Abstract
Marine hydrocarbon-degrading bacteria perform a fundamental role in the biodegradation of crude oil and its petrochemical derivatives in coastal and open ocean environments. However, there is a paucity of knowledge on the diversity and function of these organisms in deep-sea sediment. Here we used stable-isotope probing (SIP), a valuable tool to link the phylogeny and function of targeted microbial groups, to investigate polycyclic aromatic hydrocarbon (PAH)-degrading bacteria under aerobic conditions in sediments from Guaymas Basin with uniformly labeled [13C]-phenanthrene (PHE). The dominant sequences in clone libraries constructed from 13C-enriched bacterial DNA (from PHE enrichments) were identified to belong to the genus Cycloclasticus. We used quantitative PCR primers targeting the 16S rRNA gene of the SIP-identified Cycloclasticus to determine their abundance in sediment incubations amended with unlabeled PHE and showed substantial increases in gene abundance during the experiments. We also isolated a strain, BG-2, representing the SIP-identified Cycloclasticus sequence (99.9% 16S rRNA gene sequence identity), and used this strain to provide direct evidence of PHE degradation and mineralization. In addition, we isolated Halomonas, Thalassospira, and Lutibacterium sp. with demonstrable PHE-degrading capacity from Guaymas Basin sediment. This study demonstrates the value of coupling SIP with cultivation methods to identify and expand on the known diversity of PAH-degrading bacteria in the deep-sea.
Keywords: Guaymas Basin, hydrocarbon degradation, stable isotope probing, polycyclic aromatic hydrocarbons (PAHs), Cycloclasticus, deep-sea, marine environment
Introduction
In deep-sea naturally oil-laden marine sediments, such as cold seeps, hydrocarbon-degrading microorganisms contribute importantly to the diagenesis, and biological transformation of hydrocarbons. Since microorganisms, in particular oil-degrading bacteria, are the foundation of natural bioremediation processes and protagonists in the removal of hydrocarbon contaminants (Head et al., 2006; Yakimov et al., 2007), identifying these types of bacteria in the deep-sea is a first step to understanding their role in the mineralization of hydrocarbons in these environments and how they would respond to oil from the overlying water column. As evidenced during the Deepwater Horizon blowout, oil in surface waters of the Gulf of Mexico reached the seafloor through the formation and subsequent vertical sedimentation of marine oil snow (MOS; Chanton et al., 2015), which is oil-enriched mucilaginous particulates that had formed within 1–2 weeks of the blowout (Passow et al., 2012). This process can introduce large quantities of oil to the seafloor where it can have acute and lasting impacts to benthic ecosystems. Microbial degradation of hydrocarbons in deep petroleum reservoirs is well-documented (Orphan et al., 2000; Aitken et al., 2004; Jones et al., 2008; Bennett et al., 2013), and hydrocarbon-degrading bacteria in coastal environments have been investigated extensively (Yakimov et al., 2007). However, the deep-sea has received considerably less attention in this respect since the first published reports on hydrocarbon biodegradation in the deep-sea in the 1970s (Schwarz et al., 1974a,b). With exploration and production for oil in deeper water provinces having accelerated in recent years, there is a need to improve our understanding of the diversity and catabolic potential of oil-degrading bacteria in deep-sea sediments.
A model system for studying the diversity and evolution of hydrocarbon-degrading bacteria in the deep-sea exists in the Guaymas Basin (Teske et al., 2014). Located at approximately 2000 m water depth on the seabed in the Gulf of California, this submarine spreading center is characterized by hydrocarbon seeps, hydrothermal plumes, and hot springs. The high temperatures (up to 200–300°C) in sub-surface Guaymas sediments lead to the pyrolysis of organic material in these organic-rich sediments (3–12% [wt/wt] near the sediment surface; Lanza-Espino and Soto, 1999), as well as to the production of significant quantities of petroleum hydrocarbons in the deep subsurface sediments (Peter et al., 1991). The vent fluids, which are laden with petrochemicals, migrate upward to the sediment surface, thus providing a natural model system for studying the microbiology of deep-sea hydrocarbon-degrading communities. These communities are largely responsible for recycling of the hydrocarbons in highly active sediments at Guaymas (Pearson et al., 2005).
Stable-isotope probing (SIP) has been used successfully on environmental samples to identify a microbial group(s) of interest based on their ability to assimilate the stable isotope, thereby being able to link the phylogenetic identity of an organism to its function (Dumont and Murrell, 2005). The technique therefore has promise to explore the diversity of hydrocarbon-degrading bacteria in natural environments, and to link this phenotype to phylogenetic identity. Few reports have, however, applied SIP with hydrocarbon substrates to the study of hydrocarbon-degraders in ocean systems (Gutierrez et al., 2011, 2013b; Mishamandani et al., 2014; Sauret et al., 2014). Recently, the application of SIP has uncovered sulfate-reducing bacteria of the Desulfosarcina/Desulfococcus clade as key players in the anaerobic degradation of alkane hydrocarbons at deep marine seeps, including the Guaymas Basin (Kleindienst et al., 2014). Here, we studied PAH-degrading bacteria in sediment cores collected from the Guaymas Basin – a model system for studying the diversity of these organisms in the deep-sea (Teske et al., 2014) and where aromatic/naphthenic hydrocarbon levels (including PAHs) can constitute up to 30% of the oil (Bazylinski et al., 1988). For this, DNA-SIP and cultivation-based methods were used to identify PAH-degrading bacteria in the surficial sediment environment of Guaymas at ~2000 m depth below the sea surface in order to expand current knowledge on the diversity of hydrocarbon-degrading microbial communities in Guaymas Basin oil-rich sediments.
Materials and Methods
Field Samples
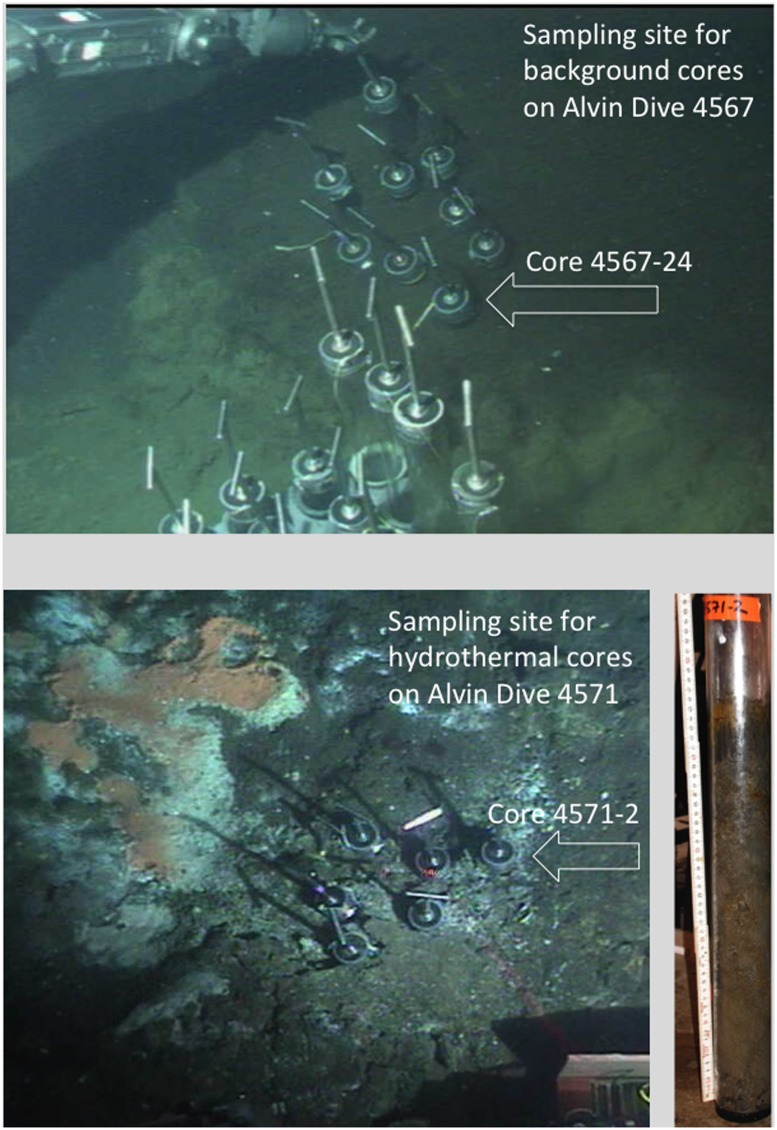
Samples were collected in 2009 on R/V Atlantis cruise AT15-56 by push coring with the submersible Alvin. Core 4567-24 was collected on November 28, 2009 from cold non-hydrothermal sediment with an in situ temperature of +4 to 5°C throughout the sediment core, no free sulfide, and no overlying bacterial mat, at a water depth of 2011 m at 27°0.542′N, 111°24.488′W. Core 4571-2 was collected on December 2, 2009 from a site with oil-rich sediments next to a well-developed Beggiatoa mat, at a water depth of 2007 m at 27°0.388′N, 111°24.560′W (Figure 1). This core was characterized by high porewater sulfide concentrations in the range of 2–4.5 mM, and an in situ temperature gradient of ca. 10°C at the surface to near 50°C at 40 cm depth, as measured with Alvin’s Heatflow probe (McKay et al., 2012). Cores were brought to the surface, immediately transferred and kept in a cold room +4°C, then sectioned by depth. At the time of collection, aliquots of the core samples 0–4 cm below seafloor were stored in sterile Falcon tubes and kept at +4°C for subsequent use within 2 weeks in enrichment, mineralization, degradation, and SIP experiments (described below).
