C. elegans transition zone structures are dispensable for axoneme assembly but are required for cell–matrix interactions during neurite extension, revealing an unexpected role for the transition zone in cell adhesion.
Abstract
Cilia are cellular projections that perform sensory and motile functions. A key ciliary subdomain is the transition zone, which lies between basal body and axoneme. Previous work in Caenorhabditis elegans identified two ciliopathy-associated protein complexes or modules that direct assembly of transition zone Y-links. Here, we identify C. elegans CEP290 as a component of a third module required to form an inner scaffolding structure called the central cylinder. Co-inhibition of all three modules completely disrupted transition zone structure. Surprisingly, axoneme assembly was only mildly perturbed. However, dendrite extension by retrograde migration was strongly impaired, revealing an unexpected role for the transition zone in cell adhesion.
Introduction
The cilium is an evolutionarily ancient structure found in most eukaryotic lineages (Hodges et al., 2010). Projecting from the cell surface, cilia mediate sensory perception and developmental signaling. A subset of cilia is also motile, enabling movement of cells and fluids. Cilia dysfunction has been linked to a spectrum of human disorders, known as ciliopathies (Fliegauf et al., 2007), ranging from disorders affecting a single type of cilium to broad-based disorders affecting multiple tissues and organs.
All cilia arise from a centriole-derived basal body, connected to the plasma membrane via transition fibers. The axonemal microtubules constituting the core of the cilium are extensions of basal body microtubules. Immediately distal to the basal body lies the transition zone, distinguished by several conserved features, including a central cylinder or apical ring that lies internal to the microtubule doublets and Y-links connecting axonemal microtubules to the ciliary membrane (Reiter et al., 2012). The ciliary membrane is contiguous with the plasma membrane but is compositionally distinct, as is the underlying cytoplasm, suggesting that structures at the ciliary base (the transition fibers and/or transition zone) control access to the ciliary compartment (Rosenbaum and Witman, 2002).
During ciliogenesis, centrioles dock to a membrane, either to a vesicle in the cytoplasm that fuses with the plasma membrane (Sorokin, 1962) or to the plasma membrane itself. Docking requires the transition fibers (Schmidt et al., 2012), which in vertebrates are derived from appendages present on mature centrioles. Finally, motor-driven intraflagellar transport (IFT; Cole et al., 1998) extends the ciliary axoneme. While the molecular mechanisms underlying axoneme extension are comparatively well understood, less is known about basal ciliary structures and their role in the early stages of ciliogenesis (Reiter et al., 2012). For the transition zone, proteomic approaches in vertebrates identified three distinct ciliopathy-associated multiprotein complexes or modules: the MKS, NPHP-1,-4,-8 (NPHP) and NPHP-5,-6 (CEP290) modules (Garcia-Gonzalo et al., 2011; Sang et al., 2011; Chih et al., 2012). These are supported by genetic analyses in Caenorhabditis elegans, which have established independent assembly pathways for two modules (MKS and NPHP; Williams et al., 2011). It is currently unclear where these modules fit within the transition zone structure or how it assembles during ciliogenesis. Further, while the transition zone clearly functions as a ciliary gate, there is some uncertainty as to what extent it is also required for axoneme assembly, with loss of transition zone components in vertebrates resulting in loss of cilia in some tissues but not others (Garcia-Gonzalo et al., 2011; Sang et al., 2011; Chih et al., 2012).
The nematode C. elegans has emerged as a major experimental model to study centrioles and cilia. A key feature of C. elegans is that cilia are limited to the dendritic endings of postmitotic sensory neurons and are dispensable for viability and fertility, which facilitates loss-of-function studies (Inglis et al., 2007). Here, we take advantage of this experimental model to dissect the assembly and function of the transition zone. We identify a C. elegans homologue of CEP290 and show it to be a component of a third, independently targeted transition zone module required to form the central cylinder, which acts as an inner scaffolding structure for transition zone assembly. Simultaneous inhibition of all three modules completely disrupts transition zone structure. Surprisingly, axoneme assembly is not significantly impaired. Instead, perturbing the transition zone disrupts cell–matrix interactions during dendrite extension, revealing an unexpected role for the transition zone in cell adhesion.
Results and discussion
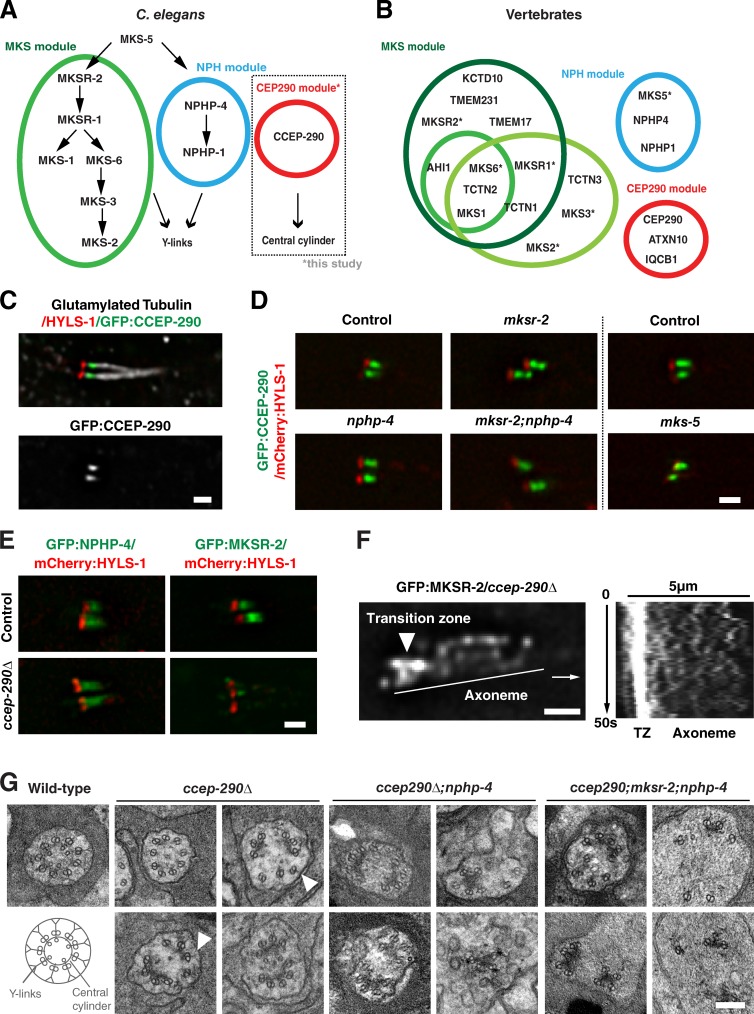
Increasing evidence points to the transition zone being organized into multiple protein complexes or modules with distinct functions. Best characterized are the MKS and NPHP modules, which are composed of proteins mutated in the ciliopathies Meckel syndrome and nephronophthisis. Previous work in C. elegans showed these to be recruited independently of each other and function redundantly in assembly of transition zone Y-links (Fig. 1 A; Williams et al., 2011). Proteomic analysis in vertebrates identified a potential third module including the key ciliopathy protein CEP290 (Fig. 1 B; Sang et al., 2011). Reciprocal BLAST searches identified a C. elegans homologue encoded by the predicted gene Y47G6A.17, which we named CCEP-290 for C. elegans CEP290 (Fig. S1, A and B). Phylogenetic analysis found CEP290 to be conserved across all major eukaryotic phyla, supporting a central role for this protein at the transition zone (Fig. S1 C; Hodges et al., 2010).
