Abstract
Despite the availability of deep-sequencing techniques, genomic and transcriptomic data remain unevenly distributed across phylogenetic groups. For example, reptiles are poorly represented in sequence databases, hindering functional evolutionary and developmental studies in these lineages substantially more diverse than mammals. In addition, different studies use different assembly and annotation protocols, inhibiting meaningful comparisons. Here, we present the “Reptilian Transcriptomes Database 2.0,” which provides extensive annotation of transcriptomes and genomes from species covering the major reptilian lineages. To this end, we sequenced normalized complementary DNA libraries of multiple adult tissues and various embryonic stages of the leopard gecko and the corn snake and gathered published reptilian sequence data sets from representatives of the four extant orders of reptiles: Squamata (snakes and lizards), the tuatara, crocodiles, and turtles. The LANE runner 2.0 software was implemented to annotate all assemblies within a single integrated pipeline. We show that this approach increases the annotation completeness of the assembled transcriptomes/genomes. We then built large concatenated protein alignments of single-copy genes and inferred phylogenetic trees that support the positions of turtles and the tuatara as sister groups of Archosauria and Squamata, respectively. The Reptilian Transcriptomes Database 2.0 resource will be updated to include selected new data sets as they become available, thus making it a reference for differential expression studies, comparative genomics and transcriptomics, linkage mapping, molecular ecology, and phylogenomic analyses involving reptiles. The database is available at www.reptilian-transcriptomes.org and can be enquired using a wwwblast server installed at the University of Geneva.
Keywords: reptiles, transcriptomes, deep sequencing, deep sequencing, squamates, turtles, turtles, Archosauria
Introduction
The fields of genomics and transcriptomics maintain their fast development thanks to the improvement of sequencing technologies and associated bioinformatic tools for assembly and annotation. The availability of deep-sequencing techniques that produce hundreds of thousands to millions of reads of variable sizes, at only a fraction of the time and cost of Sanger sequencing, is leading to the accumulation of data in an increasing number of species. Despite these technological advances, sequencing data remain unevenly distributed across phylogenetic groups; for example, 50 of the 72 vertebrate genomes currently available in Ensembl belong to mammals (Flicek et al. 2014), most of them originating from the “Mammals Genome Project” (Lindblad-Toh et al. 2011), even though mammals comprise only 8% of the total number of vertebrate species. Current sequencing initiatives aim at a better coverage of eukaryotic lineages with, for example, the “Genome 10k Project,” the “i5k initiative,” and the “959 Nematode Genomes” proposing to sequence the genomes of 10,000 vertebrate, 5,000 insects and 959 nematode species, respectively (Genome 10K Community of Scientists 2009; Kumar et al. 2012; i5K Consortium 2013).
Although the availability of genome sequence data in a broad range of species would greatly facilitate functional evolutionary and developmental studies, these proposals will require a considerable amount of time to yield high-quality genomic resources. To overcome the high cost and difficulties of de novo genome sequencing and annotation, analyzing transcriptomes is an appealing alternative: Less sequencing data and computational resources are required. To generate a reference data set of annotated genes of a nonmodel species, one can pool RNA from multiple tissues, developmental stages and individuals for the preparation of complementary DNA (cDNA) libraries (Ekblom and Galindo 2011), thus sequencing a maximum number of genes. Special attention should be given to the selection of tissues for mRNA extraction to ensure that a high number of transcripts is expressed and sequenced (Ramskold et al. 2009). The transcriptome can be further enriched by the normalization of the cDNA libraries, as it equalizes the abundance of expressed transcripts by decreasing the proportion of the most highly expressed mRNAs, hence, facilitating the identification of poorly transcribed genes (Zhulidov et al. 2004).
Several freely available and efficient de novo transcriptome assemblers are available (e.g., Trans-Abyss [Robertson et al. 2010], Oases [Schulz et al. 2012], and Trinity [Grabherr et al. 2011]) that take into account the particularities of transcriptome sequence data sets, such as the variation in sequencing depth due to the variable abundance of transcripts and the presence of multiple transcript variants because of alternative splicing (Martin and Wang 2011). Annotation of the assembled transcripts is probably the most difficult step. Indeed, as functional analyses of transcripts are rarely undertaken in a systematic fashion, the identification of genes relies heavily on homology inference based on similarity analyses against already characterized sequences from other species. Even though the most widely used program for this task is the Basic Local Alignment Search Tool (BLAST; Altschul et al. 1990), a variety of protocols (type of BLAST, search parameters, databases inquired) are used in published studies, thus hindering the comparison of results. Furthermore, the sequence data provided with a specific publication vary extensively from none to raw deep-sequencing reads to annotated full transcripts. The only current database that integrates large-scale comparative genomic analyses within a single analytical and phylogenetic framework is Ensembl (Kersey et al. 2014). Yet, only high-quality fully sequenced genomes are incorporated in Ensembl, creating the necessity to generate smaller databases that target insufficiently represented lineages.
Among the most underrepresented vertebrate groups in terms of genomic data is the paraphyletic Class Reptilia. With more than 10,000 described species (http://www.reptile-database.org, last accessed June 10, 2015), only 12 genomes have been fully sequenced with varying quality: The Anole lizard Anolis carolinensis (Alfoldi et al. 2011), the Chinese softshell turtle Pelodiscus sinensis and the Green sea turtle Chelonia midas (Wang et al. 2013), the Western painted turtle Chrysemys picta (Shaffer et al. 2013), the Chinese alligator Alligator sinensis (Wan et al. 2013), the American alligator Alligator mississippiensis, the gharial Gavialis gangeticus and the saltwater crocodile Crocodylus porosus (St John et al. 2012), the Burmese python Python molurus (Castoe et al. 2013), the King cobra Ophiophagus hannah (Vonk et al. 2013), the speckled rattlesnake Crotalus mitchellii (Gilbert et al. 2014), and the corn snake Pantherophis guttatus (Ullate-Agote et al. 2015). According to the Genome 10k project, only nine additional reptilian genomes are in sequencing progress, still a low number to efficiently analyze the variety of developmental characters and evolutionary novelties encountered in this highly diverse group.
The aim of this study is to build the second version of the “Reptilian Transcriptomes Database” (http://www.reptilian-transcriptomes.org, last accessed June 10, 2015; Tzika et al. 2011), providing a high-quality and extensive annotation of transcriptomes and genomes from species that cover the major lineages of the Class Reptilia. We generated new transcriptomic data from two species, of interest for evolutionary developmental studies: The leopard gecko, Eublepharis macularius, and the corn snake, Pa. guttatus. For both species, Roche 454 and Illumina reads were sequenced from normalized cDNA libraries of multiple adult tissues (brain, kidneys, testes) and various embryonic stages. We also gathered published reptilian transcriptomic/genomic data sets, selecting representatives of the four extant orders (fig. 1): 1) Four Squamata, including two snakes (P. molurus [Castoe et al. 2011] and Thamnophis elegans [Schwartz et al. 2010]) and two lizards (Chalcides ocellatus [Brandley et al. 2012] and Chamaeleo chamaeleon [Bar-Yaacov et al. 2013]); 2) the single living tuatara species, Sphenodon punctatus (Miller et al. 2012); 3) three Crocodilia species (Cr. porosus [St John et al. 2012], Al. mississippiensis [Kunstner et al. 2010], and G. gangeticus); and 4) one Testudines (Chr. picta [Shaffer et al. 2013]).
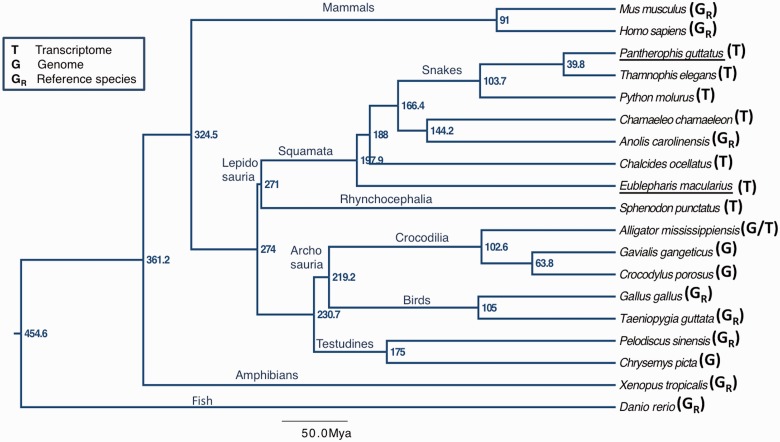
Fig. 1.—
Chronogram among the selected reptilian and reference species used for annotation. The letters between parentheses after the species names indicate the data type (T, transcriptome; G, genome; GR, genome of a reference species). The underlined species were newly sequenced in our laboratory for this study. The tree topology and divergence times are based on the “TimeTree of Life” estimates (Hedges et al. 2006).
