Schwann cells degrade myelin after injury by a novel form of selective autophagy, myelinophagy, which is positively regulated by the JNK/c-Jun pathway and is defective in the injured central nervous system.
Abstract
Although Schwann cell myelin breakdown is the universal outcome of a remarkably wide range of conditions that cause disease or injury to peripheral nerves, the cellular and molecular mechanisms that make Schwann cell–mediated myelin digestion possible have not been established. We report that Schwann cells degrade myelin after injury by a novel form of selective autophagy, myelinophagy. Autophagy was up-regulated by myelinating Schwann cells after nerve injury, myelin debris was present in autophagosomes, and pharmacological and genetic inhibition of autophagy impaired myelin clearance. Myelinophagy was positively regulated by the Schwann cell JNK/c-Jun pathway, a central regulator of the Schwann cell reprogramming induced by nerve injury. We also present evidence that myelinophagy is defective in the injured central nervous system. These results reveal an important role for inductive autophagy during Wallerian degeneration, and point to potential mechanistic targets for accelerating myelin clearance and improving demyelinating disease.
Introduction
In peripheral nerves, myelin breakdown, or demyelination, is a universal outcome of a remarkably wide range of conditions that involve disturbance to Schwann cells or the nerve environment, whether due to genetic or acquired disease, toxicity, or nerve transection/crush. It has also become clear from studies on cut nerves that, perhaps surprisingly, Schwann cells themselves have the ability to turn against their own myelin and initiate myelin breakdown, in addition to being able to call on macrophages for myelin phagocytosis (Hirata and Kawabuchi, 2002). The maintenance of healthy myelin and normal nerve function depends on tight control of this intrinsic potential for myelin destruction. In contrast to Schwann cells, the myelin-forming cells of the central nervous system (CNS), oligodendrocytes, appear to be unable to digest myelin, a feature that has been linked to poor regenerative ability of CNS tissue (Brosius Lutz and Barres, 2014).
In spite of the central position of myelin breakdown in Schwann cell biology and pathology, the cellular and molecular mechanisms that make Schwann cell–mediated myelin digestion possible have not been established. While earlier authors were often cautious about myelin breakdown mechanisms (Holtzman and Novikoff, 1965), more recent literature frequently invokes phagocytosis as the mechanism by which Schwann cells digest their myelin after nerve transection/crush. But this notion is problematic. This is because phagocytosis is a process by which cells ingest cell-extrinsic material, but myelin is initially an intrinsic Schwann cell component, being an integral part of the Schwann cell membrane. Furthermore, there is no evidence that myelin separates from Schwann cells during the first, Schwann cell–dependent phase of myelin breakdown (see further below in the Introduction), although this would be required if myelin were to be phagocytosed by Schwann cells. Rather, in a process requiring actin polymerization, the myelin sheath breaks up into intracellular oval-shaped myelin segments that gradually fragment into smaller intracellular debris (Jung et al., 2011b). In the present work, we have examined the mechanism by which Schwann cells initiate digestion of intracellular myelin using nerve transection as a model for demyelination.
Schwann cells possess an unusual degree of phenotypic plasticity, and nerve transection triggers a large-scale transformation of the myelin and nonmyelin (Remak) cells of undisturbed nerves to form the repair (Bungner) Schwann cells of injured nerves (Arthur-Farraj et al., 2012; Brosius Lutz and Barres, 2014; Jessen et al., 2015). A major component of this cellular reprogramming is the removal of myelin. In the first phase of myelin clearance, the Schwann cells themselves break down 40–50% of the myelin during the first 5–7 d after injury (Perry et al., 1995). Subsequently, macrophages that invade injured nerves play the major role in myelin breakdown by phagocytosis in conjunction with antibodies and complement. It is likely that Schwann cells take part in phagocytosis of myelin debris during this second phase of myelin clearance (Hirata and Kawabuchi, 2002; Ramaglia et al., 2008; Vargas et al., 2010; Dubový et al., 2013). The importance of the initial Schwann cell–mediated phase of demyelination is underscored by the observation that 7 d after cutting, myelin is cleared normally from the nerves of CCR2−/− mice, although macrophages do not accumulate significantly in injured nerves in this mutant (Niemi et al., 2013).
Macroautophagy is an inducible degradation system by which cells break down their own organelles and large macromolecules. Autophagy involves the formation of an isolation membrane that extends around cytoplasmic cargo to form an autophagosome, which transfers cargo to the lysosome for degradation (Rubinsztein et al., 2012). During starvation, autophagic degradation of cytoplasmic constituents provides a protective mechanism for energy release. In addition, specialized forms of autophagy mediate the delivery of specific cargo to the autophagosome, including intracellular pathogens (xenophagy; Levine et al., 2011), cellular organelles (mitophagy, ribophagy; Kiel, 2010), and storage vesicles such as lipid droplets (lipophagy; Singh et al., 2009).
While it has been understood for a long time that the first phase of myelin digestion in injured nerves is performed by Schwann cells, the cellular mechanism that enables Schwann cells to destroy their own myelin has not been established. Most nerve pathology involves breakdown of the myelin sheath, although demyelination occurs most acutely after injury, as part of the cellular reprogramming from myelin cells to repair cells. Therefore, understanding the molecular mechanisms of Schwann cell–mediated demyelination in injured nerves potentially has broader implications. Here, we present evidence that autophagy, a mechanism by which many cells digest their intrinsic cellular components, plays a central role in Schwann cell myelin breakdown. We show that nerve injury triggers strong activation of Schwann cell autophagy, find myelin debris in autophagosomes, and demonstrate a strong requirement for autophagy in myelin digestion, revealing a novel form of selective autophagy of the myelin sheath, myelinophagy.
Results
Autophagy is induced in Schwann cells in response to nerve injury
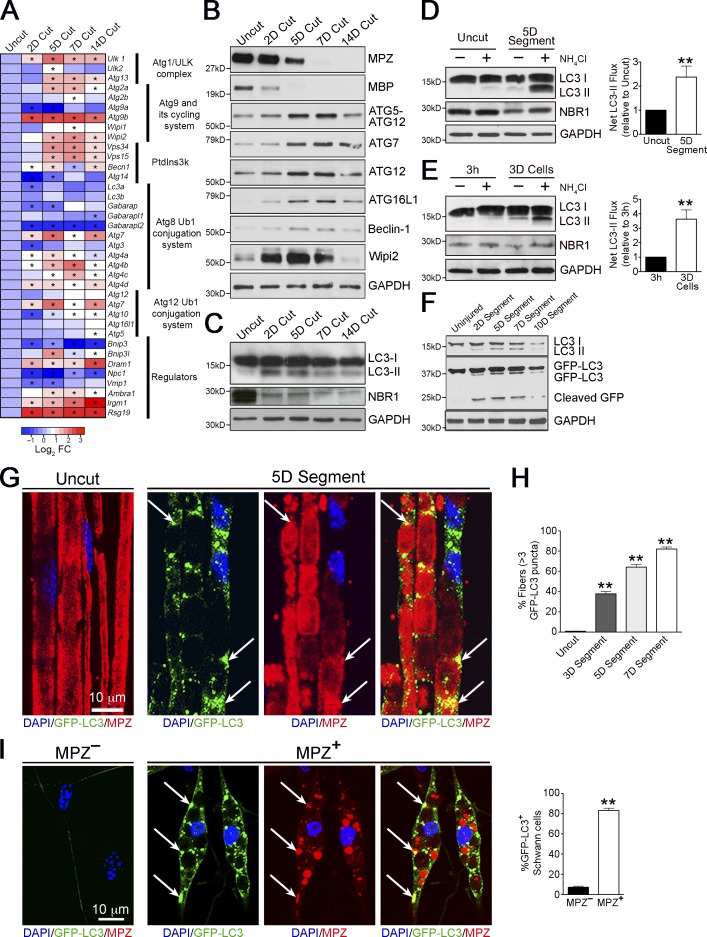
Transcriptional regulation of autophagy-related genes coincides with autophagy induction in several systems (Klionsky et al., 2012). To examine whether autophagy is involved in myelin degradation, we analyzed expression of autophagy-related genes broadly classified into functional subgroups comprising the core autophagy machinery and its main regulators (Feng et al., 2014). Sciatic nerves were transected and the distal stump was analyzed over the time course of demyelination. We found a strong induction of several autophagy-related genes during demyelination, notably members of the ULK complex and Atg9 cycling system, and Atg7, all essential for autophagosome formation (Fig. 1 A and Fig. S1). Concurrent with the degradative decline of myelin proteins MPZ and MBP, we observed an increase, compared with uninjured levels, of several ATG proteins including ATG7, components of the ATG16L complex, Wipi2, and Beclin1 in injured nerves, which is consistent with a role for autophagy in Wallerian degeneration (Fig. 1 B; quantified in Fig. S2).
Figure 1.
Autophagy is activated during Wallerian degeneration. (A) Heat map showing qPCR analysis of changes in expression of autophagy-related genes in cut nerves in vivo at different time points (e.g., 2D Cut = 2 d after nerve cut) relative to control uncut nerves. Data are expressed as log2 fold change relative to uncut nerves (red-blue color scale). Significant changes (P < 0.05) are denoted by asterisks within the heat map. Bar graphs depicting fold changes of all the genes examined are shown in Fig. S1. (B and C) Western blotting showing expression of selected autophagy-related proteins (B), and LC3 (LC3-I, 18 kD; LC3-II, 16 kD) and NBR1 (C), in uncut nerves and cut nerves in vivo at different time points. Myelin proteins MPZ and MBP are used as controls to show demyelination after nerve cut. GAPDH is used as a loading control. Quantification of Western blots is shown in Fig. S2, A and B, respectively. (D and E) Western blots showing that LC3-II and NBR1 accumulate after a 3-h treatment with the lysosomal blocker NH4Cl in nerve segments maintained in vitro (D), and dissociated Schwann cell cultures (E). Graphs show significantly higher net LC3-II flux (D) in demyelinating nerves (5 d in vitro [5D]) compared with freshly isolated nerves (Uncut), and demyelinating Schwann cells (E) cultured for 3 d (3D) compared with freshly plated cells cultured for 3 h (3h). Data are presented as mean ± SEM (error bars) from a minimum of three independent experiments. **, P < 0.01. A quantification of NBR1 levels is shown in Fig. S2 (C and D). (F) Western blots showing expression of endogenous LC3 and GFP-LC3 in nerve segments from GFP-LC3 mice maintained in vitro at different time points. Cleaved GFP bands demonstrate a transient increase in autophagic flux after nerve injury. Quantification of Western blots is shown in Fig. S2 E. (G–I) GFP-LC3 puncta in nerves from GFP-LC3 mice. (G) The immunolabeled micrographs show the accumulation of GFP-LC3 puncta in teased nerve segments maintained in vitro for 5 d compared with control teased uncut nerves. MPZ labels myelin. Individual MPZ and GFP-LC3 labeling for 5D nerve segments are shown to illustrate colocalization of MPZ+ myelin debris and GFP-LC3+ autophagosomes (arrows). (H) Graph shows quantification of fibers positive for GFP-LC3 puncta in nerves cultured for 3, 5, and 7 d. n = 3 mice for each time point. Data are presented as mean ± SEM (error bars). **, P < 0.01 (cultured segments relative to uncut nerves). (I) Immunolabeling showing elevated expression of GFP-LC3 puncta in dissociated Schwann cells degrading myelin, shown by the presence of MPZ+ myelin compared with that seen in MPZ– Schwann cells. Individual MPZ and GFP-LC3 labeling for MPZ+ Schwann cells are shown to illustrate colocalization of MPZ+ myelin debris and GFP-LC3+ autophagosomes (arrows). The graph shows quantification of the number of GFP-LC3+ Schwann cells that are MPZ+ or MPZ–. Data are presented as mean ± SEM (error bars) from a minimum of three independent experiments with a minimum of 60 cells analyzed per experiment. **, P < 0.01 (MPZ+ cells relative to MPZ– cells).