Figure 1.
CCEP-290 belongs to a third transition zone module required for assembly of the central cylinder. (A) Transition zone modules in C. elegans, based on genetic interactions and localization interdependencies (Huang et al., 2011; Williams et al., 2011; this study). (B) Modules in vertebrates, based on proteomic studies (green, red, blue: Sang et al., 2011; dark green: Chih et al., 2012; light green: Garcia-Gonzalo et al., 2011). *, C. elegans names used for ease of comparison. HGNC names: B9D1/MKSR1, B9D2/MKSR2, TMEM216/MKS2, TMEM67/MKS3, RPGRIP1L/MKS5, CC2D2A/MKS6. (C) Immunofluorescence micrographs of phasmid (tail) cilia of worms expressing GFP:CCEP-290 and stained for HYLS-1 and glutamylated tubulin. (D and E) Localization interdependencies between CCEP-290 and MKS/NPHP module components MKS-5, NPHP-4, and MKSR-2. Panels show phasmid cilia in worms coexpressing mCherry:HYLS1 and GFP:CCEP-290/MKSR-2/NPHP-4 in wild-type and transition zone mutants, as indicated. (F) MKSR-2 at transition zones is reduced due to dispersal along the ciliary axoneme. A still image and kymograph from a time-lapse sequence of GFP:MKSR-2 in ccep-290Δ mutant phasmids is shown (see also Video 1). The line indicates the kymograph axis. (G) Transmission electron micrographs of amphid transition zones in wild-type, ccep-290Δ, ccep-290Δ;nphp-4, and ccep-290;mksr-2;nphp-4 mutants. While a central cylinder is not apparent and transition zones are fragmented, Y-links (arrowheads) are still occasionally present in ccep-290Δ mutants. Inner singlet microtubule numbers are reduced (3.8 ± 1.5, n = 25 wild-type; 1.6 ± 1.4, n = 21 ccep-290Δ; t test; P < 0.0001), potentially due to loss of the central cylinder to which they normally attach. Transition zone structures are completely lost in ccep-290;mksr-2;nphp-4 triple mutants. Bars: (C, D, and E) 1 µm; (F) 5 µm; (G) 200 nm.
Generation of a single-copy GFP transgene under control of endogenous regulatory sequences revealed expression exclusively in ciliated neurons in a manner dependent on the transcription factor DAF-19 (Fig. S1 D), which serves as a master regulator of ciliogenesis in C. elegans (Swoboda et al., 2000). Consistent with localization to transition zones, CCEP-290 was found distal to the basal body marker HYLS-1 (Dammermann et al., 2009) but proximal to the axoneme positive for polyglutamylated tubulin (Fig. 1 C). Comparisons by high-resolution two-color fluorescence imaging (Wan et al., 2009) using HYLS-1 as a point of reference showed this localization to be distinct from that of the MKS and NPHP module components MKSR-2 and NPHP-4. Whereas MKSR-2 and NPHP-4 localized to the transition zone periphery, consistent with localization to Y-links or the ciliary membrane, CCEP-290 occupied a more central position (Fig. S2, A and B). CCEP-290 recruitment was unaffected in mutants of MKS-5, MKSR-2, and NPHP-4, the most upstream components of the MKS and NPHP assembly pathways (Fig. 1 D). CCEP-290 therefore independently targets to the transition zone.
Since C. elegans neurons are refractory to RNAi, we obtained an in-frame deletion mutant, ccep-290(tm4927), hereafter referred to as ccep-290, affecting the N terminus of the protein (Fig. S1 E). We also generated a complete gene deletion, ccep-290Δ, using CRISPR/Cas9-mediated genome editing (Fig. S1 F). Both mutants, as well as two additional C-terminal truncation mutants, were found to be viable and fertile, with no apparent morphological defects (Fig. S1 G and not depicted). Examining other transition zone proteins in ccep-290Δ mutants, we observed no change in localization of NPHP-4. In contrast, MKSR-2 at transition zones appeared much reduced (Fig. 1 E), which is consistent with recent findings in Drosophila melanogaster (Basiri et al., 2014). Localization of downstream components of the MKS pathway was also perturbed (Fig. S2 C). Closer inspection revealed that MKSR-2 still targeted to cilia (70.1 ± 16.0% of controls, t test, P < 0.0001). However, most of the protein (61.8 ± 16.4%) was now found dispersed along the axoneme, with MKSR-2 particles moving in a manner characteristic of diffusion (Fig. 1 F and Video 1). Instead of targeting MKSR-2 as expected for an upstream component in the MKS assembly pathway, CCEP-290 therefore tethers MKSR-2 to the transition zone.