FIGURE 1.
In situ still photographs of sampling sites during Alvin dives 4567 and 4571, obtained with the Alvin frame-grabber system (http://4dgeo.whoi.edu/alvin). The (Top) image shows the benthic sediment without microbial mats or hydrothermal features sampled during dive 4567; the (Bottom) image shows the microbial mats and sulfur precipitates on the sediment surface that reveal hydrothermal influence. Right, photo of core 4571-2 after shipboard retrieval; the reddish spots are oil droplets in the sediment core.
SIP Incubations
Prior to commencing SIP incubations, mineralization assays using 14C-labeled naphthalene (NAP), phenanthrene (PHE), anthracene (ANT), pyrene (PYR), fluoranthene (FLU), and benz[a]anthracene (BaA) were performed on the collected sediment samples to determine which of these hydrocarbons would warrant SIP with their 13C-labeled counterparts. [9-14C]-PHE (8.3 mCi mmol-1), [1, 2, 3, 4, 4a, 9a-14C]-ANT (17.3 mCi mmol-1), [4, 5, 9, 10-14C]-PYR (61 mCi mmol-1), [3-14C]-FLU (45 mCi mmol-1), and [U-14C]-NAP (17.8 mCi mmol-1) were from Sigma–Aldrich (St Louis, MO, USA). [5, 6-14C]-BaA (54.6 mCi mmol-1) was obtained from Chemsyn Science Laboratories (Lenexa, KS, USA). Mineralization assays were conducted in sterile 40-ml amber-glass EPA vials, each containing a 14C-labeled test compound (to 20,000 d.p.m., except for PYR which was added to 3,000 d.p.m.) and 2.5 mg of the respective unlabeled test compound in 4.5 ml of ONR7a medium (Dyksterhouse et al., 1995). For the inoculum, 0.5 g of wet sediment sample was inoculated into the vials. Killed controls were prepared by adding 85% phosphoric acid to pH of ≤1 prior to inoculation. All treatments were conducted in triplicate. For the CO2 trap, a sterile glass test tube (12 mm × 75 mm) containing a piece of filter paper saturated with 60 ml of 2 M KOH was inserted into each vial. The vials were sealed with foil-covered Teflon-lined caps and incubated with shaking (100 r.p.m.) at 4°C or 21°C in order to determine which incubation temperature would be most suitable for SIP. The filter paper from each vial was removed daily and the captured 14C from any 14CO2 respired was counted on a Packard (Meriden, CT, USA) Tri-Carb liquid scintillation analyzer (model 1900TR). The KOH-saturated filter paper from each vial was replaced at each sampling point for the course of the experiment. The percentage of 14C mineralized for each compound was calculated by subtracting the triplicate values for the acidified controls from those of the experimental and then dividing by the total d.p.m. of 14C added.
Stable-isotope probing incubations were performed using 125-ml sterilized glass screw-top Erlenmeyer flasks with caps that were lined with aluminum foil to prevent sorption of hydrocarbons. Each flask contained 18 ml of ONR7a medium and 2 g of sediment slurry from core 4567-24. [U-13C]-PHE was synthesized by methods described elsewhere (Zhang et al., 2011). Five sets of replicate flasks were prepared and run in parallel. Duplicate flasks were prepared with 1 mg of [U-13C]-PHE for the SIP incubation, and a second set of duplicates was prepared with 1 mg of unlabeled PHE. To determine the endpoint of each SIP experiment, the mineralization of [U-14C]-PHE was measured in triplicate flasks by liquid scintillation counting of 14CO2 trapped in KOH-soaked filter paper over time, as described above. An additional set of triplicate flasks was used to monitor the disappearance of the unlabeled PHE by HPLC; samples were periodically taken from these flasks for DNA extraction and subsequent measurement of the abundance of target organisms identified through SIP. Triplicate flasks of acid-inhibited controls (pH ≤ 2) containing unlabeled PHE were prepared by adding ca. 0.7 ml of 85% phosphoric acid. All flasks were incubated on an orbital shaker (250 r.p.m.; 21°C) in the dark. At the endpoint of each SIP incubation – defined as the time when the extent of mineralization of the 14C-labeled substrate began to approach an asymptote – whole DNA from the total volume in the duplicate flasks amended with the [U-13C]-PHE and the corresponding duplicate set with unlabeled PHE was extracted as previously described (Tillett and Neilan, 2000).
Caesium Chloride (CsCl) Gradient Ultracentrifugation and Identification of 13C-Enriched DNA
To separate 13C-enriched and unenriched DNA, total extracted DNA from each sample was added to caesium chloride (CsCl) solutions (1.72 g ml-1) for isopycnic ultracentrifugation and gradient fractionation, as previously described (Jones et al., 2011). Five microliters of purified Escherichia coli DNA (ca. 40 ng ml-1) was added and mixed into each tube as an internal standard of unlabeled DNA prior to ultracentrifugation. Denaturing gradient gel electrophoresis (DGGE) was then performed on each fraction to visualize the separation of DNA. For this, PCR amplification of each fraction was carried out with primers 63f-GC (Marchesi et al., 1998) and 517r (Muyzer et al., 1993) using a PCR program as described by Yu and Morrison (2004). PCR products were confirmed on a 1.5% (w/v) agarose gel alongside a HindIII DNA ladder (Invitrogen, Carlsbad, CA, USA). DGGE was performed using 6.5% acrylamide gels containing a denaturant range of 30–60% (100% denaturant contains 7.0 M urea and 40% molecular-grade formamide). After electrophoresis for 16 h at 60°C and 60 V, gels were stained with ethidium bromide at 1:25 000 dilutions for 15 min. Gel image colors were inverted, adjusted for contrast, and cropped to only the regions displaying bands with the GNU Image Manipulation Program (GIMP; version 2.6.8).
16S rRNA Gene Libraries of 13C-Enriched DNA
To identify PHE-degrading bacteria, a 16S rRNA gene clone library comprising 96 clones was prepared from the 13C-enriched DNA fractions (Singleton et al., 2006) using general bacterial primers 27f and 1492r for PCR amplification, followed by partial sequencing with primer 27f (Wilmotte et al., 1993) at the Beckman Coulter Genomics sequencing facility (Danvers, MA, USA). The 13C-enriched heavy DNA fractions were selected based on the DGGE evidence, which is discussed below. After excluding vector sequences, poor-quality reads and chimeras, the clone sequences were grouped into operational taxonomic units (OTUs) based on applying a 97% sequence identity cutoff. Using the complete linkage clustering and dereplicate tools available at the Pyrosequencing Pipeline tool of RDP-II (Cole et al., 2009), representative sequences were selected to represent dominant OTUs identified in each of the libraries. Near-complete 16S rRNA gene sequences for the represented sequences were obtained at the University of North Carolina at Chapel Hill Genome Analysis Facility. Sequences were edited and assembled using the program Sequencher 4.8 (Gene Codes Corp., Ann Arbor, MI, USA). The BLASTN search program and RDP-II (Maidak et al., 1999) were used to check for close relatives and phylogenetic affiliation.
