Abstract
The importance of tissue oxygenation has led to a great interest in methods for imaging pO2 in vivo. Electron paramagnetic resonance imaging (EPRI) provides noninvasive, near absolute 1 mm-resolved 3D images of pO2 in the tissues and tumors of living animals. Current EPRI image reconstruction methods tend to be time consuming and preclude real-time visualization of information. Methods are presented to significantly accelerate the reconstruction process in order to enable real-time reconstruction of EPRI pO2 images. These methods are image reconstruction using graphics processing unit (GPU)-based 3D filtered back-projection and lookup table parameter fitting. The combination of these methods leads to acceleration factors of over 650 compared to current methods and allows for real-time reconstruction of EPRI images of pO2 in vivo.
INTRODUCTION
Studies have shown that the oxygenation status of tumors has many important prognostic implications (1). Low oxygen concentration (pO2), or hypoxia, has been found to result in increased resistance to radiation therapy for cancer cells (2,3) and in human patients there is strong correlation between electrode measurements of hypoxia and radiotherapy treatment failure (4). Hypoxic tumors have increased resistance to chemotherapy (5), faster tumor growth caused by abnormal proliferation (6), and higher potential for metastatic progression (7).
Due to these effects associated with hypoxia, there is an interest in methods for probing/imaging tissue pO2. Advances in such methods are detailed in the literature (8–11). Electron paramagnetic resonance imaging (EPRI) has proven to be a robust modality for imaging tissue pO2. EPRI provides noninvasive, highly-resolved, both spatially (~1mm3 voxels) and in pO2 (1-3 torr), near absolute quantitative 3D images of in vivo pO2 (12–16). Low frequency (250MHz) EPRI excitation (comparable to those for 6T MRI) provide penetration and, therefore, imaging deep into tissue (>7cm). Standard EPRI image acquisition time is currently on the order of 10 minutes.
EPRI can be efficiently implemented as a tomographic imaging modality. EPR signals acquired under the conditions of applied magnetic field gradients can be viewed from a mathematical standpoint as Radon transformation projections acquired in a 3D spherical geometry. The inversion of the Radon transformation can be applied to recover a 3D image from these projections. Various reconstruction methods can be used. One of the common analytic (those that provide explicit expressions in the form of analytic functions for the solution) reconstruction methods is filtered back-projection (FBP). For practical application, the analytic algorithm must be discretized to handle discrete data and the resulting sampled FBP is an approximation to the analytic solution.
This article describes an improvement to pulse EPRI methodology. A single 3D EPRI image provides the spin distribution in a sample. To acquire the pO2 distribution, a series of 3D images are acquired with different pulse sequence parameters, which are chosen to monitor the evolution of spin magnetization as a function of time, the process of spin-lattice relaxation. The spin-lattice relaxation rate is then obtained in every voxel of a 3D image by fitting this evolution to a known function of time. The local spin-lattice relaxation rate is proportional to pO2 and therefore provides a quantitative and near absolute measure of pO2 at each voxel (17). 3D image reconstruction is a part of pO2 image processing and one of the major contributors to the pO2 image processing time.
Conventional, “multi-stage”, reconstruction of 3D tomographic EPRI images has been limited to particular projection schemes. This is a computationally efficient but highly restrictive method (18,19). In particular, this method does not allow correct image reconstruction from an incomplete or other arbitrary projection set. The most flexible FBP implementation for reconstruction of 3D images is a “single-stage” method, which involves direct back-projection of each projection through the entire 3D image. This method is computationally intensive and relatively slow on a conventional CPU. In this work, we present an implementation of single-stage reconstruction for EPRI using a graphics processing unit (GPU), which provides both versatility to reconstruct images from arbitrary projection distributions and speed. GPUs have been used in many imaging applications to improve reconstruction speed. In fact, the first non-graphics GPU application was to accelerate FBP for CT image reconstruction (20). GPUs have also been used for many other image reconstruction applications (21).
An iterative fitting method can be used to characterize the exponential signal intensity decay rate measuring pO2 at each voxel. However, this is a time consuming process. An alternative fitting method is described here, which implements a predefined lookup table to match the observed signal decay to ideal signal decay curves. This lookup table method may result in modestly reduced accuracy but is significantly faster. In certain circumstances, sacrificing image accuracy for convenience and speed is a favorable tradeoff. This is particularly true if offline image reconstruction will regain that accuracy. The method utilized in this work is similar to that used in magnetic resonance fingerprinting (22).
Conventional personal computer central processor unit (CPU)-based reconstruction and optimization-based parameter fitting are both time consuming processes and represent bottlenecks for the speed with which EPRI pO2 images can be reconstructed. Using these methods, flexible real-time EPRI oxygen image reconstruction is extremely difficult. This work focuses on new methods and implementations of image reconstruction routines providing reconstruction speed enhancements, which enable a more flexible, real-time EPRI reconstruction.
Real-time image reconstruction has been implemented in several imaging modalities, such as MRI (23, 24) and CT (25). In vivo imaging is often time sensitive, especially for human subjects. Real-time reconstruction, allowing for visualization of the buildup of an image as projection data are acquired, can help to reduce scanning time by resolving certain issues early. Such issues can include errors in setup of either the subject or imaging equipment, subject motion, imaging equipment failure, etc. Real-time reconstruction can allow an operator to potentially identify a particular region of interest (ROI) earlier during imaging. Furthermore, as the temporal resolution of EPRI continues to improve, real-time reconstruction will be necessary to eventually enable real-time visualization of in vivo pO2 distributions, essentially analogous to fluoroscopic x-ray imaging.
This paper discusses the simultaneous implementation of GPU-based FBP reconstruction to more rapidly generate 3D images and the use of a lookup table for rapid fitting of pO2 related parameters. The combination of these two methods results in multiple orders of magnitude image reconstruction speed enhancement, which provides the capability to reconstruct an updated image as each successive projection is acquired. This enables real-time reconstruction of EPRI pO2 images.
