Highlight
Metabolomics was shown to be a powerful tool for both quantifying intracellular compounds and qualitatively assessing biochemical pathways. This approach underlined metabolic routes involved in the synthesis of biofuel-relevant oils.
Key words: Alternative crop, erucic acid, GC-MS, jet fuel, LC-MS/MS, metabolomics, oilseed, pennycress, plant metabolism, Thlaspi arvense L., triacylglycerols.
Abstract
Pennycress (Thlaspi arvense L.), a plant naturalized to North America, accumulates high levels of erucic acid in its seeds, which makes it a promising biodiesel and industrial crop. The main carbon sinks in pennycress embryos were found to be proteins, fatty acids, and cell wall, which respectively represented 38.5, 33.2, and 27.0% of the biomass at 21 days after pollination. Erucic acid reached a maximum of 36% of the total fatty acids. Together these results indicate that total oil and erucic acid contents could be increased to boost the economic competitiveness of this crop. Understanding the biochemical basis of oil synthesis in pennycress embryos is therefore timely and relevant to guide future breeding and/or metabolic engineering efforts. For this purpose, a combination of metabolomics approaches was conducted to assess the active biochemical pathways during oil synthesis. First, gas chromatography–mass spectrometry (GC-MS) profiling of intracellular metabolites highlighted three main families of compounds: organic acids, amino acids, and sugars/sugar alcohols. Secondly, these intermediates were quantified in developing pennycress embryos by liquid chromatography–tandem mass spectrometry (LC-MS/MS) in multiple reaction monitoring mode. Finally, partitional clustering analysis grouped the intracellular metabolites that shared a similar pattern of accumulation over time into eight clusters. This study underlined that: (i) sucrose might be stored rather than cleaved into hexoses; (ii) glucose and glutamine would be the main sources of carbon and nitrogen, respectively; and (iii) glycolysis, the oxidative pentose phosphate pathway, the tricarboxylic acid cycle, and the Calvin cycle were active in developing pennycress embryos.
Introduction
Petroleum is the largest energy source in the USA, accounting for 28% of all energy consumed in 2013 (www.eia.gov). The fact that petroleum-based fuels will eventually be depleted requires the development of renewable fuels. Indeed, biofuel production has attracted considerable research attention in both developing and industrialized countries (Demirbas, 2009). In addition to its ability to mitigate the approaching shortage of petroleum, renewable energy has the additional environmental benefit of being a low contributor of greenhouse gases (Kim and Dale, 2005). Biofuel is a renewable fuel that can be produced from plant biomass components such as oil, starch, and cell wall. In the USA, the most common biofuels, such as ethanol and biodiesel, are currently produced from corn, soybean, and other high cost commodity crops (Kim and Dale, 2005; Demirbas, 2009; Moser et al., 2009a ). In fact, it seems crucial to address that the biofuel industry should not use crops with valuable food applications. However, the availability of other suitable bioenergy plants, such as sugarcane, is limited to certain geographies and climates. Taking these challenges into consideration, the biofuel industry is in need of alternative crops that meet the following criteria: (i) a favourable biomass composition for biofuel production; (ii) the ability to grow in a variety of soils and climates; and (iii) have no competition with food crops. The alternative bioenergy crops that have been studied so far include, but are not restricted to, Crambe (Li et al., 2012), Camelina (Frohlich and Rice, 2005), Brassica carinata (Bouaid et al., 2009), Miscanthus (Robson et al., 2013), and sugarcane (Hojilla-Evangelista et al., 2013).
Field pennycress (Thlaspi arvense L.; Supplementary Fig. S1 available at JXB online) is a winter annual that grows widely across temperate regions of North America and the southern hemisphere (Warwick et al., 2002). It has been identified as an oilseed crop that could be a suitable source for biofuel (Vaughn et al., 2005; Hojilla-Evangelista et al., 2013). Indeed, pennycress is a member of the Brassicaceae family and is adapted to a wide range of climate conditions (Vaughn et al., 2005; Cermak et al., 2013; Hojilla-Evangelista et al., 2013). It germinates in the autumn and grows slowly during the winter months. Following the flowering period in the spring, pennycress seeds can be harvested before summer crops are planted (Fan et al., 2013). Thus, pennycress is capable of growing in a rotation with commodity crops without displacing them (Isbell, 2009; Phippen and Phippen, 2012; Cermak et al., 2013). The potential average yield of pennycress seeds is 1500kg ha–1, which is equivalent to 600–1200 l ha–1 of oil in comparison with 450 and 420–640 l ha–1 in the cases of soybean and camelina oils, respectively (Boateng et al., 2010; Phippen and Phippen, 2012). Therefore, pennycress has been studied as an alternative crop that can be used for biofuel. Harvested pennycress seeds contain ~36% oil of which ~94% are unsaturated fatty acids that confer specific physico-chemical properties to pennycress oil. The most abundant unsaturated fatty acid is erucic acid [(Z)-docos-13-enoic acid], a monounsaturated fatty acid with 22 carbons. Pennycress oil has been shown to be suitable for biodiesel production due to its high cetane number of 59.8 and excellent low temperature properties (Moser et al., 2009a ). These characteristics meet the US biodiesel standard ASTM D6751. Furthermore, results from a life cycle assessment revealed that renewable fuels produced from pennycress oil, in combination with hydrogenation, deoxygenation, isomerization, and hydrocracking reactions, could qualify as a biomass-derived diesel according to the Renewable Fuels Standard (RFS2) (Fan et al., 2013). Therefore, further increases in oil accumulation and erucic acid level by breeding and/or metabolic engineering will ensure pennycress economical viability as a dedicated bioenergy crop. Understanding the biochemical pathways involved in oil synthesis in pennycress is hence timely to guide future crop improvement efforts.
In plants, different pathways in central metabolism are responsible for allocating the carbon skeletons, reducing power and energy required for fatty acid synthesis. Underlying the pathways that are actively involved in erucic acid synthesis in pennycress requires a relatively new discipline known as metabolomics (Cocuron et al., 2014). As an alternative to genomics, transcriptomics, and proteomics, metabolomics plays a pivotal role in investigating genotype–phenotype relationships by quantitative profiling of metabolites in a given organism (Ogura et al., 2013). The strength of metabolomics lies in the fact that chemical compounds serve as a direct signature of biochemical activity, unlike genes and proteins which are prone to a variety of modifications. As of today, two major approaches have been commonly used in metabolomics; untargeted and targeted (Patti et al., 2012). The untargeted approach, known as metabolite fingerprinting, involves the profiling of all compounds present, whereas the targeted approach refers to the quantitative measurement of specific intermediates within given metabolic pathways (Ogura et al., 2013). On the one hand, metabolite fingerprinting can be conducted with gas chromatography–mass spectrometry (GC-MS) (Fiehn, 2008) and/or liquid chromatography–mass spectrometry (LC-MS) which are powerful analytical techniques to unravel the metabolic state of a given organism. On the other hand, obtaining quantitative information on metabolites involved in core biochemical pathways becomes possible with an approach of targeted metabolomics. However, a challenge in accomplishing such a task relies on the choice of the instruments that are capable of detecting and quantifying low concentrations of intermediates that are of interest. Among all the instruments commonly used in metabolomics, liquid chromatography–tandem mass spectrometry (LC-MS/MS) has been given special emphasis due to its high accuracy and sensitivity (Bajad et al., 2006; Luo et al., 2007; Cocuron et al., 2014). LC-MS/MS combines two main modules: liquid chromatography and mass spectrometry. In liquid chromatography, a column separates metabolites according to their chemical properties. Afterwards, these separated compounds undergo electrospray ionization (ESI), producing specific parent/daughter ions that are in turn detected by a triple-quadrupole mass spectrometer in multiple reaction monitoring (MRM) mode (Luo et al., 2007; Cocuron et al., 2014). In previous studies, LC-MS/MS has been shown to be a powerful tool for separating and quantifying known intermediates of central metabolic pathways including glycolysis, the oxidative pentose phosphate pathway (OPPP), and the tricarboxylic acid (TCA) cycle (Koubaa et al., 2013; Cocuron and Alonso, 2014; Cocuron et al., 2014). Therefore, targeted metabolomics studies should highlight which pathways are metabolically active during fatty acid synthesis through the quantification of signature metabolites using LC-MS/MS.