Real-Time Quantitative PCR
To quantify genes of the most dominant OTU, primers for real-time quantitative PCR (qPCR) were developed using the Probe Design and Probe Match tools of ARB, as previously described (Gutierrez et al., 2011). Primer specificity was confirmed with the Probe Check tool of RDP-II. The optimal annealing temperature of each primer pair was determined using an Eppendorf (Hauppauge, NY, USA) Mastercycler gradient thermal cycler. The template for these reactions, and for the construction of respective standard curves for quantitative PCR, was a plasmid containing a representative sequence that had been linearized using PstI (New England BioLabs, Ipswich, MA, USA) and purified using the QIAquick nucleotide removal kit (Qiagen, Valencia, CA, USA). The qPCR primer pairs, their amplification efficiency (Pfaffl, 2001), optimal annealing temperature, detection limit and RDP hits are shown in Table 1. To confirm the fractions from the DGGE profiles that corresponded to unlabeled DNA, the abundance of the E. coli 16S rRNA genes was quantified in each fraction using E. coli primers ECP79f (5′-GAAGCTTGCTTCTTTGCT-3′) and ECR620r (5′-GAGCCCGGGGATTTCACA-3′) and a qPCR program with an annealing temperature of 55°C and an extended extension step of 45 s (Sabat et al., 2000).
Table 1.
Quantitative PCR primers developed and used in this study.
| Target OTU | Primer name | Primer sequence (5′→3′) | Tm (°C)a | qPCR standardb | Amplicon length | Amplification efficiencyc | Detection limitd | RDP hitse |
|---|---|---|---|---|---|---|---|---|
| 1 | Cyc-467f | AACCTTAGGCCCTGACGT | 57 | Phenanthrene 1 | 128 | 1.68 | 21 | 81 |
| Cyc-577r | TGTTTAACCGCCTACGCG | 83 (68) |
aEmpirically determined PCR annealing temperature.
bRepresentative clone sequence from which plasmid DNA was used to generate a standard curve. The plasmid was linearized with PstI.
cAmplification efficiency (Pfaffl, 2001) with operational taxonomic unit (OTU)-specific primers.
dDetection limit of the qPCR assay expressed as number of 16S rRNA gene copies per milliliter of culture.
eNumber of sequences returned by the Ribosomal Database Project II release 10.18 (Cole et al., 2009; excluding sequences from this study) with no mismatches to primer pairs. Value in parentheses is the total hits that the primer pair targets.
Purified DNA from time-series incubations with unlabeled hydrocarbon was quantified using a NanoDrop ND-3300 fluorospectrometer (Thermo, Waltham, MA, USA) and the Quant-iT Picogreen double-stranded DNA (dsDNA) kit (Invitrogen). As duplicates of the separated 12C- and 13C-labeled incubations for each of the three SIP incubations displayed similar distributions of DNA in the fractions, as well as similar DGGE profiles, only the replicate incubation whose fractions contained the highest total amount of DNA was used for further analyses. SIP-identified sequences were quantified in each separated SIP fraction using triplicate reactions by qPCR, as described previously (Singleton et al., 2006). Single reactions were performed on DNA extracted from each of the triplicate samples from the time series incubations containing unlabeled hydrocarbon.
Isolation of Phenanthrene-Degrading Strains and their Mineralization of 14C Phenanthrene
The oil-contaminated surface (0–4 cm) core samples (4571-2 and 4567-24) from the Guaymas Basin were used to isolate bacteria capable of degrading polycyclic aromatic hydrocarbons (PAHs). For this, PHE was used as a representative growth substrate for PAH-degrading organisms. Samples (5 μl) of sediment were streaked directly onto ONR7a agar plates that were then sprayed with PHE dissolved in acetone (ca. 5% w/v) as the sole source of carbon and energy (Kiyohara et al., 1982). The acetone volatilizes off immediately in the process of spraying, leaving behind a thin layer of the PHE on the agar surface. Agar plates were stored in closed plastic bags in the dark at room temperature for up to 4 weeks. Colonies forming clearing zones were picked and subcultured onto fresh ONR7a agar medium amended with the PHE until pure cultures were obtained prior to storage in glycerol (30% v/v) at -80°C.
The potential of the strains to mineralize 14C-labeled PHE was determined as described above. For preparation of inocula for these experiments, each strain was grown in ONR7a liquid medium amended with Na-pyruvate (0.1% w/v) and the cell biomass washed several times with fresh ONR7a prior to use.
16S rRNA Gene Sequencing and Phylogenetic Analysis
Total genomic DNA of isolated strains was recovered using the method of Tillett and Neilan (2000). The 16S rRNA genes were amplified by PCR with primers 27f (Wilmotte et al., 1993) and 1492r (Lane, 1991). The resulting product was then cloned into the plasmid PCR4-TOPO using the TOPO-TA cloning kit for sequencing (Invitrogen, Carlsbad, CA, USA). The insert was sequenced with primers M13f, M13r, 338f, and 338r (Amann et al., 1990); 907f (Lane et al., 1985); and 907r (Wilmotte et al., 1993) at the University of North Carolina Genome Analysis Facility. Sequences were assembled using the program Sequencher 4.8 (Gene Codes Corp., Ann Arbor, MI, USA). The consensus sequence was submitted to GenBank and checked for close relatives and phylogenetic affiliation using the BLAST search program and RDP-II (Maidak et al., 1999). The search results were used as a guide for tree construction. Additional related 16S rRNA sequences identified from the BLASTN and RDP-II search were retrieved from GenBank.
The 16S rRNA sequences of the isolated strains and SIP-identified sequence were aligned using CLUSTAL_X (Thompson et al., 1994) with the identified close relatives. A neighbor-joining tree was constructed with bootstrapped replication (1000 times) and Zymobacter palmae (D14555) was used as an outgroup.
Nucleotide Sequence Accession Numbers
The 16S rRNA gene sequence of Cycloclasticus SIP clone PHE1 and the isolated strains Cycloclasticus sp. strain BG-2, Halomonas sp. strain BG-3a, Thalassospira sp. strain BG-3b, and Lutibacterium sp. strain BG-4 were deposited with GenBank under accession numbers KF875697, KF875699, KM404161, KM404162, and KM404163, respectively.
Results and Discussion
Exposure of Sediment Samples to Labeled and Unlabeled PAHs
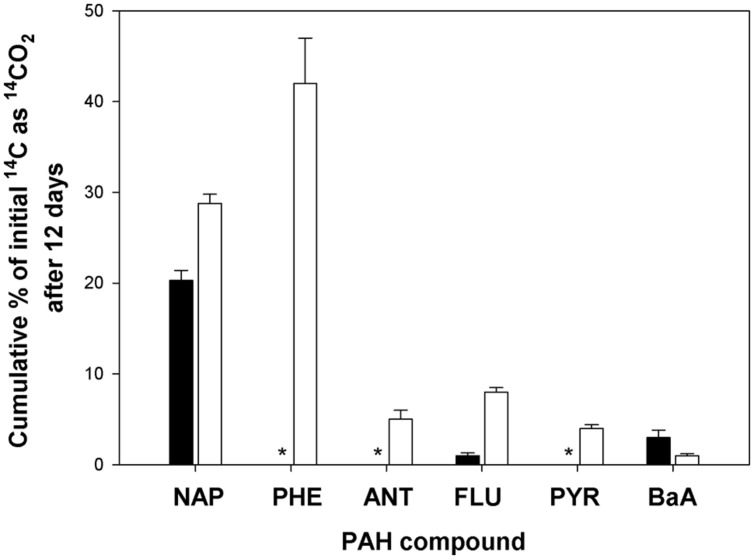
We determined the potential of the bacterial community in the two surface sediment core samples (4571-2 and 4567-24) to mineralize various 14C-labeled PAHs (NAP, PHE, ANT, FLU, PYR, or BaA), since these hydrocarbons have been shown to be present in oily surficial sediment samples at Guaymas (Bazylinski et al., 1988). This was important to thereby inform our choice of the hydrocarbon(s) that would be most suitable for obtaining sufficient incorporation of the 13C into biomass, including DNA, since mineralization of a substrate can be suggestive of growth on that substrate. 14C-hydrocarbon incubations conducted at 4°C with each of the six hydrocarbons and the two sediment samples yielded very low levels of mineralization (<0.5% mineralized of total hydrocarbon; data not shown). As shown in Figure 2, 14C incubations at 21°C using the 4571-2 sediment as inoculum revealed that significant levels of NAP had been mineralized (20.3 ± 1.1% cumulative 14CO2 captured of total initial 14C), whereas low mineralization levels (<3.5% cumulative 14CO2 captured of total initial 14C) were measured for FLU and BaA. PHE, ANT, and PYR were not mineralized by the bacterial community in the 4571-2 sediment sample. Conversely, all six of these PAHs were mineralized by the 4567-24 sediment sample at 21°C, with highest levels of cumulative 14CO2 captured from PHE (42.0 ± 7.0%) and NAP (28.8 ± 1.0%) of total initial 14C for each of these compounds. Whilst the oil-rich core 4571-2 was expected to have yielded higher mineralization levels than the quite oxidized 4567-24 sediment core, the converse which was measured may be attributed to the microbial community of core 4567-24 having been more amenable to aerobic conditions than the sulfide-adapted microbial inhabitants of core 4571-2. Cultured strains and uncultured clones of aromatic-degrading sulfate reducing bacteria, mainly belonging to the Desulfobacteraceae, have been described from Guaymas sediments (Phelps et al., 1998; Kniemeyer et al., 2003), and most likely play a significant role in the anaerobic oxidation and complete degradation of aromatic hydrocarbons at Guaymas. For SIP, achieving sufficient incorporation of the labeled carbon under short incubation times is desirable in order to minimize the potential for cross-feeding (see below). Hence, based on the significant mineralization of PHE measured with the 4567-24 sediment sample at 21°C, SIP experiments were subsequently conducted using this PAH, sediment sample and incubation temperature.
