CENP-C promotes kinetochore targeting of other constitutive centromere–associated network (CCAN) subunits by directly interacting with the four-subunit CCAN subcomplex CENP-HIKM and spatially organizing the localization of all other CCAN subunits downstream of CENP-A.
Abstract
Kinetochores are multisubunit complexes that assemble on centromeres to bind spindle microtubules and promote faithful chromosome segregation during cell division. A 16-subunit complex named the constitutive centromere–associated network (CCAN) creates the centromere–kinetochore interface. CENP-C, a CCAN subunit, is crucial for kinetochore assembly because it links centromeres with the microtubule-binding interface of kinetochores. The role of CENP-C in CCAN organization, on the other hand, had been incompletely understood. In this paper, we combined biochemical reconstitution and cellular investigations to unveil how CENP-C promotes kinetochore targeting of other CCAN subunits. The so-called PEST domain in the N-terminal half of CENP-C interacted directly with the four-subunit CCAN subcomplex CENP-HIKM. We identified crucial determinants of this interaction whose mutation prevented kinetochore localization of CENP-HIKM and of CENP-TW, another CCAN subcomplex. When considered together with previous observations, our data point to CENP-C as a blueprint for kinetochore assembly.
Introduction
Accurate and timely chromosome segregation in mitosis and meiosis is essential for cellular and organismal viability. During mitosis, sister chromatids generated upon DNA replication retain cohesion until they are bioriented on the mitotic spindle. Release of sister chromatid cohesion at the metaphase-to-anaphase transition allows the spindle to separate the sister chromatids into two genetically identical daughter cells (Cheerambathur and Desai, 2014).
Kinetochores mediate the attachment of sister chromatids to spindle microtubules. These large structures, consisting of several copies of ∼30 core subunits, become established on a segment of specialized chromatin named the centromere, whose main hallmark is the presence of the histone H3 variant CENP-A (or CenH3; Fig. 1 A; McAinsh and Meraldi, 2011; Fukagawa and Earnshaw, 2014). At low resolution, kinetochores appear as laminar structures, with an outer plate receiving the ends of spindle microtubules, and an inner plate adjacent to dense centromeric chromatin (Santaguida and Musacchio, 2009; Cheeseman, 2014). The outer kinetochore plate hosts the KMN (Knl1 complex, Mis12 complex, Ndc80 complex) network, a 10-subunit assembly that plays a crucial role as a receptor for microtubules (Cheeseman et al., 2006; DeLuca et al., 2006). The inner kinetochore hosts the constitutive centromere–associated network (CCAN), a complex of at least 16 different CENPs (centromeric proteins), several of which were originally identified in the CENP-A interactome of vertebrates (McCleland et al., 2004; Obuse et al., 2004; Foltz et al., 2006; Izuta et al., 2006; Okada et al., 2006; Hori et al., 2008).
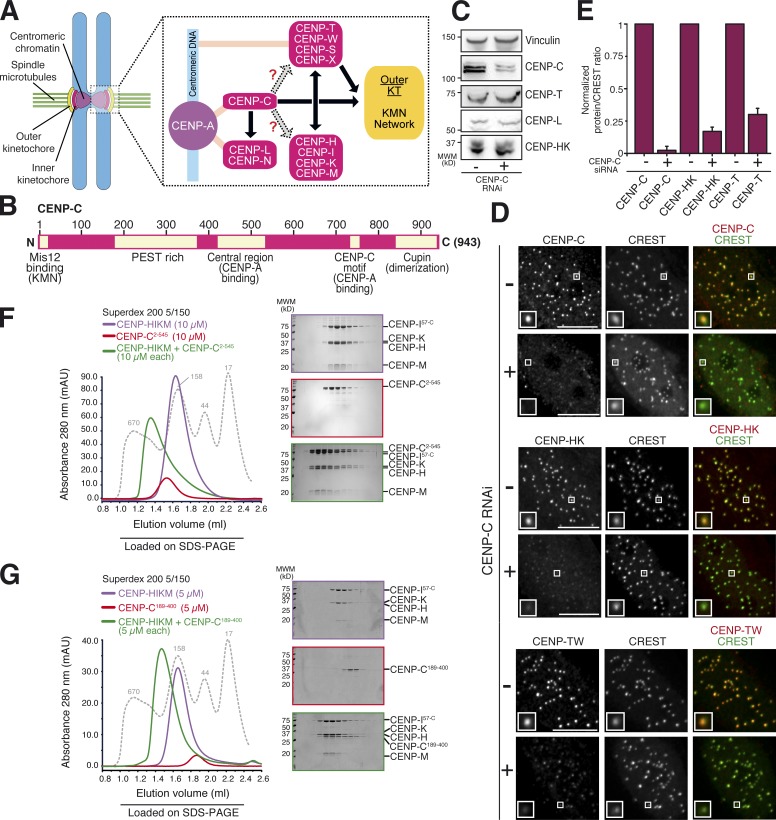
Figure 1.
Depletion of CENP-C abolishes localization of CCAN to kinetochores. (A) Schematic kinetochore organization. CCAN subunits are shown in hot pink. Light pink lines indicate presumed contacts with centromeric chromatin. Black arrows indicate direct and well-established interactions. Gray arrows represent interactions that are still uncharacterized and that were characterized in this study. For instance, whether localization of CENP-STWX to inner kinetochores requires CENP-C is controversial. Direct interaction of CENP-STWX to CENP-HIKM has been demonstrated recently and reflects in codependency for kinetochore localization. Both CENP-STWX and CENP-C contribute to recruitment of outer kinetochore components. (B) Schematic representation of human CENP-C depicting some of the protein’s crucial domains. N, N terminus; C, C terminus. (C) Whole cell protein extracts from Flp-In T-REx HeLa cells treated with control or CENP-C siRNAs were run on SDS-PAGE and immunoblotted for the indicated kinetochore proteins. Vinculin served as a loading control. MWM, molecular weight marker. (D) Representative images showing kinetochore levels of CENP-C, CENP-TW (with an antibody raised against the CENP-TW complex), and CENP-HK (with an antibody raised against the CENP-HK complex) in Flp-In T-REx HeLa cells upon treatment with control and CENP-C siRNA in interphase. Kinetochores were visualized with CREST sera. Upon CENP-C siRNA, CENP-C, CENP-HK, and CENP-TW were displaced from kinetochores. Bars, 10 µm. Magnification = 630×. Insets show a magnification of the white boxed areas that capture one or more kinetochores in each panel. (E) Quantifications are expressed as normalized protein/CREST fluorescence intensity ratios from the experiment in D. Graphs and bars indicate means ± SEM of three independent experiments for CENP-C, CENP-HK, and CENP-TW. (F) SEC elution profile shows CENP-C2–545 (∼62 kD) elutes earlier than predicted based on its molecular mass, consistent with an unfolded structure. The modest height of the CENP-C peak is caused by low abundance of aromatic amino acids. CENP-C2–545 and CENP-HIKM, both at 10 µM, form a stable, apparently stoichiometric complex on an analytical SEC. Shown is the typical outcome of at least five experiments. (G) SEC elution profile of CENP-C189–400 (∼24 kD) and of its combination with CENP-HIKM, both at 5 µM. Also CENP-C189–400 and CENP-HIKM form a stoichiometric complex and coelute in analytical SEC. Shown is the typical outcome of at least three experiments. The gray dotted lines show the elution profile of globular markers of known molecular masses, as indicated.
A subset of CCAN subunits, including CENP-N, the CENP-TW complex, and CENP-C, have been recognized as potential “founders” of kinetochore assembly downstream of CENP-A (Fig. 1 A; Hori et al., 2008, 2013; Carroll et al., 2009, 2010; Gascoigne et al., 2011; Guse et al., 2011; Kato et al., 2013). In vitro, CENP-N has been shown to interact directly with CENP-A nucleosomes (Carroll et al., 2009). Chl4, the CENP-N orthologue in Saccharomyces cerevisiae, additionally binds tightly to Iml3 (orthologous to CENP-L; Guo et al., 2013; Hinshaw and Harrison, 2013). Although functionally poorly characterized, the CENP-LN complex has been shown to bind directly to CENP-C and to require it for kinetochore localization, at least in yeast (Tanaka et al., 2009; Hinshaw and Harrison, 2013). In vertebrates, the CENP-LN complex has been shown to contribute to localization of downstream CCAN subunits, including those in the CENP-HIKM complex (Carroll et al., 2009, 2010).
CENP-T and CENP-W are histone fold domain–containing proteins and form a tightly interacting complex (Foltz et al., 2006; Hori et al., 2008). They also interact with two additional histone fold domain proteins named CENP-S and CENP-X (Amano et al., 2009; Nishino et al., 2012). It has been proposed that the CENP-STWX complex may form a specialized centromeric nucleosome-like structure (Nishino et al., 2012), but a recent study suggested that these proteins bind preferentially to internucleosomal linker DNA (Takeuchi et al., 2014). Importantly, the N-terminal region of CENP-T interacts directly, in a phosphorylation-dependent manner, with the Ndc80 subcomplex in the KMN network at the outer kinetochore (Bock et al., 2012; Schleiffer et al., 2012; Malvezzi et al., 2013; Nishino et al., 2013), thus contributing to outer kinetochore assembly. When expressed ectopically on a noncentromeric chromosome site, the N-terminal region of CENP-T supported outer kinetochore assembly independently of other inner kinetochore subunits (Gascoigne et al., 2011; Hori et al., 2013).
