. Med. Virol. 2008;1227:1222–1227. doi: 10.1002/jmv.21193. [DOI] [PubMed] [Google Scholar]
- 14.Yamashita T., Sakae K., Kobayashi S., Ishihara Y., Miyake T., Mubina A., Isomura S. Isolation of cytopathic small round virus (Aichi virus) from Pakistani children and Japanese travelers from Southeast Asia. Microbiol. Immunol. 1995;39:433–435. doi: 10.1111/j.1348-0421.1995.tb02225.x. [DOI] [PubMed] [Google Scholar]
- 15.Yang S., Zhang W., Shen Q., Yang Z., Zhu J., Cui L., Hua X. Aichi virus strains in children with gastroenteritis, China. Emerg. Infect. Dis. 2009;15:1703–1705. doi: 10.3201/eid1510.090522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaikkonen S., Räsänen S., Rämet M. Aichi virus infection in children with acute gastroenteritis in Finland. Epidemiol. Infect. 2010;138:1166–1171. doi: 10.1017/S0950268809991300. [DOI] [PubMed] [Google Scholar]
- 17.Le Guyader F.S., le Saux J.-C., Ambert-Balay K., Krol J., Serais O., Parnaudeau S., Giraudon H., Delmas G., Pommepuy M., Pothier P., et al. Aichi virus, norovirus, astrovirus, enterovirus, and rotavirus involved in clinical cases from a French oyster-related gastroenteritis outbreak. J. Clin. Microbiol. 2008;46:4011–4017. doi: 10.1128/JCM.01044-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh D.-Y., Silva P.A., Hauroeder B., Diedrich S., Cardoso D.D.P., Schreier E. Molecular characterization of the first Aichi viruses isolated in Europe and in South America. Arch. Virol. 2006;151:1199–1206. doi: 10.1007/s00705-005-0706-7. [DOI] [PubMed] [Google Scholar]
- 19.Reuter G., Boldizsár A., Papp G., Pankovics P. Detection of Aichi virus shedding in a child with enteric and extraintestinal symptoms in Hungary. Arch. Virol. 2009;154:1529–1532. doi: 10.1007/s00705-009-0473-y. [DOI] [PubMed] [Google Scholar]
- 20.Räsänen S., Lappalainen S., Kaikkonen S., Hämäläinen M., Salminen M., Vesikari T. Mixed viral infections causing acute gastroenteritis in children in a waterborne outbreak. Epidemiol. Infect. 2010;138:1227–1234. doi: 10.1017/S0950268809991671. [DOI] [PubMed] [Google Scholar]
- 21.Sdiri-Loulizi K., Gharbi-Khélifi H., de Rougemont A., Chouchane S., Sakly N., Ambert-Balay K., Hassine M., Guédiche M.N., Aouni M., Pothier P. Acute Infantile Gastroenteritis Associated with Human Enteric Viruses in Tunisia. J. Clin. Microbiol. 2008;46:1349–1355. doi: 10.1128/JCM.02438-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sdiri-Loulizi K., Hassine M., Gharbi-Khelifi H., Sakly N., Chouchane S., Guediche M.N., Pothier P., Aouni M., Ambert-Balay K. Detection and Genomic Characterization of Aichi Viruses in Stool Samples from Children in Monastir, Tunisia. J. Clin. Microbiol. 2009;47:2275–2278. doi: 10.1128/JCM.00913-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamashita T., Sakae K., Ishihara Y., Isomura S., Utagawa E. Prevalence of newly isolated, cytopathic small round virus (Aichi strain) in Japan. J. Clin. Microbiol. 1993;31:2938–2943. doi: 10.1128/jcm.31.11.2938-2943.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goyer M., Aho L.-S., Bour J.-B., Ambert-Balay K., Pothier P. Seroprevalence distribution of Aichi virus among a French population in 2006–2007. Arch. Virol. 2008;153:1171–1174. doi: 10.1007/s00705-008-0091-0. [DOI] [PubMed] [Google Scholar]
- 25.Ribes J.M., Montava R., Téllez-Castillo C.J., Fernández-Jiménez M., Buesa J. Seroprevalence of Aichi Virus in a Spanish Population from 2007 to 2008. Clin. Vaccine Immunol. 2010;17:545–549. doi: 10.1128/CVI.00382-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sdiri-Loulizi K., Hassine M., Bour J.-B., Ambert-Balay K., Mastouri M., Aho L.-S., Gharbi-Khelifi H., Aouni Z., Sakly N., Chouchane S., et al. Aichi virus IgG seroprevalence in Tunisia parallels genomic detection and clinical presentation in children with gastroenteritis. Clin. Vaccine Immunol. 2010;17:1111–1116. doi: 10.1128/CVI.00059-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamashita T., Ito M., Tsuzuki H. Identification of Aichi virus infection by measurement of immunoglobulin responses in an enzyme-linked immunosorbent assay. J. Clin. Microbiol. 2001;39:4178–4180. doi: 10.1128/JCM.39.11.4178-4180.