Mio, a conserved member of the SEACAT/GATOR2 complex, is required for concentration of active Plk1 and Aurora kinases at spindle poles and for subsequent spindle formation and chromosome segregation.
Abstract
Coordination of cell growth and proliferation in response to nutrient supply is mediated by mammalian target of rapamycin (mTOR) signaling. In this study, we report that Mio, a highly conserved member of the SEACAT/GATOR2 complex necessary for the activation of mTORC1 kinase, plays a critical role in mitotic spindle formation and subsequent chromosome segregation by regulating the proper concentration of active key mitotic kinases Plk1 and Aurora A at centrosomes and spindle poles. Mio-depleted cells showed reduced activation of Plk1 and Aurora A kinase at spindle poles and an impaired localization of MCAK and HURP, two key regulators of mitotic spindle formation and known substrates of Aurora A kinase, resulting in spindle assembly and cytokinesis defects. Our results indicate that a major function of Mio in mitosis is to regulate the activation/deactivation of Plk1 and Aurora A, possibly by linking them to mTOR signaling in a pathway to promote faithful mitotic progression.
Introduction
The Nup107–160 complex (Nup107 complex) is an evolutionarily conserved nucleoporin subcomplex that plays a crucial role in nuclear pore complex (NPC) assembly, mRNA export, and cell differentiation (Boehmer et al., 2003; Harel et al., 2003; Walther et al., 2003; González-Aguilera and Askjaer, 2012). A small fraction of the Nup107 complex localizes to kinetochores from early prophase to late anaphase (Belgareh et al., 2001). Efficient depletion of the Nup107 complex component Seh1 from mammalian cells causes chromosome alignment and segregation defects (Zuccolo et al., 2007) by altering the centromeric localization of the chromosomal passenger complex (Platani et al., 2009).
During mitosis, a signaling network involving the kinases Aurora A, Polo-like kinase 1 (Plk1), and CDK1/Cyclin B and their counteracting phosphatases controls the localization and function of various components of the mitotic spindle (Carmena et al., 2009; Rieder, 2011). Aurora A kinase localizes on centrosomes and spindle pole microtubules from late S phase throughout mitosis, where it plays a role in mitotic entry, centrosome maturation and separation, and bipolar spindle formation and function (Barr and Gergely, 2007; Carmena et al., 2009; Hochegger et al., 2013). Aurora A substrates include TPX2 (Kufer et al., 2002), TACC3 (Giet et al., 2002; Barros et al., 2005), Ajuba (Hirota et al., 2003), Eg5 (Giet et al., 1999), and HURP (Yu et al., 2005; Wong et al., 2008).
Plk1 is a critical regulator of mitosis that regulates centrosome maturation, kinetochore–microtubule attachment, and cleavage furrow ingression (Petronczki et al., 2008; Bruinsma et al., 2012; Zitouni et al., 2014). Spindle pole localization of Plk1 controls recruitment of pericentrin and γ-tubulin complexes to centrosomes (Lane and Nigg, 1996; Casenghi et al., 2003; Lee and Rhee, 2011) and has also been implicated in centrosome disjunction and separation (Bruinsma et al., 2012). Centrosomal Plk1 additionally controls spindle positioning and orientation by regulating binding of the dynein–dynactin complex to its cortical targeting factors Numa and LGN (Kiyomitsu and Cheeseman, 2012). During prometaphase, Plk1 localization at kinetochores is required for chromosome alignment and faithful chromosome segregation (Elowe et al., 2007; Liu et al., 2012; Maia et al., 2012).
Mitotic activity of Aurora A and Plk1 kinases is controlled by a balance of phosphorylation and dephosphorylation in time and space. Aurora A activation depends on the autophosphorylation of Thr288 in its activation loop, which occurs primarily at centrosomes (Littlepage et al., 2002; Zorba et al., 2014) and on TPX2-mediated localization and activation on spindle microtubules (Kufer et al., 2002; Bayliss et al., 2003; Eyers and Maller, 2003, 2004; Tsai et al., 2003). Aurora A/Bora activates Plk1 at centrosomes in late G2/prophase via phosphorylation of its activation loop at Thr210 (Macůrek et al., 2008; Seki et al., 2008).
Mammalian target of rapamycin (mTOR) is a serine/threonine protein kinase involved in cell proliferation, cell size regulation, transcription, and cytoskeletal regulation in response to a variety of input signals (Harris and Lawrence, 2003; Jacinto and Hall, 2003; Wullschleger et al., 2006). Two mTOR complexes have been identified in mammalian cells mTORC1 and mTORC2 (Guertin and Sabatini, 2007). The mTORC1 complex contains the regulatory protein raptor and, by regulating the phosphorylation of p70S6 kinase and 4E-binding protein 1 (4EBP1), controls their downstream functions in protein translation, cell growth, and cell proliferation (Loewith et al., 2002). mTORC2 contains the regulatory subunit rictor and is involved in regulation of the actin cytoskeleton (Jacinto et al., 2004). Almost all documented mTOR functions take place during interphase, although the mTORC1 complex has been implicated in mitotic entry in fission yeast through the stress MAPK pathway (Petersen and Nurse, 2007). mTORC1 activation requires Rag-GTPases, two regulators of which have recently been identified: the SEACAT/GATOR1 and 2 subcomplexes (Panchaud et al., 2013b).
Here, we have identified a mitotic role for Mio, a highly conserved member of the SEACAT/GATOR2 complex, which regulates Rag-1 and mTOR signaling (Bar-Peled et al., 2013; Panchaud et al., 2013a). We show that Mio is required for spindle formation and subsequent chromosome segregation and for the proper concentration of active Plk1 and Aurora A kinases at centrosomes and spindle poles. Our results suggest that Mio may regulate the activation/deactivation of Plk1 and Aurora A, possibly by linking them to mTOR signaling in a pathway that promotes faithful mitotic progression.
Results
Nucleoporin Seh1 interacts with Mio, WDR24, and WDR59, components of the SEACAT/GATOR2 complex, in mammalian cells
Seh1 is a member of the Nup107 complex that functions in NPC assembly and localizes to kinetochores in mitosis (Belgareh et al., 2001). To better define its biochemical interactions in mammalian cells, we isolated GFP-Seh1 both from asynchronous and mitotically arrested HeLaGFPSeh1 cells (Platani et al., 2009) and identified interacting proteins by SILAC (stable isotope labeling by amino acids in cell culture) mass spectrometry (Ong and Mann, 2007). This analysis identified all known Seh1-interacting proteins, including components of the Nup107 complex (Nup107, 98, 133, 160, and 85) and SEACAT/GATOR2 complexes (Mios, WDR24, and WDR59; Fig. 1 A and Fig. S1 A; Dokudovskaya et al., 2011; Bar-Peled et al., 2013; Panchaud et al., 2013a). Interestingly, Seh1 interacts with the SEACAT/GATOR2 complex equally in interphase and mitosis, whereas its interaction with the rest of the Nup107 complex is stronger during mitosis.
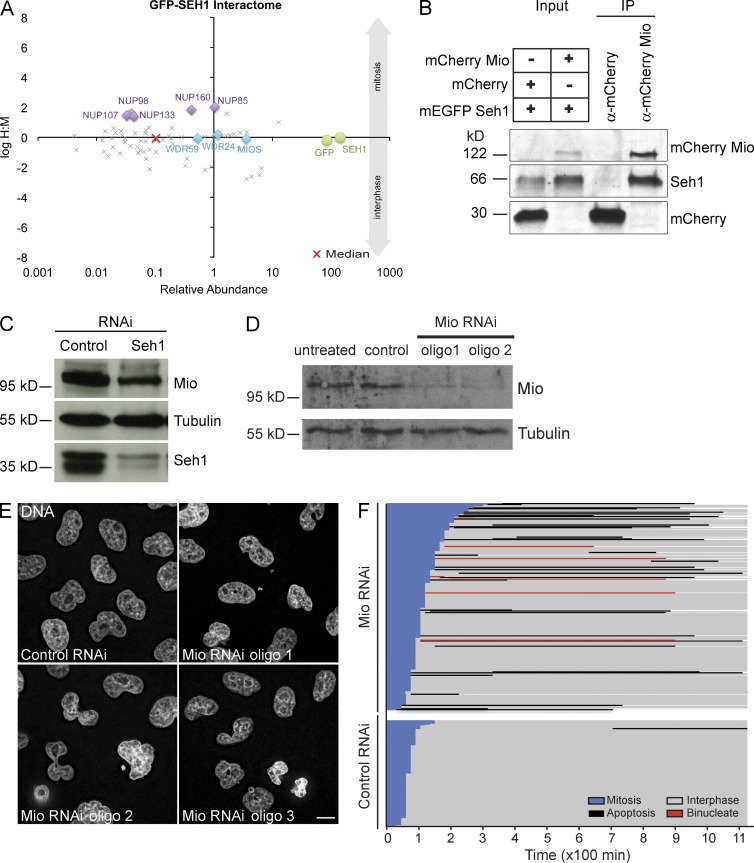
Figure 1.
Seh1 interacts with the SEACAT complex both in interphase and mitosis. (A) Seh1 complexes were purified from a cell line stably expressing GFP-Seh1, HeLaEGP-SEH1. Cells were differentially labeled with isotopic amino acids by growth in SILAC media before immunopurification of GFP-Seh1 complexes. Where indicated, cells were arrested in mitosis using 200 ng/ml nocodazole for 14 h. Immunopurified GFP-Seh1 complexes were analyzed by quantitative mass spectrometry. Plot of log ratio H/M versus relative abundance (summarized peptide intensities normalized by molecular weight) for all quantified proteins is shown. Bait protein GFP-Seh1 (green circles) and the interactors Nup107 complex (purple diamonds) and SEACAT/GATOR2 complex (blue diamonds) are highlighted. This nonbiased quantitative experiment was performed twice in asynchronous cell populations and once in mitotically arrested cells, and top hits were chosen for targeted follow-up validation by immunoprecipitation/Western blot analysis using specific antibodies. (B) Mio interacts with Seh1 in mammalian cells. Immunoprecipitates (IP) are shown of total protein extracts from HeLa cells transfected with mCherry, mCherry-Mio, and EGFP-Seh1. Immunoprecipitation was performed using α-RFP binder, and Seh1 was detected using α-Seh1 antibody. (C) Immunoblots of HeLa cell lysates treated with siRNAs corresponding to negative control and Seh1 (probed using α-Mio and α-Seh1) show that depletion of Seh1 can affect Mio stability. (D) Immunoblots of HeLa cell lysates treated with siRNAs corresponding to negative control and Mio (probed using α-Mio) show efficient depletion of Mio protein with different siRNA oligos 48 h after transfection. (C and D) Tubulin served as a loading control. (E) Abnormal nuclear morphology detected after Mio depletion. HeLa cells transfected with control and Mio siRNA oligos for 48 h were stained with DAPI to reveal the DNA and nuclear morphology. (F) Mio depletion increases mitotic length, binucleation, and apoptosis. HeLa cells transfected with control or Mio siRNAs were incubated for 30 h before phase-contrast imaging for a further 19 h. Images were acquired every 15 min. Image sequences were then analyzed tracking individual control (n = 132) and Mio-depleted cells (n = 289) to determine their behavior. Fate profiles of live control or Mio-depleted cells are shown as a function of time. Bars represent total time spent at different cell cycle stages for individual cells. Starting point T = 0 is NEBD. Bar, 10 µm.
We confirmed the Mio–Seh1 interaction using recombinant mCherry-Mio expressed in mammalian cells. Seh1 coimmunoprecipitated with mCherry-Mio protein but not with the mCherry control (Fig. 1 B). Furthermore, levels of endogenous Mio protein were reduced in HeLa cells upon Seh1 depletion (Fig. 1 C). Because Seh1 overexpression does not increase Mio protein levels (Senger et al., 2011), this suggests that Seh1 promotes Mio’s stability in mammalian cells. Although Seh1 is a nucleoporin, depletion of Mio resulted in no detectable changes in NPC structure or assembly that could be detected by mAb414 staining against FG repeat nucleoporins. The nuclear rim localization of endogenous Seh1 also appeared not to be affected (Fig. S1 B).
Mio depletion causes mitotic defects
Having previously described a requirement for Seh1 for mitotic progression (Platani et al., 2009), we next explored the role of Mio in mitotic spindle assembly and chromosome segregation. RNAi depletion of Mio resulted in an increase in the number of binucleate cells, micronucleation, and aberrant nuclear morphology with a variety of different oligos against Mio (Fig. 1, D and E; and see the following paragraphs). To better define the Mio depletion phenotype in living cells, we used time-lapse phase-contrast microscopy. Cells transfected with control siRNA underwent mitosis with normal timing, and cytokinesis was completed within 2 h followed by a period in interphase (Fig. 1 F). Apoptotic events were rarely seen (1.6%), indicating that phototoxicity is minimal under our imaging conditions. Mio-depleted cells remained in mitosis and cytokinesis for extended periods of time before either furrow ingression resulted in binucleation, a period in interphase, or apoptosis (which was observed both during mitosis and interphase; Fig. 1 F). Mio is therefore required for normal mitotic progression. Interestingly, Mio depletion did not result in an increase in the fraction of cells in mitosis (Fig. 2 A). Therefore, the increased time that cells spend in mitosis must be counterbalanced by a decrease in mitotic entry.