Fluctuations in levels of lipidated LC3 (LC3 II) are frequently used to determine autophagy induction (Klionsky et al., 2012). We analyzed levels of LC3 II in response to injury in vivo and observed a clear increase in its levels, peaking at 5 d and decreasing thereafter. Conversely, levels of the autophagy substrate NBR1 (Klionsky et al., 2012) showed a clear decrease in injured nerves (Fig. 1 C). p62, another autophagy substrate, could not be used since we found its levels to be transcriptionally regulated by nerve injury, unlike NBR1 (unpublished data). To confirm that this increase reflected an induction of autophagy, we examined autophagic flux in nerve segments maintained in vitro. In this established model of Schwann cell demyelination, short nerve segments are cultured in vitro, leading to progressive myelin breakdown, similar to the changes seen in vivo. Myelin breakdown is mainly performed by Schwann cells because invading macrophages are absent (Fernandez-Valle et al., 1995). We found that treatment with the lysosome inhibitor NH4Cl led to a strong accumulation of LC3 II and NBR1 in demyelinating nerve segments cultured in vitro for 5 d (5D Cut), but not in freshly isolated nerve segments (Uncut). Net LC3 II flux calculations confirmed the significantly higher autophagic flux in the demyelinating segments, compared with uncut segments (Fig. 1 D). To confirm these findings, we examined autophagy flux in dissociated Schwann cell cultures. In this model, highly pure myelinating Schwann cells, obtained from sciatic nerves of 8–10 d old rats, are plated and cultured over a period of time, leading to progressive myelin degradation in the cells. We found an accumulation of LC3 II and NBR1 in demyelinating cultures but not in freshly plated cells. These findings were confirmed by net LC3 II calculations (Fig. 1 E). Collectively, this indicates that up-regulation of LC3 II and breakdown of the autophagy substrate NBR1 after injury is a result of a strong and rapid autophagy induction, concurrent with the onset of myelin degradation.
To show that autophagy was induced specifically in demyelinating Schwann cells, we used mice expressing a GFP-tagged LC3 transgene, a model widely used to monitor autophagy in vivo (Mizushima et al., 2004). In cultured nerves from these mice, we observed the appearance of cleaved GFP bands, indicative of autophagic breakdown of cargo (Klionsky et al., 2012), during active demyelination (2, 5, and 7 d) but not when demyelination is essentially complete (10 d; Fig. 1 F). This confirmed our findings that autophagy is induced during active demyelination at 2–7 d after injury and down-regulated thereafter.
Uninjured myelinated fibers from these mice contained few or no GFP-LC3 punctate structures. In contrast, numerous GFP-LC3 puncta, representing autophagosomes (Klionsky et al., 2012), were associated with degenerating myelin in nerve segments maintained in vitro for 3–7 d (Fig. 1, G and H). The lysosomal marker LAMP1 has been shown to be associated with degenerating myelin fragments 3 and 7 d after nerve injury (Jung et al., 2011a). Similar results were obtained with dissociated Schwann cell cultures, where numerous GFP-LC3 puncta were observed in actively demyelinating Schwann cells (MPZ+), whereas myelin-negative cells were largely negative for GFP-LC3 puncta (Fig. 1 I). Together this indicates that in individual cells, autophagosome formation correlates with active myelin digestion.
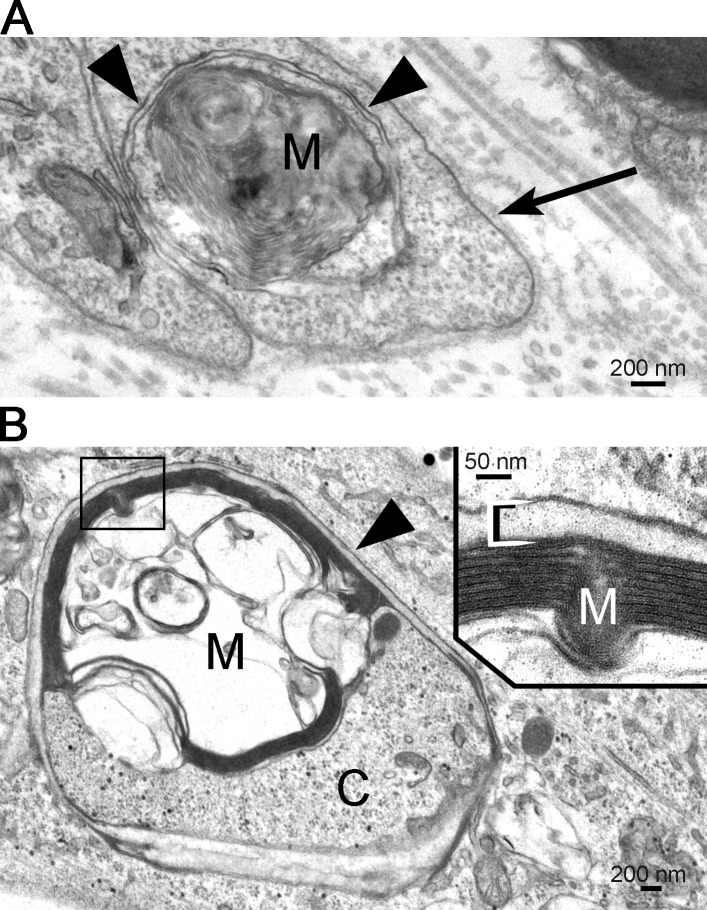
To confirm that the Schwann cell autophagy machinery is selectively targeted to myelin during active digestion, electron microscopy of cultured nerve segments and Schwann cell cultures were performed. We found myelin debris clearly enclosed in double membrane vesicles typical of autophagosomes (Fig. 2). These observations show that autophagy is activated during Wallerian degeneration, specifically in demyelinating Schwann cells.
Figure 2.
Schwann cell autophagosomes enclose degraded myelin. Electron micrographs show the presence of structures typical of autophagosomes surrounding myelin debris. (A) Myelin debris (M) are surrounded by the double membrane of an autophagosome (arrowheads) within a Schwann cell in a nerve piece. Note the basal lamina surrounding the Schwann cell processes (arrow) and the collagen fibrils outside the basal lamina. (B) Myelin debris (M) and cytoplasm (C) are enclosed by an autophagosome (arrowhead) in a cultured Schwann cell. The myelin is shown at high magnification in the inset.
Pharmacological inhibition of autophagy impairs demyelination
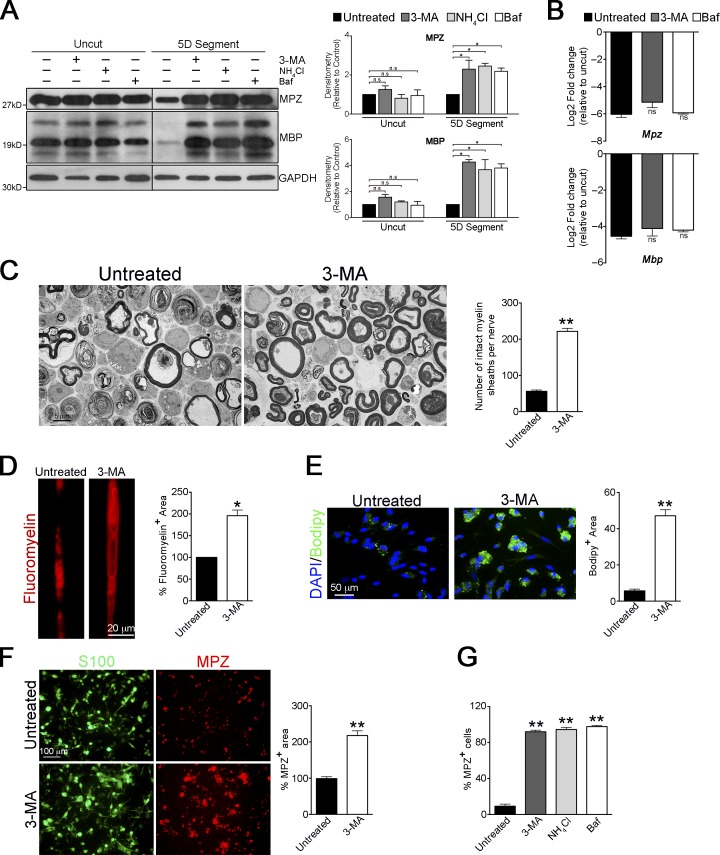
If Schwann cells degrade myelin by autophagy, autophagy inhibitors should reduce myelin breakdown. We therefore cultured nerve segments with the widely used autophagy inhibitors 3-methyladenine (3-MA) and Bafilomycin A1 (Klionsky et al., 2012). The lysosomal inhibitor NH4Cl was included for comparison. These inhibitors strikingly prevented the degradation of the myelin proteins MPZ and MBP in nerve segments maintained in vitro for 5 d but not in uncut nerve segments, indicating that autophagy is required for myelin protein breakdown (Fig. 3 A). This was not due to transcriptional changes, as we did not find significant differences in mRNA levels in nerve segments treated with these autophagy inhibitors (Fig. 3 B). Histologically, we found that 3-MA reduced myelin collapse in cultured nerves compared with control nerves, in which most sheaths appeared as collapsed whorls (Fig. 3 C).
Figure 3.
Pharmacological block of autophagy prevents myelin degradation. (A) Western blot showing a block in degradation of the myelin proteins MPZ and MBP in nerve segments maintained in vitro for 5 d and treated with different pharmacological inhibitors: untreated, lysosomal inhibitor, NH4Cl, and autophagy inhibitors 3-MA and bafilomycin A1 (Baf). No such effect is seen in freshly isolated nerve segments treated with these inhibitors for 3 h. The black line indicates that intervening lanes have been spliced out. Graphs show densitometric quantification of Western blots. Data are presented as mean ± SEM (error bars) from three independent experiments. n.s., nonsignificant; *, P < 0.05. (B) qPCR analysis showing no significant differences (n.s.) in mRNA levels of Mpz and Mbp in nerve segments maintained in vitro for 5 d and treated with 3-MA and Baf, compared with untreated segments. Data are expressed as log2 fold change in 5D cultured nerve segments relative to uncut nerves. n = 3 for each condition. Data are presented as mean ± SEM (error bars). (C) Electron micrographs showing abundant intact myelin sheaths in nerve segments cultured in vitro for 4 d in the presence of 3-MA, compared with untreated cultures. The graph shows a quantification of the number of intact myelin sheaths in untreated segments and segments treated with 3-MA. (D) Teased fibers from untreated and 3-MA treated nerve segments (5 d) stained with FluoroMyelin red. The graph shows a quantification of the myelin fluorescent area. (E) Immunolabeling showing a block in the degradation of lipids (Bodipy+ cells) in dissociated Schwann cell cultures after 3 d of 3-MA treatment. Graph shows quantification of Bodipy+ area. (F) 3-MA blocks myelin protein degradation in dissociated Schwann cell cultures. Immunolabeling showing a block in degradation of MPZ in dissociated Schwann cells (S100+) cultured for 5 d in the presence of 3-MA. The graph shows MPZ+ area in dissociated cultures treated with 3-MA. (G) Graph showing the number of MPZ+ Schwann cells in dissociated cultures treated for 5 d with 3-MA, bafilomycin, and NH4Cl. (C–G) Data are presented as mean ± SEM (error bars) from three independent experiments with a minimum of 10 picture frames analyzed per condition/experiment. *, P < 0.05; **, P < 0.01 (treated cells relative to untreated controls).
