Rpgrip1l regulates proteasomal activity at the basal body via Psmd2 and thereby controls ciliary signaling.
Abstract
Mutations in RPGRIP1L result in severe human diseases called ciliopathies. To unravel the molecular function of RPGRIP1L, we analyzed Rpgrip1l−/− mouse embryos, which display a ciliopathy phenotype and die, at the latest, around birth. In these embryos, cilia-mediated signaling was severely disturbed. Defects in Shh signaling suggested that the Rpgrip1l deficiency causes an impairment of protein degradation and protein processing. Indeed, we detected a cilia-dependent decreased proteasomal activity in the absence of Rpgrip1l. We found different proteasomal components localized to cilia and identified Psmd2, a component of the regulatory proteasomal 19S subunit, as an interaction partner for Rpgrip1l. Quantifications of proteasomal substrates demonstrated that Rpgrip1l regulates proteasomal activity specifically at the basal body. Our study suggests that Rpgrip1l controls ciliary signaling by regulating the activity of the ciliary proteasome via Psmd2.
Introduction
Primary cilia act as antenna-like structures, transducing signals from the environment to control essential cellular processes such as proliferation, apoptosis, migration, and differentiation. Cilia consist of three different compartments—the basal body (BB), the transition zone (TZ), and the axoneme. The BB is a modified centriole from which the ciliary microtubule-based framework, the axoneme, grows out. The intermediate region between the BB and the axoneme is called the TZ. Mutations in the gene RPGRIP1L lead to a broad spectrum of severe human diseases, which are based on cilia dysfunction commonly summarized as ciliopathies (Arts et al., 2007; Delous et al., 2007; Wolf et al., 2007; Zaghloul and Katsanis, 2010). Rpgrip1l is an evolutionary conserved gene encoding a protein that is located at the base of cilia (Arts et al., 2007; Delous et al., 2007; Vierkotten et al., 2007; Garcia-Gonzalo et al., 2011; Williams et al., 2011; Mahuzier et al., 2012). Rpgrip1l−/− mouse embryos display a ciliopathy phenotype with defects in numerous organs and die, at the latest, around birth (Vierkotten et al., 2007; Besse et al., 2011; Gerhardt et al., 2013).
Protein degradation is an essential process to ensure proper cellular signaling and hence a normal development of an organism (Liu et al., 2012). In eukaryotes, ∼80–90% of all proteins are degraded by the 26S proteasome (Lilienbaum, 2013), which functions as catalytic component of the ubiquitin–proteasome system that consists of 19S and 20S subunits. Proteins become phosphorylated as well as ubiquitinated and bind to the 19S regulatory complex. Subsequently, they are degraded by the multiple peptidase activities containing 20S subunit (Coux et al., 1996). In addition to controlling protein degradation, the proteasome is able to cleave one or more peptide bonds of a target protein to modify its activity, an event known as proteolytic processing. As a result of its importance for the degradation or processing of proteins, the ubiquitin–proteasome system is involved in the regulation and proper functioning of numerous signaling pathways. For example, the proteasome is responsible for the degradation of β-Catenin, the mediator of canonical Wnt signaling, as well as for the processing of Gli3, an important component of the Sonic hedgehog (Shh) pathway. In case of Shh signaling, the presence of the ligand Shh induces the activation of Gli3-190 which then switches on the transcription of Shh target genes. In the absence of Shh, Gli3-190 is proteolytically processed to Gli3-83, the predominant repressor of Shh target gene transcription (Wang et al., 2000). Previously, different studies described an association between proteasomal degradation and ciliary proteins. For example, the loss of Bbs4, Bbs7, and Ofd1 results in a decreased proteasomal activity, respectively (Gerdes et al., 2007; Gascue et al., 2012; Liu et al., 2014). Furthermore, proteasomal degradation is involved in ciliogenesis and the intraflagellar transport (Guo et al., 2011; Zalli et al., 2012; Kasahara et al., 2014).
Interestingly, a link between primary cilia and the autophagy–lysosome system, another protein degradation mechanism, has been found (Pampliega et al., 2013; Tang et al., 2013). Autophagy is characterized by the formation of autophagosomes, double-membrane vesicles. These autophagosomes enclose their targets and then fuse their external membrane with lysosomes to degrade their cargo. Targets of autophagy can be long-lived or aggregated proteins as well as intracellular pathogens and cellular organelles (Farré et al., 2008; Kraft et al., 2008; Orvedahl et al., 2011; Rott et al., 2011; Wang and Klionsky, 2011; Thurston et al., 2012; Lilienbaum, 2013). It was shown that ciliary signaling induces autophagy (Pampliega et al., 2013).
Analyzing the molecular background of ciliopathies based on Rpgrip1l deficiency in this study, we uncovered that Rpgrip1l regulates proteasomal activity. However, it does not control total cellular proteasomal activity per se but exclusively at primary cilia. Our data reveal the localization of proteasomal components at the ciliary base and a decreased activity of the ciliary proteasome in the absence of Rpgrip1l. We show that Rpgrip1l interacts with Psmd2, a proteasomal 19S subunit component, at the ciliary TZ. These observations are consistent with the hypothesis that Rpgrip1l realizes the regulation of proteasomal activity at the ciliary base via Psmd2.
Results
Rpgrip1l is located at the ciliary TZ in G0 phase and at centrosomes during mitosis
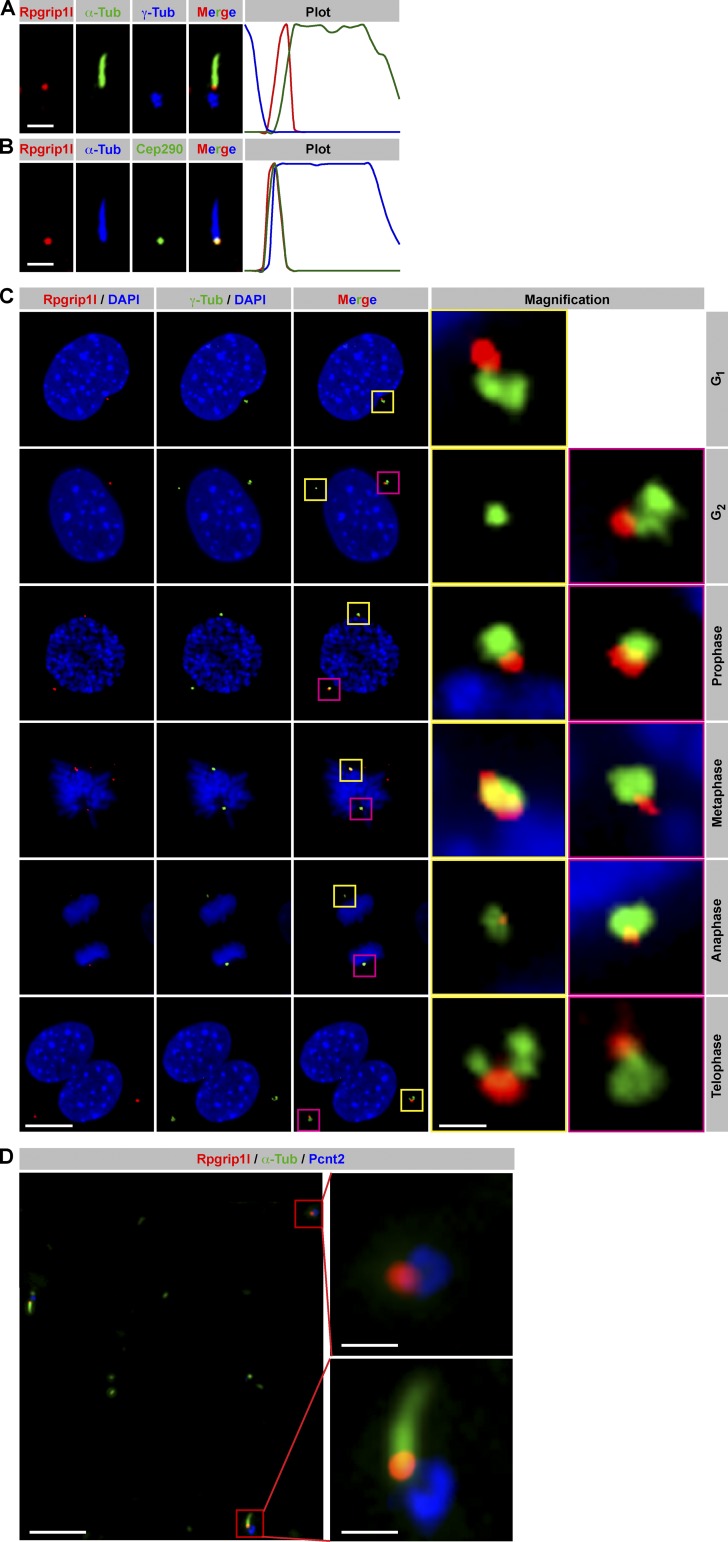
Originally, Rpgrip1l has been shown to be a BB protein (Arts et al., 2007; Delous et al., 2007; Vierkotten et al., 2007). More recent reports describe a more detailed localization at the ciliary TZ (Garcia-Gonzalo et al., 2011; Williams et al., 2011; Mahuzier et al., 2012). We choose mouse embryonic fibroblasts (MEFs) as in vitro system for analyzing Rpgrip1l function. In these primary cells, Rpgrip1l is located at the TZ during G0 phase (Fig. 1, A and B) and at both centrosomes during mitosis (Fig. 1 C). In embryonic day 12.5 (E12.5) limb cilia, Rpgrip1l is also present at the TZ and at centrosomes (Fig. 1 D). This means that in vivo studies of limbs, a well-known system for studying functional mechanisms, can be used to complement the data of MEFs.
Figure 1.
Rpgrip1l localizes at the ciliary TZ in G0 phase and at centrosomes during mitosis. (A–C) Immunofluorescence on MEFs (isolated from E12.5 embryos). (A and B) Plots indicate fluorescent intensities of each channel along the cilium from base (left) to tip (right). (A) The ciliary axoneme is marked by acetylated α-tubulin (α-Tub) and the BB by γ-tubulin. (B) The ciliary axoneme is marked by acetylated α-tubulin (α-Tub), and the TZ is marked by Cep290. (C) Centrosomes are marked by γ-tubulin, and cell nuclei are marked by DAPI. Insets illustrate higher magnifications of boxed regions. (D) Immunofluorescence on E12.5 murine limbs. The ciliary axoneme is marked by acetylated α-tubulin and centrosomes by Pcnt2. Bars: (A and B) 1 µm; (C) 10 µm; (D, overview) 5 µm; (D, magnification) 1 µm.
Rpgrip1l controls ciliary length
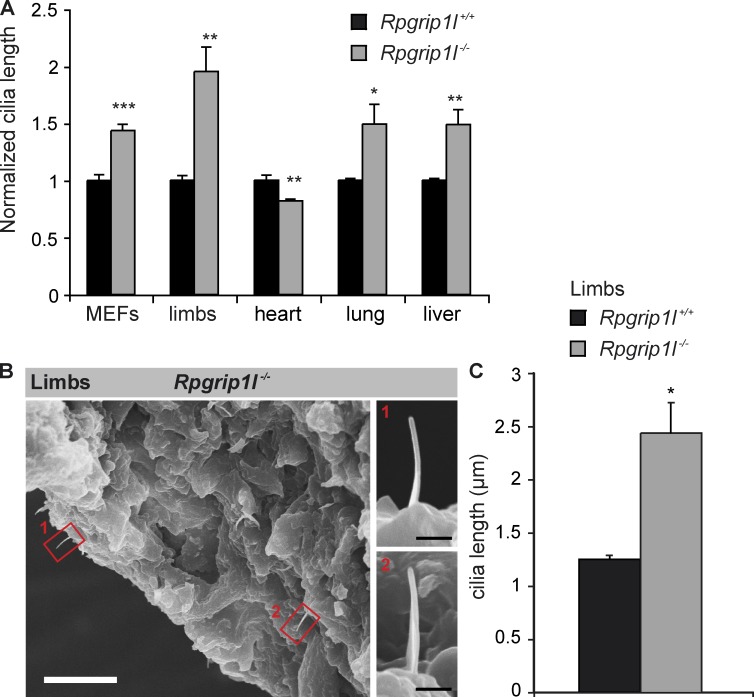
The absence of particular TZ proteins results in differences in ciliary length (Tammachote et al., 2009; Cui et al., 2011; Dowdle et al., 2011; Garcia-Gonzalo et al., 2011). Such alterations are indicative for ciliary signaling defects (Cui et al., 2011; Dowdle et al., 2011; Garcia-Gonzalo et al., 2011; Ishikawa and Marshall, 2011; Larkins et al., 2011). We measured the length of cilia in Rpgrip1l−/− MEFs and in different organs of Rpgrip1l−/− mouse embryos by using immunofluorescence. Although the cilia in Rpgrip1l−/− MEFs, limbs, livers, and lungs are significantly longer, the length of cardiac cilia is significantly reduced (Fig. 2 A). We evaluated the quality of our fluorescence-based ciliary length measurements by comparing these results with previously published scanning EM (SEM) data. The absolute value we measured for wild-type (WT) limb cilia is 1.25 ± 0.05 µm (standard error of the mean; Fig. 2 C), which correlates with SEM quantifications (Haycraft et al., 2005; Moon et al., 2014). Moreover, we performed SEM analyses to quantify the length of Rpgrip1l−/− limb cilia. The elongation of limb cilia, in the absence of Rpgrip1l, was verified by these SEM experiments (Fig. 2, B and C).
Figure 2.
