Summary
Using high-density genetic markers, gene flow is identified from diploid Miscanthus sinensis to tetraploid M. sacchariflorus in Japan, in contrast to genetic isolation between these species in China.
Key words: Biomass crop, cross-ploidy introgression, hybridization, Poaceae, polyploidy, population genetics, RAD-seq.
Abstract
Unilateral introgression from diploids to tetraploids has been hypothesized to be an important evolutionary mechanism in plants. However, few examples have been definitively identified, perhaps because data of sufficient depth and breadth were difficult to obtain before the advent of affordable high-density genotyping. Throughout Japan, tetraploid Miscanthus sacchariflorus and diploid Miscanthus sinensis are common, and occasionally hybridize. In this study, 667 M. sinensis and 78 M. sacchariflorus genotypes from Japan were characterized using 20 704 SNPs and ten plastid microsatellites. Similarity of SNP genotypes between diploid and tetraploid M. sacchariflorus indicated that the tetraploids originated through autopolyploidy. Structure analysis indicated a gradient of introgression from diploid M. sinensis into tetraploid M. sacchariflorus throughout Japan; most tetraploids had some M. sinensis DNA. Among phenotypically M. sacchariflorus tetraploids, M. sinensis ancestry averaged 7% and ranged from 1–39%, with introgression greatest in southern Japan. Unexpectedly, rare (~1%) diploid M. sinensis individuals from northern Japan were found with 6–27% M. sacchariflorus ancestry. Population structure of M. sinensis in Japan included three groups, and was driven primarily by distance, and secondarily by geographic barriers such as mountains and straits. Miscanthus speciation is a complex and dynamic process. In contrast to limited introgression between diploid M. sacchariflorus and M. sinensis in northern China, selection for adaptation to a moderate maritime climate probably favoured cross-ploidy introgressants in southern Japan. These results will help guide the selection of Miscanthus accessions for the breeding of biomass cultivars.
Introduction
High-resolution analyses of population structure, which have been enabled by second-generation sequencing technologies, can provide new insights into the processes of speciation in plants and facilitate crop improvement by guiding marker-trait association studies and identifying groups to test for heterotic combinations. Polyploidy is a primary driver of evolution in flowering plants (Adams and Wendel, 2005; Barabaschi et al., 2012), and it has long been recognized that polyploidization of amphidiploids can result in new, genetically isolated species in a single event (Winge, 1917; Stebbins, 1959). Additionally, introgression between plant species plays an important role in local adaptation and speciation (Rieseberg et al., 2003; Arnold, 2004; Arnold et al., 2008). In contrast to the genetic isolation typically observed between populations of different ploidies, Stebbins (1971) noted that introgression across ploidy levels can also occur in plants. Moreover, Stebbins (1971) postulated that introgression of genes between diploid and tetraploid populations would usually flow preferentially from diploids to tetraploids (via unreduced gametes and/or triploid bridges) and these gene movements could have large evolutionary consequences (Petit et al., 1999). To date, however, few examples of gene flow from diploids to tetraploids have been reported for wild plants (Kim et al., 2008; Wang et al., 2014), but prior technological limitations in the ability to detect small introgressions in a large sampling of genotypes and populations may have contributed to a lack of reports. The extent to which polyploidy has contributed to speciation or, in contrast, limited differentiation of populations via gene exchange, can now be explored in detail with analyses of population structure using high marker densities.
Miscanthus is a genus of perennial grasses native to east Asia and Oceania, and includes polyploid and diploid species that are able to hybridize. A close relative of sugarcane (Saccharum hybrids), Miscanthus is also useful in its own right as a lignocellulosic biomass crop, and as a popular ornamental in European and North American gardens. However, there has been little or no effort to domesticate Miscanthus in its native lands. M. sacchariflorus (Maxim.) Hack. and M. sinensis Andersson are among the most widely distributed and divergent species within Miscanthus sensu stricto and are the parent species of the biomass crop M.×giganteus (Hodkinson et al., 2002a,b ; Clifton-Brown et al., 2008; Sacks et al., 2013). M. sinensis has a caespitose form, prefers aerobic soils especially on hilly sites that are infrequently disturbed by grazing or fire, and is typically diploid with a monoploid genome size of about 2.5–2.8 pg (Clifton-Brown et al., 2008; Rayburn et al., 2009; Sacks et al., 2013; Li et al., 2013; Jiang et al., 2013; Moon et al., 2013; Chae et al., 2014). In contrast, M. sacchariflorus has a spreading rhizomatous habit, prefers riparian environments, and can be diploid or tetraploid with a monoploid genome size of about 2.1–2.3 pg (Rayburn et al., 2009; Li et al., 2013; Moon et al., 2013; Chae et al., 2014). Though the natural ranges of M. sinensis and M. sacchariflorus overlap from ~29° N to 43° N, M. sinensis is distributed further south to at least ~18° N in Hainan, whereas M. sacchariflorus is distributed further north to ~50° N in eastern Russia (Sacks et al., 2013). Thus, M. sinensis and M. sacchariflorus are well differentiated phylogenetically, morphologically, and ecologically.
Throughout Japan, diploid M. sinensis and tetraploid M. sacchariflorus are common, and although they typically occupy different niches, sympatric populations occur. Though both diploid and tetraploid M. sacchariflorus have been found in mainland Asia (Yan et al., 2012; Li et al., 2013; Moon et al., 2013), an extensive survey in Japan reported only tetraploids (Hirayoshi et al. 1957).
Hodkinson and Renvoize (2001) defined the nothospecies M.×giganteus J.M. Greef and Deuter ex Hodkinson and Renvoize (syn. M. ogiformis Honda if awns present; Honda, 1939) as a hybrid between M. sacchariflorus and M. sinensis. In 1935, a single triploid M.×giganteus genotype was exported from Japan to Denmark (Greef et al., 1997; Głowacka et al., 2014). This M.×giganteus genotype has become an important crop for the emerging lignocellulosic bioenergy industry in Europe and North America owing to its high yield, low input requirements, low risk of invasiveness, high rate of photosynthesis at low temperatures, and broad adaptation (Barney and Ditomaso, 2008; Pyter et al., 2009; Somerville et al., 2010; Purdy et al., 2013). Subsequent to its initial introduction to Europe, a few additional triploid M.×giganteus genotypes have been found growing in situ in Japan (Adati and Shiotani, 1962), and others have been obtained by germinating seed collected from wild plants of M. sinensis (Hirayoshi et al., 1957) or M. sacchariflorus (Nishiwaki et al., 2011; Dwiyanti et al., 2013) from locations where both species grew sympatrically.
In mainland Asia, where diploid M. sacchariflorus is common, it naturally crosses with M. sinensis to produce homoploid hybrids that have previously been named M. purpurascens or M. sinensis var. purpurascens (Jiang et al., 2013; Chae et al., 2014; Głowacka et al., 2014). These diploid interspecific hybrids backcross infrequently with M. sinensis but do not form a hybrid swarm (Jiang et al., 2013; Clark et al., 2014). However, the extent of genetic exchange between M. sinensis and M. sacchariflorus in Japan, where M. sacchariflorus is thought to be exclusively tetraploid, is unknown beyond the occasional production of sterile triploid M.×giganteus hybrids (Hirayoshi et al., 1957; Adati and Shiotani, 1962).
In addition to the discovery of new M.×giganteus genotypes in nature, human-directed crosses between diploid M. sinensis and tetraploid M. sacchariflorus can be made intentionally, utilizing germplasm with desired traits and exploiting the genetic diversity of these obligate-outcrossing species to maximize heterosis. Previous efforts to breed new triploid genotypes of M.×giganteus by Hirayoshi et al. (1960), and the release of ‘Nagara’ in 2006 by M. Deuter of Tinplant (Klein Wanzleben, Germany) indicate that this approach is viable. Recently, more than 30 new triploid M.×giganteus genotypes have been bred at the University of Illinois and field evaluations of these have begun. Crucial to the success of breeding new biomass cultivars of M.×giganteus will be an in-depth understanding of genetic diversity and population structure for M. sinensis and M. sacchariflorus to guide the selection of parental genotypes for combining ability, adaptation, and novel alleles.