Lipids constitute 70–80% of peripheral myelin (Chrast et al., 2011), and lipid droplets can serve as autophagic cargo (Singh et al., 2009). Therefore, we examined lipid degradation in the presence of 3-MA. Nerve segments stained with FluoroMyelin, a dye for myelin lipids, showed a significantly greater fluorescent area after 3-MA treatment compared with controls, indicating a reduction in myelin lipid degradation in the absence of autophagosome formation (Fig. 3 D).
Finally, we examined the effects of pharmacological inhibition of autophagy specifically in demyelinating Schwann cells, using dissociated cell cultures. We found that inhibition of autophagy led to a notable reduction in myelin protein and lipid breakdown (Fig. 3, E–G), similar to that seen in nerve segments.
These results show that in injured nerves, Schwann cell autophagy is a major contributor to degradation of myelin proteins and lipids and to the collapse of myelin architecture.
Genetic inactivation of autophagy
The results above (Fig. 3) suggest that autophagy inhibition leads to an impairment in myelin breakdown. However, since chemical modulators cannot be guaranteed to be entirely specific, we analyzed the role of autophagy in myelin breakdown using genetic ablation of autophagy function. For this, we generated mice that lack ATG7, an essential autophagy gene, specifically in Schwann cells. Mice carrying an Atg7loxP allele (Komatsu et al., 2005) were crossed with mice that express Cre recombinase in Schwann cells only (Mpz-Cre; Feltri et al., 2002) to generate Mpz-Cre+;Atg7loxP/loxP (Atg7 cKO) mice (Fig. S3, A–C).
The mutant mice were born and survived normally. Notably, the uninjured adult peripheral nerves of Atg7 conditional knockout (cKO) mice were indistinguishable from wild-type (WT) control littermates. Sensorimotor function, histological appearance, G ratio, the levels of myelin proteins, and lipid composition were all normal (Fig. S3, D–G; see, however, Jang et al., 2015). This is in line with our findings above (Fig. 1), that autophagy is not significantly activated in mature uncut nerves.
Nerve homogenates from uninjured and injured ATG7 cKO mice showed strongly reduced ATG7, ATG5-ATG12, and LC3 II in uninjured and cut nerves, indicating impairment of autophagy. The residual proteins are likely to be expressed by cells other than Schwann cells present in the nerve (Fig. S4, A–C).
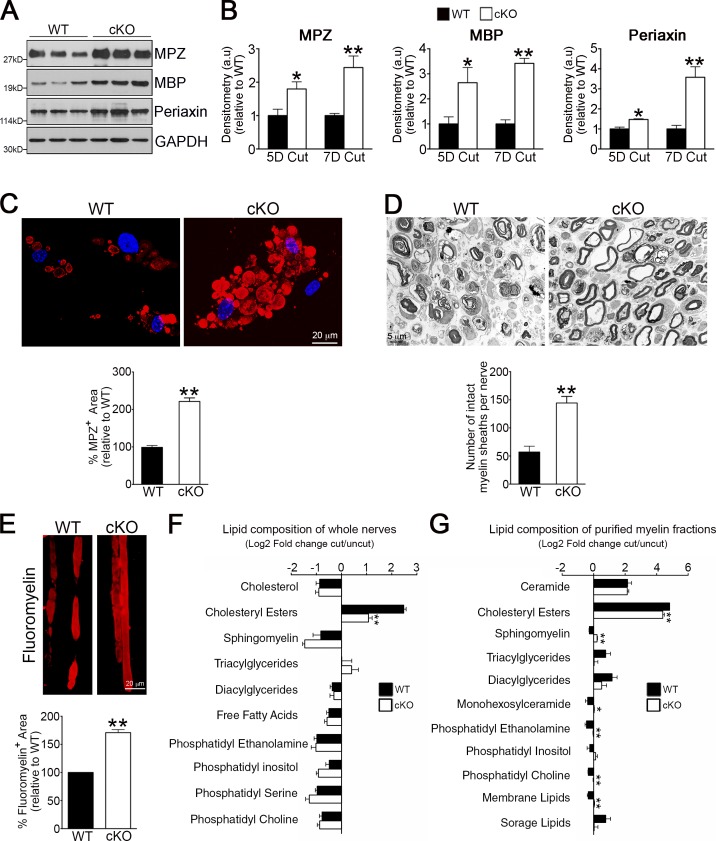
Striking differences were found when myelin breakdown was compared in Atg7 cKO and WT nerves after injury. A significant reduction was seen in injury-induced breakdown of myelin proteins in transected nerves in vivo (Fig. 4, A and B), in cultured nerve segments (Fig. S4 D), and in Schwann cell cultures where Atg7 cKO cells typically retained bloated “cauliflower” morphology for many days due to the persistence of undigested myelin (Fig. 4 C). In transected Atg7 cKO nerves, there was also a striking preservation of noncollapsed myelin (Fig. 4 D), confirming previous results using the autophagy blocker 3-MA (Fig. 3 C).
Figure 4.
Genetic inactivation of autophagy retards myelin degradation in vivo. (A) Western blot showing higher levels of the myelin proteins MPZ, MBP, and Periaxin in 5 d cut nerves from Atg7 cKO mice compared with WT controls. (B) Densitometric analysis of Western blots showing higher levels of myelin proteins in 5 and 7 d cut nerves from Atg7 cKO mice compared with WT controls. For each comparison, the value for cKO is normalized to that seen in WT. n = a minimum of three mice for the genotype/time point. Data are presented as mean ± SEM (error bars). *, P < 0.05; **, P < 0.01 (Atg7 cKO relative to WT). (C) Immunolabeling showing MPZ+ myelin inclusions in cultured WT Schwann cells and cells from Atg7 cKO nerves that often show bloated “cauliflower” morphology. The graph shows quantification of MPZ+ area. Data are presented as mean ± SEM (error bars) from three independent experiments with a minimum of 60 cells analyzed per condition. **, P < 0.01 (Atg7 cKO relative to WT). (D) Electron micrographs showing several intact myelin sheath profiles in 5 d cut nerves from Atg7 cKO mice. The graph shows a quantification of the number of intact myelin sheaths. Data are presented as mean ± SEM (error bars) from three independent experiments with a minimum of 18 picture frames analyzed per condition/experiment. **, P < 0.01 (Atg7 cKO relative to WT). (E) Teased fibers of 5 d cut nerves from WT and Atg7 cKO mice stained with FluoroMyelin red to show myelin. The graph shows a quantification of the myelin fluorescent area. Data are presented as mean ± SEM (error bars) from three independent experiments with a minimum of 10 picture frames analyzed per condition/experiment. **, P < 0.01 (Atg7 cKO relative to WT). (F) The lipid composition of whole sciatic nerves from 5 d cut WT and Atg7 cKO mice expressed as log2 fold change compared with uncut nerves. n = a minimum of four mice for each genotype. Data are presented as mean ± SEM (error bars). **, P < 0.01. (G) The lipid composition of purified myelin obtained from sciatic nerves from 5 d cut WT and Atg7 cKO mice, expressed as log2 fold change compared with uncut nerves. The individual lipid species detected by UPLC were grouped in distinct lipid classes as shown in the graph. “Membrane lipids” refers to all lipid species detected that are the major structural lipids in the eukaryotic membrane, including phosphatidylethanolamines, phosphatidylcholines, and phosphatidylinositols, and “storage lipids” include triacylglycerides and cholesteryl esters. See Table S2 for changes in levels of individual lipid species in WT and Atg7 cKO mice. n = 3 mice for each genotype. Data are presented as mean ± SEM (error bars). *, P < 0.05; **, P < 0.01 (Atg7 cKO relative to WT).
Next, we analyzed whether myelin lipid degradation was also affected in cut nerves from the Atg7 cKO mice. Using FluoroMyelin histochemistry, we found that degradation of myelin lipid was reduced in Atg7 cKO nerves (Fig. 4 E). Second, using TLC-based screening, we examined the lipid composition of uncut and 5 d cut nerves from Atg7 cKO and WT mice. This revealed that up-regulation of cholesteryl esters, which are by-products of myelin lipid breakdown (Goodrum et al., 1994), was markedly lower in Atg7 cKO nerves than in WT ones, indicating slower myelin breakdown (Fig. 4 F). Finally, to differentiate between myelinophagy and the generic lipophagy that likely takes place in whole nerve extracts, we purified myelin from uncut and 5 d cut nerves, and used UPLC to reveal how the lipid composition of myelin changes during the 5 d degradation period. This showed that the typical down-regulation or up-regulation of a large proportion of lipid species seen in WT myelin was notably impaired in Atg7 cKO myelin and confirmed the difference in cholesteryl esters seen previously (Fig. 4 G and Table S2). These results demonstrate that inhibition of autophagy results in aberrant breakdown of the lipid component of the myelin sheath.