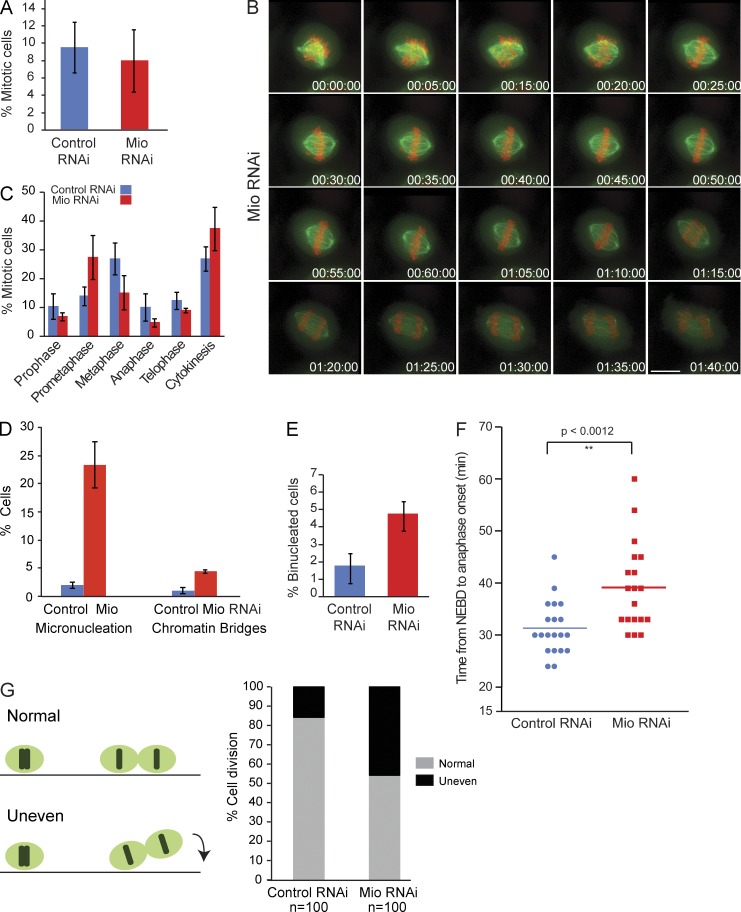
Figure 2.
Mio is required for mitotic progression. (A) Mitotic index of control (blue bar) and Mio-depleted cells (red bar) 48 h after transfection (n = 3). (B) Depletion of Mio causes spindle assembly and cytokinesis defects. Selected maximum intensity projections from time-lapse images show the mitotic and cytokinesis defects. HeLa cell line stably expressing EGFP-tubulin (green) and mRFP-H2B (red), HeLaEGFPTubulin-mRFPH2B, was transfected with Mio siRNA oligos. Images were collected every 5 min at 46 h for a period of 120 min. Numbers indicate time in hours/minutes/seconds. (C) Quantitation of different mitotic stages for control (blue bars) and Mio-depleted cells (red bars; n = 3). (D) Quantitation of micronucleation and chromatin bridges in control and Mio-depleted cells. Error bars represent SD. (E) Binucleation index of control (blue bar) and Mio-depleted cells (red bar) 48 h after transfection (n = 3). (F) Mitotic progression scatter plots of anaphase onset with NEBD as T = 0 in control (blue) and Mio siRNA-treated cells (red) from live cell videos. Statistical significance was determined by a two-tailed, unpaired t test. (G) Quantitation of normal and misoriented cell divisions in cells treated as in B (n = 3). Bar, 10 µm.
More detailed live cell imaging of cells stably expressing mRFP-H2B and EGFP–α-tubulin or EGFP-H2B alone revealed that the kinetics of bipolar spindle formation was abnormal in Mio-depleted cells, although chromosomes eventually aligned on what appeared to be a metaphase plate (Fig. 2 B and Fig. S2 D). Phenotypic analysis of mitotic progression in Mio-depleted cultures showed an increase in prometaphase cells (from 13 to 28%; Fig. 2 C) with spindles often in closer proximity to one side of the cell. During mitotic exit, chromosome segregation defects, including increased chromatin bridges and micronucleation, were observed (Fig. 2 D). Cytokinesis also appeared to be delayed (the percentage of cells in cytokinesis increased from 26 to 37%; Fig. 2 C). In 6.9% of cells, no furrow ingression was observed followed by either fusion of chromatin masses or production of binucleate cells (the latter increased twofold relative to control-depleted cells; Fig. 2 E). Whereas control cells progressed to anaphase soon after metaphase chromosome alignment (31.35 ± 5.2 min), Mio depletion resulted in a prolongation of the period from nuclear envelope breakdown (NEBD) to anaphase onset (40.5 ± 9.4 min; Fig. 2 F). In addition, Mio-depleted cells showed an uneven timing of daughter cell adhesion to the substratum (Fig. 2 G), a characteristic of uneven cell division, as orientation of the cell division plane is controlled by the orientation of the mitotic spindle relative to the cell cortex.
Mio depletion results in spindle and centrosomal defects
The previous observations suggested a potential defect in spindle formation and/or chromosome capture after Mio depletion. However, Mio-depleted cells displayed no obvious defects in kinetochore–microtubule interactions, checkpoint proteins, or checkpoint bypass, at least as reported by Bub1 and Sgo1 staining and the ability of cells to undergo mitotic arrest in nocodazole (Fig. S2, A–C). We therefore looked in detail at centrosome-mediated spindle formation.
Closer inspection of the distribution of α-tubulin and the centrosomal markers γ-tubulin and pericentrin in Mio-depleted cells revealed an increase in several spindle defects. These included abnormal tilt of the spindle axis, disrupted or unfocused spindle poles, monopolar spindles, displacement of the spindle from the cell center toward the cell cortex, and failure of centrosomes to localize at both spindle poles (Fig. 3 A). Most depleted cells showed one or a combination of the aforementioned phenotypes.
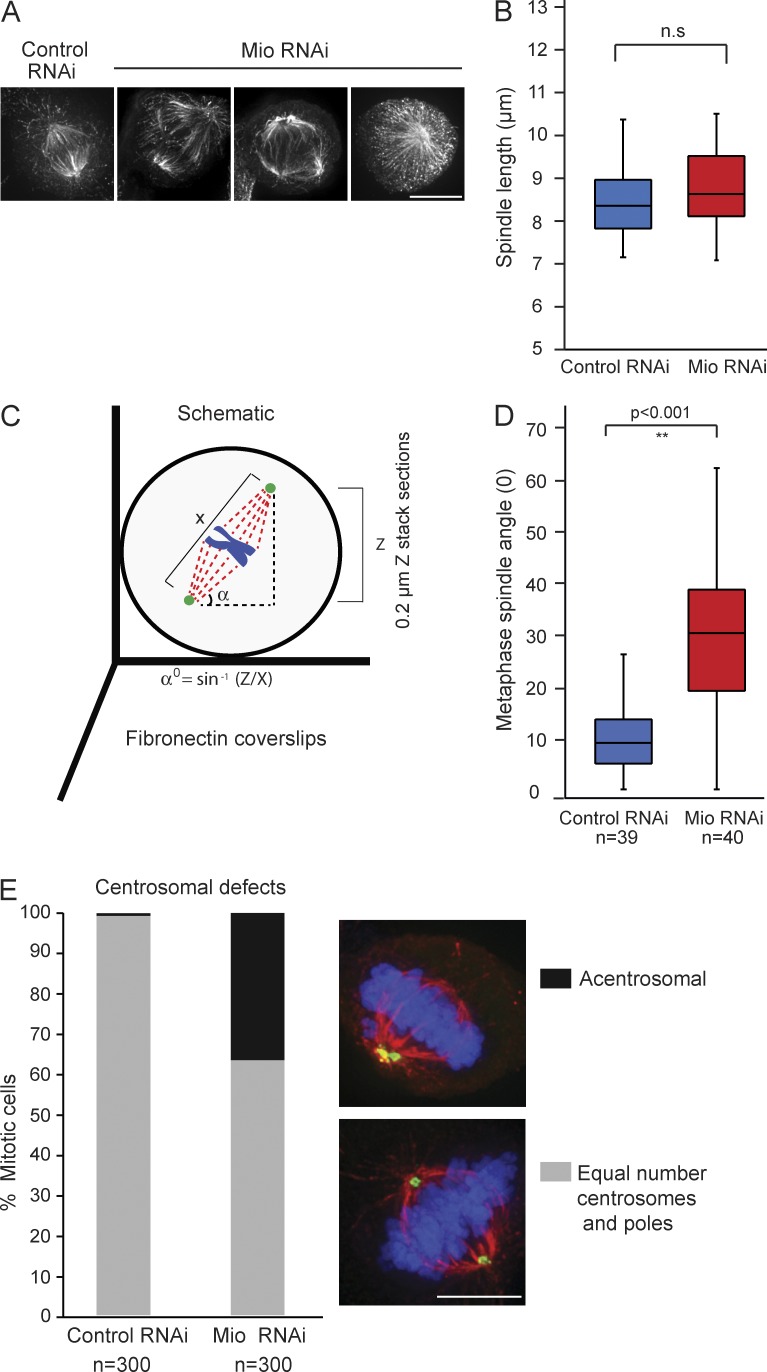
Figure 3.
Mio depletion leads to spindle misorientation and centrosomal defects. (A) Abnormal spindle morphology detected after Mio depletion. HeLa cells were transfected with control and Mio siRNA oligos for 48 h and stained with α-tubulin. (B) Quantification of metaphase spindle length between control (blue) and Mio-depleted cells (red) shows no significant change in the length of the mitotic spindle. (C) Schematic depicting the spindle angle (α) measurement relative to the fibronectin substratum. (D) Quantification of metaphase spindle angles between control (blue) and Mio-depleted cells (red) showing a significant increase of >20o of spindle angle. n = 40 from three experiments. (E) Quantification of mitotic metaphase cells with an unequal number of centrosomes and spindle poles. 3D maximum intensity projections of representative metaphase cells immunostained with α-pericentrin (green), α-tubulin (red), and DNA (blue) are shown on the right. 300 cells per condition (n = 3). Bars, 10 µm.
To measure the orientation of the mitotic spindle in relation to the substratum, 3D image stacks were acquired for Mio-depleted cells grown on fibronectin-coated coverslips. In control RNAi cells grown on these coverslips, the metaphase spindle axis tends to align parallel to the substratum. Although spindle length was normal in Mio-depleted cells (Fig. 3 B), the spindle azimuth, as judged by the metaphase angle α, was significantly increased (Fig. 3, C and D). In addition, Mio-depleted cells showed an increase in bipolar, monoastral spindles, with one pole lacking γ-tubulin or pericentrin (Fig. 3 E).
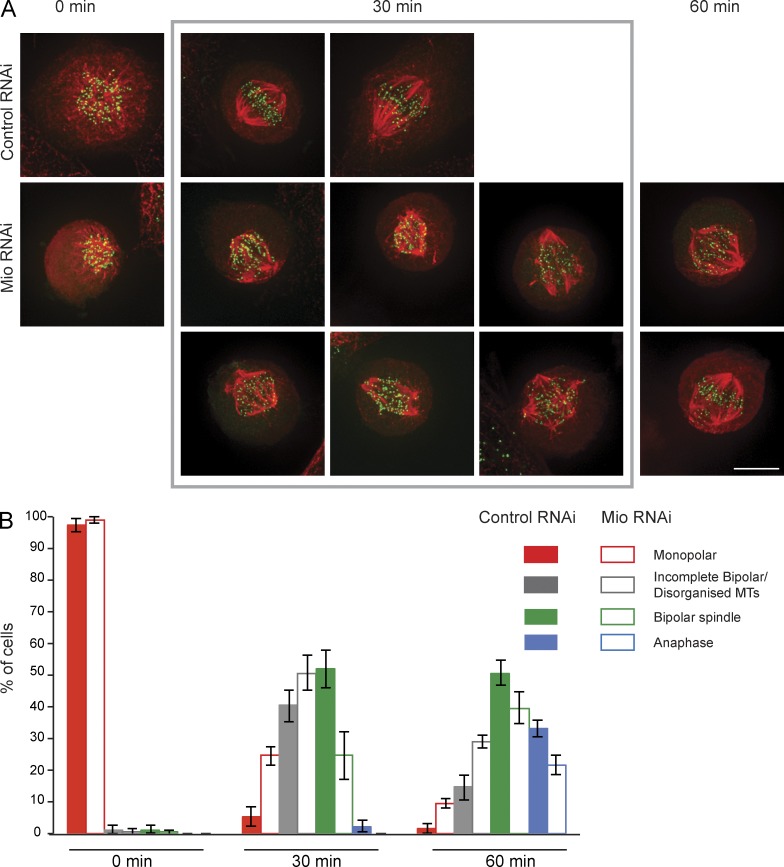
To allow a quantitative measurement of spindle assembly and chromosome alignment, we examined the recovery of Mio-depleted cells from Monastrol, a kinesin Eg5 inhibitor that arrests mitotic cells in prometaphase with monopolar spindles without affecting microtubule dynamics (Kapoor et al., 2000). At 30 min after Monastrol washout, control mitotic cells had rapidly formed a bipolar spindle, with only 40% of cells still having a disorganized spindle. In contrast, at 30 min after Monastrol washout, Mio-depleted cells had a pronounced delay in bipolar spindle formation, with 76% of cells having monopolar or incomplete bipolar spindles (Fig. 4).
Figure 4.
Bipolar spindle formation is delayed in Mio-depleted cells. (A) Control and Mio-depleted cells were arrested with Eg5 inhibitor Monastrol for 3 h. The drug was washed out with fresh medium, and cells were fixed at the indicated time points. Cells were immunostained with α-tubulin (red) and α-ACA (green). (B) Quantitation of spindle and chromosome alignment status in control and Mio-depleted cells at the indicated time points after Monastrol release. 300 cells/time point (n = 3). Bar, 10 µm.
Micronuclei and lagging chromosomes can occur as a result of kinetochore–microtubule misattachments, either due to defects in error correction mechanisms, microtubule dynamics, or spindle formation. After observing defects in spindle formation and micronuclei in Mio-depleted cells, we decided to look at kinetochore–microtubule attachments in detail. We observed no changes in Survivin or Aurora B levels at centromeres of Mio-depleted cells (Fig. S3, A and B), and cold-stable kinetochore fibers were also present, suggesting that the kinetochore attachment site is not disrupted (Fig. S3 C).
Plk1 and Aurora A are misregulated after Mio depletion
To characterize the mechanism underlying the various defects in Mio-depleted mitotic cells, we focused on Plk1 and Aurora A. Both of these mitotic kinases have numerous important roles in spindle formation and centrosome separation (Lens et al., 2010).