CENP-C (943 residues in humans) was among the first human centromere proteins to be discovered (Earnshaw and Rothfield, 1985; Saitoh et al., 1992). Sequence analysis predicts that CENP-C is mostly intrinsically disordered. Near its N terminus, CENP-C contains a binding site for the Mis12 subcomplex of the KMN network (Fig. 1 B; Przewloka et al., 2011; Screpanti et al., 2011; Hornung et al., 2014). Additionally, CENP-C contains two related short motifs that direct it to the CENP-A nucleosome, an interaction believed to be compatible with CENP-N binding (Milks et al., 2009; Carroll et al., 2010; Guse et al., 2011; Kato et al., 2013). Thus, CENP-C, like CENP-T, has the potential to bridge the underlying centromeric chromatin with the outer kinetochore. Finally, the C-terminal region of CENP-C has been shown to mediate dimerization and to promote interactions with CENP-A deposition machinery (Sugimoto et al., 1997; Cohen et al., 2008; Trazzi et al., 2009; Moree et al., 2011; Dambacher et al., 2012; Unhavaithaya and Orr-Weaver, 2013).
The precise localization dependencies of the centromere proximal proteins CENP-C, CENP-N, and CENP-TW are controversial and so is their role in the recruitment of additional CCAN subunits, including those in a four-subunit subcomplex consisting of the CENP-H, CENP-K, CENP-I, and CENP-M subunits (the CENP-HIKM complex), which we have recently characterized (Basilico et al., 2014). It has been shown that depletion of CENP-C in HeLa cells leads to mislocalization of the subunits of the CENP-HIKM complex and of CENP-T (Carroll et al., 2010; Basilico et al., 2014), suggesting that both localize downstream of CENP-C. Furthermore, CENP-T and the CENP-HIKM subunits were shown to be codependent for kinetochore localization (Hori et al., 2008; Gascoigne et al., 2011; Basilico et al., 2014). Other studies, however, did not report effects from CENP-C depletion on kinetochore localization of CENP-I or CENP-T (Liu et al., 2006; Gascoigne et al., 2011). It was therefore proposed that CENP-T may encode a CENP-C–independent pathway of kinetochore assembly downstream of CENP-A (Gascoigne et al., 2011). Here, we provide strong mechanistic insight on the role of CENP-C and identify CENP-C as a blueprint of kinetochore assembly and as the protein that spatially organizes the localization of all other CCAN subunits downstream of CENP-A.
Results and discussion
Depletion of CENP-C disrupts CCAN organization
We used a protocol previously established in our laboratory (Basilico et al., 2014) to achieve substantial CENP-C depletion by RNAi in HeLa cells (Fig. 1 C). By Western blotting, the overall cellular levels of several additional CCAN subunits, including CENP-T, CENP-L, and CENP-HK, appeared unaffected (Fig. 1 C). Conversely, immunofluorescence experiments demonstrated that the kinetochore levels of CENP-C, CENP-HK, and CENP-TW were strongly reduced in CENP-C–depleted cells (Fig. 1, D and E). These experiments confirm evidence from our and other laboratories that CENP-TW complex localization to kinetochores requires CENP-C (Carroll et al., 2010; Basilico et al., 2014; Kim and Yu, 2015; Krizaic et al., 2015; Logsdon et al., 2015; Tachiwana et al., 2015). Our evidence that CENP-C is required for correct localization of CENP-HK is also consistent with previous studies (Milks et al., 2009; Carroll et al., 2010; Gascoigne et al., 2011). These results, when considered together with evidence that the subunits of the CENP-HIKM complex are required for localization of CENP-TW (Hori et al., 2008; Basilico et al., 2014), suggest that the CENP-TW complex may not create a CENP-C–independent pathway of kinetochore assembly downstream of CENP-A (Gascoigne et al., 2011).
CENP-HIKM and CENP-C interact directly
To investigate whether the observed dependency of the CENP-HIKM complex on CENP-C reflected a direct interaction, we built recombinant versions of CENP-HIKM (Basilico et al., 2014) and CENP-C. We did not observe binding of CENP-HIKM to CENP-C constructs encompassing the majority of the C-terminal region (CENP-C632–943; unpublished data). Conversely, recombinant CENP-C2–545 (Fig. S1 A) coeluted in a stoichiometric complex with the CENP-HIKM complex from a Superdex 200 size-exclusion chromatography (SEC) column, which separates proteins based on size and shape (Fig. 1 F).
CENP-C2–545 contains a Mis12 binding motif (Przewloka et al., 2011; Screpanti et al., 2011), a region rich in proline, glutamate, serine, and threonine (PEST in single letter amino acid code) and therefore referred to as “PEST rich” (Cohen et al., 2008), and a kinetochore targeting domain recently shown to be responsible for a specific interaction with the CENP-A nucleosome (Fig. 1 B; Yang et al., 1996; Song et al., 2002; Fachinetti et al., 2013; Kato et al., 2013). We asked whether the PEST domain was involved in CENP-HIKM binding. Indeed, recombinant CENP-C189–400 (essentially limited to the PEST domain; Fig. S1 B) was sufficient to interact with CENP-HIKM in the SEC assay (Fig. 1 G). Thus, CENP-C and CENP-HIKM bind directly, and the PEST region of CENP-C is sufficient for this interaction in vitro.
Characterization of the CENP-C–CENP-HIKM interaction
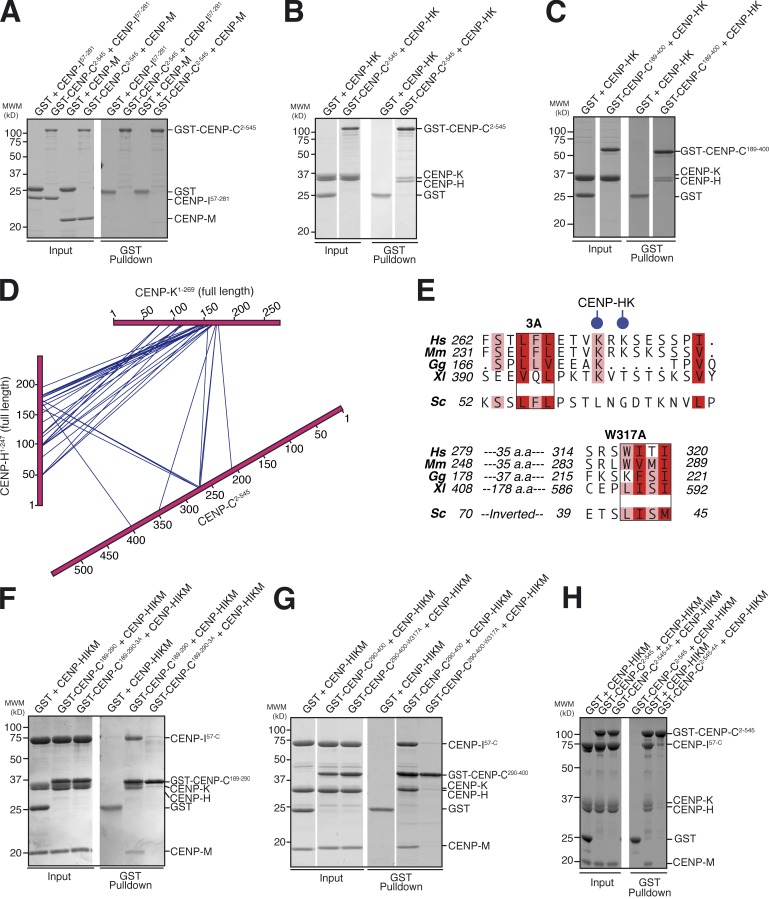
Next, we tried to identify which subunits of the CENP-HIKM complex might be involved in the interaction with CENP-C. To this end, we immobilized a GST–CENP-C2–545 fusion protein to glutathione beads and used it as bait in pull-down assays with CENP-M, the CENP-HK complex, and CENP-I57–281 (full-length CENP-I could not be produced because it is not soluble in the absence of CENP-M and CENP-HK; Basilico et al., 2014). Neither CENP-M nor CENP-I57–281 bound the GST–CENP-C2–545 bait (Fig. 2 A). In contrast, the CENP-HK complex bound the GST–CENP-C2–545 bait (Fig. 2 B) and also the GST–CENP-C189–400 bait (Fig. 2 C), indicating that the CENP-HK subcomplex is sufficient for a tight interaction with CENP-C. We conclude that the PEST-rich domain of CENP-C and the CENP-HK subunits of the CENP-HIKM complex are the main determinants of the interaction of CENP-C with the CENP-HIKM complex.