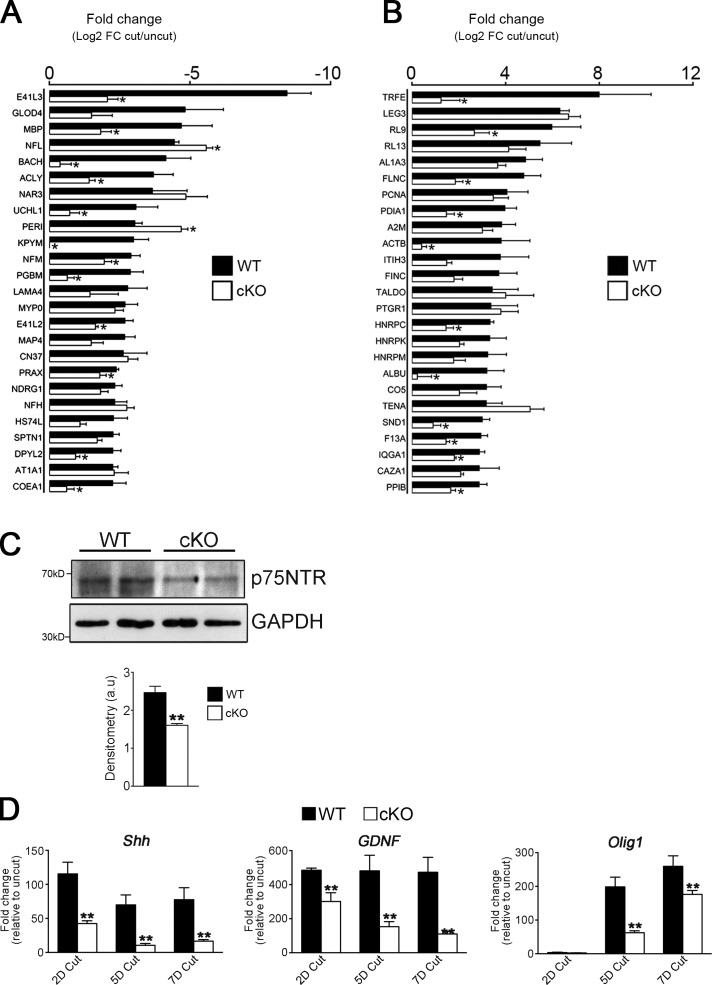
After injury, myelin and nonmyelin (Remak) Schwann cells undergo a c-Jun–dependent reprogramming to form repair Schwann cells (Arthur-Farraj et al., 2012). We examined whether this important cell type conversion was altered in Atg7 cKO nerves using global proteomic analysis. Injury (5 d cut nerves) resulted in substantial changes in the proteomic profile, involving both up- and down-regulation (Fig. S4 E), as described previously (Jiménez et al., 2005). Notably, we found that 12 of the 25 most strongly down-regulated proteins after nerve cut in WT nerves were down-regulated to a significantly lesser degree in Atg7 cKO mice (Fig. 5 A). Conversely, 11 of the 25 most strongly up-regulated proteins in WT nerves were up-regulated to a significantly lesser degree in Atg7 cKO mice (Fig. 5 B). To further confirm that generation of repair Schwann cells was affected in the Atg7 cKO mice, we examined expression of markers associated with these cells after injury (Arthur-Farraj et al., 2012). We found that the expression of p75NTR protein, and of the Shh, GDNF, and Olig1 genes, was significantly reduced in the Atg7 cKO mice (Fig. 5, C and D).
Figure 5.
Genetic inactivation of autophagy retards the generation of repair cells after injury in vivo. (A and B) Graph showing fold change of the top 25 down-regulated (A) and up-regulated (B) proteins in 5 d cut nerves relative to uninjured nerves in WT and Atg7 cKO mice from proteomics analysis (Fig. S4 E). n = 5 mice for each genotype. Data are presented as mean ± SEM (error bars). *, P < 0.05 (Atg7 cKO relative to WT). (C) Western blot showing lower levels of the repair Schwann cell marker p75NTR in 5 d cut nerves from Atg7 cKO mice compared with WT controls. The graph shows densitometric analysis of Western blots. n = 3 mice for each genotype. Data are presented as mean ± SEM (error bars). **, P < 0.01. (D) qPCR analysis showing significantly lower levels of the mRNA levels of the repair Schwann cell markers Shh, GDNF, and Olig1 in 2, 5, and 7 d cut nerves from Atg7 cKO mice compared with WT controls. Data are expressed as fold change in cut nerves relative to uncut nerves. n = 3 mice for each genotype/time point. Data are presented as mean ± SEM (error bars). **, P < 0.01 (Atg7 cKO relative to WT).
These data indicate that activation of autophagy is part of the normal generation of repair Schwann cells after injury.
Myelinophagy is mTOR independent, driven by JNK/c-Jun, and reduced in the injured CNS
Because the control of myelin breakdown is a central issue in nerve injury and pathology, we studied the regulation of myelinophagy, examining the mTOR and JNK/c-Jun pathways, a mouse model of demyelinating neuropathy, and autophagy in the injured CNS.
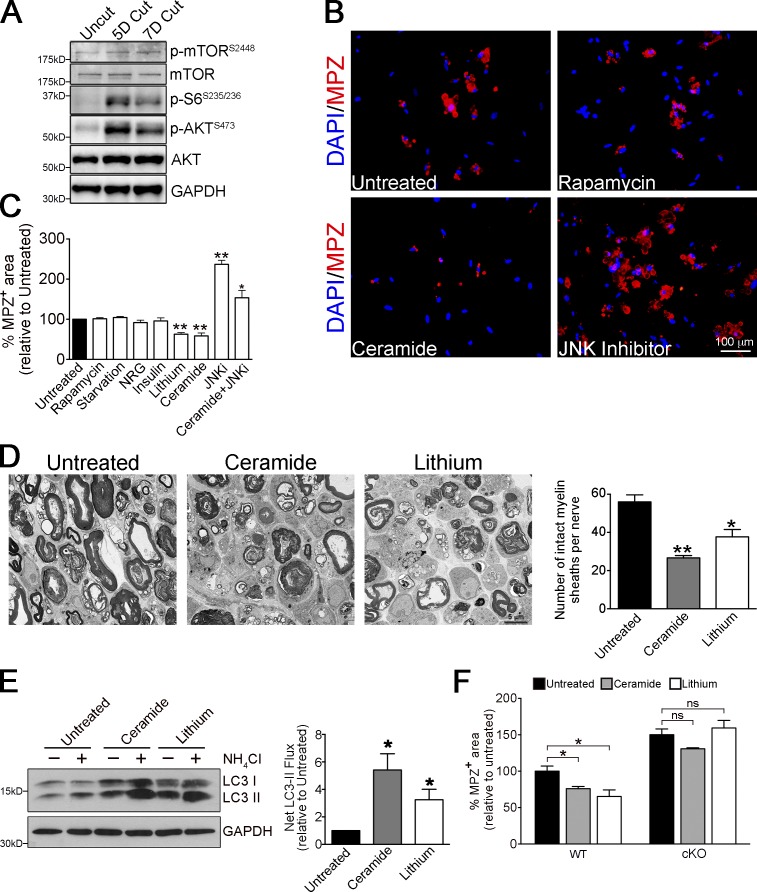
In starvation autophagy, the best-studied model of autophagy regulation, autophagy is activated by reduction in the activity of the autophagy inhibitor mTOR (Ravikumar et al., 2010). Therefore, we examined levels of pmTOR, and pAKT and pS6, which are upstream and downstream markers for mTOR, respectively. While uninjured nerves showed low levels of pmTOR, pS6, and pAKT, these were significantly up-regulated at 5 and 7 d after transection (Fig. 6 A). In addition, neither mTOR activation by starvation (serum deprivation) nor mTOR inhibition by rapamycin altered the rate of myelin breakdown in Schwann cell cultures (Fig. 6, B and C). This was confirmed by analysis of myelin sheath collapse and measurements of autophagy flux in cultured nerve segments treated with rapamycin (Fig. S5, B and C). Treatment with insulin or neuregulin, stimulators of mTOR, also failed to affect myelin protein breakdown (Fig. 6 C). Together this shows that down-regulation of mTOR does not provide the signal for activation of Schwann cell autophagy.
Figure 6.
Myelinophagy is mTOR independent and promoted by lithium and ceramide. (A) Western blot showing increased expression of pmTOR, p-S6, and p-AKT in cut WT nerves. Quantification of Western blots is shown in Fig. S5 A. (B) Immunolabeling showing regulation of MPZ breakdown by rapamycin, ceramide, or JNK inhibitor SP600125 in Schwann cell cultures (from P8 mice animals, treated 3 d in vitro). Myelin breakdown is unchanged by rapamycin, reduced by ceramide, and increased by JNK inhibitor compared with untreated cultures. (C) Graph showing MPZ+ myelin area in Schwann cell cultures (as in Fig. 6 B) after different treatments. Data are presented as mean ± SEM (error bars) from three independent experiments with a minimum of 200 cells analyzed per condition/experiment. *, P < 0.05; **, P < 0.01 (treated cells relative to untreated controls). (D) Electron micrographs showing fewer intact myelin sheaths in nerve segments maintained in vitro for 4 d in the presence of ceramide and lithium, compared with control cultures. The graph shows quantification of the number of intact myelin sheaths in control segments and segments treated with ceramide and lithium. Data are presented as mean ± SEM (error bars) from three independent experiments with a minimum of 10 picture frames analyzed per condition/experiment. *, P < 0.05; **, P < 0.01 (treated cells relative to untreated controls). (E) Western blot showing increased LC3 II accumulation in nerve segments maintained in vitro for 3 d and treated with ceramide or lithium in the presence or absence of NH4Cl (3 h treatment). The graph shows increased net LC3 II flux after ceramide and lithium treatment compared with control cultures. Data are presented as mean ± SEM (error bars) from three independent experiments. *, P < 0.05 (treated cells relative to untreated controls). (F) Graph showing quantification of MPZ+ area in control Schwann cell cultures (WT) and cultures in which autophagy was blocked (Atg7 cKO cultures). The increased myelin degradation of MPZ seen after treatment with ceramide and lithium in WT dissociated Schwann cell cultures is blocked in Atg7 cKO cultures. See Fig. S5 D for pictures of immunolabeling. Data are presented as mean ± SEM (error bars) from three independent experiments with a minimum of 10 picture frames analyzed per condition/experiment. *, P < 0.05; ns, not significant (treated cells relative to untreated controls).
In contrast, two alternative autophagy inducers reported to function through mTOR-independent pathways, lithium (LiCl) and ceramide (Ravikumar et al., 2010), accelerated the rate of myelin protein breakdown both in dissociated Schwann cells (Fig. 6, B and C) and in cultured nerve segments (Fig. 6 D). Notably, we found that LiCl and ceramide significantly increased autophagy flux in nerve segments maintained in vitro (Fig. 6 E). Strikingly, we found that the acceleration of myelin degradation in Schwann cell cultures treated by these two compounds was effectively blocked when autophagy was impaired by using Atg7 cKO Schwann cells (Fig. 6 F) or by using 3-MA (Fig. S5 D; quantification not shown). Collectively, these results show that Schwann cell autophagy is regulated by mTOR-independent pathways.
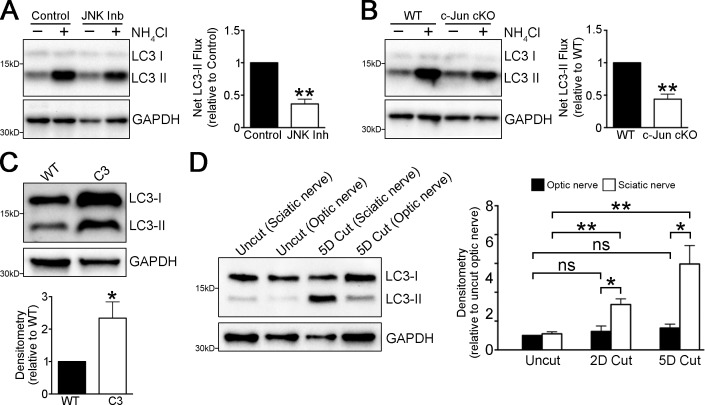
Ceramide has been proposed to act via the JNK/c-Jun pathway to promote autophagy (Ravikumar et al., 2010). We reported previously that c-Jun is a master regulator of the Schwann cell response to injury, and that myelin protein and lipid degradation are delayed in c-Jun cKO mice (Arthur-Farraj et al., 2012). Accordingly, treatment with JNK inhibitor SP600125 strongly inhibited myelin protein breakdown in Schwann cell cultures (Fig. 6, B and C). To test whether the JNK/c-Jun pathway regulated myelinophagy, we examined autophagic flux in nerves from WT mice treated with SP600125 and nerves from WT and c-Jun cKO mice. Autophagic flux was significantly reduced in SP600125-treated and c-Jun cKO nerves (Fig. 7, A and B). This indicates that the JNK/c-Jun pathway is involved in driving Schwann cell autophagy in injured nerves.