METHODS
Pulsed EPRI Imaging Model
Pulse EPRI methods developed in our laboratory have been detailed elsewhere (26). A brief description follows. Electron spin echo (ESE) R2 imaging was our first approach (27). More recently, inversion recovery electron spin echo (IRESE) R1 imaging has been implemented, which reduced confounding spin probe concentration dependent self-broadening and therefore enabled essentially absolute pO2 imaging (17). The pulse EPRI signal comes from an exogenous triarylmethyl (trityl) spin probe with an unpaired electron centrally located on the molecule that is remarkably difficult to reduce (16, 28). Both R1 and R2 imaging methods provide oxygen information. The electron relaxation rates (R1 and R2) are proportional to pO2 and therefore directly report local pO2. Linear magnetic field gradients are applied to spatially encode an object. Gradients are applied along different directions in a 3D spherical geometry, producing radial k-space projections, which, when Fourier transformed, provide spatial projections. An inverse radon transform, such as FBP, from a set of spatial projections obtained from different angular views can reconstruct a 3D image of spin density. A series of 3D images is acquired at different times during magnetic relaxation to observe average relaxation rates from each image voxel. Fitting the signal relaxation rates at each voxel to known exponential functions governing the relaxation (refer to Eq. (6)), produces a 3D parametric image with pO2 information from each voxel.
3D FBP Algorithm
Without derivation, the FBP algorithm (29) is described in Eq. (1)–(5). Here, f(x, y, z) is the 3D object being imaged; p(t, φ, θ) is a 1D spatial projection at angle (φ, θ); g(t, φ, θ) is the filtered 1D projection; and h(t) is the unit impulse response for the parabolic filter. In Eq. (2), t is the location of a point (x, y, z) on a given projection at some angle (φ, θ). In Eq. (4), the operator F–1{–} represents the inverse Fourier transform. In practice, these functions and variables are discretized, resulting in a sampled FBP algorithm that provides an approximation of the analytic solution.
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
GPU Implementation of FBP
The GPU is a processing unit specialized to rapidly execute many specific operations in parallel for accelerated creation of images intended for output to a display. The back-projection process in FBP is an analogous process and can take advantage of the excellent parallel computing abilities of the GPU using compute unified device architecture (CUDA) techniques. The GPU card used was a commercially available GeForce GTX 760 with compute capability 3.0, global memory 4GB, and 1152 cores or streaming processors.
The GPU consists of a streaming multiprocessor (SM), which is comprised of many streaming processors (SPs). The GeForce GTX 760 has 6 SMs, each containing 192 SPs. To utilize the acceleration capability of the GPU for the time consuming back-projection process, a kernel function executing the necessary operations for back-projection must be written in CUDA C. The GPU runs the kernel function on a grid, which is a set of blocks, each consisting of a set of threads. A thread is a single execution of the kernel function with a specific index. Each thread uses its index to select the processed data and execution unit so that all threads may cooperatively process the entire data set in parallel.
An efficient parallelization strategy involves assigning a calculation for each specific voxel to an individual thread. In general, blocks and grids can be arranged into 1D, 2D, or 3D structures. For a reconstructed image matrix of size [N, N, N], the thread block is chosen to be a 1D structure of size [N] and the grid is chosen to be a 2D structure of size [N, N]. As such, the kernel function then performs the parallelized back-projection process so that the proper value or values from each projection point or points are back-projected onto each voxel. This is done for all voxels simultaneously. The results of the image reconstruction on the GPU are then retrieved.
R1/R2 Hybrid Imaging
Equation (6) below shows the function describing the EPR signal (S) as a function of inversion delay time (T) and echo delay time (τ), as well as spin probe concentration (A), R1 relaxation, and R2 relaxation. While R1 imaging is the preferred EPRI technique, both R1 and R2 imaging have advantages and disadvantages.
For R2 imaging, signal is not a function of inversion delay time because no inversion is used (equivalent to T = ∞), which results in an exponential decay as seen in the right side of the curve in Fig. 1. The disadvantage of R2 imaging is that accuracy of reported pO2 is decreased by self-broadening effects for higher concentrations of spin probe (17). The advantage of R2 imaging is that it provides an absolute measure of the voxel amplitude, A, which is a measure of spin probe concentration.
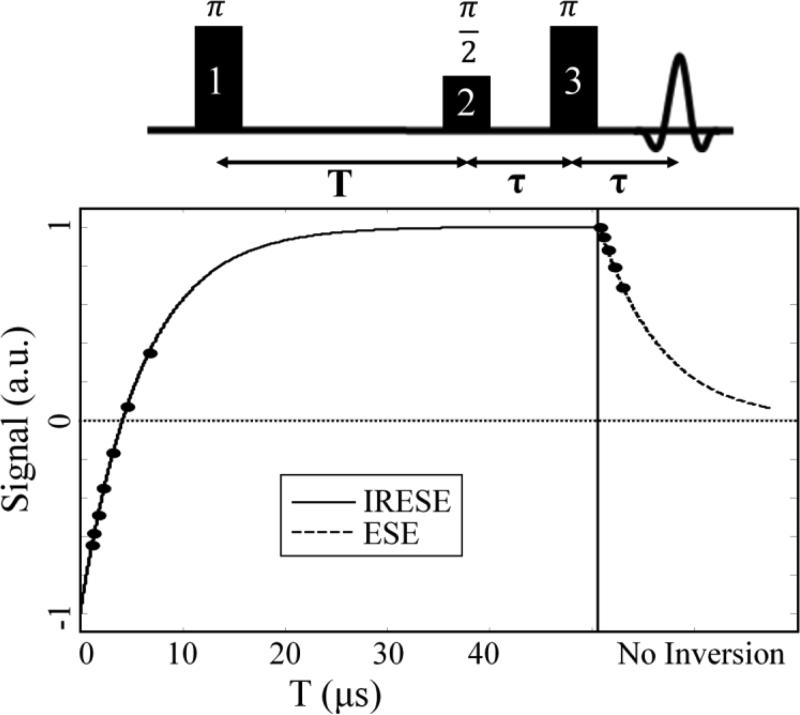
Figure 1.
Diagram of pulse sequence used for EPRI is shown at top of figure. For ESE pulse sequences, only pulses 2 and 3 are used, whereas for IRESE pulse sequences all three pulses are used. At the end of the pulse sequence the echo can be read out after a delay (τ). The expected shape of the magnetic relaxation curve at the 12 different time points used for R1/R2 imaging is also plotted.
For R1 imaging, signal is not a function of echo delay (held constant). This results in exponential recovery as seen in the left side of the curve in Fig. 1. The advantage of using R1 imaging is that it ameliorates the detrimental self-broadening effects present in R2 imaging, thereby providing an absolute measure of pO2. The disadvantage is that A measured using R1 imaging is not a true measure of A because it has a confounding dependence on the R2 relaxation. It is useful to have an accurate measurement of both spin probe concentration and pO2. This is why a hybrid R1/R2 imaging methodology is introduced (Fig. 1), where both IRESE and ESE pulse sequences are used to measure both R1 and R2, and subsequently use Eq. (6) to simultaneously fit A, R1, and R2.
| (6) |
Standard R2 imaging uses 5 values for τ (logarithmically spaced from 0.730μs to 3.000μs) to obtain a fit. For R2 imaging, the ESE pulse sequence consists of a π/2 pulse, followed by a delay (τ), then a π pulse and the same delay (τ) before the echo is read out. This is the pulse sequence shown in Fig. 1 if only pulses 2 and 3 are used.