In this work, both qualitative and quantitative approaches were combined to understand the biochemical basis of oil synthesis in pennycress embryos by: (i) analysing the biomass accumulation that determined the main carbon sinks; (ii) conducting a metabolomic profiling study using GC-MS to identify the main classes of metabolites present in pennycress embryos; and (iii) quantifying intracellular compounds involved in central metabolism through LC-MS/MS.
Materials and methods
Chemicals
Metabolite standards, 3 N methanolic HCl, and toluene were purchased from Sigma. [U-13C]Glucose, [U-13C]glycine, and [U-13C]fumarate were obtained from Isotec. Potassium hydroxide (KOH), methylene chloride, ethoxyamine hydrochloride, and MSTFA+1% TMCS (N-methyl-N-trimethylsilytrifluoroacetamide plus 1% trimethylchlorosilane), solvents for GC-MS and LC-MS/MS, were purchased from Fisher Scientific. Gibberellins (GA4/GA7) and Murashige and Skoog basal salt were ordered from PhytoTechnology Laboratories.
Plant growth
Pennycress seeds of the Ames 30982 accession were obtained from the North Central Regional Plant Introduction Station. The seeds were germinated on plates prior to transfer to pots (Supplementary Fig. S1 at JXB online). Briefly, the seeds were sterilized for 5min with 50% bleach in a 2ml tube and rinsed with sterile water a total of four times. Then, the seeds were placed between two aseptic Whatman papers in a 100×15mm glass Petri dish. Sterile Murashige and Skoog salt medium containing 1mM G4/G7 gibberellins, pH 6.0, was added and the plate was sealed with parafilm. Seeds were allowed to germinate for 3–5 d at 22 °C. Finally, the germinated kernels were transferred to pots (14 cm×14 cm×18cm), and grown in a growth chamber at 22 °C under a constant light intensity of 200 μmol m–2 s–1 and a 16h/8h day/night cycle. Upon emergence of the first pair of true leaves, the plants were transferred to a cold room (4 °C) for 3 weeks. The light intensity and day/night cycle were 100 μmol m–2 s–1 and 10h/14h, respectively. This step was crucial in ensuring that plants flowered later on. The plants were then placed back into their initial growth chamber and allowed to grow until maturity. The pennycress flowers were hand pollinated and tagged every day in order to study the embryo metabolism at different developmental stages.
Biomass extraction
Oil, proteins, and starch were sequentially extracted as previously described (Cocuron et al., 2014). A 1:5 dilution was applied to the fatty acid methyl ester (FAME) samples. The remaining pellet after oil, protein, and starch extraction was considered to represent the cell wall.
Biomass quantification
Oil
Oil content was determined by GC-MS. FAMEs were analysed using a Thermo Trace 1310 gas chromatograph coupled to an ISQ single quadrupole mass spectrometer. FAME derivatives were separated using an Omegawax 250 capillary (30 m×0.25 mm×0.25 μm) column from Supelco at a constant flow rate of 1.4ml min–1. Helium was used as the carrier gas. The GC conditions were as follows: initial temperature was set to 170 °C and held for 30 s. The oven temperature was then raised to 245 °C at 100 °C min–1 and held for 8.75min. The injection temperature was fixed at 225 °C and the injection mode set to split with a split ratio of 10. For the MS analysis, the mass spectra were acquired using electron impact (EI) ionization in positive ion mode. The ion source and the interface temperatures were set to 200 °C and 250 °C, respectively. GC-MS data were acquired and processed using Xcalibur software. FAME derivatives were identified using the NIST 11 library and neat FAME standards purchased from Sigma.
Proteins, starch, and cell wall
Proteins and starch were quantified following the steps previously described (Cocuron et al., 2014). Cell wall was estimated by subtracting oil, protein, and starch content from the total dry weight (DW).
Metabolite extraction
Metabolites were extracted from pennycress embryos at six different stages [11, 13, 15, 17, 19, and 21 days after pollination (DAP)] using boiling water, as previously described (Cocuron et al., 2014). Prior to extraction, 500, 500, and 1000 nmol of [U-13C]glucose, [U-13C]glycine, and [U-13C]fumarate were added, respectively, as internal standards. The hot water extraction was used for the untargeted and targeted metabolomics studies.
GC-MS analysis of intracellular metabolites
Derivatization
Extracted and lyophilized metabolites were derivatized as previously described (Koek et al., 2006) with minor modifications. Briefly, 200 μl of methylene chloride was added and the samples were dried under a stream of nitrogen. This step was repeated twice. Then, 100 μl of pyridine was added to the vials along with 50 μl of a 56mg ml–1 ethoxyamine hydrochloride solution in pyridine. Samples were flushed with nitrogen for 10 s, resuspended using a vortex, and incubated at 40 °C for 90min in a dry bath. Finally, 350 μl of MSTFA+1% TMCS reagent was added to the samples which were flushed with nitrogen for 10 s and incubated at 40 °C for 50min.
GC-MS analysis
Alkylsilyl derivates were analysed using a Thermo Trace 1310 gas chromatograph coupled to an ISQ single quadrupole mass spectrometer. Alkylsilyl derivates were separated using a TG-5MS capillary (30 m×0.25 mm×0.50 μm) column from Thermo Scientific at a constant flow rate of 1.4ml min–1. Helium was used as the carrier gas. The GC conditions were as follows: initial temperature was set to 70 °C and held for 5min. The oven temperature was then raised to 235 °C at 3 °C min–1. A second ramp was applied at a rate of 6 °C min–1 to reach a final temperature of 320 °C which was held for 5min. The injection temperature was fixed at 240 °C and the injection mode was set to split with a split ratio of 3.6. For the MS analysis, the mass spectra were acquired using EI ionization in positive ion mode. The ion source and the interface temperatures were set to 300 °C and 325 °C, respectively. GC-MS data were acquired and processed using Xcalibur software. Alkylsilyl derivatives were identified using the NIST 11 library.
LC-MS/MS quantification of intracellular metabolites
After lyophilization, extracts were resuspended in 500 μl of nanopure water and vortexed. A 200 μl aliquot of sample was loaded onto a 0.2 μm nanosep MF centrifugal device in order to quantify the sugars. The remaining 300 μl was transferred to a 3kDa Amicon Ultra 0.5ml filtering device for the quantification of amino acids, phosphorylated compounds, and organic acids. The samples were spun at 14 000 g for 45min at 4 °C. The intracellular metabolites were separated and quantified as previously described (Cocuron et al., 2014) with minor modifications.
Sugars and sugar alcohols
A 15 μl aliquot of extract was diluted in a LC-MS/MS vial containing 975 μl of acetonitrile/water (60:40) solution, and 10 μl of the diluted sample was injected onto the LC-MS/MS column.