Figure 7.
Regulation of myelinophagy. (A and B) Graph showing reduced LC3 II accumulation in nerve segments (A) maintained in vitro for 5 d and treated with JNK inhibitor in the presence and absence of NH4Cl (3 h treatment) and in nerve segments (B) maintained in vitro for 5 d from cJun cKO mice compared with WT mice, in the presence and absence of NH4Cl (3 h treatment). Graphs show reduced net LC3 II flux. Data are presented as mean ± SEM (error bars) from three independent experiments. **, P < 0.01 (treated cells relative to untreated cells; cJun cKO relative to WT). (C) Western blots showing elevated LC3 II levels in uninjured C3 nerves compared with WT nerves. The graph shows densitometric quantification of blots. n = 3 mice for each genotype. Data are presented as mean ± SEM (error bars). *, P < 0.05 (C3 mice relative to WT). (D) Western blots showing that LC3 II levels in optic nerves are substantially lower than in sciatic nerve 3 and 5 d after cut. The graph shows densitometric quantification of blots. Data are presented as mean ± SEM (error bars) from three independent experiments. *, P < 0.05 (sciatic nerves relative to optic nerves); **, P < 0.01 (cut sciatic nerves relative to uncut nerves); n.s., not significant (cut optic nerves relative to uncut nerves).
Previously, we showed aberrant up-regulation of Schwann cell c-Jun in human demyelinating neuropathies and in the C3 mouse model of the most common hereditary demyelinating neuropathy, Charcot-Marie-Tooth disease (CMT1A; Hutton et al., 2011; Verhamme et al., 2011; Hantke et al., 2014). We therefore postulated that C3 nerves might also show aberrant up-regulation of autophagy even in the absence of nerve injury. We found that compared with control nerves of the same age (8 mo), C3 nerves showed significant up-regulation of LC3 II (Fig. 7 C). This finding awaits further investigation, but it points to aberrant autophagy induction in response to myelin instability and suggests a common mechanism for Schwann cell–mediated demyelination in injury and disease.
Poor axonal regeneration in the CNS is in large part caused by the inability of oligodendrocytes to break down myelin effectively (Brosius Lutz and Barres, 2014). We therefore examined autophagy in the injured CNS. In the optic nerve, we found that LC3 II levels were not significantly elevated after injury, in contrast to the sciatic nerve (Fig. 7 D). This suggests that the failure of the CNS to clear myelin is associated with reduced activation of autophagy
Discussion
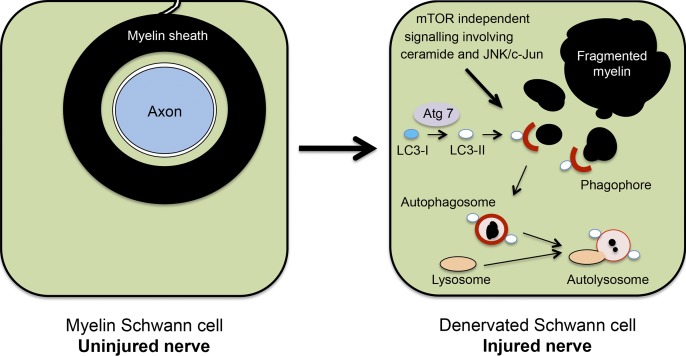
Myelin destruction in injured nerves has long been recognized to take place in two phases: initially, myelin breakdown by the Schwann cells themselves, followed by myelin phagocytosis performed mostly by hematogenous macrophages (Brosius Lutz and Barres, 2014). Phagocytosis has often been suggested to be the mechanism by which Schwann cells digest their myelin, in spite of the fact that myelin is initially an intrinsic Schwann cell component (Holtzman and Novikoff, 1965; Jung et al., 2011b). While Schwann cells may well assist macrophages in phagocytosing myelin debris at later times after injury, we show here that autophagy, a mechanism by which a wide variety of cells break down their own intrinsic cellular components, is an important mechanism by which Schwann cells initiate the degradation of myelin proteins and lipids after nerve injury (Fig. 8).
Figure 8.
Outline of myelinophagy. (left) A transverse section through a myelin Schwann cell in an uninjured nerve. Note that the myelin sheath is in direct continuity with the Schwann cell membrane and an integral component of the Schwann cell. (right) A myelin Schwann cell after nerve injury and axonal degeneration. Note that the myelin sheath has broken up into myelin fragments lying in the Schwann cell cytoplasm. The proposed role of autophagy in digesting these fragments is illustrated.
Autophagy-associated genes and autophagic flux were strikingly induced in transected nerves, and myelin-containing double-membrane autophagosomes were found in demyelinating Schwann cells. Genetic inhibition of autophagy inhibited myelin protein and lipid breakdown in injured nerves, and similar results were obtained with pharmacological autophagy inhibitors. When the bulk of myelin breakdown was accomplished, Schwann cells down-regulated autophagy, and myelin-negative cells were largely negative for GFP-LC3 autophagosomes, indicating a cell-autonomous mechanism for reduction of autophagy once myelin is digested. Collectively, these observations indicate that after injury of peripheral nerves, myelin proteins and lipids are selectively targeted for degradation by activation of myelin autophagy in Schwann cells.
This injury-induced Schwann cell myelinophagy is mTOR-independent. Namely, it is not activated by suppression of mTOR, the classical mechanism for autophagy activation, as we found that mTOR was activated, rather than suppressed, in transected nerves (Kroemer et al., 2010; Ravikumar et al., 2010). mTOR-independent autophagy has been described in other systems, and can be stimulated by lithium and ceramide (Ravikumar et al., 2010), both of which also promoted myelin autophagy in Schwann cells. The mechanism for activation of autophagy by mTOR suppression also exists in Schwann cells, however, because mTOR inhibition by starvation or rapamycin activates autophagy in Trembler J mice, where it functions to clear PMP22 myelin protein aggregates and improve myelination (Rangaraju et al., 2010).
Our data show that activation of autophagy is an important component of the reprogramming of Schwann cells that is triggered by nerve cut (Jessen et al., 2015). In injured nerves, we see activation of mTOR, a pathway that drives major biosynthetic processes and accelerates catabolism and growth. These functions are likely to be required during the dynamic Schwann cell response to injury, which, in addition to myelin breakdown, involves major reorganization of the molecular and morphological profile of myelin and nonmyelin (Remak) cells to generate repair (Bungner) Schwann cells (Arthur-Farraj et al., 2012). The coexistence of autophagy to promote removal of redundant myelin and mTOR activation therefore makes biological sense.
The JNK1/c-Jun pathway stimulates autophagy in several cell types, and ceramide stimulates autophagy in part by activating this pathway (Pattingre et al., 2009). In line with this, we find that pharmacological or genetic inhibition of JNK1/c-Jun reduces Schwann cell autophagic flux. Because nerve injury rapidly activates JNK1/c-Jun in Schwann cells (Parkinson et al., 2008), it is likely that the delay in myelin clearance seen when the JNK1/c-Jun pathway is inhibited pharmacologically or genetically is in part mediated by defective autophagy (Mirsky et al., 2008; Arthur-Farraj et al., 2012). Another potential link between JNK1/c-Jun and autophagy is seen in uninjured nerves of C3 mice that model CMT1A demyelinating neuropathy. We found that in these nerves, high Schwann cell c-Jun expression (Hantke et al., 2014) is accompanied by elevated LC3 II, which is consistent with autophagy activation.
Myelin breakdown in the CNS is ineffective. In line with this, we found a marked difference between the injured optic and sciatic nerves, as LC3 II levels, a key indicator of autophagy, were not significantly elevated in the injured optic nerve in contrast to the sciatic. It is intriguing that elevation of c-Jun in cut optic nerves is also minimal relative to that seen in Schwann cells (Vaudano et al., 1996), although the relevance of this to myelin breakdown and regeneration remains to be established.
In conclusion, we have identified myelinophagy as a major mechanism for Schwann cell–mediated myelin breakdown in injured nerves. We also provide evidence that myelin autophagy is aberrantly regulated in demyelinating peripheral neuropathy, and defective in CNS glia after injury. Identifying an endogenous Schwann cell mechanism for the myelin breakdown that takes place when nerves are cut has broader implications, since activation of autophagy can now be considered to be a plausible candidate mechanism for myelin breakdown in other situations. This includes acquired and inherited demyelinating disease and the age-related degeneration of myelin. Therefore, the recognition of this mechanism in Schwann cells potentially provides novel targets for manipulating demyelination in injury and disease. It also throws new light on the remarkable absence of myelin breakdown by oligodendrocytes. It would be important in future studies to test whether modulation of myelinophagy improves repair in both the injured peripheral nervous system and CNS, and prevents myelin breakdown in demyelinating neuropathies.
Materials and methods
Animals
Animal experiments conformed to UK Home Office guidelines under the supervision of UCL Biological Services, and to the Spanish/European regulations on animal use under the supervision of the institutional committee at CIC bioGUNE (AAALAC-accredited facility) and the Instituto de Neurociencias de Alicante.
Atg7fl/fl mice were made by insertion of loxP sequences and a stop codon within introns 13 and 14 of the atg7 gene (Komatsu et al., 2005). N. Mizushima (The University of Tokyo, Tokyo, Japan) allowed us to use the Atg7fl/fl mice and GFP-LC3 transgenic mice for these studies, and S. Tooze (Cancer Research UK, London, England, UK) and A.K. Simon (University of Oxford, Headington, Oxford, England, UK) provided these mice.
Mpz-Cre mice express Cre recombinase under the control of a mouse mpz promoter and regulatory sequences on chromosome 15. This transgene contains the complete mouse MPZ gene with 6 kb of promoter, in which the ATG start of translation has been mutated and substituted with Cre, and is expressed in Schwann cells, but not in other cell types (Feltri et al., 2002). Mpz-Cre mice were provided by L. Feltri and L. Wrabetz (State University of New York at Buffalo, Buffalo, NY).
These mice were crossed with Atg7fl/fl mice to generate the Mpz-Cre+;Atg7loxP/loxP mice to obtain Schwann cells deficient in atg7.
Genotyping
DNA for genotyping was extracted from ear or tail samples using the Hot Sodium Hydroxide and Tris method (HotSHot; Truett et al., 2000). The ear or tail samples were incubated in alkaline solution (25 mM NaOH and 0.2 mM disodium EDTA, pH 12) for 1 h at 95°C, cooled, and then mixed with neutralizing buffer (40 mM Tris-HCl, pH 5). 1 or 2 µl of this reaction mixture was used for a 25-µl PCR reaction.