FIGURE 2.
Cumulative 14CO2 recovered from incubations with [14C]-naphthalene (NAP), phenanthrene (PHE), anthracene (ANT), fluoranthene (FLU), pyrene (PYR), or benz[a]anthracene (BaA) by surface (0–4 cm) sediment cores 4571-2 (solid bar) and 4567-24 (open bar) during incubation at 21°C for 12 days, as compared to acid-killed controls. Bars are the averages and SD from triplicate incubations. *values for cumulative 14CO2 recovered were equal or below that of the respective control incubations.
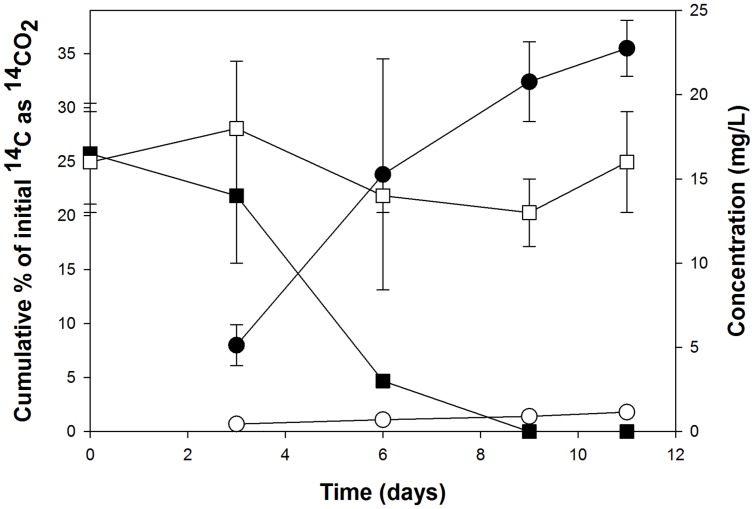
Careful attention must be employed in the design and execution of SIP in order for it to yield interpretable, unambiguous results. One of the main challenges in SIP is obtaining sufficient incorporation of the 13C into biomass, which in the case for DNA-SIP, its enrichment into DNA. Whilst the extent of labeling can be increased with longer incubation times, this can lead to the 13C becoming distributed among other members of the microbial community – i.e., those not necessarily directly capable of metabolizing the isotopically labeled substrate – by cross-feeding on 13C-labeled metabolic byproducts, intermediates, or dead cells (Lueders et al., 2004). To avert this, we had set up several 12C and 14C incubations that ran in parallel to the 13C incubations in order to tractably measure the degradation (by GC-MS) and mineralization (by scintillation counting) of the PHE to help guide our selection of the point at which to terminate the 13C incubations (endpoint of experiment) whereby sufficient 13C incorporation had been achieved with minimal cross-feeding. As shown in Figure 3, complete removal of the PHE occurred after day 9, whereas mineralization of the 14C substrate appeared to reach an asymptote by day 11. Based on these results, the endpoint selected for extraction of DNA from 13C incubations was 11 days. DNA extractions were performed on each of the duplicate 13C incubations for subsequent isopycnic ultracentrifugation to isolate the 13C-enriched ‘heavy’ DNA for analysis.
FIGURE 3.
Cumulative 14CO2 recovered from the incubations with 14C-labeled PHE (circles) that were run in parallel to the stable-isotope probing (SIP) incubations, and the respective removal of this polycyclic aromatic hydrocarbon (PAH) in incubations with the corresponding unlabeled substrates as measured by HPLC (squares). Each data point is the mean of results from triplicate flasks ± SD. Filled symbols represent live (uninhibited) cultures; open symbols represent acid-inhibited controls. Some error bars are smaller than the symbol.
All live (non-acid treated) incubations were observed to produce a rusty-yellowish coloration after 3 days. This is suggestive of the extracellular accumulation of an oxidized intermediate(s) from the metabolism of PHE, as has been observed previously for the accumulation of 1-hydroxy-2-naphthoate during PHE degradation (Stringfellow and Aitken, 1994).
Identification of 13C-Labeled 16S rRNA Genes
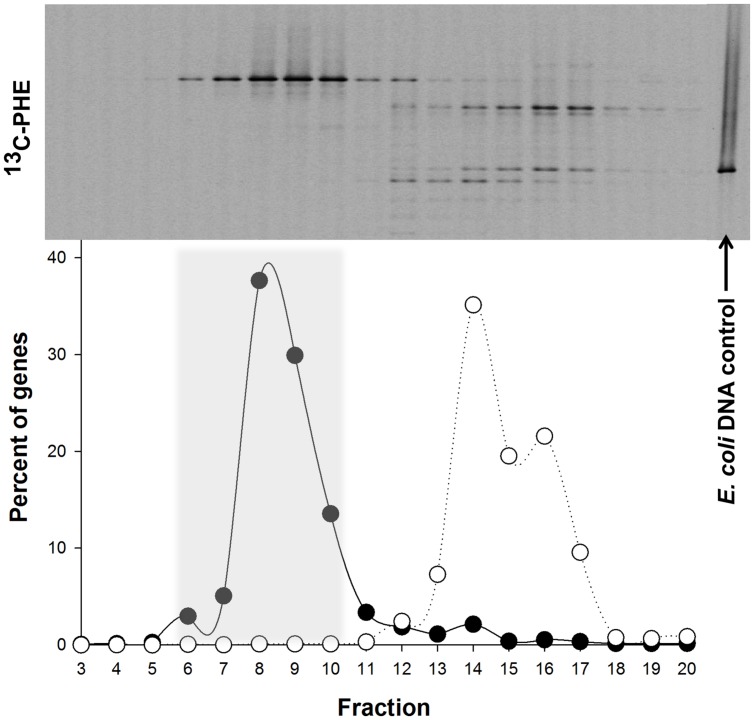
Denaturing gradient gel electrophoresis analysis of the fractions derived from the labeled and unlabeled incubations showed clear evidence of isotopic enrichment of DNA in 13C-PHE incubations, separation of 13C-labeled and unlabeled DNA, and different banding patterns between the 13C-enriched and unenriched DNA fractions (Figure 4). One band in particular was especially dominant in fractions containing 13C-enriched DNA. For the 13C-incubation shown in Figure 4, fractions 6–10 were combined and used in the generation of the 16S rRNA gene clone library. Fractions 4–8 of the duplicate gradient were combined in a similar fashion (data not shown). After excluding vector sequences, poor sequence reads, chimeras, and singleton sequences, the clone library constructed from pooled 13C-enriched DNA comprised 68 sequences. Of these 68 sequences, 3 OTUs were identified of which two were singleton sequences affiliated to Marinobacterium and Propionibacterium and not further analyzed. OTU-1, designated SIP clone PHE1, which comprised the majority (95%) of the 68 sequences (>99% sequence identity), was found affiliated to the genus Cycloclasticus.
FIGURE 4.
Distribution of the ‘heavy’ and ‘light’ DNA in separated SIP fractions. The top of the panel shows the denaturing gradient gel electrophoresis (DGGE) profiles of bacterial PCR products from separated [13C]-PHE fractions with decreasing densities from left to right. The position of unlabeled Escherichia coli DNA, which was used as an internal control in all three isopycnic centrifugations, is shown on the right. The distribution of qPCR-quantified 16S rRNA gene sequences in fractions from [13C]-PHE incubations is shown below the DGGE image for Cycloclasticus (●) and for E. coli (○). Fractions 6–10 (shaded area) were determined to represent 13C heavy DNA and were combined for further analysis. Gene abundance in a fraction are presented as a percentage of the total bacterial 16S rRNA genes quantified in the displayed range of fractions. DGGE banding patterns for a given fraction are aligned with the corresponding gene abundance data below.