Standard R1 imaging uses 8 values for T (the first 7 are logarithmically spaced from 0.430μs to 6.000μs; the final point has no inversion, i.e. T = 0.000μs) and a fixed τ value (0.730μs) to obtain a fit. For R1 imaging the IRESE pulse sequence consists of an inversion π pulse, followed by a delay (T) for longitudinal magnetization recovery, and an ESE pulse sequence to read out the echo. To simulate infinite T, no inversion pulse is used (pulse 1, π pulse) for the final point.
Since no inversion is used for R2 points, the final R1 point and the initial R2 point are equivalent. Therefore, for R1/R2 hybrid imaging, 12 points (as opposed to the combined 13 R1 and R2 points) are used in total for the fit (Fig. 1). T values used for the first 8 points are those used for standard R1 imaging and, for the final 4 points, no inversion is used (i.e., T = 0.000μs). The τ values for the final 5 points are those used for standard R2 imaging and, for the first 7 points, τ = 0.730μs. Acquiring more points (12 as opposed to 8 or 5) results in a R1/R2 image with lower signal to noise ratio (SNR) compared to a R1 image of the same time duration (SNR for R1/R2 image is 80% that of a R1 image). With this tradeoff in SNR, R1/R2 imaging provides images with accurate measures of both pO2 and spin probe concentration.
Lookup Table Fitting Algorithm
Currently, voxel-by-voxel curve fitting is performed using built in Matlab™ (Mathworks, MA) functions. For R1/R2 imaging, the difference between acquired data points and a curve calculated from the fitting function given in Eq. (6), with guesses for the fit parameters (A, R1, and R2) is iteratively minimized using the “fminsearch” function in Matlab™. Unconstrained nonlinear optimization is used to iteratively update the fit parameters to best fit the data (30). This method can be time consuming (see Table 4). This fitting method will be referred to as the FMS method.
Table 4.
Comparison of fit times for the different fitting methods. Acceleration factor is compared to standard FMS method.
| FMS | LUT (0.001 μs−1) | LUT (0.01 μs−1) | |
|---|---|---|---|
| Fit Time (s) | 282 | 0.264 | 0.035 |
| Acceleration Factor | – | 1068 | 8057 |
An alternative fitting method uses a lookup table to match acquired data to curves generated using different fit parameter values. The curve generated from known parameters that provides the closest match gives an approximation of the corresponding parameters for the acquired data. This can also be referred to as using a dictionary. Previous studies have used such methods for parameter mapping in MRI applications. Complete dictionaries (number of dictionary elements equal to data dimensionality) can be generated using independent component analysis or principal component analysis (31). Over-complete dictionaries have been used as well (32, 33). These methods however are relatively complicated and dictionary generation and fitting can be time consuming (on the order of tens of minutes).
In this work, a less complicated and faster lookup table (LUT) fitting method is used, similar to other previously used methods (22, 34). The algorithm consists of two main parts: lookup table generation and data fitting. To generate the necessary lookup tables, parameters from the data to be fit are determined: number of time points, τ and T values used for each time point, and number of time points corresponding to either R1 or R2 portions of the data.
A linear exponential decay fit is used for both R1 and R2 determination (Eq. (7) and (8), respectively). For R1/R2 imaging, R1 and R2 lookup tables are generated and used separately. The range and resolution of R2 values (∝ pO2) suitable for the data are determined for R2 lookup table generation. Here, the R2 values used were linearly spaced from ΔR2 to 1.61 μs−1, with ΔR2 equal to 0.001, 0.01, or 0.1 μs−1. This range corresponds to a pO2 range of 0-150 torr, which includes values observed in previous EPRI experiments. Unit vectors are necessary for the lookup table entries because degree of matching between data and lookup table entries is determined based on dot product amplitude.
Unit vector R2 lookup table entries, L̂R2, are generated using Eq. (7), (derived from Eq. (6) with A = 1 and T = ∞). The R1 lookup table is produced in the same way. The range of R1 values (∝ pO2) used is the same as that used for the R2 values. Equation (8) shows the exponential used to generate the vectors that are normalized to be unit vectors and used as the entries in the R1 lookup table, L̂R1.
| (7) |
| (8) |
Following lookup table generation, R2 is determined first. The R2 portion of the data is selected (final 5 data points) and its dot products with all R2 lookup table entries are computed. The entry resulting in the maximum dot product is found and used to determine the corresponding R2 value.
A is then determined. Again, only the R2 portion of the data is used. From Eq. (6) it can be seen that, for ideal data, A is equal to the R2 data points, SR2,i, divided by the corresponding factor accounting for exponential decay due to R2 relaxation. Ideally, this quotient would be constant for all data points. However, due to noise, this is not necessarily true and A is approximately determined for non-ideal data by finding this quotient for each of the 5 data points and taking the average, as in Eq. (9), where N is the number of R2 data points (e.g., 5), SR2.i is the ithe entry in the R2 data vector, τ delay associated with this ith R2 data point, and the denominator represents the corresponding factor accounting for exponential signal decay due to R2 relaxation.
| (9) |
The final step is to determine R1. Similar to the R2 fitting process, only the R1 portion of the data is considered. Equation (10) shows how the R1 data is conditioned to the functional form in Eq. (8), where is the original R1 portion of the data and is the conditioned data. The dot products of with all R1 lookup table entries are then computed. The entry resulting in the maximum dot product determines the corresponding R1 value. It should be noted that the coefficient with a value of 2 in Eq. (6) is for an ideal inversion pulse that rotates the magnetization by exactly π. However, in practice there are spatial inhomogeneities in the applied pulse, resulting in imperfect magnetization inversion, causing this coefficient to deviate from the ideal value of 2 based on spatial location. However, by conditioning the affected R1 data as in Eq. (10), the true coefficient is retained as a scaling of the magnitude of the data vector corresponding to a specific spatial location. This additional scalar multiplicative factor will not alter which lookup table entry corresponds to the maximum dot product with as this is a relative comparison.
