Abstract
Venom peptides from predatory organisms are a resource for investigating evolutionary processes such as adaptive radiation or diversification, and exemplify promising targets for biomedical drug development. Terebridae are an understudied lineage of conoidean snails, which also includes cone snails and turrids. Characterization of cone snail venom peptides, conotoxins, has revealed a cocktail of bioactive compounds used to investigate physiological cellular function, predator-prey interactions, and to develop novel therapeutics. However, venom diversity of other conoidean snails remains poorly understood. The present research applies a venomics approach to characterize novel terebrid venom peptides, teretoxins, from the venom gland transcriptomes of Triplostephanus anilis and Terebra subulata. Next-generation sequencing and de novo assembly identified 139 putative teretoxins that were analyzed for the presence of canonical peptide features as identified in conotoxins. To meet the challenges of de novo assembly, multiple approaches for cross validation of findings were performed to achieve reliable assemblies of venom duct transcriptomes and to obtain a robust portrait of Terebridae venom. Phylogenetic methodology was used to identify 14 teretoxin gene superfamilies for the first time, 13 of which are unique to the Terebridae. Additionally, basic local algorithm search tool homology-based searches to venom-related genes and posttranslational modification enzymes identified a convergence of certain venom proteins, such as actinoporin, commonly found in venoms. This research provides novel insights into venom evolution and recruitment in Conoidean predatory marine snails and identifies a plethora of terebrid venom peptides that can be used to investigate fundamental questions pertaining to gene evolution.
Keywords: venomics, venom evolution, Terebridae, teretoxins, transcriptomics, Conoidea
Introduction
Venom is widely spread throughout the animal kingdom, mostly as a foraging adaptation, such as in most predatory mammals, snakes, spiders, scorpions, cephalopods, and gastropods, but also as a defensive mechanism as in some lizards, fishes, echinoderms, and insects (Casewell et al. 2013). Animal venoms are among the most complex biochemical natural secretions known and comprise a mixture of bioactive compounds often referred to as toxins (Norton and Olivera 2006; Vonk et al. 2013; von Reumont et al. 2014). Despite their complexity, there is a high degree of convergence throughout the animal kingdom in the basic molecular structure and targets of venom toxins, which include most major physiological pathways and tissues accessible by blood (Escoubas and King 2009; Casewell et al. 2013). These features make venom an extremely successful evolutionary innovation, whose components are ideal candidates for drug discovery and therapeutic development (Fry and Wüster 2004; Twede et al. 2009; Puillandre and Holford 2010). Despite their great potential as model systems for a diverse array of biological areas, including molecular evolution (Duda and Palumbi 1999, 2000; Vonk et al. 2013), functional convergence (Fry et al. 2009), drug discovery (Escoubas and King 2009; Koh and Kini 2012), or structural biology (Tsetlin 1999; Terlau and Olivera 2004; Dutertre and Lewis 2010), most venomous animals remain understudied. However, in the postgenomic era, the concept of a model system is rapidly evolving and venomous organisms, such as the Terebridae, are an attractive option for investigating gene evolution, particularly in venomics research.
With the decreasing costs and increasing efficiency of next-generation sequencing (NGS) techniques, molecular and functional genomic studies enable venomous taxa, such as the predatory snails of the Conoidea superfamily, to become model organisms in the drug discovery arena (fig. 1). The globally distributed Conoidea, which includes Conidae (∼800 species), Terebridae (∼400 species), and Turridae (∼3,000 species), is one of the most diverse groups of venomous organisms in the marine realm and the enormous variety of conoidean venom peptide toxins greatly outnumber that of snakes, a pharmaceutical industry favorite due to ease of collection and quantity of available venom (Escoubas and King 2009). The Conoidea, divided into 16 families, have been perfecting the art of the hunt for over 50 Myr (Bandyopadhyay et al. 2006; Puillandre et al. 2008; Bouchet et al. 2011). A notable example of cone snail venom characterization is the discovery and development of the analgesic therapeutic ziconotide (Prialt, Jazz Pharmaceuticals) (Miljanich 1997, 2004; Olivera 2000). Given their potential, cone snails and conotoxins have been investigated for several decades, but represent only a fraction of the species richness found in the larger Conoidean superfamily. Characterization of the monophyletic Terebridae, an understudied and very diverse lineage of Conoidea, would identify venom peptides distinct from cone snails that can be used to study molluscan species and venom diversification, as well as to provide new compounds for biomedical drug discovery and development (Holford, Puillandre, Terryn, et al. 2009).