Primers for genotyping Atg7loxP/loxP were 5′-TGGCTGCTACTTCTGCAATGATGT-3′ and 5′-CAGGACAGAGACCATCAGCTCCAC-3′ (1,500-bp band for WT allele and 550-bp band flox allele; Komatsu et al., 2005). The primers for the Mpz-cre transgene are 5′-GCTGGCCCAAATGTTGCTGG-3′ and 5′-CCACCACCTCTCCATTGCAC-3′ (480-bp band; Feltri et al., 2002).
Models to study Schwann cell myelin breakdown
We used three models to study Schwann cell myelin breakdown in response to nerve injury. (1) After nerve transection (6–8 wk old mice), which results in axonal degeneration, the distal stump was most commonly cut into further 2–3-mm segments and left in situ, most commonly for 3–5 d, before examination. (2) After nerve transection (6–8 wk old mice), the distal stump was removed, cut into 3–5 mm segments, and maintained in tissue culture most commonly for 3–5 d before analysis (nerve segment experiments). (3) Schwann cell cultures were generated by enzymatic dissociation of postnatal day 10 (P10) nerves (that are already extensively myelinated).
Surgical nerve injury
Sciatic nerve surgeries were done as described previously (Woodhoo et al., 2009). The right sciatic nerve was exposed and transected at the sciatic notch (distal stump). Skin was closed using veterinary autoclips. The transected nerve was excised for analysis at various time points. Contralateral uninjured sciatic nerves were used as controls for Western blotting, immunohistochemistry, or electron microscopy analysis.
Optic nerve surgeries were done as described previously (Parrilla-Reverter et al., 2009). The left optic nerve transection was performed intracranially, 3 mm from the posterior pole of the eye. Contralateral uninjured optic nerves were used as controls for Western blotting.
Sciatic nerve segments and Schwann cell culture
Sciatic nerves from adult mice were dissected and placed in L15 medium on ice. The epineurium and superfluous connective tissue were removed using forceps. Desheathed nerves were then cut into 5-mm segments, transferred to DMEM supplemented with 5% FBS, and incubated at 37°C/5% CO2. Segments were removed from culture for analysis at various time points, either by Western blotting, teased fiber preparation, or dissociation.
For dissociated Schwann cells, nerves were transferred to a dissociating enzyme solution containing 10 µg/ml collagenase in Kreb’s Ca2+ and Mg2+ free medium plus 0.25% trypsin and placed at 37°C/5% CO2 for 30 min. Nerves were triturated gently before the addition of 10% FBS/DMEM and centrifuged at 1,000 rpm for 10 min. The cells were plated as drops on coverslips coated with poly-l-lysine and laminin. Cells were incubated at 37°C/5% CO2 and allowed to adhere overnight (16 h).
Autophagy specific inhibitors used were: ammonium chloride at 10–20 mM, 3-MA at 100 mM, protease inhibitor cocktail at 1:1,000, Bafilomycin A1 at 1 nM, and rapamycin at 50 ng/ml (Sigma-Aldrich).
Immunohistochemistry
Cultured nerve segments were fixed for 2 h at RT in 4% paraformaldehyde/PBS, and dissociated Schwann cell cultures were fixed at RT in 4% paraformaldehyde for 10 min. Cultured nerve segments were subjected to teasing before immunolabeling. Individual nerve fibers were gently teased and separated using forceps and 26 gauge needles, and plated onto Superfrost glass slides (Menzel-Glasër).
Teased fibers and dissociated cells were treated for 10 min in 100% ice-cold methanol before immunolabeling. Blocking solution (10% fetal bovine serum, 1.8% lysine, and 0.02% sodium azide in PBS) was used before incubation with primary antibodies overnight followed by secondary antibodies for 1 h (1:500, Alexa Fluor 488 [Molecular Probes] or Cy3 [Jackson ImmunoResearch Laboratories, Inc.]). Samples were stained with the nuclear probe DAPI (1:1,000; Thermo Fisher Scientific) to visualize nuclei and were mounted in fluorescent mounting medium (Citifluor).
Immunofluorescent cells or teased nerves were visualized at room temperature using either an Optiphot-2 or an Eclipse E800 fluorescent microscope (both from Nikon) equipped with a Plan Fluor 20×/0.50 NA or 40×/1.30 NA oil objective lens. Images were captured using a digital camera (DXM1200; Nikon) and ACT-1 acquisition software (Nikon).
Confocal microscopy was performed at room temperature on a confocal microscope (TCS SPE; Leica) controlled with Leica LAS AF software and equipped with violet (405 nm laser diode), blue (argon, 488 nm), yellow (561 nm solid state Laser), and red (633 nm HeNe laser) excitation laser lines. A 63× oil-immersion objective lens (ACS Apochromat 63.0× 1.30 NA oil; Leica) was used for analysis. All confocal images represent the maximum projection from total stacks. Dissociated cells were typically imaged using a maximum projection of 20 stacks; teased fibers were typically imaged using a maximum projection of 30 stacks.
For illustration purposes, images were cropped and brightness/contrast were adjusted using Photoshop (CS6; Adobe); the same adjustments were applied to every pixel in each RGB channel. For data analysis, ImageJ software (National Institutes of Health) was used.
Antibodies
The following antibodies were used: ATG5 (rabbit; Western blot [WB] 1:1,000; Cell Signaling Technology), ATG7 (rabbit; WB 1:1,000; Cell Signaling Technology), ATG16L1 (rabbit; WB 1:500; Cosmobio), Beclin-1 (rabbit; WB 1:1,000; Cell Signaling Technology), GAPDH (rabbit; WB 1:4,000; Sigma-Aldrich), LC3B (rabbit; WB 1:1,000; Sigma-Aldrich), GFP (rabbit; immunofluorescence [IF] 1:1,000; WB 1:1,000; Invitrogen), Wipi2 (mouse; WB 1:1,000; AbD Serotec), MBP (mouse; WB 1:1,000, IF 1:1,000; Covance), NBR1 (mouse; WB 1:1,000; Abcam), mTOR (rabbit; WB 1:1,000; Cell Signaling Technology), p-mTORS2448 (rabbit; WB 1:1,000; Cell Signaling Technology), MPZ (chicken; WB 1:1,000, IF 1:1,000; Aves Labs), Periaxin (affinity-purified rabbit polyclonal antibody against a full-length fusion protein of rat Periaxin [Gillespie et al., 1994]; a gift from P. Brophy, University of Edinburgh, Edinburgh, Scotland, UK; WB 1:5,000), pAKT (rabbit; WB 1:2,000; Cell Signaling Technology), pS6 (rabbit; WB 1:1,000; Cell Signaling Technology), S100 (rabbit; IF 1:1,000; Dako), and F4/80 (rat; IF 1:50; Bio-Rad Laboratories).
Electron microscopy
Samples were fixed in 2.5% glutaraldehyde/2% paraformaldehyde in 0.1 M cacodylate buffer, pH 7.4, overnight at 4°C. Samples were post-fixed with 1% OsO4, embedded in Agar 100 epoxy resin, and sectioned at 70 nm. Transverse ultrathin sections of tibial nerve at 3 mm from the cut site or uncut nerve were mounted on film and 18–20 photographs were taken at 2,500× using an electron microscope (1010; Jeol) with Gatan software and analyzed with ImageJ software.
For detection of autophagosomes, a modified version of the electron microscopy protocol was used. In brief, cells or nerve segments were fixed in fixative solution (2.5% glutaraldehyde/2% paraformaldehyde in 0.1 M cacodylate buffer, pH 7.4, overnight at 4°C), and then osmicated in 1% osmium tetroxide and 1.5% potassium ferrocyanide (added to increase membrane resolution) for 45 min at 4°C. This was followed by standard EM protocol and visualization.
RNA isolation and quantitative PCR
RNA was isolated with TRIzol (Invitrogen), and its concentration and integrity were determined. Quantitative PCR (qPCR) was performed using a ViiA 7 Real-Time PCR System (Life Technologies). Ct values were normalized to the housekeeping expression (GAPDH). Primer sequences are described in Table S1.
Lipid analysis of whole nerves
Nerves (3 mg) from five WT and five Atg7 cKO mice (uncut and cut nerves) were homogenized in ice-cold PBS and lipids were extracted from homogenates as described previously (Bligh and Dyer, 1959). In brief, lipids were exhaustively extracted from homogenates using a mixture of chloroform and methanol to form a miscible system. This was diluted with chloroform and water to separate the homogenate into two layers: a chloroform layer containing all the lipids and a methanolic layer containing all the nonlipids. A purified lipid extract was obtained by isolating the chloroform layer.
The lipid extracts were separated by thin layer chromatography and quantified by optical densitometry as detailed previously (Ruiz and Ochoa, 1997). In brief, lipids were first separated by one-dimensional thin-layer chromatography using an EDTA-impregnated chromatoplate. This was followed by acquisition and processing of an image of the TLC plate using Molecular Imager and Quantity One software (Bio-Rad Laboratories). To get the amount of each lipid, the integrated optical densities of the individual spots were interpolated in the corresponding lipid standard curve. Finally, the results were normalized per gram of tissue.
Lipidomics analysis of purified myelin
Myelin was purified from uncut and 5 d cut sciatic nerve extracts by density gradient centrifugation, based on previously published protocols (Larocca and Norton, 2007). In brief, the sciatic nerves were cut into pieces with a razor blade on a cold glass plate and the small nerve segments were suspended in 0.27 M sucrose solution containing 20 mM Tris-Cl buffer, pH 7.45, followed by homogenization with a homogenizer (Precellys 24; Precellys). The connective tissue was eliminated by passing the homogenate through layers of cheese cloth. The tissue suspension was then layered over 0.83 M sucrose solution and centrifuged for 45 min at 82,000 g, 4°C. The crude myelin fraction, which forms at the 0.27 M/0.83 M sucrose interface, was then recovered, subjected to hypoosmotic shock, and centrifuged to remove microsomal contamination.
Lipid profiling of purified myelin fractions from control and cut nerves from three WT and three cKO mice were performed at OWL Metabolomics. Ultraperformance liquid chromatography/electrospray/mass spectrometry–based platforms were used for optimal profiling of glycerolipids, cholesteryl esters, sphingolipids, and glycerophospholipids as described previously (Barr et al., 2012; Varela-Rey et al., 2014). In brief, purified myelin samples were mixed with water (4°C) and chloroform/methanol at −20°C (overall water/chloroform/methanol, 1:3:4, vol/vol/vol) in 1.5-ml microtubes. The extraction solvent was spiked with metabolites not detected in unspiked purified myelin extracts. After brief vortex mixing, the samples were incubated for 1 h at −20°C. Then, extracts were mixed with water (ammonium hydroxide, pH 9) and after brief vortex mixing, an additional incubation (1 h at −20°C) of the samples was performed. After centrifugation at 16,000 g for 15 min, the organic phase was collected and the solvent was removed under vacuum. The dried extracts were then reconstituted in acetonitrile/isopropanol (50:50; vol/vol), centrifuged (16,000 g for 5 min), and transferred to vials for UPLC-ESI-MS analysis (Acquity UPLC hyphenated to Xevo G2 Q-ToF; Waters). All data were processed using the TargetLynx application manager for MassLynx 4.1 software (Waters). Normalization factors were calculated for each metabolite by dividing their intensities in each sample by the recorded intensity of an appropriate internal standard.