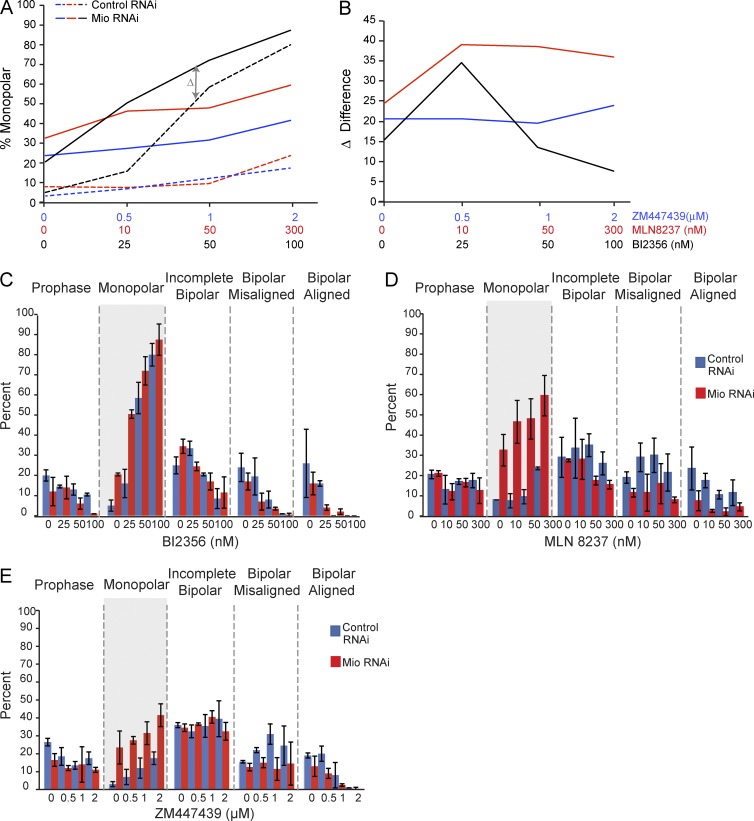
Limiting titration of the specific Plk1 inhibitor BI2356 revealed a synthetic interaction with Mio depletion. In Mio-depleted cells, lower concentrations of BI2536 caused a synergistic increase in the percentage of cells with monopolar spindles relative to the same drug treatment of control RNAi cells (Fig. 5, A–C). Thus, cells depleted of Mio assembled monopolar spindles at concentrations of the drug that did not appreciably affect cells after the control RNAi. This was seen most clearly in Fig. 5 B, which plots Δ, the difference in the percentage of monopolar spindles after drug treatments with or without Mio RNAi. A positive slope of Δ indicates a synthetic response, whereas a horizontal line indicates no increase in drug sensitivity after Mio RNAi. It is important to note that after an initial strong synthetic response, the Δ curve for Plk1 inhibition decreases because both the control and Mio-depleted samples are approaching a limit of 90% monopolar cells.
Figure 5. .
Mio-depleted cells show sensitivity to Plk1 and Aurora A inhibition. (A) Quantitation of monopolar spindles in control (dotted lines) and Mio-depleted cells (solid lines) 46 h after siRNA transfection followed by 1.5-h incubation into the indicated concentrations of ZM447439, BI2356, and MLN8237. (B) The difference (Δ) in monopolar spindle percentage between control and Mio-depleted cells at each indicated drug concentration. (C–E) Mitotic profile of control and Mio-depleted cells 48 h after siRNA transfection followed by 1.5-h incubation into the indicated concentrations of BI2356 (C), MLN8237 (D), and ZM447439 (E). (A–E) 300 cells per condition/drug concentration (n = 3). Error bars represent SD. The highlighted gray areas of the monopolar spindles mitotic stage are used for the graphs in A and B.
Limiting titration of the Aurora A inhibitor MLN8237 also showed a significant, albeit weaker, synthetic interaction (Fig. 5, A, B, and D). In contrast, ZM447439 treatment resulted in an almost horizontal line for Δ, as both control and Mio-depleted cells responded to the Aurora B inhibition with the same efficiency over the range of ZM447439 concentrations tested (Fig. 5, A, B, and E).
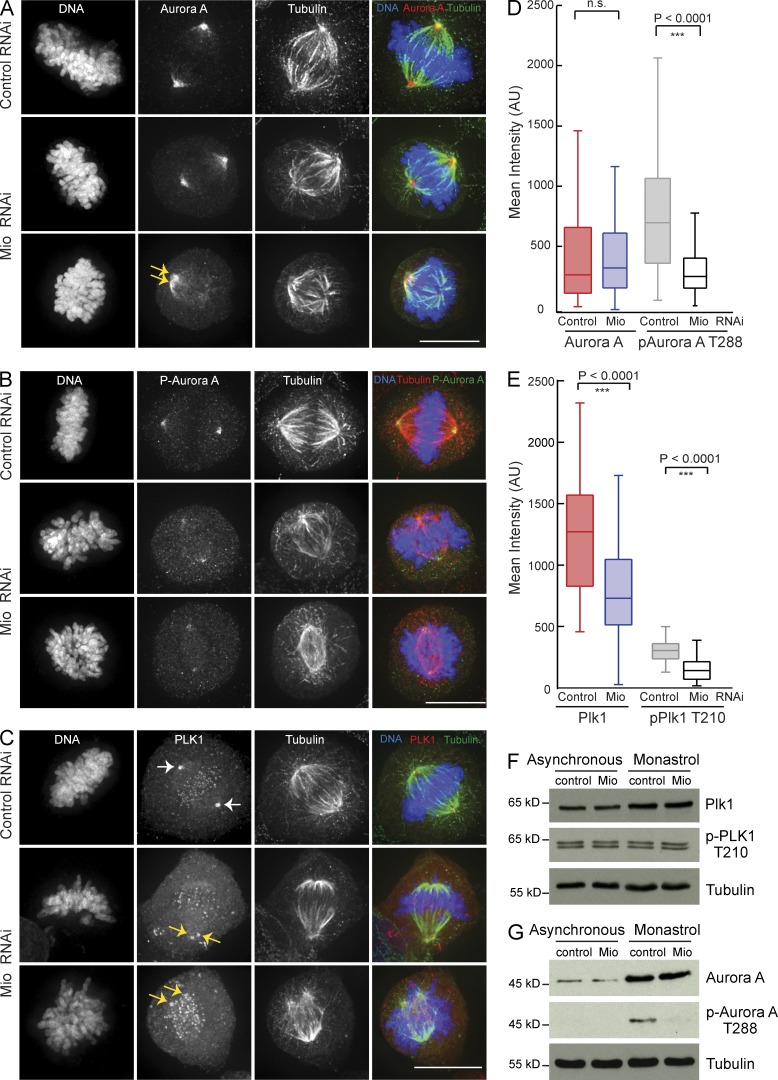
Depletion of Mio had significant effects on the activation of Aurora A and Plk1 at centrosomes. For example, although we observed no changes in total Aurora A localization or the amount on centrosomes and spindles (Fig. 6 A), the level of active Aurora A (Aurora AT288ph, which is phosphorylated at Thr288 on the activation loop) on centrosomes was significantly lower in Mio-depleted cells than in RNAi control cells (Fig. 6, B and D). We confirmed this by immunoblotting (Fig. 6 G). Rescue experiments on HeLa si02ResGFP-Mio cells, where the endogenous Mio was depleted using si02, rescued the prometaphase delay phenotype and restored Aurora AT288ph levels at spindle poles (Fig. S1, C–E). Depletion of all Mio using si01 gave the expected prometaphase delay and reduced Aurora AT288ph signal.
Figure 6.
Plk1 and Aurora A are misregulated after Mio depletion. (A) Control and Mio-depleted cells were fixed and immunostained with α–Aurora A (red), α-tubulin (green), and DNA (blue). Arrows point to centrosomes. (B) Control and Mio-depleted cells were fixed and immunostained with α-p–Aurora AT288 (green), α-tubulin (red), and DNA (blue). (C) Control and Mio-depleted cells were fixed and immunostained with α-Plk1 (red), α-tubulin (green), and DNA (blue). Arrows point to centrosomes. (D) Quantification of Aurora A and p–Aurora AT288 levels at spindle poles in control and Mio-depleted cells. Depletion of Mio reduces p–Aurora AT288 levels, whereas Aurora A is mostly retained. (E) Quantification graph of PLK1 and p-PLK1T210 levels at centrosomes in control and Mio-depleted cells. Both Plk1 and p-Plk1T120 levels at centrosomes are reduced. (D and E) Fluorescence intensities are in arbitrary units (AU). Error bars represent SD. (F and G) Immunoblots of HeLa cell lysates treated with siRNAs corresponding to negative control and Mio from asynchronous and Monastrol-arrested cells (probed using α-PLK1, α–p-PLK1T210, α–Aurora A, and α-p–Aurora AT288) show reduction of Aurora AT288ph signal upon Mio depletion, whereas total Aurora A, Plk1, and Plk1T210ph levels appear unchanged. Tubulin serves as a loading control. n.s., not significant. Bars, 10 µm.
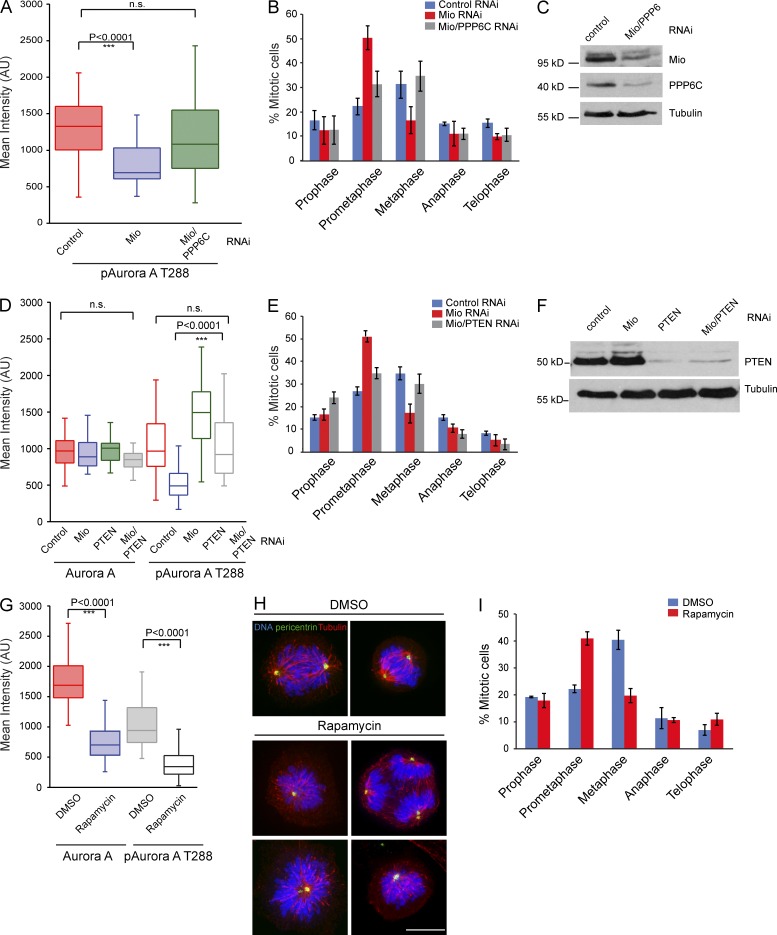
Protein phosphatase 6 regulates mitotic spindle formation by controlling the T loop phosphorylation of Aurora A bound to its activator TPX2 (Zeng et al., 2010). Thus, PPP6C depletion would enhance Aurora A activity and potentially restore the mitotic function of Mio-depleted cells. Indeed, co-depletion of Mio and PPP6C (catalytic subunit; see Fig. 10 C) rescued the prometaphase delay phenotype and restored Aurora AT288ph levels at spindle poles (see Fig. 10, A and B).
Figure 10.
Activity of mTOR is reduced in mitotic cells after Mio depletion. (A) Quantification of p–Aurora AT288 levels at spindle poles in control, Mio-depleted, and Mio/PPP6C co-depleted cells. (B) Mitotic profile of control, Mio-depleted, and Mio/PPP6C co-depleted cells 48 h after siRNA transfection. 300 cells per condition (n = 3). (C and F) Immunoblots of HeLa cell lysates treated with siRNAs corresponding to negative control, Mio, Mio with PPP6C, PTEN, and Mio/PTEN from asynchronous cells (probed using α-Mio, α-PPP6C, and α-PTEN) show efficient depletion of Mio, PPP6C, and PTEN. Tubulin serves as a loading control. (D) Quantification of Aurora A and p–Aurora AT288 levels at spindle poles in control, Mio-depleted, and Mio/PTEN co-depleted cells. PTEN co-depletion with Mio rescues the p–Aurora AT288 levels reduction. (E) Mitotic profile of control, Mio-depleted, and Mio and PTEN co-depleted cells 48 h after siRNA transfection. 300 cells per condition (n = 3). (G) Quantification of Aurora A and p–Aurora AT288 levels at spindle poles in control DMSO- and rapamycin-treated cells. (A, D, and G) Fluorescence intensities are in arbitrary units (AU). (H) Control DMSO- and rapamycin-treated cells were fixed and immunostained with α-pericentrin (green), α-tubulin (red), and DNA (blue). (I) Mitotic profile of control DMSO- and rapamycin-treated cells after 4-h incubation ( n = 3). Error bars represent SD. n.s., not significant. Bar, 10 µm.
Plk1 localization and activity also showed a strong response to Mio depletion. Centrosomal levels of both total Plk1 and active PLK1T210ph were reduced (Fig. 6, C and E; and Fig. S5 C). In contrast, Plk1 localization to centrosomes and kinetochores was not affected (Fig. 6 C). In immunoblots, total Plk1 and active PLK1T210ph protein levels in asynchronous and Monastrol-arrested mitotic cells appeared to be unaffected in Mio-depleted cells (Fig. 6 F). It thus appears that the reduction in Plk1 protein levels and activity is restricted to centrosomes.