Figure 2.
Characterization of the CENP-C–CENP-HIKM interaction. (A) GST pull-down assay of GST–CENP-C2–545 with CENP-M or CENP-I57–281. CENP-C does not bind to CENP-M or CENP-I57–281 in this assay. (B) GST pull-down assay of GST–CENP-C2–545 with CENP-HK. (C) GST pull-down assay of GST–CENP-C189–400 with CENP-HK. In B and C, CENP-C2–545 and CENP-C189–400, respectively, bind to CENP-HK. (D) Summary of cross-links within the CENP-C2–545–CENP-HK complex. Intermolecular cross-links are shown in blue. Intramolecular cross-links are not shown but are listed in Table S1. CENP-HK binds to CENP-C within its PEST-rich domain. (E) Within the boxed regions are two conserved hydrophobic motifs in CENP-C189–400. In the 3A mutant, L265, F266A, and L267A were mutated to alanine. The 4A mutant combines 3A with W317A. Two Lys residues found to cross-link with CENP-HK are shown with blue circles. Budding yeast Mif2 is shown for comparison. Curiously, the regions conserved in Mif2 seem to be inverted. Hs, Homo sapiens; Mm, Mus musculus; Gg, Gallus gallus; Xl, X. laevis; Sc, S. cerevisiae. Red and pink boxes indicate positions in the CENP-C sequence that are either fully conserved (per residue class, e.g., hydrophobic and aromatic) or conserved in at least four in five sequences, respectively. (F) GST–CENP-C189–290-3A was unable to bind HIKM in a GST pull-down assay, in contrast to wild-type CENP-C189–290. (G) GST–CENP-C290–400-W317A was unable to bind HIKM in a GST pull-down assay, in contrast to wild-type GST–CENP-C290–400. (H) GST pull-down assay of GST–CENP-C2–545 shows binding to HIKM, whereas GST–CENP-C2–545-4A is effectively impaired in HIKM binding. White lines indicate that intervening lanes have been spliced out. MWM, molecular weight marker.
Further characterization of the CENP-C–CENP-HK interaction
To further characterize the interaction of CENP-C with CENP-HK, we subjected the CENP-C2–545–CENP-HK complex to cross-linking with the bifunctional reagent BS2G (bis[sulfosuccinimidyl]glutarate), which cross-links the primary amines of lysine side chains within a distance compatible with the length of the cross-linker (7.7 Å; Maiolica et al., 2007; Herzog et al., 2012). Subsequent mass spectrometry analysis identified numerous cross-links between CENP-C and CENP-HK, the majority of which mapped to the PEST domain of CENP-C (Fig. 2 D and Table S1), in agreement with the binding assays discussed in the previous paragraph.
Low average representation of hydrophobic residues is a defining feature of intrinsically disordered proteins. Within such proteins, however, binding sites for interacting partners often coincide with short stretches of hydrophobic and aromatic amino acids. We noticed that two closely spaced lysine residues (K271 and K273), which were identified as hotspots in the cross-linking experiments, flank a Leu-Phe-Leu motif (residues 265–267) that appears to be conserved in evolution (Fig. 2 E). A second conserved hydrophobic/aromatic motif (residues 317–320) was also identified. Interestingly, both the Leu-Phe-Leu motif and the second motif appear to be present in Mif2p, the S. cerevisiae homologue of CENP-C, but in Mif2p their relative position is swapped.
To test a possible role of the CENP-C hydrophobic motifs in the interaction with the CENP-HIKM complex, we initially divided the CENP-C PEST region in two separate constructs of approximately similar length (CENP-C residues 189–290 and 290–400) each containing only one of the two hydrophobic motifs. As immobilized GST fusion proteins, both CENP-C constructs were sufficient for an interaction with the CENP-HIKM complex in pull-down assays (Fig. 2, F and G), suggesting that the binding site for CENP-HK on CENP-C is extended, as already suggested by the cross-linking experiments.
We therefore tested the effects of mutations in the individual hydrophobic motifs of CENP-C. To this end, we created a triple alanine mutant of residues Leu265, Phe266, and Leu267 (henceforth referred to as “3A” mutant) on the GST–CENP-C189–290 construct (GST–CENP-C189–290-3A) as well as a single alanine mutant at residues Trp317 (W317A) on the CENP-C290–400 construct (CENP-C290–400-W317A). Both mutants prevented the interaction of their cognate CENP-C segment with CENP-HIKM, clearly implicating the mutated CENP-C residues in the interaction (Fig. 2, F and G). On the other hand, mutations in a highly conserved Glu-Phe-Ile-Ile-Asp motif at residues 302–306 of CENP-C did not interfere with the interaction with CENP-HK (Fig. S2, A and B).
We then combined the mutations (henceforth referred to as “4A” mutant) in the context of the GST–CENP-C2–545 construct (GST–CENP-C2–545-4A) and tested their effects on CENP-HIKM binding. Although neither mutation grossly perturbed CENP-HIKM binding in isolation (unpublished data), their combination disrupted CENP-HIKM binding (Fig. 2 H), confirming the importance of both hydrophobic motifs in the CENP-C PEST region on the interaction with CENP-HIKM. In SEC experiments, binding of CENP-C2–545-3A or CENP-C2–545-W317A to CENP-HIKM was partially impaired, whereas the combination of mutations in the CENP-C2–545-4A mutant entirely prevented binding to CENP-HIKM (Fig. S1, C–F), demonstrating that the mutations have additive negative effects on the physical interaction of the CENP-HIKM complex and CENP-C. The interaction of CENP-C2–545-4A with the Mis12 complex (Petrovic et al., 2010, 2014; Screpanti et al., 2011), which involves the N-terminal region of CENP-C (Fig. 1 B), appeared unaltered in comparison to wild-type CENP-C2–545 (Fig. S3, A–E).
The CENP-C–CENP-HIKM interaction promotes CENP-HIKM localization to kinetochores
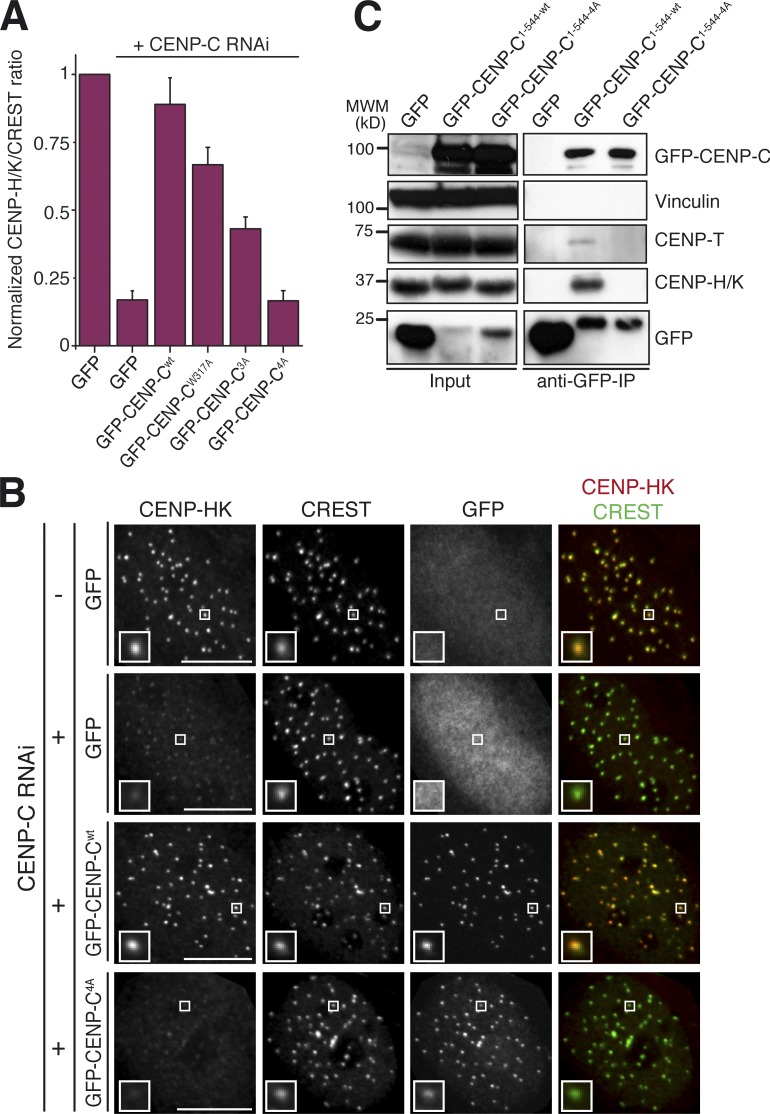
We asked whether the CENP-HIKM binding interface of CENP-C is important for kinetochore localization of CCAN subunits in HeLa cells. For this, we generated inducible stable cell lines that expressed full-length wild-type GFP–CENP-C, or the 3A, W317A, or 4A mutants (Fig. S3, F and G). After depletion of endogenous CENP-C by RNAi and induction of transgene expression, the levels of CENP-HK at kinetochores were quantified (Fig. 3 A). Expression of GFP–CENP-Cwt largely rescued the kinetochore levels of CENP-HK observed in control cells. GFP–CENP-C3A and GFP–CENP-CW317A, on the other hand, only produced a partial rescue of the kinetochore levels of CENP-HK (Fig. 3 A), in agreement with their partial retention of the interaction with CENP-HIKM observed in SEC experiments (Fig. S1, C–F). Conversely, GFP–CENP-C4A was unable to produce significant rescue of the CENP-HK levels (Fig. 3, A and B), in line with the biochemical experiments.