| (10) |
Simulated Model
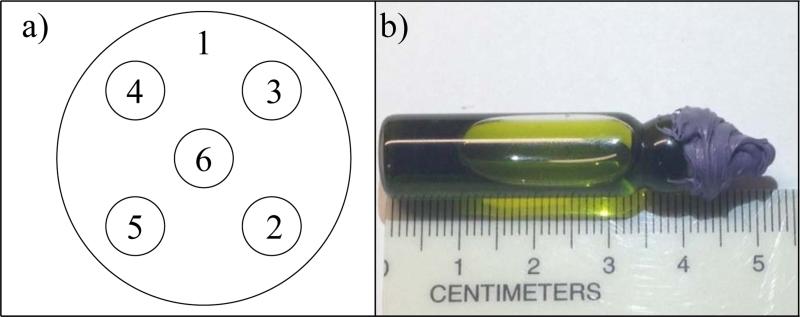
Simulated experiments with and without noise are used to compare the accuracy of different fitting methods. A mathematical phantom consisting of six spheres: five non-overlapping spheres of equal size (0.5cm radius) embedded into a larger sphere (2.5cm radius). Fig. 2a shows a slice through the 3D mathematical phantom.
Figure 2.
(a) 2D slice through the 3D mathematical used to generate simulated projections. The spherical regions are labelled. (b) Photograph of the 1.0 mM trityl bottle phantom used for imaging.
The model used allows for arbitrary choices of A, R1, and R2. The parameters used for simulated experiments are shown in Table 1. The R1 values used correspond to an approximate pO2 range of 2 to 25 torr. To avoid unwanted inaccuracies from reconstruction artifacts, a large number of projections were used for simulated experiments (6368, equal solid angle distribution). Projections were calculated using the Radon transform. Reconstructed images were 64×64×64 voxels, corresponding to a 10×10×10 cm3 field of view.
Table 1.
Parameters used for generation of simulated data from the digital phantom
| Phantom Region | A (a.u.) | R1 (μs−1) | R2 (μs−1) |
|---|---|---|---|
| 1 | 0.06 | 0.33 | 0.67 |
| 2 | 0.02 | 0.20 | 0.29 |
| 3 | 0.04 | 0.22 | 0.33 |
| 4 | 0.08 | 0.25 | 0.40 |
| 5 | 0.10 | 0.29 | 0.50 |
| 6 | 0.15 | 0.40 | 1.00 |
Noisy data were simulated by adding random Gaussian noise to projections. Noise levels in experimental data were used to determine the magnitude of the simulated noise. Noise levels corresponding to 60, 10, 5, 2, and 1 minute images were used. Projection SNR corresponding to these imaging times for 6368 projection images with 12 time points was 29.17, 21.39, 18.38, 14.40, and 11.39 dB respectively. SNR in dB was calculated using Eq. (11), where S is signal and σN is the standard deviation of the noise.
| (11) |
Phantom Experiments
A bottle phantom was imaged to test the new methods on experimental data. This phantom was a flat-bottomed borosilicate glass cylinder of 9.5mm inner diameter filled with a 1mM solution of trityl as shown in Fig. 2b. The sample was deoxygenated using a multiple-cycle freeze-pump-thaw technique and flame sealed to ensure internal pO2 of ~0 torr.
Images were standard R1/R2, 10 minute images. Images were reconstructed from 208 projections with equal solid angle spacing onto 64×64×64, 100×100×100, or 128×128×128 voxel image matrices with a physical field of view of . The standard time points for R1/R2 imaging discussed in the R1/R2 Hybrid Imaging subsection of the Methods section were used (12 time points).
RESULTS AND DISCUSSION
GPU Accelerated 3D Reconstruction
The speeds of three different 3D image reconstruction algorithms were compared to evaluate the advantage of GPU-based reconstruction. These three algorithms were: GPU-based single-stage 3D FBP (GS), CPU-based single-stage 3D FBP (CS), and CPU-based multi-stage 3D FBP (CM). A bottle phantom was imaged and reconstructed onto 643, 1003, and 1283 image matrices, as described previously.
It can be seen in Tables 2 and 3 that the GS method enables faster image reconstruction compared to the other two methods. For a 1283 reconstructed object, the CS method required 83 seconds to reconstruct one 3D image. The CM method required 1.108 seconds for reconstruction. However, the GS method was the fastest, requiring only 0.4 seconds for reconstruction, including both computing time and time to transfer data back to the CPU. If only computing time is considered, the GS reconstruction method requires 0.022 seconds, which is 3772 times faster than the CS method and 50.4 times faster than the CM method.
Table 2.
Comparison of time required for the back-projection process for 3D image reconstruction using three different algorithms. The process on the GPU can be split into 2 portions: compute time and transfer time.
| Image Matrix Size (voxels) | GPU Single-Stage (GS) | CPU Multi-Stage (CM) | CPU Single-Stage (CS) | ||
|---|---|---|---|---|---|
| Compute Time (s) | Transfer Time (s) | Total Reconstruction Time (s) | Reconstruction Time (s) | Reconstruction Time (s) | |
| 643 | 0.011 | 0.049 | 0.060 | 0.203 | 13.0 |
| 1003 | 0.016 | 0.229 | 0.245 | 0.624 | 44.0 |
| 1283 | 0.022 | 0.378 | 0.400 | 1.108 | 83.0 |
Table 3.
Acceleration factors for GPU based single-stage reconstruction versus previously used CPU based reconstruction methods.
| Image Matrix Size (voxels) | GS vs. CM not considering transfer time | GS vs. CM considering transfer time | GS vs. CS not considering transfer time | GS vs. CS considering transfer time |
|---|---|---|---|---|
| 643 | 18.4 | 3.40 | 1181 | 216 |
| 1003 | 39.0 | 2.55 | 2750 | 180 |
| 1283 | 50.4 | 2.70 | 3772 | 207 |
| Average Result | 35.9 | 2.88 | 2568 | 201 |
For a more comprehensive comparison, the three average reconstruction speeds for different reconstructed image sizes were compared. Table 3 shows that, on average, the GPU-based reconstruction provides acceleration of the 3D reconstruction by a factor of 2.88 compared to the CM method and 201 compared to the CS method.