In a previous study, a broad survey of M. sinensis genetic diversity with accessions primarily from China, Korea and Japan was conducted, and six groups were identified, including one each in northern Japan (northern Honshu and Hokkaido) and southern Japan (Clark et al., 2014). It was also found that nearly all of the ornamental cultivars of M. sinensis grown in the USA were derived from southern Japan. However, there has not yet been a population genetic study with sufficient resolution to observe how the genetic structure of M. sinensis in Japan was affected by geographic features such as straits and mountain ranges. A limitation of the previous study (Clark et al., 2014) was that only 131 wild-collected M. sinensis genotypes from Japan were able to be evaluated, with only 34 of those from central and southern Japan, and no Japanese M. sacchariflorus. Thus, the current study was conducted to provide an in-depth understanding of M. sinensis population structure in Japan as it relates to geography, and to establish a baseline understanding of M. sacchariflorus diversity in Japan, whereas the previous study was a broad East Asia-wide assessment of relationships among M. sinensis populations. Very little is known about the genetic structure of M. sacchariflorus, including the relationship between its diploid and tetraploid forms, and the amount of introgression with M. sinensis, if any. Indeed, there is longstanding disagreement about whether tetraploid M. sacchariflorus is allo-, segmental-, or auto-polyploid (Adati, 1958, 1959; Adati and Shiotani, 1962; Takahashi and Shibata, 2002; Chae et al., 2014). Thus, the objectives of this study were to (i) detect spatial genetic structure of Japanese M. sinensis and M. sacchariflorus, (ii) more precisely identify the genetic origins within Japan of ornamental and US naturalized M. sinensis, and (iii) assess the degree of hybridization and introgression between M. sinensis and M. sacchariflorus in Japan, and determine the ploidy of any hybrids.
Materials and methods
Plant materials and genotyping
In total, 1513 genotypes of Miscanthus were studied. Focus was especially placed on 667 M. sinensis genotypes from 202 accessions, and 78 M. sacchariflorus genotypes from 53 accessions, collected from the wild in Japan and studied for the first time here (i.e. the Japan dense-sampling set; Table 1, Supplementary dataset S1). Germplasm from the Japan dense-sampling set was collected as seed and/or clonal propagules in 1996 and from 2007–2011. Each seed accession was a bulk collection from between one and 50 mother plants, whereas each clonal accession came from a single individual. In addition to the 255 accessions (745 individuals) from the Japan dense-sampling set, we also studied 622 M. sinensis and four M. floridulus (Labill.) Warb. ex K. Schum. & Lauterb. accessions (one genotype per accession) primarily from China, Korea, and Japan, 11 M. sacchariflorus from China and Korea, and eight M. sinensis × M. sacchariflorus F1 hybrids from China that we evaluated previously (i.e. the region-wide set; Supplementary dataset S1; Clark et al., 2014), in order to understand relationships among accessions from Japan in a regional context. The Japanese Miscanthus accessions were also compared to 79 diploid M. sinensis or M. sinensis×M. sacchariflorus ornamental cultivars available in the USA, 42 naturalized M. sinensis genotypes from 13 accessions collected in the USA, one diploid M. sacchariflorus ornamental cultivar, and the triploid biomass cultivar M.×giganteus ‘Illinois’ (Supplementary dataset S1).
Table 1.
Origins of Miscanthus accessions genotyped in the present study
| Island | Species | Seed only | Clonal only | Clonal + seed |
|---|---|---|---|---|
| Hokkaido | M. sinensis | 91 | ||
| M. sacchariflorus | 2 | 3 | ||
| Honshu | M. sinensis | 88 | 3 | |
| M. sacchariflorus | 3 | 32 | ||
| Shikoku | M. sinensis | 5 | ||
| Kyushu | M. sinensis | 14 | 1 | |
| M. sacchariflorus | 1 | 6 | ||
| Total | M. sinensis | 198 | 4 | 6 |
| M. sacchariflorus | 6 | 41 | 6 |
Restriction site-associated DNA sequencing (RAD-seq) and plastid genotyping were performed using methods described previously (Clark et al., 2014). For RAD-seq genotyping, a PstI–MspI digestion was used to sequence tags adjacent to PstI sites, and 95 barcoded samples were multiplexed into each of ten libraries. Each library was run in one lane on a HiSeq 2000 (Illumina, San Diego, California, USA) for 100bp single-end reads at the University of Illinois Roy J. Carver Biotechnology Center DNA Sequencing Unit. All sequencing data has been deposited in the NCBI Sequence Read Archive, BioProject ID PRJNA261699. All samples were also genotyped with ten plastid microsatellite markers (de Cesare et al., 2010; Jiang et al., 2012) scored by electrophoresis on an ABI 3730 (Applied Biosystems, now part of Thermo Fisher Scientific, Waltham, Massachusetts, USA) followed by allele calling in STRand (Toonen and Hughes, 2001).
Genetic data analysis
The UNEAK pipeline in TASSEL 3.0.162 (Lu et al., 2013) was used to call single nucleotide polymorphism (SNP) genotypes from RAD-seq data using a minimum call rate of 0.5 and a minimum minor allele frequency of 0.01. In addition to the 745 individuals from the Japan dense-sampling set, 645 individuals from the region-wide set, 42 US naturalized genotypes, and 81 cultivars (Clark et al., 2014) were included in the SNP-calling pipeline, yielding 20 704 SNPs after removing SNPs that appeared heterozygous in one or more doubled haploid lines. Though polyploidy represents a challenge for SNP-calling, the UNEAK pipeline was designed to distinguish paralogues in polyploids and the use of doubled haploid M. sinensis lines further enabled this differentiation (Clark et al., 2014).
SNPs were analysed with the software Structure 2.3.4 (Falush et al., 2003) to identify new genetic groups, assign individuals to previously identified groups (Clark et al., 2014), and detect admixture and hybridization between species (see also Supplementary Materials and methods). Structure Harvester (Earl and VonHoldt, 2011) was used to determine the best number of clusters (K). To determine the origins of ornamental and naturalized accessions of M. sinensis available in the US, the USEPOPINFO and PFROMPOPFLAGONLY options were used. To determine the power of Structure to detect hybridization, analyses were conducted on groups of simulated hybrid individuals using individuals from the dataset as parents, and on a simulated population of individuals from the common ancestor of M. sinensis and M. sacchariflorus. Principal components analysis performed with adegenet (Jombart and Ahmed, 2011) was also used to compare and validate the results from Structure.
The R package mmod (Winter, 2012) was used to calculate the differentiation statistic Jost’s D (Jost, 2008) using the 20 704 RAD-seq SNPs between pairs of groups as identified by discriminant analysis of principal components (DAPC; Jombart et al., 2010). Jost’s D was calculated individually for each locus, then averaged across loci. Nei’s D (expected heterozygosity) was calculated for the same groups using allele frequencies calculated by the glMean function in adegenet (Jombart and Ahmed, 2011). To control for differences in group size, for each genetic group, 500 jack-knifed subgroups containing 100 individuals each were used to calculate Nei’s D, and the mean and standard error were calculated across jack-knifed replicates. F ST was calculated in the R package pegas (Paradis, 2010) to determine the differentiation of each Japanese M. sinensis genetic group from Japanese M. sinensis as a whole.
Spatial principal components analysis (sPCA), implemented in the R package adegenet (Jombart et al., 2008) was used to identify spatial patterns in genetic variation of M. sinensis across the major islands of Japan using RAD-seq SNPs. The R package ade4 (Chessel et al., 2004) was used to plot the results.
A haplotype network was generated from all ten chloroplast microsatellite markers, as in Clark et al. (2014). Any individuals with missing data were removed from the haplotype network analysis, leaving 731 M. sinensis and M. sacchariflorus individuals from 252 accessions from the Japan dense-sampling set.
Flow cytometry
Flow cytometry was performed using a protocol modified from Rayburn et al. (2009). Flow cytometry was used to determine the nuclear DNA content of all M. sacchariflorus and M.×giganteus individuals for which live plants were available (72 out of 78 from the Japan dense-sampling set, plus two from Korea and nine from China from the region-wide set), as well as a sample of 32 M. sinensis individuals from the Japan dense-sampling set.