These results are consistent with the fact that Plk1 is activated at centrosomes during G2 by Aurora A complexed with its cofactor Bora (Macůrek et al., 2008; Seki et al., 2008).
Localization of HURP and MCAK is altered upon Mio depletion
Both Aurora A and Plk1 are regulatory kinases. We therefore sought to identify downstream effectors that might be responsible for the spindle and mitotic abnormalities observed after Mio depletion and the consequent loss of active Aurora A and Plk1 from centrosomes.
One likely target is HURP, whose binding to microtubules is regulated by Aurora A phosphorylation (Yu et al., 2005; Wong et al., 2008). HURP is part of a Ran-dependent complex required for chromatin-dependent microtubule nucleation (Koffa et al., 2006; Silljé et al., 2006). In control RNAi cells, HURP was localized to spindle microtubules in a band transverse to the spindle axis that was maximal in the vicinity of chromosomes. This localization is consistent with the regulation of HURP by Ran-GTP.
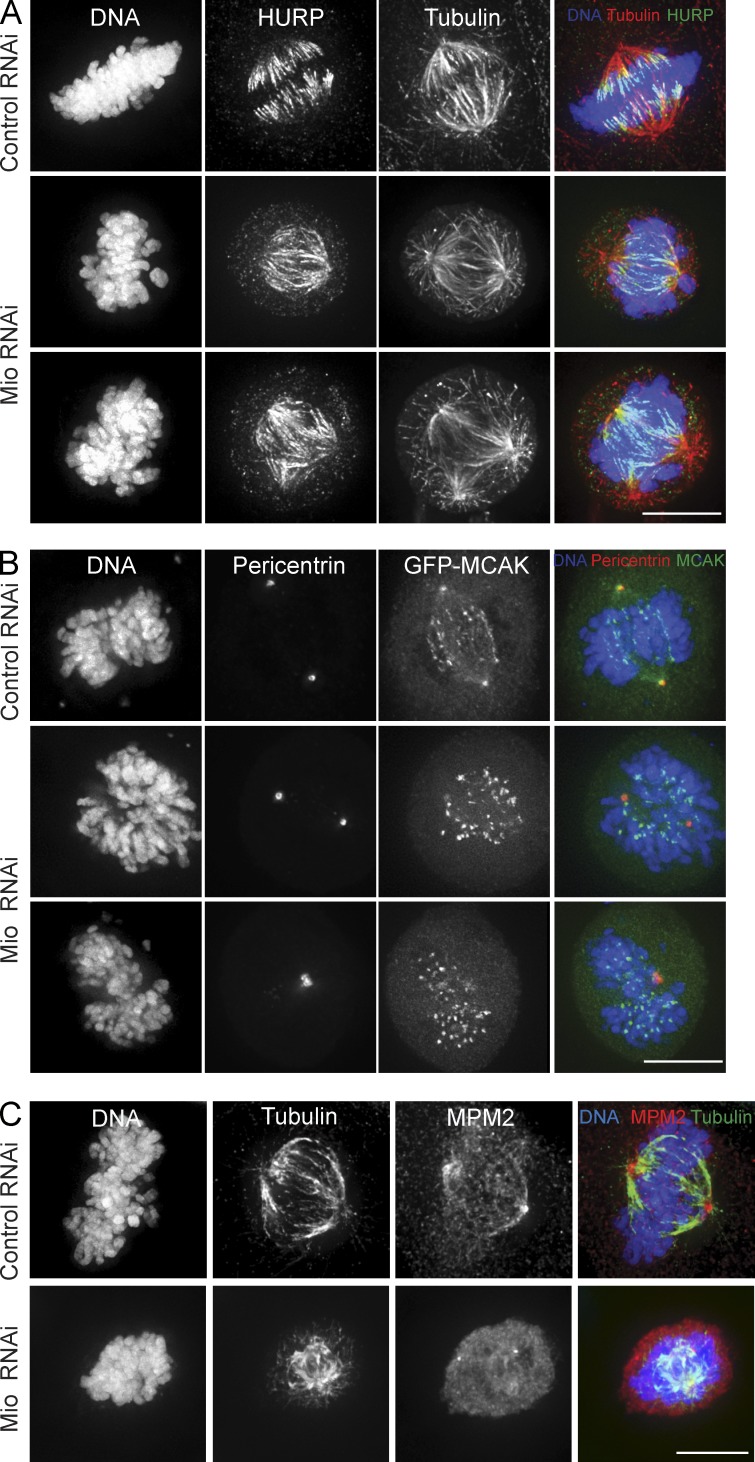
The distribution of HURP on the spindle was dramatically altered upon Mio depletion. The protein was then evenly distributed throughout the spindle, and the chromosome-proximal characteristic band was lost (Fig. 7 A). A similar mislocalization of HURP was previously seen in cells treated with selective Aurora A inhibitors or depleted of the Aurora A–activating partner TPX2 (Kesisova et al., 2013).
Figure 7.
Spindle assembly factors HURP and MCAK are misregulated after Mio depletion. (A) Control and Mio-depleted cells were fixed and immunostained with α-HURP (green), α-tubulin (red), and DNA (blue). (B) Control and Mio-depleted cells transfected with GFP-MCAK (green) were fixed and immunostained with α-pericentrin (red) and DNA (blue). (C) Control and Mio-depleted cells were fixed and immunostained with α-MPM2 (red), α-tubulin (green), and DNA (blue). Bars, 10 µm.
This effect of Mio depletion on HURP appears to be quite specific. TPX2 is a partner of HURP in a complex that contributes to Ran-dependent bipolar spindle formation (Koffa et al., 2006). No changes were observed in either localization or abundance of TPX2 in Mio-depleted cells (Fig. S4, A and C). TPX2 is an important activator of Aurora A (Kufer et al., 2002; Tsai et al., 2003; Özlü et al., 2005), and these observations therefore suggest that the decrease in the levels of active Aurora AT288ph is likely not due to altered TPX2 protein levels. Aurora A still interacted with TPX2 after Mio depletion (Fig. S4 C). Kinase assays on TPX2–Aurora A complexes confirmed a reduction of Aurora A kinase activity toward TPX2 and histone H3 (Fig. S4 D).
Localization of a second important Aurora A substrate was also changed after Mio RNAi. MCAK is a microtubule-depolymerizing kinesin that has an essential role in chromosome segregation by controlling kinetochore–microtubule attachments (Tanenbaum et al., 2011). MCAK is localized to inner centromeres, kinetochores, and spindle poles of mitotic cells, and its subcellular localization and activity are controlled by phosphorylation (Andrews et al., 2004). Importantly, the spindle pole association of MCAK is dependent on Aurora A phosphorylation (Zhang et al., 2008).
The level of centrosomal MCAK was dramatically reduced in cells exposed to Mio RNAi (Fig. 7 B). In contrast, the pool of kinetochore-associated MCAK appeared unaffected by Mio depletion. In cells exposed to control RNAi, GFP-MCAK localized to both kinetochores and spindle poles as previously described (Fig. 7 B; Davis et al., 1983; Lane and Nigg, 1996; Logarinho and Sunkel, 1998). The level of centrosomal MPM2 staining (Davis et al., 1983) was dramatically reduced in cells exposed to Mio RNAi (Fig. 7 C), confirming a reduction of Plk1 activity at the centrosomes (Lane and Nigg, 1996; Logarinho and Sunkel, 1998).
These effects of Mio depletion were specific for HURP and MCAK and were not a result of a general disruption of the centrosome. It has been suggested that Plk1 activity also contributes to centrosomal localization of the plus end–directed bipolar kinesin Eg5 before mitotic entry (Bertran et al., 2011; Smith et al., 2011). Eg5 localized normally to centrosomes in Mio-depleted cells (Fig. S4 B; Lane and Nigg, 1996). Thus, interphase CDK and Plk1 activities in Mio-depleted cells are sufficient to load Eg5 on centrosomes before mitotic entry (Davis et al., 1983; Logarinho and Sunkel, 1998).
NuMA, LGN, and the dynein–dynactin motor complex function at the cell cortex, where they help to generate pulling forces on astral microtubules and direct spindle rotation (Woodard et al., 2010; Kiyomitsu and Cheeseman, 2012). Both NuMA and the dynactin subunit p150 localized normally to spindles and spindle poles in Mio-depleted cells (Fig. S5). However, cortical levels of p150 were slightly increased near the spindle pole (Fig. S5 B). This was the exact opposite from the expected cortical p150 localization at the cortex furthest away from the pole. The above observation is consistent with the hypothesis that spindle pole–localized active Plk1 regulates dynactin complex dissociation from the cell cortex during spindle orientation.
Mitotic progression is impaired in the absence of the Mio Ring/PHD domain
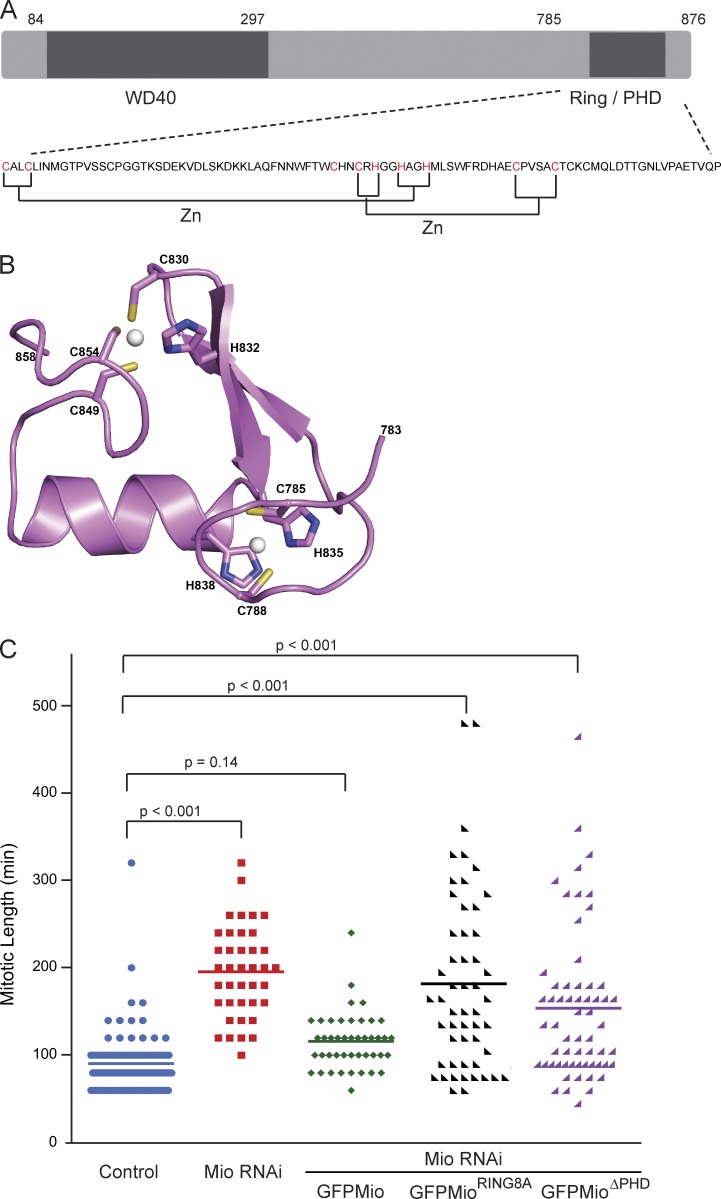
Mio is highly conserved from yeast to mammals (Iida and Lilly, 2004). Its N-terminal domain contains several WD40 repeats that are likely to be involved in protein–protein interaction (Smith et al., 1999). Mio also has a C-terminal domain with a pattern of conserved histidine and cysteine residues resembling a RING/PHD domain (Fig. 8 A). Such RING/PHD domains frequently mediate E3 ligase activity (Deshaies and Joazeiro, 2009). We used the Protein Homology/Analogy Recognition Engine version 2.0 (Phyre2) server (Kelley and Sternberg, 2009) to model Mio’s RING/PHD domain structure on the crystal structure of Ring E3 ubiquitin ligase RNF8 bound to a UBC13/MMS2 heterodimer (Protein Data Bank accession no. 4ORH). Cysteine and histidine residues that are predicted to coordinate the two zinc atoms and help maintain the domain structure are highlighted (Fig. 8, A [red] and B [labels]). Based on this model, we decided to create two Mio mutants: a C-terminal RING/PHD deletion (ΔPHD) missing the last 91 amino acids and a combination of eight specific point mutations (C785, C788, C830, H832, H835, H838, C849, and C854) that convert predicted zinc-binding cysteine and histidine residues to alanine. This would be predicted to disrupt the folding of the Ring domain (Fig. 8 B).
Figure 8.
Mitotic progression is impaired in the absence of the Ring/PHD domain. (A) Schematic diagram showing the domain structure of Mio and the sequence of its C-terminal Ring/PHD domain. The predicted cysteines and histidines involved in zinc coordination are labeled in red. (B) Cartoon representation of predicted 3D molecular structure of Mio C-terminal Ring/PHD domain showing the mutated residues in stick form. Zinc ions are shown in ball representation (as white spheres). (C) Mitotic progression scatter plots with NEBD as T = 0 in control (blue), Mio siRNA-treated cells with and without (red) expression of siRNA-resistant versions of wild-type GFP-Mio (green), GFP-MioRing8A (black), and GFP-MioΔPHD (purple) from live cell videos. n = 214 cells from two independent experiments. Statistical significance was determined by an unpaired Student’s t test.
We performed rescue experiments to evaluate the importance of the RING/PHD domain in mitotic progression (Fig. 8 C). These experiments also serve as overall controls showing that the delay in mitotic progression, observed after Mio depletion, is not due to off-target effects of the RNAi. Stable cell lines expressing siRNA-resistant wild-type or mutant Mio (si02ResGFP-MioWT, si02ResGFP-MioRING8A, and si02ResGFP-MioΔPHD) were depleted of endogenous Mio. The wild-type transgene efficiently rescued the increase in mitotic timing (Fig. 8 C) and the convoluted nuclear shape observed upon Mio depletion. In contrast, neither the Ring8A nor ΔPHD mutants were able to fully rescue these phenotypes. Thus, despite our inability to demonstrate an E3 ubiquitin ligase activity for purified Mio (unpublished data), the Mio C-terminal RING/PHD region clearly is required for normal mitotic progression.