Amino acids
A 20 μl aliquot of extract was added to a vial containing 880 μl of nano-pure water and 100 μl of 10mM hydrochloric acid, and 10 μl of the diluted sample was injected onto the column.
Phosphorylated compounds and organic acids
A 20 μl aliquot of sample was diluted in 180 μl nanopure water, and 20 μl was injected onto the column.
Statistical analyses
Two-tailed, type 3 Student’s tests (t-test) were performed considering as statistically significant P-values <0.05. Clustering analyses were performed using MetaboAnalyst v2.5 (Xia et al., 2009, 2012), a free online software (www.metaboanalyst.ca). Briefly, for each metabolite, the quantities across different developmental stages were divided by the highest one. Then, the relative values were uploaded in MetaboAnalyst using the format of samples in row (unpaired). Finally, the K-means partitional clustering was performed by the software.
Results
Biomass accumulation in developing pennycress embryos
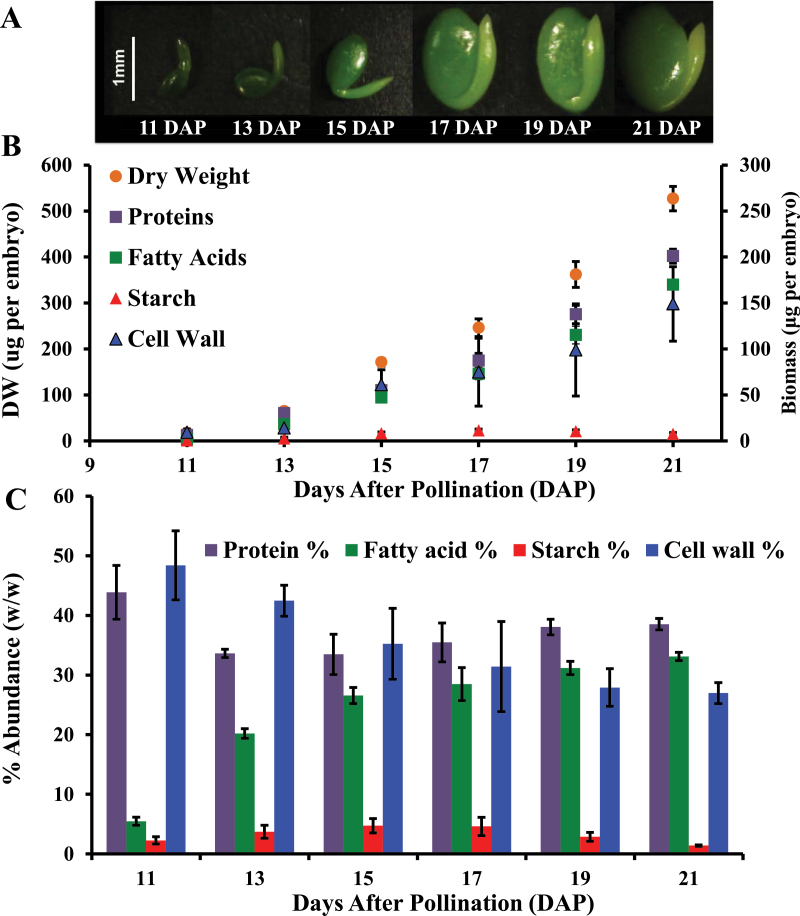
Biomass components are the final products of central metabolism, and their relative abundance reflects the allocation of carbon by primary metabolic pathways. In order to characterize the main carbon sinks and their accumulation rates, pennycress embryos were dissected at different stages (Fig. 1A) and then dried prior to biomass sequential extraction (Cocuron et al., 2014). Fatty acids, proteins, starch, and cell wall were quantified as described in the Materials and methods. A pennycress embryo grew on average 50.2 μg (DW d–1 (R 2=0.97), accumulating fatty acids, protein, cell wall, and starch with the rates of 16.8 (R 2=0.94), 19.3 (R 2=0.95), 12.8 (R 2=0.95), and 1.9 μg d–1 (R 2=0.97), respectively (Fig. 1B). The protein:fatty acid ratio in pennycress embryos dropped from 8.0 at 11 DAP to 1.2 at 21 DAP, indicating an increase in oil accumulation (Fig. 1C). Fatty acid composition varied across developmental stages to reach a steady state at 15 DAP. Indeed, linoleic acid (C18:2) was found to be the most abundant at 11 DAP (33.8±1.5) whereas erucic acid (C22:1) was under the limit of detection. Then, at 19 DAP, erucic acid became the most abundant fatty acid, reaching a plateau at 36% (Supplementary Fig. S2 at JXB online).
Fig. 1.
Biomass composition of pennycress embryos at different stages of development. (A) Pictures of the embryos at different stages of development under a dissecting microscope. (B) Biomass accumulation rate of pennycress embryos. The orange circles, purple squares, green squares, red triangles, and blue triangles, respectively, represent the dry weight, the amounts of protein, fatty acid, starch, and cell wall accumulating in a pennycress embryo (n=4 biological replicates). (C) Biomass abundance in pennycress embryo. The purple, green, red, and blue bars are associated, respectively, with the percentage (w/w) of protein, fatty acid, starch, and cell wall characterizing a single embryo. Error bars are the SD of four biological replicates.
Metabolite profiling in pennycress embryos
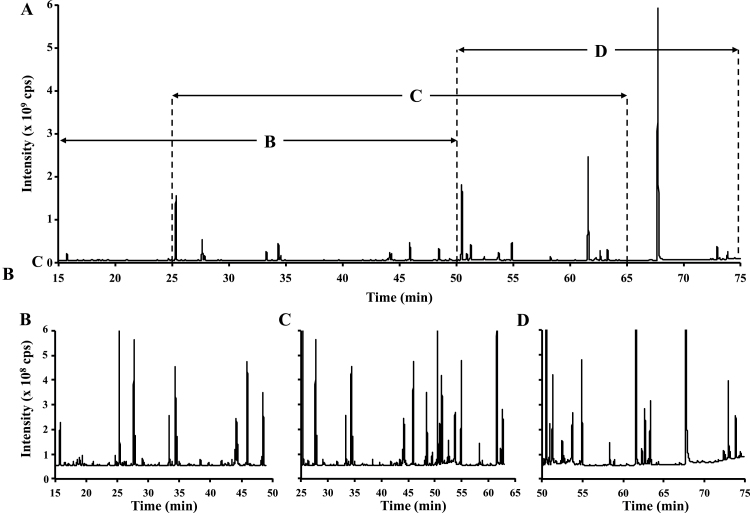
During the developmental process, embryos produce a wide variety of metabolites in a temporal fashion as a result of changes in their metabolism. Metabolite profiling, also known as untargeted metabolomics, enables the detection of intracellular compounds at a given time. Through this approach one can gain qualitative (rather than quantitative) information about specific classes of intermediates accumulating at the same time as the synthesis of a product of interest. In this study, metabolite profiling was used to characterize all the compounds that were present during fatty acid synthesis. For this purpose, intracellular metabolites were extracted from 17 DAP pennycress embryos with cold methanol:chloroform:water (MCW 2.5:1:1, v:v:v) (Fiehn, 2006) or boiling water (Alonso et al., 2010b ) and then were chemically modified with MSTFA+1% TMCS (Koek et al., 2006). Through the comparison between GC-MS profiles of the derivatized metabolites, boiling water was shown to be the most suitable method, enabling the detection of 385 peaks versus 344 for MCW (data not shown). A total of 112 peaks out of 385 were assigned with a probability ≥50% using the NIST 11 library (Fig. 2; Supplementary Table S1 at JXB online). The identification of the detected peaks qualitatively showed the presence of three main classes of metabolites (sugars, amino acids, and organic acids), and to a lesser extent, alkaloids, polyamines, phosphorylated metabolites, and free fatty acids (Fig. 2; Supplementary Table S1).