Results
Major groupings, admixture, and hybridization of Miscanthus based on SNP data
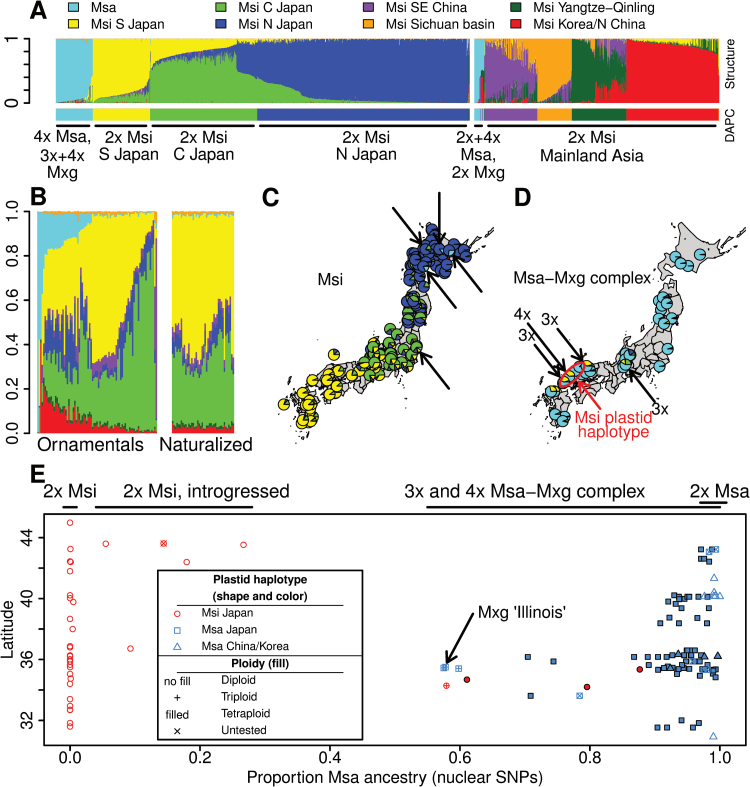
Structure analysis of the Japan dense-sampling set identified K=4 (three M. sinensis and one M. sacchariflorus) as the most reproducible estimate (Supplementary Fig. S1). Thus, the high density sampling in this study enabled identification of three M. sinensis groups in Japan (northern, central, and southern, hereafter called N, Central, and S Japan when referring to genetic clusters as opposed to geographic regions; Fig. 1A, C), where previous low density sampling had identified only two groups (northern and southern). A combined analysis of the Japan dense-sampling set with the region-wide set at K=8 identified the seven genetic groups from the previous study (six M. sinensis and one M. sacchariflorus; Clark et al., 2014) plus the one additional M. sinensis group identified in the analysis of the Japan dense-sampling set (Fig. 1A, C). The first principal component of the SNP data was strongly correlated with M. sacchariflorus ancestry identified by Structure (r 2=0.99; Supplementary Fig. S2A), and Structure runs on simulated hybrids indicated that even highly backcrossed (BC5) individuals could be distinguished from the parent species (Supplementary Fig. S2B, Table S1).
Fig. 1.
Structure and DAPC results using 20 704 nuclear SNPs. Msi=Miscanthus sinensis, Msa=M. sacchariflorus, Mxg=M.×giganteus. (A) Bar plot of Q values (proportion ancestry estimated in Structure) for 745 individuals from the Japan dense-sampling set and 645 individuals from the previously published region-wide set (Clark et al., 2014). Each of five runs included 253 individuals from the Japan-dense set (one per accession) plus all 645 individuals from the region-wide set; mean Q values are shown for individuals that were present in more than one run. Each of the eight groups is represented by a different colour. The narrower bottom bar indicates DAPC group assignments. (B) Mean Q values for 81 ornamental individuals and 42 naturalized individuals from the USA, when the parameters USEPOPINFO and PFROMPOPFLAGONLY were used in Structure to assign ancestry from native populations. (C) Map of Q values for Msi individuals in Japan, including 667 from the Japan-dense set and 128 from the region-wide set. Five individuals with Msa ancestry 6–27%, including four diploids and one of undetermined ploidy, are indicated with arrows. (D) Map of Q values for 78 Msa–Mxg complex individuals from Japan, all from the Japan dense-sampling set. Four individuals with Msi ancestry 39–42% are indicated with arrows, and the ploidy determined by flow cytometry is indicated; all other individuals shown were tetraploid except for six of undetermined ploidy. The red ellipse indicates the sampling area for all four Msa–Mxg individuals with an Msi plastid haplotype (other individuals within the ellipse have an Msa plastid haplotype). (E) Latitude vs Q values for 89 native-collected Msa–Mxg complex individuals, Mxg ‘Illinois’ (assuming origin in Yokohama, Japan; indicated with an arrow), 28 random Msi individuals that were subjected to flow cytometry, and five Msi individuals with Msa ancestry >5%. Colour and shape of symbols in (E) are used redundantly to indicate plastid haplotype and collection location, and fill is used to indicate ploidy, with filled points outlined in black to make them more easily visible.
Based on admixture estimates, M. sinensis genotypes in Japan were strongly isolated from each of the other five groups identified (Fig. 1A, Supplementary Dataset S1). Isolation of M. sinensis from M. sacchariflorus in Japan was especially strong. Only 9 of the 667 phenotypically M. sinensis genotypes evaluated had <99% M. sinensis ancestry. Unexpectedly, however, four diploid individuals from Hokkaido and one from Ibaraki (central Honshu) had hybrid ancestry >5% from M. sacchariflorus (27%, 18%, 14%, 6%, and 9% respectively), and were part of seed accessions that were otherwise non-hybrid (EBI-2009-02c, Koike-05a, EBI-2008-46c, EBI-2008-37e, JA55-2c; Supplementary dataset S1). For EBI-2009-02c, intermediate morphological characteristics were observed between M. sacchariflorus and M. sinensis, including axillary branching, which is characteristic of M. sacchariflorus, and trichomes on the abaxial surface of leaves, which is characteristic of M. sinensis (Fig. 2). Among the Japanese genotypes with ≥99% M. sinensis ancestry, only 39 out of 795 had less than 95% Japanese ancestry. Most of the non-Japanese admixture observed for M. sinensis from Japan was with the southeast (SE) China M. sinensis group (Fig. 1A, Supplementary Dataset S1).
Fig. 2.
Photographs of EBI-2009-02c, an M. sinensis×M. sacchariflorus individual grown from seed collected in Hokkaido, Japan. Ancestry of EBI-2009-02c according to Structure was ~73% M. sinensis from N Japan and ~27% M. sacchariflorus (Fig. 1A, C, E). Its plastid haplotype was commonly found among M. sinensis in Hokkaido (haplotype C, Fig. 4). (A) Close-up showing axillary branching and long internodes, which are characteristic of M. sacchariflorus. (B) Broader view, with more branching visible. (C–E) Abaxial leaf surface of three plants, showing presence or absence of trichomes. Scale is identical in C–E. (C) Non-hybrid M. sinensis displaying trichomes (arrow), which is typical for this species. (D) EBI-2009-02c, with trichomes (arrow). (E) Diploid non-hybrid M. sacchariflorus, with a glabrous phenotype that is typical of this species.
Of the three Japanese M. sinensis genetic groups (as identified by DAPC), the N Japan group was the least diverse and the Central Japan group was the most diverse, both in terms of plastid haplotypes and nuclear SNPs (Table 2). Among the three Japanese M. sinensis groups, S Japan was the most differentiated from the others based on F ST, and Central Japan was the least differentiated (Table 2). Pairwise Jost’s D between DAPC groups revealed that the S Japan group was more closely related to the SE China group than to the N Japan group (Table 3). Of the three M. sinensis groups in Japan, S Japan was the most closely related to each mainland Asia group, and N Japan was the most distantly related (Table 3).
Table 2.
Diversity statistics for Japanese groups of M. sinensis (Msi) and the M. sacchariflorus–M.×giganteus complex (Msa–Mxg)
For Msi, the Gini-Simpson index and Nei’s D were estimated using 500 jack-knifed groups containing 100 individuals from each Msi group, and standard errors are calculated across jack-knifed replicates. For Msa–Mxg, the Gini-Simpson index was estimated without jack-knifing owing to small sample size, and Nei’s D was not calculated because of expected bias from the SNP-mining method (most individuals used for SNP mining in UNEAK were Msi). F ST indicates differentiation of each Japanese Msi group from the other two, and the standard error is given across loci.
| Group | Number of individuals | Number of plastid haplotypes | Gini-Simpson index, plastid haplotypes | Nei’s D, nuclear SNPs | F ST, nuclear SNPs |
|---|---|---|---|---|---|
| N Japan Msi | 446 | 16 | 0.4533±0.0022 | 0.13055±0.00003 | 0.0421±0.0004 |
| C Japan Msi | 226 | 19 | 0.7747±0.0011 | 0.13460±0.00002 | 0.0238±0.0003 |
| S Japan Msi | 122 | 17 | 0.7606±0.0002 | 0.13217±0.00001 | 0.0455±0.0005 |
| All Japan Msi | 794 | 38 | 0.7911±0.0006 | 0.14247±0.00001 | |
| Japan Msa–Mxg | 78 | 19 | 0.8340±0.0375 |
Table 3.