Figure 3.
The CENP-C4A mutant abrogates rescue of CENP-HK localization to kinetochores. (A) Quantification of immunofluorescence experiment in fixed Flp-In T-REx HeLa cells depleted of endogenous CENP-C (where indicated) and expressing the indicated GFP constructs. The kinetochore levels of CENP-HK were measured and normalized to CREST. Graphs and bars indicate means ± SEM of three independent experiments. (B) Representative images of the dataset quantified in A and documenting the levels of CENP-HK protein in cells expressing GFP–CENP-C or GFP–CENP-C4A. Bars, 10 µM. Magnification = 630×. Insets show a magnification of the white boxed areas that capture one or more kinetochores in each panel. (C) GFP–CENP-C1–544 but not the GFP–CENP-C1–544-4A mutant coimmunoprecipitates CENP-HK and CENP-TW. α-GFP coimmunoprecipitation analysis was performed on protein extracts from cycling Flp-In T-REx HeLa cells expressing GFP, GFP–CENP-C, or GFP–CENP-C4A from an inducible promoter. Total protein extracts (Input) and immunoprecipitates (α-GFP co-IP) were separated by SDS-PAGE and blotted with the indicated antibodies. Vinculin served as loading control. IP, immunoprecipitation.
These results were supported by immunoprecipitation experiments in which we used GFP–CENP-C1–544-wt or GFP–CENP-C1–544-4A as baits (we did not use full-length CENP-C for these experiments to avoid dimerization with endogenous CENP-C via the CENP-C C-terminal domain). GFP–CENP-C1–544-wt efficiently pulled down CENP-HK, whereas GFP–CENP-C1–544-4A failed to do so (Fig. 3 C). Overall, the remarkable consistency of the results of experiments in vitro and in vivo allows us to conclude that CENP-HIKM binds directly to CENP-C and that this interaction is required for kinetochore localization of CENP-HIKM.
CENP-TW localization to kinetochores depends on HIKM
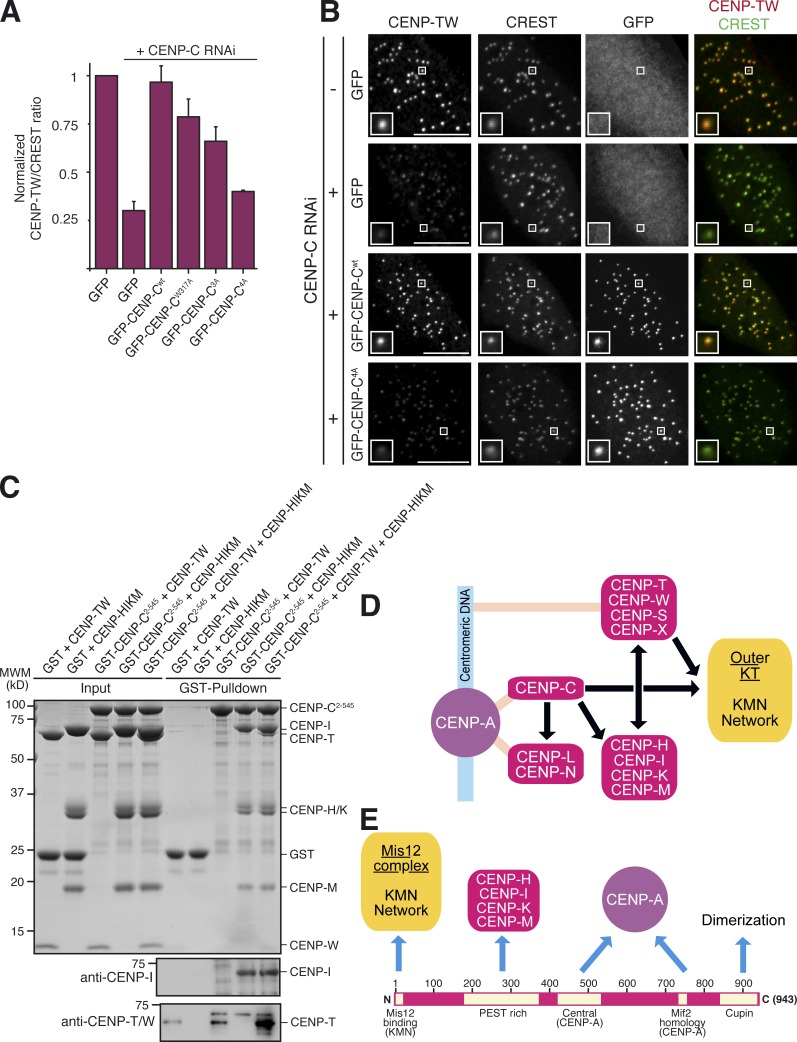
In Fig. 1 (D and E), we demonstrated that kinetochore localization of CENP-TW depends on CENP-C. Kinetochore localization of CENP-TW had been previously shown to depend also on the CENP-HIKM complex (Hori et al., 2008; Basilico et al., 2014). Thus, we analyzed how CENP-C mutants impairing CENP-HIKM localization to kinetochores impacted kinetochore localization of CENP-TW. As for CENP-HK, wild-type CENP-C fully rescued kinetochore localization of CENP-TW, but there was a progressive reduction of CENP-TW kinetochore levels when CENP-C mutants were expressed (Fig. 4, A and B). In agreement with these observations, we readily identified CENP-TW in precipitates of GFP–CENP-C1–544-wt but not in precipitates of GFP–CENP-C1–544-4A (Fig. 3 C).
Figure 4.
CENP-C4A abrogates rescue of CENP-TW localization to kinetochores. (A) Quantification of immunofluorescence experiment in fixed Flp-In T-REx HeLa cells depleted of endogenous CENP-C (where indicated) and expressing the indicated GFP constructs. The kinetochore levels of CENP-TW were measured and normalized to CREST. Graphs and bars indicate mean ± SEM of three independent experiments. (B) Representative images of the dataset quantified in A and documenting the levels of CENP-TW protein in cells expressing GFP–CENP-C or GFP–CENP-C4A. Bars, 10 µM. Magnification = 630×. (C) GST–CENP-C2–545 does not bind directly to CENP-TW in a GST pull-down assay. Bands were visualized with Coomassie brilliant blue staining. (left) Only when HIKM is included in the assay, CENP-TW is incorporated into the complex. Because CENP-I and CENP-T comigrate on the SDS-PAGE gel shown, 10% of the pull-down samples were separated on a new SDS-PAGE and subjected to Western blotting with either a CENP-T or CENP-I antibody. (D) Recasting of the kinetochore assembly scheme based on our new results. (E) The CENP-C sequence contains a linear array of motifs whose succession appears to correlate with the order of kinetochore assembly from the CENP-A nucleosome toward the outer kinetochore. N, N terminus; C, C terminus.
GST–CENP-C2–545 did not interact with CENP-TW in a pull-down assay with recombinant proteins (Fig. 4 C). In contrast, CENP-TW was pulled down by GST–CENP-C2–545 when CENP-HIKM was included in the reaction (Fig. 4 C), in agreement with our previous results showing a direct interaction of CENP-HIKM with CENP-TW (Basilico et al., 2014). The result was confirmed by Western blot analysis of immunoprecipitated fractions (Fig. 4 C). Although these data argue that CENP-HIKM is necessary for kinetochore recruitment of CENP-TW (Basilico et al., 2014), the interaction of CENP-HIKM with CENP-TW in vitro appears to be relatively low affinity in comparison with the very robust interaction of CENP-HIKM with CENP-C. This predicts that CENP-TW may require additional contacts for efficient kinetochore targeting. Collectively, the localization and interaction results, including the observation that CENP-HIKM and CENP-TW are direct interaction partners (Basilico et al., 2014), support the conclusion that both CENP-HIKM and CENP-TW depend on CENP-C for kinetochore localization and that CENP-TW does not bind tightly to CENP-C, at least in the absence of other interaction partners.