Fig. 2.
Metabolite profiling of pennycress embryos at 17 DAP. (A) GC-MS chromatogram of 17 DAP pennycress embryos obtained after MSTFA derivatization. Enlarged chromatogram areas depicting the main classes of compounds, (B) amino acids, (C) organic acids, and (D) sugars found in pennycress embryos. The NIST 11 library was used to assign the different peaks.
Comparative metabolomics analyses of developing pennycress embryos
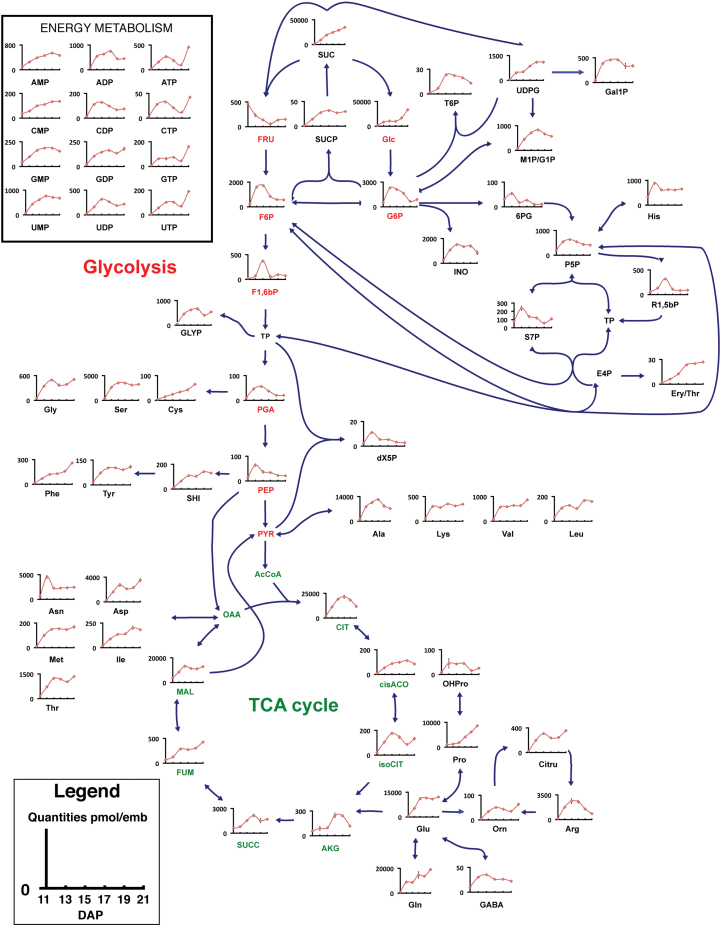
For the purpose of quantifying the compounds characterized by metabolite profiling, boiling water extraction was performed on pennycress embryos harvested at different stages of development. Extracted compounds were then analysed by LC-MS/MS and quantified according to 13C-labelled internal standards as well as standard curves generated for each metabolite. The percentage recovery of this method was previously determined in plant tissues for each metabolite (Cocuron et al., 2014). Intermediates from glycolysis, the OPPP, the TCA cycle, and the Calvin cycle were measured by LC-MS/MS, indicating that all these pathways are active in developing pennycress embryos (Fig. 3; Supplementary Table S2 at JXB online).
Fig. 3.
Metabolic map of pennycress embryos at different stages of development. Values are expressed in pmol per embryo and are the average ±SD of three biological replicates from embryos harvested at 11, 13, 15, 17, 19, and 21 DAP. SUC, sucrose; FRU, fructose; GLC, glucose; INO, inositol; GLY, glycerol; Ery/Thr, erythritol/threitol; Ala, alanine; Arg, arginine; Asn, asparagine; Asp, aspartate; Cys, cysteine; Lys, lysine; Gln, glutamine; Glu, glutamate; Gly, glycine; His, histidine; OHPro, hydroxyproline; Leu, leucine; Ile, isoleucine; Met, methionine; Phe, phenylalanine; Pro, proline; Ser, serine; Thr, threonine; Tyr, tyrosine; Val, valine; GABA, 4-aminobutyric acid; Orn, ornithine; Citru, citrulline; T6P, trehalose 6-phosphate; UDPG, UDP-glucose; SUCP, sucrose 6-phosphate; G1P, glucose 1-phosphate; M1P/G1P, mannose 1-phosphate/glucose 1-phosphate; F6P, fructose 6-phosphate; G6P, glucose 6-phosphate; 6PG, 6-phosphogluconic acid; P5P, pentose 5-phosphate; R1,5-bP, ribulose 1,5-bisphosphate; S7P, sedoheptulose 7-phosphate; E4P, eryhtrose 4-phosphate; F1,6bP, fructose 1,6-bisphosphate; GLYP, glycerol-phosphates; TP, triose phosphates; PGA, 2–3 phosphoglycerates; dX5P, deoxyxylulose 5-phosphate; PEP, phosphoenolpyruvate; SHI, shikimate; PYR, pyruvate; AcCoA, acetyl-CoA; CIT, citrate; cisACO, cis-aconitate; isoCIT, isocitrate; AKG, α-ketoglutarate; SUCC, succinate; FUM, fumarate; MAL, malate; OAA, oxaloacetate. Metabolites coloured in red and green correspond to glycolysis and the TCA cycle, respectively.
Sugars are the principal source of carbon provided by the mother plant to the embryos (Schwender and Ohlrogge, 2002; Sriram et al., 2004; Goffman et al., 2005; Alonso et al., 2007, 2010a ; Allen et al., 2009; Lonien and Schwender, 2009). Sucrose and glucose were quantified as the main free sugars in developing pennycress embryos. Their levels increased by 33-fold, from 1033.2±54.3 pmol to 33479.1±1031.6 pmol per embryo for glucose, and from 1161.7±130.0 pmol to 34963.7±112.4 pmol per embryo for sucrose (Fig. 3; Supplementary Table S2 at JXB online). The main sugar alcohols were found to be sorbitol and inositol, with respective levels of 949.1±281.5 pmol and 844.8±155.5 pmol per embryo at 21 DAP (Fig. 3; Supplementary Table S2).
Plant embryos not only receive free amino acids as the source of nitrogen but also produce their own for protein biosynthesis (Schwender and Ohlrogge, 2002; Goffman et al., 2005; Alonso et al., 2007, 2010a; Allen et al., 2009; Lonien and Schwender, 2009). The total amino acid content underwent a 10-fold increase in pennycress embryos between 11 and 21 DAP. Alanine, asparagine, aspartate, glutamate, glutamine, proline, and serine were the seven most abundant amino acids across different developmental stages. Indeed, they represented between 83% and 90% of the total amino acids (Fig. 3; Supplementary Table S2 at JXB online). Besides serine that is synthesized from 3-phosphoglycerate, the six others are all produced from organic acids, at the level of the TCA cycle (Fig. 3). The TCA cycle is also important for generating reducing power (FADH2 and NADH) that can be used for biomass synthesis and/or for ATP production by oxidative phosphorylation. Malate and citrate, which are involved in fatty acid synthesis and elongation, respectively (Fatland et al., 2000; Nikolau et al., 2000; Alonso et al., 2010a ; Baud and Lepiniec, 2010), reached 94% of the total organic acids at 13 DAP (Fig. 3; Supplementary Table S2).