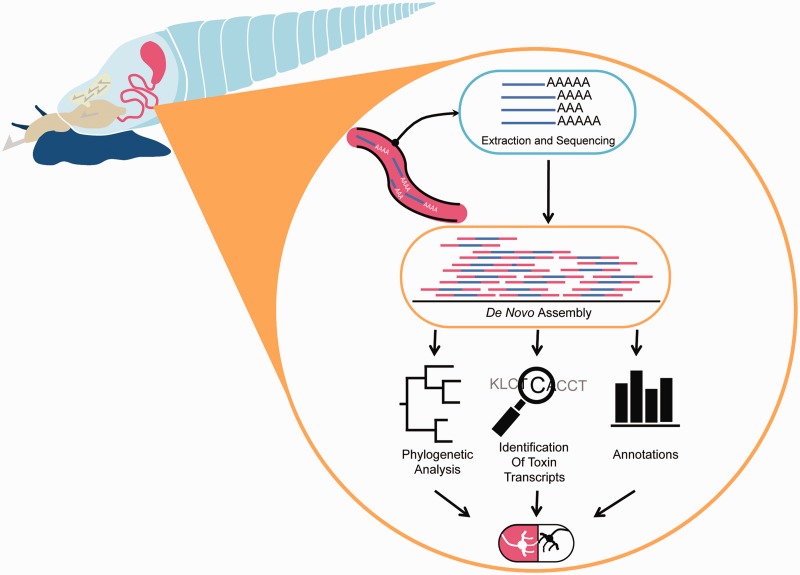
Fig. 1.—
From venom to drugs. Depiction of strategy for identifying venom peptides in conoidean snails using NGS. The venom duct is dissected from conoidean snails and mRNA is extracted. Extracted mRNA is sequenced and de novo assembled. Assembled sequences are then annotated for a variety of comparative analyses such as phylogenetic reconstruction, identification of putative toxins, and assignment of GO function. When combined, comparative analyses can identify lineages that produce bioactive compounds with potential biomedical application for drug development (the pill at the bottom illustrates therapeutic potential).
Two terebrid species, Triplostephanus anilis (Röding, 1798) and Terebra subulata (Linnaeus, 1767), were selected for venom duct transcriptome characterization using NGS (fig. 2). Both species belong to a lineage of venomous terebrids that has been identified as clade C in a recent phylogenetic reconstruction of the Terebridae (Castelin et al. 2012). Terebrids are vermivorous (worm hunting) and certain lineages, similar to cone snails, use a sophisticated venom apparatus to inject a cocktail of peptide toxins to rapidly immobilize their prey. The conoidean venom apparatus includes a convoluted tubular venom gland with a muscular bulb, propulsing the venomous secretion. Conoidea have evolved a peculiar mechanism of using marginal radular teeth for stabbing the prey, and in some groups the latter are modified in hypodermic needles to inject venom into the prey (Taylor et al. 1993; Kantor and Taylor 2000; Holford, Puillandre, Modica, et al. 2009; Holford, Puillandre, Terryn, et al. 2009; Castelin et al. 2012). Not all terebrids have a venom apparatus, and at least three different hunting physiologies are described for this family (Miller 1970). Recent studies have facilitated the identification of terebrid lineages that produce venom by correlating the molecular phylogeny of the Terebridae to the evolution of its venom apparatus (Holford, Puillandre, Modica, et al. 2009; Holford, Puillandre, Terryn, et al. 2009; Castelin et al. 2012). Using this biodiversity derived discovery approach, Tr. anilis and Te. subulata were selected for venom characterization, as they are representatives of a clade that has a similar venom apparatus to that of cone snails and produce venom peptides to subdue their prey.