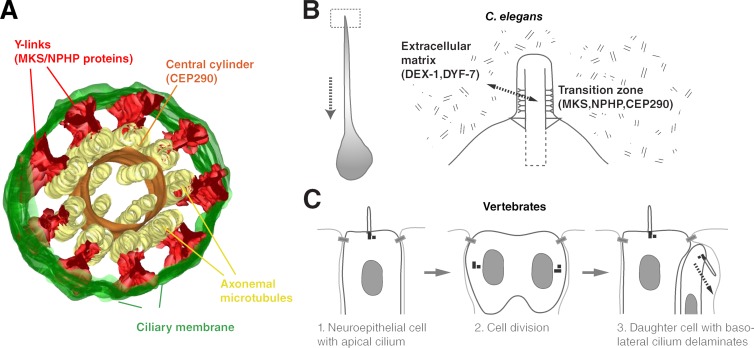
Examination of the C. elegans transition zone by serial-section electron microscopy reveals two prominent ultrastructural features: Y-links connecting axonemal microtubules to the ciliary membrane and the central cylinder to which axonemal outer doublet and inner singlet microtubules are attached (Fig. 1 G; Perkins et al., 1986). Tomographic reconstructions show both structures to be contiguous along the longitudinal axis of the transition zone, forming elongated sheets (Video 2; see also Fig. 4 A). Previous work in C. elegans had implicated MKS and NPHP proteins in assembly of Y-links. Loss of these connectors results in detachment and dilation of the ciliary membrane, while leaving the central cylinder and associated axonemal microtubules intact (Williams et al., 2011). The same role had been ascribed to CEP290 in Chlamydomonas reinhardtii, although Y-links were still observed, albeit rarely, in cep290 mutants (Craige et al., 2010). In contrast, loss of CCEP-290 in C. elegans ccep-290Δ mutants resulted in fragmentation of the central cylinder, disrupting the radially symmetric array of axonemal microtubules (Fig. 1 G). Unlike in MKS/NPHP mutants, there was no detachment of the ciliary membrane and Y-links could still be found in some sections (arrowheads). ccep-290Δ;nphp-4 mutants displayed an intermediate phenotype, with 5/11 transition zones appearing fragmented and the remainder fully disorganized. Finally, disruption of all three modules in ccep-290;mksr-2;nphp-4 triple mutants resulted in a complete loss of transition zone structures, with axonemal microtubule doublets dissociated from each other and the ciliary membrane (Fig. 1 G). These results are consistent with our high-resolution localization data and highlight the distinct roles of CCEP-290 and MKS/NPHP proteins at the transition zone, with CCEP-290 serving as a core component of the central cylinder, which appears to act as an inner scaffold for transition zone assembly, while MKS/NPHP proteins function in assembly of peripheral Y-links.
Figure 4.
Molecular architecture of the transition zone and function in cell adhesion. (A) 3D reconstruction model of the transition zone in C. elegans, highlighting key features. See also Video 2. Based on protein localization and mutant phenotypes, CCEP-290 is an essential component of the central cylinder (this study), whereas MKS and NPHP module components function in assembly of Y-links (Williams et al., 2011). Transition zone mutants display no apparent defects in axoneme assembly. However, ciliary gating and cell adhesion are compromised. (B) The dendrite of C. elegans amphid and phasmid neurons forms by retrograde extension, with the cell body moving backward while the dendritic tip remains in place. The transition zone mediates tip anchorage via interactions with the extracellular matrix. (C) Positioning of the primary cilium determines cell fate in vertebrate neuroepithelium. Cells with apically positioned cilia maintain their position at the apical adherens junction belt, whereas cells with basolateral cilia delaminate (Wilsch-Bräuninger et al., 2012; Paridaen et al., 2013). Differential signaling, but also mechanical anchorage by the cilium, could explain this result.
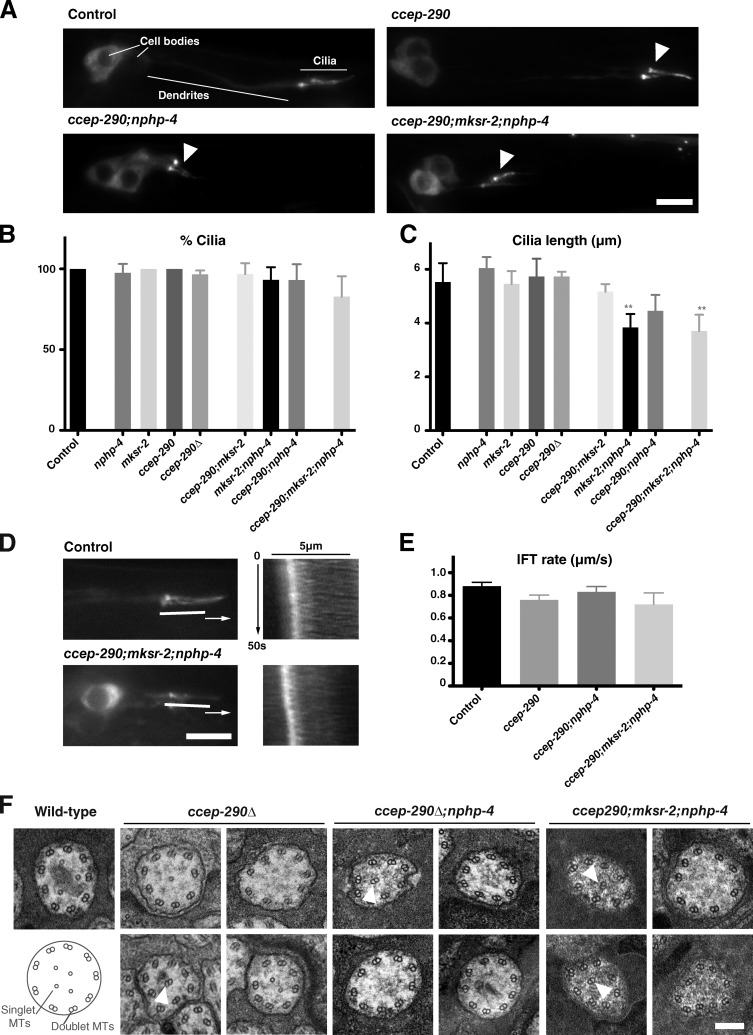
We next examined the consequences of transition zone perturbations on cilia assembly. A commonly used method to assess cilia integrity in C. elegans is the dye-fill assay, which monitors uptake of the lipophilic dye DiI (Inglis et al., 2007). This dye is taken up by a subset of ciliated neurons (12 amphid neurons in the head, 4 phasmid neurons in the tail) through their exposed ciliated endings and accumulates in their cell bodies. Defective dye-filling (Dyf) is often indicative of compromised cilia structure. As previously reported for other transition zone mutants (Williams et al., 2008, 2011), ccep-290 mutants individually did not display significant dye-fill defects. However, double mutants with either nphp-1 or nphp-4 were strongly Dyf. In contrast, no genetic interactions were detected with mksr-2. Dye-filling was restored by expression of the appropriate GFP transgene, confirming specificity of the mutant phenotypes and functionality of our GFP reporters (Fig. S1 H; Fig. S2, D and E; and not depicted). These results would appear to support a role for the transition zone in cilia assembly. However, direct visualization of phasmid cilia using the IFT marker CHE-11:GFP failed to reveal any major defects. Most strikingly, ∼83% of cilia were found to remain in ccep-290;mksr-2;nphp-4 triple mutants in which all three transition zone modules are inhibited (Fig. 2, A and B). Cilia lengths (Fig. 2 C) and IFT rates (Fig. 2 D, E) were also largely normal. Ciliary ultrastructure was also unaffected with the exception of occasional displaced doublet microtubules, likely reflecting disorganization within the transition zone (Fig. 2 F). We conclude that loss of transition zone structures has only mild effects on axoneme assembly and organization in C. elegans.