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pham N.T.K., Trinh Q.D., Nguyen T.A., Dey S.K., Phan T.G., Hoang L.P., Khamrin P., Maneekarn N., Okitsu S., Mizuguchi M., et al. Development of genotype-specific primers for differentiation of genotypes A and B of Aichi viruses. J. Virol. Methods. 2009;156:107–110. doi: 10.1016/j.jviromet.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 29.Verma H., Chitambar S.D., Gopalkrishna V. Circulation of Aichi virus genotype B strains in children with acute gastroenteritis in India. Epidemiol. Infect. 2011;139:1687–1691. doi: 10.1017/S0950268810002979. [DOI] [PubMed] [Google Scholar]
- 30.Drexler J.F., Baumgarte S., de Souza Luna L.K., Eschbach-Bludau M., Lukashev A.N., Drosten C. Aichi virus shedding in high concentrations in patients with acute diarrhea. Emerg. Infect. Dis. 2011;17:1544–1548. doi: 10.3201/eid1708.101556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitajima M., Haramoto E., Phanuwan C., Katayama H. Prevalence and genetic diversity of Aichi viruses in wastewater and river water in Japan. Appl. Environ. Microbiol. 2011;77:2184–2187. doi: 10.1128/AEM.02328-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lodder W.J., Rutjes S.A., Takumi K., de Roda Husman A.M. Aichi virus in sewage and surface water, the Netherlands. Emerg. Infect. Dis. 2013;19:1222–1230. doi: 10.3201/eid1908.130312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lukashev A.N., Drexler J.F., Belalov I.S., Eschbach-Bludau M., Baumgarte S., Drosten C. Genetic variation and recombination in Aichi virus. J. Gen. Virol. 2012;93:1226–1235. doi: 10.1099/vir.0.040311-0. [DOI] [PubMed] [Google Scholar]
- 34.Nielsen A.C.Y., Gyhrs M.L., Nielsen L.P., Pedersen C., Böttiger B. Gastroenteritis and the novel picornaviruses aichi virus, cosavirus, saffold virus, and salivirus in young children. J. Clin. Virol. 2013;57:239–242. doi: 10.1016/j.jcv.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 35.Kitajima M., Hata A., Yamashita T., Haramoto E., Minagawa H., Katayama H. Development of a reverse transcription-quantitative PCR system for detection and genotyping of Aichi viruses in clinical and environmental samples. Appl. Environ. Microbiol. 2013;79:3952–3958. doi: 10.1128/AEM.00820-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hata A., Katayama H., Kojima K., Sano S., Kasuga I., Kitajima M., Furumai H. Effects of rainfall events on the occurrence and detection efficiency of viruses in river water impacted by combined sewer overflows. Sci. Total Environ. 2014;468:757–763. doi: 10.1016/j.scitotenv.2013.08.093. [DOI] [PubMed] [Google Scholar]
- 37.Hata A., Katayama H., Kitajima M., Visvanathan C., Nol C., Furumai H. Validation of internal controls for extraction and amplification of nucleic acids from enteric viruses in water samples. Appl. Environ. Microbiol. 2011;77:4336–4343. doi: 10.1128/AEM.00077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alcalá A., Vizzi E., Rodríguez-Díaz J., Zambrano J.L., Betancourt W., Liprandi F. Molecular detection and characterization of Aichi viruses in sewage-polluted waters of Venezuela. Appl. Environ. Microbiol. 2010;76:4113–4115. doi: 10.1128/AEM.00501-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sdiri-Loulizi K., Hassine M., Aouni Z., Gharbi-Khelifi H., Sakly N., Chouchane S., Guédiche M.N., Pothier P., Aouni M., Ambert-Balay K. First molecular detection of Aichi virus in sewage and shellfish samples in the Monastir region of Tunisia. Arch. Virol. 2010;155:1509–1513. doi: 10.1007/s00705-010-0744-7. [DOI] [PubMed] [Google Scholar]
- 40.Hansman G.S., Oka T., Li T.-C., Nishio O., Noda M., Takeda N. Detection of human enteric viruses in Japanese clams. J. Food Prot. 2008;71:1689–1695. doi: 10.4315/0362-028x-71.8.1689. [DOI] [PubMed] [Google Scholar]
- 41.Sinclair R.G., Choi C.Y., Riley M.R., Gerba C.P. Pathogen surveillance through monitoring of sewer systems. Adv. Appl. Microbiol. 2008;65:249–269. doi: 10.1016/S0065-2164(08)00609-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamashita T., Adachi H., Hirose E., Nakamura N., Ito M., Yasui Y., Kobayashi S., Minagawa H. Molecular detection and nucleotide sequence analysis of a new Aichi virus closely related to canine kobuvirus in sewage samples. J. Med. Microbiol. 2014;63:715–720. doi: 10.1099/jmm.0.070987-0. [DOI] [PubMed] [Google Scholar]