Pairwise Jost’s D of Japanese M. sinensis groups
Mean and standard error were calculated across 20 704 SNP loci. Colour names correspond to colours in Fig. 1.
| C Japan | S Japan | Korea, N China (red) | SE China (purple) | Yangtze-Qinling (dark green) | Sichuan (orange) | Msa (cyan) | |
|---|---|---|---|---|---|---|---|
| N Japan (blue) | 0.0169±0.0003 | 0.0313 ± 0.0006 |
0.0571±0.0009 | 0.0492±0.0008 | 0.0597±0.0009 | 0.0743±0.0011 | 0.1224±0.0017 |
| C Japan (light green) | 0.0127±0.0003 | 0.0403±0.0007 | 0.0326±0.0006 | 0.0435±0.0008 | 0.0592±0.0010 | 0.1080±0.0016 | |
| S Japan (yellow) | 0.0337±0.0006 | 0.0294±0.0006 | 0.0395±0.0007 | 0.0567±0.0010 | 0.1058±0.0016 |
Ornamental M. sinensis and M. sinensis×M. sacchariflorus accessions from the USA had, on average, 39% ancestry to the S Japan genetic group, 32% to the Central Japan group, 11% to the N Japan group, 6% to the Korea/N China group, and 7% to the M. sacchariflorus group (Fig. 1B). Naturalized M. sinensis accessions collected in the USA had more uniform Q values among individuals than ornamental cultivars, and most of their ancestry was from S and Central Japan. Although M. sacchariflorus ancestry was negligible (0.7%) within the naturalized USA accessions, they did have 2.6% ancestry from the Korea/N China group, whereas native Japanese M. sinensis only had 0.2% ancestry from the Korea/N China group.
For all M. sacchariflorus studied, including those from Japan, China and Korea, a single group was identified via Structure analysis (Fig 1A). Nine individuals from S Japan with M. sacchariflorus phenotypes (six of which were collected as clones and three as seeds) had hybrid ancestry (Q values) >20% from M. sinensis, three of which were triploid and five tetraploid (ploidy was undetermined for one individual owing to loss of the plant; Fig. 1A, D, E, Table 4); thus, these individuals were probably F1 and BC1 interspecific hybrids (i.e. M.×giganteus). Only 11 of the 69 phenotypically M. sacchariflorus Japanese genotypes that were confirmed to be tetraploid had ≥98% of their nuclear alleles from M. sacchariflorus, with the remaining accessions having M. sinensis ancestry (predominantly Japanese) ranging from 2–39%, with a mean of 7% and median of 5% (Fig. 1A and E). Thus, recurrent backcrossing of hybrid individuals probably produced the observed gradient of M. sinensis introgression into M. sacchariflorus (Fig. 1E). For phenotypically M. sacchariflorus individuals with <20% M. sinensis ancestry, latitude was negatively correlated with M. sinensis ancestry (Fig. 1E), indicating that introgression was more frequent in southern Japan than northern Japan. In contrast to the frequent introgression of M. sinensis genes into tetraploid M. sacchariflorus in Japan, seven diploid and three tetraploid M. sacchariflorus from China each had ≥98% M. sacchariflorus ancestry. However, two M. sacchariflorus from Korea had only 92–94% M. sacchariflorus ancestry, with most of the remainder from the Korea/N China M. sinensis group.
Table 4.
F1 and BC1 M.×giganteus collected from the wild in Japan
M.×giganteus ‘Illinois’ is included for comparison. Msa=M. sacchariflorus. Proportion Msa ancestry=Q value estimated by Structure.
| Accession | Type | Prefecture | Proportion Msa ancestry | Ploidy | Plastid haplotype group |
|---|---|---|---|---|---|
| JM11-006 | Clone | Yamaguchi | 0.796 | 4× | B |
| JA52a | Seed | Fukuoka | 0.784 | NA (dead plant) | Msa |
| Gifu-2010-020d | Seed | Gifu | 0.744 | 4× | Msa |
| JM11-002 | Clone | Fukuoka | 0.709 | 4× | Msa |
| Gifu-2010-014a | Seed | Gifu | 0.705 | 4× | Msa |
| JM11-013 | Clone | Shimane | 0.611 | 4× | B |
| JM11-031 | Clone | Tottori | 0.598 | 3× | Msa |
| JM11-010 | Clone | Yamaguchi | 0.579 | 3× | B |
| Gifu-2010–025 | Clone | Gifu | 0.578 | 3× | Msa |
| ‘Illinois’ | Clone | Kanagawa | 0.575 | 3× | Msa |
Spatial analysis of M. sinensis SNP data
If M. sinensis individuals were sorted by Q value, the values changed abruptly in several regions of the bar plot, suggesting barriers to gene flow (Fig. 1A). Spatial principal components analysis of nuclear SNP data indicated the geographical locations and relative strengths of these barriers to gene flow for M. sinensis in Japan (Fig. 3). The first three eigenvectors with positive spatial autocorrelation were chosen for analysis, based on a screeplot of genetic variance vs spatial autocorrelation (Supplementary Fig. S3). The first eigenvector, which had by far the highest variance (Fig. 3A; 12.7% of genetic variation between sites), represented a genetic gradient north to south in Japan, as well as differentiation of the region to the southwest of the Noto Peninsula. The second eigenvector, representing 2.7% of the genetic variation between sites, revealed central Honshu as the most divergent region, and a steep genetic cline near the Japanese Alps (Fig. 3B). The third eigenvector, representing 1.4% of genetic variation between sites, showed a gradient from east to west (Fig. 3C). None of these three eigenvectors revealed genetic structure within Hokkaido despite the large sample size in that region.
Fig. 3.
Spatial principal components analysis (sPCA) of M. sinensis in Japan using 5359 SNPs across 782 individuals from 205 collection sites. (A–C) Interpolation of scores of lag vectors of the first three eigenvectors produced by sPCA. Scores are represented the darkness of greyscale pixels. The percentage of genetic variation between sites explained by each of the three eigenvectors is indicated. (This figure is available in colour at JXB online.)
Plastid microsatellites
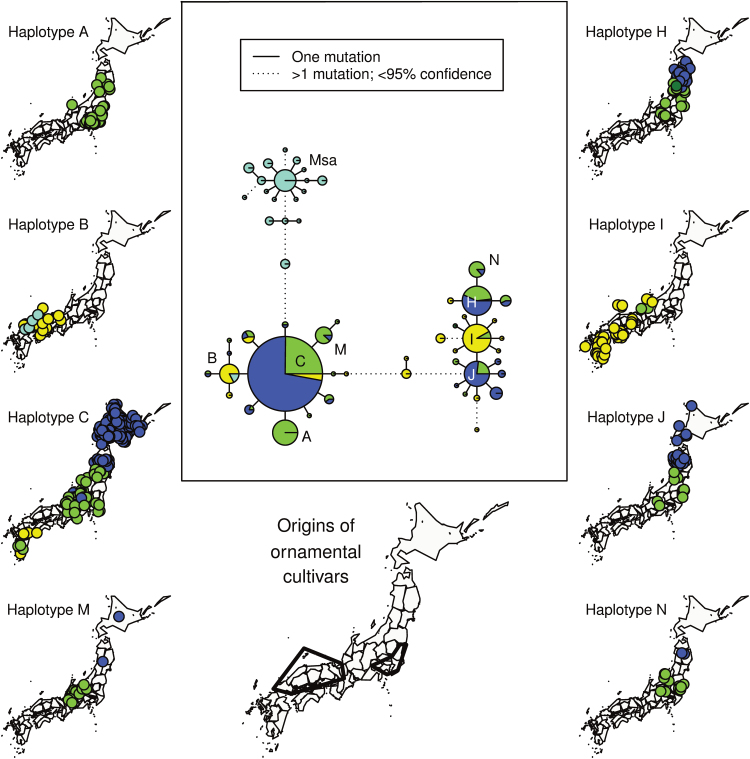
Across the Japan dense-sampling set of 745 individuals (255 accessions), 57 unique plastid haplotypes were identified. From these, a haplotype network was calculated (Fig. 4), which included a sub-network of haplotypes specific to M. sacchariflorus and two major sub-networks of haplotypes common in M. sinensis. The topology of the haplotype network (Fig. 4) was slightly different from the previously published network (Clark et al., 2014) owing to homoplasy of microsatellite alleles, absence of haplotypes present only in mainland Asia, and/or differences in haplotype frequency. Strong geographic structure was seen among the major M. sinensis haplotypes (Fig. 4), including haplotypes A and B which are common among ornamental cultivars available in the USA and Europe (Clark et al., 2014). Of the 78 phenotypically M. sacchariflorus individuals (53 accessions) genotyped, four did not have a plastid haplotype that was part of the M. sacchariflorus sub-network, but instead had haplotype B, an M. sinensis haplotype common in Shikoku and southern Honshu where those four accessions were collected (Figs. 1D, E and 4) and not found anywhere else in Asia (Clark et al., 2014). These four interspecific hybrid individuals with M. sinensis plastids were collected along the west coast of Chūgoku (Fig. 1D) and 58–88 % of their nuclear DNA was from M. sacchariflorus with the remainder from M. sinensis; one individual was triploid and the others were tetraploid. Though M. sinensis plastids were found introgressed into M. sacchariflorus, M. sacchariflorus plastids were not found introgressed into M. sinensis.