Active mTOR levels are reduced at centrosomes in mitotic cells upon Mio depletion
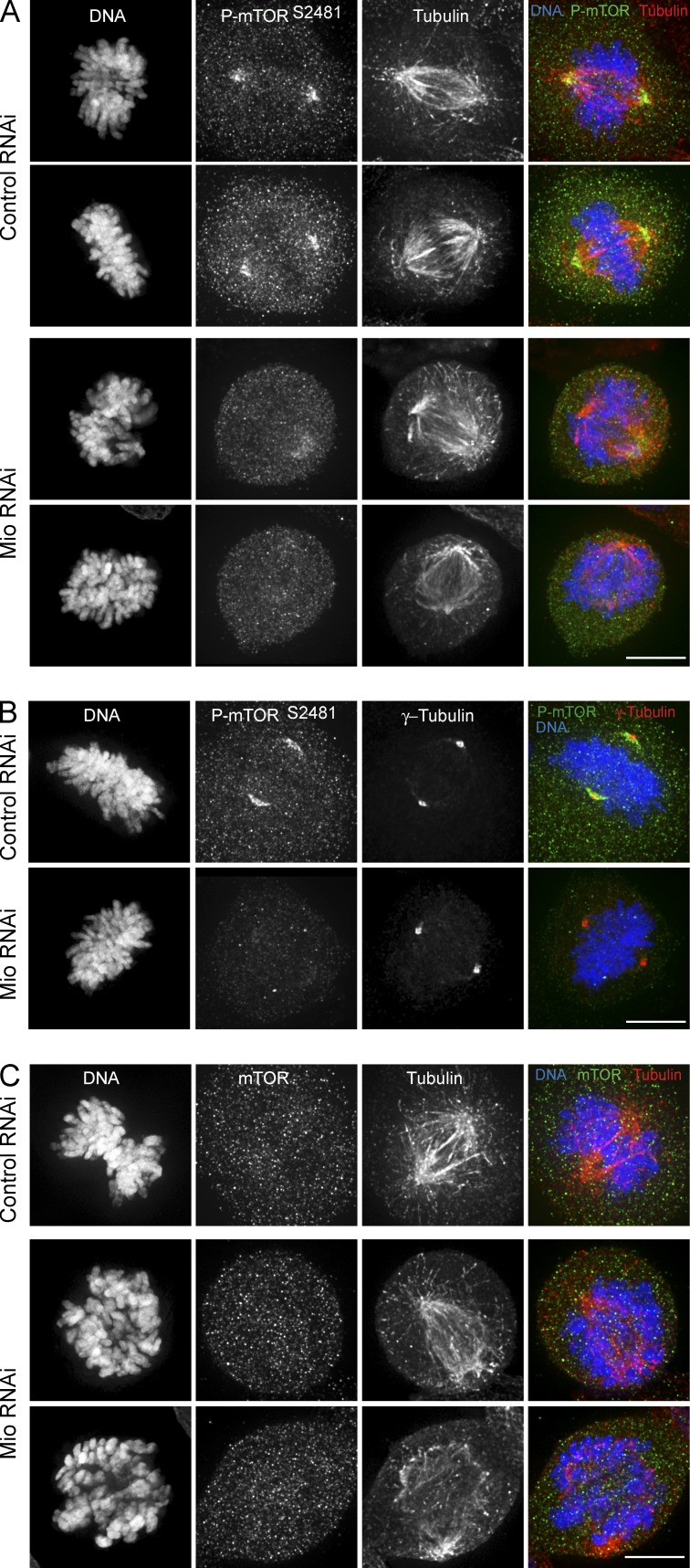
Mio is a highly conserved member of the SEACAT/GATOR2 complex, antagonizing GATOR1, a known inhibitor of Rag-1 GTPase, and hence promoting mTOR signaling (Bar-Peled et al., 2013; Panchaud et al., 2013a). We therefore decided to look directly at levels of active mTOR in cells after Mio depletion. Upon its activation, mTOR is phosphorylated on several sites, including Ser2481 (Peterson et al., 2000). mTOR Ser2481 autophosphorylation correlates with mTOR catalytic activity (Soliman et al., 2010). Although levels of total mTOR appeared unaffected by Mio depletion (Fig. 9 C), levels of active mTOR (mTORS2481ph) on centrosomes and spindles were much lower in Mio-depleted cells than in control RNAi cells (Fig. 9, A and B).
Figure 9.
Activity of mTOR is reduced in mitotic cells after Mio depletion. (A) Control and Mio-depleted cells were fixed and immunostained with α–p-mTOR (Ser2481; green), α-tubulin (red), and DNA (blue). (B) Control and Mio-depleted cells were fixed and immunostained with α–p-mTOR (Ser2481; green), γ-tubulin (red), and DNA (blue). (C) Control and Mio-depleted cells were fixed and immunostained with α-mTOR (green), α-tubulin (red), and DNA (blue). Bars, 10 µm.
We next examined phosphorylation of known mTORC1 substrates as a direct measure of mTORC1 activity. Amino acid withdrawal leads to rapid mTORC1 inactivation (Sancak et al., 2008). Mio depletion strongly inhibited the amino acid–induced activation of mTORC1 as monitored by levels of S6K1 and 4EBP1 phosphorylation (Fig. S4 F). Rescue experiments on HeLa si02ResGFP-Mio cells where only the endogenous Mio was depleted using si02 (si01 targets both endogenous and exogenous constructs) restored both mTORS2481ph levels at spindle poles and p70S6KT389ph phosphorylation (Fig. S4, E and F).
If Mio depletion disrupts mitotic progression by inhibiting mTOR, activation of the kinase by an alternative pathway might be expected to at least partially rescue the Mio depletion phenotype. PTEN (phosphatase and tensin homologue) dephosphorylates phosphoinositide-3,4,5-triphosphate (PIP3), a potent activator of AKT kinase signaling (Maehama and Dixon, 1998). mTOR is a key downstream target of AKT (Sabatini, 2006); thus, PTEN inactivation results in increased mTOR activity. Co-depletion of Mio and PTEN showed a partial rescue of mitotic progression and increased Aurora AT288ph levels at spindle poles (Fig. 10, D–F).
To determine by independent method that inhibition of mTOR can explain the mitotic defects observed upon Mio depletion, we developed a protocol that enabled us to inhibit the kinase without blocking cell cycle progression. Rapamycin, when bound to intracellular protein FKBP12, inhibits mTORC1 kinase activity (Guertin and Sabatini, 2009). Rapamycin treatment for 4 h resulted in an increase of prometaphase cells that also showed a decrease of both Aurora and Aurora AT288ph levels at spindle poles (Fig. 10, G–I), reinforcing the idea that reduced mTORC1 activity causes the mitotic defects observed upon Mio depletion.
We conclude that Mio is required for maintenance of active mTOR levels in mitosis.
Discussion
Mio is a component of the SEACAT/GATOR2 complex, which has been reported to be involved in mTOR regulation by the Rag-GTPase (Bar-Peled et al., 2013; Panchaud et al., 2013a). Here, we confirm that Mio interacts with Seh1, a member of the SEACAT/GATOR2 and Nup107 nucleoporin complexes, both in interphase and mitosis. Our experiments identified roles for Mio in timely mitotic progression, centrosome separation, spindle formation, and orientation/positioning in human mitotic cells. Indeed, 35% of mitotic cells observed after Mio depletion had a bipolar, monoastral structure (i.e., they lacked a centrosome and astral microtubules at one of the two spindle poles). Functional analysis revealed that Mio plays a critical role in activation of the essential mitotic kinases, Aurora A and Plk1, at spindle poles. We could find no role for Mio in NPC assembly or structure, suggesting that the mitotic phenotypes of Mio depletion may reflect the loss of SEACAT/GATOR2 regulation of mTOR kinase signaling. Thus, our experiments reveal a previously unsuspected role for mTOR in mitotic regulation.
Centrosome separation and bipolar spindle formation both require Plk1 and Aurora A activity. Mio depletion caused a marked reduction of active Aurora AT288ph without changes in the localization or levels of total Aurora A or TPX2. Spindle length was normal after Mio depletion, which is consistent with the interaction between Aurora A and TPX2 being unaffected (Bird and Hyman, 2008) and suggesting that the reduction of Aurora AT288ph levels observed upon Mio depletion was not a result of the loss of TPX2-mediated activation. Despite the reduced activity of Aurora A and defects in centrosome separation after Mio depletion, bipolar spindle formation still occurred, possibly as a result of microtubule nucleation from chromosomes via the Ran-GTP pathway (Caudron et al., 2005; Kaláb et al., 2006; Kalab and Heald, 2008).
Aurora A activates centrosomal Plk1 through phosphorylation of Thr210 (Macůrek et al., 2008; Seki et al., 2008; Carmena, 2012; Bruinsma et al., 2014). Active Plk1 controls entry into mitosis and also promotes centrosome maturation and separation (Bruinsma et al., 2012; Zitouni et al., 2014). Plk1 localization influences its activity and vice versa (Kishi et al., 2009). Upon Mio depletion, levels of total Plk1 and active Plk1T210ph were reduced at centrosomes. As a result, Mio-depleted cells were hypersensitive to Plk1 inhibition as detected by synthetic interaction with the Plk1 inhibitor BI2536, resulting in an increase in monopolar spindles even at low drug concentrations. The reduction in active Plk1 levels in Mio-depleted cells may result in a partial inhibition of mitotic entry and may explain the apparent paradox that Mio-depleted cells exhibit a substantial delay in mitotic progression without an accompanying increase in mitotic index.
Mio depletion from HeLa cells causes multiple mitotic defects such as a lengthening of the interval between NEBD and anaphase onset, spindle assembly defects including an increase in the number of bipolar monoastral spindles, a prometaphase delay often associated with monopolar spindles, and a delay or failure in cytokinesis. Interestingly, in Drosophila melanogaster, where Mio has been reported to play a role in oocyte maturation, the microtubule-dependent translocation of centrioles from the anterior to the posterior of stage 1 oocyte nuclei is defective, as is the microtubule-dependent translocation of RNA-binding protein Orb (Iida and Lilly, 2004). Thus, the involvement of Mio with microtubule function is conserved across species.
Aurora A regulates mitotic spindle formation at least partially as part of a multiprotein subunit complex containing HURP, TPX2, and Eg5 kinesin (Koffa et al., 2006). HURP is a microtubule stabilizer (Silljé et al., 2006; Santarella et al., 2007) that localizes mainly to kinetochore microtubules and requires Aurora A phosphorylation for its microtubule binding (Yu et al., 2005; Wong et al., 2008). TPX2 binds to and activates Aurora A on the mitotic spindle (Kufer et al., 2002; Giubettini et al., 2011). Eg5 is required for centrosome separation after NEBD (Kapitein et al., 2005; Smith et al., 2011; Mardin and Schiebel, 2012). Our analysis of mitotic spindles after Mio depletion showed a dramatic change in HURP localization, with the protein spreading throughout the spindle, as previously seen upon Aurora A kinase inhibition (Kesisova et al., 2013). Indeed, we observed a decrease in Aurora A activation (detected as a decrease in Aurora A phosphorylated on its activation loop at Thr288) on Mio-depleted mitotic spindles. Eg5 and TPX2 localization, on the other hand, appeared to be unaffected. Thus, defects in Eg5 localization to centrosomes and microtubules are unlikely to explain the bipolar, monoastral spindles observed in 35% of Mio-depleted mitotic cells.
The kinesin-13 family microtubule depolymerase MCAK associates with centromeres and growing microtubule ends, where it plays a role in correction of kinetochore–microtubule misattachments from prophase to anaphase. MCAK also localizes to spindle poles, where it is required for the assembly and dynamics of the mitotic spindle. MCAK localization at the centromere and its depolymerase activity are regulated by Aurora B (Andrews et al., 2004; Kline-Smith et al., 2004; Lan et al., 2004; Knowlton et al., 2006). In contrast, MCAK localization at the poles is regulated by Aurora A activity (Zhang et al., 2008). In Mio-depleted cells, we observed a reduction of MCAK localization at the poles, whereas MCAK’s centromeric localization appeared to be unaffected. Interference with MCAK activity specifically at centrosomes could partly explain the spindle assembly defects observed upon Mio depletion.
mTOR plays key roles in several cellular processes through interactions with other proteins to form mTOR complexes 1 and 2 (mTORC1/2). mTORC1 positively regulates cell growth, proliferation, and the cell cycle, whereas mTORC2 plays key roles in cell survival, metabolism, and actin organization (Laplante and Sabatini, 2009; Wang and Proud, 2011). Recent studies have suggested a link between mTORC1 and mitosis. mTORC1 complex components have been shown to associate with mitotic spindles in mouse oocytes (Kogasaka et al., 2013), and overexpression of mTORC1 results in impairment of spindle formation and aneuploidy (Astrinidis et al., 2006). In yeast, Aurora A promotes cell division during recovery from mTOR-mediated cell cycle arrest by driving spindle pole body recruitment of Polo (Hálová and Petersen, 2011), while at the same time overexpression of Plk1 increases the phosphorylation of mTORC1 substrates (Renner et al., 2010), suggesting the existence of a molecular and functional link between Plk1 mitotic kinase, mTOR signaling, and spindle formation.
Here, we show for the first time that mTORC1 is an important regulator of mitotic spindle assembly. Our results reveal a link between Mio and SEACAT/GATOR2 complex (Bar-Peled et al., 2013; Panchaud et al., 2013a) regulation of mTORC1 kinase and the spatiotemporal activation of Aurora A and Plk1 at centrosomes during mitosis. The function of Mio in the SEACAT/GATOR2 complex is not known. Although we have been unable to confirm any E3 enzymatic activity for purified Mio in vitro, the presence of a RING/PHD domain in Mio suggests that the protein might control the stability/interaction of key components of the mTOR signaling pathway. Indeed, the inability of Mio RING/PHD domain mutants to rescue mitotic progression suggests that the RING/PHD domain does play a role in Mio function during mitosis. Future studies will focus on the role of Mio in the SEACAT/GATOR2 complex and the mechanisms by which this complex links mTOR signaling to the regulation of Aurora A and Plk1 at mitotic centrosomes.