- 43.Di Martino B., di Profio F., Ceci C., di Felice E., Marsilio F. Molecular detection of Aichi virus in raw sewage in Italy. Arch. Virol. 2013;158:2001–2005. doi: 10.1007/s00705-013-1694-7. [DOI] [PubMed] [Google Scholar]
- 44.Kitajima M., Iker B.C., Pepper I.L., Gerba C.P. Relative abundance and treatment reduction of viruses during wastewater treatment processes—Identification of potential viral indicators. Sci. Total Environ. 2014;488–489:290–296. doi: 10.1016/j.scitotenv.2014.04.087. [DOI] [PubMed] [Google Scholar]
- 45.Cantalupo P.G., Calgua B., Zhao G. Raw sewage harbors diverse viral populations. MBio. 2011;2:e00180-11. doi: 10.1128/mBio.00180-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ng T.F.F., Marine R., Wang C., Simmonds P., Kapusinszky B., Bodhidatta L., Oderinde B.S., Wommack K.E., Delwart E. High variety of known and new RNA and DNA viruses of diverse origins in untreated sewage. J. Virol. 2012;86:12161–12175. doi: 10.1128/JVI.00869-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prevost B., Lucas F.S., Goncalves A., Richard F., Moulin L., Wurtzer S. Large scale survey of enteric viruses in river and waste water underlines the health status of the local population. Environ. Int. 2015;79:42–50. doi: 10.1016/j.envint.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 48.Rosario K., Nilsson C., Lim Y.W., Ruan Y., Breitbart M. Metagenomic analysis of viruses in reclaimed water. Environ. Microbiol. 2009;11:2806–2820. doi: 10.1111/j.1462-2920.2009.01964.x. [DOI] [PubMed] [Google Scholar]
- 49.Bibby K., Viau E., Peccia J. Viral metagenome analysis to guide human pathogen monitoring in environmental samples. Lett. Appl. Microbiol. 2011;52:386–392. doi: 10.1111/j.1472-765X.2011.03014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bibby K., Peccia J. Identification of viral pathogen diversity in sewage sludge by metagenome analysis. Environ. Sci. Technol. 2013;47:1945–1951. doi: 10.1021/es305181x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vilariño M.L., le Guyader F.S., Polo D., Schaeffer J., Kröl J., Romalde J.L. Assessment of human enteric viruses in cultured and wild bivalve molluscs. Int. Microbiol. 2009;12:145–151. [PubMed] [Google Scholar]
- 52.Benabbes L., Ollivier J., Schaeffer J., Parnaudeau S., Rhaissi H., Nourlil J., le Guyader F.S. Norovirus and other human enteric viruses in moroccan shellfish. Food Environ. Virol. 2013;5:35–40. doi: 10.1007/s12560-012-9095-8. [DOI] [PubMed] [Google Scholar]
- 53.Betancourt W.Q., Kitajima M., Wing A.D., Regnery J., Drewes J.E., Pepper I.L., Gerba C.P. Assessment of virus removal by managed aquifer recharge at three full-scale operations. J. Environ. Sci. Heal. Part A. 2014;49:1685–1692. doi: 10.1080/10934529.2014.951233. [DOI] [PubMed] [Google Scholar]
- 54.Wei J., Jin Y., Sims J.T., Kniel K.E. Fate of human enteric viruses during dairy manure-based composting. J. Food Prot. 2010;73:1543–1547. doi: 10.4315/0362-028x-73.8.1543. [DOI] [PubMed] [Google Scholar]
- 55.Kingsley D.H., Chen H., Hoover D.G. Inactivation of selected picornaviruses by high hydrostatic pressure. Virus Res. 2004;102:221–224. doi: 10.1016/j.virusres.2004.01.030. [DOI] [PubMed] [Google Scholar]
- 56.Kingsley D.H., Li X., Chen H. Temperature effects for high-pressure processing of Picornaviruses. Food Environ. Virol. 2014;6:58–61. doi: 10.1007/s12560-013-9131-3. [DOI] [PubMed] [Google Scholar]
- 57.Cromeans T., Park G.W., Costantini V., Lee D., Wang Q., Farkas T., Lee A., Vinjé J. Comprehensive comparison of cultivable norovirus surrogates in response to different inactivation and disinfection treatments. Appl. Environ. Microbiol. 2014;80:5743–5751. doi: 10.1128/AEM.01532-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.U.S. Environmental Protection Agency . Federal Register. U.S. Environmental Protection Agency; Washington, DC, USA: 2009. pp. 51850–51862. [Google Scholar]