While the CM method is significantly faster than the CS method and nearly as fast as the GS method, it is restrictively specialized only for the case of equal linear angle spaced projections. Other projection distributions can be reconstructed using the CM method by interpolating onto equal linear angle spaced projections. This works well for distributions such as equal solid angle spaced projections, but for arbitrary projection distributions this may create appreciable artifacts in the reconstructed image. To accurately reconstruct images from arbitrary projection distributions, single-stage reconstruction is necessary. However, the CS method is relatively slow. The GS method presented in this paper is preferable to both methods. It offers accurate reconstruction from arbitrary projection sets using single-stage reconstruction and takes advantage of the highly parallelizable nature of the FBP algorithm to provide extremely fast image reconstruction using the GPU.
Lookup Table Accelerated Fitting
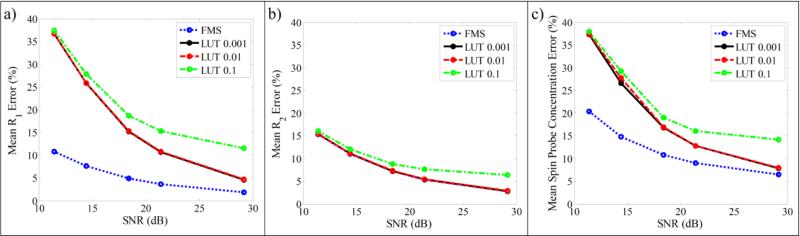
A comparison of the accuracy of fits for R1, R2, and spin probe concentration using different fitting methods is shown in Fig. 3. The error for the tested fitting methods (LUT method with lookup table parameter resolution of 0.1, 0.01, and 0.001 μs−1, as well as the FMS method) asymptotically decreases towards the error level of the noiseless data as SNR increases. Fig. 3 shows that the fit accuracy using the LUT method depends on parameter resolution for the lookup table. Over the range of SNR values used here, lookup tables with R1 and R2 resolution of 0.01 and 0.001 μs−1 result in essentially equivalent accuracy independent of SNR (see red and black curves, respectively in Fig. 3, which essentially lie on top of one another), while resolution of 0.1 μs−1 gives lower accuracy. This implies that, for this application, 0.01 μs−1 resolution may be sufficient.
Figure 3.
Plots of mean error in the fitted parameters versus SNR of simulated projection data, using different fitting methods. (a) Mean R1 error, (b) mean R2 error, and (c) mean spin probe concentration error. Note curves for LUT 0.01 and 0.001 are nearly equivalent.
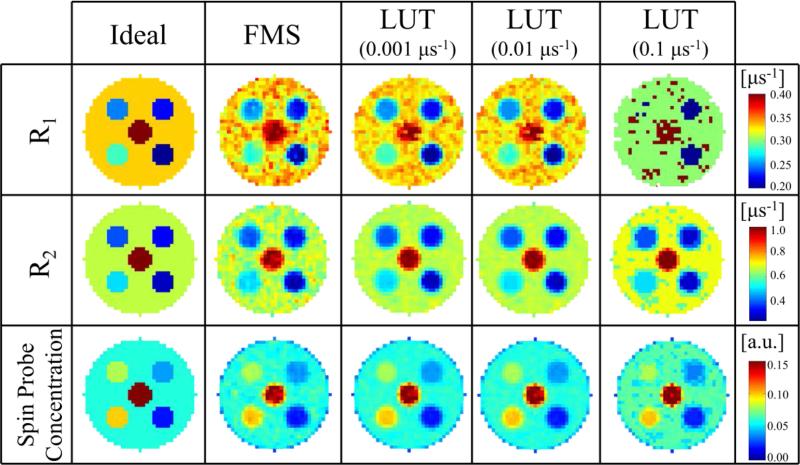
Fig. 4 provides a visual comparison of images reconstructed from the mathematical phantom described in the Methods section, with noise equivalent to a 10 minute image (SNR = 21.39 dB), using different fitting methods. The ideal data shown are the mathematical phantom discretized onto the same 64×64×64 matrix as the reconstructed images. It can be seen that, when a lookup table with sufficient parameters resolution is used, the LUT method provides similar results to the FMS method. However, lookup tables with decreased parameter resolution eventually fail to appropriately fit the data, which is most apparent in the R1 image fit using the LUT method with 0.1 μs−1 resolution.
Figure 4.
Visual comparison of results using different fitting methods. Simulated images using the mathematical phantom are reconstructed using the GS method and either the FMS method or the LUT method with different resolution lookup tables. Noise equivalent to a 10 minutes image (i.e., SNR = 21.39dB) is added and 12 time points are used for the fit. The ideal images are the discretized mathematical phantom.
For simulated 10 minute images with 12 time points, average R1 error using the LUT method with 0.01 μs−1 resolution is 10.5% compared to 3.62% for the FMS method, whereas the R2 error for the two methods is nearly equivalent (5.4% for 0.01 μs−1 resolution LUT method; 5.3% for FMS method). The spin probe concentration error is 12.82% for the 0.01 μs−1 resolution LUT method and 9.04% for the FMS method. Fig. 3 shows that, as SNR increases and fit errors for the various methods approach that of the noiseless cases, the accuracy for both the 0.01 μs−1 resolution LUT method and the FMS method approach similar values (1.39%, 1.61%, 5.8% vs. 0.63%, 1.13%, 5.14% for the R1, R2, and spin probe concentration error using the 0.01 μs−1 resolution LUT method vs. using the FMS method respectively).
While accuracy may decrease when using the LUT fitting method, fitting speed increases significantly. Table 4 shows that, when using 0.01 μs−1 resolution LUT method there is nearly a 4 order of magnitude acceleration compared to the FMS method. If the application required a lookup table with higher parameter resolution, the acceleration factor is just over 3 orders of magnitude using the 0.001 μs−1 resolution LUT method compared to the FMS method. Tradeoffs in accuracy may be acceptable for real-time imaging applications where only a rough idea of the image is required. Following an experiment, higher-quality images with increased accuracy can be produced using other, more robust, yet time intensive methods.
In the future, lookup tables incorporating multiple parameters into a single lookup table, rather than using the multiple lookup table algorithm described in the methods, may provide improved accuracy. For the current application however, such multi-parameter lookup tables became unacceptably large.
Real-Time pO2 Image Reconstruction
To achieve real-time reconstruction, a new, updated image must be reconstructed as each new projection is acquired. Current standard R1/R2 EPRI oxygen images acquire full sets of 208 projections for 12 different time points in 10 minutes. Each projection is acquired at all of the time points before the next projection is acquired. Therefore, for a standard 10 minute, 208 projection image, each projection is acquired in 2.88 seconds. Accordingly, for real-time image reconstruction, reconstruction speeds of at least 2.88 seconds/image are necessary. Using standard reconstruction methods this is very difficult. Accelerations of both 3D image reconstruction and fitting of voxel-by-voxel signal relaxation are necessary. Table 5 summarizes the total pO2 image reconstruction times when using different combinations of the 3D FBP implementations and fitting methods described above.