Rpgrip1l deficiency affects ciliary length. (A and B) MEFs were isolated from E12.5 WT and Rpgrip1l−/− embryos. (A) Comparison of WT and Rpgrip1l−/− ciliary length. Embryonic organs were separated from E12.5 embryos. At least 20 cilia per embryo were analyzed. n values (n refers to the number of embryos) are as follows: MEFs: WT, n = 4; Rpgrip1l−/−, n = 5; limbs: WT, n = 4; Rpgrip1l−/−, n = 5; hearts: WT, n = 4; Rpgrip1l−/−, n = 5; lungs: WT, n = 4; Rpgrip1l−/−, n = 4; livers: WT, n = 4; Rpgrip1l−/−, n = 4. Error bars show standard error of the mean. (B) Elongation of E12.5 Rpgrip1l−/− limb cilia are confirmed in comparison to fluorescence based cilia length measurement (10 cilia; SEM: 2.88 ± 0.14 µm; immunofluorescence: 2.44 ± 0.265 µm). Magnifications (termed as 1 and 2) show two examples of Rpgrip1l−/− limb cilia. Bars: (overview) 10 µm; (magnification) 1 µm. (C) Absolute values of the quantified ciliary length of WT (n = 4 embryos) and Rpgrip1l−/− (n = 5 embryos) limbs at E12.5 based on immunofluorescence. Rpgrip1l-negative cilia (2.44 ± 0.265 µm [standard error of the mean]) are significantly longer than WT cilia (1.25 µm ± 0.05 µm [standard error of the mean]). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Rpgrip1l regulates proteasomal activity at the ciliary base
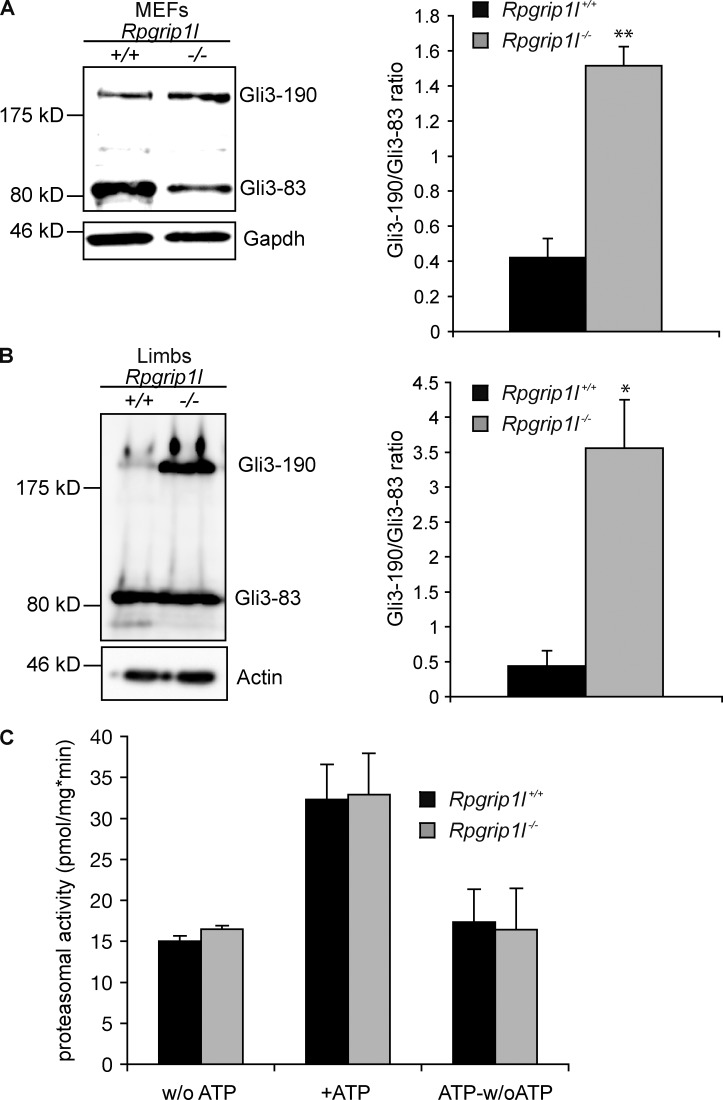
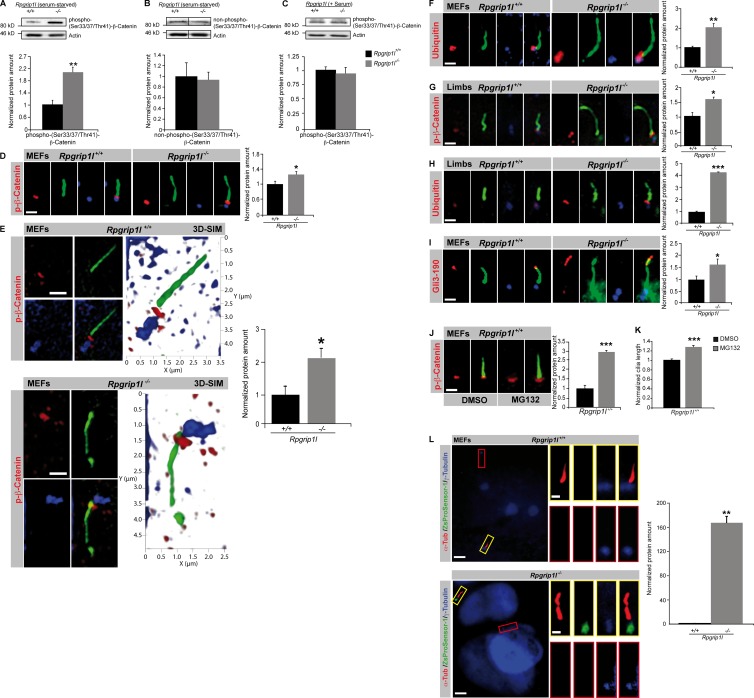
The observations of significant elongation of cilia in our in vitro and in vivo model systems led us to the question if ciliary signaling is affected in these systems. Previously, we found an elevated Gli3-190/Gli3-83 ratio in Rpgrip1l−/− mouse embryos as a result of a reduced proteolytic processing of Gli3 (Vierkotten et al., 2007). Consistent with this suspicion, the Gli3-190/Gli3-83 ratios are also elevated in Rpgrip1l−/− MEFs and isolated limbs (Fig. 3, A and B). Because previous studies showed that the deficiency of other TZ proteins affects events upstream of Gli3 in the Shh signal cascade (Garcia-Gonzalo et al., 2011) and since Smo regulates Gli3 modification via the Evc–Evc2 complex (Dorn et al., 2012), we analyzed the amount of Smo and Evc in cilia of SAG (Shh agonist)-treated Rpgrip1l−/− MEFs. We could not detect any alteration in the amount of either protein in cilia of Rpgrip1l−/− MEFs (Fig. S1, A and B) making it likely that Rpgrip1l affects Shh signaling at the level of Gli3. In search of the cause for the increased Gli3-190/Gli3-83 ratio, we investigated proteasomal activity. However, changes in overall cellular proteasomal activity were not detected in Rpgrip1l−/− MEFs (Fig. 3 C). Because Gli3-processing is dependent on cilia (Haycraft et al., 2005; Besse et al., 2011), we could not exclude that proteasomal activity is reduced exclusively at cilia. Potentially, an alteration of proteasomal degradation specific at cilia is undetectable in measurements of general proteasomal substrates. To test this hypothesis, we measured the amount of phospho-(S33/37/T41)-β-Catenin, a well-known proteasomal substrate (Aberle et al., 1997; Hart et al., 1999; Kitagawa et al., 1999; Latres et al., 1999; Liu et al., 1999; Winston et al., 1999). A previous study showed a localization of phospho-(S33/37/T41)-β-Catenin at the ciliary base (Corbit et al., 2008), suggesting that this is the location of its degradation. In total lysates of serum-starved Rpgrip1l−/− MEFs (on average, 82% of all cells had cilia), the amount of phospho-(S33/37/T41)-β-Catenin is significantly increased in comparison to serum-starved WT MEFs (on average, 88.6% of all cells produced cilia; Fig. 4 A), whereas non–phospho-(S33/37/T41)-β-Catenin is unaltered (Fig. 4 B). In total lysates of non–serum-starved Rpgrip1l−/− MEFs (on average, 4% of all cells possessed cilia), the amount of phospho-(S33/37/T41)-β-Catenin is not changed in comparison to non–serum-starved WT MEFs (on average, 6.67% of all cells had cilia; Fig. 4 C), demonstrating that the degradation of phospho-(S33/37/T41)-β-Catenin is cilia dependent. The amount of phospho-(S33/37/T41)-β-Catenin is significantly increased at the ciliary base of Rpgrip1l−/− MEFs (Fig. 4 D). To confirm these data, which were collected by using standard resolution microscopy, we also measured the amount of phospho-(S33/37/T41)-β-Catenin using super-resolution microscopy. 3D structured illumination microscopy (SIM; Gustafsson et al., 2008; Schermelleh et al., 2008) analyses confirmed the finding of significantly elevated levels of phospho-(S33/37/T41)-β-Catenin at the ciliary TZ in Rpgrip1l−/− MEFs (Fig. 4 E).
Figure 3.
Rpgrip1l deficiency leads to elongation of limb cilia and an increase of the Gli3-190/Gli3-83 ratio but does not alter overall cellular proteasomal activity. (A and B) Western blot analysis of MEFs and E12.5 limbs (WT: n = 3 embryos; Rpgrip1l−/−: n = 3 embryos, respectively). Gapdh (A) and Actin (B) serve as loading controls. Evaluation of the Gli3-190/Gli3-83 ratio depicts a significant increase in the Rpgrip1l−/− situation of 3.6-fold (MEFs, A) and 8.1-fold (limbs, B). (C) Proteasomal activity was measured in total cell lysates of MEFs isolated from E12.5 WT (n = 8) and Rpgrip1l−/− embryos (n = 4). No alteration is detectable in Rpgrip1l−/− MEFs in comparison to WT MEFs either without ATP (activity of the 20S proteasomal subunit) or after ATP addition (activity of the 20S proteasomal subunit and 26S proteasome together). There is also no alteration in the activity of the 26S proteasome alone (ATP-w/o ATP), demonstrating that the activity of the 19S proteasomal subunit is unchanged in the absence of Rpgrip1l. Error bars show standard error of the mean. *, P < 0.05; **, P < 0.01.
Figure 4.
Rpgrip1l deficiency causes impaired proteasomal activity at primary cilia. (A–F and I–L) MEFs were isolated from E12.5 WT and Rpgrip1l−/− embryos. (A and B) Western blot analysis of WT and Rpgrip1l−/− MEF lysates (n = 4 embryos, respectively). (C) Western blot analysis of WT and Rpgrip1l−/− MEF lysates (n = 3 embryos, respectively). (A–C) Actin serves as a loading control. (A) Phospho-(S33/37/T41)-β-Catenin is significantly increased in serum-starved Rpgrip1l−/− MEF lysates (82% of all cells had cilia; in serum-starved Rpgrip1l+/+ MEF lysates, 88.67% of all cells possessed cilia) but not in non–serum-starved Rpgrip1l−/− MEF lysates (4% of all cells displayed cilia; in non–serum-starved Rpgrip1l+/+ MEF lysates, 6.67% of all cells carried cilia; C). (B) Non–phospho-(S33/37/T41)-β-Catenin is unaltered in serum-starved Rpgrip1l−/− MEF lysates. Black lines indicate that intervening lanes have been spliced out. (D–F, I, and L) Immunofluorescence on MEFs of E12.5 WT and Rpgrip1l−/− embryos (both genotypes: p-β-Catenin: n = 5; p-β-Catenin (3D-SIM, n = 3; Ubiquitin, n = 4; Gli3-190, n = 6; ZsProSensor-1, n = 3; n refers to the number of embryos, respectively). Per embryo, 15 cilia were quantified for p-β-Catenin, 10 cilia were quantified for p-β-Catenin (3D-SIM) and for Ubiquitin, and 20 cilia were quantified for Gli3-190. (G and H) Immunofluorescence on limbs of E12.5 WT and Rpgrip1l−/− embryos (n = 3 embryos, respectively). Per embryo, 20 cilia were quantified for p-β-Catenin and Ubiquitin. All quantified proteins are shown in red (D–J), the ciliary axoneme is marked by acetylated α-tubulin (green; D–J), and the BB is marked by γ-tubulin (blue; D–F, H, and I) or by Pcnt2 (blue; G). (J and K) Immunofluorescence on MEFs of WT embryos (n = 4). 25 cilia per embryo were used for phospho-(S33/37/T41)-β-Catenin and cilia length quantification. (L) Proteasome activity assay on WT and Rpgrip1l−/− MEFs. Cilia are marked by acetylated α-tubulin (α-Tub), and centrosomes/basal bodies are marked by γ-tubulin. Colored squares mark cilia with basal bodies (yellow squares) as well as centrosomes (red squares), which are presented magnified. The green ZsProSensor-1 protein signal is exclusively detected at the ciliary base in Rpgrip1l−/− MEFs. Error bars show standard error of the mean. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Bars, 1 µm.
If proteasomal degradation occurs at the ciliary base, ubiquitinated proteins should accumulate there. We used an antibody specifically recognizing an epitope of ubiquitin proteins and detected an accumulation at cilia of WT MEFs. In Rpgrip1l−/− MEFs, there are significantly more ubiquitinated proteins at the ciliary base (Fig. 4 F) consistent with a decrease of proteasomal activity in the absence of Rpgrip1l. In line with these findings in our in vitro model, the amount of phospho-(S33/37/T41)-β-Catenin and Ubiquitin is also significantly elevated at the ciliary base of Rpgrip1l−/− limbs (Fig. 4, G and H). Additionally, the ciliary amount of Gli3-190 is significantly elevated in Rpgrip1l−/− MEFs (Fig. 4 I). To test whether the elevation of the different proteins at cilia of Rpgrip1l−/− MEFs could be caused by a dysfunction of the proteasome, we treated WT MEFs with the proteasome inhibitor MG132 (Rock et al., 1994) and expected to provoke a situation comparable to the Rpgrip1l−/− state. After MG132 treatment, the amount of phospho-(S33/37/T41)-β-Catenin is significantly increased at the ciliary base of WT MEFs (Fig. 4 J). Furthermore, MG132 treatment results in a significant elongation of MEF cilia (Fig. 4 K), mimicking the increased ciliary length of Rpgrip1l−/− MEFs (Fig. 2 A). In a parallel approach, we tested whether a fluorescence reporter system could be used to evaluate whether the activity of the proteasome is decreased in the absence of Rpgrip1l. In general, these reporters are rapidly diffusing and do not accumulate at specific subcellular localizations. Therefore, they may not be ideal for measuring proteasomal activity at the ciliary base. However, differential proteasomal activity in the nuclear and cytoplasmic compartments of MBECs and HepG2 cells using the Proteasome Sensor Vector has been previously reported (Zheng et al., 2006). This vector encodes the ZsProSensor-1 protein, a fusion of a GFP with a degradation domain, which targets the protein for rapid degradation by the proteasome. We transfected the Proteasome Sensor Vector into WT as well as Rpgrip1l−/− MEFs and detected the green fluorescence signal at 88.9% of all analyzed Rpgrip1l−/− MEF cilia (n = 16 out of 18 cilia) but never at WT cilia (n = 0 out of 37 cilia; Fig. 4 L), demonstrating a diminished activity of the proteasome. Consequently, the amount of ZsProSensor-1 is significantly elevated at the ciliary base of Rpgrip1l−/− MEFs (Fig. 4 L). Interestingly, the accumulation of the green fluorescence signal in Rpgrip1l−/− MEFs was observed exclusively at cilia and not at centrosomes or other subcellular locations (Fig. 4 L).