Fig. 4.
Haplotype network of Miscanthus based upon ten plastid microsatellite markers, and maps of sampling locations of the most common haplotypes. Circle area in the network is proportional to the number of unique accessions with each haplotype, and pie slice area is proportional to the number of individuals in each DAPC group as determined by nuclear SNPs (Fig. 1A, lower bar, with corresponding colours). Msa indicates the sub-network belonging to M. sacchariflorus. The rest of the network is found in M. sinensis. The eight most common haplotypes in M. sinensis are indicated with letters. Haplotypes A, B, C, H, I, and J correspond to identically named haplotypes from Clark et al. (2014). Probable geographic origins of ornamental M. sinensis cultivars (bottom, centre) were determined by the presence of haplotypes A, B, and I and the absence of other haplotypes.
DNA content
All tested individuals that had >60% M. sacchariflorus ancestry were tetraploid, with the exception of seven diploids from China (Fig. 1E). As expected, all tested M. sinensis individuals were diploid (Fig. 1E). Of the five interspecific hybrids that phenotypically resembled M. sinensis and had M. sinensis plastids, but had 6–27% M. sacchariflorus ancestry based on nuclear SNPs, four were determined to be diploid (Fig. 1E), and the fifth individual died before it could be tested.
Discussion
Introgression of M. sinensis DNA into tetraploid M. sacchariflorus
Though previous studies have also identified triploid hybrids between tetraploid M. sacchariflorus and diploid M. sinensis from wild populations in Japan (Hirayoshi et al., 1957, 1960; Adati and Shiotani, 1962; Hodkinson et al., 2002c; Nishiwaki et al., 2011; Dwiyanti et al., 2013) and even one interspecific tetraploid hybrid (Dwiyanti et al., 2013), this is the first study to establish that introgression of M. sinensis DNA into tetraploid M. sacchariflorus is common in Japan, resulting in a tetraploid population that has a continuous gradient of M. sinensis nuclear genetic ancestry ranging up to 39%. Only 16% of the phenotypically M. sacchariflorus tetraploids from Japan had ≥98% M. sacchariflorus ancestry, whereas all nine of the M. sacchariflorus from China that were studied (seven diploids and two tetraploids) exceeded this threshold. Similar to the unintrogressed M. sacchariflorus from China, ≥99% of M. sinensis from Japan had ≥99% M. sinensis ancestry (Fig. 1A, E). Moreover, the amount of introgression of M. sinensis DNA into M. sacchariflorus was negatively correlated with latitude (Fig. 1E), and is highest where flowering times of M. sinensis and M. sacchariflorus have the greatest overlap and where alleles for adaptation to a warm climate would be expected to have the greatest benefit to M. sacchariflorus. Thus, the tetraploid Miscanthus in Japan, which had been considered to be allo- or autopolyploid M. sacchariflorus, are in fact predominantly a hybrid swarm derived from autotetraploid M. sacchariflorus and diploid M. sinensis. In contrast to the tetraploid hybrid swarm that we identified in Japan, Jiang et al. (2013) reported that hybrids between diploid M. sacchariflorus and diploid M. sinensis in China had approximately equal genetic contributions from both parents, and they did not find evidence of introgression from one species into the other. Another notable contrast between the tetraploid interspecific hybrids in Japan and diploid interspecific hybrids in China is that the tetraploids in Japan are phenotypically most similar to the M. sacchariflorus parent, including the development of long rhizomes, whereas the interspecific diploids in China are phenotypically most similar to the M. sinensis parent, including caespitose habit. Such differences in the growth characteristics of the hybrids would be expected to have substantial effects on their adaptation and competitiveness.
This study is also the first to report M. sinensis plastids introgressed into wild-collected tetraploid M. sacchariflorus from Japan, with about 4% of the phenotypically M. sacchariflorus tetraploids having M. sinensis plastids in nuclear genetic backgrounds that ranged from 61–88% M. sacchariflorus. Consistent with the findings of M. sinensis plastids introgressed into wild M. sacchariflorus tetraploids from Japan, Hirayoshi et al. (1960) produced a triploid and a tetraploid progeny from a purposeful cross between diploid M. sinensis var. condensatus as the female parent and tetraploid M. sacchariflorus as the male parent. In China, wild-collected hybrids between diploid M. sacchariflorus and diploid M. sinensis were also found to have plastids from either parent (Jiang et al., 2013; Clark et al., 2014).
Cytogenetic evidence has resulted in conflicting reports as to whether tetraploid M. sacchariflorus is allopolyploid or autopolyploid, although the most modern studies suggest autopolyploidy (Adati and Shiotani, 1962; Takahashi and Shibata, 2002). The present study also does not support the allopolyploid hypothesis. If the M. sacchariflorus in Japan were allotetraploid, derived from diploid M. sacchariflorus and diploid M. sinensis, it would be expected that at least half their ancestry would be from M. sinensis, but this was not observed, as M. sinensis ancestry was typically <20% and never greater than 39% (Fig 1E). Moreover, it was found that the tetraploid M. sacchariflorus from Japan co-clustered with the diploid and tetraploid M. sacchariflorus from China and Korea. Thus, tetraploid M. sacchariflorus in Japan probably originated via autopolyploidization of diploid M. sacchariflorus, but subsequent and ongoing crosses with diploid M. sinensis have resulted in a predominantly interspecific hybrid population of tetraploids in Japan. Given that both diploid and tetraploid hybrids between M. sacchariflorus and M. sinensis are typically fertile and have normal meioses (Hirayoshi et al., 1960; Jiang et al., 2013; Clark et al., 2014), their genomes may not be sufficiently differentiated to result in allopolyploid speciation of hybrids. Whereas polyploidization of amphidiploids is typically considered a speciation event, polyploidization of M. sacchariflorus has facilitated considerable introgression of genes from diploid M. sinensis in Japan (i.e. brought the nascent differentiating genomes of M. sacchariflorus and M. sinensis back together). Thus, for M. sacchariflorus and M. sinensis, speciation seems to be an ongoing, lengthy, and dynamic process, rather than a single discrete event.
Adaptive advantages of interspecific tetraploids to a warming climate provide a possible explanation for the absence of diploid M. sacchariflorus in Japan. Temperatures and flora in much of Japan around the last glacial maximum were similar to those of contemporary inland eastern Russia, where diploid M. sacchariflorus is common but M. sinensis is absent or rare (Winkler and Wang, 1993; Adams and Faure, 1997; Ray and Adams, 2001). When the climate subsequently warmed and M. sinensis migrated from a refuge in southeast Asia to Japan ~14 000 years before present (Clark et al., 2014), the cold-adapted M. sacchariflorus in Japan may have benefited from the introgression of M. sinensis genes that conferred adaptation to warmer environments. Moreover, introgression would have preferentially favoured fitness of a tetraploid M. sacchariflorus–M.×giganteus complex over diploid M. sacchariflorus and diploid interspecific hybrids, owing to the tetraploid hybrids’ competitive rhizomatous habit combined with heterosis and adaptation to a warming climate, thus possibly explaining why diploid M. sacchariflorus is absent or exceedingly rare in Japan today. This hypothesis is consistent with the prediction of Stebbins (1971) that cross-ploidy introgressions have played a large role in ecological adaptation. Japan’s maritime climate, lacking extreme temperatures, may have driven the M. sacchariflorus conversion from diploid to tetraploid more completely than in mainland Asia. Observations from a field trial located in southern Illinois at the Dixon Springs Experiment Station (37.4° N; USDA hardiness zone 6/7) indicate that some diploid M. sacchariflorus are unadapted to warm temperate environments (e.g. by flowering and going dormant many months before the end of the growing season), whereas all tested tetraploid M. sacchariflorus from Japan are well-adapted to such environments. However, the range of putatively diploid M. sacchariflorus in China extends as far south as 28°N (Sacks et al., 2013). As subsequent analyses of population structure allow the identification of the closest living diploid M. sacchariflorus relatives of Japanese tetraploid M. sacchariflorus, it will be possible to more fully test this hypothesis by comparing the adaptation of these diploid M. sacchariflorus to that of their induced tetraploids, their diploid and tetraploid progeny from crosses with diploid M. sinensis from southern Japan, and natural tetraploid M.×giganteus and M. sacchariflorus genotypes. Additionally, if the hypothesis is correct, we expect to see introgression from M. sinensis into M. sacchariflorus press northward as the climate warms.