Figure 2.
Ciliogenesis is largely normal in transition zone mutants. (A) Phasmid cilia in wild-type and transition zone mutants visualized by the IFT marker CHE-11:GFP (arrowheads). (B and C) Quantitation of phasmid cilia number per animal (B, n > 50 animals) and length (C, n > 25 cilia) based on CHE-11:GFP. Error bars indicate the 95% confidence intervals. Asterisks indicate statistically significant difference to wild-type (t test, P < 0.01). (D) Stills and kymographs from time-lapse sequences of IFT in phasmids of wild-type and transition zone triple mutants expressing CHE-11:GFP. (E) Anterograde IFT rates in the middle segment of wild-type and transition zone mutant phasmids (n > 40 particles per strain). Error bars indicate the 95% confidence interval. (F) Transmission electron micrographs of amphid cilia at the level of the middle segment of the axoneme in wild-type, ccep-290Δ, ccep-290Δ;nphp-4, and ccep-290;mksr-2;nphp-4 mutants. Axonemes were essentially normal except for occasional displaced doublet microtubules (arrowheads). Doublet microtubule number was not significantly affected (8.7 ± 1.3 2×MTs/cilium, n = 38 wild type; 7.7 ± 1.8 2×MTs/cilium, n = 27 triple mutant; t test, NS). Bars: (A and D) 5 µm; (F) 200 nm.
While cilia assembly was largely unaffected in transition zone mutants, neuronal morphology was strongly perturbed. This was most clearly seen in phasmids, where dendrites collapsed almost entirely, with cilia found immediately adjacent to the cell body (Fig. 2 A). This phenomenon was previously reported in certain combinations of transition zone mutants (Williams et al., 2008, 2011). Loss of contact with the external environment rather than ciliogenesis defects could explain the observed lack of dye-filling, and dendrite lengths do correlate with dye-fill phenotypes (compare Figs. 3 C and S2 E). Complete dendrite collapse was not observed in amphids (Fig. S3, C and D). However, cilia frequently failed to extend into the channel formed by socket and sheath glia (Fig. S3, A and B).
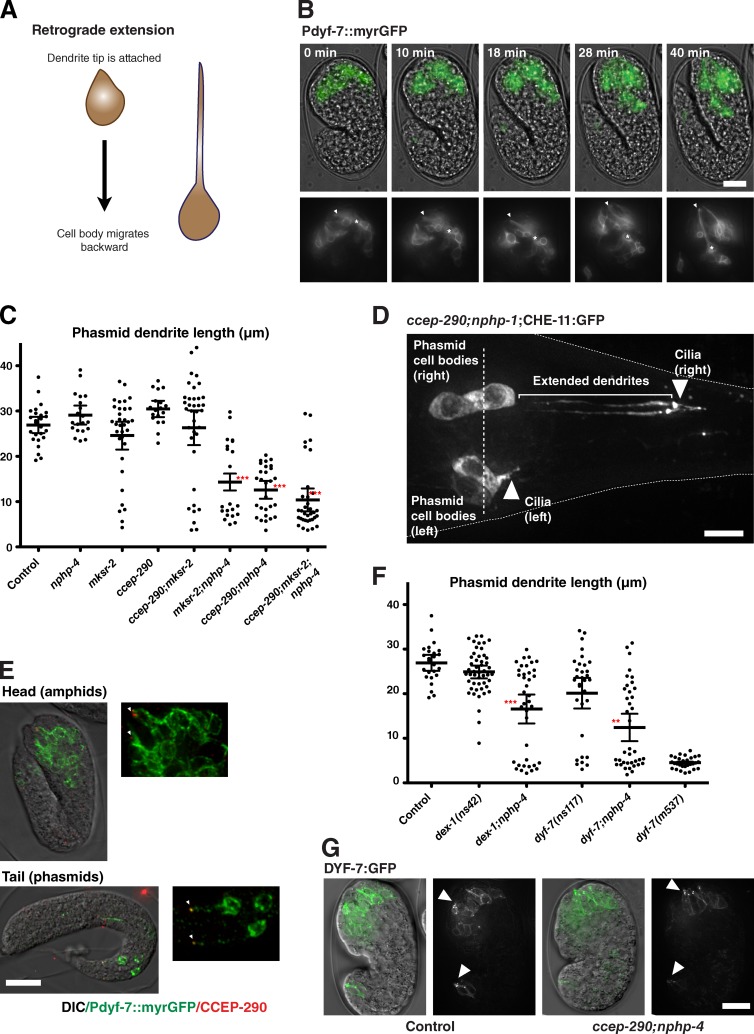
Figure 3.
A role for the transition zone in dendrite attachment. (A) Schematic of dendrite formation by retrograde extension in C. elegans amphids. (B) Stills from time-lapse movie of embryo expressing myristoylated GFP in amphid neurons undergoing retrograde migration. Overlay of GFP signal is shown on transmitted light images of the embryo. Insets show GFP only. Note that the position of dendritic tips (arrowhead) appears fixed as cell bodies (asterisk) move. See also Video 3. (C) Quantitation of phasmid dendrite lengths in wild-type and transition zone mutants at the L4 stage, using CHE-11:GFP. Each dot on the scatter plot represents a single dendrite, with bars indicating average and 95% confidence intervals. n > 20 dendrites per condition. Asterisks indicate statistically significant differences to wild-type (t test; ***, P < 0.001). (D) Tail of a ccep-290;nphp-1 mutant animal expressing CHE-11:GFP. While dendrite extension has failed in both phasmids on the left hand side, right-hand-side phasmids have formed normally. Note that cell bodies are positioned similarly on both sides (broken line). (E) Immunofluorescence micrographs of amphids and phasmids of late embryos expressing myristoylated GFP and LAP:CCEP-290 stained for CCEP-290 with antibody to S-tag. CCEP-290 is located at the tip of the elongating dendrite (arrowheads). (F) Phasmid dendrite lengths in combinations of dex-1/dyf-7 and nphp-4 mutants, quantified as in C. Wild-type data are shown for comparison. ns42 and ns117 are hypomorphic alleles of dex-1 and dyf-7, dyf-7(m537) closer to a loss of function (Heiman and Shaham, 2009). Asterisks indicate statistically significant differences to dex-1/dyf-7 single mutants (t test; ***, P < 0.001; **, P < 0.01). (G) Still images of wild-type and ccep-290;nphp-4 mutant embryos expressing DYF-7:GFP. Overlay of GFP signal on transmitted light images of the embryo. Note the enrichment of DYF-7:GFP at the tips of elongating dendrites (arrowheads). Bars: (D) 5 µm; (B, E, and G) 10 µm. Insets in E are magnified 2×.