Based on these observations, it appears that Rpgrip1l regulates proteasomal activity in a cilia-dependent manner. Because Rpgrip1l localizes to the ciliary TZ, it is possible that it controls proteasomal activity directly at primary cilia. To our knowledge, the existence of a ciliary proteasome (a proteasome that localizes and functions at cilia) has been assumed (Wen et al., 2010) but not proven until now.
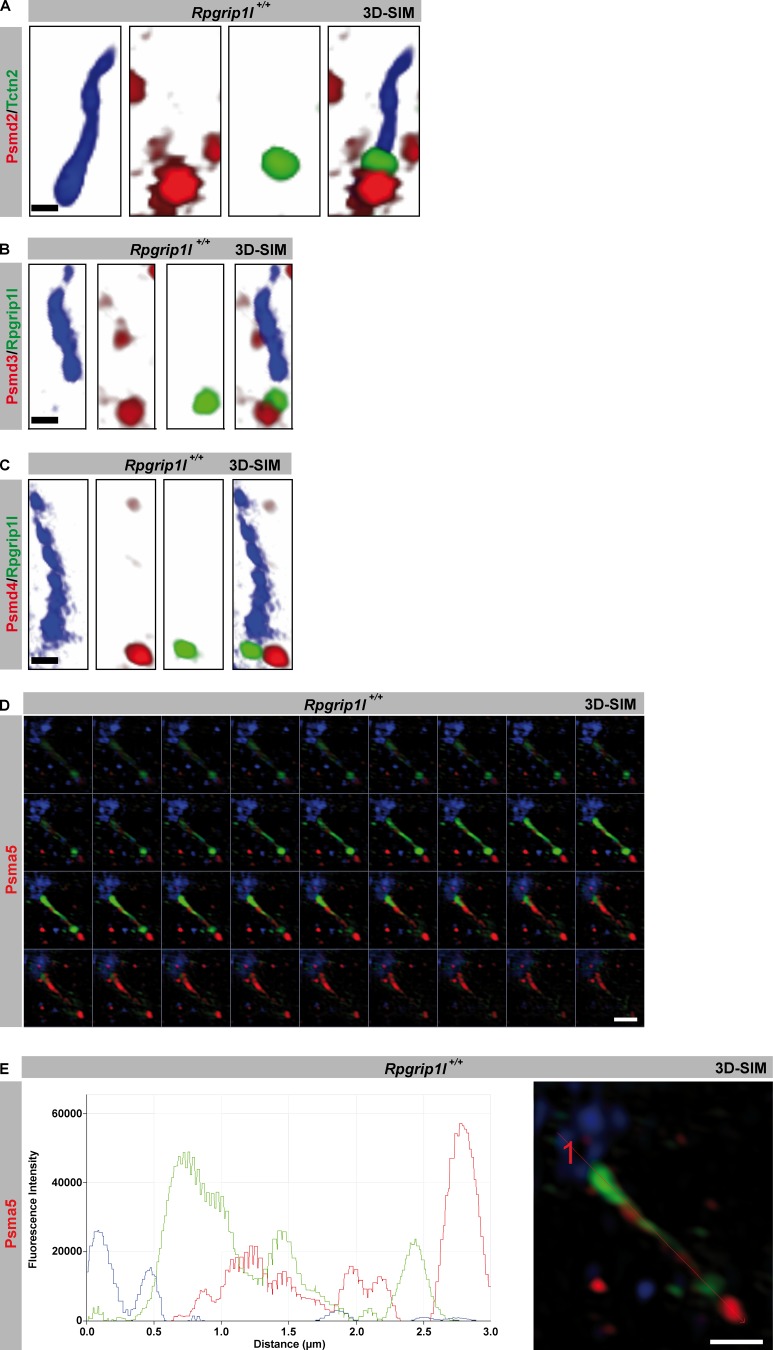
A prerequisite for a proteasome to function at the base of cilia is the presence of components of the 19S and 20S proteasomal subunits. Psmd2, Psmd3, and Psmd4, which are components of the 19S proteasomal subunit, are found at the BB of WT MEF cilia by using 3D-SIM (Fig. 5, A–C), supporting the assumed existence of a ciliary proteasome. Psma5, which is a component of the 20S proteasomal subunit is detected at the ciliary base (BB as well as TZ) and along the axoneme of WT MEFs (Fig. 5, D and E), indicating a transport of Psma5 through the cilium. Assuming a transport of Psma5 along the cilium, we propose the BB as the location were the ciliary proteasome localizes and functions. This is the most likely scenario because we never detected components of the 19S proteasomal subunit along the axoneme of MEF cilia.
Figure 5.
Components of the S19 and S20 proteasomal subunits localize at primary cilia. (A–E) Immunofluorescence on MEFs of E12.5 WT and Rpgrip1l−/− embryos. Higher resolution was achieved by using SIM. The ciliary axoneme is marked by acetylated α-tubulin (blue). The TZ is marked by Tctn2 (A) or Rpgrip1l (B and C). (D and E) The ciliary axoneme is marked by acetylated α-tubulin (green) and the BB by γ-tubulin (blue). Psma5 is shown in red. (D) Z-stack of the single optical sections from E with the appropriate plot of the depicted representative image. Bars: (A–C) 0.5 µm; (D and E) 1 µm.
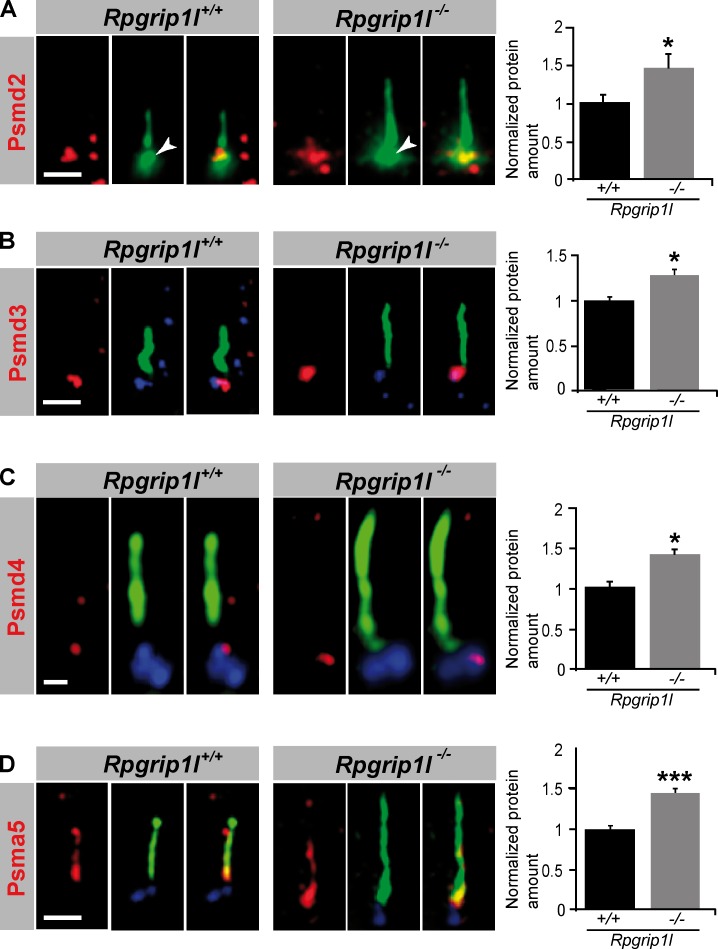
It has been reported that inhibition of the proteasome leads to an accumulation of proteasomal subunits at the centrosome (Fabunmi et al., 2000). So the question arises whether proteasomal components accumulate in a significantly higher amount at the ciliary base of Rpgrip1l−/− MEFs. All analyzed proteasomal components, Psmd2, Psmd3, Psmd4 and Psma5, were significantly increased at the ciliary base in Rpgrip1l−/− MEFs (Fig. 6, A–D), underscoring the likelihood that Rpgrip1l deficiency affects proteasomal activity at primary cilia.
Figure 6.
Rpgrip1l deficiency results in an accumulation of proteasomal subunit components at the base of cilia. (A–D) Immunofluorescence on MEFs of E12.5 WT and Rpgrip1l−/− embryos (both genotypes: Psmd2, n = 7 embryos; Psmd3, n = 3 embryos; Psmd4, n = 3 embryos; Psma5, n = 5 embryos). At least 10 cilia per embryo were used for Psmd2, Psmd3, and Psmd4 quantification, respectively, and 20 cilia per embryo were used for Psma5 quantification. The ciliary axoneme is marked by acetylated α-tubulin (green). The BB is marked by Pcnt (green; white arrowheads; A) or γ-tubulin (blue; B–D). Error bars show standard error of the mean. *, P < 0.05; ***, P < 0.001. Bars: (A, B, and D) 1 µm; (C) 0.5 µm.
Rpgrip1l interacts with Psmd2, a component of the proteasomal 19S subunit
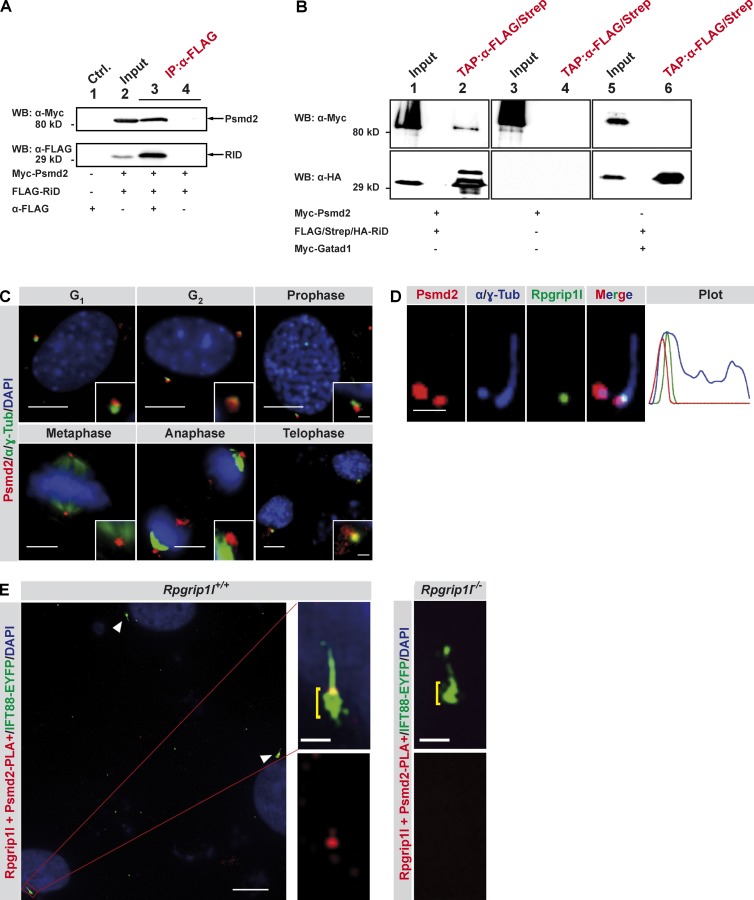
To unravel how Rpgrip1l regulates the activity of the proteasome, we searched for novel interaction partners of Rpgrip1l by performing a yeast two-hybrid screen. Among a total of six proteins, we found Psmd2 as a potential interaction partner (Fig. S1 C). We used the tagged RPGR-interacting domain (RID) of Rpgrip1l (the full-length Rpgrip1l could not be stably expressed) and a tagged full-length Psmd2 for transient overexpression in NIH3T3 and HEK293T cells. Using coimmunoprecipitation and tandem affinity purification assays, we confirmed the interaction of Rpgrip1l with Psmd2 (Fig. 7, A and B).
Figure 7.
Rpgrip1l interacts with the 19S proteasomal subunit component Psmd2. (A) Coimmunoprecipitation experiments in NIH3T3 cells. Myc-tagged Psmd2 full-length protein and FLAG-tagged RID were transiently overexpressed and tested for interaction by coimmunoprecipitation from total cell lysates. Immunoprecipitation assays were performed by using an anti-FLAG antibody. Myc-Psmd2 coimmunoprecipitated with FLAG-RID (lane 3). Ctrl., control; IP, immunoprecipitation; WB, Western blot. (B) Myc-tagged Psmd2 full-length protein and FLAG/Strep/HA-tagged RID domain of Rpgrip1l were transiently overexpressed in HEK293T cells and tested for interaction by tandem affinity purification (TAP) tag experiments from total cell lysates. FLAG/Strep/HA-tagged RID was immunopurified using anti-FLAG beads, eluted with FLAG-protein, and recaptured on anti-Strep beads. The final eluation was performed by using biotin. After this sequential purification, we identified Myc-Psmd2 (lane 2) but not the unrelated protein Gatad1 (lane 6) in addition to purified FLAG/Strep/HA-RID. (C–E) Immunofluorescence on MEFs isolated from WT or Rpgrip1l−/− E12.5 embryos (n = 3 embryos, respectively). (C) Centrosomes are marked by γ-tubulin as well as acetylated α-tubulin (α-Tub), and cell nuclei were marked by DAPI. Insets illustrate higher magnifications. (D) The ciliary axoneme and the BB are marked by acetylated α-tubulin and by γ-tubulin, respectively. The plot shows fluorescence intensities of the depicted representative image. (E) In situ proximity ligation assay (in situ PLA) on MEFs. Cell nuclei are marked by DAPI, and the ciliary axoneme are marked by transiently transfected Ift88-EYFP. Additional accumulation of Ift88-EYFP at the ciliary base is highlighted by yellow brackets. White arrowheads point to cilia. Bars: (C, all images; and D, overview) 10 µm; (D and E, magnifications) 1 µm.