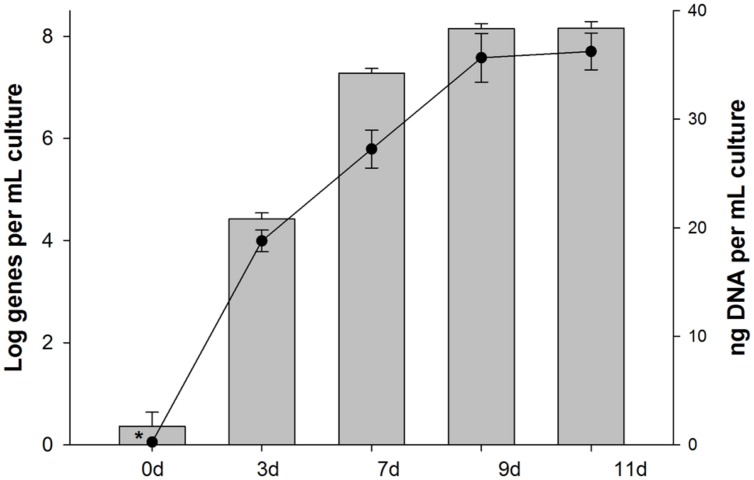
During incubations of the 4567-24 sediment sample with unlabeled PHE in parallel with the SIP incubation, the abundance of 16S rRNA genes for SIP clone PHE1 (OTU-1) increased by several orders of magnitude (Figure 5), thus providing further confirmation of its enrichment on PHE as a growth substrate. By day 3 of the PHE enrichment, the gene abundance increased by over three orders of magnitude, coinciding with the time-frame for the initial stages of disappearance, and mineralization of this compound (Figure 3). By day 9, the gene abundance had increased by ca. six orders of magnitude, coinciding with the almost complete disappearance and high mineralization rate of the PHE. The increase in gene abundance coincided with an increase in the total concentration of DNA, an indicator of cell growth. Collectively, the low bacterial diversity identified in the heavy DNA fractions, which is almost exclusively represented by the Cycloclasticus OTU (SIP clone PHE1), and the dramatic increase in the abundance of these organisms in the incubations with unlabeled PHE, strongly supports that this OTU was solely responsible for degradation of the PAH. In addition, since growth of these organisms coincided with PAH disappearance and the appearance of their 16S rRNA genes only in the most heavily enriched 13C-DNA fractions of incubations containing the labeled substrate, their presence in clone libraries was unlikely due to cross-feeding on a PAH metabolite. We cannot, however, disregard the possibility that other bacterial taxa in the Guaymas 4567-24 sediment sample also possessed the capacity to degrade PHE or its metabolites – they were merely not strongly represented in the most highly 13C-enriched fractions analyzed. However, as discussed below, we also isolated PHE-degrading strains from this sediment sample that are affiliated to other genera. In previous pyrosequencing analyses of the bacterial diversity of Guaymas Basin sediments, Cycloclasticus related sequences composed 0.12% of the average bacterial community at nearby sites, suggesting that representatives of the clade may be poised to act in this oily habitat (Biddle et al., 2012).
FIGURE 5.
Abundance of 16S rRNA genes of Cycloclasticus (SIP clone PHE1) in samples collected from incubations with unlabeled PHE that were run in parallel during SIP. Bars are the averages and SD of results from triplicate qPCRs measuring the abundance of group-specific 16S rRNA genes. Circles are the means and standard deviations of triplicate measurements of the total mass of DNA per sample. Bars or data points with asterisks represent values with one or more readings below the quantification limit of the assay and are presented as the largest possible value for that point.
Most studies that have reported the identification of Cycloclasticus in marine sediments have been conducted in shallow waters. Using PCR-DGGE and 16S rRNA analysis, Cui et al. (2008) identified a Cycloclasticus ribotype in a deep sub-surface sediment core collected at 3542 m depth from the mid Atlantic Ridge and posited these organisms to play a key role in PHE degradation. However, since the authors were unable to isolate a representative of these organisms, likely owing to their poor cultivability, this precluded the opportunity to directly determine the potential of these organisms to degrade PAHs. In a study by Goetz and Jannasch (1993), a number of aerobic obligate aromatic-degrading bacteria were isolated from Guaymas sediment that, although not phylogenetically characterized by 16S rRNA sequencing, represent good candidates as possible members of the genus Cycloclasticus. Our use of DNA-SIP with a 13C-labeled PAH demonstrates the power of this molecular tool in uncovering the diversity of PAH-degraders in deep-sea sediment where they may be a minority portion of the overall community, but that may contribute significantly to the degradation of oil hydrocarbons.
Isolation and Molecular Analysis of PAH-Degrading Strains
Polycyclic aromatic hydrocarbon-degrading bacteria were isolated from surface (0–4 cm) sediment core sample 4567-24 with enrichment on PHE-sprayed agar plates. Several colonies surrounded by clearing zones grew out on the plates, which is indicative of their capacity to utilize the PHE as a sole source of carbon and energy. Subsequent purification and sequencing identified four morphologically distinct isolates designated strains BG-2, BG-3a, BG-3b, and BG-4. Phylogenetic analysis based on 16S rRNA gene sequences indicated that the strains, respectively, belong to the genus Cycloclasticus, Halomonas, Thalassospira, and Lutibacterium. Members comprising each of these genera have been described with PAH-degrading qualities, whilst members of Cycloclasticus are recognized as obligate degraders of PAHs (Yakimov et al., 2007). They have been commonly found in marine coastal sediments (Dyksterhouse et al., 1995; Geiselbrecht et al., 1998), in surface sediment of the west Pacific (Wang et al., 2008) and, as noted above, in deep sediment cores of the mid Atlantic Ridge (Cui et al., 2008). Strains of Halomonas and Thalassospira were enriched with crude oil and individual species and mixtures of PAH substrates from the mid Atlantic Ridge (Cui et al., 2008). Though the ability of these four isolates to directly utilize hydrocarbons as a sole source of carbon and energy was not evaluated, their occurrence in deep-sea sediments and enrichment on oil hydrocarbons suggests that these organisms may play a dominant role in the degradation of oil hydrocarbons in deep-sea surficial sediments.