Table 5.
Summary of total pO2 image reconstruction times when using different combinations of 3D FBP implementations and fitting methods.
| Reconstruction Method (3D FBP + Fitting) | Minimum Reconstruction Time (s) | Maximum Reconstruction Time (s) |
|---|---|---|
| CM + FMS | – | 284 |
| CM + LUT 0.001 | – | 2.70 |
| CM + LUT 0.01 | – | 2.47 |
| CS + FMS | 283 | 503 |
| CS + LUT 0.001 | 1.33 | 221 |
| CS + LUT 0.01 | 1.10 | 221 |
| GS + LUT 0.001 | 0.85 | 0.98 |
| GS + LUT 0.01 | 0.62 | 0.76 |
Minimum reconstruction time is for reconstruction from a single projection. Maximum reconstruction time is for reconstruction from the full 208 projections. The CM method requires reconstruction from a full projection set and therefore only one reconstruction time is reported. Numbers shown next to LUT denote parameter resolution used in μs−1.
Using the CM method for 3D reconstruction and FMS method for fitting, requires 0.203 seconds per 3D reconstruction at each time point for standard 64×64×64 R1/R2 EPRI pO2 images, plus 282 seconds for fitting. In this case, 3D reconstruction is relatively fast while fitting limits the overall reconstruction speed. Using the LUT fitting method and CM reconstruction method, pO2 image reconstruction time is faster than projection acquisition speed. However, multi-stage reconstruction (e.g., the CM method) cannot handle arbitrary projection sets and therefore is not a viable candidate for real-time reconstruction from incomplete sets of projections during acquisition.
Single-stage reconstruction, however, can be used for any arbitrary set of projections and therefore can be used to reconstruct images from incomplete projection sets, but the current implementation on the CPU is restrictively slow. 3D image reconstruction speed is proportional to the number of projections in the image. Total pO2 image reconstruction time for single-stage methods can be calculated as the time to back-project each projection multiplied by both the total number of projections used and the number of time points used (e.g., 12 for standard R1/R2 imaging) plus the time to fit the spectral dimension. When using a GPU, time to transfer 3D images between GPU and CPU must be added as well. When using the CS method and FMS method, both 3D reconstruction and fitting can be time consuming and real-time pO2 image reconstruction is not possible. If the CS method and LUT method are used, for smaller numbers of projections, reconstruction is fast enough to keep up with data acquisition; however as imaging progresses, real-time pO2 image reconstruction cannot be achieved.
The methods presented in this paper (GPU-based single-stage 3D reconstruction and lookup table fitting), when used in concert, provide the versatility of single-stage reconstruction and the speed necessary for real-time pO2 image reconstruction. Using the GS method and 0.001 μs−1 resolution LUT method results in reconstruction speed acceleration by up to a factor of 511 compared to current methods. Using the GS method and 0.01 μs−1 resolution LUT method results in reconstruction speed acceleration by up to over a factor of 650 compared to current methods.
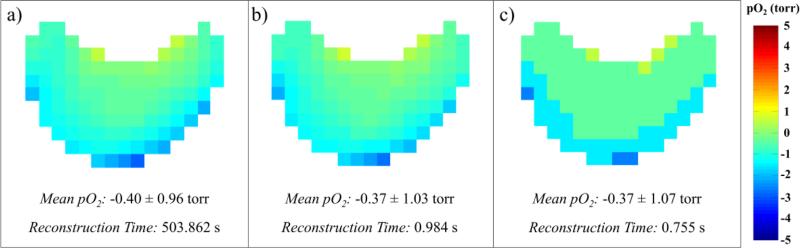
Fig. 5 shows that, if a lookup table with appropriate resolution is used, the accelerated methods for EPRI pO2 image reconstruction presented provide essentially the same result as previously used methods. Only very subtle differences can be visually discerned between the image reconstructed using the CS+FMS methods and that reconstructed using the GS+LUT with 0.001 μs−1 resolution (average voxel absolute pO2 difference was 0.08 torr). Using a 0.01 μs−1 resolution lookup table still provides a reasonable image, however the effect of decreased lookup table resolution can clearly be observed as a decrease in pO2 resolution of the image (average voxel absolute pO2 difference was 0.28 torr). With these accelerated reconstruction speeds, the new methods enable real-time reconstruction of pO2 images for imaging times as short as 3.4 minutes when using 0.001 μs−1 resolution or 2.6 minutes when using 0.01 μs−1 resolution lookup tables. As imaging times decrease, reconstruction speed must increase accordingly to enable real-time reconstruction. More powerful, higher-end GPU cards exist that could increase reconstruction speeds. However, GPU-based single-stage 3D reconstruction and lookup table fitting together provide EPRI pO2 image reconstruction accelerations sufficient to enable real-time reconstruction for current imaging applications.
Figure 5.
Axial slice through EPRI pO2 image of bottle phantom showing meniscus in trityl solution. (a) Image reconstructed using previous methods: CS+FMS. (b) Image reconstructed using accelerated methods: GS+LUT 0.001 μs−1 resolution. (c) Image reconstructed using accelerated methods: GS+LUT 0.01 μs−1 resolution.
CONCLUSIONS
Methods providing EPRI pO2 image reconstruction acceleration are presented. The acceleration is sufficient to enable real-time pO2 image reconstruction. It should be emphasized that any sacrifice in reconstruction quality that is associated with the techniques discussed in this work can be recovered post acquisition with more standard reconstruction techniques, for example, avoiding the loss of pO2 resolution with the LUT technique.
Real-time reconstruction is advantageous for different reasons. For example, for in vivo imaging applications, particularly in the patient setting, procedures are time sensitive. Real-time image reconstruction can help operators recognize and resolve issues with an EPRI scan early and limit wasted time if the operator must wait for imaging to be completed to discover these issues. For example, if an image becomes corrupted during imaging, (due to improper setup of the subject and/or imaging equipment, subject motion, imaging equipment failure, etc.) real-time reconstruction allows detection of such issues prior to scan completion. Real-time reconstruction can also identify a region of interest (ROI) earlier during imaging.