Proteasomes have been detected in the nucleus, in the microsomes, in the cytosol, at the centrosomes and at the base of cilia (Wigley et al., 1999; Brooks et al., 2000; Gerdes et al., 2007). Interestingly, both Rpgrip1l and Psmd2, are located at centrosomes during mitosis (Fig. 1 C and Fig. 7 C). Because the mother centriole functions as ciliary BB, this finding indicates that their interaction might occur at the ciliary base. Immunofluorescence on ciliated MEFs reveals a partial overlap of Rpgrip1l and Psmd2 staining at the TZ (Fig. 7 D), indicating that Psmd2 localizes mainly at the BB but also partially at the TZ. This identifies the TZ as a potential Rpgrip1l–Psmd2 interaction site. To further investigate this question, we used a technique called in situ proximity ligation assay (PLA) in which an interaction is detected via a fluorescence signal (Söderberg et al., 2006). We tested its applicability for this approach by investigating the already known interaction of Rpgrip1l with Nphp4 in the TZ. In this assay, a PLA signal can be observed at the TZ of WT MEF cilia in 100% of the cases, whereas we did not detect a PLA signal at Rpgrip1l−/− MEF cilia (Fig. S1 D). Analyzing the interaction of Rpgrip1l with Psmd2, the PLA signal is always present at the TZ of WT MEF cilia, but we never detected a PLA signal at Rpgrip1l−/− cilia (Fig. 7 E), indicating that Rpgrip1l and Psmd2 interact at the ciliary TZ of MEFs. Although Rpgrip1l and Psmd2 are observed at the centrosomes in the course of mitosis (Fig. 1 C and Fig. 7 C), we detected the Rpgrip1l/Psmd2 PLA signal exclusively in the presence of cilia (Fig. 7 E) but not at centrosomes (Fig. S1 F). These findings suggest that Rpgrip1l regulates the activity of a cilia-specific proteasome directly at the TZ of primary cilia and that this regulation may be realized via Psmd2.
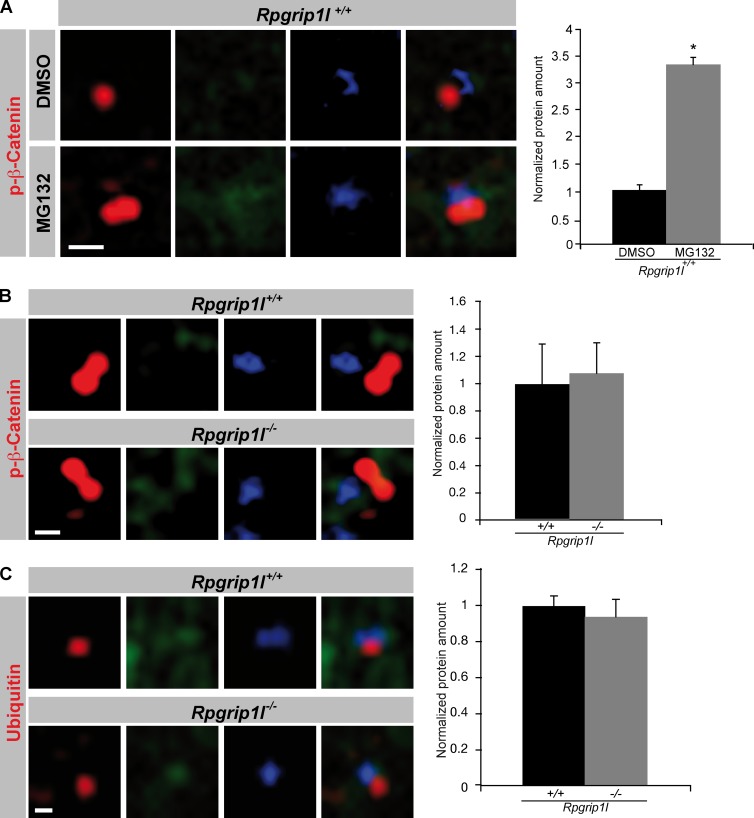
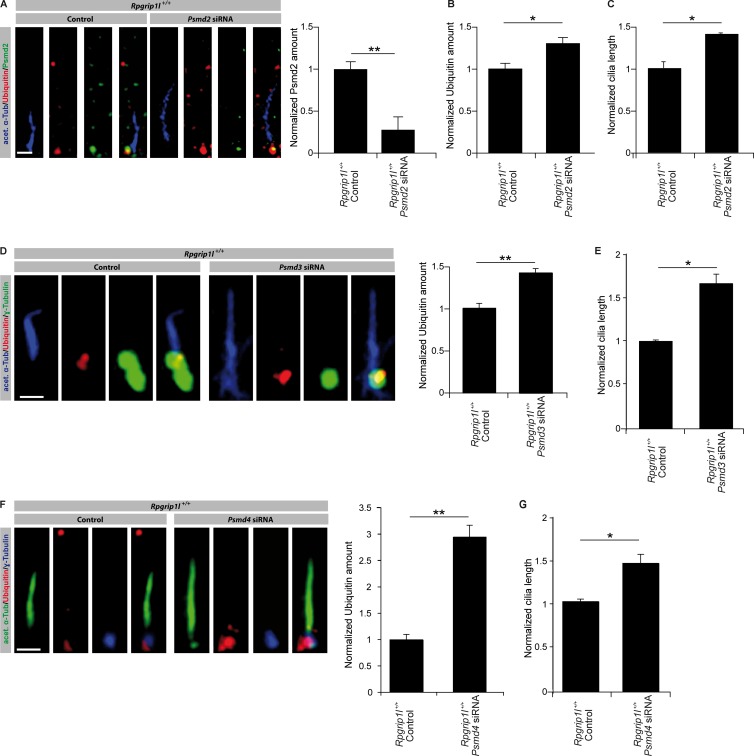
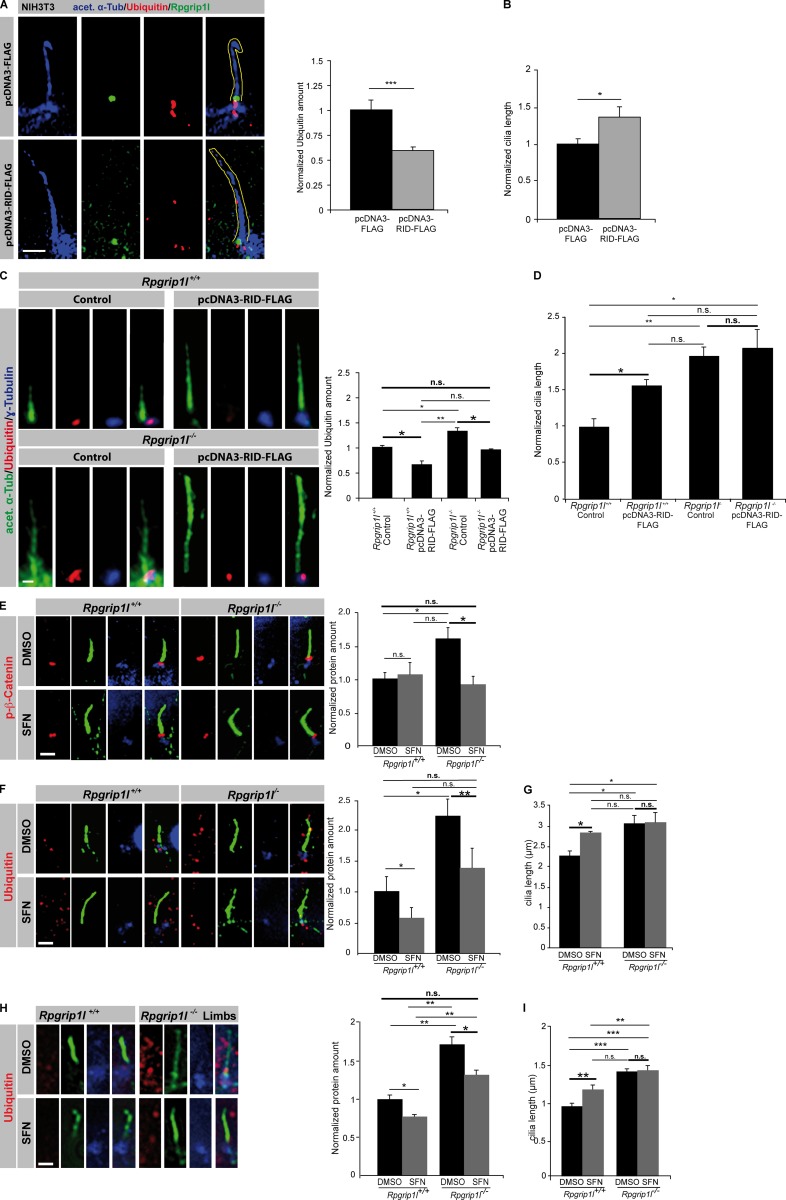
Because in MEFs, the interaction of Rpgrip1l and Psmd2 seems to take place only at the ciliary TZ, but Rpgrip1l is also located at centrosomes during mitosis, the question arises whether Rpgrip1l also regulates the activity of the centrosomal proteasome. Proteasomal degradation at the centrosome has previously been reported for ubiquitinated proteins (Fabunmi et al., 2000; Puram et al., 2013). Using MG132, an inhibitor of proteasomal activity, we were able to show that the centrosomal (pericentriolar) proteasome is also responsible for the degradation of phospho-(S33/37/T41)-β-Catenin (Fig. 8 A). In contrast to the situation at the ciliary base, the amount of phospho-(S33/37/T41)-β-Catenin and ubiquitinated proteins is unaltered at the centrosomes of Rpgrip1l−/− MEFs (Fig. 8, B and C). These results correspond to the finding that the green fluorescence signal of the Proteasome Sensor Vector exclusively accumulates at the ciliary base in the absence of Rpgrip1l (Fig. 4 L), indicating that Rpgrip1l only affects the activity of the ciliary proteasome in MEFs. Consequently, our data reveal a regulation of proteasomal activity by Rpgrip1l exclusively at primary cilia and most likely via the interaction with Psmd2. To evaluate whether Rpgrip1l deficiency affects the activity of the 26S proteasome by affecting the function of the 19S proteasomal subunit, the amount of ubiquitinated proteins was determined at the ciliary base of MEFs in which several components of the 19S proteasomal subunit (Psmd2, Psmd3, or Psmd4) were knocked down. In MEFs, Psmd2, Psmd3, and Psmd4 knockdowns result in a significantly higher amount of ubiquitinated proteins at the ciliary base (Fig. 9, A, B, D, and F; and Fig. S2, A and B), mimicking the Rpgrip1l−/− status. This result suggests that Rpgrip1l regulates the activity of the ciliary proteasome by affecting the 19S proteasomal subunit. Remarkably, knockdowns of all these 19S proteasomal subunit components lead to significant ciliary elongation in MEFs (Fig. 9, C, E, and G). Considering that Rpgrip1l interacts with Psmd2 by means of its RID domain, we tested whether Rpgrip1l overexpression increases the activity of the ciliary proteasome by transfecting the RID domain of Rpgrip1l into NIH3T3 cells as well as into WT MEFs. We quantified the amount of Ubiquitin at the ciliary base as well as ciliary length. The amount of Ubiquitin was significantly decreased at the ciliary base of RID-transfected NIH3T3 cells and WT MEFs (Fig. 10, A and C), indicating that the ciliary proteasome is regulated by Rpgrip1l via its interaction with Psmd2 at the ciliary TZ. Interestingly, cilia were significantly longer after RID transfection (Fig. 10, B and D). To exclude that these effects are caused by an overexpression of any type of protein, we also transfected a control protein (Wnk2) into NIH3T3 cells and detected no alteration in Ubiquitin amount or cilia length (Fig. S2, C and D). Because the transfection of the RID domain leads to an increased activity of the ciliary proteasome, we also transfected the RID domain into Rpgrip1l−/− MEFs to elevate the activity of the ciliary proteasome. Indeed, the ciliary amount of Ubiquitin was restored in these MEFs (Fig. 10 C). However, ciliary length of Rpgrip1l−/− MEFs was unaltered after transfection (Fig. 10 D), implying that the RID domain of Rpgrip1l is essential for the regulation of the ciliary proteasome but is dispensable for the regulation of cilia length. Consequently, another part of the Rpgrip1l protein is needed to control the length of cilia.
Figure 8.
Proteasomal activity is unaltered at Rpgrip1l−/− centrosomes. (A–C) Immunofluorescence on MEFs isolated from E12.5 WT and Rpgrip1l−/− embryos (WT: p-β-Catenin (treated with DMSO or MG132): n = 3 embryos; both genotypes: p-β-Catenin and Ubiquitin: n = 3 embryos, respectively). The ciliary axoneme is marked by acetylated α-tubulin (green) and the centrosomes (basal bodies in case of ciliary presence) by γ-tubulin (blue). All quantified proteins are shown in red. An axonemal-like green staining is not visible, demonstrating that the blue staining marks centrosomes. (A) After treatment of WT MEFs with the proteasome inhibitor MG132, the amount of phospho-(S33/37/T41)-β-Catenin is significantly increased at the centrosome. (B and C) The amounts of phospho-(S33/37/T41)-β-Catenin and Ubiquitin are unaltered at the centrosome of Rpgrip1l−/− MEFs. (A–C) Per embryo, 20 cilia were used in the quantifications. Error bars show standard error of the mean. *, P < 0.05. Bars, 0.5 µm.
Figure 9.
Knockdown of the 19S proteasomal subunit components Psmd2, Psmd3, and Psmd4 reduce the activity of the ciliary proteasome. (A–G) Immunofluorescence on MEFs of E12.5 WT embryos (both genotypes: n = 3 embryos). (A) The ciliary axoneme is marked by acetylated α-tubulin (acet. α-Tub). Transfection of MEFs with siRNA against Psmd2 and quantification of the ciliary Psmd2 amount (A) and the ciliary Ubiquitin amount (B). (C) Measurement of cilia length after treatment with siRNA against Psmd2. (D and E) Transfection of MEFs with siRNA against Psmd3. (D) Quantification of Ubiquitin amount at cilia. The ciliary axoneme is marked by acetylated α-tubulin (acet. α-Tub)and the BB by γ-tubulin. (E) Measurement of cilia length. (F and G) Transfection of MEFs with siRNA against Psmd4. (F) Quantification of Ubiquitin amount at cilia. The ciliary axoneme is marked by acetylated α-tubulin (acet. α-Tub) and the BB by γ-tubulin. (G) Measurement of cilia length. Error bars show SEM. *, P < 0.05; **, P < 0.01. Bars: (A) 0.5 µm; (D and F) 1 µm.