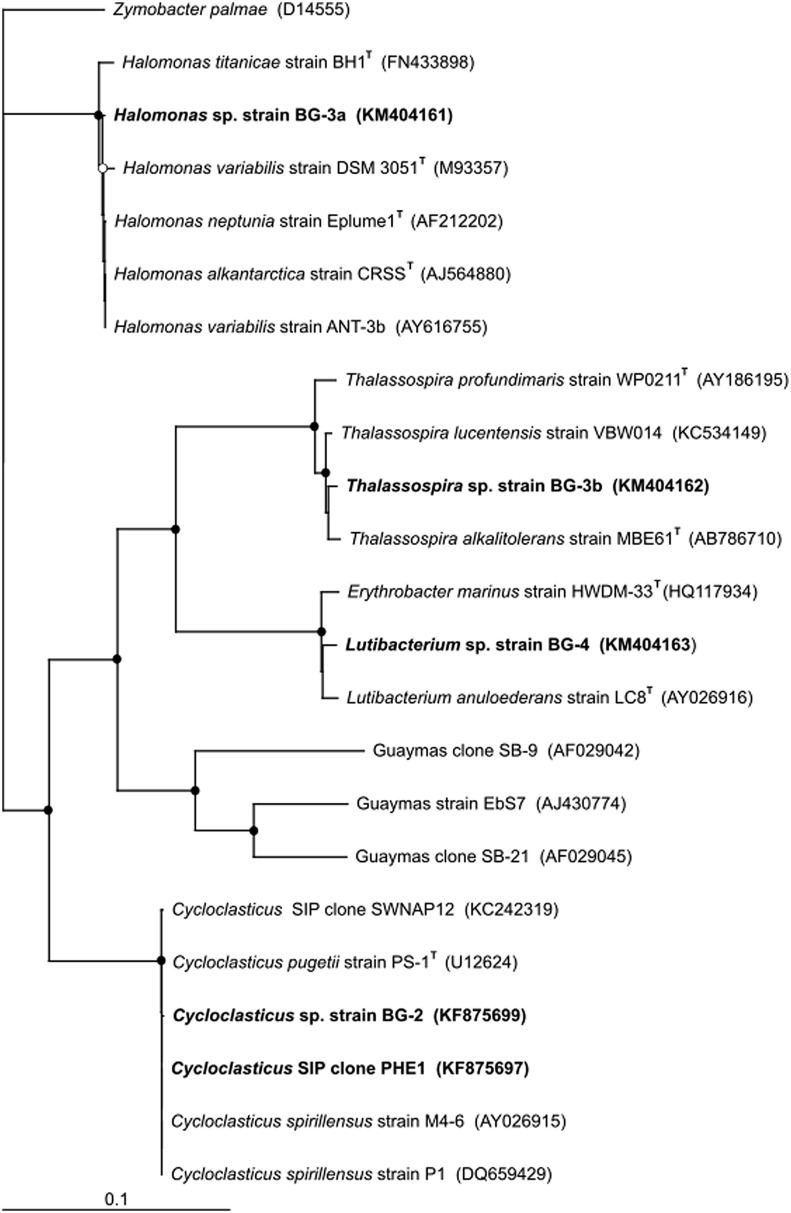
The near-complete 16S rRNA gene sequence (>1400 bp) of each of these PHE-degrading strains were compared with related GenBank sequences, including sequences from studies investigating hydrocarbon-degrading bacteria in deep-sea sediments (Figure 6). From a BLAST analysis, the highest level (99.9%) of sequence identity for strain BG-2 was to Cycloclasticus sp. strain P1 isolated from deep-sea sediment of the West Pacific at 2682 m water depth (Wang et al., 2008), Cycloclasticus spirillensus strain M4-6 isolated from marine macrofaunal burrow sediments of Lowes Cove in Maine, USA (Chung and King, 2001), and to Cycloclasticus sp. clone SWNAP12 which was identified in 13C-enriched DNA of a SIP enrichment of a surface oil slick sample collected during the Deepwater Horizon oil spill (Gutierrez et al., 2013b). The next closest cultivated relative to BG-2 was C. pugetii strain PS-1T (99.7% sequence identity) isolated from marine sediment of Puget Sound (Dyksterhouse et al., 1995). Notably, isolated strain BG-2 shared 99.7% 16S rRNA gene sequence identity with SIP clone PHE1. Strain BG-3a was most closely related to the type strains Halomonas alkaliantarctica strain CRSST (Poli et al., 2007; 99.4% sequence identity), H. neptunia strain Eplume1T (Kaye et al., 2004; 99.3% sequence identity), and the exopolysaccharide (EPS)-producer H. variabilis strain ANT-3b (Pepi et al., 2005; 99.3% sequence identity). The next closest cultivated relatives to BG-3a were H. titanicae strain BH1T (Sanchez-Porro et al., 2010) and H. variabilis strain DSM 3051T (Arahal et al., 2002; 98.8% sequence identity). Strain BG-3b shared 99.4% sequence identity to Thalassospira lucentensis strain VBW014 (Rajasabapathy et al., 2014), and to the type strains T. alkalitolerans strain MBE#61T (Tsubouchi et al., 2014) and T. profundimaris strain WP0211T (Liu et al., 2007) with 99.1% and 99.3% sequence identity, respectively. Strain BG-4 was most closely related to the type strains Lutibacterium anuloederans strain LC8T (Chung and King, 2001) and Erythrobacter marinus strain HWDM-33T (Jung et al., 2012) with 98.7 and 98.6% sequence identity, respectively. In a previous deep sequencing survey of bacterial diversity at Guaymas basin, Halomonas sp. accounted for 0.1% of sequences, whereas Thalassospira and Lutibacterium were not detected (Biddle et al., 2012). This highlights the importance of using enrichment [cultivation-independent (DNA-SIP) and/or cultivation-dependent] methods to uncover minority taxa that may not be identified by solely sequencing surveys, and to linking these organisms to a particular function – in this case the degradation of hydrocarbons.
FIGURE 6.
Phylogenetic tree of SIP clone PHE1 and of the isolated strains from Guaymas Basin surface (0–4 cm) sediment shown alongside closely related sequences and type strains from GenBank. Guaymas strain EbS7 and Guaymas clones SB-9 and SB-21 are included as representatives of sulfate-reducing bacteria identified at Guaymas Basin that are capable of oxidizing aromatic hydrocarbons. Aerobic PAH-degrading Cycloclasticus P1, isolated from deep-sea sediments of the West Pacific, and other strains of this genus isolated from shallower sediment locations (strains M4-6 and PS-1T), are also included. The tree was constructed using the neighbor-joining algorithm. Nodes with bootstrap support (1000 replications) of at least 65% (○) and 90% (●) are marked. Accession numbers of all sequences are given in parentheses. The scale bar indicates the difference of number of substitutions per site.
Whilst members of the genus Cycloclasticus are recognized as obligate degraders of PAHs (Yakimov et al., 2007), only some members comprising the genera Halomonas, Thalassospira, and Lutibacterium have been reported as non-obligate degraders of PAHs (Chung and King, 2001; Melcher et al., 2002; Kodama et al., 2008; Zhao et al., 2010; Gutierrez et al., 2013a). The ability of these isolated strains (BG-2, BG-3a, BG-3b, BG-4) to grow on and degrade PHE was confirmed on ONR7a liquid medium amended with the PAH as the sole carbon source (data not shown). The ability of the strains to mineralize PHE was assessed by measuring for the liberation of 14CO2 from 14C-labeled PHE. After 12 days of incubation, significant levels (P < 0.05) of the PAH was mineralized by strain BG-2 (11.7 ± 0.7%), strain BG-3a (5.1 ± 2.3%), strain BG-3b (8.4 ± 3.5%), and strain BG-4 (16.7 ± 4.0%) of total added 14C-labeled compound when compared to respective acid-killed controls (data not shown). During these mineralization experiments, a notable feature observed with Cycloclasticus sp. strain BG-2 was the production of a rusty-yellow coloration in the medium after a few days, as similarly observed in the SIP incubations (see above).
To the best of our knowledge, this study represents the first application of SIP to identify bacterial taxa with the ability to degrade PAHs in deep-sea sediments. In particular, this work has added to our current knowledge on the diversity of hydrocarbon-degrading bacteria in Guaymas Basin sediments – an environment with abundant sediment surface hydrocarbons (Goetz and Jannasch, 1993). In summary, the dominant representation of Cycloclasticus in the 13C-enriched clone library and recognition of members in this genus as obligate degraders of PAHs (Yakimov et al., 2007) suggests that Cycloclasticus may play an important role in the degradation of these compounds at Guaymas Basin. Using DNA-SIP and cultivation-based methods, we have shown that these organisms and other hydrocarbon-degraders (Halomonas, Thalassospira, and Lutibacterium) co-exist in Guaymas surface sediments. We posit that the activities of these organisms are likely to have a significant influence on the natural attenuation of PAHs and other hydrocarbon constituents of the crude oil that is a natural feature to this deep-sea vent site.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by a Marie Curie International Outgoing Fellowship (PIOF-GA-2008-220129) within the Seventh European Community Framework Programme. Sampling at Guaymas Basin was supported by the U.S. National Science Foundation (grant OCE-0647633) to AT. Partial support was also provided by the National Institute of Environmental Health Sciences (grant # 5 P42 ES005948). We thank Stephen Richardson for assistance with HPLC analysis. Finally, we would like to also thank the two reviewers for their valuable comments during the preparation of the manuscript.