Version 2 of the LANE runner software (Tzika et al. 2011) was implemented to annotate the selected and newly sequenced assemblies. LANE runner 2.0 integrates 1) iterative BLAST+ searches (Camacho et al. 2009) against multiple databases, 2) Reciprocal Best BLAST Hits (RBBH) identification for homology assessment, and 3) consensus sequence building to assemble sequences exhibiting the same annotation. This approach allowed us to annotate 50–70% of the transcripts from each data set, a higher percentage than in previously published transcriptomic studies. Using reference data sets, such as the Core Eukaryotic Genes Mapping Approach (CEGMA) (Parra et al. 2007), we show that combining transcriptomes of different tissues and developmental stages substantially increases the completeness of the assembled transcriptomes.
Using the annotated transcripts, we built concatenated protein alignments of single-copy families (up to 86,153 amino acids per species after quality trimming) and inferred maximum-likelihood phylogenetic trees that support the positions of turtles (Tzika et al. 2011) and the tuatara as sister groups of Archosauria and Squamata, respectively. The Reptilian Transcriptomes 2.0 resource is the largest transcriptomic database for reptiles to date and covers a large proportion of their diversity, including representatives of the four extant orders (Squamata, Rhynchocephalia, Crocodilia, and Testudines). This resource will be continuously updated to include selected new transcriptomic data as they become available, thus making it a reference for differential expression studies, comparative genomics/transcriptomics, linkage mapping, molecular ecology, and phylogenomic studies involving reptiles. The Reptilian Transcriptomes Database 2.0 is publicly available (www.reptilian-transcriptomes.org) for download and can be enquired through LANE runner 2.0 using a wwwblast server installed at the University of Geneva.
Materials and Methods
Transcriptomes Sequencing and Assembly
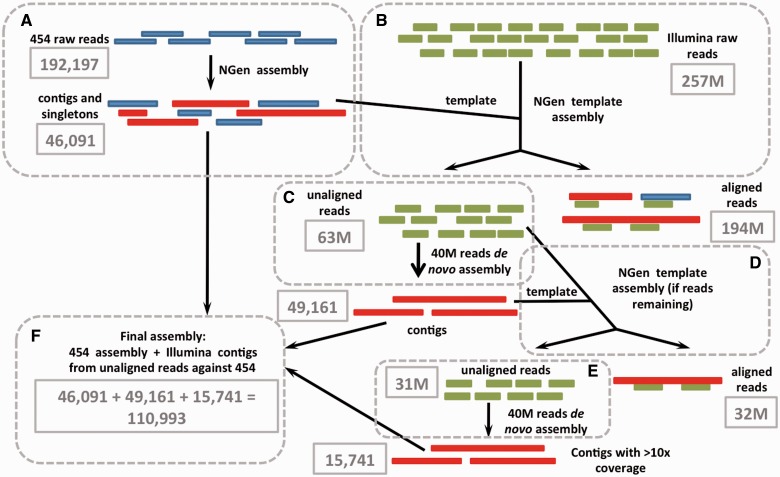
We collected samples from individuals of our established Pa. guttatus and E. macularius colonies. Animal housing and samplings were performed in accordance with the Swiss animal welfare regulation (permit number 1008/3421/0). Normalized libraries (using the Trimmer-2 cDNA normalization kit; Evrogen) were prepared from 1) adult organs (testes, kidneys, and brain), and 2) embryos at two or three developmental stages (E10, E30, and E47 for Pa. guttatus and E8 and E24 for E. macularius). Each library was sequenced with the “454” (half-plate) and the Illumina (one lane, 100-base paired-end reads) technologies. For Pa. guttatus, we also included in the analyses our previously published vomeronasal organ (VNO) transcriptome (Brykczynska et al. 2013). We designed an assembly pipeline (incorporating SeqMan NGen v11.0; DNASTAR) presented in figure 2 to exclude redundancy among the “454” and Illumina contigs. Briefly, it comprises the following steps (detailed description in the supplementary methods, Supplementary Material online): 1) Assembly of the “454” reads (fig. 2A), 2) alignment of all the Illumina reads to the “454” assembly (fig. 2B), and 3) de novo and template assembly of the nonaligned Illumina reads (fig. 2C–E) in subsets of 40 million reads (due to computational resources limitation). The final assembly (fig. 2F) comprises the “454” assembly (contigs and singletons) and the Illumina contigs. Remaining adaptor sequences at the extremities or within contigs and singletons are removed with LANE runner. For each species, we assembled a mix of all cDNA libraries as well as each library (adults, embryos, or VNO) separately. To assess whether the mix assemblies are good representatives of the individual data sets, we compared the former with the latter by performing a template transcriptome assembly in NGen. Default parameters were used for template assemblies (with “other” as sequencing technology) except for the “Minimum Match Percentage” parameter that was set to 80. The mix assemblies were used for subsequent annotation. The “454” raw reads of the Al. mississippiensis brain transcriptome (Kunstner et al. 2010) were assembled with NGen, as well.
Fig. 2.—
Assembly pipeline used to combine the “454” (in blue) and Illumina (in green) reads into nonredundant contigs (in red). Framed values correspond to those obtained for the E. macularius mix data set (multiple developmental stages and multiple adult organs), provided as an example. Dashed boxes delineate major steps of the assembly: (A) The “454” reads are de novo assembled, (B) the Illumina reads are aligned to the “454” assembly, and (C–E) iterative building of nonredundant Illumina contigs. The final assembly (F) includes both the “454” assembly and the Illumina contigs.
Annotation of Transcriptomes
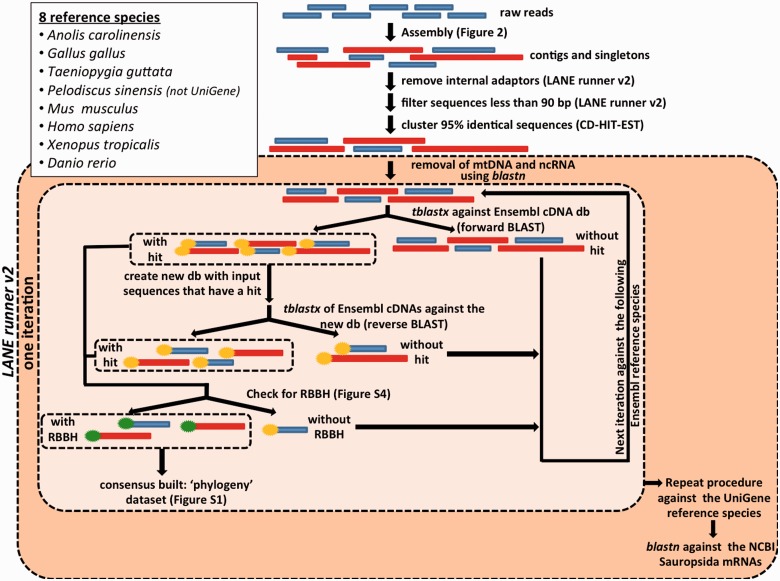
We annotated the transcriptomes and/or genomes of the following representatives of the four extant reptilian orders: 1) Six Squamata, including three snakes (P. molurus, Pa. guttatus, and T. elegans) and three lizards (E. macularius, Cham. chamaeleon, and C. ocellatus); 2) three Crocodilia (G. gangeticus, Cr. porosus, and Al. mississippiensis); 3) the single living Rhynchocephalia (S. punctatus); and 4) one Testudines (Chr. picta). Table 1 contains detailed information on the data sets and figure 1 shows the phylogenetic relationships among these species together with the reference species used for annotation (topology and divergence times based on the “Timetree of Life” estimates; Hedges et al. 2006). During our study, additional reptilian transcriptomes and genomes became available (e.g., Margres et al. 2013; Vonk et al. 2013; Wang et al. 2013); however, they either correspond to species closely related to the ones already selected or the transcriptomes originate from very specialized organs (e.g., the venom glands of snakes), in which case there is a high abundance of tissue-specific transcripts (e.g., coding for venom toxins) rather than a wide coverage of the species’ transcriptome. Similarly to the Reptilian Transcriptome 1.0 (Tzika et al. 2011), the annotation in the Reptilian Transcriptomes 2.0 is based on iterative BLAST searches, but with three major modifications: 1) The identification of RBBH to obtain a higher quality annotation (Altenhoff and Dessimoz 2009; Dalquen and Dessimoz 2013); 2) the use of the improved BLAST+ (release 2.2.28; Camacho et al. 2009), instead of the wwwblast web server; and 3) the use of Clustal Omega, instead of MUSCLE, as the multiple sequence alignment software for consensus building. The software LANE runner (Tzika et al. 2011) was upgraded (version 2.0) to accommodate these modifications. An overview of the annotation pipeline is presented in figure 3 and supplementary figure S1, Supplementary Material online, and a detailed description of the process is given in the supplementary methods and figure S2, Supplementary Material online.