Figure 10.
Transfection of the RID domain and SFN treatment increase proteasomal activity and rescue the decreased activity of the ciliary proteasome caused by Rpgrip1l deficiency. (A and B) Immunofluorescence on NIH3T3 cells. Per control and per transfected cells, 20 cilia were used for quantification. The ciliary axoneme is marked by acetylated α-tubulin (acet. α-Tub). (A) Yellow lines encircle the shape of cilia. (C–I) The most important significance comparisons are written in bold with larger asterisks as are larger not significant comparisons. (C and D) Immunofluorescence on MEFs of E12.5 WT and Rpgrip1l−/− embryos (both genotypes, n = 3 embryos). At least 10 cilia per embryo were used for quantification. The ciliary axoneme is marked by acetylated α-tubulin (acet. α-Tub) and the BB by γ-tubulin. (E–G) Immunofluorescence on MEFs isolated from E12.5 WT and Rpgrip1l−/− embryos (both genotypes: p-β-Catenin and Ubiquitin [treated with DMSO or SFN], n = 3 embryos, respectively). Per embryo, 20 cilia were used in the quantifications. The ciliary axoneme is marked by acetylated α-tubulin (green), and the BB is marked by γ-tubulin (blue). All quantified proteins are shown in red. (H and I) Whole embryo culture treatment of embryos isolated on E12.5 and incubated for 5 h in 20 µmol SFN (both genotypes: Ubiquitin [treated with DMSO or SFN], n = 3 embryos, respectively]). Per embryo, 20 cilia were used in the quantifications. The ciliary axoneme is marked by acetylated α-tubulin (green), and the BB is given by γ-tubulin (blue). Bars, 1 µm. Error bars show standard error of the mean. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Because Psmd2 is a 19S proteasomal subunit, and hence, Rpgrip1l may have only a direct regulatory impact on the 19S subunit, it could be possible that a drug-induced activation of the 20S proteasomal subunit leads to an increase of proteasomal activity in Rpgrip1l−/− MEFs. We treated WT and Rpgrip1l−/− MEFs with sulforaphane (SFN), which activates the catalytic proteasomal 20S subunit (Kwak et al., 2007) and detected a reduction of proteasomal substrates at the ciliary base to a WT level (Fig. 10, E and F), indicating that SFN treatment rescues the activity of the ciliary proteasome in Rpgrip1l−/− MEFs.
However, ciliary length is not recovered in Rpgrip1l−/− MEFs after SFN treatment (Fig. 10 G). Interestingly, like MG132 treatment, SFN treatment leads to a significant elongation of WT cilia (Fig. 10 G). To exclude that the findings of the SFN treatment experiments are only phenomenona in cell culture, we performed the treatment in ex vivo–cultured mouse embryos. In this experiment, the activity of the ciliary proteasome is also rescued by SFN treatment, because the Ubiquitin amount at the ciliary base in Rpgrip1l−/− embryos decreased to WT level after SFN treatment (Fig. 10 H). Like in the in vitro SFN experiment, ciliary length is significantly elevated in WT embryos and remains significantly increased in Rpgrip1l−/−, SFN-treated embryos (Fig. 10 I).
Discussion
Mutations in the gene RPGRIP1L result in various ciliopathies in humans (Zaghloul and Katsanis, 2010), accentuating it as a proper candidate gene to examine the underlying molecular dysfunction. Using the mouse as a model, we show that the absence of Rpgrip1l leads to an impairment of the ciliary proteasome, which is located at the BB of cilia. Because proteasomal function is essential for proper development and function of multiple organs (Rubinsztein, 2006; Breusing et al., 2009; Wang and Robbins, 2014), our findings broach the issue of disturbed protein degradation as a cause of ciliopathies.
In the Rpgrip1l−/− state, we detected a decrease of proteasomal activity at the ciliary base in vitro and in vivo (Fig. 4) but not at the centrosome (Fig. 8, B and C), indicating that the regulation of the proteasome via Rpgrip1l is cilia dependent. It might be that the ciliary and the centrosomal proteasome are one and the same. But, we are able to demonstrate that Rpgrip1l regulates exclusively the activity of the ciliary proteasome.
Recently, it was shown that the ciliopathy-associated proteins Bbs4 and Ofd1 are essential for the proteasomal degradation or processing of proteins involved in paracrine signaling (Liu et al., 2014). Our study confirms the discovery of ciliary base proteins as regulators of proteasomal activity and hence cilia-mediated signaling. Additionally, our data complete these findings by revealing that the ciliopathy-associated protein Rpgrip1l does not control proteasomal activity in general but specifically the activity of the ciliary proteasome. Alternatively, it was reported that the TZ protein Nphp4 affects the ubiquitin conjugation system via its interaction with Jade-1, a novel ubiquitin ligase (Borgal et al., 2012). This may indicate that the elevated amounts of proteins, which we detect at the ciliary base, could also result from defective ubiquitination. But, in contrast to this possibility, we detected an accumulation of ubiquitin at the ciliary base (Fig. 4, F and H), arguing for a proteasomal defect rather than for a ubiquitination defect. Additionally, this result also speaks against a phosphorylation defect and indicates that the elevated amount of phospho-(S33/37/T41)-β-Catenin results from impaired proteasomal activity. Thus, although Rpgrip1l and Nphp4 interact with each other (Arts et al., 2007; Delous et al., 2007) and function in the same TZ module (Czarnecki and Shah, 2012), the impact of these two proteins on protein degradation is different. Although Nphp4 has an effect on the labeling of proteins destined to become degraded by the proteasome, Rpgrip1l affects the activity of the ciliary proteasome most likely by interacting with the proteasomal 19S subunit component Psmd2 at the ciliary TZ (Fig. 7). It was shown before that Psmd2 is able to regulate proteasomal activity (Matsuyama et al., 2011). We confirmed this finding in WT MEFs by using RNAi because knockdown of Psmd2 results in a significantly decreased proteasomal activity at the ciliary base (Fig. 9, A and B). Remarkably, the presence of Psmd2 at the ciliary base does not depend on Rpgrip1l (Fig. 6 A), indicating that Rpgrip1l controls the activity of the ciliary proteasome not by anchoring Psmd2 to cilia. This is a major difference to the regulation of proteasomal activity by Bbs4 and Ofd1. Though Bbs4 and Ofd1 also interact with proteasomal components, Bbs4 and Ofd1 deficiency leads to a decreased amount of these proteasomal components (Liu et al., 2014). Thus, Bbs4 and Ofd1 are essential for the composition of the proteasome. Moreover, Bbs4 and Ofd1 are thought to control proteasomal activity within the whole cell (Liu et al., 2014), whereas Rpgrip1l is the first protein for which it is demonstrated that it specifically and exclusively regulates the activity of the proteasome at the ciliary base.
Interestingly, Bbs4 recruitment is disturbed in autophagy-deficient cells (Tang et al., 2013). Consequently, if the ciliary localization of Bbs4 is essential for proper proteasomal activity, it is possible that impaired autophagy leads to a reduced proteasomal activity. In contrast, Ofd1 is degraded by autophagy (Tang et al., 2013). Because it is unknown whether an elevated amount of Ofd1 results in an increased proteasomal activity, it is difficult to evaluate if disturbed autophagy really provokes a reduced proteasomal activity. Until now, an association between autophagy and Rpgrip1l has not been described, but studying this potential connection would help to unravel the relationship between cilia, proteasome, and autophagy.
A potential association of Rpgrip1l with proteasomal activity has been found before. Mahuzier et al. (2012) showed that Rpgrip1l knockdown results in a reduction of Dsh at the ciliary base in MDCK cells. Because Dsh is degraded by the proteasome, these data imply a negative regulation of the ciliary proteasome by Rpgrip1l in MDCK cells and hence the opposite of our results. Previous data and our cilia length measurements implicate that Rpgrip1l deficiency affects cilium biogenesis and stability depending on the cell type. For example, cilia are absent in the node and the neural tube (Vierkotten et al., 2007; Besse et al., 2011), shorter in the heart and the cochlea (Fig. 2 A; Mahuzier et al., 2012; Gerhardt et al., 2013) and longer in the limbs, the lung, the liver, and in MEFs (Fig. 2 A). Thus, it is possible that the differences in the study by Mahuzier et al. (2012) and the work presented here reflect differences in the molecular mechanisms between cilia of MEFs and MDCK cells. The phenomenon that the absence of Rpgrip1l leads to longer cilia in some organs and to shorter cilia in other organs is difficult to explain, because it is not known whether the composition of the ciliary proteome is different across organs. It was shown before that loss of one single protein (Mks1 and Tctn1, respectively) leads to ciliary shape defects in some organs, but not in all (Weatherbee et al., 2009; Garcia-Gonzalo et al., 2011), indicating the existence of organ-specific cilia formation mechanisms.
According to our data, a decreased activity of the ciliary proteasome is a potential cause of ciliopathies. Thus, it will be indispensable to uncover the detailed molecular mechanisms underlying the regulation of the ciliary proteasome by Rpgrip1l. The outcome of these future studies could be the prerequisite for new therapies to treat ciliopathies. In this context, treatment of Rpgrip1l−/− MEFs and limbs (ex vivo embryo culture) with SFN seems to result in a nearly restored activity of the ciliary proteasome (Fig. 10, E, F, and H), suggesting that enhancement of the proteasomal 20S subunit activity rescues proteasomal function even in the absence of Rpgrip1l. It was shown previously that SFN treatment of different cells increases the expression of 20S proteasomal subunit components (Kwak et al., 2007; Ramirez and Singletary, 2009) and that SFN realizes this effect by activating Nrf2 (nuclear factor erythroid 2–related factor 2) signaling (Kwak et al., 2003), a signaling pathway known to promote transcriptional antioxidant response (Rushmore et al., 1991; Zhang et al., 1992; Itoh et al., 1997; Li et al., 2012). Because we found a higher amount of Psma5, a component of the 20S proteasomal subunit, in cilia of Rpgrip1l−/− MEFs, why up-regulating the expression of 20S proteasomal subunits via application of SFN is able to compensate the reduced proteasomal activity in Rpgrip1l−/− cells remains an open and difficult question. Possibly, SFN is able to affect the activity of the ciliary proteasome via another, yet unknown way. Consequently, future studies will be necessary to understand the effect of SFN on proteasomal activity in the absence of Rpgrip1l.
Formerly, an association between proteasomal degradation and the intraflagellar transport was suggested (Guo et al., 2011), making it likely that proteasomal function regulates ciliary length. Interestingly, cilia of WT MEFs are significantly longer after MG132 and SFN treatment (Fig. 4 K and Fig. 10 G) and also ciliary length of WT limbs is significantly elevated after SFN treatment (Fig. 10 I), meaning that proteasomal activation and inhibition increases ciliary length. But although the activity of the ciliary proteasome is almost normalized in SFN-treated Rpgrip1l−/− MEFs and limbs, their cilia are not shorter after SFN treatment (Fig. 10, G and I), indicating that another factor is responsible for the ciliary elongation in the absence of Rpgrip1l.
It was reported in a previous study that proteasomal dysfunction is only one contributor to the pathology of ciliopathies (Liu et al., 2014). Our data confirm this perception, because increased ciliary length is not (only) caused by reduced proteasomal activity in the absence of Rpgrip1l. But nevertheless in the case of some organs (e.g., the forebrain), the severe defects caused by Rpgrip1l deficiency are based on the impairment of proteasomal activity (Besse et al., 2011). The consequences of proteasomal dysfunction for different paracrine signaling pathways have been shown recently (Liu et al., 2014). Because mutations in RPGRIP1L result in many different severe human ciliopathies (Zaghloul and Katsanis, 2010), our analyses make an important contribution to how ciliary proteins control proteasomal activity. To our knowledge, it was never shown before that a ciliary protein regulates specifically and exclusively the ciliary proteasome and that this regulation is realized by an interaction with a proteasomal component in a qualitative rather than in a quantitative level. The interaction of Rpgrip1l with the 19S proteasomal subunit component Psmd2 indicates that Rpgrip1l controls the activity of the ciliary proteasome by regulating the activity of the 19S proteasomal subunit. Consequently, knockdowns of different components of the 19S proteasomal subunit result in a decreased activity of the ciliary proteasome (Fig. 9, A, B, D, and F; and Fig. S2, A and B), resembling the Rpgrip1l−/− status (Fig. 4 D). Rpgrip1l does not ensure the accurate localization of Psmd2 to the ciliary base but regulates the activity of the ciliary proteasome most likely via its interaction with Psmd2. How Rpgrip1l affects Psmd2 function in detail, e.g., via triggering a conformational alteration, has to be elucidated in the future and will give novel mechanistical insights into the molecular background underlying severe human ciliopathies.
Materials and methods
Ethics statement and animal husbandry
All mice (Mus musculus) used in this study were on the C3H background and kept under standard housing conditions with a 12/12-h dark–light cycle and with food and water ad libitum. All animal procedures were performed in accordance with National Institutes of Health guidelines and with local and state regulations for research with animals.
Mouse strains
We used Rpgrip1l+/− mice for breeding and mated Rpgrip1l+/− male and female mice to obtain WT and Rpgrip1l−/− mouse embryos. Rpgrip1l+/− mice were generated as follows: A Rpgrip1l-null allele was produced by using homologous recombination. The targeting construct was designed to introduce a PGKneo cassette replacing exon 4 and 5 of the Rpgrip1l coding sequence. The linearized vector was electroporated into R1 embryonic stem cells, and after G418 selection, all clones were screened by Southern blot analysis. Targeted embryonic stem cells were used to create chimeras that passed the Rpgrip1l mutation on to their progeny. F1 animals were intercrossed to derive homozygous mutant F2 newborns and embryos by timed mating. For genotyping, the following primers were used: KORpgrip1l_forward, 5′-GGCCTCCCCTTTGTCAT-3′; KOneo_forward, 5′-ACGAGTTCTTTCTGAGGGGATC-3′; and KORpgrip1l_reverse, 5′-CAGCTTTCCTTGTGTCTCTACTT-3′.