References
- Aitken M. M., Jones D. M., Larter S. R. (2004). Anaerobic hydrocarbon biodegradation in deep subsurface oil reservoirs. Nature 431 291–294. 10.1038/nature02922 [DOI] [PubMed] [Google Scholar]
- Amann R. I., Binder B. J., Olson R. J., Chisholm S. W., Devereux R., Stahl D. A. (1990). Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56 1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arahal D. R., Ludwig W., Schleifer K. H., Ventosa A. (2002). Phylogeny of the family Halomonadaceae based on 23S and 16S rDNA sequence analyses. Int. J. Syst. Evol. Microbiol. 52 241–249. [DOI] [PubMed] [Google Scholar]
- Bazylinski D. A., Farrington J. W., Jannasch H. W. (1988). Hydrocarbons in surface sediments from a Guaymas Basin hydrothermal vent site. Org. Geochem. 12 547–558. 10.1016/0146-6380(88)90146-5 [DOI] [Google Scholar]
- Bennett B., Adams J. J., Gray N. D., Sherry A., Oldenburg T. B. P., Huang H., et al. (2013). The controls on the composition of biodegraded oils in the deep subsurface – Part 3. The impact of microorganisms distribution on petroleum geochemical gradients in biodegraded petroleum reservoirs. Org. Geochem. 56 94–105. 10.1016/j.orggeochem.2012.12.011 [DOI] [Google Scholar]
- Biddle J. F., Cardman Z., Mendlovitz H., Albert D. B., Lloyd K. G., Boetius A., et al. (2012). Anaerobic oxidation of methane at different temperature regimes in Guaymas Basin hydrothermal sediments. ISME J. 6 1018–1031. 10.1038/ismej.2011.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanton J., Zhao T., Rosenheim B. E., Joye S., Bosman S., Brunner C., et al. (2015). Using natural abundance radiocarbon to trace the flux of petrocarbon to the seafloor following the Deepwater Horizon oil spill. Environ. Sci. Technol. 49 847–854. 10.1021/es5046524 [DOI] [PubMed] [Google Scholar]
- Chung W. K., King G. M. (2001). Isolation, characterization, and polyaromatic hydrocarbon degradation potential of aerobic bacteria from marine macrofaunal burrow sediments and description of Lutibacterium anuloederans gen. nov., sp. nov., and Cycloclasticus spirillensus sp. nov. Appl. Environ. Microbiol. 67 5585–5592. 10.1128/AEM.67.12.5585-5592.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J. R., Wang Q., Cardenas E., Fish J., Chai B., Farris R. J., et al. (2009). The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37 D141–D145. 10.1093/nar/gkn879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z., Lai Q., Dong C., Shao Z. (2008). Biodiversity of polycyclic aromatic hydrocarbon-degrading bacteria from deep sea sediments of the Middle Atlantic Ridge. Environ. Microbiol. 10 2138–2149. 10.1111/j.1462-2920.2008.01637.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont M., Murrell J. (2005). Stable isotope probing – linking microbial identity to function. Nat. Rev. Microbiol. 3 499–504. 10.1038/nrmicro1162 [DOI] [PubMed] [Google Scholar]
- Dyksterhouse S. E., Gray J. P., Herwig R. P., Cano Lara J., Staley J. T. (1995). Cycloclasticus pugetii gen. nov., sp.nov., an aromatic hydrocarbon-degrading bacterium from marine sediments. Int. J. Syst. Bacteriol. 45 116–123. 10.1099/00207713-45-1-116 [DOI] [PubMed] [Google Scholar]
- Geiselbrecht A. D., Hedlund B. P., Tichi M. A., Staley J. T. (1998). Isolation of marine polycyclic aromatic hydrocarbon (PAH)-degrading Cycloclasticus strains from the Gulf of Mexico and comparison of their PAH degradation ability with that of Puget Sound Cycloclasticus strains. Appl. Environ. Microbiol. 64 4703–4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz F. E., Jannasch H. W. (1993). Aromatic hydrocarbon-degrading bacteria in the petroleum-rich sediments of Guaymas Basin hydrothermal vent site: Preference for aromatic carboxylic acids. Geomicrobiol. J. 11 1–18. 10.1080/01490459309377928 [DOI] [Google Scholar]
- Gutierrez T., Berry D., Yang T., Mishamandani S., McKay L., Teske A., et al. (2013a). Role of bacterial exopolysaccharides (EPS) in the fate of the oil released during the Deepwater Horizon oil spill. PLoS ONE 8:e67717 10.1371/journal.pone.0067717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez T., Singleton D. R., Berry D., Yang T., Aitken M. D., Teske A. (2013b). Hydrocarbon-degrading bacteria enriched by the Deepwater Horizon oil spill identified by cultivation and DNA-SIP. ISME J. 7 2091–2104. 10.1038/ismej.2013.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez T., Singleton D. R., Aitken M. D., Semple K. T. (2011). Stable isotope probing of an algal bloom to identify uncultivated members of the Rhodobacteraceae associated with low-molecular-weight polycyclic aromatic hydrocarbon degradation. Appl. Environ. Microbiol. 77 7856–7860. 10.1128/AEM.06200-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head I. M., Jones D. M., Röling W. F. (2006). Marine microorganisms make a meal of oil. Nat. Rev. Microbiol. 4 173–182. 10.1038/nrmicro1348 [DOI] [PubMed] [Google Scholar]
- Jones D. M., Head I. M., Gray N. D., Adams J. J., Rowan A. K., Aitken C. M., et al. (2008). Crude-oil biodegradation via methanogenesis in subsurface petroleum reservoirs. Nature 451 176–180. 10.1038/nature06484 [DOI] [PubMed] [Google Scholar]
- Jones M. D., Singleton D. R., Sun W., Aitken M. D. (2011). Multiple DNA extractions coupled with stable-isotope probing of anthracene-degrading bacteria in contaminated soil. Appl. Environ. Microbiol. 77 2984–2991. 10.1128/AEM.01942-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y.-T., Park S., Oh T.-K., Yoon J.-H. (2012). Erythrobacter marinus sp. nov., isolated from seawater. Int. J. Syst. Evol. Microbiol. 62 2050–2055. 10.1099/ijs.0.034702-0 [DOI] [PubMed] [Google Scholar]
- Kaye J. Z., Márquez M. C., Ventosa A., Baross J. A. (2004). Halomonas neptunia sp. nov., Halomonas sulfidaeris sp. nov., Halomonas axialensis sp. nov. and Halomonas hydrothermalis sp. nov.: halophilic bacteria isolated from deep-sea hydrothermal-vent environments. Int. J. Syst. Evol. Microbiol. 54 499–511. 10.1099/ijs.0.02799-0 [DOI] [PubMed] [Google Scholar]
- Kiyohara H., Nagao K., Yana K. (1982). Rapid screen for bacteria degrading water-insoluble, solid hydrocarbons on agar plates. Appl. Environ. Microbiol. 43 454–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleindienst S., Herbst F.-A., Stagars M., von Netzer F., von Bergen M., Seifert J., et al. (2014). Diverse sulphate-reducing bacteria of the Desulfosarcina/Desulfococcus clade are the key alkane degraders at marine seeps. ISME J. 8 2029–2044. 10.1038/ismej.2014.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniemeyer O., Fischer T., Wilkes H., Glöckner F. O., Widdel F. (2003). Anaerobic degradation of ethylbenzene by a new type of marine sulfate-reducing bacterium. Appl. Environ. Microbiol. 69 760–768. 10.1128/AEM.69.2.760-768.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama Y., Stiknowati L. I., Ueki A., Ueki K., Watanabe K. (2008). Thalassospira tepidiphila sp. nov., a polycyclic aromatic hydrocarbon-degrading bacterium isolated from seawater. Int. J. Syst. Evol. Microbiol. 58 711–715. 10.1099/ijs.0.65476-0 [DOI] [PubMed] [Google Scholar]
- Lane D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Sequencing Techniques in Bacterial Systematics, eds Stackebrandt E., Goodfellow M. (New York, NY: John Wiley & Sons; ), 115–175. [Google Scholar]
- Lane D. J., Pace B., Olsen G. J., Stahl D. A., Sogin M. L., Pace N. R. (1985). Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. U.S.A. 82 6955–6959. 10.1073/pnas.82.20.6955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanza-Espino G., Soto L. A. (1999). Sedimentary geochemistry of hydrothermal vents in Guaymas Basin, Gulf of California, Mexico. Appl. Geo. Chem. 14 499–510. 10.1016/s0883-2927(98)00064-x [DOI] [Google Scholar]
- Liu C., Wu Y., Li L., Ma Y., Shao Z. (2007). Thalassospira xiamenensis sp. nov. and Thalassospira profundimaris sp. nov. Int. J. Syst. Evol. Microbiol. 57 316–320. 10.1099/ijs.0.64544-0 [DOI] [PubMed] [Google Scholar]