Table 1.
Reptilian Transcriptomes and Genomes Considered for Annotation
| Species | Vernacular Name | Data Type | Source | Sequencing/Assembly | Size |
|---|---|---|---|---|---|
| Chalcides ocellatus | Ocellated skink | cDNA | Brandley et al. (2012) | Illumina assembly from uterus | 300,966 contigs |
| Sphenodon punctatus | Tuatara | cDNA | Miller et al. (2012) | Illumina assembly from embryos | 32,911 contigs |
| Python molurus bivittatus | Burmese python | cDNA | Castoe et al. (2011) | “454” assembly from liver and heart | 37,245 contigs |
| Chamaeleo chamaeleon | Common chameleon | cDNA | Bar-Yaacov et al. (2013) | SOLiD assembly multitissue | 164,692 contigs |
| Thamnophis elegans | Garter snake | cDNA | Schwartz et al. (2010) | “454” assembly multitissue and multi-individual | 188,940 contigs and singletons |
| Chrysemys picta | Painted turtle | gDNA | Shaffer et al. (2013) | Genome assembly 3.0.1 | 6,080 scaffolds |
| Gavialis gangeticus | Gharial | gDNA | St John et al. (2012) | Draft genome assembly | 47,351 scaffolds |
| Alligator mississippiensis | American alligator | gDNA | St John et al. (2012) | Draft genome assembly | 8,897 scaffolds |
| Crocodylus porosus | Saltwater crocodile | gDNA | St John et al. (2012) | Draft genome assembly | 23,365 scaffolds |
| Alligator mississippiensis | American alligator | cDNA | Kunstner et al. (2010) | Brain “454” reads | 438,029 reads |
| Pantherophis guttatus | Corn snake | cDNA | Brykczynska et al. (2013) | VNO “454” and Illumina libraries | 343,062 and 54.4M reads from “454” and Illumina |
| Pantherophis guttatus | Corn snake | cDNA | This study | Adults testes, brain and kidneys “454” and Illumina paired-end libraries | 135,630 and 145M reads from “454” and Illumina |
| Pantherophis guttatus | Corn snake | cDNA | This study | Embryonic “454” and Illumina paired-end libraries | 45,417 and 129.8M reads from “454” and Illumina |
| Eublepharis macularius | Leopard gecko | cDNA | This study | Adults testes, brain and kidneys “454” and Illumina paired-end libraries | 112,760 and 128M reads from “454” and Illumina |
| Eublepharis macularius | Leopard gecko | cDNA | This study | Embryonic “454” and Illumina paired-end libraries | 79,437 and 129.8M reads from “454” and Illumina |
Fig. 3.—
Outline of the transcriptome annotation pipeline. All steps included in the outer dash-framed box are performed in LANE runner. The steps of a single iteration (i.e., using one reference species) are grouped in the inner dashed frame. The reference species iteratively considered for annotation are listed in the inset. Query sequences having a hit are indicated with a yellow mark and those having an RBBH with a green mark.
Consensus Sequences
Consensus sequences were built using LANE runner 2.0 as follows: 1) The best match of each input sequence is identified considering the ordered criteria of e-value, match length, and % identity; 2) input sequences matching with the same database sequence are aligned using Clustal Omega (Sievers and Higgins 2014); 3) a majority consensus is built, where the database sequence is used only as an anchoring point to define the relative positions of the input sequences; and 4) the consensus is named after the database sequence. When several input sequences match with a single database sequence, we obtain a “one-to-many consensus.” When a single input sequence matches a database sequence, it is labeled as a “one-to-one” consensus, where no alignment is necessary and the input sequence takes the name of the database sequence. Contrary to LANE runner v1 (Tzika et al. 2011) that used MUSCLE (Edgar 2004), LANE runner 2.0 uses Clustal Omega which is better adapted to the alignment of small fragments against a long reference sequence.
Two data sets were generated and used to build consensus sequences (fig. 3 and supplementary fig. S1, Supplementary Material online). The “phylogeny data set” consisted of the RBBH against the Ensembl cDNA and the UniGene databases. The “annotation data set” combined 1) the phylogeny data set, 2) the “NCBI Sauropsida mRNAs” BLASTn results, and 3) the non-RBBH Ensembl and UniGene database matches. The phylogeny data set corresponds to a high-quality annotation that can be used to build phylogenetic trees, whereas the annotation data set maximizes the number of annotated transcripts in the species investigated.
Genome Annotation
We developed a simple method to partially annotate the following genomes for which annotation is not yet available in the literature or public databases: Al. mississippiensis, G. gangeticus, Cr. porosus (St John et al. 2012), and Chr. picta (Shaffer et al. 2013). The pipeline is outlined in supplementary figure S3, Supplementary Material online, with Chrysemys as an example. First, we used BioMart (Smedley et al. 2009) to extract the exons, as well as the 5′ and 3′ 1,000-base flanks of Pelodiscus Ensembl cDNAs. Then, we performed an NGen assembly where the Chrysemys genome was used as template and the Pelodiscus exons and flank sequences as reads (default NGen parameters, “Other” as sequencing technology, and Minimum Match Percentage set to 80). Using the “mpileup” option of samtools (Li et al. 2009), we extracted the genome segments that align to the Pelodiscus exons and flanks. These sequences are potential exons (including 3′- and 5′- untranslated regions [UTRs]) of the Chrysemys genome that we want to annotate. To assemble the potential exons into transcripts, a BLASTn search was performed against the Pelodiscus cDNA database. The BLASTn search settings were the same as for the mtDNA comparison (see supplementary methods, Supplementary Material online), without considering a minimum match length threshold to permit the alignment of even the shortest potential exons. As in our annotations of transcriptomes, consensus sequences were built for the exons that had a hit against the cDNA database (supplementary fig. S3, Supplementary Material online). The same procedure was used for annotation of the genomes of the three crocodilian species except that both Gallus and Pelodiscus exons were aligned to each crocodilian genome, and both Gallus and Pelodiscus cDNA databases were used in the BLASTn search. Annotated genes were restricted to a phylogeny data set by keeping only: 1) Consensus sequences named after a Gallus transcript and greater than 90 bp for the crocodilian species, and 2) consensus sequences greater than 90 bp for Chr. picta.
Annotation of the Pa. guttatus and E. macularius Individual Libraries
For Pa. guttatus and E. macularius, we separately sequenced cDNA libraries from different organs and embryonic developmental stages. For each of the two species, all reads from all libraries were pooled before performing assembly and annotation (figs. 2 and 3). We then investigated, with a tBLASTx search (same settings as for the Ensembl cDNA searches), the contribution of the individual libraries to the annotation data set and to the “orphans” generated with the mixed annotation.