Proteomic analysis
Protein from five WT and five cKO samples (both uncut and cut nerves) was extracted in a buffer containing 7 M urea, 2 M thiourea, and 4% CHAPS. After in-solution tryptic digestion, peptide mixtures obtained from the digestions were separated by on-line NanoLC (nLC) and analyzed using electrospray tandem mass spectrometry. Peptide separation was performed on a nanoACQUITY UPLC system (Waters) connected to a mass spectrometer (LTQ Orbitrap XL ETD; Thermo Fisher Scientific).
Progenesis LC-MS software (Nonlinear Dynamics) was used for differential protein expression analysis. Raw files were imported into the program, and one of the samples was selected as a reference run to which the precursor masses in all the other samples were aligned. Abundance ratios between the run to be aligned and the reference run were calculated for all features at given retention times. These values were then logarithmized and the software, based on the analysis of the distribution of all ratios, automatically calculated a global scaling factor. Once normalized, samples were grouped in the corresponding experimental category and compared. Differences between groups were only considered for peptide abundances with an ANOVA p-value <0.05 and a ratio >1.5 in either direction. A peak list containing the differential peptides was generated for each comparison, and searched against a database using the Mascot Search engine (www.matrixscience.com). Only proteins with at least two peptides passing the FDR <1% filter were considered for further analysis. Finally, only proteins with ANOVA P < 0.05 and a ratio >1.5 in either direction were considered as significantly deregulated. To calculate total protein volumes, searches with unfiltered data were also performed following the same approach. The fold change between cut nerves and their respective control uncut nerves were then calculated for each replicate.
Western blotting
Nerves from surgical transection or cultured nerve segments were snap-frozen in liquid nitrogen and transferred to tubes containing 10–12 10B lysing matrix beads and 100 µl of lysis buffer (5 M urea, 2.5% SDS, 50 mM Tris, and 30 mM NaCl). Samples were placed in a Fastprep homogenizer for 45 s at level 6, chilled on ice, and then subjected to a repeat homogenization at the same setting. Lysates were then centrifuged at 4°C at 10,000 rpm for 2 min. Supernatants were transferred to new tubes and centrifuged again at the same settings. New supernatants were collected and stored at −80°C.
Cultured Schwann cells were extracted by cell scraping. Cells on 35-mm tissue culture dishes were washed twice with PBS before addition of 100 µl of lysis buffer (T-PER Tissue Protein Extraction Reagent; Thermo Fisher Scientific). Cells were scraped with a plastic cell lifter and the lysate was collected. Lysates were then centrifuged at 4°C at 10,000 rpm for 2 min. Supernatants were transferred to new tubes and centrifuged again at the same settings. New supernatants were collected and stored at −80°C.
Between 5 µg and 40 µg of protein samples were loaded onto acrylamide gels and run alongside prestained standard molecular weight markers (PageRuler prestained protein ladder; Thermo Fisher Scientific) to enable identification of band sizes. Gels were run using the mini Protean II gel electrophoresis apparatus (Bio-Rad Laboratories). Gels were transferred to nitrocellulose (Hybond ECL; Amersham Biosciences) or PVDF (Immobilon-P; EMD Millipore) membranes using the Bio-Rad Laboratories semi-dry system.
Membranes were blocked in 5% fat-free milk powder (Sigma-Aldrich) in PBS for 1 h (at room temperature) and subsequently incubated overnight at 4°C in primary antibodies diluted in 5% milk/PBS on a slow rotator (Gallenkamp). HRP-conjugated secondary antibodies (1:2,000) were used (Jackson ImmunoResearch Laboratories) and membranes were developed with ECL reagent (GE Healthcare). Experiments were repeated at least three times with fresh samples and representative pictures are shown. Densitometric quantification of Western blots was performed using ImageJ analysis software or Image Lab 4.1 (Bio-Rad Laboratories). The measurements were normalized to loading controls (GAPDH or β-Actin), and for each experiment, at least three different biological replicates were used.
Behavioral tests
All experiments were performed according to UK Home Office guidelines. Four mice from each genotype were tested and at least three measurements from each mouse were taken to obtain mean values.
Walking beam
Mice walked along a wooden beam 1 m long, 20 cm high, and 0.5 cm wide to assess sensorimotor function. Foot slips and beam falls were counted over the middle distance of 70 cm. A performance score was given as follows: 0 and 1 foot slips = a score of 1; 2–5 foot slips = a score of 2; >5 foot slips or at least 1 beam fall = a score of 3. Mice were first accustomed to the beam in training sessions on two days that included walking on wider beams of 2 cm and 1.2 cm. At the end of the second day, mice were scored on the 0.5 cm beam.
Sciatic function index (SFI)
Paw prints were taken from 6-wk-old mice to calculate the SFI. The SFI formula correlates mouse paw prints with nerve function, as described previously (Inserra et al., 1998). The hind limb paws of the mice were painted on the plantar surface with black paint. Mice then walked down a 54 × 5-cm wooden corridor lined with 200 gsm highly absorbent paper (Goldline). Prints were subsequently scanned (Perfection 3100; Epson) to generate digital paw prints. Toe spread (the distance between the first and fifth toes) and print length (the distance between the third toe and the heel) were measured using ImageJ, and Excel (Microsoft) was used to convert the measurements into SFI values using an established formula (Inserra et al., 1998). The SFI formula is: SFI = 118.9 (ETS − NTS/NTS) − 51.2 (EPL − NPL/NPL) − 7.5, where ETS is experimental toe spread, NTS is normal toe spread, EPL is experimental paw length, and NPL is normal paw length. An SFI value of −100 represents complete loss of function.
Hanging wire test
Masking tape was placed around the perimeter of a cage lid. Mice were placed individually on top of the lid, which was then slowly turned around with the mouse hanging on upside down by all four legs. Mice were allowed to hang for 2 min. Latency until fall was noted, unless mice remained on the lid for the full 2 min.
Net LC3 II flux
Net LC3 II flux was calculated by analyzing LC3 II turnover by Western blotting in the presence or absence of lysosomal inhibitors. In brief, net LC3 II flux was calculated by subtracting the densitometry value of normalized (relative to GAPDH) LC3 II in the sample treated with lysosomal inhibitor by the value in the control sample (untreated). A minimum of three biological replicate experiments were performed for net LC3 II flux calculations.
Statistics
Values are given as means ± SEM (n = 3 unless otherwise stated). Statistical significance was analyzed with the Student’s t test and p-values <0.05 were considered significant.
Online supplemental material
Fig. S1 shows qPCR analysis of autophagy-related genes from Fig. 1 A. Fig. S2 shows densitometric quantification of Western blots from Fig. 1. Fig. S3 shows that uninjured Atg7 cKO nerves are normal using behavioral and biochemical analyses. Fig. S4 shows that Atg7 cKO nerves have reduced autophagy activation after nerve injury, and that this retards myelin degradation. Fig. S5 shows that myelinophagy is mTOR-independent, and is regulated by ceramide and lithium. Table S1 shows the list of primers used for qPCR analysis. Table S2 shows changes in levels of individual lipid species after nerve injury in ATG7 WT and cKO mice as determined by UPLC analysis of purified myelin. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201503019/DC1.
Supplementary Material
Acknowledgements
We thank Jane Pendjiky for help with imaging and Mark Turmaine for help with electron microscopy. The authors thank Dr. Paolo Salomoni and Dr. Harold Jefferies for helpful discussions, and Dr. Marta Agudo, Dr. Eloisa Herrera, and Dr. Hugo Cabedo for training us in optic nerve surgery. We thank OWL Metabolomics for lipid profiling and the Genome Analysis and Proteomics platforms at CIC bioGUNE.
This work was funded by a Wellcome Trust Program grant to K.R. Jessen and R. Mirsky. The research leading to these results has received funding from the European Community’s Seventh Framework Program (FP7/2007-2013) under grant agreement No. HEALTH-F2-2008-201535. This work was funded by grant PI12/00005, co-financed by the ISCIII-Subdirección General de Evaluación and Fondo Europeo de Desarrollo Regional (FEDER), Fundación Científica de la Asociación Española Contra el Cáncer (Cancer Infantil 2011), Departamento de Educación Política Lingüística y Cultura del Gobierno Vasco (PI2013-46), and the Ministerio de Economía y Competitividad–Plan Nacional de I+D+I 2008-2011 Subprograma Ramón y Cajal RYC2010-06901 to A. Woodhoo. The other co-authors are supported by the following grants: NIH AT-1576 (to M.L. Martínez-Chantar and J.M. Mato), SAF 2011-29851 (to J.M. Mato), Sanidad Gobierno Vasco 2012 (2011111106; to M. Varela-Rey), ETORTEK-2011 (to M.L. Martínez-Chantar and A.M. Aransay), Educacion Gobierno Vasco 2011 (to M.L. Martínez-Chantar), and FIS PI11/01588 (to M.L. Martínez-Chantar). CIBERehd is funded by the Instituto de Salud Carlos III. The Proteomics Platform at CIC bioGUNE lab is a member of Proteored, PRB2-ISCIII, and is supported by grant PT13/0001 of the PE I+D+i 2013-2016 (http://www.idi.mineco.gob.es/portal/site/MICINN/menuitem.7eeac5cd345b4f34f09dfd1001432ea0/?vgnextoid=83b192b9036c2210VgnVCM1000001d04140aRCRD), funded by ISCIII and FEDER.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- 3-MA
- 3-methyladenine
- cKO
- conditional knockout
- CNS
- central nervous system
- P
- postnatal day
- qPCR
- quantitative PCR
- WT
- wild type
References
- Arthur-Farraj P.J., Latouche M., Wilton D.K., Quintes S., Chabrol E., Banerjee A., Woodhoo A., Jenkins B., Rahman M., Turmaine M., et al. 2012. c-Jun reprograms Schwann cells of injured nerves to generate a repair cell essential for regeneration. Neuron. 75:633–647. 10.1016/j.neuron.2012.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr J., Caballería J., Martínez-Arranz I., Domínguez-Díez A., Alonso C., Muntané J., Pérez-Cormenzana M., García-Monzón C., Mayo R., Martín-Duce A., et al. 2012. Obesity-dependent metabolic signatures associated with nonalcoholic fatty liver disease progression. J. Proteome Res. 11:2521–2532. 10.1021/pr201223p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh E.G., and Dyer W.J.. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917. 10.1139/o59-099 [DOI] [PubMed] [Google Scholar]