Antibodies
We used the following primary antibodies: rabbit anti-actin (A2066; Sigma-Aldrich), mouse anti-FLAG (200472; Agilent Technologies), mouse anti-Gapdh (G8795; Sigma-Aldrich), goat anti-Gli3 (AF3690; R&D systems), mouse anti-HA (MMS-101P; Covance), rabbit anti-Hprt (ab10479; Abcam), rabbit anti-Myc (sc-789; Santa Cruz Biotechnology, Inc.), rabbit anti-phospho-(S33/37/T41)-β-Catenin (9561; Cell Signaling Technology), rabbit anti–nonphospho-(S33/37/T41)-β-Catenin (4270; Cell Signaling Technology), rabbit anti-Psma5 (PA1-1962; Thermo Fisher Scientific), mouse anti-Pcnt (611814; BD), goat anti-Pcnt2 (sc-28145; Santa Cruz Biotechnology, Inc.), goat anti-Psmd2 (IMG-3751; Imgenex), rabbit anti-Psmd3 (S2824; Sigma-Aldrich), rabbit anti-Psmd4 (14899-1-AP; Proteintech), rabbit anti-Smo (ab72130; Abcam), mouse anti–acetylated α-tubulin (T6793; Sigma-Aldrich), mouse anti–γ-tubulin (T6557; Sigma-Aldrich), and rabbit anti-Ubiquitin (U5379; Sigma-Aldrich). The antibodies to Cep290 as well as Nphp4 are gifts provided by S. Saunier (Institut National de la Santé et de la Recherche Médicale, Université Paris Descartes, Paris, France). The rabbit polyclonal antibody against Cep290 was generated against the murine sequence (amino acids 330–360 and amino acids 700–735). The anti-Nphp4 antibody was raised in rabbits by injection of a recombinant His-tagged human Nphp4 fragment (amino acids 673–1,032). The antibody against Evc is a gift provided by R. Rohatgi (Stanford University School of Medicine, Stanford, CA). The rabbit anti-Evc antibody was generated against the mouse sequence (amino acids 461–1,005). The polyclonal antibody against Rpgrip1l was generated by immunizing rabbits with a GST-Rpgrip1l fusion protein encompassing the murine Rpgrip1l-RID domain (amino acids 1,060–1,264) by Pineda antibody services. The polyclonal antibody against Gli3-190 was generated by immunizing rabbits with a His-Gli3 fusion protein encompassing the mouse Gli3-C terminal region (3,473–4,806 bp) by Pineda antibody services. Antibodies were affinity purified with the antigen coupled to Ni-NTA agarose (QIAGEN). Antibody specificity is confirmed by Western blotting and immunofluorescence analyses (Fig. S3, Fig. S4, and Fig. S5).
Cell culture, transfection, and drug treatment
MEFs were isolated from single E12.5 embryos after performing standard procedures. Nphp4-negative MEFs are a gift provided by C. Vesque and S. Schneider-Maunoury (both Institut de Biologie Paris Seine-Biologie du Développement, Sorbonne Universités, Paris, France). MEFs, NIH3T3 (MEF cell line), and HEK293T (human embryonic kidney) cells were grown in DMEM supplemented with 10% FCS, 1:100 (vol/vol) l-glutamine (Gibco), 1:100 (vol/vol) sodium pyruvate (Gibco), 1:100 (vol/vol) nonessential amino acids (Gibco), and 1:100 (vol/vol) penicillin/streptomycin (Gibco) at 37°C and 5% CO2. For induction of ciliogenesis, MEFs were grown to confluency and serum starved with medium containing 0.5% FCS for ≥24 h.
The transfection of MEFs, NIH3T3 and HEK293T cells with the particular constructs used in different experiments was performed by using Lipofectamine 2000 (Invitrogen). The RID domain of murine Rpgrip1l (aa 1,076–1,264) was cloned into the Flag-tagged pcDNA3 vector (Invitrogen) and expressed under the control of the cytomegalovirus (CMV) promotor. Murine full-length Wnk2 was cloned into the Myc-tagged pCMV vector (Takara Bio Inc.). Both plasmids were separately transfected into NIH3T3 cells. Each transfection experiment has been repeated once.
Cells were treated for 6 h with 20 µM MG132 (ALX-260-092; Enzo Life Sciences) or for 20 h with 5 µM SFN (ALX-350-230-M010; Enzo Life Sciences). As a solvent control, cells were treated for the same time with 0.1% DMSO, respectively. To stimulate Shh signaling, cells were treated for 24 h with 200 nM SAG (EMD Millipore). To ensure ciliary absence in the in situ PLA experiment (Fig. S1 E), MEFs were treated with 10 µg/ml nocodazole (M1404; Sigma-Aldrich) and incubated for 3 h in 4°C. All treatment experiments were repeated once.
Coimmunoprecipitation
Tagged Psmd2 full-length protein and the RID domain of Rpgrip1l were transiently overexpressed in NIH3T3 cells. After 24 h of expression, cells were lysed on ice in lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, and 0.25% Na-deoxycholate) supplemented with complete protease inhibitor cocktail (Roche) and PMSF (Sigma-Aldrich). Lysates were precleared with protein G–agarose (EMD Millipore) overnight followed by an overnight incubation with anti-FLAG or anti-Myc protein G–agarose. Afterward, beads with bound protein complexes were washed in lysis buffer, taken up in 4× sample buffer (50 mM Tris-HCl, pH 6.8, glycerol, 2% SDS, 20% β-mercaptoethanol, and bromophenol blue), and heated for 10 min at 95°C. Beads were precipitated by centrifugation. Supernatant was run on a SDS-PAGE gel and then immunoblotted. Coimmunoprecipitation experiments were repeated three times.
Cryosections
Embryos were fixed in 4% PFA and incubated in 30% sucrose (in PBS) overnight at 4°C. On the following day, they were embedded in Tissue-Tek O.C.T. Compound (Miles Laboratories, Inc.) and then stored at −80°C. Transverse cryostat sections (7 µm in thickness) were prepared.
Ex vivo embryo culture
All solutions were preheated to 37°C, and isolation was performed at this temperature. Embryos at the embryonic stage E12.5 were isolated at 37°C retaining the connection between embryo and placenta, respectively (Zeeb et al., 2012), and then transferred in pairs into whole-embryo culture bottles containing preincubated DMEM (Gibco) and FBS (Mycoplex PAA). Bottles were incubated with 20-µm SFN or 0.1% DMSO as a vector control for 5 h at 95% O2 and 5% CO2 atmosphere and 37°C. Viability of the embryos was checked, extraembryonic tissue was removed, and the limbs were fixed for 90 min with 4% PFA at room temperature. The PFA was replaced by 30% PBS-Sucrose solution and the limbs incubated at 4°C overnight. On the next day, the limbs were embedded in Tissue-Tek O.C.T. Compound. The ex vivo embryo culture experiment was repeated once.
Image processing
Image acquisition and data analysis were performed at room temperature using a microscope (Imager.A2; Carl Zeiss), 100×, NA 1.46 oil immersion objective lens (Carl Zeiss), a monochrome charge-coupled device camera (AxioCam MRm; Carl Zeiss), and the AxioVision Rel. 4.8 software package (Carl Zeiss).
3D-SIM was performed on a ELYRA PS.1 system (Carl Zeiss) equipped with an electron-multiplying charge-coupled device (iXON DU-885; Andor Technology) with 1,004 × 1,002 pixels at room temperature. Z stacks were taken using a 100× α-Plan Apochromat oil immersion objective with an NA of 1.46. To generate structured illumination, a grid pattern is projected onto the image plane in five different positions and at five different modulation angles to obtain high frequency information within the low frequency information captured by the optical system. For the Dylight405-, Dylight488-, Cy3-channel back computation of the lower frequencies, Fourier transformation was performed using the ZEN Structured Illumination Processing tool (Carl Zeiss) to increase the resolution in the final image. Fluorochromes used were Dylight405, Dylight488, Cy3, DAPI, EGFP, and ZsGreen.
Immunofluorescence
For immunofluorescence on MEFs, cells were plated on coverslips, and after reaching confluency, they were serum starved for ≥24 h. Cells were fixed with 4% PFA (for stainings with the antibodies to Rpgrip1l, Nphp4, Cep290, phospho-(S33/37/T41)-β-Catenin, Evc, Smo, and Gli3-190), methanol (for stainings with the antibody against Psmd2 and Psmd4), methanol/acetone (for stainings with the antibodies to Ubiquitin and Psmd3), or 4% PFA/methanol (for stainings with the antibody against Psma5), respectively. Fixed cells were rinsed three times with PBS after permeabilization with PBS/0.5% Triton X-100 for 10 min. After three washes with PBS, they were blocked for ≥10 min at room temperature in PBST (PBS/0.1% Triton X-100) containing 10% donkey serum. Diluted primary antibodies in block were incubated overnight at 4°C. After three washing steps with PBST, incubation with fluorescent secondary antibody diluted in block was performed at room temperature for 1 h followed by several washings and subsequent embedding with Mowiol optionally containing DAPI (Merck).
For immunofluorescence on cryosections, cryosections were washed with PBS and permeabilized with PBS/0.5% Triton X-100. Blocking was performed with 10% FCS in PBS/0.1% Triton X-100. The sections then were incubated with the primary antibodies diluted in blocking solution overnight at 4°C. After three washing steps, they underwent an incubation in the secondary antibody (diluted in blocking solution) for 2 h and then were washed again. Finally, they were embedded in Mowiol containing DAPI (Merck). All immunofluorescence experiments were repeated at least one time.
In situ PLA
The in situ PLA (PLA or DUOLINK) was performed according to the manufacturer’s manual (OLink Bioscience) and was used to detect the subcellular localization of the interaction between Rpgrip1l and Psmd2. First, Rpgrip1l and Psmd2 are recognized by specific primary antibodies. Subsequently, PLA probes consisting of species-specific secondary antibodies, each with a unique short DNA strand attached to it, bind to the primary antibodies. The addition of two other circle-forming DNA oligonucleotides leads to an interaction of the DNA strands if Rpgrip1l and Psmd2 are in close proximity (<40 nm). After joining of the two added oligonucleotides by enzymatic ligation, a rolling circle amplification using a polymerase starts. The amplified DNA is visualized by fluorescent-labeled complementary oligonucleotide probes. The cilium in the in situ PLA experiments was marked by the product of a transiently transfected IFT88-EYFP construct (Fig. 7 E and Fig. S1 D), which was a gift provided by D.R. Beier (Center for Developmental Biology and Regenerative Medicine, University of Washington School of Medicine, Seattle, WA). The IFT88-EYFP fusion protein was present in the pJAG368 vector and expressed under the control of the SV40 promoter. The centrosome in the in situ PLA experiment was marked by the product of a transiently transfected Ofd1-EYFP construct (Fig. S1, E and F), which was a gift provided by B. Franco (Telethon Institute of Genetics and Medicine, Naples, Campania, Italy). The Ofd1 full-length sequence was inserted into the pcDNA3x(+)MyEGFP vector and expressed under the control of the CMV promoter. The PLA experiments were repeated once.
Measurement of proteasomal activity in whole-cell lysates
Measurement of proteasomal activity was performed according to Reinheckel et al. (1998) with a few modifications. Cells were washed twice with PBS. After addition of lysis buffer (250 mM sucrose, 25 mM Hepes, 10 mM magnesium chloride, 1 mM EDTA, and 1.7 mM DTT), cells were lysed as a result of repeated freeze–thaw cycles. To differentiate between 20S proteasome activity (ATP independent) and 26S proteasome activity (ATP dependent), an assay in the presence of ATP and one with depleted ATP were performed simultaneously. ATP was depleted using 0.1 mg/ml hexokinase (Sigma-Aldrich) and 15 mM 2-deoxyglucose (Sigma-Aldrich). In the parallel incubation, ATP (Sigma-Aldrich) was added with a 2 mM final concentration. Afterward, cell lysates were incubated for 30 min at 37°C with 225 mM Tris buffer, pH 7.8, containing 45 mM potassium chloride, 7.5 mM magnesium acetate, 7.5 mM magnesium chloride, and 1 mM DTT and the fluorogenic peptide suc-LLVY-MCA (7-amino-4-methylcoumarin; Sigma-Aldrich). The fluorogenic peptide is degraded by the chymotrypsin-like protease of the proteasome into the degradation product MCA (Sigma-Aldrich). The MCA liberation measurement was performed with 360-nm excitation and 460-nm emission. As a standard for quantification, free MCA in different concentrations was used. Quantification of proteasomal activity in whole cell lysates was repeated one time.
Quantitations of immunofluorescence intensities
Intensity of ciliary protein staining was quantified using ImageJ (National Institutes of Health). For quantifications of the axoneme-associated protein Gli3-190, we used the area marked by acetylated α-tubulin and quantified the average pixel intensity to take the cilia length into account thereby making the data of WT and Rpgrip1l−/− cilia comparable. For ciliary base protein intensities, we selected the region labeled by γ-tubulin and the area in-between the γ-tubulin staining and the proximal part of the acetylated α-tubulin staining (for TZ proteins) and measured the total pixel intensity. In the case of the Psmd2 knockdown experiment, γ-tubulin was not used, but a triple staining with antibodies to acetylated α-tubulin, Psmd2, and Ubiquitin was performed (Fig. 9 A). To measure the amounts of Psmd2 and Ubiquitin, we determined the greatest distance of γ-tubulin to the proximal part of acetylated α-tubulin in all quantifications we performed by using γ-tubulin and acetylated α-tubulin. This was 1.5 µm. Afterward, we used a square of 1.5 × 1.5 µm to quantify the amount of Ubiquitin and Psmd2 at the ciliary base. To get rid of the ratio of unspecific (background) staining, we subtracted the mean value of the average pixel intensity (in the case of axoneme-associated proteins) or of the total pixel intensity (for all BB or TZ proteins) of three neighboring regions free from specific staining.
SEM
Freshly dissected limbs from E12.5 embryos were fixed in 2.5% glutaraldehyde. After three washes with 0.1 M cacodylate buffer, the limbs were broken in liquid nitrogen, dehydrated through a graded series of acetone, and dried chemically using hexamethylsilazane. Samples were then mounted on stubs followed by gold sputter coating. Cilia were imaged using a scanning electron microscope (Leo 1430 VP; Carl Zeiss). The SEM experiment has been repeated once.
Figure preparation
Figure preparation was performed by using Photoshop 7 or CS4 (Adobe), and collages of the all images for figure preparation were arranged using Illustrator CS4 (Adobe).