Materials and methods
Cell culture
HeLa Kyoto cells were grown in DMEM supplemented with 10% FCS, 0.2 mM l-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin. Cell lines stably expressing EGFP-Mio, EGFP-Mio Ring mutant, and EGFP-Mio-ΔPHD were generated using the HeLa Flp-In cell line (Klebig et al., 2009) and Flp-in system vectors (Invitrogen) and were maintained in the aforementioned media with an additional 200 µg/ml hygromycin.
The HeLaEGFP-H2B Kyoto cell line was generated by transfecting human H2B coding sequence fused to 3′ of EGFP (Takara Bio Inc.) to HeLa Kyoto cells and was maintained in G418 (EGFP-Seh1 cDNA were previously described [Platani et al., 2009]), and the HeLa KyotomRFP-H2B GFP-tubulin cell line was generated for this study by transfecting mRFP-H2B and GFP–α-tubulin cDNA to HeLa Kyoto cell lines and was maintained in G418.
RNAi and transfection experiments
cDNA transfections were performed using X-treme Gene (Roche). RNAi experiments were performed using annealed siRNA oligos (Ambion and QIAGEN) using HiPerFect reagent (QIAGEN). Full sequences of siRNA oligos are provided in Table S1.
Microscopy and image analysis
For immunofluorescence, 3D datasets were acquired using a camera (CoolSnap HQ cooled CCD; Photometrics) on a microscope (DeltaVision Spectris; Applied Precision). Optical sections were acquired every 0.2 µm, and 3D datasets were deconvolved using the constrained iterative algorithm (Swedlow et al., 1997; Wallace et al., 2001) implemented in the SoftWoRx software (Applied Precision). For live cell imaging, cells were grown on glass-bottomed Lab-Tek dishes (Thermo Fisher Scientific), transfected with appropriate siRNAs, and maintained in a humidified 37°C chamber in a CO2-independent phenol red–free DMEM (Invitrogen). Images were collected using a 100×/1.4 NA PlanApo objective lens with optical sections every 0.5 µm and a 20×/0.4 PlanFL Ph1 objective. Images were loaded into Photoshop (Adobe) or OMERO (Open Microscopy Environment) and adjusted for display (Allan et al., 2012).
Quantification of Plk1 and p-Plk1 on ∼80 cells per condition was performed as follows: deconvolved images were imported into OMERO (Swedlow et al., 2009), and segmentation of centrosomal foci (pericentrin FITC, reference channel) was performed using the Otsu method, implemented in Matlab (MathWorks). These segmentation masks were then used to calculate intensities for these bodies in Plk1 and p-Plk1 channels, and output was imported into Excel (Microsoft) for plotting. Signal within these volumes was quantified, background corrected, and represented as mean fluorescent intensity/pixel. Quantification of Aurora A and P–Aurora A on ∼210 cells per condition was performed directly on their respective channel. That is, deconvolved images were imported into OMERO, and Otsu segmentation of the spindle and centrosomal signal was performed on the FITC channel (Aurora A and P–Aurora A) staining. The signal within these volumes was quantified, background corrected, and represented as fluorescence intensity/pixel.
cDNAs, antibodies, and immunofluorescence
All fixation, permeabilization, and immunostaining were performed at room temperature. Cells grown on coverslips were fixed in a 3.7% formaldehyde/PBS solution for 10 min and permeabilized in PBS–0.5% Triton X-100 for 10 min. Cells were blocked in 10% normal donkey serum for 1 h at room temperature before antibody incubations. For γ-tubulin staining, cells were fixed in 100% methanol for 10 min and rehydrated in PBS for 20 min before staining. For Bub1, MPM2, and Sgo1 staining, a prepermeabilization step with 0.1% Triton X-100 in Pipes-Hepes-EGTA-MgCl2 buffer for 1 min was included followed by PFA fixation. Antibodies used in this study are listed as follows. Antibody against human Mio was raised in rabbits against the C-terminal part of the protein corresponding to amino acids 720–876 expressed as a His-tagged protein. The antibody was purified from serum using the corresponding protein immobilized on Affigel beads (Bio-Rad Laboratories). The other antibodies used are tyrosinated tubulin YL12 (rat; Abcam), α-tubulin (mouse, DMIA; Sigma-Aldrich), PLK1 (mouse; EMD Millipore), p-Plk1T210 (mouse; Abcam), Aurora A (mouse; BD), P–Aurora A T288 (rabbit; Abcam), Eg5 (rabbit; Novus Biologicals), GFP (rabbit; Life Technologies), ACA (human; Antibodies Incorporated), mAb414 (mouse; Covance), pericentrin (rabbit; Abcam), γ-tubulin (mouse; Sigma-Aldrich), Hurp (rabbit; Abcam), Numa (rabbit; Abcam), TPX2 (mouse; Abcam), p150 (mouse; BD), Aurora B (rabbit; Abcam), Survivin (rabbit; Novus Biologicals), Bub1 (mouse; Abcam), Sgo1 (mouse; Abcam), mTOR and p-mTOR2481 (rabbit; Cell Signaling Technology), p70S6K (Thr389, antibody kit, rabbit; Cell Signaling Technology), PTEN (rabbit, D4.3; Cell Signaling Technology), 4EBP1 (53H11, rabbit; Cell Signaling Technology), 4EBP1 (Ser65, rabbit; Cell Signaling Technology), MPM2 (mouse; EMD Millipore), PPP6C (rabbit; Bethyl Laboratories, Inc.), SAPK/JNK (56G8, rabbit; Cell Signaling Technology), SAPK/JNK (Thr183/Tyr185, rabbit; Cell Signaling Technology), p38 MAPK (D13E1, rabbit; Cell Signaling Technology), p38 MAPK (Thr180/Tyr182; rabbit, Cell Signaling Technology), p44/42 MAPK and p44/42 Thr202/Tyr204 (137F5, rabbit; Cell Signaling Technology), and Seh1 (rabbit, generated against full-length histidine-tagged human protein; Platani et al., 2009). All affinity-purified donkey secondary antibodies (labeled either with FITC, Alexa Fluor 488, TRITC, Alexa Fluor 594, or Cy5) were purchased from Jackson ImmunoResearch Laboratories, Inc. Peroxidase-conjugated donkey anti–rabbit and goat anti–mouse antibodies were purchased from Roche.
Monastrol (EMD Millipore) was used at 100 µM. ZM447439 (Tocris Bioscience) was used at 0.5, 1, and 2 µM; MLN8237 (Selleckchem) was used at 10, 50, and 300 nM; BI2356 (Selleckchem) was used at 25, 50, and 100 nM for 1.5 h; and rapamycin (Sigma-Aldrich) was used at 200 nM for 4 h followed by recovery in normal medium.
For spindle length measurements, cells were stained with centromeric marker (ACA) to identify clear metaphase plates and centrosome marker (pericentrin). Pole to pole measurements were made from single sections of 3D datasets using OMERO. Only spindles parallel to the plane of focus were used to avoid artifacts that can be created by spindle rotation into the plane of focus. Spindle rotation measurements of the metaphase spindle with respect to the fibronectin substratum were performed as previously described (Toyoshima and Nishida, 2007). In brief, cells that showed a congressed metaphase plate were stained with γ-tubulin or pericentrin to mark the poles and Hoechst 33342 to mark DNA. Stacks of 3D images at 0.2-µm steps were acquired, and the poles’ distances in X, Y, and Z were determined using OMERO. The spindle angle was calculated using inverse trigonometry.
For analysis of cold-stable microtubules, cells were incubated in ice-cold media for 10 min before incubation in ice-cold Pipes-Hepes-EGTA-MgCl2 buffer/0.5% Triton X-100 for 2 min. Fixation was performed in ice-cold methanol for 2 min, followed by washing with PBS twice and final rehydration in fresh PBS for 15 min.
Mammalian expression plasmids of EGFP-Mio, EGFP-Mio-ΔPHD (1–2,353 bp), and Cherry-Mio were cloned using pEGFP-C1 and mCherry-C1 vectors. Mio cDNA was amplified with Phusion polymerase (New England Biolabs, Inc.) from total HeLa RNA extracted with TRIzol (Invitrogen). Site-directed mutagenesis was performed using the QuikChange XL system (Agilent Technologies) according to the manufacturer’s protocol.
siRNA-resistant GFP-Mio, GFP-Mio-Ring mutant, and GFP-Mio-ΔPHD cDNA was generated by site-directed mutagenesis carrying silent nucleotide mutations (in bold letters: 5′-GCCGCGACCTTGCTATCCATA-3′) at positions 150, 153, 156, 159, 162, and 165, respectively.
Isolation of mCherry-Mio
mCherry and mCherry-Mio complexes were isolated from 3 × 10–cm dishes of asynchronous HeLa Kyoto cells 28 h after transfection with the corresponding cDNA constructs. Cells were washed twice in ice-cold PBS and collected and lysed in 0.6 ml lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.5% NP-40, and protease inhibitors). Extracts were left on ice for 30 min before centrifugation at 20,000 g for 10 min at 4°C. Equilibrated RFP trap beads (ChromoTek) were added to cleared cell lysates and rotated at 4°C for 2 h. The beads were washed four times with wash buffer (50 mM Tris, pH 7.5, 150 mM NaCl, and 0.05% NP-40) before sample buffer addition, SDS-PAGE, and Western blotting.
Isolation of Aurora A–TPX2 complexes and kinase assays
Aurora A–TPX2 complexes were isolated from 6 × 10–cm dishes of HeLa cells at ∼60–70% confluency as described in Zeng et al. (2010). In brief, cells were arrested in mitosis with 100 ng/ml nocodazole for 16 h, lysed in 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, protease inhibitors, and phosphatase inhibitor cocktail 2 (Sigma-Aldrich), and left on ice for 30 min before centrifugation at 20,000 g for 20 min at 4°C. Equilibrated protein G–Sepharose beads (GE Healthcare) together with either 2 µg of control mouse IgG or TPX2 antibody were added to the cell lysates and rotated at 4°C for 2 h. The beads were washed in lysis buffer and TBS before sample buffer addition, SDS-PAGE, and Western blotting. For in vitro kinase assays, isolated Aurora A–TPX2 complexes were incubated for 30 min at 30°C in a 20-µl solution of 50 mM Tris-HCl, pH 7.4, 50 mM KCl, 10 mM MgCl2, 20 mM β-glycerolphosphate, 15 mM EGTA, 100 µM ATP, 0.5 µl γ-[32P]ATP solution, and 1 µg Histone H3 substrate (New England Biolabs, Inc.) before the addition of sample buffer. Reactions were analyzed by SDS-PAGE and autoradiography.
Amino acid starvation and stimulation
HeLa cells grown in RPMI were washed in PBS, and starvation was performed by incubating the cells for 50–60 min in amino acid–free RPMI without serum. Cells were then stimulated for 20 min by the addition of RPMI containing a 2× concentrated solution of amino acids. After stimulation, the final concentration of amino acids in the media was the same as in the original RPMI medium.
Mass spectrometry
For Seh1 interactome mapping in interphase and mitosis by quantitative affinity purification/mass spectrometry, 10 × 15–cm dishes of cells were differentially encoded by growth in SILAC DMEM supplemented with 10% dialyzed FBS and 100 U/ml penicillin/streptomycin and containing either ”light” l-arginine and l-lysine (HeLaGFP), “medium” l-arginine–13C and l-lysine 4,4,5,5-D4 (HeLaGFP-Seh1/interphase), or “heavy” l-arginine–13C/15N and l-lysine–13C/15N (HeLaGFP-Seh1/mitosis) for 7 d. Equal protein amounts of whole cell extract were incubated with GFP-TrapA (ChromoTek) affinity resin for 1 h at 4°C, the beads were washed in ice-cold radio immunoprecipitation assay buffer and combined, and the proteins were eluted for gel separation, trypsin digestion, and liquid chromatography–tandem mass spectrometry analysis on a mass spectrometer system (LTQ-Orbitrap; ThermoElectron) as previously described (Prévost et al., 2013). In brief, for efficient elution of bound proteins, a bead equivalent volume of 1% SDS was added, heated at 95°C for 10 min, and then a 4× volume of dH2O was added. The matrix was mixed, and the solution was removed and reduced to the original bead-equivalent volume using a speedvac. Proteins were reduced and alkylated by the addition of 10 mM DTT at 95°C for 2 min followed by the addition of 50 mM iodoacetamide at room temperature for 30 min. Sample buffer was added, and proteins were separated on 10% Bis-Tris gels. Gels were Coomassie stained and destained overnight before excision of 10 gel slices. Peptides resulting from in-gel digestion with trypsin (Promega) were extracted from the gel slices. An aliquot of each tryptic digest was analyzed by liquid chromatography–electrospray ionization–tandem mass spectrometry on a mass spectrometer with a nanospray source (LTQ Orbitrap XL hybrid; Thermo Fisher Scientific) and an HPLC (UltiMate 3000 RSLC nano; Dionex). The system was controlled by Xcalibur software version 2.0.7 (Thermo Fisher Scientific). Peptides were loaded onto a trap column (C18 CapTrap; Michrom) for 5 min at 15 µl/min and then were eluted over a 60-min gradient of 3–45% acetonitrile with 0.1% formic acid at 0.3 µl/min onto a 10-cm column with integrated emitter tip (Picofrit; New Objective) packed with 5 µm Zorbax SBC18 (Agilent Technologies) and nanosprayed into the mass spectrometer. Mass spectrometry scans were acquired in the Orbitrap module, and MS2 scans were acquired in the ion trap module using data-dependent acquisition of the top five ions from each MS scan. Database searching against the human UniProt database and quantitation were performed using MaxQuant version 1.2.7. Protein groups and peptide lists were analyzed in Excel (Microsoft).