The new understanding that most of the tetraploid phenotypically M. sacchariflorus in Japan are in fact backcross hybrids between M. sacchariflorus and M. sinensis with variable degrees of introgression from M. sinensis also leads to an interesting question of nomenclature. Hodkinson and Renvoize (2001) defined the hybrid between M. sacchariflorus and M. sinensis as the nothospecies M.×giganteus. The International Code of Nomenclature for algae, fungi, and plants (McNeill et al., 2012) further indicates that a nothotaxa includes all filial and backcross individuals that are recognizably derived from the defined parental taxa. Molecular markers have allowed pure M. sacchariflorus to be distinguished from F1 and backcross hybrids with M. sinensis, though phenotypically even F1 triploid M.×giganteus can be difficult to distinguish from tetraploid M. sacchariflorus. Thus, many of the tetraploid genotypes in Japan that look like M. sacchariflorus phenotypically may be most accurately referred to as M.×giganteus. Perhaps it would be most accurate to refer to this group in Japan as an M. sacchariflorus–M.×giganteus complex. Nomenclature details aside, researchers should be cognizant of the complex nature of the tetraploid Miscanthus populations in Japan.
For the development of biomass cultivars, the new triploid F1 M.×giganteus accessions that were identified here and imported into the USA will be immediately useful in field trials to compare their performance to the current agronomic standard, M.×giganteus ‘Illinois’. Though M. sacchariflorus is the maternal parent of ‘Illinois’ (Hodkinson et al., 2002c), the results here indicate that M. sinensis can be the maternal parent with similar probability. Crosses to create new M.×giganteus can therefore be performed in either direction, and maternal cytoplasmic effects on the performance of new hybrids should be investigated. Also, the degree of introgression from M. sinensis should now be taken into account when selecting a parent from the tetraploid M. sacchariflorus–M.×giganteus complex for crosses with diploid M. sinensis and evaluating the performance of their progeny. As traits of interest are mapped on the M. sinensis and M. sacchariflorus genomes in the near future, it is possible that a greater genetic dosage from one parent species or the other will be desired for particular genes or genomic regions, in which case particular tetraploid M. sacchariflorus–M.×giganteus accessions may be selected as parents based on which regions of the M. sinensis genome they do or do not possess.
Introgression of M. sacchariflorus DNA into diploid M. sinensis
Although much less frequent than introgression of diploid M. sinensis into tetraploid M. sacchariflorus, introgression of M. sacchariflorus DNA into M. sinensis was also observed, particularly in Hokkaido (Figs. 1A, C, E and 2). The M. sinensis individual with the greatest amount of M. sacchariflorus DNA, EBI-2009-02c, was collected in Rikubetsu, the coldest place in Japan, where Miscanthus is rare (T. Yamada, personal observation). Hybridization of M. sinensis and M. sacchariflorus to produce diploid progeny that could backcross to diploid M. sinensis is difficult to explain, given that no endemic diploid M. sacchariflorus are known to exist in Japan. Recent importation of diploid M. sacchariflorus from China is one possible explanation for the presence of interspecific diploid progeny. Another possibility is the production of highly rare monoploid gametes from tetraploid M. sacchariflorus or triploid M.×giganteus. However, M.×giganteus from the south is unlikely to have contributed to the ancestry of these diploid M. sinensis× M. sacchariflorus hybrids, which were found in the north, because the diploid hybrids did not have any ancestry from the S Japan M. sinensis group. Moreover, in northern Japan tetraploid M. sacchariflorus typically flower much later than M. sinensis. Lastly, the possibility must be considered that endemic populations of diploid M. sacchariflorus either recently existed in northern Japan and have been lost, or exist currently but have remained undetected, and these diploid M. sacchariflorus crossed with M. sinensis.
Spatial genetic structure in M. sinensis
Spatial principal components analysis revealed several distinct genetic clines in M. sinensis, which reflect different demographic processes and their relative importance in shaping the population structure of M. sinensis in Japan. The largest eigenvector by far indicated a cline from south to north (Fig. 3A), which could reflect progressive founder effects as M. sinensis migrated from southeast China and colonized Japan (Clark et al., 2014). The largest eigenvector also corresponds to geographic distance from Korea and the Ryukyu islands, with which ongoing genetic exchange is probably taking place. The region to the southwest of the Noto Peninsula was also distinguished by the first eigenvector, perhaps because this maritime region is isolated by mountains and ocean from other nearby maritime regions. Consistent with the Structure analysis identifying a distinct central Japan M. sinensis group (Fig. 1C), the second eigenvector distinguished central Honshu from northern and southern Japan (Fig. 3B). The Japanese Alps have been a barrier to gene flow, as indicated by a steep cline in this region for the second eigenvector (Fig. 3B) and by greater genetic differentiation for the S Japan group than the other Japanese M. sinensis groups to the north of these mountains (Table 2). The combination of the first and second eigenvectors gave a similar pattern to a genetic cline previously found across southern Japan and Korea in M. sinensis (Slavov et al., 2013) and is also similar to the pattern that was seen with Structure (Fig. 1A). The third eigenvector showed a cline east to west in central and southern Japan (Fig. 3C), suggesting that there may be gene flow along the coasts that bypasses the clines seen in the first two eigenvectors. Most of the genetic variation between sites (83.2%) remained unexplained by these three eigenvectors, indicating that obligate outcrossing and wind dispersal of seed and pollen, in combination with the relatively recent colonization of Japan by M. sinensis (within the past ~14,000 years; Clark et al., 2014) resulted in an unstructured pattern of allele frequencies at most loci.
The plastid results were consistent with those of previous studies (Shimono et al., 2013; Clark et al., 2014), which indicated the presence of two major groups of plastid haplotypes in Japan that probably correspond to two or more colonization events. Additionally, it was found that the eight most common haplotypes for M. sinensis in Japan (four from each of the two major groups) all had well-defined geographic ranges (Fig. 4), indicating strong barriers to seed flow. The Tsugaru Strait is one such barrier, given that the Hokkaido population almost exclusively had haplotype C, despite haplotypes H and J being common nearby in northern Honshu. Kyushu is similarly isolated; the only major haplotypes found there are C and I, despite haplotype B being common nearby in southern Honshu and Shikoku. The Japanese Alps also seem to block seed flow, given that haplotypes A, H, J, M, and N were only found north of the mountain range, whereas haplotypes B and I were only found south of it. Cytoplasmic markers can exhibit stronger population structure than nuclear markers owing to undergoing a higher rate of genetic drift as a result of having a smaller effective population size (McCauley, 1995).
Origins of ornamental cultivars and naturalized M. sinensis in the USA
In this study, geographic resolution was added to the previous finding that ornamental and naturalized M. sinensis in the USA originated from southern Japan (Clark et al., 2014). The Q values (Fig. 1B) and the geographic distribution of plastid haplotypes (Fig. 4) for the ornamental cultivars indicated that there were multiple introductions to the USA but from two small areas (Fig. 4) of east-central Japan (eastern parts of Kantō and Chūbu) and south-western Japan (Chūgoku and western Kansai). The Q values among the naturalized populations in the USA were less varied than Q values among ornamental cultivars, indicating that these populations originated from a small subset of ornamental cultivars. These findings on the origins of the ornamental cultivars in the USA and Europe are consistent with historical documentation that the Yokohama Nursery Company played an important role in distributing Japanese plants, including Miscanthus, internationally during the late 1800s and early 1900s (Galloway, 1907, see entry 10524; http://www.nal.usda.gov/exhibits/speccoll/exhibits/show/nursery-and-seed-trade-catalog/japanese-nursery-and-seed-trad, last accessed 5 January 2015; http://www.yokohamaueki.co.jp/ayumi/index.html, last accessed 5 January 2015). The ornamental M. sinensis×M. sacchariflorus ‘Purpurascens’ was a likely ancestor of many of the other ornamental cultivars, given that its M. sinensis ancestry is from the Korea/N China genetic group (red, Fig. 1A, B), and that ornamental cultivars with ancestry from the Korea/N China cluster tended to have a similar amount of ancestry from M. sacchariflorus (Fig. 1B). Among ornamentals and US naturalized accessions with negligible M. sacchariflorus ancestry, a small but significant amount of Korea/N China ancestry was present in some genotypes, possibly indicating purifying selection to remove M. sacchariflorus genomic regions, given that this pattern of admixture was rare in the native range.