Proteasome activity assay at the ciliary base
The activity of the ciliary proteasome was evaluated using WT and Rpgrip1l−/− MEFs transiently transfected with the Proteasome Sensor Vector (Takara Bio Inc.). This vector encodes the so-called ZsProSensor-1 protein, a fusion of a bright GFP with a degradation domain that targets the protein for rapid degradation by the proteasome. If the green fluorescence accumulates, proteasomal activity is decreased. If there is no accumulation of green fluorescence, the proteasome is functioning normally.
An accumulation of the ZsProSensor-1 protein was observed exclusively at the ciliary base of Rpgrip1l−/− MEFs (Fig. 4 L), although fluorescence proteasome reporter proteins are usually rapidly diffusing. Potentially, it accumulates in this way because the ZsProSensor-1 is a specific degradation motif of ornithine decarboxylase that has been fused to the fluorescent ZsGreen protein. This fusion protein is degraded by the proteasome without requiring prior ubiquitination. Hence, a trapping of polyubiquitinated proteins by, for example, receptors localized at the ciliary base cannot be an explanation for the accumulation of the ZsProSensor-1 at the base of Rpgrip1l−/− cilia. Ornithine decarboxylase has been shown to be proteasomally degraded with the help of its interaction with AZ1 (Zhang et al., 2003). In turn, AZ1 interacts with Aurora A and is essential for Aurora A degradation by the proteasome (Lim and Gopalan, 2007). Interestingly, Aurora A localizes to the ciliary BB (Pugacheva et al., 2007) and potentially is degraded by the ciliary proteasome. Maybe at least partially, ornithine decarboxylase is also degraded by the ciliary proteasome and caused by the reduced activity of the ciliary proteasome in the absence of Rpgrip1l, the ZsProSensor-1 protein accumulates at the cilium. The proteasome activity assay at the ciliary base was repeated once.
RNAi experiments
A pool of ON-TARGET plus siRNA against Psmd2 (J-042377-05, J-042377-06, J-042377-07, and J-042377-08; GE Healthcare), a pool of 27mer siRNA duplexes against Psmd3 (SR416821; OriGene), and a pool of FlexiTube Gene Solution siRNAs against Psmd4 (GS19185; QIAGEN) were transfected into WT MEFs. 200 pmol of each siRNA were used. Each RNAi study was repeated once.
Statistical data
Data are presented as means ± SEM. Two-tailed Student’s t test was performed for all compared data by using Excel (Microsoft). A P < 0.05 was considered to be statistically significant (one asterisk), a P < 0.01 was regarded as statistically very significant (two asterisks), and a P < 0.001 was considered to be highly significant statistically (three asterisks).
Tandem affinity purification
HEK293T cells were transfected with constructs that encoded the RID domain of Rpgrip1l fused to FLAG/Strep/HA-tags and that encoded full-length Psmd2 fused to a Myc-tag. After 24 h, transfected cells were lysed in lysis buffer containing 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, protease inhibitor cocktail (Roche), and PMSF (Sigma-Aldrich) for 5 min at 4°C and sedimentated at 10,000 g. Afterward, the supernatant was incubated for 2 h at 4°C with M2 agarose FLAG-beads (Sigma-Aldrich), affinity purified by using Micro Bio-Spin Chromatography Columns (Bio-Rad Laboratories, Inc.), and eluted with 300 µg/ml FLAG peptide (produced at the Biological Medical Research Center of the Heinrich Heine University Düsseldorf) in TBS (150 mM NaCl, 3 mM KCl, and 25 mM Tris-HCl, pH 7.4). The second purification was realized by incubation of the eluates with Strep-Tactin Superflow (IBA) on Micro Bio-Spin Chromatography Columns (Bio-Rad Laboratories, Inc.). Protein complexes were eluated with 2.5 mM/ml biotin (Sigma-Aldrich) in Strep buffer (100 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 1 mM EDTA). To concentrate the proteins, the eluate was incubated in StrataClean resin (Agilent Technologies) beads overnight at 4°C. After centrifugation for 5 min at 4°C and subsequent resuspending of the beads, the supernatant as well as the beads were heated at 95°C for 10 min. Then, the samples were separated by SDS-PAGE and immunoblotted. Tandem affinity purification experiments were repeated three times.
Western blotting
Whole-cell lysates were obtained by lysis with radioimmunoprecipitation buffer (150 mM sodium chloride, 50 mM Tris-HCl, pH 7.4, 0.1% sodium deoxycholate, and 1 mM EDTA). Protein content was measured by the Bradford method, and samples were normalized. 20 mg of total protein was separated by SDS-PAGE on polyacrylamide gels (10%) and transferred to a polyvinylidene fluoride membrane (Bio-Rad Laboratories, Inc.). The membrane was probed with antibodies against Psmd2, Psmd3, Psma5, Myc, phospho-(S33/37/T41)-β-Catenin, nonphospho-(S33/37/T41)-β-Catenin, HA, or FLAG. Anti-actin, anti-Gapdh, and anti-Hprt antibodies were used as loading controls. Proteins were detected with secondary antibodies conjugated to horseradish peroxidase (RPN4201 and RPN4301) and the ECL detection kit (both GE Healthcare). Visualization of protein bands was realized by LAS-4000 mini (Fujifilm). Band intensities were measured by using ImageJ (National Institutes of Health).
Yeast two-hybrid screen
The screen was performed by using a Gal4-based yeast two-hybrid system (BD) and a mouse E11.5 cDNA library (BD). As host, the yeast strain AH109 was used, which carried the HIS3 (histidine), ADE2 (adenine), MEL1 (α-galactosidase), and LacZ (β-galactosidase) reporter genes. Interactions were investigated by evaluation of reporter gene activation via growth on selective media (HIS3 and ADE2 reporter genes), α-galactosidase colorimetric plate assays (MEL1 reporter gene), and β-galactosidase colorimetric filter lift assays (LacZ reporter gene). The assessment of the putative protein interactions was realized on the basis of growth and selective media and staining in α- and β-galactosidase activity assays. The yeast two-hybrid screen was repeated twice.
Online supplemental material
Fig. S1 shows that the ciliary amount of Smo and Evc is unaltered in Rpgrip1l−/− MEFs, a list of potential Rpgrip1l interaction partners identified by yeast two-hybrid screening, an in situ PLA displaying that the known interaction partners Rpgrip1l and Nphp4 are in close proximity (<40 nm) in the ciliary TZ, and an in situ PLA demonstrating that Rpgrip1l and Psmd2 are not in close proximity at the centrosome. The immunofluorescence experiments with the corresponding quantifications in Fig. S2 prove that treatment of WT and Rpgrip1l−/− MEFs with siRNA against Psmd3 as well as Psmd4 results in a significant reduction of Psmd3 and Psmd4 at the BB, respectively, and that the overexpression of proteins (presented by the example of Wnk2) does not always lead to an alteration of proteasomal activity at the ciliary base or of ciliary length. Fig. S3 shows antibody specificity of the anti-Gli3, anti-Gli3-190, anti-Psma5, anti-Psmd3, and anti-Psmd2 antibodies based on Western blot analyses. Fig. S4 illustrates antibody specificity in wide-field views (anti-Cep290, anti-Rpgrip1l, anti–p-β-Catenin, anti-Ubiquitin, and anti–Gli3-190 antibodies), by using Nphp4−/− MEFs and by using antigens against Smo and Evc. Fig. S5 depicts antibody specificity in wide-field views (anti-Psmd2, anti-Psmd3, anti-Psma5, and anti-Psmd4 antibodies) and by using antigens against Psmd2, Psmd3, Psma5, and Psmd4. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201408060/DC1.
Acknowledgments
The authors thank the members of the Center for Advanced Imaging at the Heinrich Heine University Düsseldorf for their support concerning the used imaging applications. Special thanks go to Kerstin Rose for generating the antibody to Gli3-190, to Yvonne Jäger, Jennifer Auchter, Katrin Götz, and the work group of Axel Gödecke for producing plasmids as well as to David R. Beier for providing the Ift88-EYFP construct and to Brunella Franco for providing the Ofd1-EYFP construct. Moreover, we are also grateful to Sophie Saunier for providing the antibodies against Cep290 and Nphp4 and to Rajat Rohatgi for providing the antibody to Evc. We express our particular gratitude to Christine Vesque and Sylvie Schneider-Maunoury for supplying Nphp4-negative MEFs as well as to Jennifer Axnick and Eckhard Lammert for giving us the possibility to perform ex vivo embryo culture experiments. Finally, we give thanks to Laura Rose and Renate Dildrop for fruitful discussion and critical reading of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (Sonderforschungsbereiche 590 and 612) to U. Rüther.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- BB
- basal body
- CMV
- cytomegalovirus
- MEF
- mouse embryonic fibroblast
- PLA
- proximity ligation assay
- RID
- RPGR-interacting domain
- SEM
- scanning EM
- SFN
- sulforaphane
- Shh
- Sonic hedgehog
- SIM
- structured illumination microscopy
- TZ
- transition zone
- WT
- wild type
References
- Aberle H., Bauer A., Stappert J., Kispert A., and Kemler R.. 1997. β-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 16:3797–3804. 10.1093/emboj/16.13.3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts H.H., Doherty D., van Beersum S.E., Parisi M.A., Letteboer S.J., Gorden N.T., Peters T.A., Märker T., Voesenek K., Kartono A., et al. 2007. Mutations in the gene encoding the basal body protein RPGRIP1L, a nephrocystin-4 interactor, cause Joubert syndrome. Nat. Genet. 39:882–888. 10.1038/ng2069 [DOI] [PubMed] [Google Scholar]
- Besse L., Neti M., Anselme I., Gerhardt C., Rüther U., Laclef C., and Schneider-Maunoury S.. 2011. Primary cilia control telencephalic patterning and morphogenesis via Gli3 proteolytic processing. Development. 138:2079–2088. 10.1242/dev.059808 [DOI] [PubMed] [Google Scholar]
- Borgal L., Habbig S., Hatzold J., Liebau M.C., Dafinger C., Sacarea I., Hammerschmidt M., Benzing T., and Schermer B.. 2012. The ciliary protein nephrocystin-4 translocates the canonical Wnt regulator Jade-1 to the nucleus to negatively regulate β-catenin signaling. J. Biol. Chem. 287:25370–25380. 10.1074/jbc.M112.385658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breusing N., Arndt J., Voss P., Bresgen N., Wiswedel I., Gardemann A., Siems W., and Grune T.. 2009. Inverse correlation of protein oxidation and proteasome activity in liver and lung. Mech. Ageing Dev. 130:748–753. 10.1016/j.mad.2009.09.004 [DOI] [PubMed] [Google Scholar]
- Brooks P., Fuertes G., Murray R.Z., Bose S., Knecht E., Rechsteiner M.C., Hendil K.B., Tanaka K., Dyson J., and Rivett J.. 2000. Subcellular localization of proteasomes and their regulatory complexes in mammalian cells. Biochem. J. 346:155–161. 10.1042/0264-6021:3460155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbit K.C., Shyer A.E., Dowdle W.E., Gaulden J., Singla V., Chen M.H., Chuang P.T., and Reiter J.F.. 2008. Kif3a constrains β-catenin-dependent Wnt signalling through dual ciliary and non-ciliary mechanisms. Nat. Cell Biol. 10:70–76. 10.1038/ncb1670 [DOI] [PubMed] [Google Scholar]
- Coux O., Tanaka K., and Goldberg A.L.. 1996. Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 65:801–847. 10.1146/annurev.bi.65.070196.004101 [DOI] [PubMed] [Google Scholar]
- Cui C., Chatterjee B., Francis D., Yu Q., SanAgustin J.T., Francis R., Tansey T., Henry C., Wang B., Lemley B., et al. 2011. Disruption of Mks1 localization to the mother centriole causes cilia defects and developmental malformations in Meckel-Gruber syndrome. Dis. Model. Mech. 4:43–56. 10.1242/dmm.006262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnecki P.G., and Shah J.V.. 2012. The ciliary transition zone: from morphology and molecules to medicine. Trends Cell Biol. 22:201–210. 10.1016/j.tcb.2012.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delous M., Baala L., Salomon R., Laclef C., Vierkotten J., Tory K., Golzio C., Lacoste T., Besse L., Ozilou C., et al. 2007. The ciliary gene RPGRIP1L is mutated in cerebello-oculo-renal syndrome (Joubert syndrome type B) and Meckel syndrome. Nat. Genet. 39:875–881. 10.1038/ng2039 [DOI] [PubMed] [Google Scholar]
- Dorn K.V., Hughes C.E., and Rohatgi R.. 2012. A Smoothened-Evc2 complex transduces the Hedgehog signal at primary cilia. Dev. Cell. 23:823–835. 10.1016/j.devcel.2012.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdle W.E., Robinson J.F., Kneist A., Sirerol-Piquer M.S., Frints S.G., Corbit K.C., Zaghloul N.A., van Lijnschoten G., Mulders L., Verver D.E., et al. 2011. Disruption of a ciliary B9 protein complex causes Meckel syndrome. Am. J. Hum. Genet. 89:94–110. 10.1016/j.ajhg.2011.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabunmi R.P., Wigley W.C., Thomas P.J., and DeMartino G.N.. 2000. Activity and regulation of the centrosome-associated proteasome. J. Biol. Chem. 275:409–413. 10.1074/jbc.275.1.409 [DOI] [PubMed] [Google Scholar]
- Farré J.C., Manjithaya R., Mathewson R.D., and Subramani S.. 2008. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev. Cell. 14:365–376. 10.1016/j.devcel.2007.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo F.R., Corbit K.C., Sirerol-Piquer M.S., Ramaswami G., Otto E.A., Noriega T.R., Seol A.D., Robinson J.F., Bennett C.L., Josifova D.J., et al. 2011. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat. Genet. 43:776–784. 10.1038/ng.891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascue C., Tan P.L., Cardenas-Rodriguez M., Libisch G., Fernandez-Calero T., Liu Y.P., Astrada S., Robello C., Naya H., Katsanis N., and Badano J.L.. 2012. Direct role of Bardet-Biedl syndrome proteins in transcriptional regulation. J. Cell Sci. 125:362–375. 10.1242/jcs.089375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes J.M., Liu Y., Zaghloul N.A., Leitch C.C., Lawson S.S., Kato M., Beachy P.A., Beales P.L., DeMartino G.N., Fisher S., et al. 2007. Disruption of the basal body compromises proteasomal function and perturbs intracellular Wnt response. Nat. Genet. 39:1350–1360. 10.1038/ng.2007.12 [DOI] [PubMed] [Google Scholar]