Dendrite extension in amphids has been shown to occur by retrograde extension, whereby the cell body migrates backward while the dendritic tip remains anchored in place (Heiman and Shaham, 2009; Fig. 3, A and B; and Video 3). Failure of dendrite extension in transition zone mutants could reflect a failure of cell migration or dendrite anchorage. The stochastic nature of the phenotype and bilaterally symmetric organization of phasmids allowed us to distinguish between these possibilities. In cases where one set of phasmids displayed collapsed dendrites, their cell bodies were positioned opposite their counterparts with normally extended dendrites (Fig. 3 D), indicating that cell migration occurred normally while dendrite attachment was defective. Interestingly, dendrite lengths are bimodally distributed, either normal or fully collapsed (Fig. 3 C). Thus, attachment only occurs at the tip of the dendrite. If that attachment is lost, the dendrite collapses entirely.
Cilia could contribute to neuronal morphology in a variety of ways, including by signaling. However, dendrite extension occurs before cilia are fully formed (Sulston et al., 1983; Fujiwara et al., 1999) and was normal in IFT mutants in which axoneme assembly was inhibited (Snow et al., 2004; Fig. S3 H). Because signaling requires an intact cilium, this would support a direct anchoring role rather than an indirect signaling one, mediated by some structure at the ciliary base. A unique feature of C. elegans is that centrioles degenerate early during ciliogenesis (Dammermann et al., 2009) and do not persist at the base of mature cilia (Perkins et al., 1986). While transition zone proteins are present at the time of dendrite elongation (Fig. 3 E), centriole degeneration is well underway at this time (Fig. S3 I), making it unlikely that centrioles or centrosomes contribute to dendrite anchorage. Finally, loss of the basal body protein HYLS-1 only mildly perturbed dendrite extension in a manner synergistic with loss of NPHP-4 (Fig. S3 H), likely reflecting transition zone disorganization in hyls-1 mutants. Our results are therefore most consistent with a direct anchoring function mediated by the transition zone.
Dendrite extension in PQR neurons has been reported to occur in the canonical, anterograde manner by growth cone crawling (Kirszenblat et al., 2011). It is therefore not surprising that dendrites in PQR neurons are unaffected (Fig. S3, E and F). In contrast, both amphid and phasmid dendrites form by retrograde extension. Why the more severe phenotype in phasmids? We speculate that this is due to cooperativity in attachment between neurons sharing the same channel. Indeed, dendrites of the two neurons sharing each phasmid channel always succeeded or failed to extend together, whereas there was no correlation between dendrite extension on the left and right sides of the animal (65% displayed collapsed dendrites on the left; 63% on the right; 41% both sides, the expected probability for two independent events; n = 46 animals). With 13 transition zones per amphid compared with two per phasmid, cooperative attachment could make amphid dendrites more resistant to collapse. Differences in cell morphology may also contribute. It is notable in this context that failure of dendrite extension in phasmids, unlike in amphids (Heiman and Shaham, 2009), is not accompanied by failure of sheath glia extension (Fig. S3 G).
For a structure at the dendritic tip to provide anchorage during retrograde extension, it must interact with neighboring cells or other immobile structures. For amphids, that structure is the extracellular matrix, composed in part of the tectorin-related proteins DEX-1 and DYF-7. Loss of either protein results in loss of anchorage and dendrite collapse (Heiman and Shaham, 2009). This was also true for the strong loss-of-function mutant dyf-7(m537) in phasmids (Fig. 3 F), indicating that dendrites are secured to the extracellular material as in amphids. This adhesive matrix remains in place in transition zone mutants (Fig. 3 G). If transition zones and tectorins function together in cell adhesion, we should be able to observe genetic interactions between mutants in both structures. Indeed, hypomorphic mutants of dex-1 and dyf-7 were found to synergize with mutations in nphp-4, which alone do not result in dendrite phenotypes (Fig. 3, C and F). Thus, transition zones mediate dendrite attachment via interactions with the extracellular matrix.
In summary, our work has helped clarify the molecular architecture and function of the ciliary transition zone (see also Fig. 4, A and B). By combining perturbation of CCEP-290, which affects the central cylinder, with previously described perturbations that affect the Y-links (Williams et al., 2011), we were able to completely disrupt transition zone structure. Our finding, that axoneme assembly remains relatively normal when the transition zone is lost, argues that there is no fundamental requirement for the transition zone in axoneme assembly. The picture has been less clear in vertebrates, where loss of transition zone components has been reported to result in ciliary defects in some tissues but not others (Garcia-Gonzalo et al., 2011; Sang et al., 2011; Chih et al., 2012). One possible explanation for this context dependence is that it relates to transition zone function in ciliary gating (Craige et al., 2010). Perturbed entry of ciliary components could affect cilia assembly in some cases but not others. Alternatively, loss or impairment of transition zone structures could affect cilia stability. As previously shown for basal bodies in Tetrahymena thermophila (Bayless et al., 2012), cilia motility or fluid flow over nonmotile cilia results in mechanical stresses that could damage cilia unless stabilized by structures at their base. In less challenging environments, such as C. elegans cilia protected inside the cuticle of the worm, impaired stability might have less deleterious consequences.
