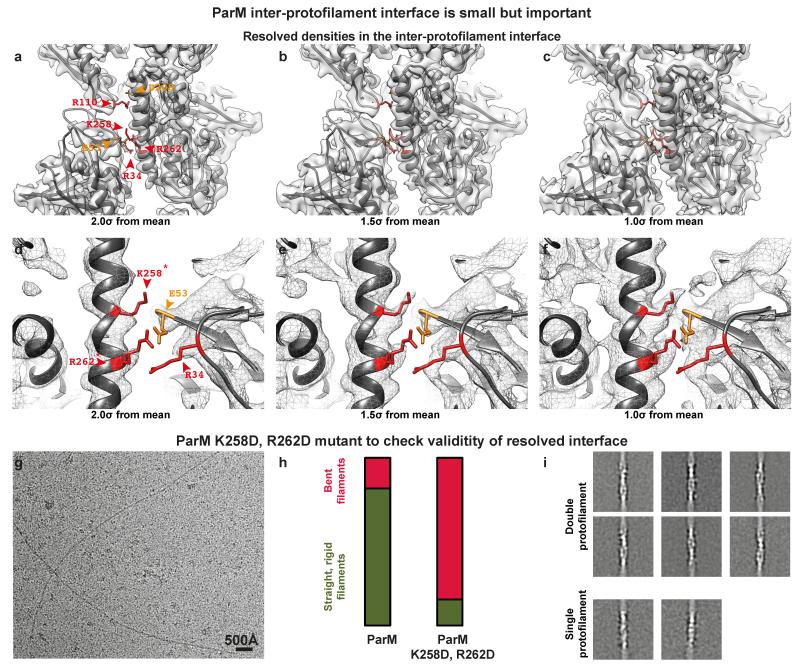
Extended Data Figure 3. The ParM inter-protofilament interface is small but important.
(a) Cryo-EM density for the ParM+AMPPNP filament is shown at an isosurface contour level of 2.0 σ from the mean value. Overlaid on the density, refined atomic coordinates from REFMAC are additionally displayed as grey ribbons. Residues forming salt bridges at the inter-protofilament interface are highlighted. (b) The same figure as a), except the cryo-EM density shown at an isosurface contour level of 1.5 σ from the mean. (c) 1.0 σ from the mean. (d-f) A magnified view of the primary salt-bridged interface consisting of charged residues that form the ParM inter-protofilament interface. The cryo-EM density is shown as a mesh at three different contour levels to demonstrate resolved side chain densities. Positively charged residues are highlighted in red while negatively charged residues are highlighted in orange. (g) Two residues (K258 and R262) that were the best resolved (marked with an * in d), were mutated to aspartic acid to test the importance of this inter-protofilament interface. A cryo-EM image of this mutant protein assembled with AMPPNP is shown. A much higher concentration of the protein was required to obtain filaments on cryo-EM grids (Supplementary Methods). This experiment was repeated four times. (h) Randomly selected cryo-EM images of ParM+AMPPNP and ParM(K258D, R262D)+AMPPNP were used to count occurrences of straight and bent filaments by visual inspection. The results of this quantification are shown as a percentage bar diagram. For the ParM protein, 82% of all filaments were classified as straight, while 18% were bent (n=345). Using exactly the same classification criteria, only 15% of the filaments were found to be straight and 85% of the filaments were bent (n=45) for the ParM(K258D, R262D) mutant protein. (i) Reference-free class averages show that most of the ParM(K258D, R262D) filaments are made up of double protofilaments like wild-type ParM. Some class averages show evidence of bending. A few class averages show that single protofilaments were present in the sample (lower panels). However, the double mutation destabilises the entire ParM filament, making filament formation an unfavourable reaction, illustrating that even though the inter-protofilament interface is small, it is critical for ParM filament formation.