Conclusions
The CCAN has been implicated in the fascinating mechanism of new CENP-A deposition during the cell cycle, a process that is at the basis of epigenetic inheritance of centromeres (Fukagawa and Earnshaw, 2014). Moreover, the inner kinetochore is directly involved in recruiting, shaping, and functionally influencing the outer kinetochore (Schittenhelm et al., 2007; Ribeiro et al., 2010; Gascoigne et al., 2011; Przewloka et al., 2011; Screpanti et al., 2011; Matson et al., 2012; Hori et al., 2013; Hornung et al., 2014; Suzuki et al., 2014). Finally, the CCAN has been implicated in interactions with the microtubule plus end, thus possibly complementing the function of the outer kinetochore in chromosome alignment (McClelland et al., 2007; Amaro et al., 2010; Dumont et al., 2012). Understanding the physical organization of the inner kinetochore is therefore an important goal of current kinetochore research. Here, we have identified the PEST domain of CENP-C as an interaction domain for the CENP-HIKM complex and implicated four conserved residues of CENP-C in this interaction. Mutation of these residues impairs binding of CENP-C to CENP-HIKM (but not to the Mis12 complex) in vitro to undetectable levels and leads to a complete loss of CENP-HIKM kinetochore localization in vivo. The same mutations on CENP-C also lead to a dramatic loss of CENP-TW from kinetochores, which is consistent with our previous observation that CENP-TW binds the CENP-HIKM complex directly (Basilico et al., 2014). N-terminal or C-terminal tagging of CENP-C interfered with kinetochore recruitment of the Mis12 complex or with stable incorporation of CENP-C to kinetochores, respectively, preventing us from testing rigorously the effects of the four CENP-C mutations on kinetochore recruitment of the Mis12 complex (unpublished data). However, our results in vitro and previous evidence (Basilico et al., 2014; Kim and Yu, 2015) argue that depletion of HIKM subunits is not sufficient for robust displacement of the Mis12 complex from kinetochores.
Our results are consistent with a model in which CENP-C occupies the first position downstream of CENP-A in a single pathway of kinetochore assembly (Fig. 4 D). Our data argue that CENP-TW positions itself in this pathway, together with other CCAN subunits, downstream of CENP-C, in agreement with several other studies (Carroll et al., 2010; Basilico et al., 2014; Kim and Yu, 2015; Krizaic et al., 2015; Logsdon et al., 2015; Tachiwana et al., 2015). Claims that CENP-T and CENP-C establish independent pathways of kinetochore recruitment downstream from CENP-A (Gascoigne et al., 2011; Rago et al., 2015) may reflect differences in the levels of RNAi-based depletion or in the details of the localization analysis. Several CCAN subunits have been implicated in the deposition of new CENP-A required for centromere propagation (see for instance Okada et al., 2009; Moree et al., 2011; Fachinetti et al., 2013). This is a potential confounding factor because the localization pattern of inner kinetochore proteins cannot be solely viewed as a representation of a static set of protein–protein interactions. Interference with new CENP-A deposition during each cell cycle will eventually affect indirectly the pattern of localization of inner kinetochore subunits, regardless of their direct physical interactions. Be that as it may, our conclusion that CENP-T is positioned downstream of CENP-C is not inconsistent with the identified role of CENP-T in regulating outer kinetochore assembly through an interaction with the Ndc80 complex and possibly other outer kinetochore components (Gascoigne et al., 2011; Bock et al., 2012; Schleiffer et al., 2012; Nishino et al., 2012, 2013; Malvezzi et al., 2013; Kim and Yu, 2015; Rago et al., 2015). The conclusion, however, implies that such role of CENP-TW in kinetochore assembly occurs downstream of CENP-C (Fig. 4 D).
Dissection of CENP-C is bringing to light a linear organization of binding motifs within a largely structurally disordered protein (Fig. 4 E). Specifically, the N-terminal region and the central domain have been shown to interact with the outer kinetochore and the centromere, respectively (Przewloka et al., 2011; Screpanti et al., 2011; Kato et al., 2013; Hornung et al., 2014), whereas the C-terminal region contains binding sites for proteins involved in new CENP-A deposition and CENP-C dimerization (Sugimoto et al., 1997; Cohen et al., 2008; Trazzi et al., 2009; Moree et al., 2011; Dambacher et al., 2012; Unhavaithaya and Orr-Weaver, 2013). We now report that between the outer kinetochore binding site and the CENP-A binding site is a binding site for the CCAN subunits CENP-HIKM. This linear organization of motifs (Fig. 4 E) justifies our interpretation that CENP-C may be a blueprint that determines the correct positioning of kinetochore subunits along the inner to outer kinetochore axis (Suzuki et al., 2014). In the future, it will be important to elucidate the entire network of interactions of CCAN subunits around the CENP-C scaffold through biochemical reconstitution and in vivo validation.
Materials and methods
Expression and purification of CENP-C constructs
From a codon-optimized cDNA synthesized by GeneArt (Life Technologies) encoding the human CENP-C sequence, different CENP-C constructs have been subcloned in pGEX-6P-2rbs (GenBank accession code KM817768), a modified pGEX-6P vector (GE Healthcare; the vector utilizes an inducible tac promoter), as a 3′ fusion to the sequence encoding GST. Mutant CENP-C constructs were created by site-directed mutagenesis using the QuikChange kit (Agilent Technologies). All constructs and mutants were sequence verified. The expression and purification procedure was the same for all CENP-C constructs and mutants. The parental pGEX-6P-2rbs vector expressing CENP-C2–545 was a gift of S. Jeganathan (Max-Planck Institute of Molecular Physiology, Dortmund, Germany). Escherichia coli C41 (DE3) cells harboring vectors expressing CENP-C constructs or CENP-C mutant constructs were grown in Terrific Broth at 37°C to an OD600 of 0.8–1, when 0.2 mM IPTG was added and the culture was grown at 20°C for ∼15 h. Cell pellets were resuspended in lysis buffer (50 mM Hepes, pH 7.5, 500 mM NaCl, 10% glycerol, and 2 mM β-mercaptoethanol), lysed by sonication, and cleared by centrifugation at 48,000 g at 4°C for 30 min. The cleared lysate was applied to glutathione Sepharose 4 Fast Flow beads (GE Healthcare) preequilibrated in lysis buffer, incubated at 4°C for 2 h, washed with 70 column volumes of lysis buffer and either subjected to an overnight cleavage reaction with 3C protease to separate CENP-C constructs from GST or eluted in a lysis buffer supplemented with 20 mM glutathione. A 5-ml HiTrap Heparin HP column (GE Healthcare) was preequilibrated in 20 mM Hepes, pH 7.5, 200 mM NaCl, 5% glycerol, and 2 mM β-mercaptoethanol. The eluate from glutathione beads was adjusted to a final salt concentration of 200 mM NaCl, loaded onto the Heparin column, and eluted with a linear gradient of 200–1,200 mM NaCl in 20 bed column volumes. Fractions containing CENP-C were concentrated and loaded onto a Superdex 200 SEC column (GE Healthcare) preequilibrated in SEC buffer (20 mM Hepes, pH 7.5, 300 mM NaCl, 2.5% glycerol, and 2 mM Tris(2-carboxyethyl)phosphine [TCEP]). Fractions containing CENP-C were concentrated, flash frozen in liquid nitrogen, and stored at −80°C.
GST pull-down assays
All GST pull-down experiments were performed using preblocked GSH Sepharose beads in pull-down buffer (10 mM Hepes, pH 7.5, 200 mM NaCl, 0.05% Triton X-100, 2.5% glycerol, and 1 mM TCEP). GST–CENP-C various constructs always served as bait at a 1-µM concentration, whereas potential binding partners served as prey at a 3-µM concentration. The bait was loaded to 12-µl preblocked beads, before the prey was added. At the same time, 1 µg of each protein was added into Laemmli sample loading buffer for the input gel. The reaction volume was added up to 40 µl with buffer and incubated at 4°C for 1 h, rotating. Beads were spun down at 500 g for 3 min. The supernatant was removed, and beads were washed twice with 250 µl buffer. The supernatant was removed completely, and samples were boiled in 15 µl Laemmli sample loading buffer and run on a 14% SDS-PAGE gel. Bands were visualized with Coomassie brilliant blue staining.
Preblocking of GSH Sepharose beads
750 µl GSH Sepharose beads were washed twice with 1 ml washing buffer (20 mM Hepes, pH 7.5, and 200 mM NaCl) and incubated in 1 ml blocking buffer (20 mM Hepes, pH 7.5, 500 mM NaCl, and 500 µg/µl BSA) overnight at 4°C rotating. Beads were washed 5× with 1 ml washing buffer and resuspended in 500 µl washing buffer to have a 50/50 slurry of beads and buffer.
Analytical SEC migration shift assays
Analytical SEC experiments were performed on a calibrated Superdex 200 5/150 column (GE Healthcare). All samples were eluted under isocratic conditions at 4°C in SEC buffer (20 mM Hepes, pH 7.5, 300 mM NaCl, 2.5% glycerol, and 2 mM TCEP) at a flow rate of 0.2 ml/min. Elution of proteins was monitored at 280 nm. 100-µl fractions were collected and analyzed by SDS-PAGE and Coomassie blue staining. To detect the formation of a complex, proteins were mixed at the indicated concentrations in 50 µl, incubated for at least 2 h on ice, and then subjected to SEC.