Results
Assembly of the Corn Snake, Leopard Gecko, and American Alligator Transcriptomes
For Pa. guttatus and E. macularius, we used the “454” and Illumina technologies to sequence a multitissue normalized library (kidney, testes, and brain) from three adult individuals, as well as one normalized library from embryos of various developmental stages. In addition, the VNO reads that we sequenced for Pa. guttatus (Brykczynska et al. 2013) were included in the analysis. The number of “454” reads obtained for each library varies between 45,000 and 343,000 (table 2), with an average read length of 219–401 bp. The number of Illumina 100-base paired-end reads ranged from 128 millions (M) in E. macularius (adult data set) to 145M in Pa. guttatus (adult). The individual (adults, embryos, VNO) and mixed (all reads) data sets obtained with the “454” technology were assembled using NGen (fig. 2A). The percentage of reads discarded due to quality filtering and trimming varied from 0.6% to 25.8% (table 2). The same approach was used to assemble the Al. mississippiensis brain transcriptome “454” reads (Kunstner et al. 2010; SRR029332). The “454” assembly was then used as template to remove redundant Illumina reads (fig. 2B). Depending on the data set, 48–75% Illumina reads matched with the “454” assembly, this percentage being higher for the E. macularius than for the Pa. guttatus assemblies. Forty millions of the remaining unaligned Illumina reads were then de novo assembled (fig. 2C). This assembly was used as a template for all unaligned Illumina reads (fig. 2D). Yet unassembled Illumina reads were de novo assembled by batches of 40M (fig. 2E). The final assemblies consisted of the “454” contigs and singletons and the Illumina contigs. LANE runner 2.0 was used to remove adaptor sequences within the contigs. More than 95% of contigs/singletons of the individual data sets aligned to the mix assembly sequence (supplementary table S2, Supplementary Material online), thus the latter was used for subsequent annotation.
Table 2.
NGen Assembly Workflow Statistics
|
Pantherophis guttatus |
Eublepharis macularius |
||||||
|---|---|---|---|---|---|---|---|
| Adults | Embryos | VNO | Mix | Adults | Embryos | Mix | |
| Number of plates | 1 | 1 | 1 | 3 | 1 | 1 | 2 |
| 454 reads | 135,630 | 45,417 | 343,062 | 524,109 | 112,760 | 79,437 | 192,197 |
| 454 discarded | 8,557 (6.3%) | 4,374 (9.6%) | 1,912 (0.6%) | 15,071 (2.9%) | 29,056 (25.8%) | 4,466 (5.6%) | 33,522 (17.4%) |
| 454 contigs | 17,570 | 6,133 | 38,666 | 45,955 | 10,635 | 6,595 | 17,876 |
| 454 singletons | 27,826 | 17,069 | 56,632 | 98,265 | 26,200 | 13,434 | 28,215 |
| Av. contig length | 556 | 499 | 447 | 497 | 393 | 545 | 712 |
| Greater than 500 bp | 9,585 | 2,951 | 12,369 | 17,075 | 3,192 | 3,439 | 13,122 |
| Greater than 1 Kb | 1,471 | 343 | 1,325 | 3,667 | 265 | 595 | 3,404 |
| 454 assembly | 45,396 | 23,202 | 95,298 | 144,220 | 36,835 | 20,029 | 46,091 |
| Number of lanes | 0.5 | 0.5 | 0.5 | 1.5 | 0.5 | 0.5 | 1 |
| Illumina reads | 145M | 129.8M | 54.4M | 329.2M | 128M | 129.8M | 257.8M |
| Aligned to 454 | 92M (63%) | 62.8M (48%) | 32.3M (59%) | 216.9M (66%) | 82M (64%) | 94.8M (73%) | 194.2M (75%) |
| Illumina contigs | 63,227 | 86,371 | 28,453 | 134,457 | 50,737 | 36,254 | 64,902 |
| Av. contig length | 423 | 394 | 575 | 396 | 410 | 454 | 389 |
| Greater than 500 bp | 20,805 | 25,892 | 14,891 | 39,300 | 15,131 | 13,668 | 18,577 |
| Greater than 1 Kb | 2,738 | 3,570 | 2,652 | 3,589 | 953 | 2,097 | 1,831 |
| Number iterations | 1 | 2 | 1 | 3 | 1 | 1 | 2 |
| Final assembly | 108,623 | 109,573 | 123,751 | 278,677 | 87,572 | 56,283 | 110,993 |
| After adaptor removal | 108,678 | 109,589 | 124,012 | 279,699 | 87,703 | 56,302 | 111,237 |
Note.—The two first shaded rows correspond to the total number of contigs/singletons obtained from "454" and Illumina reads, and the third shaded row corresponds to the total number of contig/singletons after removal of adaptors.
Annotation of the Transcriptomes
Along with these three newly assembled reptilian transcriptomes (corn snake, leopard gecko, and American alligator), another five transcriptomes, originating from different tissues and developmental stages, were annotated: Cham. chamaeleon, T. elegans, P. molurus, S. punctatus, and C. ocellatus (fig. 1 and table 1). First, we preprocessed the assemblies by 1) removing sequences of ≤ 90 nucleotides length and 2) clustering ≥ 95% identical sequences using CD-HIT-EST (Li and Godzik 2006; Fu et al. 2012) by maintaining only the longest sequence from each cluster (fig. 3).
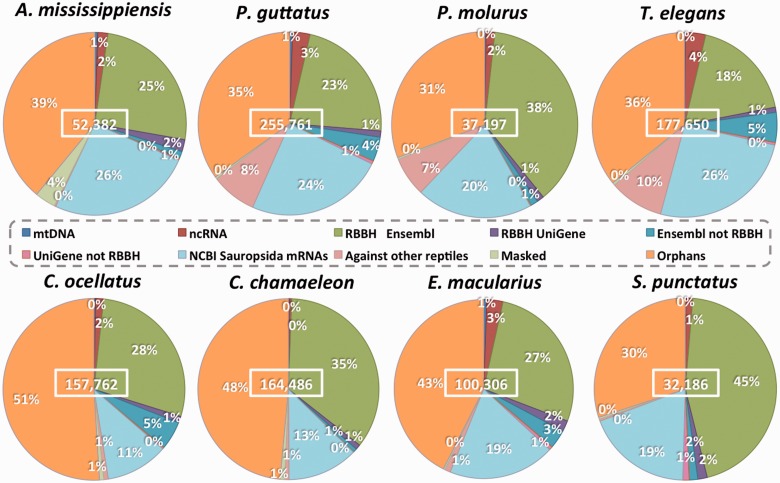
Our annotation pipeline (fig. 3 and supplementary fig. S1, Supplementary Material online) consists of iterative BLAST searches and identification of RBBH for a higher quality result, all steps being performed with the updated version of LANE runner (version 2.0). The filtered contigs and singletons of each reptilian transcriptome were compared with a BLASTn search to 1) the mitochondrial genome of the corresponding species or that of a closely related one and 2) a database including Ensembl v73 ncRNA sequences from eight reference species: A. carolinensis, Gallus gallus, Taeniopygia guttata, Pe. sinensis, Mus musculus, Homo sapiens, Xenopus tropicalis, and Danio rerio (hereafter named Anolis, Gallus, Taeniopygia, Pelodiscus Mus, Homo, Xenopus and Danio, to distinguish them as reference species from the annotated ones). Less than 1% of contigs and singletons corresponded to mtDNA sequences and only 1–4% to ncRNA (fig. 4). RBBH were identified with tBLASTx searches against the Ensembl v73 coding cDNA and “UniGene November 2013” databases of each reference species (fig. 3; see Supplementary Material online for details). Second, the sequences still nonannotated by this iterative process were aligned with tBLASTx against an “NCBI (National Center for Biotechnology Information) November 2013” database containing mRNA sequences from Sauropsida. The annotated sequences were divided into two data sets: 1) An annotation data set comprising all annotated sequences (with and without RBBH) and 2) a high quality phylogeny data set including only the annotated sequences with a RBBH. In a last round of BLAST searches, the nonannotated contigs/singletons of each reptilian species were compared with the annotation data set and the nonannotated sequences of the other seven reptilian transcriptomes. Finally, the remaining sequences were masked using RepeatMasker to check whether their lack of annotation was due to the presence of repetitive elements. The sequences lacking annotation after all these processes are called orphans (supplementary fig. S1, Supplementary Material online).