Conclusions
In Japan, speciation between tetraploid M. sacchariflorus and diploid M. sinensis is an ongoing and dynamic process, with gene exchange occurring in both directions but asymmetrically in favour of diploid to tetraploid. Tetraploidy seems to have promoted introgression of genes from diploid M. sinensis into tetraploid M. sacchariflorus in Japan to a greater extent than gene exchange between sympatric diploid M. sinensis and diploid M. sacchariflorus populations in China. These conclusions are consistent with the theory of Stebbins (1971) that unilateral introgressive hybridization across ploidy levels can play an important role in plant evolution. The M.×giganteus–M. sacchariflorus complex in Japan is expected to be an outstanding resource for developing new biomass cultivars.
To develop improved biomass cultivars of Miscanthus, it will be desirable to genetically map agronomic traits in M. sinensis and tetraploid M. sacchariflorus, and identify the best parents of each species for breeding new triploid M.×giganteus cultivars. Artificial backcrossed populations, derived from crosses between tetraploid M.×giganteus and tetraploid M. sacchariflorus, will be useful for elucidating the role of M. sinensis genes introgressed into a tetraploid M. sacchariflorus genetic background. Experiments with such introgressants will provide insights into their possible selective advantage to wild populations of M. sacchariflorus, as well as their potential utility for breeding biomass cultivars. Additionally, an understanding of how the degree of M. sinensis introgression in tetraploid M. sacchariflorus–M.×giganteus complex genotypes affects heterosis of triploid M.×giganteus hybrids would be useful for breeding bioenergy cultivars. The Miscanthus cultivars that are ultimately developed for the bioenergy industry are likely to be complex hybrids possessing traits from multiple species and geographic regions.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Choice of K in Structure analysis.
Figure S2. Validation of Structure for detecting introgression.
Figure S3. Screeplot from sPCA.
Table S1. Expected and observed Q values from Structure runs on simulated hybrid individuals.
Supplementary Dataset S1. A Microsoft Excel file containing a table of collection data and genetic results by individual, as well as a table providing the sequences of 289 pairs of RAD-tags that are highly diagnostic of M. sinensis vs M. sacchariflorus.
Acknowledgments
We thank Melina Salgado, Megan Swanson, Kasia Dubiel, Yingying Zheng, Jack Cygan, and Travis Hurt for assistance with plant propagation, tissue collection, DNA extraction, and PCR. PstI adapters were provided by Pat Brown. We thank Jessica Petersen, Stephen Downie, Katy Heath, and two anonymous reviewers for providing valuable feedback on an earlier version of this manuscript. This work was supported by the Office of Science - Biological and Environmental Research, US Department of Energy [Project ID 0017582] and by the Energy Biosciences Institute. Lindsay Clark is grateful for support by a travel award from the US Department of Energy, Office of Science, Office of Basic Energy Sciences (BES) and the Office of Biological and Environmental Research (BER), DE-FOA-0000995.
References
- Adams JM, Faure H. 1997. Preliminary vegetation maps of the world since the last glacial maximum: an aid to archaeological understanding. Journal of Archaeological Science 24, 623–647. [Google Scholar]
- Adams KL, Wendel JF. 2005. Polyploidy and genome evolution in plants. Current Opinion in Plant Biology 8, 135–141. [DOI] [PubMed] [Google Scholar]
- Adati S. 1958. Studies on the genus Miscanthus with special reference to the Japanese species suitable for breeding purposes as fodder crops. Bulletin of the Faculty of Agriculture, Mie University 17, 1–112. [Google Scholar]
- Adati S. 1959. Cytogenetics of Japanese wild forage Miscanthus species. Proceedings of the X International Congress of Genetics, August 20–27, 1958, McGill University, Montreal, Canada. Toronto: University of Toronto Press. [Google Scholar]
- Adati S, Shiotani I. 1962. The cytotaxonomy of the genus Miscanthus and its phylogenic status. Bulletin of the Faculty of Agriculture, Mie University 25, 1–24. [Google Scholar]
- Arnold ML. 2004. Transfer and origin of adaptations through natural hybridization: were Anderson and Stebbins right? The Plant Cell 16, 562–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold ML, Cornman RS, Martin NH. 2008. Hybridization, hybrid fitness and the evolution of adaptations. Plant Biosystems 142, 166–171. [Google Scholar]
- Barabaschi D, Guerra D, Lacrima K, Laino P, Michelotti V, Urso S, Valè G, Cattivelli L. 2012. Emerging knowledge from genome sequencing of crop species. Molecular Biotechnology 50, 250–266. [DOI] [PubMed] [Google Scholar]
- Barney JN, Ditomaso JM. 2008. Nonnative species and bioenergy: are we cultivating the next invader? BioScience 58, 64–70. [Google Scholar]
- De Cesare M, Hodkinson TR, Barth S. 2010. Chloroplast DNA markers (cpSSRs, SNPs) for Miscanthus, Saccharum and related grasses (Panicoideae, Poaceae). Molecular Breeding 26, 539–544. [Google Scholar]
- Chae WB, Hong SJ, Gifford JM, Rayburn AL, Sacks EJ, Juvik JA. 2014. Plant morphology, genome size, and SSR markers differentiate five distinct taxonomic groups among accessions in the genus Miscanthus . GCB Bioenergy 6, 646–660. [Google Scholar]
- Chessel D, Dufour AB, Thioulouse J. 2004. The ade4 package - I : One-table methods. R News 4, 5–10. [Google Scholar]
- Clark LV, Brummer JE, Głowacka K, et al. 2014. A footprint of past climate change on the diversity and population structure of Miscanthus sinensis . Annals of Botany 114, 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton-Brown J, Chiang Y-C, Hodkinson TR. 2008. Miscanthus: genetic resources and breeding potential to enhance bioenergy production. In: Vermerris W, ed. Genetic improvement of bioenergy crops. Berlin: Springer, 273–294. [Google Scholar]
- Dwiyanti MS, Rudolph A, Swaminathan K, et al. 2013. Genetic analysis of putative triploid Miscanthus hybrids and tetraploid M. sacchariflorus collected from sympatric populations of Kushima, Japan. BioEnergy Research 6, 486–493. [Google Scholar]
- Earl DA, VonHoldt BM. 2011. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4, 359–361. [Google Scholar]
- Falush D, Stephens M, Pritchard JK. 2003. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 164, 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway BT. 1907. Seeds and plants imported during the period from December, 1903, to December, 1905; inventory no. 11, nos.9897 to 16796. United States Department of Agriculture Bureau of Plant Industry-Bulletin no. 97. Washington: United States. Bureau of Plant Industry. [Google Scholar]
- Głowacka K, Clark LV, Adhikari S, et al. 2014. Genetic variation in Miscanthus× giganteus and the importance of estimating genetic distance thresholds for differentiating clones. GCB Bioenergy doi: 10.1111/gcbb.12166. [Google Scholar]
- Greef J, Deuter M, Jung C, Schondelmaier J. 1997. Genetic diversity of European Miscanthus species revealed by AFLP fingerprinting. Genetic Resources and Crop Evolution 44, 185–195. [Google Scholar]
- Hirayoshi I, Nishikawa K, Hakura A. 1960. Cyto-genetical studies on forage plants (VIII) 3×- and 4×- hybrid arisen from the cross Miscanthus sinensis var. condensatus× Miscanthus sacchariflorus . Research Bulletin of the Faculty of Agriculture, Gifu University 12, 82–88. [Google Scholar]
- Hirayoshi I, Nishikawa K, Kubono M, Murase T. 1957. Cyto-genetical studies on forage plants (VI): On the chromosome number of Ogi (Miscanthus sacchariflorus). Research Bulletin of the Faculty of Agriculture, Gifu University 8, 8–13. [Google Scholar]