A second major conclusion of our work is that the transition zone is required for adhesion to the extracellular matrix to allow dendrite extension. While cilia have not previously been implicated in cell adhesion, signs of crosstalk have been observed for many years. Thus, NPHP proteins localize to cell–cell and cell–matrix junctions (Benzing et al., 2001) and have been found to be required for epithelial morphogenesis (Delous et al., 2009). Conversely, junctional proteins have been found at cilia (Fan et al., 2004; Sfakianos et al., 2007). Cilia have been linked to many developmental processes in vertebrates and we speculate that cilia-mediated adhesion contributes to some of these processes. For example, during development of the cerebral cortex, delamination of neural precursors from the apical adherens belt is an important determinant of cell fate. These precursors are ciliated, and it has been shown that the position of the primary cilium determines whether a daughter cell delaminates after mitosis (Wilsch-Bräuninger et al., 2012; Paridaen et al., 2013; Fig. 4 C). Importantly, cilia repositioning precedes any change in polarity and is therefore the driving force behind loss of attachment. While it is possible that cilia-dependent signaling underlies this phenomenon, we suggest an alternative explanation: a direct, mechanical role for the cilium in cell adhesion. As in C. elegans, analysis of IFT mutants (impairing cilia-dependent signaling but not transition zone function) could distinguish between these possibilities.
Materials and methods
Sequence analysis
CEP290 orthologues were identified by reciprocal BLAST analysis using human CEP290 as the starting point. Where direct comparisons failed to identify a clear homologue, indirect searches were performed using less divergent related species as intermediates. Orthology relationships were confirmed using an algorithm optimized for coiled-coil proteins (Kuhn et al., 2014). Conserved motifs within CEP290 were identified by Meme (http://meme-suite.org/) and sequences were aligned using MAFFT within Jalview (http://www.jalview.org). Maximum likelihood phylogenetic trees were constructed using IQ-TREE under the LG+G model (Le and Gascuel, 2008). Clade supports were assessed by ultrafast bootstrap (UFBoot) analysis (Minh et al., 2013).
C. elegans strains and culture conditions
Strains used are listed in Table S1. ccep-290 mutants were obtained from the C. elegans Knockout Consortium/NBRP-Japan (tm4927) and the Million Mutation Project (gk415029 and gk107339). The ccep-290Δ mutant was generated by Cas9-induced homologous recombination (Dickinson et al., 2013), replacing the entire ccep-290 coding region and 5′ and 3′ UTR with the selectable marker unc-119. Detailed information for other mutants is available from WormBase (http://www.wormbase.org). Mutants were outcrossed 6× to N2 wild-type before phenotypic analysis and introduction of fluorescent markers. Transgenic strains expressing fluorescently tagged CCEP-290, HYLS-1, MKSR-2, and NPHP-4 were generated by cloning the corresponding genomic loci, including 5′ and 3′ regulatory sequences and N-terminal LAP-GFP/mCherry into the appropriate targeting vector and Mos1-mediated transposition (Frøkjaer-Jensen et al., 2008). Strains expressing GFP:CCEP-290, DYF-7:GFP, MKS-2:GFP, MKS-6:GFP, MKSR-1:GFP, and Pnas-31:mCherry/OSM-6:GFP from extrachromosomal arrays were generated by coinjection with the marker plasmid pRF4. All strains were maintained at 23°C.
Immunofluorescence and fixed imaging
Immunofluorescence experiments were performed as described previously (Oegema et al., 2001) using Cy3/Cy5-labeled affinity-purified rabbit polyclonal antibodies against HYLS-1 (Dammermann et al., 2009) and SAS-4 (Dammermann et al., 2004), as well as unlabeled mouse monoclonal antibodies against polyglutamylated tubulin (GT335; provided by C. Janke, Institut Curie, Paris, France). Residual fluorescence signal was used to detect GFP/mCherry-tagged protein in fixed embryos. Mouse monoclonal antibodies against S-peptide (MA1-981; Thermo Fisher Scientific) were used to detect LAP-tagged CCEP-290 in embryos also expressing other GFP fusions. In brief, worms were permeabilized by freeze crack, fixed in −20°C methanol for 20 min, rehydrated in PBS, blocked for 20 min in AbDil (PBS, 2% BSA, 0.1% Triton X-100), incubated with primary antibodies (1 µg/ml HYLS-1/SAS-4/MA1-981, 4 µg/ml GT335) in AbDil for 1.5 h, washed with PBST (PBS, 0.1% Triton X-100), incubated with secondary antibodies (15 µg/ml Cy2/Cy3/Cy5-labeled donkey anti-mouse antibody; Jackson ImmunoResearch Laboratories, Inc.) in AbDil for 1 h, washed with PBST, and incubated with 1 µg/ml Hoechst 33342 in PBS for 5 min before mounting in 0.5% p-phenylenediamine, 20 mM Tris, pH 8.8, and 90% glycerol. 3D wide-field datasets were acquired using a 100× 1.4 NA Super Plan-Apochromat lens on a microscope (DeltaVision; Applied Precision) equipped with a 7-Color SSI module and a cooled charge-coupled device (CCD) camera (CoolSNAP-HQ2; Photometrics), computationally deconvolved using the enhanced ratio constrained iterative deconvolution algorithm, and maximum-intensity projected in SoftWorx (Applied Precision), before being imported into Photoshop (Adobe) for panel preparation. No nonlinear gamma corrections were performed during image processing.
Live imaging
Where possible, worms expressing fluorescent markers were examined without fixation, anaesthetized with 10 mM tetramisole, and mounted on 2% agarose pads. Imaging was performed at room temperature on the set-up described above. For examination of neuronal morphology and localization interdependencies, 3D wide-field datasets were acquired, deconvolved, and processed as described above. Exposure conditions and scaling were kept constant when comparing signal across mutant backgrounds. At least 10 animals were examined per condition. Dendrite lengths (from cell body to ciliary base) and cilia dimensions (base to tip) were measured in MetaMorph on strains expressing CHE-11:GFP. For examination of protein localization at high spatial resolution, L4 larvae coexpressing GFP-tagged transition zone proteins and mCherry:HYLS-1 were imaged with the additional use of a 1.6× optovar, and deconvolved image stacks were analyzed in MetaMorph. Where phasmid cilia lay flat within the image plane, the dimensions of GFP and mCherry signals were measured using the linescan function and averaged over >20 cilia per strain. Potential shifts between GFP and mCherry signals due to chromatic aberration were excluded by imaging a strain coexpressing the same protein in both colors. For time-lapse imaging of neurite extension, single-plane GFP and transmitted light images were acquired every 2 min in embryos expressing myristoylated GFP under a pan-neuronal promoter, using low-power LED illumination to avoid photodamage. For analysis of IFT and GFP:MKSR-2 trafficking in ccep-290Δ mutants, single-plane GFP images of phasmid cilia in L4 larvae were acquired every second. Anterograde movement rates for the middle segment were determined by single-particle tracking in MetaMorph (n = 40–140 particles per strain). Kymographs along the ciliary axis were similarly generated in MetaMorph.