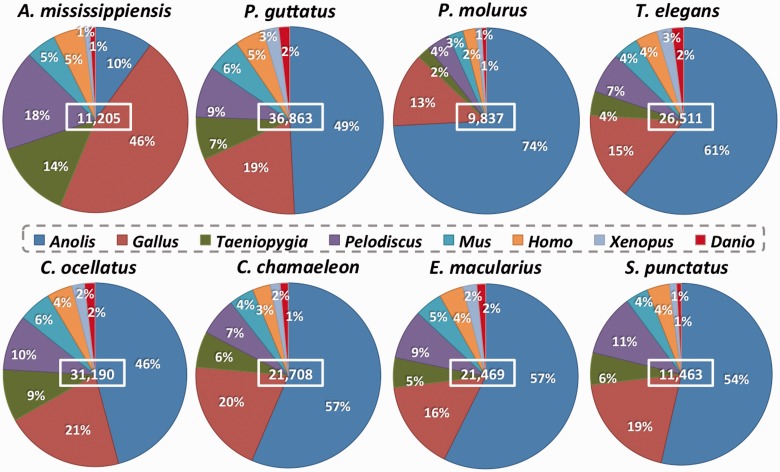
Fig. 4.—
Piecharts showing the percentage of contig/singletons annotated at each step of the pipeline. The number of input sequences for each transcriptome is indicated in the middle of each graph.
As shown in figure 4, the annotation success is high but varies among species, with the number of orphans ranging from 30% in S. punctatus to 51% in C. ocellatus. Compared with the Reptilian Transcriptome v1, this percentage is improved (i.e., reduced), in part due to the BLAST search step against the NCBI Sauropsida mRNAs, which include transcripts of reptilian species closely related to those under study here. This step indeed resulted in the annotation of 11 to 26% of the filtered contigs and singletons, the percentage being highest for the transcriptomes of snakes and Al. mississippiensis. On the other hand, the UniGene database provided little improvement in the annotation (1 to 3% of the total), probably because of its high overlap with the Ensembl cDNA database. The percentage of input sequences with a RBBH varies from 19 to 47%, with the greatest proportions found in S. punctatus and P. molurus transcriptomes.
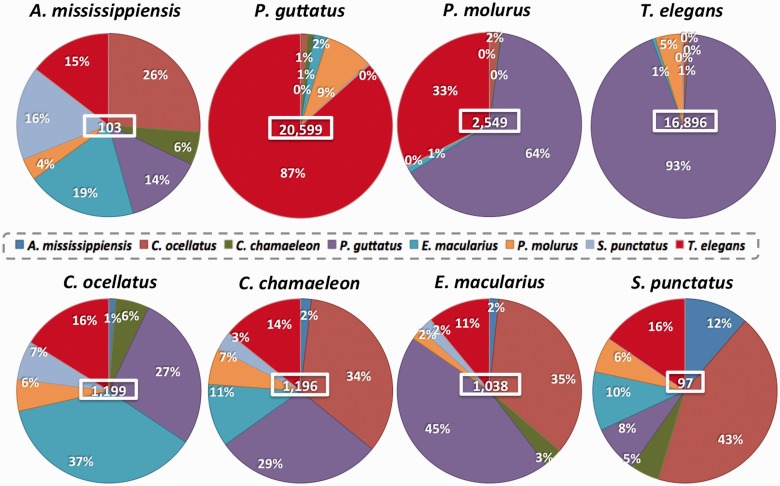
The BLAST searches among the reptilian transcriptomes substantially improved the snakes’ annotation (figs. 4 and 5), as 7 to 10% of their contigs/singletons matched with other transcriptomes, mainly those of other snakes. In particular, 93% of the T. elegans sequences still nonannotated after BLAST searches against Ensembl, UniGene and NCBI Sauropsida mRNAs, matched a Pa. guttatus sequence (fig. 5). Similar results are obtained for the reverse comparison (87% of the nonannotated Pa. guttatus sequences matched T. elegans). These results strongly suggest that these sequences represent snake-specific transcripts. For the other transcriptomes, only a small number (<1,200) of contigs/singletons were annotated, probably because none of these reptiles is more closely related to each others than to one of the Archosauria or Squamata reference species (Anolis and Gallus) (fig. 1). Finally, very few repetitive elements were identified, with the highest percentage of masked sequences in Al. mississippiensis (4%) and ≤1% for all the other species.
Fig. 5.—
Piecharts showing the percentage of nonannotated contigs/singletons that match with the other annotated reptilian transcriptomes. The total number of hits is indicated in the middle of each graph.
Even though the number of input sequences annotated here from each reptilian species is greater compared with the Reptilian Transcriptome v1 and other transcriptomic studies, the percentage of orphans is still substantial, which can be due to a combination of factors such as 1) the absence of full genome sequence data from the particular species for extensive annotation, 2) the presence of sequencing or assembly artifacts, 3) the presence of genomic contaminations in cDNA libraries, 4) the large evolutionary distance of the eight reptilian species from the reference species (fig. 1)—Anolis being the most closely related reference species to Squamata (144–197 Myr) and to S. punctatus (271 Ma), whereas Gallus is the most closely related to Al. mississippiensis (219 Myr)—and 5) the presence of real orphans (also known as taxonomically restricted genes), which lack sequence similarity or even homology to other species’ genes and, depending on the organism, may correspond to 10–20% of all genes (Khalturin et al. 2009; Tautz and Domazet-Loso 2011).
In an attempt to rule out some of these hypotheses, we compared the orphans with available reptilian genomes. We mapped all but 87 contigs of the 11,435 P. molurus orphans, 98.1% of the 89,329 Pa. guttatus orphans, and 95% of the 20,441 Al. mississippiensis orphans to their respective genomes using NGen. We obtained similar results when performing a BLASTn search against the genome of each corresponding species. Thus, at least for these three species, the vast majority of nonannotated contigs/singletons are not artifacts but must be either real transcripts or genomic DNA contaminations (introns, intergenic regions). Furthermore, we aligned the Pa. guttatus and T. elegans orphans to the two snake genomes available: P. molurus (100 Myr) and the king cobra O. hannah (44 Myr). Twenty-six percent out of 63,631 T. elegans orphans and 30% out of 89,329 Pa. guttatus orphans were aligned to the P. molurus genome, whereas these percentages greatly increased (68% and 74%) for the comparison with the more closely related O. hannah genome. These results provide further support that these orphans are real transcripts.
Finally, we compared the quality statistics of the annotated contigs/singletons and the orphans (supplementary table S3, Supplementary Material online) with a two-sample t-test. In all transcriptomes, the average length of the annotated sequences is significantly greater than that of the orphans (P value < 0.001). This measure remains significant even if we consider separately the average length of contigs and singletons. The contig coverage (average number of reads per contig) is also significantly higher for the annotated than for the orphan contigs (P value < 0.001). For example, in P. molurus and T. elegans, there is almost a 5-fold coverage difference between annotated contigs and orphans.
Consensus Sequences Statistics
Following the BLAST searches, we used LANE runner 2.0 to assemble “consensuses” (see Materials and Methods for details). When a single input sequence matches a database sequence, it is labeled as a “one-to-one consensus” and the input sequence takes the name of the database sequence. When multiple contigs/singletons match the same database sequence, LANE runner 2.0 performs Clustal Omega (Sievers and Higgins 2014) alignments to define the relative positions of the input sequences and assembles them into a “one-to-many consensus.”
Overall, 20–39% of the consensuses are “one-to-many” (supplementary table S4, Supplementary Material online) with an average transcript coverage of 18–40%. In the case of the Ensembl phylogeny data set, we observe a greater percentage of “one-to-many consensuses” (28–54%) and a large mean coverage of these transcripts (19–39%). To investigate whether the coverage of transcripts in all our analyzed data sets is 3′- or 5′-biased, we split the “one-to-many consensuses” against Anolis genes into three parts: The 3′ 30%, the middle 40%, and the 5′ 30%. We observed that in the 3′- and 5′-ends, the percentage of gaps ranged from 64% to 90% versus 49–72% in the middle. As we extended the Anolis cDNAs with 1,000-bases upstream and downstream of the CDS (in order to include the missing UTRs), the lower coverage of these regions suggests that some of them are not part of the real mRNAs or are highly variable between distantly related species.