Projection acquisition schemes have been developed that provide approximately uniform sampling throughout the imaging process. Such acquisition schemes enable reconstruction of useful intermediate images from incomplete projection data, which closely approximate the final image reconstructed from complete projection data (35, 36). For EPRI data acquired using these robust techniques, the real-time reconstruction presented in this work allows for the visualization of the buildup of an oxygen image as data are acquired.
Studies suggest that both chronic and transient states of hypoxia may exist in tumors and that they may have differing physiological and clinical implications (11). This dynamic nature of the in vivo oxygen environment has led to increased interest in time resolved measurements and visualization of pO2 in order to help clarify the differences between chronic and transient hypoxia. To this end, efforts are being made to increase EPRI temporal resolution (37–41).
As EPRI image acquisition times approach real-time acquisition, the real-time image reconstruction discussed herein will eventually enable real-time visualization of temporal pO2 fluctuations. This could allow for real-time differentiation of chronically hypoxic regions and transiently hypoxic regions. One potential future approach is a sliding window or view-sharing technique, which could provide quasi-real-time pO2 imaging based on rapid reconstruction of images from temporally overlapping projection subsets throughout acquisition. Such methods have been used for real-time CT (25) and MRI (34,42) applications. In addition to visualizing transient hypoxia, future real-time EPRI pO2 imaging could enable image guided treatment adaptation based on spatiotemporally heterogeneous tumor pO2, EPRI-guided biopsies, and real-time subject positioning and motion monitoring.
The methods presented in this work provide EPRI pO2 image reconstruction acceleration factors of over 650. This enables real-time reconstruction of noninvasively acquired EPRI images of in vivo oxygen concentrations for the first time.
ACKNOWLEDGMENTS
This work was supported by grants from the NIH (P41 EB002034 and R01 CA98575). The authors are very grateful for the insight provided by Dr. Xiaochuan Pan and his research group in helping to develop and refine this work.
REFERENCES
- 1.Overgaard J. Hypoxic radiosensitization: adored and ignored. J Clin Oncol. 2007;25(26):4066–4074. doi: 10.1200/JCO.2007.12.7878. [DOI] [PubMed] [Google Scholar]
- 2.Schwarz G. Uber Desensibilisierung gegen Rontgen- und Radiumstrahlen. Munchner Medizinische Wochenschrift. 1909;56:1217–1218. [Google Scholar]
- 3.Hall EJ. Radiobiology for the Radiologist. 5th Ed. Lippincott Williams & Wilkins; Philadelphia: 2000. [Google Scholar]
- 4.Hockel M, Schlenger K, Aral B, Mitze M, Schaffer U, Vaupel P. Association between tumor hypoxia and malignant progression in advanced cancer of the uterine cervix. Cancer Res. 1996;56(19):4509–4515. [PubMed] [Google Scholar]
- 5.Shannon AM, Bouchier-Hayes DJ, Condron CM, Toomey D. Tumour hypoxia, chemotherapeutic resistance and hypoxia-related therapies. Cancer Treat Rev 2003. 2003;29(4):297–307. doi: 10.1016/s0305-7372(03)00003-3. [DOI] [PubMed] [Google Scholar]
- 6.Carmeliet P, Dor Y, Herbert JM, Fukumura D, Brusselmans K, Dewerchin M, Neeman M, Bono F, Abramovitch R, Maxwell P, Koch CJ, Ratcliffe P, Moons L, Jain RK, Collen D, Keshert E, Keshet E. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature. 1998;394(6692):485–490. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- 7.Rofstad EK. Microenvironment-induced cancer metastasis. Int J Radiat Biol. 2000;76(5):589–605. doi: 10.1080/095530000138259. [DOI] [PubMed] [Google Scholar]
- 8.Dewhirst MW, Klitzman B, Braun RD, Brizel DM, Haroon ZA, Secomb TW. Review of methods used to study oxygen transport at the microcirculatory level. Int J Cancer. 2000;90(5):237–255. [PubMed] [Google Scholar]
- 9.Zhao DW, Jiang L, Mason RP. Measuring changes in tumor oxygenation. Methods Enzymol. 2004;386:378–418. doi: 10.1016/S0076-6879(04)86018-X. [DOI] [PubMed] [Google Scholar]
- 10.Tatum JL. Hypoxia: Importance in tumor biology, noninvasive measurement by imaging, and value of its measurement in the management of cancer therapy. Int J Rad Biol. 2006;82(10):699–757. doi: 10.1080/09553000601002324. [DOI] [PubMed] [Google Scholar]
- 11.Bayer C, Vaupel P. Acute versus chronic hypoxia in tumors: Controversial data concerning time frames and biological consequences. Strahlenther Onkol. 2012;188(7):616–627. doi: 10.1007/s00066-012-0085-4. [DOI] [PubMed] [Google Scholar]
- 12.Elas M, Ahn KH, Parasca A, Barth ED, Lee D, Haney C, Halpern HJ. Electron paramagnetic resonance oxygen images correlate spatially and quantitatively with Oxylite oxygen measurements. Clin Cancer Res. 2006;12(14 Pt 1):4209–4217. doi: 10.1158/1078-0432.CCR-05-0446. [DOI] [PubMed] [Google Scholar]
- 13.Epel B, Sundramoorthy SV, Mailer C, Halpern HJ. A versatile high speed 250-MHz pulse imager for biomedical applications. Concept Magn Reson B Magn Reson Eng. 2008;33B(3):163–176. doi: 10.1002/cmr.b.20119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Epel B, Sundramoorthy SV, Barth ED, Mailer C, Halpern HJ. Comparison of 250 MHz electron spin echo and continuous wave oxygen EPR imaging methods for in vivo applications. Med Phys. 2011;38(4):2045–2052. doi: 10.1118/1.3555297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Epel B, Haney CR, Hleihel D, Wardrip C, Barth ED, Halpern HJ. Electron paramagnetic resonance oxygen imaging of a rabbit tumor using localized spin probe delivery. Med Phys. 2010;37(6):2553–2559. doi: 10.1118/1.3425787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elas M, Williams BB, Parasca A, Mailer C, Pelizzari CA, Lewis MA, River JN, Karczmar GS, Barth ED, Halpern HJ. Quantitative tumor oxymetric images from 4D electron paramagnetic resonance imaging (EPRI): Methodology and comparison with blood oxygen level-dependent (BOLD) MRI. Magn Reson Med. 2003;49(4):682–691. doi: 10.1002/mrm.10408. [DOI] [PubMed] [Google Scholar]