Transmission electron microscopy and electron tomography
L4-stage worms were fixed in 2.5% glutaraldehyde in 0.1 mol/liter sodium phosphate buffer, pH 7.4, overnight at 4°C. Samples were postfixed in 0.5–2% osmium tetroxide in the same buffer, washed in ddH2O, dehydrated in a graded series of ethanol, and embedded in Agar100 resin. 70-nm serial sections were poststained with aqueous uranyl acetate and lead citrate and examined with a Morgagni 268D microscope (FEI) equipped with an 11-megapixel Morada CCD camera (Olympus-SIS) and operated at 80 kV. At least five animals were examined per condition. For electron tomography, 200-nm sections were examined with a microscope (Tecnai G2 20; FEI) equipped with a CCD camera (Eagle 4k HS; FEI) and operated at 200 kV. Single axis tilt series were acquired using SerialEM (Mastronarde, 2005) by stepwise tilting the sample on average from −60° to +60° in 1° increments. Tomogram reconstruction with 10-nm gold beads as fiducials and model generation was performed using the IMOD software package (Kremer et al., 1996).
Online supplemental material
Fig. S1 details the identification and initial characterization of C. elegans CEP290, including sequence alignments, expression analysis, and phenotypic characterization of ccep-290 mutants. Fig. S2 presents a high-resolution analysis of transition zone protein localization and localization interdependencies, as well as an analysis of genetic interactions between transition zone mutants as revealed by the dye-fill assay. Fig. S3 presents a further characterization of dendrite extension, including phenotypes in other ciliated neurons, glial cell morphology, and contribution of other ciliary structures. Table S1 lists C. elegans strains used in this study. Table S2 provides accession numbers of CEP290 orthologues. Video 1 shows MKSR-2 particle movement along the ciliary axoneme in a ccep-290 mutant. Video 2 shows an electron tomogram and 3D reconstruction model of the C. elegans transition zone. Video 3 shows dendrite extension by retrograde migration in the C. elegans embryo. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201501013/DC1.
Acknowledgements
We thank Karen Oegema for comments on the manuscript, the Vienna Biocenter C. elegans community, and members of the Dammermann laboratory for discussions; the Caenorhabditis Genetics Center, Max Heiman (Boston Children’s Hospital, Boston, MA), Carsten Janke, Michel Leroux (Simon Fraser University, Burnaby BC, Canada), Shohei Mitani (Tokyo Women’s Medical University, Tokyo, Japan), and Bradley Yoder (The University of Alabama at Birmingham, Birmingham, AL) for strains and reagents, Bui Quang Minh for help with bioinformatics, and the Campus Science Support Facilities GmbH (CSF) EM facility for technical assistance.
This work was supported by start-up funding from Max F. Perutz Laboratories and grants Y597-B20 and P24296-B20 from the Austrian Science Fund (FWF) to A. Dammermann, as well as a predoctoral fellowship from the Boehringer Ingelheim Fonds to C. Schouteden.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CCD
- charge-coupled device
- IFT
- intraflagellar transport
References
- Basiri M.L., Ha A., Chadha A., Clark N.M., Polyanovsky A., Cook B., and Avidor-Reiss T.. 2014. A migrating ciliary gate compartmentalizes the site of axoneme assembly in Drosophila spermatids. Curr. Biol. 24:2622–2631. 10.1016/j.cub.2014.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayless B.A., Giddings T.H. Jr., Winey M., and Pearson C.G.. 2012. Bld10/Cep135 stabilizes basal bodies to resist cilia-generated forces. Mol. Biol. Cell. 23:4820–4832. 10.1091/mbc.E12-08-0577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzing T., Gerke P., Höpker K., Hildebrandt F., Kim E., and Walz G.. 2001. Nephrocystin interacts with Pyk2, p130(Cas), and tensin and triggers phosphorylation of Pyk2. Proc. Natl. Acad. Sci. USA. 98:9784–9789. 10.1073/pnas.171269898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chih B., Liu P., Chinn Y., Chalouni C., Komuves L.G., Hass P.E., Sandoval W., and Peterson A.S.. 2012. A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat. Cell Biol. 14:61–72. 10.1038/ncb2410 [DOI] [PubMed] [Google Scholar]
- Cole D.G., Diener D.R., Himelblau A.L., Beech P.L., Fuster J.C., and Rosenbaum J.L.. 1998. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 141:993–1008. 10.1083/jcb.141.4.993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craige B., Tsao C.C., Diener D.R., Hou Y., Lechtreck K.F., Rosenbaum J.L., and Witman G.B.. 2010. CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J. Cell Biol. 190:927–940. 10.1083/jcb.201006105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A., Müller-Reichert T., Pelletier L., Habermann B., Desai A., and Oegema K.. 2004. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell. 7:815–829. 10.1016/j.devcel.2004.10.015 [DOI] [PubMed] [Google Scholar]
- Dammermann A., Pemble H., Mitchell B.J., McLeod I., Yates J.R. III, Kintner C., Desai A.B., and Oegema K.. 2009. The hydrolethalus syndrome protein HYLS-1 links core centriole structure to cilia formation. Genes Dev. 23:2046–2059. 10.1101/gad.1810409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delous M., Hellman N.E., Gaudé H.M., Silbermann F., Le Bivic A., Salomon R., Antignac C., and Saunier S.. 2009. Nephrocystin-1 and nephrocystin-4 are required for epithelial morphogenesis and associate with PALS1/PATJ and Par6. Hum. Mol. Genet. 18:4711–4723. 10.1093/hmg/ddp434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D.J., Ward J.D., Reiner D.J., and Goldstein B.. 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods. 10:1028–1034. 10.1038/nmeth.2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S., Hurd T.W., Liu C.J., Straight S.W., Weimbs T., Hurd E.A., Domino S.E., and Margolis B.. 2004. Polarity proteins control ciliogenesis via kinesin motor interactions. Curr. Biol. 14:1451–1461. 10.1016/j.cub.2004.08.025 [DOI] [PubMed] [Google Scholar]
- Fliegauf M., Benzing T., and Omran H.. 2007. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 8:880–893. 10.1038/nrm2278 [DOI] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C., Davis M.W., Hopkins C.E., Newman B.J., Thummel J.M., Olesen S.P., Grunnet M., and Jorgensen E.M.. 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40:1375–1383. 10.1038/ng.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M., Ishihara T., and Katsura I.. 1999. A novel WD40 protein, CHE-2, acts cell-autonomously in the formation of C. elegans sensory cilia. Development. 126:4839–4848. [DOI] [PubMed] [Google Scholar]