Phosphorylated metabolites are key intermediates of glycolysis, OPPP, and the Calvin cycle. Therefore, measuring these compounds is essential to assess central metabolism. The major phosphorylated compounds were found to be glucose 6-phosphate, fructose 6-phosphate, and pentose 5-phosphates at 11 DAP, with respective levels of 536.4±30.5, 231.6±44.7, and 206.1±22.6 pmol per embryo, and remained high during the development of the embryo (Fig. 3; Supplementary Table S2 at JXB online). At 21 DAP, UDP-glucose became the most abundant phosphorylated metabolite (1112.5±66.5 pmol per embryo) along with glucose 1-phosphate/mannose 1-phosphate (569.3±30.1 pmol per embryo); these are major precursors for cell wall biosynthesis. Glycerol phosphate levels increased by 7-fold between 11 and 21 DAP; this metabolite provides the glycerol part of the triacylglycerols.
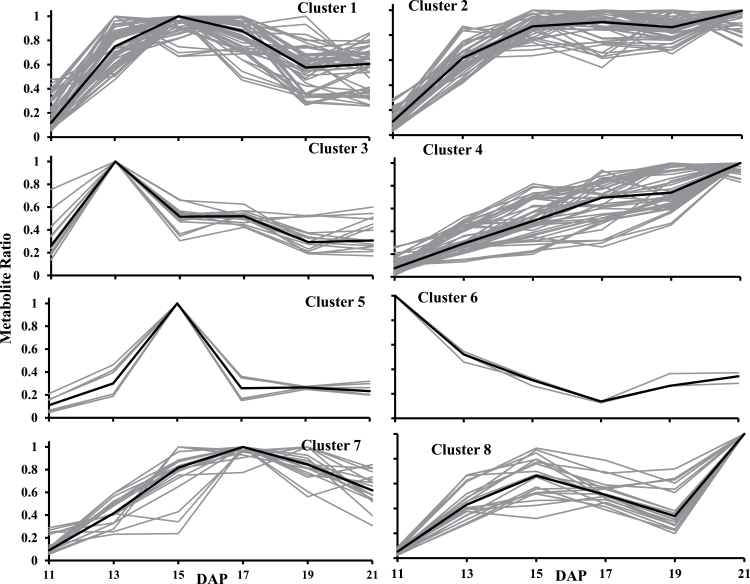
In order to group metabolites that share a similar pattern of accumulation over different stages of development, a partitional clustering analysis was performed using MetaboAnalyst (Xia et al., 2009, 2012). Intracellular metabolites in developing pennycress embryos were found to gather in eight clusters (Fig. 4; Table 1). The majority of compounds in cluster 1 increased from 11 to 15 DAP and then decreased; 33% of the phosphorylated metabolites grouped into this cluster. The intermediates from cluster 2, among which nine were amino acids, rapidly increased to reach a plateau at 15 DAP. Clusters 3, 5, and 7 peaked at 13, 15, and 17 DAP, respectively, and then decreased. Interestingly, fructose 1,6-bisphosphate and ribulose 1,5-bisphosphate grouped in cluster 7, and four of the organic acids grouped in cluster 5. Metabolites gradually accumulating during embryos development were gathered in cluster 4; the main sugars (glucose and sucrose), several of the most abundant amino acids (glutamine and proline), and phosphorylated compounds (UDP-glucose) were found in this cluster. Fructose was the only metabolite steadily decreasing over time, and therefore was separated from all the other intermediates (cluster 6). Finally, all the nucleotide triphosphates grouped together in cluster 8; these had a peak at 15 DAP and then reached an optimum at 21 DAP.
Fig. 4.
Metabolite clustering of pennycress embryos across different developmental stages. Metabolites were clustered using MetaboAnalyst v2.5. The black lines represent median intensities of corresponding clusters that were obtained from K-means analysis.
Table 1.
Clusters of metabolites in developing pennycress embryos
Metabolites were clustered using MetaboAnalyst v2.5.
| Cluster | Metabolites | |||
|---|---|---|---|---|
| Sugars and sugar alcohols | Amino acids | Phosphorylated compounds | Organic acids | |
| 1 | INO | Arg, Ala, His, GABA, OHPro | PGA, F6P, G6P, ADP, IMP, CDP, Gal1P, GLYP, UDP, P5P | isoCIT |
| 2 | Glu, Gly, Lys, Met, Ser, Thr, Tyr, Val | GDP, GMP, Man6P, UMP, SUCP | MAL, CIT | |
| 3 | Asn | 6PG, dX5P, S7P, PEP | ||
| 4 | Glc, Sorb, Ery/Thr, pentitols, SUC | Cys, Phe, Pro, Gln, Ile | UDPG, CMP, AMP, | transACO, FUM, SHI |
| 5 | F1,6bP, R1,5bP | |||
| 6 | FRU | |||
| 7 | T6P, G1P | AKG, cisACO, CIT, SUCC | ||
| 8 | Asp, Orn | ATP, CTP, GTP, UTP | ||
For abbreviations, see the legend of Fig. 3.
Abbreviations: DAP, days after pollination; DW, dry weight; FAMEs, fatty acid methyl esters; MCW, methanol:chloroform:water; MRM, multiple reaction monitoring; MSTFA, N-methyl-N-trimethylsilytrifluoroacetamide; TMCS, trimethylchlorosilane; OPPP, oxidative pentose-phosphate pathway; TCA, tricarboxylic acid.
Discussion
Pennycress naturally accumulates high levels of erucic acid in its embryos, which makes it a promising biodiesel and industrial crop (Moser et al., 2009a, b ). Understanding the biochemical basis of oil synthesis in pennycress embryos is therefore relevant to guide future breeding and/or metabolic engineering efforts. In plants, fatty acid synthesis occurs predominantly in plastids and requires carbon (acetyl-CoA), energy (ATP), and reducing power (NADH and NADPH), which are provided in situ by the activity of central metabolism (Hills, 2004; Baud and Lepiniec, 2010). The main carbon sinks in pennycress embryos were found to be proteins, fatty acids, and cell wall, which represented 38.5, 33.2, and 27.0%, respectively of the biomass at 21 DAP (Fig. 1). In comparison, the embryos of other Brassicaceae, such as Arabidopsis thaliana and Physaria fendleri, accumulate up to 40% and 55% (w/w) of oil (Lonien and Schwender, 2009; Cocuron et al., 2014); these levels could be potentially achieved in pennycress too. Erucic acid reached its highest level of ~36% of the total fatty acids in pennycress embryos at 19 DAP (Supplementary Fig. S2 at JXB online). According to the results above, future crop improvement might involve the increase of: (i) the carbon flow towards oil synthesis; and (ii) the elongation of oleic acid (C18:1) to erucic acid (C22:1). Indeed, recent genetic manipulations successfully enhanced the percentage of erucic acid in crambe (Li et al., 2012), and would be a promising approach for pennycress.