Cross-linking analysis
CENP-C2–545–CENP-H–CENP-K complex was cross-linked with isotope-labeled disuccinimidyl suberate and digested with Lys-C and trypsin after quenching with ammonium bicarbonate. Cross-linked peptides were enriched using SEC, analyzed by liquid chromatography coupled to tandem mass spectrometry, and identified by the search algorithm, xQuest. Cross-linking, mass spectrometry analysis, and database searching were performed as previously described (Herzog et al., 2012).
Visualization of the cross-links was performed by converting the raw data (in form of Excel spreadsheets) to the GEXF (Graph Exchange XML Format) data format using custom shell scripts (supplemental material). The data were then imported into the Gephi software that was modified to allow simultaneous calculation and display of curved and straight connectors (i.e., intra- and intermolecular cross-links). The Gephi graph was exported as an Illustrator file (Adobe) for final processing.
Cell culture and transfection
Parental Flp-In T-REx HeLa cells used to generate stable doxycycline-inducible cell lines were a gift from S. Taylor (University of Manchester, Manchester, England, UK). They were grown at 37°C in the presence of 5% CO2 in DMEM (PAN Biotech) supplemented with 10% tetracycline-free FBS (Invitrogen) and 2 mM l-glutamine (PAN-Biotech). Flp-In T-REx HeLa cells (Tighe et al., 2004) and maintained in DMEM supplemented with 10% tetracycline-free FBS, 2 mM l-glutamine, 250 µg/ml hygromycin (Invitrogen), and 4 µg/ml blasticidin (Invitrogen). GFP–CENP-C fusions were expressed by addition of 25 ng/ml doxycycline (Sigma-Aldrich) for 24 or 48 h. For CENP-C silencing, we used a single siRNA (target sequence: 5′-GGAUCAUCUCAGAAUAGAA-3′ obtained from Sigma-Aldrich), targeting the coding region of endogenous CENP-C mRNA. Transfections were performed with HyPerFect (QIAGEN) according to the manufacturer’s instructions. To obtain an efficient depletion of CENP-C, 60 nM siRNA was transfected 3× within 72 h. In the last 24 h, 25 ng/ml doxycycline was added to induce GFP–CENP-C construct expression. The siRNA did not affect expression of GFP-CENP. Phenotypes were analyzed 96 h after the first siRNA addition, and protein depletion was monitored by Western blotting or immunofluorescence.
Mammalian plasmids
CENP-C constructs were created by cDNA subcloning in pcDNA5/FRT/TO-EGFP-IRES vector, a modified version of pcDNA5/FRT/TO vector (Invitrogen). pcDNA5/FRT/TO vector (Invitrogen) is a tetracycline-inducible expression vector designed for use with the Flp-In T-REx system. It carries a hybrid human cytomegalovirus/TetO2 promoter for high-level, tetracycline-regulated expression of the target gene. The control plasmid for EGFP expression was created by PCR amplifying the EGFP sequence from pEGFP-C1 (Takara Bio Inc.) and cloning it into the pcDNA5/FRT/TO vector previously modified to carry an internal ribosomal entry site (IRES) sequence to obtain the pcDNA5/FRT/TO EGFP-IRES vector (Petrovic et al., 2010). All plasmids used in the study for mammalian expression (Fig. 3, B and C; and Fig. 4 B) were derived from the pCDNA5/FRT/TO-EGFP-IRES and used for genomic integration and expression of human CENP-C proteins. To create all N-terminally tagged EGFP fusions, we amplified CENP-C full-length or 1–544 fragments by PCR from a full-length human RNAi-resistant CENP-C cDNA synthesized by GeneArt (Life Technologies) and subcloned them in frame with the EGFP tag into the pcDNA5/FRT/TO EGFP-IRES vector using the restriction sites BamHI and XhoI. Mutant CENP-C constructs were created by site-directed mutagenesis using the QuikChange kit (Agilent Technologies). All constructs were sequence verified.
Immunoprecipitation and immunoblotting
Cycling cells were harvested by trypsinization and lysed by incubation in lysis buffer (75 mM Hepes, pH 7.5, 150 mM KCl, 1.5 mM EGTA, 1.5 mM MgCl2, 10% glycerol, 0.075% NP-40, 90 U/ml Benzonase [Sigma-Aldrich], and protease inhibitor cocktail [Serva]) at 4°C for 15 min followed by sonication and centrifugation. Extracts were precleared with a mixture of protein A–Sepharose (CL-4B; GE Healthcare) and protein G–Sepharose (rec-Protein G-Sepharose 4B; Invitrogen) at 4°C for 1 h. Subsequently, extracts were incubated with GFP-Traps (ChromoTek) at 4°C for 3–4 h. Immunoprecipitates were washed with lysis buffer, resuspended in Laemmli sample buffer, boiled, and analyzed by Western blotting using 12% NuPAGE gels (Life Technologies). The following antibodies were used: anti-GFP (in house–made rabbit polyclonal antibody raised against full-length GFP from Aequorea victoria, full length; 1:4,000), anti-Vinculin (mouse monoclonal, clone hVIN-1; 1:15,000; V9131; Sigma-Aldrich), anti–α-tubulin (mouse monoclonal, T9026; Sigma-Aldrich), anti–CENP-TW (in house–made rabbit polyclonal antibody SI0882, raised against the full-length protein complex; 1:800; a gift of S. Jeganathan, Max Planck Institute of Molecular Physiology, Dortmund, Germany), anti–CENP-C (rabbit polyclonal antibody SI410 raised against residues 23–410 of human CENP-C; 1:1,200; Trazzi et al., 2009), anti–CENP-HK (rabbit polyclonal antibody SI0930 raised against the full-length human CENP-HK complex; 1:1,000), anti–CENP-I (rabbit polyclonal antibody raised against residues 57–281 of human CENP-I), and anti–CENP-L (rabbit polyclonal, 17007–1-AP; Acries Antibodies). Secondary antibodies were affinity-purified anti–mouse (Amersham, part of GE Healthcare) and anti–rabbit (Amersham) conjugated to horseradish peroxidase (1:10,000). After incubation with ECL Western blotting system (GE Healthcare), images were acquired with ChemiBIS 3.2 (DNR Bio-Imaging Systems). Levels were adjusted with ImageJ (National Institutes of Health) and Photoshop (Adobe), and images were cropped accordingly
Immunofluorescence and quantification
Flp-In T-REx HeLa cells were grown on coverslips precoated with 0.01% poly-l-lysine (Sigma-Aldrich). Cells were fixed with PBS/PHEM (Pipes, Hepes, EGTA, and MgCl2)-paraformaldehyde 4% followed by permeabilization with PBS/PHEM–Triton X-100 0.5%. The following antibodies were used for immunostaining: anti–CENP-TW (in house–made rabbit polyclonal antibody SI0882, raised against the full-length protein complex; 1:800), CREST/anticentromere antibodies (human autoimmune serum, 1:100; Antibodies, Inc.), anti–CENP-C (SI410; 1:1,000), anti–CENP-HK (SI0930; 1:800). Rodamine red–conjugated, DyLight 405–conjugated secondary antibodies were purchased from Jackson ImmunoResearch Laboratories, Inc. Alexa Fluor 647–labeled secondary antibodies were obtained from Invitrogen. Coverslips were mounted with Mowiol mounting media (EMD Millipore). All experiments were imaged at room temperature using the spinning-disk confocal microscopy of a 3i Marianas system (Intelligent Imaging Innovations) equipped with a microscope (Axio Observer Z1; Carl Zeiss), a confocal scanner unit (CSU-X1; Yokogawa Electric Corporation), Plan Apochromat 63× or 100×/1.4 NA objectives (Carl Zeiss), and camera (Orca Flash 4.0 sCMOS; Hamamatsu Photonics) and converted into maximal intensity projections TIFF files for illustrative purposes. Quantification of kinetochore signals was performed on unmodified z series images using Imaris 7.3.4 software (Bitplane). After background subtraction, all signals were normalized to CREST, and values obtained for control cells were set to 1. Quantifications are based on three independent experiments in which 5–10 cells were analyzed.
Online supplemental material
Fig. S1 shows the SEC profiles of two CENP-C segments described in the Results and discussion, as well as SEC profiles demonstrating the compound effects of mutations within the PEST region of CENP-C on the CENP-C–CENP-HIKM interaction. Fig. S2 shows that a conserved Glu-Phe-Ile-Ile-Asp motif of CENP-C is not required for the interaction of CENP-C with CENP-HK. Fig. S3 shows that CENP-C2–545 binds the Mis12 complex in SEC experiments even if mutated in the interaction site for CENP-HIKM and loading controls for Figs. 3 and 4 (F and G). Table S1 contains the entire list of intra- and intermolecular cross-links and is provided online as an Excel file. Supplemental material also includes a ZIP file that provides custom scripts used to convert raw cross-linking data from an Excel spreadsheet to the GEXF. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201412028/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201412028.dv.