- Hodkinson T, Chase M, Lledó D, Salamin N, Renvoize SA. 2002. a Phylogenetics of Miscanthus, Saccharum and related genera (Saccharinae, Andropogoneae, Poaceae) based on DNA sequences from ITS nuclear ribosomal DNA. Journal of Plant Research 115, 381–392. [DOI] [PubMed] [Google Scholar]
- Hodkinson TR, Chase MW, Renvoize SA. 2002. b Characterization of a genetic resource collection for Miscanthus (Saccharinae, Andropogoneae, Poaceae) using AFLP and ISSR PCR. Annals of Botany 89, 627–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodkinson TR, Chase MW, Takahashi C, Leitch IJ, Bennet MD, Renvoize SA. 2002. c. The use of DNA sequencing (ITS and trnL-F), AFLP, and fluorescent in situ hybridization to study allopolyploid Miscanthus (Poaceae). American Journal of Botany 89, 279–286. [DOI] [PubMed] [Google Scholar]
- Hodkinson TR, Renvoize S. 2001. Nomenclature of Miscanthus xgiganteus (Poaceae). Kew Bulletin 56, 759–760. [Google Scholar]
- Honda M. 1939. Nuntia ad Floram Japoniae. XXXVIII. The Botanical Magazine 53, 99–101. [Google Scholar]
- Jiang J-X, Wang Z-H, Tang B-R, Xiao L, Ai X, Yi Z-L. 2012. Development of novel chloroplast microsatellite markers for Miscanthus species (Poaceae). American Journal of Botany 99, e230–e233. [DOI] [PubMed] [Google Scholar]
- Jiang J, Zhu M, Ai X, Xiao L, Deng G, Yi Z. 2013. Molecular evidence for a natural diploid hybrid between Miscanthus sinensis (Poaceae) and M. sacchariflorus . Plant Systematics and Evolution 299, 1367–1377. [Google Scholar]
- Jombart T, Ahmed I. 2011. adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27, 3070–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T, Devillard S, Balloux F. 2010. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genetics 11, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T, Devillard S, Dufour a-B, Pontier D. 2008. Revealing cryptic spatial patterns in genetic variability by a new multivariate method. Heredity 101, 92–103. [DOI] [PubMed] [Google Scholar]
- Jost L. 2008. G ST and its relatives do not measure differentiation. Molecular Ecology 17, 4015–4026. [DOI] [PubMed] [Google Scholar]
- Kim M, Cui M-L, Cubas P, Gillies A, Lee K, Chapman MA, Abbott RJ, Coen E. 2008. Regulatory genes control a key morphological and ecological trait transferred between species. Science 322, 1116–1119. [DOI] [PubMed] [Google Scholar]
- Li X, Hu D, Luo M, Zhu M, Li X, Luo F, Li J, Yan J. 2013. Nuclear DNA content variation of three Miscanthus species in China. Genes and Genomics 35, 13–20. [Google Scholar]
- Lu F, Lipka AE, Glaubitz J, Elshire R, Cherney JH, Casler MD, Buckler ES, Costich DE. 2013. Switchgrass genomic diversity, ploidy, and evolution: novel insights from a network-based SNP discovery protocol. PLoS Genetics 9, e1003215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley DE. 1995. The use of chloroplast DNA polymorphism in studies of gene flow in plants. Trends in Ecology and Evolution 10, 198–202. [DOI] [PubMed] [Google Scholar]
- McNeill J, Barrie FR, Buck WR, et al. 2012. International code of nomenclature for algae, fungi and plants (Melbourne code): adopted by the Eighteenth International Botanical Congress Melbourne, Australia, July 2011. Königstein: Koeltz Scientific Books. [Google Scholar]
- Moon Y-H, Cha Y-L, Choi Y-H, Yoon Y-M, Koo B-C, Ahn J-W, An G-H, Kim J-K, Park K-G. 2013. Diversity in ploidy levels and nuclear DNA amounts in Korean Miscanthus species. Euphytica 193, 317–326. [Google Scholar]
- Nishiwaki A, Mizuguti A, Kuwabara S, et al. 2011. Discovery of natural Miscanthus (Poaceae) triploid plants in sympatric populations of Miscanthus sacchariflorus and Miscanthus sinensis in southern Japan. American Journal of Botany 98, 154–159. [DOI] [PubMed] [Google Scholar]
- Paradis E. 2010. pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics 26, 419–420. [DOI] [PubMed] [Google Scholar]
- Petit C, Bretagnolle F, Felber F. 1999. Evolutionary consequences of diploid-polyploid hybrid zones in wild species. Trends in Ecology and Evolution 14, 306–311. [DOI] [PubMed] [Google Scholar]
- Purdy SJ, Maddison AL, Jones LE, Webster RJ, Andralojc J, Donnison I, Clifton-Brown J. 2013. Characterization of chilling-shock responses in four genotypes of Miscanthus reveals the superior tolerance of M.× giganteus compared with M. sinensis and M. sacchariflorus . Annals of Botany 111, 999–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyter R, Heaton E, Dohleman F, Voigt T, Long S. 2009. Agronomic experiences with Miscanthus× giganteus in Illinois, USA. In: Mielenz JR, ed. Methods in Molecular Biology. Biofuels: methods and protocols. Totowa: Humana Press, 41–52. [DOI] [PubMed] [Google Scholar]
- Ray N, Adams J. 2001. A GIS-based vegetation map of the world at the last glacial maximum (25,000–15,000 BP). Internet Archaeology doi: 10.11141/ia.11.2 [Google Scholar]
- Rayburn AL, Crawford J, Rayburn CM, Juvik JA. 2009. Genome size of three Miscanthus species. Plant Molecular Biology Reporter 27, 184–188. [Google Scholar]
- Rieseberg LH, Raymond O, Rosenthal DM, Lai Z, Livingstone K, Nakazato T, Durphy JL, Schwarzbach AE, Donovan LA, Lexer C. 2003. Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301, 1211–1216. [DOI] [PubMed] [Google Scholar]
- Sacks EJ, Juvik JA, Lin Q, Stewart JR, Yamada T. 2013. The gene pool of Miscanthus species and its improvement. In: Paterson AH, ed. Genomics of the Saccharinae. New York: Springer, 73–101. [Google Scholar]
- Shimono Y, Kurokawa S, Nishida T, Ikeda H, Futagami N. 2013. Phylogeography based on intraspecific sequence variation in chloroplast DNA of Miscanthus sinensis (Poaceae), a native pioneer grass in Japan. Botany 91, 449–456. [Google Scholar]
- Slavov G, Robson P, Jensen E, et al. 2013. Contrasting geographic patterns of genetic variation for molecular markers vs. phenotypic traits in the energy grass Miscanthus sinensis . GCB Bioenergy 5, 562–571. [Google Scholar]
- Somerville C, Youngs H, Taylor C, Davis SC, Long SP. 2010. Feedstocks for lignocellulosic biofuels. Science 329, 790–792. [DOI] [PubMed] [Google Scholar]
- Stebbins GL. 1959. The role of hybridization in evolution. Proceedings of the American Philosophical Society 103, 231–251. [Google Scholar]
- Stebbins GL. 1971. Chromosomal evolution in higher plants. London: Edward Arnold. [Google Scholar]
- Takahashi C, Shibata F. 2002. Analysis of Miscanthus sacchariflorus and M. sinensis chromosomes by fluorescence in situ hybridization using rDNA and total genomic DNA probes. Chromosome Science 6, 7–11. [Google Scholar]
- Toonen RJ, Hughes S. 2001. Increased throughput for fragment analysis on an ABI PRISM (R) automated sequencer using a membrane comb and STRand software. Biotechniques 31, 1320–1324. [PubMed] [Google Scholar]
- Wang N, Borrell JS, Bodles WJA, Kuttapitiya A, Nichols RA, Buggs RJA. 2014. Molecular footprints of the Holocene retreat of dwarf birch in Britain. Molecular Ecology 23, 2771–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winge Ö. 1917. The chromosomes. Their number and importance. Comptes Rendus des Travaux du Laboratoire Carlsberg 13, 131–275. [Google Scholar]
- Winkler MG, Wang PK. 1993. The late-quaternary vegetation and climate of China. In: Wright HE, ed. Global climates since the last glacial maximum. Minneapolis: University of Minnesota Press, 221–264. [Google Scholar]
- Winter DJ. 2012. MMOD: an R library for the calculation of population differentiation statistics. Molecular Ecology Resources 12, 1158–1160. [DOI] [PubMed] [Google Scholar]
- Yan J, Chen W, Luo F, et al. 2012. Variability and adaptability of Miscanthus species evaluated for energy crop domestication. GCB Bioenergy 4, 49–60. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.