Metabolomics emerged as a powerful tool to assess the metabolic state of a given organism/tissue (Patti et al., 2012), specifically quantifying key intermediate compounds involved in primary pathways (Huck et al., 2003; Bajad et al., 2006; Luo et al., 2007; Alonso et al., 2010b ; Koubaa et al., 2013; Cocuron and Alonso, 2014; Cocuron et al., 2014). In this study, intracellular metabolites were extracted from pennycress embryos using boiling water, which has been shown to be the most efficient method to extract water-soluble compounds from various biological sources (microorganisms, mammalian cells, plant tissues, etc.) with the maximum recovery (Alonso et al., 2010a ; El Rammouz et al., 2010; Cocuron et al., 2014). Two metabolomic approaches were applied in this work to fingerprint the physiological activities of pennycress embryos. The first one, untargeted metabolomics, is a purely qualitative method: it has been widely used to assess the global metabolite profile of a sample and to detect novel entities (Koek et al., 2006; Patti et al., 2012; Wolfender et al., 2013; Macel et al., 2014; Mie et al., 2014). In this study, metabolite fingerprinting and the main classes of intermediates in pennycress embryos were determined using GC-MS and a structural database (Fig. 2). The second approach is more targeted and relies on the selective quantification of key intracellular metabolites. The levels of intermediates may be expressed as relative (Rolletschek et al., 2011; Borisjuk et al., 2013) or absolute values (Urakami et al., 2010; Cocuron et al., 2014; Wu et al., 2014); with absolute quantities offering the possibility to draw comparisons between different metabolites, in various tissues, conditions, etc. High-throughput LC-MS/MS methods have been recently developed and validated to separate and quantify the intermediates and precursors for plant biomass synthesis: amino acids, sugars/sugar alcohols, phosphorylated compounds, and organic acids, which represent ~100 metabolites (Alonso et al., 2010b ; Koubaa et al., 2013; Cocuron and Alonso, 2014; Cocuron et al., 2014). These methods have been applied here to compare intracellular metabolite levels in pennycress embryos at different stages of development (Fig. 3; Supplementary Table S2 at JXB online). It is important to note that the targeted metabolomics study presented here was performed with the future objective to carry out flux analysis. In the plant field, it is the norm to describe units on a per embryo basis for flux analysis, and in the future it will be useful in comparing the net accumulation of intermediate metabolites with the carbon fluxes through the metabolic pathways. However, metabolite quantities have also been reported here per mg DW (Supplementary Table S3) in order to facilitate the comparison with other plants, organs, etc.
In this study, metabolomics was used to probe the activity of central metabolic pathways and to determine their implication in fatty acid synthesis. First, the main intracellular sugars and amino acids were found to be sucrose, glucose, and glutamine (Fig. 3; Supplementary Table S2 at JXB online), which were grouped into cluster 4 (Fig. 4; Table 1); these are respectively common sources of carbon and nitrogen for developing plant embryos (Schwender and Ohlrogge, 2002; Sriram et al., 2004; Goffman et al., 2005; Alonso et al., 2007, 2010a ; Allen et al., 2009; Lonien and Schwender, 2009). In developing pennycress embryos, fructose was the only metabolite in cluster 6 (Fig. 4; Table 1), with its level decreasing over time. This observation, together with the high levels of sucrose and glucose, indicate that sucrose might be stored rather than cleaved into hexoses via the invertase. In agreement with this study, targeted metabolomics on the embryos of another Brassicaceae, Physaria fendleri, also reported the accumulation of sucrose across developmental stages (Cocuron et al., 2014). A variety of integrative functions have been suggested for sucrose storage in plant embryos, including modulation of gene expression, protein turnover, and a trigger to induce storage pathway (Weber et al., 1997; Farrar et al., 2000; Borek and Nuc, 2011). In earlier studies, storage activity was shown to occur in both avocado and field bean embryos when the sucrose level increased (Weber et al., 1997; Sanchez-Romero et al., 2002). Additionally, sucrose storage is involved in the acquisition of desiccation tolerance during seed development and maturation (Businge et al., 2013). Sugars received by the embryos are metabolized in the cytosol, supplying carbon skeletons to biomass synthesis (Hills, 2004; Baud and Lepiniec, 2010). Secondly, the main organic acids were malate and citrate (Fig. 3; Supplementary Table S2 at JXB online) which have been shown to provide acetyl-CoA for fatty acid synthesis and elongation, respectively (Fatland et al., 2000; Nikolau et al., 2000; Alonso et al., 2010a ; Baud and Lepiniec, 2010). Thirdly, the presence of ribulose 1,5-bisphosphate (Fig. 3; Supplementary Table S2), a metabolite specific to the Calvin cycle, and the green colour of the embryos (Fig. 1) indicate that they are photosynthetically active between 11 and 21 DPA. It has been observed that in green seeds, light energy can be used by chloroplasts to generate ATP and NADPH (Browse and Slack, 1985; Ohlrogge et al., 2004; Schwender et al., 2004, 2006; Goffman et al., 2005). Therefore, photosynthesis might provide part of the energy and reductant necessary for fatty acid production in developing pennycress embryos. Furthermore, ribulose 1,5-bisphosphate is the substrate of the ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCo). High activities of this enzyme have been measured in Brassicaceae embryos (King et al., 1998; Ruuska et al., 2004) where it has been shown to fix the CO2 released by the pyruvate dehydrogenase, increasing the efficiency of carbon use (Schwender et al., 2004). Interestingly ribulose 1,5-bisphosphate uniquely clustered with fructose 1,6-bisphosphate (Fig. 4; Table 1), which indicates that a large portion of the fructose 1,6-bisphosphate might be produced by the Calvin cycle. Finally mitochondrial respiration and the OPPP usually are the two other pathways generating ATP and NADPH, respectively. The level of free amino acids was found to be higher than that of their direct precursors from glycolysis, the TCA cycle, and the OPPP (Fig. 3; Supplementary Table S2), revealing a high flow of carbon through these pathways. Besides photosynthesis, oxidative phosphorylation and the OPPP might be a significant source of energy and reductant for oil synthesis in developing pennycress embryos.
To date, there is only one other quantitative metabolomics study which was conducted on developing embryos of Physaria fendleri (Cocuron et al., 2014). The major developmental difference between Physaria and pennycress embryos was the rate of DW accumulation being twice faster in pennycress. The comparison of the intracellular compound levels between same stage embryos (i.e. 17 and 27 DAP for pennycress and Physaria, respectively) highlighted major biochemical and metabolic differences. First, Physaria embryos synthesized more oil (55% versus 33%; w/w). Secondly, besides sucrose, the levels of the other major free sugars were higher whereas hexose-phosphates were lower in Physaria, suggesting a faster glycolytic flow in pennycress embryos. Thirdly, the amounts of all the organic acids were lower in Physaria by a factor 6–55, which may indicate a slower TCA cycle. Finally, the intermediates of the OPPP (6-phosphogluconate, sedoheptulose 7-phosphate, and pentose-phosphates) and the Calvin cycle (ribulose 1,5-bisphosphate) were found to be higher in pennycress embryos, suggesting a larger flow of carbon through these pathways. The metabolomics study hence revealed the occurrence of key pathways involved in oil production in pennycress embryos. However, the relative contribution of each of these pathways to the synthesis of fatty acids (in terms of carbon skeletons, energy, and reductant), and the potential bottlenecks can only be determined by measuring the in vivo metabolic fluxes (Alonso et al., 2010a ; Dieuaide-Noubhani and Alonso, 2014).
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Thlaspi arvense L. plant anatomy.
Figure S2. Fatty acid composition in developing pennycress embryos.
Table S1. Untargeted metabolomics analysis of pennycress embryos at 17 DAP.
Table S2. Targeted metabolomics analyses of pennycress embryos at different developmental stages.
Table S3. Metabolite levels in developing pennycress embryos expressed in pmol mg DW–1.