Acknowledgments
We thank Giuseppe Ossolengo of the antibody facility at the European Institute of Oncology for help with antibody production, Ingrid Vetter for help with the preparation of Fig. 2 D, Sadasivam Jeganathan for the anti–CENP-TW antibody and the CENP-C2–545 construct, Stefano Maffini and Anna De Antoni for help in the development of the CENP-C RNAi protocol, Doro Vogt for preparation of purified CENP-TW protein, and the members of the Musacchio laboratory for helpful discussions.
A. Musacchio acknowledges funding by the European Union’s seventh Framework Program European Research Council agreement KINCON and the Integrated Project MitoSys. F. Herzog is supported by the Bavarian Research Center of Molecular Biosystems and by a Ludwigs Maximilian excellent junior grant.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CCAN
- constitutive centromere–associated network
- IRES
- internal ribosomal entry site
- SEC
- size-exclusion chromatography
- TCEP
- Tris(2-carboxyethyl)phosphine
References
- Amano M., Suzuki A., Hori T., Backer C., Okawa K., Cheeseman I.M., and Fukagawa T.. 2009. The CENP-S complex is essential for the stable assembly of outer kinetochore structure. J. Cell Biol. 186:173–182. 10.1083/jcb.200903100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaro A.C., Samora C.P., Holtackers R., Wang E., Kingston I.J., Alonso M., Lampson M., McAinsh A.D., and Meraldi P.. 2010. Molecular control of kinetochore-microtubule dynamics and chromosome oscillations. Nat. Cell Biol. 12:319–329. 10.1038/ncb2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basilico F., Maffini S., Weir J.R., Prumbaum D., Rojas A.M., Zimniak T., De Antoni A., Jeganathan S., Voss B., van Gerwen S., et al. 2014. The pseudo GTPase CENP-M drives human kinetochore assembly. eLife. 3:e02978 10.7554/eLife.02978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock L.J., Pagliuca C., Kobayashi N., Grove R.A., Oku Y., Shrestha K., Alfieri C., Golfieri C., Oldani A., Dal Maschio M., et al. 2012. Cnn1 inhibits the interactions between the KMN complexes of the yeast kinetochore. Nat. Cell Biol. 14:614–624. 10.1038/ncb2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C.W., Silva M.C., Godek K.M., Jansen L.E., and Straight A.F.. 2009. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat. Cell Biol. 11:896–902. 10.1038/ncb1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C.W., Milks K.J., and Straight A.F.. 2010. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 189:1143–1155. 10.1083/jcb.201001013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheerambathur D.K., and Desai A.. 2014. Linked in: formation and regulation of microtubule attachments during chromosome segregation. Curr. Opin. Cell Biol. 26:113–122. 10.1016/j.ceb.2013.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M. 2014. The kinetochore. Cold Spring Harb. Perspect. Biol. 6:a015826 10.1101/cshperspect.a015826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., and Desai A.. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997. 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Cohen R.L., Espelin C.W., De Wulf P., Sorger P.K., Harrison S.C., and Simons K.T.. 2008. Structural and functional dissection of Mif2p, a conserved DNA-binding kinetochore protein. Mol. Biol. Cell. 19:4480–4491. 10.1091/mbc.E08-03-0297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambacher S., Deng W., Hahn M., Sadic D., Fröhlich J., Nuber A., Hoischen C., Diekmann S., Leonhardt H., and Schotta G.. 2012. CENP-C facilitates the recruitment of M18BP1 to centromeric chromatin. Nucleus. 3:101–110. 10.4161/nucl.18955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., Gall W.E., Ciferri C., Cimini D., Musacchio A., and Salmon E.D.. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 127:969–982. 10.1016/j.cell.2006.09.047 [DOI] [PubMed] [Google Scholar]
- Dumont S., Salmon E.D., and Mitchison T.J.. 2012. Deformations within moving kinetochores reveal different sites of active and passive force generation. Science. 337:355–358. 10.1126/science.1221886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W.C., and Rothfield N.. 1985. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 91:313–321. 10.1007/BF00328227 [DOI] [PubMed] [Google Scholar]
- Fachinetti D., Folco H.D., Nechemia-Arbely Y., Valente L.P., Nguyen K., Wong A.J., Zhu Q., Holland A.J., Desai A., Jansen L.E., and Cleveland D.W.. 2013. A two-step mechanism for epigenetic specification of centromere identity and function. Nat. Cell Biol. 15:1056–1066. 10.1038/ncb2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E., Black B.E., Bailey A.O., Yates J.R. III, and Cleveland D.W.. 2006. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8:458–469. 10.1038/ncb1397 [DOI] [PubMed] [Google Scholar]
- Fukagawa T., and Earnshaw W.C.. 2014. The centromere: chromatin foundation for the kinetochore machinery. Dev. Cell. 30:496–508. 10.1016/j.devcel.2014.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne K.E., Takeuchi K., Suzuki A., Hori T., Fukagawa T., and Cheeseman I.M.. 2011. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 145:410–422. 10.1016/j.cell.2011.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q., Tao Y., Liu H., Teng M., and Li X.. 2013. Structural insights into the role of the Chl4-Iml3 complex in kinetochore assembly. Acta Crystallogr. D Biol. Crystallogr. 69:2412–2419. 10.1107/S0907444913022397 [DOI] [PubMed] [Google Scholar]
- Guse A., Carroll C.W., Moree B., Fuller C.J., and Straight A.F.. 2011. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 477:354–358. 10.1038/nature10379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog F., Kahraman A., Boehringer D., Mak R., Bracher A., Walzthoeni T., Leitner A., Beck M., Hartl F.U., Ban N., et al. 2012. Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science. 337:1348–1352. 10.1126/science.1221483 [DOI] [PubMed] [Google Scholar]
- Hinshaw S.M., and Harrison S.C.. 2013. An Iml3-Chl4 heterodimer links the core centromere to factors required for accurate chromosome segregation. Cell Reports. 5:29–36. 10.1016/j.celrep.2013.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., Amano M., Suzuki A., Backer C.B., Welburn J.P., Dong Y., McEwen B.F., Shang W.H., Suzuki E., Okawa K., et al. 2008. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 135:1039–1052. 10.1016/j.cell.2008.10.019 [DOI] [PubMed] [Google Scholar]
- Hori T., Shang W.H., Takeuchi K., and Fukagawa T.. 2013. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J. Cell Biol. 200:45–60. 10.1083/jcb.201210106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung P., Troc P., Malvezzi F., Maier M., Demianova Z., Zimniak T., Litos G., Lampert F., Schleiffer A., Brunner M., et al. 2014. A cooperative mechanism drives budding yeast kinetochore assembly downstream of CENP-A. J. Cell Biol. 206:509–524. 10.1083/jcb.201403081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izuta H., Ikeno M., Suzuki N., Tomonaga T., Nozaki N., Obuse C., Kisu Y., Goshima N., Nomura F., Nomura N., and Yoda K.. 2006. Comprehensive analysis of the ICEN (Interphase Centromere Complex) components enriched in the CENP-A chromatin of human cells. Genes Cells. 11:673–684. 10.1111/j.1365-2443.2006.00969.x [DOI] [PubMed] [Google Scholar]
- Kato H., Jiang J., Zhou B.R., Rozendaal M., Feng H., Ghirlando R., Xiao T.S., Straight A.F., and Bai Y.. 2013. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science. 340:1110–1113. 10.1126/science.1235532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., and Yu H.. 2015. Multiple assembly mechanisms anchor the KMN spindle checkpoint platform at human mitotic kinetochores. J. Cell Biol. 208:181–196. 10.1083/jcb.201407074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizaic I., Williams S.J., Sánchez P., Rodríguez-Corsino M., Stukenberg P.T., and Losada A.. 2015. The distinct functions of CENP-C and CENP-T/W in centromere propagation and function in Xenopus egg extracts. Nucleus. 6:133–143. 10.1080/19491034.2014.1003509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.T., Rattner J.B., Jablonski S.A., and Yen T.J.. 2006. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J. Cell Biol. 175:41–53. 10.1083/jcb.200606020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon G.A., Barrey E.J., Bassett E.A., DeNizio J.E., Guo L.Y., Panchenko T., Dawicki-McKenna J.M., Heun P., and Black B.E.. 2015. Both tails and the centromere targeting domain of CENP-A are required for centromere establishment. J. Cell Biol. 208:521–531. 10.1083/jcb.201412011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiolica A., Cittaro D., Borsotti D., Sennels L., Ciferri C., Tarricone C., Musacchio A., and Rappsilber J.. 2007. Structural analysis of multiprotein complexes by cross-linking, mass spectrometry, and database searching. Mol. Cell. Proteomics. 6:2200–2211. 10.1074/mcp.M700274-MCP200 [DOI] [PubMed] [Google Scholar]