Mio 3D structure prediction
The modeling of the Mio protein C-terminal sequence was performed using Phyre2 (Kelley and Sternberg, 2009). The predicted 3D molecular structure model of the Mio protein C-terminal sequence was created using a molecular visualization program (PyMOL version 1.5.0.4; Schrödinger, LLC).
Statistical analysis
To assess statistical significance, we determined the t test for equal or unequal variances. A p-value of <0.05 was considered to be statistically significant.
Online supplemental material
Fig. S1 shows Seh1 interactome and rescue experiments using wild-type GFP-Mio. Figs. S2 and S3 show that spindle assembly checkpoints or centromeric localization of chromosome passenger complex are not affected after Mio depletion. Micrographs of cold-stable microtubule assays are also included. Fig. S4 shows spindle assembly factors TPX2 and Eg5 localization together with Aurora A kinase assays on Aurora A–TPX2 complexes after Mio depletion. Fig. S5 shows Numa and p150 spindle localization together with p-Plk1 localization. Table S1 contains a list of siRNA duplexes used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201410001/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201410001.dv.
Acknowledgments
We would like to thank J.R. Swedlow for GFP-MCAK and GFP–-α-tubulin, Patrick Meraldi for the HeLa Kyoto Flp-In cell line, Paola Vagnarelli for mRFP-H2B, and David Kelly for initial microscopy assistance (Centre Optical Instrumentation Laboratory, Wellcome Trust Centre for Cell Biology, Edinburgh, Scotland, UK).
M. Platani was supported by a Wellcome Trust Project grant (086194/Z/08/Z), W.C. Earnshaw is supported by a Wellcome Trust Principal Research Fellow grant (073915), M. Porter is supported by a Wellcome Trust grant (095931/Z/11/Z), and A.A. Jeyaprakash acknowledges support from the Wellcome Trust for the Wellcome Trust Career Development grant (095822) and from the European Commission for the Marie Curie Career Integration Grant (FP7). L. Trinkle-Mulcahy holds a Canadian Institutes of Health Research New Investigator Award.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- mTOR
- mammalian target of rapamycin
- NEBD
- nuclear envelope breakdown
- NPC
- nuclear pore complex
- PTEN
- phosphatase and tensin homologue
References
- Allan C., Burel J.M., Moore J., Blackburn C., Linkert M., Loynton S., Macdonald D., Moore W.J., Neves C., Patterson A., et al. . 2012. OMERO: flexible, model-driven data management for experimental biology. Nat. Methods. 9:245–253. 10.1038/nmeth.1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews P.D., Ovechkina Y., Morrice N., Wagenbach M., Duncan K., Wordeman L., and Swedlow J.R.. 2004. Aurora B regulates MCAK at the mitotic centromere. Dev. Cell. 6:253–268. 10.1016/S1534-5807(04)00025-5 [DOI] [PubMed] [Google Scholar]
- Astrinidis A., Senapedis W., and Henske E.P.. 2006. Hamartin, the tuberous sclerosis complex 1 gene product, interacts with polo-like kinase 1 in a phosphorylation-dependent manner. Hum. Mol. Genet. 15:287–297. 10.1093/hmg/ddi444 [DOI] [PubMed] [Google Scholar]
- Bar-Peled L., Chantranupong L., Cherniack A.D., Chen W.W., Ottina K.A., Grabiner B.C., Spear E.D., Carter S.L., Meyerson M., and Sabatini D.M.. 2013. A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 340:1100–1106. 10.1126/science.1232044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr A.R., and Gergely F.. 2007. Aurora-A: the maker and breaker of spindle poles. J. Cell Sci. 120:2987–2996. 10.1242/jcs.013136 [DOI] [PubMed] [Google Scholar]
- Barros T.P., Kinoshita K., Hyman A.A., and Raff J.W.. 2005. Aurora A activates D-TACC-Msps complexes exclusively at centrosomes to stabilize centrosomal microtubules. J. Cell Biol. 170:1039–1046. 10.1083/jcb.200504097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss R., Sardon T., Vernos I., and Conti E.. 2003. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol. Cell. 12:851–862. 10.1016/S1097-2765(03)00392-7 [DOI] [PubMed] [Google Scholar]
- Belgareh N., Rabut G., Baï S.W., van Overbeek M., Beaudouin J., Daigle N., Zatsepina O.V., Pasteau F., Labas V., Fromont-Racine M., et al. . 2001. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J. Cell Biol. 154:1147–1160. 10.1083/jcb.200101081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertran M.T., Sdelci S., Regué L., Avruch J., Caelles C., and Roig J.. 2011. Nek9 is a Plk1-activated kinase that controls early centrosome separation through Nek6/7 and Eg5. EMBO J. 30:2634–2647. 10.1038/emboj.2011.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A.W., and Hyman A.A.. 2008. Building a spindle of the correct length in human cells requires the interaction between TPX2 and Aurora A. J. Cell Biol. 182:289–300. 10.1083/jcb.200802005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehmer T., Enninga J., Dales S., Blobel G., and Zhong H.. 2003. Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proc. Natl. Acad. Sci. USA. 100:981–985. 10.1073/pnas.252749899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruinsma W., Raaijmakers J.A., and Medema R.H.. 2012. Switching Polo-like kinase-1 on and off in time and space. Trends Biochem. Sci. 37:534–542. 10.1016/j.tibs.2012.09.005 [DOI] [PubMed] [Google Scholar]
- Bruinsma W., Macurek L., Freire R., Lindqvist A., and Medema R.H.. 2014. Bora and Aurora-A continue to activate Plk1 in mitosis. J. Cell Sci. 127:801–811. 10.1242/jcs.137216 [DOI] [PubMed] [Google Scholar]
- Carmena M. 2012. Abscission checkpoint control: stuck in the middle with Aurora B. Open Biol. 2:120095 10.1098/rsob.120095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M., Ruchaud S., and Earnshaw W.C.. 2009. Making the Auroras glow: regulation of Aurora A and B kinase function by interacting proteins. Curr. Opin. Cell Biol. 21:796–805. 10.1016/j.ceb.2009.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casenghi M., Meraldi P., Weinhart U., Duncan P.I., Körner R., and Nigg E.A.. 2003. Polo-like kinase 1 regulates Nlp, a centrosome protein involved in microtubule nucleation. Dev. Cell. 5:113–125. 10.1016/S1534-5807(03)00193-X [DOI] [PubMed] [Google Scholar]
- Caudron M., Bunt G., Bastiaens P., and Karsenti E.. 2005. Spatial coordination of spindle assembly by chromosome-mediated signaling gradients. Science. 309:1373–1376. 10.1126/science.1115964 [DOI] [PubMed] [Google Scholar]
- Davis F.M., Tsao T.Y., Fowler S.K., and Rao P.N.. 1983. Monoclonal antibodies to mitotic cells. Proc. Natl. Acad. Sci. USA. 80:2926–2930. 10.1073/pnas.80.10.2926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R.J., and Joazeiro C.A.. 2009. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78:399–434. 10.1146/annurev.biochem.78.101807.093809 [DOI] [PubMed] [Google Scholar]
- Dokudovskaya S., Waharte F., Schlessinger A., Pieper U., Devos D.P., Cristea I.M., Williams R., Salamero J., Chait B.T., Sali A., et al. A conserved coatomer-related complex containing Sec13 and Seh1 dynamically associates with the vacuole in Saccharomyces cerevisiae. Mol. Cell. Proteomics. 2011 doi: 10.1074/mcp.M110.006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowe S., Hümmer S., Uldschmid A., Li X., and Nigg E.A.. 2007. Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions. Genes Dev. 21:2205–2219. 10.1101/gad.436007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyers P.A., and Maller J.L.. 2003. Regulating the regulators: Aurora A activation and mitosis. Cell Cycle. 2:286–289. 10.4161/cc.2.4.444 [DOI] [PubMed] [Google Scholar]
- Eyers P.A., and Maller J.L.. 2004. Regulation of Xenopus Aurora A activation by TPX2. J. Biol. Chem. 279:9008–9015. 10.1074/jbc.M312424200 [DOI] [PubMed] [Google Scholar]
- Giet R., Uzbekov R., Cubizolles F., Le Guellec K., and Prigent C.. 1999. The Xenopus laevis aurora-related protein kinase pEg2 associates with and phosphorylates the kinesin-related protein XlEg5. J. Biol. Chem. 274:15005–15013. 10.1074/jbc.274.21.15005 [DOI] [PubMed] [Google Scholar]
- Giet R., McLean D., Descamps S., Lee M.J., Raff J.W., Prigent C., and Glover D.M.. 2002. Drosophila Aurora A kinase is required to localize D-TACC to centrosomes and to regulate astral microtubules. J. Cell Biol. 156:437–451. 10.1083/jcb.200108135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giubettini M., Asteriti I.A., Scrofani J., De Luca M., Lindon C., Lavia P., and Guarguaglini G.. 2011. Control of Aurora-A stability through interaction with TPX2. J. Cell Sci. 124:113–122. 10.1242/jcs.075457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Aguilera C., and Askjaer P.. 2012. Dissecting the NUP107 complex: Multiple components and even more functions. Nucleus. 3:340–348. 10.4161/nucl.21135 [DOI] [PubMed] [Google Scholar]
- Guertin D.A., and Sabatini D.M.. 2007. Defining the role of mTOR in cancer. Cancer Cell. 12:9–22. 10.1016/j.ccr.2007.05.008 [DOI] [PubMed] [Google Scholar]
- Guertin D.A., and Sabatini D.M.. 2009. The pharmacology of mTOR inhibition. Sci. Signal. 2:pe24 10.1126/scisignal.267pe24 [DOI] [PubMed] [Google Scholar]
- Hálová L., and Petersen J.. 2011. Aurora promotes cell division during recovery from TOR-mediated cell cycle arrest by driving spindle pole body recruitment of Polo. J. Cell Sci. 124:3441–3449. 10.1242/jcs.083683 [DOI] [PubMed] [Google Scholar]
- Harel A., Orjalo A.V., Vincent T., Lachish-Zalait A., Vasu S., Shah S., Zimmerman E., Elbaum M., and Forbes D.J.. 2003. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol. Cell. 11:853–864. 10.1016/S1097-2765(03)00116-3 [DOI] [PubMed] [Google Scholar]
- Harris T.E., and Lawrence J.C. Jr. 2003. TOR signaling. Sci. STKE. 2003:re15. [DOI] [PubMed] [Google Scholar]
- Hirota T., Kunitoku N., Sasayama T., Marumoto T., Zhang D., Nitta M., Hatakeyama K., and Saya H.. 2003. Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell. 114:585–598. 10.1016/S0092-8674(03)00642-1 [DOI] [PubMed] [Google Scholar]
- Hochegger H., Hégarat N., and Pereira-Leal J.B.. 2013. Aurora at the pole and equator: overlapping functions of Aurora kinases in the mitotic spindle. Open Biol. 3:120185 10.1098/rsob.120185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida T., and Lilly M.A.. 2004. missing oocyte encodes a highly conserved nuclear protein required for the maintenance of the meiotic cycle and oocyte identity in Drosophila. Development. 131:1029–1039. 10.1242/dev.01001 [DOI] [PubMed] [Google Scholar]
- Jacinto E., and Hall M.N.. 2003. Tor signalling in bugs, brain and brawn. Nat. Rev. Mol. Cell Biol. 4:117–126. 10.1038/nrm1018 [DOI] [PubMed] [Google Scholar]
- Jacinto E., Loewith R., Schmidt A., Lin S., Rüegg M.A., Hall A., and Hall M.N.. 2004. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 6:1122–1128. 10.1038/ncb1183 [DOI] [PubMed] [Google Scholar]
- Kalab P., and Heald R.. 2008. The RanGTP gradient - a GPS for the mitotic spindle. J. Cell Sci. 121:1577–1586. 10.1242/jcs.005959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaláb P., Pralle A., Isacoff E.Y., Heald R., and Weis K.. 2006. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 440:697–701. 10.1038/nature04589 [DOI] [PubMed] [Google Scholar]
- Kapitein L.C., Peterman E.J., Kwok B.H., Kim J.H., Kapoor T.M., and Schmidt C.F.. 2005. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 435:114–118. 10.1038/nature03503 [DOI] [PubMed] [Google Scholar]
- Kapoor T.M., Mayer T.U., Coughlin M.L., and Mitchison T.J.. 2000. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J. Cell Biol. 150:975–988. 10.1083/jcb.150.5.975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L.A., and Sternberg M.J.. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371. 10.1038/nprot.2009.2 [DOI] [PubMed] [Google Scholar]
- Kesisova I.A., Nakos K.C., Tsolou A., Angelis D., Lewis J., Chatzaki A., Agianian B., Giannis A., and Koffa M.D.. 2013. Tripolin A, a novel small-molecule inhibitor of aurora A kinase, reveals new regulation of HURP’s distribution on microtubules. PLoS ONE. 8:e58485 10.1371/journal.pone.0058485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi K., van Vugt M.A., Okamoto K., Hayashi Y., and Yaffe M.B.. 2009. Functional dynamics of Polo-like kinase 1 at the centrosome. Mol. Cell. Biol. 29:3134–3150. 10.1128/MCB.01663-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T., and Cheeseman I.M.. 2012. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat. Cell Biol. 14:311–317. 10.1038/ncb2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebig C., Korinth D., and Meraldi P.. 2009. Bub1 regulates chromosome segregation in a kinetochore-independent manner. J. Cell Biol. 185:841–858. 10.1083/jcb.200902128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith S.L., Khodjakov A., Hergert P., and Walczak C.E.. 2004. Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol. Biol. Cell. 15:1146–1159. 10.1091/mbc.E03-08-0581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlton A.L., Lan W., and Stukenberg P.T.. 2006. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr. Biol. 16:1705–1710. 10.1016/j.cub.2006.07.057 [DOI] [PubMed] [Google Scholar]