Acknowledgements
MC was supported by an REU summer training grant to OSU (DBI-1062144). We are grateful to The Ohio State University Targeted Metabolomics Laboratory (metabolomics.osu.edu) for access to the GC-MS and LC-MS/MS equipment funded respectively by the Center for Applied Plant Sciences (CAPS) and the Translational Plant Sciences Targeted Investment in Excellence (TIE). We thank Brooke Anderson for technical help, as well as Gary Posey (Greenhouse superintendent). Enkhtuul Tsogtbaatar is grateful for support by a travel award from the US Department of Energy, Office of Science, Office of Basic Energy Sciences (BES) and the Office of Biological and Environmental Research (BER), DE-FOA-0000995.
References
- Allen DK, Ohlrogge JB, Shachar-Hill Y. 2009. The role of light in soybean seed filling metabolism. The Plant Journal 58, 220–234. [DOI] [PubMed] [Google Scholar]
- Alonso AP, Dale VL, Shachar-Hill Y. 2010. a Understanding fatty acid synthesis in developing maize embryos using metabolic flux analysis. Metabolic Engineering 12, 488–497. [DOI] [PubMed] [Google Scholar]
- Alonso AP, Goffman FD, Ohlrogge JB, Shachar-Hill Y. 2007. Carbon conversion efficiency and central metabolic fluxes in developing sunflower (Helianthus annuus L.) embryos. The Plant Journal 52, 296–308. [DOI] [PubMed] [Google Scholar]
- Alonso AP, Piasecki RJ, Wang Y, LaClair RW, Shachar-Hill Y. 2010. b Quantifying the labeling and the levels of plant cell wall precursors using ion chromatography tandem mass spectrometry. Plant Physiology 153, 915–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajad SU, Lu W, Kimball EH, Yuan J, Peterson C, Rabinowitz JD. 2006. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography–tandem mass spectrometry. Journal of Chromatography . A 1125, 76–88. [DOI] [PubMed] [Google Scholar]
- Baud S, Lepiniec L. 2010. Physiological and developmental regulation of seed oil production. Progress in Lipid Research 49, 235–249. [DOI] [PubMed] [Google Scholar]
- Boateng AA, Mullen CA, Goldberg NM. 2010. Producing stable pyrolysis liquids from the oil-seed presscakes of mustard family plants: pennycress (Thlaspi arvense L.) and camelina (Camelina sativa). Energy & Fuels 24, 6624–6632. [Google Scholar]
- Borek S, Nuc K. 2011. Sucrose controls storage lipid breakdown on gene expression level in germinating yellow lupine (Lupinus luteus L.) seeds. Journal of Plant Physiology 168, 1795–1803. [DOI] [PubMed] [Google Scholar]
- Borisjuk L, Neuberger T, Schwender J, et al. 2013. Seed architecture shapes embryo metabolism in oilseed rape. The Plant Cell 25, 1625–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouaid A, Martinez M, Aracil J. 2009. Production of biodiesel from bioethanol and Brassica carinata oil: oxidation stability study. Bioresource Technology 100, 2234–2239. [DOI] [PubMed] [Google Scholar]
- Browse J, Slack CR. 1985. Fatty-acid synthesis in plastids from maturing safflower and linseed cotyledons. Planta 166, 74–80. [DOI] [PubMed] [Google Scholar]
- Businge E, Bygdell J, Wingsle G, Moritz T, Egertsdotter U. 2013. The effect of carbohydrates and osmoticum on storage reserve accumulation and germination of Norway spruce somatic embryos. Physiologia Plantarum 149, 273–285. [DOI] [PubMed] [Google Scholar]
- Cermak SC, Biresaw G, Isbell TA, Evangelista RL, Vaughn SF, Murray R. 2013. New crop oils—properties as potential lubricants. Industrial Crops and Products 44, 232–239. [Google Scholar]
- Cocuron JC, Alonso AP. 2014. Liquid chromatography tandem mass spectrometry for measuring 13C-labeling in intermediaries of the glycolysis and pentose-phosphate pathway. Methods in Molecular Biology 1090, 131–142. [DOI] [PubMed] [Google Scholar]
- Cocuron JC, Anderson B, Boyd A, Alonso AP. 2014. Targeted metabolomics of Physeria fendleri, an industrial crop producing hydroxy fatty acids. Plant and Cell Physiology 55, 620–633. [DOI] [PubMed] [Google Scholar]
- Demirbas A. 2009. Political, economic and environmental impacts of biofuels: a review. Applied Energy 86, S108–S117. [Google Scholar]
- Dieuaide-Noubhani M, Alonso AP. 2014. Application of metabolic flux analysis to plants. Methods in Molecular Biology 1090, 1–18. [DOI] [PubMed] [Google Scholar]
- El Rammouz R, Letisse F, Durand S, Portais JC, Moussa ZW, Fernandez X. 2010. Analysis of skeletal muscle metabolome: evaluation of extraction methods for targeted metabolite quantification using liquid chromatography tandem mass spectrometry. Analytical Biochemistry 398, 169–177. [DOI] [PubMed] [Google Scholar]
- Fan JQ, Shonnard DR, Kalnes TN, Johnsen PB, Rao S. 2013. A life cycle assessment of pennycress (Thlaspi aruense L.)-derived jet fuel and diesel. Biomass & Bioenergy 55, 87–100. [Google Scholar]
- Farrar J, Pollock C, Gallagher J. 2000. Sucrose and the integration of metabolism in vascular plants. Plant Science 154, 1–11. [DOI] [PubMed] [Google Scholar]
- Fatland B, Anderson M, Nikolau BJ, Wurtele ES. 2000. Molecular biology of cytosolic acetyl-CoA generation. Biochemical Society Transactions 28, 593–595. [PubMed] [Google Scholar]
- Fiehn O. 2006. Metabolite profiling in Arabidopsis. Methods in Molecular Biology 323, 439–447. [DOI] [PubMed] [Google Scholar]
- Fiehn O. 2008. Extending the breadth of metabolite profiling by gas chromatography coupled to mass spectrometry. Trends in Analytical Chemistry 27, 261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohlich A, Rice B. 2005. Evaluation of Camelina sativa oil as a feedstock for biodiesel production. Industrial Crops and Products 21, 25–31. [Google Scholar]
- Goffman FD, Alonso AP, Schwender J, Shachar-Hill Y, Ohlrogge JB. 2005. Light enables a very high efficiency of carbon storage in developing embryos of rapeseed. Plant Physiology 138, 2269–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hills MJ. 2004. Control of storage-product synthesis in seeds. Current Opinion in Plant Biology 7, 302–308. [DOI] [PubMed] [Google Scholar]
- Hojilla-Evangelista MP, Evangelista RL, Isbell TA, Selling GW. 2013. Effects of cold-pressing and seed cooking on functional properties of protein in pennycress (Thlaspi arvense L.) seed and press cakes. Industrial Crops and Products 45, 223–229. [Google Scholar]
- Huck JHJ, Struys EA, Verhoeven NM, Jakobs C, Van der Knaap MS. 2003. Profiling of pentose phosphate pathway intermediates in blood spots by tandem mass spectrometry: application to transaldolase deficiency. Clinical Chemistry 49, 1375–1380. [DOI] [PubMed] [Google Scholar]
- Isbell TA. 2009. US effort in the development of new crops (Lesquerella, Pennycress, Coriander and Cuphea). Oleagineux Corps Gras Lipides 16, 205–210. [Google Scholar]
- Kim S, Dale BE. 2005. Life cycle assessment of various cropping systems utilized for producing biofuels: bioethanol and biodiesel. Biomass & Bioenergy 29, 426–439. [Google Scholar]
- King WA, Gready JE, Andrews TJ. 1998. Quantum chemical analysis of the enolization of ribulose bisphosphate: the first hurdle in the fixation of CO2 by Rubisco. Biochemistry 37, 15414–15422. [DOI] [PubMed] [Google Scholar]