- Malvezzi F., Litos G., Schleiffer A., Heuck A., Mechtler K., Clausen T., and Westermann S.. 2013. A structural basis for kinetochore recruitment of the Ndc80 complex via two distinct centromere receptors. EMBO J. 32:409–423. 10.1038/emboj.2012.356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson D.R., Demirel P.B., Stukenberg P.T., and Burke D.J.. 2012. A conserved role for COMA/CENP-H/I/N kinetochore proteins in the spindle checkpoint. Genes Dev. 26:542–547. 10.1101/gad.184184.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh A.D., and Meraldi P.. 2011. The CCAN complex: linking centromere specification to control of kinetochore-microtubule dynamics. Semin. Cell Dev. Biol. 22:946–952. 10.1016/j.semcdb.2011.09.016 [DOI] [PubMed] [Google Scholar]
- McCleland M.L., Kallio M.J., Barrett-Wilt G.A., Kestner C.A., Shabanowitz J., Hunt D.F., Gorbsky G.J., and Stukenberg P.T.. 2004. The vertebrate Ndc80 complex contains Spc24 and Spc25 homologs, which are required to establish and maintain kinetochore-microtubule attachment. Curr. Biol. 14:131–137. 10.1016/j.cub.2003.12.058 [DOI] [PubMed] [Google Scholar]
- McClelland S.E., Borusu S., Amaro A.C., Winter J.R., Belwal M., McAinsh A.D., and Meraldi P.. 2007. The CENP-A NAC/CAD kinetochore complex controls chromosome congression and spindle bipolarity. EMBO J. 26:5033–5047. 10.1038/sj.emboj.7601927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milks K.J., Moree B., and Straight A.F.. 2009. Dissection of CENP-C-directed centromere and kinetochore assembly. Mol. Biol. Cell. 20:4246–4255. 10.1091/mbc.E09-05-0378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moree B., Meyer C.B., Fuller C.J., and Straight A.F.. 2011. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J. Cell Biol. 194:855–871. 10.1083/jcb.201106079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T., Takeuchi K., Gascoigne K.E., Suzuki A., Hori T., Oyama T., Morikawa K., Cheeseman I.M., and Fukagawa T.. 2012. CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell. 148:487–501. 10.1016/j.cell.2011.11.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T., Rago F., Hori T., Tomii K., Cheeseman I.M., and Fukagawa T.. 2013. CENP-T provides a structural platform for outer kinetochore assembly. EMBO J. 32:424–436. 10.1038/emboj.2012.348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obuse C., Yang H., Nozaki N., Goto S., Okazaki T., and Yoda K.. 2004. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells. 9:105–120. 10.1111/j.1365-2443.2004.00705.x [DOI] [PubMed] [Google Scholar]
- Okada M., Cheeseman I.M., Hori T., Okawa K., McLeod I.X., Yates J.R. III, Desai A., and Fukagawa T.. 2006. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8:446–457. 10.1038/ncb1396 [DOI] [PubMed] [Google Scholar]
- Okada M., Okawa K., Isobe T., and Fukagawa T.. 2009. CENP-H-containing complex facilitates centromere deposition of CENP-A in cooperation with FACT and CHD1. Mol. Biol. Cell. 20:3986–3995. 10.1091/mbc.E09-01-0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic A., Pasqualato S., Dube P., Krenn V., Santaguida S., Cittaro D., Monzani S., Massimiliano L., Keller J., Tarricone A., et al. 2010. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J. Cell Biol. 190:835–852. 10.1083/jcb.201002070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic A., Mosalaganti S., Keller J., Mattiuzzo M., Overlack K., Krenn V., De Antoni A., Wohlgemuth S., Cecatiello V., Pasqualato S., et al. 2014. Modular assembly of RWD domains on the Mis12 complex underlies outer kinetochore organization. Mol. Cell. 53:591–605. 10.1016/j.molcel.2014.01.019 [DOI] [PubMed] [Google Scholar]
- Przewloka M.R., Venkei Z., Bolanos-Garcia V.M., Debski J., Dadlez M., and Glover D.M.. 2011. CENP-C is a structural platform for kinetochore assembly. Curr. Biol. 21:399–405. 10.1016/j.cub.2011.02.005 [DOI] [PubMed] [Google Scholar]
- Rago F., Gascoigne K.E., and Cheeseman I.M.. 2015. Distinct organization and regulation of the outer kinetochore KMN network downstream of CENP-C and CENP-T. Curr. Biol. 25:671–677. 10.1016/j.cub.2015.01.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro S.A., Vagnarelli P., Dong Y., Hori T., McEwen B.F., Fukagawa T., Flors C., and Earnshaw W.C.. 2010. A super-resolution map of the vertebrate kinetochore. Proc. Natl. Acad. Sci. USA. 107:10484–10489. 10.1073/pnas.1002325107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H., Tomkiel J., Cooke C.A., Ratrie H. III, Maurer M., Rothfield N.F., and Earnshaw W.C.. 1992. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell. 70:115–125. 10.1016/0092-8674(92)90538-N [DOI] [PubMed] [Google Scholar]
- Santaguida S., and Musacchio A.. 2009. The life and miracles of kinetochores. EMBO J. 28:2511–2531. 10.1038/emboj.2009.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittenhelm R.B., Heeger S., Althoff F., Walter A., Heidmann S., Mechtler K., and Lehner C.F.. 2007. Spatial organization of a ubiquitous eukaryotic kinetochore protein network in Drosophila chromosomes. Chromosoma. 116:385–402. 10.1007/s00412-007-0103-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiffer A., Maier M., Litos G., Lampert F., Hornung P., Mechtler K., and Westermann S.. 2012. CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat. Cell Biol. 14:604–613. 10.1038/ncb2493 [DOI] [PubMed] [Google Scholar]
- Screpanti E., De Antoni A., Alushin G.M., Petrovic A., Melis T., Nogales E., and Musacchio A.. 2011. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr. Biol. 21:391–398. 10.1016/j.cub.2010.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K., Gronemeyer B., Lu W., Eugster E., and Tomkiel J.E.. 2002. Mutational analysis of the central centromere targeting domain of human centromere protein C, (CENP-C). Exp. Cell Res. 275:81–91. 10.1006/excr.2002.5495 [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Kuriyama K., Shibata A., and Himeno M.. 1997. Characterization of internal DNA-binding and C-terminal dimerization domains of human centromere/kinetochore autoantigen CENP-C in vitro: role of DNA-binding and self-associating activities in kinetochore organization. Chromosome Res. 5:132–141. 10.1023/A:1018422325569 [DOI] [PubMed] [Google Scholar]
- Suzuki A., Badger B.L., Wan X., DeLuca J.G., and Salmon E.D.. 2014. The architecture of CCAN proteins creates a structural integrity to resist spindle forces and achieve proper Intrakinetochore stretch. Dev. Cell. 30:717–730. 10.1016/j.devcel.2014.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachiwana H., Müller S., Blümer J., Klare K., Musacchio A., and Almouzni G.. 2015. HJURP involvement in de novo CenH3CENP-A and CENP-C recruitment. Cell Reports. 11:22–32. 10.1016/j.celrep.2015.03.013 [DOI] [PubMed] [Google Scholar]
- Takeuchi K., Nishino T., Mayanagi K., Horikoshi N., Osakabe A., Tachiwana H., Hori T., Kurumizaka H., and Fukagawa T.. 2014. The centromeric nucleosome-like CENP-T-W-S-X complex induces positive supercoils into DNA. Nucleic Acids Res. 42:1644–1655. 10.1093/nar/gkt1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Chang H.L., Kagami A., and Watanabe Y.. 2009. CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev. Cell. 17:334–343. 10.1016/j.devcel.2009.08.004 [DOI] [PubMed] [Google Scholar]
- Tighe A., Johnson V.L., and Taylor S.S.. 2004. Truncating APC mutations have dominant effects on proliferation, spindle checkpoint control, survival and chromosome stability. J. Cell Sci. 117:6339–6353. 10.1242/jcs.01556 [DOI] [PubMed] [Google Scholar]
- Trazzi S., Perini G., Bernardoni R., Zoli M., Reese J.C., Musacchio A., and Della Valle G.. 2009. The C-terminal domain of CENP-C displays multiple and critical functions for mammalian centromere formation. PLoS ONE. 4:e5832 10.1371/journal.pone.0005832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unhavaithaya Y., and Orr-Weaver T.L.. 2013. Centromere proteins CENP-C and CAL1 functionally interact in meiosis for centromere clustering, pairing, and chromosome segregation. Proc. Natl. Acad. Sci. USA. 110:19878–19883. 10.1073/pnas.1320074110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.H., Tomkiel J., Saitoh H., Johnson D.H., and Earnshaw W.C.. 1996. Identification of overlapping DNA-binding and centromere-targeting domains in the human kinetochore protein CENP-C. Mol. Cell. Biol. 16:3576–3586. [DOI] [PMC free article] [PubMed] [Google Scholar]