- Garcia-Gonzalo F.R., Corbit K.C., Sirerol-Piquer M.S., Ramaswami G., Otto E.A., Noriega T.R., Seol A.D., Robinson J.F., Bennett C.L., Josifova D.J., et al. 2011. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat. Genet. 43:776–784. 10.1038/ng.891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M.G., and Shaham S.. 2009. DEX-1 and DYF-7 establish sensory dendrite length by anchoring dendritic tips during cell migration. Cell. 137:344–355. 10.1016/j.cell.2009.01.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges M.E., Scheumann N., Wickstead B., Langdale J.A., and Gull K.. 2010. Reconstructing the evolutionary history of the centriole from protein components. J. Cell Sci. 123:1407–1413. 10.1242/jcs.064873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Szymanska K., Jensen V.L., Janecke A.R., Innes A.M., Davis E.E., Frosk P., Li C., Willer J.R., Chodirker B.N., et al. 2011. TMEM237 is mutated in individuals with a Joubert syndrome related disorder and expands the role of the TMEM family at the ciliary transition zone. Am. J. Hum. Genet. 89:713–730. 10.1016/j.ajhg.2011.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis P.N., Ou G., Leroux M.R., and Scholey J.M.. 2007. The sensory cilia of Caenorhabditis elegans (March 8, 2007). In WormBook The C. elegans Research Community, Editors. 10.1895/wormbook.1.126.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirszenblat L., Pattabiraman D., and Hilliard M.A.. 2011. LIN-44/Wnt directs dendrite outgrowth through LIN-17/Frizzled in C. elegans neurons. PLoS Biol. 9:e1001157 10.1371/journal.pbio.1001157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., and McIntosh J.R.. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76. 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- Kuhn M., Hyman A.A., and Beyer A.. 2014. Coiled-coil proteins facilitated the functional expansion of the centrosome. PLOS Comput. Biol. 10:e1003657 10.1371/journal.pcbi.1003657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S.Q., and Gascuel O.. 2008. An improved general amino acid replacement matrix. Mol. Biol. Evol. 25:1307–1320. 10.1093/molbev/msn067 [DOI] [PubMed] [Google Scholar]
- Mastronarde D.N. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152:36–51. 10.1016/j.jsb.2005.07.007 [DOI] [PubMed] [Google Scholar]
- Minh B.Q., Nguyen M.A., and von Haeseler A.. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30:1188–1195. 10.1093/molbev/mst024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema K., Desai A., Rybina S., Kirkham M., and Hyman A.A.. 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153:1209–1226. 10.1083/jcb.153.6.1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paridaen J.T., Wilsch-Bräuninger M., and Huttner W.B.. 2013. Asymmetric inheritance of centrosome-associated primary cilium membrane directs ciliogenesis after cell division. Cell. 155:333–344. 10.1016/j.cell.2013.08.060 [DOI] [PubMed] [Google Scholar]
- Perkins L.A., Hedgecock E.M., Thomson J.N., and Culotti J.G.. 1986. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 117:456–487. 10.1016/0012-1606(86)90314-3 [DOI] [PubMed] [Google Scholar]
- Reiter J.F., Blacque O.E., and Leroux M.R.. 2012. The base of the cilium: roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Rep. 13:608–618. 10.1038/embor.2012.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum J.L., and Witman G.B.. 2002. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 3:813–825. 10.1038/nrm952 [DOI] [PubMed] [Google Scholar]
- Sang L., Miller J.J., Corbit K.C., Giles R.H., Brauer M.J., Otto E.A., Baye L.M., Wen X., Scales S.J., Kwong M., et al. 2011. Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell. 145:513–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K.N., Kuhns S., Neuner A., Hub B., Zentgraf H., and Pereira G.. 2012. Cep164 mediates vesicular docking to the mother centriole during early steps of ciliogenesis. J. Cell Biol. 199:1083–1101. 10.1083/jcb.201202126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfakianos J., Togawa A., Maday S., Hull M., Pypaert M., Cantley L., Toomre D., and Mellman I.. 2007. Par3 functions in the biogenesis of the primary cilium in polarized epithelial cells. J. Cell Biol. 179:1133–1140. 10.1083/jcb.200709111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow J.J., Ou G., Gunnarson A.L., Walker M.R., Zhou H.M., Brust-Mascher I., and Scholey J.M.. 2004. Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat. Cell Biol. 6:1109–1113. 10.1038/ncb1186 [DOI] [PubMed] [Google Scholar]
- Sorokin S. 1962. Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J. Cell Biol. 15:363–377. 10.1083/jcb.15.2.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J.E., Schierenberg E., White J.G., and Thomson J.N.. 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100:64–119. 10.1016/0012-1606(83)90201-4 [DOI] [PubMed] [Google Scholar]
- Swoboda P., Adler H.T., and Thomas J.H.. 2000. The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol. Cell. 5:411–421. 10.1016/S1097-2765(00)80436-0 [DOI] [PubMed] [Google Scholar]
- Wan X., O’Quinn R.P., Pierce H.L., Joglekar A.P., Gall W.E., DeLuca J.G., Carroll C.W., Liu S.T., Yen T.J., McEwen B.F., et al. 2009. Protein architecture of the human kinetochore microtubule attachment site. Cell. 137:672–684. 10.1016/j.cell.2009.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C.L., Winkelbauer M.E., Schafer J.C., Michaud E.J., and Yoder B.K.. 2008. Functional redundancy of the B9 proteins and nephrocystins in Caenorhabditis elegans ciliogenesis. Mol. Biol. Cell. 19:2154–2168. 10.1091/mbc.E07-10-1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C.L., Li C., Kida K., Inglis P.N., Mohan S., Semenec L., Bialas N.J., Stupay R.M., Chen N., Blacque O.E., et al. 2011. MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J. Cell Biol. 192:1023–1041. 10.1083/jcb.201012116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsch-Bräuninger M., Peters J., Paridaen J.T., and Huttner W.B.. 2012. Basolateral rather than apical primary cilia on neuroepithelial cells committed to delamination. Development. 139:95–105. 10.1242/dev.069294 [DOI] [PubMed] [Google Scholar]