- 17.Epel B, Bowman MK, Mailer C, Halpern HJ. Absolute oxygen R1e imaging in vivo with pulse electron paramagnetic resonance. Magn Reson Med. 2014;72(2):362–368. doi: 10.1002/mrm.24926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai CM, Lauterbur PC. A gradient control device for complete 3-dimensional nuclear magnetic resonance zeugmatographic imaging. J Phys E Sci Instrum. 1980;13(7):747–750. [Google Scholar]
- 19.Hyslop WB, Woods RK, Lauterbur PC. Four-dimensional spectral-spatial imaging using projection reconstruction. IEEE Trans Med Imaging. 1995;14(2):374–383. doi: 10.1109/42.387718. [DOI] [PubMed] [Google Scholar]
- 20.Cabral B, Cam N, Foran J. Accelerated volume rendering and tomographic reconstruction using texture mapping hradware. Proc Symp Volume Visualization. 1994:91–98. [Google Scholar]
- 21.Pratx G, Xing L. GPU computing in medical physics: a review. Med Phys. 2011;38(5):2685–2697. doi: 10.1118/1.3578605. [DOI] [PubMed] [Google Scholar]
- 22.Ma D, Gulani V, Seiberlich N, Liu K, Sunshine JL, Duerk JL, Griswold MA. Magnetic resonance fingerprinting. Nature. 2013;495(7440):187–192. doi: 10.1038/nature11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eggers H, Proksa R. Multiprocessor system for real-time convolution interpolation reconstruction. Proc Int Soc Magn Reson Med. 1999:95. [Google Scholar]
- 24.Sorensen TS, Atkinson D, Schaeffter T, Hansen MS. Real-time reconstruction of sensitivity encoded radial magnetic resonance imaging using a garphics processing unit. IEEE T Med Imaging. 2009;28(12):1974–1985. doi: 10.1109/TMI.2009.2027118. [DOI] [PubMed] [Google Scholar]
- 25.Xu F, Mueller K. Real-time 3D computed tomographic reconstruction using commodity graphics hardware. Phys Med Biol. 2007;52(12):3405–3419. doi: 10.1088/0031-9155/52/12/006. [DOI] [PubMed] [Google Scholar]
- 26.Seifi P. Ph.D. Thesis. University of Chicago; Dec, 2010. Scalability of time-domain electron paramagnetic resonance in vivo imaging. [Google Scholar]
- 27.Mailer C, Sundramoorthy SV, Pelizzari CA, Halpern HJ. Spin echo spectroscopic electron paramagnetic resonance imaging. Magn Reson Med. 2006;55(4):904–912. doi: 10.1002/mrm.20849. [DOI] [PubMed] [Google Scholar]
- 28.Ardenkjaer-Larsen JH, Laursen I, Leunbach I, Ehnholm G, Wistrand LG, Petersson JS, Golman K. EPR and DNP properties of certain novel single electron contrast agents intended for oximetric imaging. J Magn Reson. 1998;133(1):1–12. doi: 10.1006/jmre.1998.1438. [DOI] [PubMed] [Google Scholar]
- 29.Deans SR. The Radon transformation and some of its applications. John Wiley & Sons; New York, NY: 1983. [Google Scholar]
- 30.Lagarias JC, Reeds JA, Wright MH, Wright PE. Convergence properties of the Nelder-Mead simplex method in low dimensions. Siam J Optimiz. 1998;9(1):112–147. [Google Scholar]
- 31.Doneva M, Senegas J, Bornert P, Eggers H, Mertins A. Accelerated MR parameter mapping using compressed sensing with model-based sparsifying transform. Proc Int Soc Magn Reson. 2009:2812. [Google Scholar]
- 32.Aharon M, Elad M, Bruckstein A. K-SVD: An algorithm for designing overcomplete dictionaries for sparse representation. IEEE Trans Signal Process. 2006;54(11):4311–4322. [Google Scholar]
- 33.Doneva M, Bornert P, Eggers H, Stehning C, Senegas J, Mertins A. Compressed sensing reconstruction for magnetic resonance parameter mapping. Magn Reson Med. 2010;64(4):1114–1120. doi: 10.1002/mrm.22483. [DOI] [PubMed] [Google Scholar]
- 34.Ehses P, Seiberlich N, Ma D, Breuer FA, Jakob PM, Griswold MA, Gulani V. IR TrueFISP with a golden-ratio-based radial readout: fast quantification of T1,T2, and proton density. Magn Reson Med. 2013;69(1):71–81. doi: 10.1002/mrm.24225. [DOI] [PubMed] [Google Scholar]
- 35.Chan RW, Ramsay EA, Cunningham CH, Plewes DB. Temporal stability of adaptive 3D radial MRI using multidimensional golden means. Magn Reson Med. 2009;61(2):354–363. doi: 10.1002/mrm.21837. [DOI] [PubMed] [Google Scholar]
- 36.Redler G, Epel B, Halpern HJ. Maximally spaced projection sequencing in electron paramagnetic resonance imaging. Concepts Magn Reson Part B. 2015 doi: 10.1002/cmr.b.21280. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Redler G, Epel B, Halpern HJ. EPR image based oxygen movies for transient hypoxia. Adv Exp Med Biol. 2014;812:127–133. doi: 10.1007/978-1-4939-0620-8_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Redler G, Epel B, Halpern HJ. Principal component analysis enhances snr for dynamic electron paramagnetic resonance oxygen imaging of cycling hypoxia in vivo. Magn Reson Med. 2014;71(1):440–450. doi: 10.1002/mrm.24631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yasui H, Matsumoto S, Devasahayam N, Munasinghe JP, Choudhuri R, Saito K, Subramanian S, Mitchel JB, Krishna MC. Low-field magnetic resonance imaging to visualize chronic and cycling hypoxia in tumor-bearing mice. Cancer Res. 2010;70(16):6427–6436. doi: 10.1158/0008-5472.CAN-10-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsumoto S, Yasui H, Mitchell JB, Krishna MC. Imaging cycling tumor hypoxia. Cancer Res. 2010;70(24):10019–10023. doi: 10.1158/0008-5472.CAN-10-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Redler G. Ph.D. Thesis. University of Chicago; Jun, 2014. Enhance dynamic electron paramagnetic resonance imaging of in vivo physiology. [Google Scholar]
- 42.Uecker M, Zhang S, Frahm J. Nonlinear inverse reconstruction for real-time MRI of the human heart using undersampled radial FLASH. Magn Reson Med. 2010;63(6):1456–1462. doi: 10.1002/mrm.22453. [DOI] [PubMed] [Google Scholar]