- Koek MM, Muilwijk B, van der Werf MJ, Hankemeier T. 2006. Microbial metabolomics with gas chromatography/mass spectrometry. Analytical Chemistry 78, 1272–1281. [DOI] [PubMed] [Google Scholar]
- Koubaa M, Cocuron J-C, Thomasset B, Alonso AP. 2013. Highlighting the tricarboxilic acid cycle: liquid and gas chromatography–mass spectrometry analyses of 13C-labeled organic acids. Analytical Biochemistry 436, 151–159. [DOI] [PubMed] [Google Scholar]
- Li XY, van Loo EN, Gruber J, Fan J, Guan R, Frentzen M, Stymne S, Zhu LH. 2012. Development of ultra-high erucic acid oil in the industrial oil crop Crambe abyssinica. Plant Biotechnology Journal 10, 862–870. [DOI] [PubMed] [Google Scholar]
- Lonien J, Schwender J. 2009. Analysis of metabolic flux phenotypes for two Arabidopsis mutants with severe impairment in seed storage lipid synthesis. Plant Physiology 151, 1617–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B, Groenke K, Takors R, Wandrey C, Oldiges M. 2007. Simultaneous determination of multiple intracellular metabolites in glycolysis, pentose phosphate pathway and tricarboxylic acid cycle by liquid chromatography–mass spectrometry. Journal of Chromatography A 1147, 153–164. [DOI] [PubMed] [Google Scholar]
- Macel M, de Vos RC, Jansen JJ, van der Putten WH, van Dam NM. 2014. Novel chemistry of invasive plants: exotic species have more unique metabolomic profiles than native congeners. Ecology and Evolution 4, 2777–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mie A, Laursen KH, Aberg KM, Forshed J, Lindahl A, Thorup-Kristensen K, Olsson M, Knuthsen P, Larsen EH, Husted S. 2014. Discrimination of conventional and organic white cabbage from a long-term field trial study using untargeted LC-MS-based metabolomics. Analytical and Bioanalytical Chemistry 406, 2885–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser BR, Knothe G, Vaughn SF, Isbell TA. 2009. a Production and evaluation of biodiesel from field pennycress (Thlaspi arvense L.) oil. Energy & Fuels 23, 4149–4155. [Google Scholar]
- Moser BR, Shah SN, Winkler-Moser JK, Vaughn SF, Evangelista RL. 2009. b Composition and physical properties of cress (Lepidium sativum L.) and field pennycress (Thlaspi arvense L.) oils. Industrial Crops and Products 30, 199–205. [Google Scholar]
- Nikolau BJ, Oliver DJ, Schnable PS, Wurtele ES. 2000. Molecular biology of acetyl-CoA metabolism. Biochemical Society Transactions 28, 591–593. [PubMed] [Google Scholar]
- Ogura T, Bamba T, Fukusaki E. 2013. Development of a practical metabolite identification technique for non-targeted metabolomics. Journal of Chromatography A 1301, 73–79. [DOI] [PubMed] [Google Scholar]
- Ohlrogge JB, Ruuska SA, Schwender J. 2004. The capacity of green oilseeds to utilize photosynthesis to drive biosynthetic processes. Plant Physiology 136, 2700–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti GJ, Yanes O, Siuzdak G. 2012. Innovation: metabolomics: the apogee of the omics trilogy. Nature Reviews . Molecular Cell Biology 13, 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phippen WB, Phippen ME. 2012. Soybean seed yield and quality as a response to field pennycress residue. Crop Science 52, 2767–2773. [Google Scholar]
- Robson P, Jensen E, Hawkins S, White SR, Kenobi K, Clifton-Brown J, Donnison I, Farrar K. 2013. Accelerating the domestication of a bioenergy crop: identifying and modelling morphological targets for sustainable yield increase in Miscanthus. Journal of Experimental Botany 64, 4143–4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolletschek H, Melkus G, Grafahrend-Belau E, Fuchs J, Heinzel N, Schreiber F, Jakob PM, Borisjuk L. 2011. Combined noninvasive imaging and modeling approaches reveal metabolic compartmentation in the barley endosperm. The Plant Cell 23, 3041–3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruuska SA, Schwender J, Ohlrogge JB. 2004. The capacity of green oilseeds to utilize photosynthesis to drive biosynthetic processes. Plant Physiology 136, 2700–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Romero C, Peran-Quesada R, Barcelo-Munoz A, Pliego-Alfaro F. 2002. Variations in storage protein and carbohydrate levels during development of avocado zygotic embryos. Plant Physiology and Biochemistry 40, 1043–1049. [Google Scholar]
- Schwender J, Goffman F, Ohlrogge JB, Shachar-Hill Y. 2004. Rubisco without the Calvin cycle improves the carbon efficiency of developing green seeds. Nature 432, 779–782. [DOI] [PubMed] [Google Scholar]
- Schwender J, Ohlrogge JB. 2002. Probing in vivo metabolism by stable isotope labeling of storage lipids and proteins in developing Brassica napus embryos. Plant Physiology 130, 347–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwender J, Shachar-Hill Y, Ohlrogge JB. 2006. Mitochondrial metabolism in developing embryos of Brassica napus. Journal of Biological Chemistry 281, 34040–34047. [DOI] [PubMed] [Google Scholar]
- Sriram G, Fulton DB, Iyer VV, Peterson JM, Zhou R, Westgate ME, Spalding MH, Shanks JV. 2004. Quantification of compartmented metabolic fluxes in developing soybean embryos by employing biosynthetically directed fractional (13)C labeling, two-dimensional [(13)C, (1)H] nuclear magnetic resonance, and comprehensive isotopomer balancing. Plant Physiology 136, 3043–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urakami K, Zangiacomi V, Yamaguchi K, Kusuhara M. 2010. Quantitative metabolome profiling of Illicium anisatum by capillary electrophoresis time-of-flight mass spectrometry. Biomedical Research 31, 161–163. [DOI] [PubMed] [Google Scholar]
- Vaughn SF, Isbell TA, Weisleder D, Berhow MA. 2005. Biofumigant compounds released by field pennycress (Thlaspi arvense) seedmeal. Journal of Chemical Ecology 31, 167–177. [DOI] [PubMed] [Google Scholar]
- Warwick SI, Francis A, Susko DJ. 2002. The biology of Canadian weeds. 9. Thlaspi arvense L. (updated). Canadian Journal of Plant Science 8, 803–823. [Google Scholar]
- Weber H, Borisjuk L, Wobus U. 1997. Sugar import and metabolism during seed development. Trends in Plant Science 2, 169–174. [Google Scholar]
- Wolfender JL, Rudaz S, Choi YH, Kim HK. 2013. Plant metabolomics: from holistic data to relevant biomarkers. Current Medicinal Chemistry 20, 1056–1090. [PubMed] [Google Scholar]
- Wu X, Li N, Li H, Tang H. 2014. An optimized method for NMR-based plant seed metabolomic analysis with maximized polar metabolite extraction efficiency, signal-to-noise ratio, and chemical shift consistency. The Analyst 139, 1769–1778. [DOI] [PubMed] [Google Scholar]
- Xia J, Mandal R, Sinelnikov IV, Broadhurst D, Wishart DS. 2012. MetaboAnalyst 2.0—a comprehensive server for metabolomic data analysis. Nucleic Acids Research 40, W127–W133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Psychogios N, Young N, Wishart DS. 2009. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Research 37, W652–W660. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.