In figure 6, we show the distribution of the consensus sequences named after an Ensembl or UniGene reference species gene. For all transcriptomes, most consensus sequences are named after the first reference species (used in the iterative BLAST searches) selected to be the most closely related to the species being annotated: Gallus is used for the annotation of 46% of the Al. mississippiensis sequences and Anolis is used for 46–74% of the sequences for the other reptilian species. For all the data sets, the combination of Anolis and Gallus sequences used as reference to build consensuses ranged from 56% to 87%. In all cases, there are more consensuses named after a Pelodiscus cDNA than after a Taeniopygia one, underlying the importance of using reference species from different taxonomic groups rather than several species from the same one. The remaining four non-Sauropsida species were used as reference to build only about 10% of the consensus sequences, due to their greater evolutionary distance.
Fig. 6.—
Piecharts showing the percentage of consensus sequences annotated with each reference species in the Ensembl or UniGene databases. The total number of consensuses is indicated in the middle of each graph.
Annotation of Genomes
We additionally annotated the genomes of three Crocodilia (Al. mississippiensis, Cr. porosus, and G. gangeticus) and a turtle (Chr. picta) using Gallus and Pelodiscus exons extracted from Ensembl (supplementary fig. S3, Supplementary Material online). We selected Chr. picta because there is a preliminary gene annotation in “Ensembl Pre!” that can help us assess the efficiency and quality of our annotation pipeline. Pelodiscus exons and flanks, as well as Gallus exons were used to identify the corresponding Crocodilia exons, whereas only the Pelodiscus sequences were used for the Chr. picta genome annotation. Seventy percent of Pelodiscus sequences were aligned to the Chr. picta genome, resulting in 143,467 potential Chr. picta exons (supplementary table S5, Supplementary Material online). Almost all (99.8%) were assembled to transcripts after a BLASTn search against the Ensembl Pelodiscus coding cDNA database, resulting in 18,447 consensus sequences. To assess the quality of this annotation, the consensuses were compared with the Chr. picta preliminary gene annotation of Ensembl: 15,028 of 18,447 (81.3%) consensus sequences had a BLASTn hit (using the same settings as for the identification of mtDNA transcripts, but increasing the minimum %ID to 90% and lowering the e-value threshold to 10−5 to be more stringent), demonstrating the efficiency of our annotation pipeline.
Only 33% of the Pelodiscus and Gallus exons were aligned to the Crocodilia genomes (supplementary table S5, Supplementary Material online), probably because none of the reference species is closely related to these crocodilian genomes and, therefore, only well-conserved genes can be identified. For each Crocodilia, around 90,000 potential exons were retrieved and almost all of them (99.7–99.8) were assembled against a Gallus or a Pelodiscus coding cDNA after a BLASTn search, yielding more than 17,000 consensus sequences. Note that the percentage of “one-to-many consensuses” (67% for Crocodilia and 87% for Chr. picta) in these annotations of genomes is higher than in the annotations of transcriptomes. For the phylogeny data set (built only with sequences greater than 90 bp and, for Crocodilia, only with sequences named after a Gallus cDNA), the percentage of “one-to-many consensuses” is even higher (around 87% in the Crocodilia and 91% in Chr. picta), indicating that most of the ≤90 bp short sequences correspond to “one-to-one consensuses.” Furthermore, there are 11,709 consensuses named after the same database sequence in Al. mississippiensis, G. gangeticus and Cr. porosus, possibly corresponding to well-conserved transcripts at this taxonomic level.
Comparison with Reference Data Sets
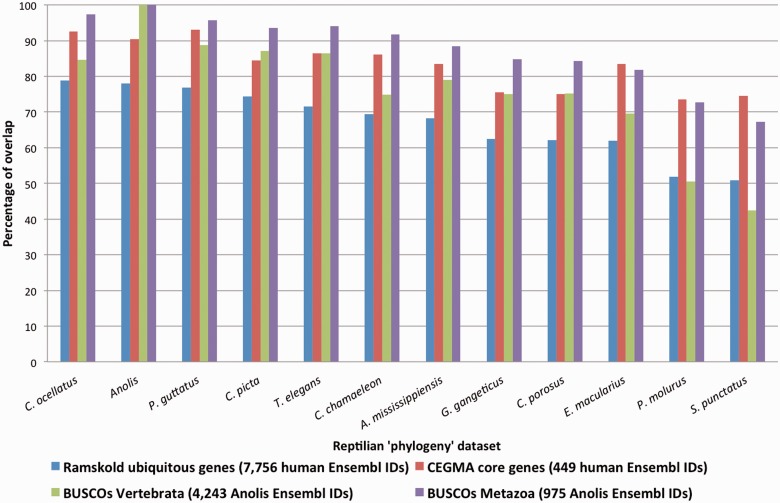
We verified the quality of our annotations by assessing their completeness against four established data sets (fig. 7): A set of ubiquitously expressed genes in human tissues and cell lines (Ramskold et al. 2009), a selection of eukaryotic genes from the CEGMA (Parra et al. 2007), and two “Benchmarking sets of Universal Single-Copy Orthologs” (BUSCOs; from OrthoDB7 [Waterhouse et al. 2013]) in Vertebrata and Metazoa. The comparisons were performed by BLASTn searches of the reference data sets against the consensuses of the phylogeny data sets. In the case of Al. mississippiensis, the results of the transcriptome and genome annotations were combined.
Fig. 7.—
Completeness of the annotated transcriptomes assessed with four reference data sets: Ramskold ubiquitously expressed genes in human (blue bars), CEGMA core human genes (red bars), OrthoDB7 BUSCOs from the vertebrate (green bars), and the metazoan (purple bars) radiation nodes. The species are ordered from higher to lower overlap with the Ramskold data set and Anolis is shown as reference.
Ramskold et al. (2009) identified 7,756 human Ensembl genes ubiquitously expressed in 16 organs and cell lines. These are mainly intracellular housekeeping genes involved in basic functions, such as metabolism, transcription or macromolecule synthesis. We identified roughly 80% of these genes in the Anolis reference genome. Note that it is unknown whether all of these genes are ubiquitously expressed in the corresponding 16 Anolis tissues/organs. We show the overlap with the Ramskold et al.’s core set of genes to be slightly higher for the C. ocellatus transcriptome annotation than for the Anolis genome (fig. 7), underlying the quality of our C. ocellatus annotation. On the other hand, S. punctatus and P. molurus presented a substantially lower percentage (about 50%) of correspondence with this Ramskold et al.’s core set of genes. For all other species investigated, the percentages of overlap were intermediate (62–77%). The best percentage among Crocodilia was found for Al. mississippiensis (68%), possibly because transcriptomic and genomic data were combined for this species.
The CEGMA data set includes a set of 456 human transcripts that are highly conserved in a wide range of eukaryotic taxa and has been previously used to assess the quality of genome annotations (Parra et al. 2007). CEGMA was built considering conserved genes in six organisms: Homo sapiens, Drosophila melanogaster, Arabidopsis thaliana, Caenorhabditis elegans, Saccharomyces cerevisiae and Schizosaccharomyces pombe, 449 of which have an Ensembl human identifier. Despite this high conservation among taxa, we identified only 90% of these genes in the Anolis reference genome (fig. 7). The high percentage (>92%) of these genes identified in Pa. guttatus and C. ocellatus indicates the high-quality annotation we achieved here. The other species exhibited an overlap with the CEGMA data set between 73% and 86% (the lowest being, again, for S. puctatus and P. molurus).
Finally, the two BUSCOs data sets, provided by OrthoDB7 (Waterhouse et al. 2013) for the Metazoan and Vertebrata radiation nodes, correspond to sequences from orthologous groups with single-copy genes present in more than 90% of the species of the corresponding radiation node. There are 41 species in the Vertebrata BUSCOs (including Gallus and Anolis) and 93 species in the Metazoan BUSCOs data sets (with 4,243 and 975 Anolis Ensembl identifiers, respectively). The Metazoan BUSCOs are a subset of the Vertebrate data set and correspond to genes conserved over a greater evolutionary period, explaining that we identify a substantially greater percentage of genes in the Metazoan (67–97%; purple columns, fig. 7) than in the vertebrate (42–89%; green columns) BUSCOs.