- Lueders T., Wagner B., Claus P., Friedrich M. (2004). Stable isotope probing of rRNA and DNA reveals a dynamic methylotroph community and trophic interactions with fungi and protozoa in oxic rice field soil. Environ. Microbiol. 6 60–72. 10.1046/j.1462-2920.2003.00535.x [DOI] [PubMed] [Google Scholar]
- Maidak B. L., Cole J. R., Parker C. T., Jr, Garrity G. M., Larsen N., Li B., et al. (1999). A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27 171–173. 10.1093/nar/27.1.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesi J. R., Sato T., Weightman A. J., Martin T. A., Fry J. C., Hiom S. J., et al. (1998). Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 64 795–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. J., MacGregor B. J., Biddle J. F., Mendlovitz H. P., Hoer D., Lipp J. S., et al. (2012). Spatial heterogeneity and underlying geochemistry of phylogenetically diverse orange and white Beggiatoa mats in Guaymas Basin hydrothermal sediments. Deep-Sea Res. Part 1 67 21–31. 10.1016/j.dsr.2012.04.011 [DOI] [Google Scholar]
- Melcher R. J., Apitz S. E., Hemmingsen B. B. (2002). Impact of irradiation and polycyclic aromatic hydrocarbon spiking on microbial populations in marine sediment for future aging and biodegradability studies. Appl. Environ. Microbiol. 68 2858–2868. 10.1128/AEM.68.6.2858-2868.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishamandani S., Gutierrez T., Aitken M. D. (2014). DNA-based stable isotope probing coupled with cultivation methods implicates Methylophaga in hydrocarbon degradation. Front. Microbiol. 5:76 10.3389/fmicb.2014.00076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G., de Waal E. C., Uitterlinden A. G. (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphan V. J., Taylor L. T., Hafenbradl D., DeLong E. F. (2000). Culture-dependent and culture-independent characterization of microbial assemblages associated with high-temperature petroleum reservoirs. Appl. Environ. Microbiol. 66 700–711. 10.1128/AEM.66.2.700-711.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passow U., Ziervogel K., Asper V., Diercks A.-R. (2012). Marine snow formation in the aftermath of the Deepwater Horizon oil spill in the Gulf of Mexico. Environ. Res. Lett. 7:035301; 10.1088/1748-9326/7/3/035301 [DOI] [Google Scholar]
- Pearson A., Seewald J. S., Eglinton T. I. (2005). Bacterial incorporation of relict carbon in the hydrothermal environment of Guaymas Basin. Geochim. Cosmochim. Acta 69 5477–5486. 10.1016/j.gca.2005.07.007 [DOI] [Google Scholar]
- Pepi M., Cesaro A., Liut G., Baldi F. (2005). An antarctic psychrotrophic bacterium Halomonas sp. ANT-3b, growing on n-hexadecane, produces a new emulsifying glycolipid. FEMS Microbiol. Ecol. 53 157–166. 10.1016/j.femsec.2004.09.013 [DOI] [PubMed] [Google Scholar]
- Peter J. M., Peltonen P., Scott S. D., Simoneit B. R. T., Kawka O. E. (1991). 14C ages of hydrothermal petroleum and carbonate in Guaymas Basin, Gulf of California: implications for oil generation, expulsion, and migration. Geology 19 253–256. [DOI] [Google Scholar]
- Pfaffl M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29 2002–2007. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps C. D., Kerkhof L. J., Young L. Y. (1998). Molecular characterization of a sulfate-reducing consortium which mineralizes benzene. FEMS Microbiol. Ecol. 27 269–279. 10.1111/j.1574-6941.1998.tb00543.x [DOI] [Google Scholar]
- Poli A., Esposito E., Orlando P., Lama L., Giordano A., de Appolonia F., et al. (2007). Halomonas alkaliantarctica sp. nov., isolated from saline lake Cape Russell in Antarctica, an alkalophilic moderately halophilic, exopolysaccharide-producing bacterium. Syst. Appl. Microbiol. 30 31–38. 10.1016/j.syapm.2006.03.003 [DOI] [PubMed] [Google Scholar]
- Rajasabapathy R., Mohandass C., Colaco A., Dastager S. G., Santos R. S., Meena R. M. (2014). Culturable bacterial phylogeny from a shallow water hydrothermal vent of Espalamaca (Faial, Azores) reveals a variety of novel taxa. Curr. Sci. 106 58–69. [Google Scholar]
- Sabat G., Rose P., Hickey W. J., Harkin J. M. (2000). Selective and sensitive method for PCR amplification of Escherichia coli 16S rRNA genes in soil. Appl. Environ. Microbiol. 66 844–849. 10.1128/AEM.66.2.844-849.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Porro C., Kaur B., Mann H., Ventosa A. (2010). Halomonas titanicae sp. nov., a halophilic bacterium isolated from the RMS Titanic. Int. J. Syst. Evol. Microbiol. 60 2768–2774. 10.1099/ijs.0.020628-0 [DOI] [PubMed] [Google Scholar]
- Sauret C., Séverin T., Vétion G., Guigue C., Goutx M., Pujo-Pay M., et al. (2014). ‘Rare biosphere’ bacteria as key phenanthrene degraders in coastal seawaters. Environ. Pollut. 194 246–253. 10.1016/j.envpol.2014.07.024 [DOI] [PubMed] [Google Scholar]
- Schwarz J. R., Walker J. D., Colwell R. R. (1974a). Deep-sea bacteria: growth and utilization of hydrocarbons at ambient and in situ pressure. Appl. Microbiol. 28 982–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz J. R., Walker J. D., Colwell R. R. (1974b). Growth of deep-sea bacteria on hydrocarbons at ambient and in situ pressure. Dev. Ind. Microbiol. 15 239–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton D. R., Sangaiah R., Gold A., Ball L. M., Aitken M. D. (2006). Identification and quantification of uncultivated Proteobacteria associated with pyrene degradation in a bioreactor treating PAH-contaminated soil. Environ. Microbiol. 8 1736–1745. 10.1111/j.1462-2920.2006.01112.x [DOI] [PubMed] [Google Scholar]
- Stringfellow W. T., Aitken M. D. (1994). Comparative physiology of phenanthrene degradation by two dissimilar pseudomonads isolated from a creosote-contaminated soil. Can. J. Microbiol. 40 432–438. 10.1139/m94-071 [DOI] [PubMed] [Google Scholar]
- Teske A., Callaghan A. V., LaRowe D. E. (2014). Biosphere frontiers of subsurface life in the sedimented hydrothermal system of Guaymas Basin. Front. Microbiol. 5:362 10.3389/fmicb.2014.00362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. (1994). CLUSTAL_X: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillett D., Neilan B. A. (2000). Xanthogenate nucleic acid isolation from cultured and environmental cyanobacteria. J. Phycol. 36 251–258. 10.1046/j.1529-8817.2000.99079.x [DOI] [Google Scholar]
- Tsubouchi T., Ohta Y., Haga T., Usui K., Shimane Y., Mori K., et al. (2014). Thalassospira alkalitolerans sp. nov. and Thalassospira mesophila sp. nov., isolated from a decaying bamboo sunken in the marine environment, and description of the genus Thalassospira. Int. J. Syst. Evol. Microbiol. 64 107–115. 10.1099/ijs.0.056028-0 [DOI] [PubMed] [Google Scholar]
- Wang B., Lai Q., Cui Z., Tan T., Shao Z. (2008). A pyrene-degrading consortium from deep-sea sediment of the West Pacific and its key member Cycloclasticus sp. P1. Environ. Microbiol. 10 1948–1963. 10.1111/j.1462-2920.2008.01611.x [DOI] [PubMed] [Google Scholar]
- Wilmotte A., Van der Auwera G., De Wachter R. (1993). Structure of the 16S ribosomal RNA of the thermophilic cyanobacterium Chlorogloeopsis HTF (‘Mastigocladus laminosus HTF’) strain PCC7518, and phylogenetic analysis. FEMS Microbiol. Lett. 317 96–100. 10.1016/0014-5793(93)81499-P [DOI] [PubMed] [Google Scholar]
- Yakimov M. M., Timmis K. N., Golyshin P. N. (2007). Obligate oil-degrading marine bacteria. Curr. Opin. Biotechnol. 18 257–266. 10.1016/j.copbio.2007.04.006 [DOI] [PubMed] [Google Scholar]
- Yu Z., Morrison M. (2004). Comparisons of different hypervariable regions of rrs genes for use in fingerprinting of microbial communities by PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 70 4800–4806. 10.1128/AEM.70.8.4800-4806.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Sangaiah R., Gold A., Ball L. M. (2011). Synthesis of uniformly 13C-labeled polycyclic aromatic hydrocarbons. Org. Biomol. Chem. 9 5431–5435. 10.1039/c0ob01107j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Wang H., Li R., Mao X. (2010). Thalassospira xianhensis sp. nov., a polycyclic aromatic hydrocarbon-degrading marine bacterium. Int. J. Syst. Evol. Microbiol. 60 1125–1129. 10.1099/ijs.0.013201-0 [DOI] [PubMed] [Google Scholar]