- Gerhardt C., Lier J.M., Kuschel S., and Rüther U.. 2013. The ciliary protein Ftm is required for ventricular wall and septal development. PLoS ONE. 8:e57545 10.1371/journal.pone.0057545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C.W., Liu G., Xiong S., Ge F., Fuse T., Wang Y.F., and Kitazato K.. 2011. The C-terminus of MIP-T3 protein is required for ubiquitin-proteasome-mediated degradation in human cells. FEBS Lett. 585:1350–1356. 10.1016/j.febslet.2011.04.015 [DOI] [PubMed] [Google Scholar]
- Gustafsson M.G., Shao L., Carlton P.M., Wang C.J., Golubovskaya I.N., Cande W.Z., Agard D.A., and Sedat J.W.. 2008. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 94:4957–4970. 10.1529/biophysj.107.120345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart M., Concordet J.P., Lassot I., Albert I., del los Santos R., Durand H., Perret C., Rubinfeld B., Margottin F., Benarous R., and Polakis P.. 1999. The F-box protein β-TrCP associates with phosphorylated β-catenin and regulates its activity in the cell. Curr. Biol. 9:207–211. 10.1016/S0960-9822(99)80091-8 [DOI] [PubMed] [Google Scholar]
- Haycraft C.J., Banizs B., Aydin-Son Y., Zhang Q., Michaud E.J., and Yoder B.K.. 2005. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 1:e53 10.1371/journal.pgen.0010053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., and Marshall W.F.. 2011. Ciliogenesis: building the cell’s antenna. Nat. Rev. Mol. Cell Biol. 12:222–234. 10.1038/nrm3085 [DOI] [PubMed] [Google Scholar]
- Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y., Oyake T., Hayashi N., Satoh K., Hatayama I., et al. 1997. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 236:313–322. 10.1006/bbrc.1997.6943 [DOI] [PubMed] [Google Scholar]
- Kasahara K., Kawakami Y., Kiyono T., Yonemura S., Kawamura Y., Era S., Matsuzaki F., Goshima N., and Inagaki M.. 2014. Ubiquitin-proteasome system controls ciliogenesis at the initial step of axoneme extension. Nat. Commun. 5:5081 10.1038/ncomms6081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M., Hatakeyama S., Shirane M., Matsumoto M., Ishida N., Hattori K., Nakamichi I., Kikuchi A., Nakayama K., and Nakayama K.. 1999. An F-box protein, FWD1, mediates ubiquitin-dependent proteolysis of β-catenin. EMBO J. 18:2401–2410. 10.1093/emboj/18.9.2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C., Deplazes A., Sohrmann M., and Peter M.. 2008. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat. Cell Biol. 10:602–610. 10.1038/ncb1723 [DOI] [PubMed] [Google Scholar]
- Kwak M.K., Wakabayashi N., Greenlaw J.L., Yamamoto M., and Kensler T.W.. 2003. Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Mol. Cell. Biol. 23:8786–8794. 10.1128/MCB.23.23.8786-8794.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak M.K., Cho J.M., Huang B., Shin S., and Kensler T.W.. 2007. Role of increased expression of the proteasome in the protective effects of sulforaphane against hydrogen peroxide-mediated cytotoxicity in murine neuroblastoma cells. Free Radic. Biol. Med. 43:809–817. 10.1016/j.freeradbiomed.2007.05.029 [DOI] [PubMed] [Google Scholar]
- Larkins C.E., Aviles G.D., East M.P., Kahn R.A., and Caspary T.. 2011. Arl13b regulates ciliogenesis and the dynamic localization of Shh signaling proteins. Mol. Biol. Cell. 22:4694–4703. 10.1091/mbc.E10-12-0994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latres E., Chiaur D.S., and Pagano M.. 1999. The human F box protein β-Trcp associates with the Cul1/Skp1 complex and regulates the stability of β-catenin. Oncogene. 18:849–854. 10.1038/sj.onc.1202653 [DOI] [PubMed] [Google Scholar]
- Li Y., Paonessa J.D., and Zhang Y.. 2012. Mechanism of chemical activation of Nrf2. PLoS ONE. 7:e35122 10.1371/journal.pone.0035122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilienbaum A. 2013. Relationship between the proteasomal system and autophagy. Int. J. Biochem. Mol. Biol. 4:1–26. [PMC free article] [PubMed] [Google Scholar]
- Lim S.K., and Gopalan G.. 2007. Antizyme1 mediates AURKAIP1-dependent degradation of Aurora-A. Oncogene. 26:6593–6603. 10.1038/sj.onc.1210482 [DOI] [PubMed] [Google Scholar]
- Liu C., Kato Y., Zhang Z., Do V.M., Yankner B.A., and He X.. 1999. β-Trcp couples beta-catenin phosphorylation-degradation and regulates Xenopus axis formation. Proc. Natl. Acad. Sci. USA. 96:6273–6278. 10.1073/pnas.96.11.6273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Urbé S., and Clague M.J.. 2012. Selective protein degradation in cell signalling. Semin. Cell Dev. Biol. 23:509–514. 10.1016/j.semcdb.2012.01.014 [DOI] [PubMed] [Google Scholar]
- Liu Y.P., Tsai I.C., Morleo M., Oh E.C., Leitch C.C., Massa F., Lee B.H., Parker D.S., Finley D., Zaghloul N.A., et al. 2014. Ciliopathy proteins regulate paracrine signaling by modulating proteasomal degradation of mediators. J. Clin. Invest. 124:2059–2070. 10.1172/JCI71898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahuzier A., Gaudé H.M., Grampa V., Anselme I., Silbermann F., Leroux-Berger M., Delacour D., Ezan J., Montcouquiol M., Saunier S., et al. 2012. Dishevelled stabilization by the ciliopathy protein Rpgrip1l is essential for planar cell polarity. J. Cell Biol. 198:927–940. 10.1083/jcb.201111009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama Y., Suzuki M., Arima C., Huang Q.M., Tomida S., Takeuchi T., Sugiyama R., Itoh Y., Yatabe Y., Goto H., and Takahashi T.. 2011. Proteasomal non-catalytic subunit PSMD2 as a potential therapeutic target in association with various clinicopathologic features in lung adenocarcinomas. Mol. Carcinog. 50:301–309. 10.1002/mc.20632 [DOI] [PubMed] [Google Scholar]
- Moon H., Song J., Shin J.O., Lee H., Kim H.K., Eggenschwiller J.T., Bok J., and Ko H.W.. 2014. Intestinal cell kinase, a protein associated with endocrine-cerebro-osteodysplasia syndrome, is a key regulator of cilia length and Hedgehog signaling. Proc. Natl. Acad. Sci. USA. 111:8541–8546. 10.1073/pnas.1323161111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A., Sumpter R. Jr., Xiao G., Ng A., Zou Z., Tang Y., Narimatsu M., Gilpin C., Sun Q., Roth M., et al. 2011. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 480:113–117. 10.1038/nature10546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampliega O., Orhon I., Patel B., Sridhar S., Díaz-Carretero A., Beau I., Codogno P., Satir B.H., Satir P., and Cuervo A.M.. 2013. Functional interaction between autophagy and ciliogenesis. Nature. 502:194–200. 10.1038/nature12639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugacheva E.N., Jablonski S.A., Hartman T.R., Henske E.P., and Golemis E.A.. 2007. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 129:1351–1363. 10.1016/j.cell.2007.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puram S.V., Kim A.H., Park H.Y., Anckar J., and Bonni A.. 2013. The ubiquitin receptor S5a/Rpn10 links centrosomal proteasomes with dendrite development in the mammalian brain. Cell Reports. 4:19–30. 10.1016/j.celrep.2013.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez M.C., and Singletary K.. 2009. Regulation of estrogen receptor α expression in human breast cancer cells by sulforaphane. J. Nutr. Biochem. 20:195–201. 10.1016/j.jnutbio.2008.02.002 [DOI] [PubMed] [Google Scholar]
- Reinheckel T., Sitte N., Ullrich O., Kuckelkorn U., Davies K.J., and Grune T.. 1998. Comparative resistance of the 20S and 26S proteasome to oxidative stress. Biochem. J. 335:637–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock K.L., Gramm C., Rothstein L., Clark K., Stein R., Dick L., Hwang D., and Goldberg A.L.. 1994. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 78:761–771. 10.1016/S0092-8674(94)90462-6 [DOI] [PubMed] [Google Scholar]
- Rott R., Szargel R., Haskin J., Bandopadhyay R., Lees A.J., Shani V., and Engelender S.. 2011. α-Synuclein fate is determined by USP9X-regulated monoubiquitination. Proc. Natl. Acad. Sci. USA. 108:18666–18671. 10.1073/pnas.1105725108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein D.C. 2006. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 443:780–786. 10.1038/nature05291 [DOI] [PubMed] [Google Scholar]
- Rushmore T.H., Morton M.R., and Pickett C.B.. 1991. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J. Biol. Chem. 266:11632–11639. [PubMed] [Google Scholar]
- Schermelleh L., Carlton P.M., Haase S., Shao L., Winoto L., Kner P., Burke B., Cardoso M.C., Agard D.A., Gustafsson M.G., et al. 2008. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 320:1332–1336. 10.1126/science.1156947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K.J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.G., and Landegren U.. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 3:995–1000. 10.1038/nmeth947 [DOI] [PubMed] [Google Scholar]
- Tammachote R., Hommerding C.J., Sinders R.M., Miller C.A., Czarnecki P.G., Leightner A.C., Salisbury J.L., Ward C.J., Torres V.E., Gattone V.H. II, and Harris P.C.. 2009. Ciliary and centrosomal defects associated with mutation and depletion of the Meckel syndrome genes MKS1 and MKS3. Hum. Mol. Genet. 18:3311–3323. 10.1093/hmg/ddp272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Lin M.G., Stowe T.R., Chen S., Zhu M., Stearns T., Franco B., and Zhong Q.. 2013. Autophagy promotes primary ciliogenesis by removing OFD1 from centriolar satellites. Nature. 502:254–257. 10.1038/nature12606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston T.L., Wandel M.P., von Muhlinen N., Foeglein A., and Randow F.. 2012. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 482:414–418. 10.1038/nature10744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierkotten J., Dildrop R., Peters T., Wang B., and Rüther U.. 2007. Ftm is a novel basal body protein of cilia involved in Shh signalling. Development. 134:2569–2577. 10.1242/dev.003715 [DOI] [PubMed] [Google Scholar]
- Wang K., and Klionsky D.J.. 2011. Mitochondria removal by autophagy. Autophagy. 7:297–300. 10.4161/auto.7.3.14502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., and Robbins J.. 2014. Proteasomal and lysosomal protein degradation and heart disease. J. Mol. Cell. Cardiol. 71:16–24. 10.1016/j.yjmcc.2013.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Fallon J.F., and Beachy P.A.. 2000. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 100:423–434. 10.1016/S0092-8674(00)80678-9 [DOI] [PubMed] [Google Scholar]
- Weatherbee S.D., Niswander L.A., and Anderson K.V.. 2009. A mouse model for Meckel syndrome reveals Mks1 is required for ciliogenesis and Hedgehog signaling. Hum. Mol. Genet. 18:4565–4575. 10.1093/hmg/ddp422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X., Lai C.K., Evangelista M., Hongo J.A., de Sauvage F.J., and Scales S.J.. 2010. Kinetics of hedgehog-dependent full-length Gli3 accumulation in primary cilia and subsequent degradation. Mol. Cell. Biol. 30:1910–1922. 10.1128/MCB.01089-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigley W.C., Fabunmi R.P., Lee M.G., Marino C.R., Muallem S., DeMartino G.N., and Thomas P.J.. 1999. Dynamic association of proteasomal machinery with the centrosome. J. Cell Biol. 145:481–490. 10.1083/jcb.145.3.481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C.L., Li C., Kida K., Inglis P.N., Mohan S., Semenec L., Bialas N.J., Stupay R.M., Chen N., Blacque O.E., et al. 2011. MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J. Cell Biol. 192:1023–1041. 10.1083/jcb.201012116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston J.T., Strack P., Beer-Romero P., Chu C.Y., Elledge S.J., and Harper J.W.. 1999. The SCFβ-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev. 13:270–283. 10.1101/gad.13.3.270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf M.T., Saunier S., O’Toole J.F., Wanner N., Groshong T., Attanasio M., Salomon R., Stallmach T., Sayer J.A., Waldherr R., et al. 2007. Mutational analysis of the RPGRIP1L gene in patients with Joubert syndrome and nephronophthisis. Kidney Int. 72:1520–1526. 10.1038/sj.ki.5002630 [DOI] [PubMed] [Google Scholar]
- Zaghloul N.A., and Katsanis N.. 2010. Functional modules, mutational load and human genetic disease. Trends Genet. 26:168–176. 10.1016/j.tig.2010.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalli D., Bayliss R., and Fry A.M.. 2012. The Nek8 protein kinase, mutated in the human cystic kidney disease nephronophthisis, is both activated and degraded during ciliogenesis. Hum. Mol. Genet. 21:1155–1171. 10.1093/hmg/ddr544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeb M., Axnick J., Planas-Paz L., Hartmann T., Strilic B., and Lammert E.. 2012. Pharmacological manipulation of blood and lymphatic vascularization in ex vivo-cultured mouse embryos. Nat. Protoc. 7:1970–1982. 10.1038/nprot.2012.120 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Talalay P., Cho C.G., and Posner G.H.. 1992. A major inducer of anticarcinogenic protective enzymes from broccoli: isolation and elucidation of structure. Proc. Natl. Acad. Sci. USA. 89:2399–2403. 10.1073/pnas.89.6.2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Pickart C.M., and Coffino P.. 2003. Determinants of proteasome recognition of ornithine decarboxylase, a ubiquitin-independent substrate. EMBO J. 22:1488–1496. 10.1093/emboj/cdg158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Ruas J.L., Cao R., Salomons F.A., Cao Y., Poellinger L., and Pereira T.. 2006. Cell-type-specific regulation of degradation of hypoxia-inducible factor 1 α: role of subcellular compartmentalization. Mol. Cell. Biol. 26:4628–4641. 10.1128/MCB.02236-05 [DOI] [PMC free article] [PubMed] [Google Scholar]