In short, and as supported by all reference data sets (fig. 7), Pa. guttatus, T. elegans, C. ocellatus and Chr. picta data sets have the most complete annotations, whereas S. punctatus and P. molurus exhibit substantially less complete annotations. These results are consistent with the source of the corresponding transcriptomes: The S. punctatus transcriptome is from a nonnormalized library of a single early stage embryo (at which developmental point a restricted panel of genes is probably expressed) and the P. molurus transcriptome is from nonnormalized libraries of heart and liver, two organs in which a high proportion of mRNAs correspond to a low number of different transcripts (Ramskold et al. 2009). The Pa. guttatus and E. macularius transcriptomes, newly sequenced for this study, also exhibit substantial differences in their annotation completeness. The Pa. guttatus transcriptome is more complete for two reasons: 1) The availability of the VNO transcriptome of Pa. guttatus doubles the number of contigs/singletons in that species, and 2) using three embryonic stages in Pa. guttatus, instead of two in E. macularius, probably enriched the transcript pool further. Note that, in general, the full length of the reference sequences was partially covered by the aligned reptilian transcripts: CEGMA mean coverage: 29–65%, Ramskold: 25–64%, BUSCOs Vertebrata: 28–69%, and BUSCOs Metazoa: 33–75%.
Large Phylogeny Inference
The phylogenetic positions of the tuatara S. punctatus and Testudines (turtles) have long been debated. The first phylogenomic analysis to investigate the question of the position of turtles (Tzika et al. 2011) supported Testudines as the sister group to Archosauria. Two subsequent phylogenomic analyses, based on nucleotide/amino-acid (Chiari et al. 2012) or ultraconserved-element alignments (Crawford et al. 2012), confirmed this hypothesis. Here, using the Reptilian Transcriptomes 2.0, consisting of a greater number of species than the first version and of better annotated transcripts, we built yet improved data sets (up to 86,153 amino acids per species after quality trimming) and inferred maximum-likelihood phylogenomic trees that support the positions of turtles (Tzika et al. 2011) and the tuatara as sister groups of Archosauria and Squamata, respectively (supplementary results, supplementary methods, table S1, and figs. S4 and S5, Supplementary Material online).
Discussion
Here, we present a pipeline for the annotation of transcriptomic and genomic data sets, which we applied to build the second version of the Reptilian Transcriptomes Database. To that end, the LANE runner software was updated with the incorporation of new functionalities that allowed us to 1) perform iterative BLAST searches using the BLAST+ program against multiple databases and reference species covering a wide evolutionary range within vertebrates, 2) identify RBBH, and 3) build consensus sequences by joining the contigs/singletons that correspond to the same reference sequence. This approach fulfilled our three objectives: 1) The production of a high-quality annotation, thanks to the identification of RBBHs, which can be used for phylogenomic analyses (phylogeny data set); 2) the generation of an exhaustive annotation resource (annotation data set); and 3) the use of a single workflow for the analysis of all data sets, independently of the species, tissues, or sequencing technology, greatly facilitating comparative analyses. This new annotation resource is the largest transcriptomic database for reptiles to date and covers a large proportion of their diversity, including representatives of the four extant orders (Squamata, Rhynchocephalia, Crocodilia, and Testudines). Furthermore, its structure facilitates the incorporation of new data sets as they are produced, making it an expansible resource rather than a static list of genes. The Reptilian Transcriptomes Database 2.0 is available at www.reptilian-transcriptomes.org.
Using our transcriptome annotation pipeline, we annotated between 50% and 70% of the nonredundant contigs and singletons for each species. For the reannotated transcriptomes, this percentage is in all cases higher than in the corresponding original publication, underlying the efficiency of our approach. For instance, Schwartz et al. (2010) annotated 25% of the garter snake T. elegans transcripts, whereas, using our workflow, we increased this value to 64%. Note, however, that the only reptilian reference at the time of that original publication was the draft genome of Anolis. But even in the case of the Cham. chamaeleon transcriptome, that was recently published, we doubled the number of annotated transcripts (from 25% to 50%) (Bar-Yaacov et al. 2013). For the newly sequenced Pa. guttatus transcriptome presented here, not only does it contain more contigs/singletons than the first version (79,688 transcripts in version 1 vs. 255,761 transcripts in version 2), but it also annotates a greater percentage of them (46% in the first vs. 65% in the second version). The better performances of our updated pipeline are due to 1) the addition of Illumina data, increasing the coverage of each transcript; 2) the sequencing of transcripts from multiple adult tissues and embryonic stages; and 3) an improvement in the annotation thanks to the use of new reference databases.
Even though we obtained a higher percentage of annotation than in previous studies, there is still a substantial proportion of orphan contigs. Our comparisons with the corresponding species genome or with genomes of closely related species suggest that these orphans are unlikely to be sequencing or assembly errors, as most show a hit to a genomic region. Therefore, there are two explanations for the lack of annotation of these sequences. First, some of these sequences may represent highly variable regions of transcripts (hence, difficult to annotate with similarity searches), either because they are not translated (UTRs) or because they do not code for functionally constrained protein domains. Second, many of these sequences are likely to be real orphans, that is, taxonomically restricted genes, which emerged recently in time, hence, lack homology to divergent species genes (Khalturin et al. 2009; Tautz and Domazet-Loso 2011). It was long assumed that such orphan genes appear due to duplication events followed by rapid divergence, but a recent study proposed an alternative mechanism of de novo emergence from noncoding DNA regions (Neme and Tautz 2013), favoring the appearance of short genes and open-reading frames. In addition, comparisons among the transcriptomes from different species of snakes in our database confirmed that a substantial proportion of orphans are taxonomically restricted genes in that lineage as 7–10% could be identified through similarity searches among snake transcriptomes. These pools of taxonomically restricted genes are therefore of particular interest because they may be involved in lineage-specific adaptations. However, until high-quality genomic data from more closely related species are available, the identification of taxonomically restricted genes remains a challenge.
Our genome annotation pipeline proved to be a simple and accurate method to identify a large proportion of genuine transcripts. Even though it is not as exhaustive as available genome prediction pipelines, which combine genome sequencing and RNA-Seq data from the same species to identify genes, our database and corresponding pipeline provide a first good draft annotation before full genome sequencing data becomes available. The high overlap (81.3%) between the Chr. picta cDNA set from Ensembl Pre! and our data set proves the efficiency of our method, especially when the annotation of closely related species is available.
We also observed a high variability, among species, in the completeness of the annotation. In the case of genome annotation, the well-conserved transcripts were more easily retrieved than the fast evolving ones. Regarding the transcriptome annotation, the variability in the quality of the results is associated with the specific approach used to generate the raw data: These transcriptomes were obtained from different tissues and using different sequencing and assembly techniques. Comparisons against reference data sets (CEGMA core genes, BUSCOs, and ubiquitously expressed genes) showed that the Pa. guttatus and C. ocellatus transcriptomes are the most complete, whereas the ones of S. punctatus and P. molurus present the lowest annotation and coverage. Moreover, even though we identified numerous transcripts (on average 35,912 per species), they cover a variable proportion (per-species average of 18–54%) of the full-length cDNAs.
Several modifications of our pipeline could yet increase the speed and quality of annotation. We could optimize the selection of reference databases as, for example, there is a great overlap between the Ensembl and UniGene databases, making comparisons with the second one highly redundant. Even more, the reliability and possible function of orphans could be analyzed by identifying open-reading frames and protein motifs/domains using databases, such as InterPro, SMART, Pfam, PANTHER, and PROSITE.
Using our transcriptome annotation, we built very large alignments of concatenated homologous protein sequences (from single-copy gene families) and generated the best- supported phylogeny to date among the five major lineages of Sauropsida. All analyses strongly support the position of turtles as the sister group of Archosauria (Tzika et al. 2011) and the position of the tuatara as the sister group of Squamata (both, long-standing questions).
In conclusion, the Reptilian Transcriptomes Database 2.0 presented here is a useful resource for comparative genomics/transcriptomics, differential expression analyses, as well as molecular ecology, developmental and evolutionary studies involving reptiles. For instance, this database can be used as a reference for RNA-Seq and in situ hybridization studies aiming to identify genes that are differentially expressed in distinct cell types, tissues or phenotypes. Such studies are not possible in the absence of a high-quality annotated transcriptome/genome from either the species of interest or a closely-related one.