- Koffa M.D., Casanova C.M., Santarella R., Köcher T., Wilm M., and Mattaj I.W.. 2006. HURP is part of a Ran-dependent complex involved in spindle formation. Curr. Biol. 16:743–754. 10.1016/j.cub.2006.03.056 [DOI] [PubMed] [Google Scholar]
- Kogasaka Y., Hoshino Y., Hiradate Y., Tanemura K., and Sato E.. 2013. Distribution and association of mTOR with its cofactors, raptor and rictor, in cumulus cells and oocytes during meiotic maturation in mice. Mol. Reprod. Dev. 80:334–348. 10.1002/mrd.22166 [DOI] [PubMed] [Google Scholar]
- Kufer T.A., Silljé H.H., Körner R., Gruss O.J., Meraldi P., and Nigg E.A.. 2002. Human TPX2 is required for targeting Aurora-A kinase to the spindle. J. Cell Biol. 158:617–623. 10.1083/jcb.200204155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W., Zhang X., Kline-Smith S.L., Rosasco S.E., Barrett-Wilt G.A., Shabanowitz J., Hunt D.F., Walczak C.E., and Stukenberg P.T.. 2004. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol. 14:273–286. 10.1016/j.cub.2004.01.055 [DOI] [PubMed] [Google Scholar]
- Lane H.A., and Nigg E.A.. 1996. Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J. Cell Biol. 135:1701–1713. 10.1083/jcb.135.6.1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M., and Sabatini D.M.. 2009. mTOR signaling at a glance. J. Cell Sci. 122:3589–3594. 10.1242/jcs.051011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., and Rhee K.. 2011. PLK1 phosphorylation of pericentrin initiates centrosome maturation at the onset of mitosis. J. Cell Biol. 195:1093–1101. 10.1083/jcb.201106093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lens S.M., Voest E.E., and Medema R.H.. 2010. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat. Rev. Cancer. 10:825–841. 10.1038/nrc2964 [DOI] [PubMed] [Google Scholar]
- Littlepage L.E., Wu H., Andresson T., Deanehan J.K., Amundadottir L.T., and Ruderman J.V.. 2002. Identification of phosphorylated residues that affect the activity of the mitotic kinase Aurora-A. Proc. Natl. Acad. Sci. USA. 99:15440–15445. 10.1073/pnas.202606599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.S., Song B., Tang J., Liu W., Kuang S., and Liu X.. 2012. Plk1 phosphorylates Sgt1 at the kinetochores to promote timely kinetochore-microtubule attachment. Mol. Cell. Biol. 32:4053–4067. 10.1128/MCB.00516-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J.L., Bonenfant D., Oppliger W., Jenoe P., and Hall M.N.. 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell. 10:457–468. 10.1016/S1097-2765(02)00636-6 [DOI] [PubMed] [Google Scholar]
- Logarinho E., and Sunkel C.E.. 1998. The Drosophila POLO kinase localises to multiple compartments of the mitotic apparatus and is required for the phosphorylation of MPM2 reactive epitopes. J. Cell Sci. 111:2897–2909. [DOI] [PubMed] [Google Scholar]
- Macůrek L., Lindqvist A., Lim D., Lampson M.A., Klompmaker R., Freire R., Clouin C., Taylor S.S., Yaffe M.B., and Medema R.H.. 2008. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature. 455:119–123. 10.1038/nature07185 [DOI] [PubMed] [Google Scholar]
- Maehama T., and Dixon J.E.. 1998. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 273:13375–13378. 10.1074/jbc.273.22.13375 [DOI] [PubMed] [Google Scholar]
- Maia A.R., Garcia Z., Kabeche L., Barisic M., Maffini S., Macedo-Ribeiro S., Cheeseman I.M., Compton D.A., Kaverina I., and Maiato H.. 2012. Cdk1 and Plk1 mediate a CLASP2 phospho-switch that stabilizes kinetochore-microtubule attachments. J. Cell Biol. 199:285–301. 10.1083/jcb.201203091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardin B.R., and Schiebel E.. 2012. Breaking the ties that bind: New advances in centrosome biology. J. Cell Biol. 197:11–18. 10.1083/jcb.201108006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S.E., and Mann M.. 2007. Stable isotope labeling by amino acids in cell culture for quantitative proteomics. Methods Mol. Biol. 359:37–52. [DOI] [PubMed] [Google Scholar]
- Özlü N., Srayko M., Kinoshita K., Habermann B., O’Toole E.T., Müller-Reichert T., Schmalz N., Desai A., and Hyman A.A.. 2005. An essential function of the C. elegans ortholog of TPX2 is to localize activated aurora A kinase to mitotic spindles. Dev. Cell. 9:237–248. 10.1016/j.devcel.2005.07.002 [DOI] [PubMed] [Google Scholar]
- Panchaud N., Péli-Gulli M.P., and De Virgilio C.. 2013a Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Sci. Signal. 6:ra42 10.1126/scisignal.2004112 [DOI] [PubMed] [Google Scholar]
- Panchaud N., Péli-Gulli M.P., and De Virgilio C.. 2013b SEACing the GAP that nEGOCiates TORC1 activation: Evolutionary conservation of Rag GTPase regulation. Cell Cycle. 12:2948–2952. 10.4161/cc.26000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J., and Nurse P.. 2007. TOR signalling regulates mitotic commitment through the stress MAP kinase pathway and the Polo and Cdc2 kinases. Nat. Cell Biol. 9:1263–1272. 10.1038/ncb1646 [DOI] [PubMed] [Google Scholar]
- Peterson R.T., Beal P.A., Comb M.J., and Schreiber S.L.. 2000. FKBP12-rapamycin-associated protein (FRAP) autophosphorylates at serine 2481 under translationally repressive conditions. J. Biol. Chem. 275:7416–7423. 10.1074/jbc.275.10.7416 [DOI] [PubMed] [Google Scholar]
- Petronczki M., Lénárt P., and Peters J.M.. 2008. Polo on the rise—from mitotic entry to cytokinesis with Plk1. Dev. Cell. 14:646–659. 10.1016/j.devcel.2008.04.014 [DOI] [PubMed] [Google Scholar]
- Platani M., Santarella-Mellwig R., Posch M., Walczak R., Swedlow J.R., and Mattaj I.W.. 2009. The Nup107-160 nucleoporin complex promotes mitotic events via control of the localization state of the chromosome passenger complex. Mol. Biol. Cell. 20:5260–5275. 10.1091/mbc.E09-05-0377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévost M., Chamousset D., Nasa I., Freele E., Morrice N., Moorhead G., and Trinkle-Mulcahy L.. 2013. Quantitative fragmentome mapping reveals novel, domain-specific partners for the modular protein RepoMan (recruits PP1 onto mitotic chromatin at anaphase). Mol. Cell. Proteomics. 12:1468–1486. 10.1074/mcp.M112.023291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner A.G., Créancier L., Dos Santos C., Fialin C., Recher C., Bailly C., Kruczynski A., Payrastre B., and Manenti S.. 2010. A functional link between polo-like kinase 1 and the mammalian target-of-rapamycin pathway? Cell Cycle. 9:1690–1696. 10.4161/cc.9.9.11295 [DOI] [PubMed] [Google Scholar]
- Rieder C.L. 2011. Mitosis in vertebrates: the G2/M and M/A transitions and their associated checkpoints. Chromosome Res. 19:291–306. 10.1007/s10577-010-9178-z [DOI] [PubMed] [Google Scholar]
- Sabatini D.M. 2006. mTOR and cancer: insights into a complex relationship. Nat. Rev. Cancer. 6:729–734. 10.1038/nrc1974 [DOI] [PubMed] [Google Scholar]
- Sancak Y., Peterson T.R., Shaul Y.D., Lindquist R.A., Thoreen C.C., Bar-Peled L., and Sabatini D.M.. 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 320:1496–1501. 10.1126/science.1157535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarella R.A., Koffa M.D., Tittmann P., Gross H., and Hoenger A.. 2007. HURP wraps microtubule ends with an additional tubulin sheet that has a novel conformation of tubulin. J. Mol. Biol. 365:1587–1595. 10.1016/j.jmb.2006.10.064 [DOI] [PubMed] [Google Scholar]
- Seki A., Coppinger J.A., Jang C.Y., Yates J.R., and Fang G.. 2008. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science. 320:1655–1658. 10.1126/science.1157425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger S., Csokmay J., Akbar T., Jones T.I., Sengupta P., and Lilly M.A.. 2011. The nucleoporin Seh1 forms a complex with Mio and serves an essential tissue-specific function in Drosophila oogenesis. Development. 138:2133–2142. 10.1242/dev.057372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silljé H.H., Nagel S., Körner R., and Nigg E.A.. 2006. HURP is a Ran-importin β-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr. Biol. 16:731–742. 10.1016/j.cub.2006.02.070 [DOI] [PubMed] [Google Scholar]
- Smith T.F., Gaitatzes C., Saxena K., and Neer E.J.. 1999. The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24:181–185. 10.1016/S0968-0004(99)01384-5 [DOI] [PubMed] [Google Scholar]
- Smith E., Hégarat N., Vesely C., Roseboom I., Larch C., Streicher H., Straatman K., Flynn H., Skehel M., Hirota T., et al. . 2011. Differential control of Eg5-dependent centrosome separation by Plk1 and Cdk1. EMBO J. 30:2233–2245. 10.1038/emboj.2011.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliman G.A., Acosta-Jaquez H.A., Dunlop E.A., Ekim B., Maj N.E., Tee A.R., and Fingar D.C.. 2010. mTOR Ser-2481 autophosphorylation monitors mTORC-specific catalytic activity and clarifies rapamycin mechanism of action. J. Biol. Chem. 285:7866–7879. 10.1074/jbc.M109.096222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swedlow J.R., Sedat J.W., and Agard D.A.. 1997. Deconvolution in optical microscopy. In Deconvolution of Images and Spectra. Jansson P.A., editor. Academic Press, New York: 284–309. [Google Scholar]
- Swedlow J.R., Goldberg I.G., Eliceiri K.W., and OME Consortium . 2009. Bioimage informatics for experimental biology. Annu. Rev. Biophys. 38:327–346. 10.1146/annurev.biophys.050708.133641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum M.E., Medema R.H., and Akhmanova A.. 2011. Regulation of localization and activity of the microtubule depolymerase MCAK. BioArchitecture. 1:80–87. 10.4161/bioa.1.2.15807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima F., and Nishida E.. 2007. Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. EMBO J. 26:1487–1498. 10.1038/sj.emboj.7601599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M.Y., Wiese C., Cao K., Martin O., Donovan P., Ruderman J., Prigent C., and Zheng Y.. 2003. A Ran signalling pathway mediated by the mitotic kinase Aurora A in spindle assembly. Nat. Cell Biol. 5:242–248. 10.1038/ncb936 [DOI] [PubMed] [Google Scholar]
- Wallace W., Schaefer L.H., and Swedlow J.R.. 2001. A workingperson’s guide to deconvolution in light microscopy. Biotechniques. 31:1076–1078. [DOI] [PubMed] [Google Scholar]
- Walther T.C., Alves A., Pickersgill H., Loïodice I., Hetzer M., Galy V., Hülsmann B.B., Köcher T., Wilm M., Allen T., et al. . 2003. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell. 113:195–206. 10.1016/S0092-8674(03)00235-6 [DOI] [PubMed] [Google Scholar]
- Wang X., and Proud C.G.. 2011. mTORC1 signaling: what we still don’t know. J. Mol. Cell Biol. 3:206–220. 10.1093/jmcb/mjq038 [DOI] [PubMed] [Google Scholar]
- Wong J., Lerrigo R., Jang C.Y., and Fang G.. 2008. Aurora A regulates the activity of HURP by controlling the accessibility of its microtubule-binding domain. Mol. Biol. Cell. 19:2083–2091. 10.1091/mbc.E07-10-1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodard G.E., Huang N.N., Cho H., Miki T., Tall G.G., and Kehrl J.H.. 2010. Ric-8A and Gi α recruit LGN, NuMA, and dynein to the cell cortex to help orient the mitotic spindle. Mol. Cell. Biol. 30:3519–3530. 10.1128/MCB.00394-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S., Loewith R., and Hall M.N.. 2006. TOR signaling in growth and metabolism. Cell. 124:471–484. 10.1016/j.cell.2006.01.016 [DOI] [PubMed] [Google Scholar]
- Yu C.T., Hsu J.M., Lee Y.C., Tsou A.P., Chou C.K., and Huang C.Y.. 2005. Phosphorylation and stabilization of HURP by Aurora-A: implication of HURP as a transforming target of Aurora-A. Mol. Cell. Biol. 25:5789–5800. 10.1128/MCB.25.14.5789-5800.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng K., Bastos R.N., Barr F.A., and Gruneberg U.. 2010. Protein phosphatase 6 regulates mitotic spindle formation by controlling the T-loop phosphorylation state of Aurora A bound to its activator TPX2. J. Cell Biol. 191:1315–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Ems-McClung S.C., and Walczak C.E.. 2008. Aurora A phosphorylates MCAK to control ran-dependent spindle bipolarity. Mol. Biol. Cell. 19:2752–2765. 10.1091/mbc.E08-02-0198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitouni S., Nabais C., Jana S.C., Guerrero A., and Bettencourt-Dias M.. 2014. Polo-like kinases: structural variations lead to multiple functions. Nat. Rev. Mol. Cell Biol. 15:433–452. 10.1038/nrm3819 [DOI] [PubMed] [Google Scholar]
- Zorba A., Buosi V., Kutter S., Kern N., Pontiggia F., Cho Y.J., and Kern D.. 2014. Molecular mechanism of Aurora A kinase autophosphorylation and its allosteric activation by TPX2. eLife. 3:e02667 10.7554/eLife.02667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccolo M., Alves A., Galy V., Bolhy S., Formstecher E., Racine V., Sibarita J.B., Fukagawa T., Shiekhattar R., Yen T., and Doye V.. 2007. The human Nup107-160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J. 26:1853–1864. 10.1038/sj.emboj.7601642 [DOI] [PMC free article] [PubMed] [Google Scholar]