- Brosius Lutz A., and Barres B.A.. 2014. Contrasting the glial response to axon injury in the central and peripheral nervous systems. Dev. Cell. 28:7–17. 10.1016/j.devcel.2013.12.002 [DOI] [PubMed] [Google Scholar]
- Chrast R., Saher G., Nave K.A., and Verheijen M.H.. 2011. Lipid metabolism in myelinating glial cells: lessons from human inherited disorders and mouse models. J. Lipid Res. 52:419–434. 10.1194/jlr.R009761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubový P., Jančálek R., and Kubek T.. 2013. Role of inflammation and cytokines in peripheral nerve regeneration. Int. Rev. Neurobiol. 108:173–206. [DOI] [PubMed] [Google Scholar]
- Feltri M.L., Graus Porta D., Previtali S.C., Nodari A., Migliavacca B., Cassetti A., Littlewood-Evans A., Reichardt L.F., Messing A., Quattrini A., et al. 2002. Conditional disruption of β 1 integrin in Schwann cells impedes interactions with axons. J. Cell Biol. 156:199–209. 10.1083/jcb.200109021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., He D., Yao Z., and Klionsky D.J.. 2014. The machinery of macroautophagy. Cell Res. 24:24–41. 10.1038/cr.2013.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Valle C., Bunge R.P., and Bunge M.B.. 1995. Schwann cells degrade myelin and proliferate in the absence of macrophages: evidence from in vitro studies of Wallerian degeneration. J. Neurocytol. 24:667–679. 10.1007/BF01179817 [DOI] [PubMed] [Google Scholar]
- Gillespie C.S., Sherman D.L., Blair G.E., and Brophy P.J.. 1994. Periaxin, a novel protein of myelinating Schwann cells with a possible role in axonal ensheathment. Neuron. 12:497–508. 10.1016/0896-6273(94)90208-9 [DOI] [PubMed] [Google Scholar]
- Goodrum J.F., Earnhardt T., Goines N., and Bouldin T.W.. 1994. Fate of myelin lipids during degeneration and regeneration of peripheral nerve: an autoradiographic study. J. Neurosci. 14:357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantke J., Carty L., Wagstaff L.J., Turmaine M., Wilton D.K., Quintes S., Koltzenburg M., Baas F., Mirsky R., and Jessen K.R.. 2014. c-Jun activation in Schwann cells protects against loss of sensory axons in inherited neuropathy. Brain. 137:2922–2937. 10.1093/brain/awu257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata K., and Kawabuchi M.. 2002. Myelin phagocytosis by macrophages and nonmacrophages during Wallerian degeneration. Microsc. Res. Tech. 57:541–547. 10.1002/jemt.10108 [DOI] [PubMed] [Google Scholar]
- Holtzman E., and Novikoff A.B.. 1965. Lysomes in the rat sciatic nerve following crush. J. Cell Biol. 27:651–669. 10.1083/jcb.27.3.651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton E.J., Carty L., Laurá M., Houlden H., Lunn M.P., Brandner S., Mirsky R., Jessen K., and Reilly M.M.. 2011. c-Jun expression in human neuropathies: a pilot study. J. Peripher. Nerv. Syst. 16:295–303. 10.1111/j.1529-8027.2011.00360.x [DOI] [PubMed] [Google Scholar]
- Inserra M.M., Bloch D.A., and Terris D.J.. 1998. Functional indices for sciatic, peroneal, and posterior tibial nerve lesions in the mouse. Microsurgery. 18:119–124. [DOI] [PubMed] [Google Scholar]
- Jang S.Y., Shin Y.K., Park S.Y., Park J.Y., Rha S.H., Kim J.K., Lee H.J., and Park H.T.. 2015. Autophagy is involved in the reduction of myelinating Schwann cell cytoplasm during myelin maturation of the peripheral nerve. PLoS ONE. 10:e0116624 10.1371/journal.pone.0116624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen K.R., Mirsky R., and Lloyd A.C.. 2015. Schwann cells: development and role in nerve repair. Cold Spring Harb. Perspect. Biol.:a020487 10.1101/cshperspect.a020487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez C.R., Stam F.J., Li K.W., Gouwenberg Y., Hornshaw M.P., De Winter F., Verhaagen J., and Smit A.B.. 2005. Proteomics of the injured rat sciatic nerve reveals protein expression dynamics during regeneration. Mol. Cell. Proteomics. 4:120–132. 10.1074/mcp.M400076-MCP200 [DOI] [PubMed] [Google Scholar]
- Jung J., Cai W., Jang S.Y., Shin Y.K., Suh D.J., Kim J.K., and Park H.T.. 2011a Transient lysosomal activation is essential for p75 nerve growth factor receptor expression in myelinated Schwann cells during Wallerian degeneration. Anat. Cell Biol. 44:41–49. 10.5115/acb.2011.44.1.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J., Cai W., Lee H.K., Pellegatta M., Shin Y.K., Jang S.Y., Suh D.J., Wrabetz L., Feltri M.L., and Park H.T.. 2011b Actin polymerization is essential for myelin sheath fragmentation during Wallerian degeneration. J. Neurosci. 31:2009–2015. 10.1523/JNEUROSCI.4537-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel J.A. 2010. Autophagy in unicellular eukaryotes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365:819–830. 10.1098/rstb.2009.0237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Abdalla F.C., Abeliovich H., Abraham R.T., Acevedo-Arozena A., Adeli K., Agholme L., Agnello M., Agostinis P., Aguirre-Ghiso J.A., et al. 2012. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 8:445–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., et al. 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169:425–434. 10.1083/jcb.200412022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G., Mariño G., and Levine B.. 2010. Autophagy and the integrated stress response. Mol. Cell. 40:280–293. 10.1016/j.molcel.2010.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larocca J.N., and Norton W.T.. 2007. Isolation of myelin. In Current Protocols in Cell Biology. Chapter 3:Unit3.25. [DOI] [PubMed] [Google Scholar]
- Levine B., Mizushima N., and Virgin H.W.. 2011. Autophagy in immunity and inflammation. Nature. 469:323–335. 10.1038/nature09782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirsky R., Woodhoo A., Parkinson D.B., Arthur-Farraj P., Bhaskaran A., and Jessen K.R.. 2008. Novel signals controlling embryonic Schwann cell development, myelination and dedifferentiation. J. Peripher. Nerv. Syst. 13:122–135. 10.1111/j.1529-8027.2008.00168.x [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Matsui M., Yoshimori T., and Ohsumi Y.. 2004. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell. 15:1101–1111. 10.1091/mbc.E03-09-0704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemi J.P., DeFrancesco-Lisowitz A., Roldán-Hernández L., Lindborg J.A., Mandell D., and Zigmond R.E.. 2013. A critical role for macrophages near axotomized neuronal cell bodies in stimulating nerve regeneration. J. Neurosci. 33:16236–16248. 10.1523/JNEUROSCI.3319-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson D.B., Bhaskaran A., Arthur-Farraj P., Noon L.A., Woodhoo A., Lloyd A.C., Feltri M.L., Wrabetz L., Behrens A., Mirsky R., and Jessen K.R.. 2008. c-Jun is a negative regulator of myelination. J. Cell Biol. 181:625–637. 10.1083/jcb.200803013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrilla-Reverter G., Agudo M., Nadal-Nicolás F., Alarcón-Martínez L., Jiménez-López M., Salinas-Navarro M., Sobrado-Calvo P., Bernal-Garro J.M., Villegas-Pérez M.P., and Vidal-Sanz M.. 2009. Time-course of the retinal nerve fibre layer degeneration after complete intra-orbital optic nerve transection or crush: a comparative study. Vision Res. 49:2808–2825. 10.1016/j.visres.2009.08.020 [DOI] [PubMed] [Google Scholar]
- Pattingre S., Bauvy C., Levade T., Levine B., and Codogno P.. 2009. Ceramide-induced autophagy: to junk or to protect cells? Autophagy. 5:558–560. 10.4161/auto.5.4.8390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry V.H., Tsao J.W., Fearn S., and Brown M.C.. 1995. Radiation-induced reductions in macrophage recruitment have only slight effects on myelin degeneration in sectioned peripheral nerves of mice. Eur. J. Neurosci. 7:271–280. 10.1111/j.1460-9568.1995.tb01063.x [DOI] [PubMed] [Google Scholar]
- Ramaglia V., Wolterman R., de Kok M., Vigar M.A., Wagenaar-Bos I., King R.H., Morgan B.P., and Baas F.. 2008. Soluble complement receptor 1 protects the peripheral nerve from early axon loss after injury. Am. J. Pathol. 172:1043–1052. 10.2353/ajpath.2008.070660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaraju S., Verrier J.D., Madorsky I., Nicks J., Dunn W.A. Jr., and Notterpek L.. 2010. Rapamycin activates autophagy and improves myelination in explant cultures from neuropathic mice. J. Neurosci. 30:11388–11397. 10.1523/JNEUROSCI.1356-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B., Sarkar S., Davies J.E., Futter M., Garcia-Arencibia M., Green-Thompson Z.W., Jimenez-Sanchez M., Korolchuk V.I., Lichtenberg M., Luo S., et al. 2010. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol. Rev. 90:1383–1435. 10.1152/physrev.00030.2009 [DOI] [PubMed] [Google Scholar]
- Rubinsztein D.C., Shpilka T., and Elazar Z.. 2012. Mechanisms of autophagosome biogenesis. Curr. Biol. 22:R29–R34. 10.1016/j.cub.2011.11.034 [DOI] [PubMed] [Google Scholar]
- Ruiz J.I., and Ochoa B.. 1997. Quantification in the subnanomolar range of phospholipids and neutral lipids by monodimensional thin-layer chromatography and image analysis. J. Lipid Res. 38:1482–1489. [PubMed] [Google Scholar]
- Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A.M., and Czaja M.J.. 2009. Autophagy regulates lipid metabolism. Nature. 458:1131–1135. 10.1038/nature07976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truett G.E., Heeger P., Mynatt R.L., Truett A.A., Walker J.A., and Warman M.L.. 2000. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques. 29:52–54: 54. [DOI] [PubMed] [Google Scholar]
- Varela-Rey M., Iruarrizaga-Lejarreta M., Lozano J.J., Aransay A.M., Fernandez A.F., Lavin J.L., Mósen-Ansorena D., Berdasco M., Turmaine M., Luka Z., et al. 2014. S-adenosylmethionine levels regulate the schwann cell DNA methylome. Neuron. 81:1024–1039. 10.1016/j.neuron.2014.01.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas M.E., Watanabe J., Singh S.J., Robinson W.H., and Barres B.A.. 2010. Endogenous antibodies promote rapid myelin clearance and effective axon regeneration after nerve injury. Proc. Natl. Acad. Sci. USA. 107:11993–11998. 10.1073/pnas.1001948107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaudano E., Campbell G., and Hunt S.P.. 1996. Change in the molecular phenotype of Schwann cells upon transplantation into the central nervous system: down-regulation of c-jun. Neuroscience. 74:553–565. 10.1016/0306-4522(96)00199-6 [DOI] [PubMed] [Google Scholar]
- Verhamme C., King R.H., ten Asbroek A.L., Muddle J.R., Nourallah M., Wolterman R., Baas F., and van Schaik I.N.. 2011. Myelin and axon pathology in a long-term study of PMP22-overexpressing mice. J. Neuropathol. Exp. Neurol. 70:386–398. 10.1097/NEN.0b013e318217eba0 [DOI] [PubMed] [Google Scholar]
- Woodhoo A., Alonso M.B., Droggiti A., Turmaine M., D’Antonio M., Parkinson D.B., Wilton D.K., Al-Shawi R., Simons P., Shen J., et al. 2009. Notch controls embryonic Schwann cell differentiation, postnatal myelination and adult plasticity. Nat. Neurosci. 12:839–847. 10.1038/nn.2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.