As additional data accumulate, we will expand and improve the Reptilian-Transcriptomes.org resource. Additional transcriptomes of other reptilian species will undoubtedly become available. The Order Squamata, which comprises 9,372 species (i.e., 95% of all reptiles), would particularly warrant further sequencing efforts. We will also add new reptilian reference species in our pipeline as the annotation of their genomes becomes available, and we will continue updating the database by 1) annotating and including new transcriptomes and 2) improving the quality and completeness of the ones that are already present in the database. Finally, as sequencing throughput and quality will continue to improve, we eventually aim at building a reptilian “phylome” (Huerta-Cepas et al. 2007) that could be incorporated into MANTiS, a software that allows users to explore and interrogate genome content and associated functional data within a phylogenetic context (Tzika et al. 2008).
Supplementary Material
Supplementary methods, results, figures S1–S5, tables S1–S5, and bibliography are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors are grateful to Adrien Debry and Rosa Afonso for maintenance of the Pa. guttatus and E. macularius colonies. Most of the computations were performed at the Vital-IT (http://www.vital-it.ch) Center for high-performance computing of the SIB (Swiss Institute of Bioinformatics) and the Baobab cluster (https://baobabmaster.unige.ch/iface/) of the University of Geneva (Switzerland). They thank the reviewers for constructive comments, which improved the manuscript. This work was supported by grants from the University of Geneva (Switzerland), the Swiss National Science Foundation (FNSNF, grant 31003A_125060), and the SystemsX.ch initiative (project EpiPhysX).
Literature Cited
- Alfoldi J, et al. 2011. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 477(7366):587–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenhoff AM, Dessimoz C. 2009. Phylogenetic and functional assessment of orthologs inference projects and methods. PLoS Comput Biol. 5(1):e1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Bar-Yaacov D, et al. 2013. The first Chameleon transcriptome: comparative genomic analysis of the OXPHOS system reveals loss of COX8 in Iguanian lizards. Genome Biol Evol. 5(10):1792–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandley MC, et al. 2012. Uterine gene expression in the live-bearing lizard, Chalcides ocellatus, reveals convergence of squamate reptile and mammalian pregnancy mechanisms. Genome Biol Evol. 4(3):394–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brykczynska U, et al. 2013. Contrasted evolution of the vomeronasal receptor repertoires in mammals and squamate reptiles. Genome Biol Evol. 5(2):389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, et al. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. [Google Scholar]
- Castoe TA, et al. 2011. A multi-organ transcriptome resource for the Burmese Python (Python molurus bivittatus). BMC Res Notes. 4:310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoe TA, et al. 2013. The Burmese python genome reveals the molecular basis for extreme adaptation in snakes. Proc Natl Acad Sci U S A. 110(51):20645–20650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiari Y, et al. 2012. Phylogenomic analyses support the position of turtles as the sister group of birds and crocodiles (Archosauria). BMC Biol. 10:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NG, et al. 2012. More than 1000 ultraconserved elements provide evidence that turtles are the sister group of archosaurs. Biol Lett. 8(5):783–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalquen DA, Dessimoz C. 2013. Bidirectional best hits miss many orthologs in duplication-rich clades such as plants and animals. Genome Biol Evol. 5(10):1800–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekblom R, Galindo J. 2011. Applications of next generation sequencing in molecular ecology of non-model organisms. Heredity (Edinb) 107(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, et al. 2014. Ensembl 2014. Nucleic Acids Res. 42(Database issue):D749–D755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, et al. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28(23):3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genome 10K Community of Scientists. 2009. Genome 10K: a proposal to obtain whole-genome sequence for 10,000 vertebrate species. J Hered. 100(6):659–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, et al. 2014. Endogenous hepadnaviruses, bornaviruses and circoviruses in snakes. Proc Biol Sci. 281(1791):20141122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29(7):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, et al. 2006. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics 22(23):2971–2972. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J, et al. 2007. The human phylome. Genome Biol. 8(6):R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- i5K Consortium. 2013. The i5K Initiative: advancing arthropod genomics for knowledge, human health, agriculture, and the environment. J Hered. 104(5):595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey PJ, et al. 2014. Ensembl Genomes 2013: scaling up access to genome-wide data. Nucleic Acids Res. 42(Database issue):D546–D552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalturin K, et al. 2009. More than just orphans: are taxonomically- restricted genes important in evolution? Trends Genet. 25(9):404–413. [DOI] [PubMed] [Google Scholar]
- Kumar S, et al. 2012. Toward 959 nematode genomes. Worm 1(1):42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunstner A, et al. 2010. Comparative genomics based on massive parallel transcriptome sequencing reveals patterns of substitution and selection across 10 bird species. Mol Ecol. 19(Suppl. 1):266–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22(13):1658–1659. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, et al. 2011. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478(7370):476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margres MJ, et al. 2013. The venom-gland transcriptome of the eastern coral snake (Micrurus fulvius) reveals high venom complexity in the intragenomic evolution of venoms. BMC Genomics 14:531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JA, Wang Z. 2011. Next-generation transcriptome assembly. Nat Rev Genet. 12(10):671–682. [DOI] [PubMed] [Google Scholar]
- Miller HC, et al. 2012. De novo sequence assembly and characterisation of a partial transcriptome for an evolutionarily distinct reptile, the tuatara (Sphenodon punctatus). BMC Genomics 13:439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neme R, Tautz D. 2013. Phylogenetic patterns of emergence of new genes support a model of frequent de novo evolution. BMC Genomics 14:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G, et al. 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23(9):1061–1067. [DOI] [PubMed] [Google Scholar]
- Ramskold D, et al. 2009. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol. 5(12):e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G, et al. 2010. De novo assembly and analysis of RNA-seq data. Nat Methods. 7(11):909–912. [DOI] [PubMed] [Google Scholar]
- Schulz MH, et al. 2012. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28(8):1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz TS, et al. 2010. A garter snake transcriptome: pyrosequencing, de novo assembly, and sex-specific differences. BMC Genomics 11:694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer HB, et al. 2013. The western painted turtle genome, a model for the evolution of extreme physiological adaptations in a slowly evolving lineage. Genome Biol. 14(3):R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Higgins DG. 2014. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol Biol. 1079:105–116. [DOI] [PubMed] [Google Scholar]
- Smedley D, et al. 2009. BioMart—biological queries made easy. BMC Genomics 10:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John JA, et al. 2012. Sequencing three crocodilian genomes to illuminate the evolution of archosaurs and amniotes. Genome Biol. 13(1):415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Domazet-Loso T. 2011. The evolutionary origin of orphan genes. Nat Rev Genet. 12(10):692–702. [DOI] [PubMed] [Google Scholar]
- Tzika AC, et al. 2008. MANTIS: a phylogenetic framework for multi-species genome comparisons. Bioinformatics 24(2):151–157. [DOI] [PubMed] [Google Scholar]
- Tzika AC, et al. 2011. Reptilian-transcriptome v1.0, a glimpse in the brain transcriptome of five divergent Sauropsida lineages and the phylogenetic position of turtles. Evodevo 2(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullate-Agote A, et al. Forthcoming 2015. The genome sequence of the corn snake (Pantherophis guttatus), a valuable resource for EvoDevo studies in squamates. Int J Dev Biol. doi: 10.1387/ijdb.150060at. [DOI] [PubMed] [Google Scholar]
- Vonk FJ, et al. 2013. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc Natl Acad Sci U S A. 110(51):20651–20656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan QH, et al. 2013. Genome analysis and signature discovery for diving and sensory properties of the endangered Chinese alligator. Cell Res. 23(9):1091–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, et al. 2013. The draft genomes of soft-shell turtle and green sea turtle yield insights into the development and evolution of the turtle-specific body plan. Nat Genet. 45(6):701–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, et al. 2013. OrthoDB: a hierarchical catalog of animal, fungal and bacterial orthologs. Nucleic Acids Res. 41(D1):D358–D365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhulidov PA, et al. 2004. Simple cDNA normalization using Kamchatka crab duplex-specific nuclease. Nucleic Acids Res. 32(3):e37. [DOI] [PMC free article] [PubMed] [Google Scholar]