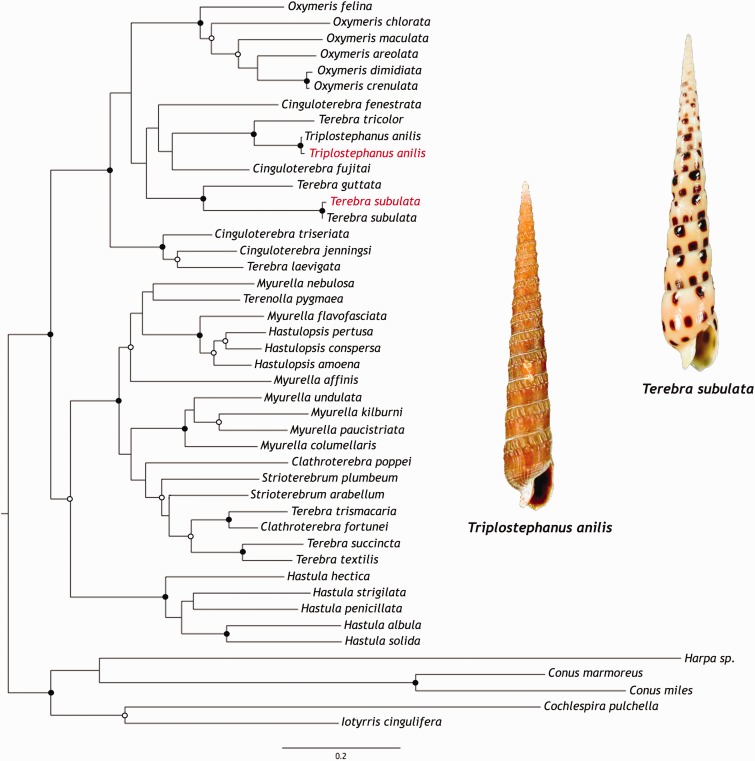
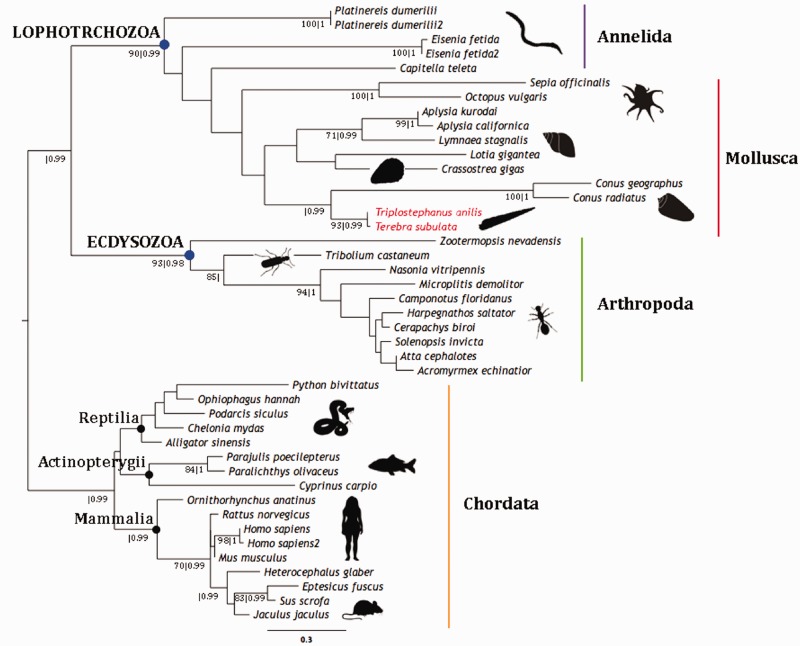
Fig. 2.—
Terebridae phylogeny. Phylogenetic reconstruction of the Terebridae using nuclear gene 18S and mitochondrial genes 12S, 16S, and COI under ML optimality criteria with GTR + G + I and 1,000 pseudoreplicates. Supported nodes indicated by closed circles (bootstrap ≥90) and open circles (bootstrap ≥70). Sequences obtained from our transcriptomes are highlighted in red and shell images of Tr. anilis and Te. subulata are also shown.
This study provides the first, to our knowledge, NGS transcriptome analysis of Terebridae venom ducts to investigate terebrid venom composition. Terebrids express a diverse array of hypervariable disulfide-rich peptide toxins, teretoxins, which come in an assortment of molecular scaffolds that are significantly different from conotoxins (Imperial et al. 2007; Puillandre and Holford 2010; Kendel et al. 2013; Anand et al. 2014). Several novel putative Tr. anilis and Te. subulata teretoxin precursors are identified, and the evolutionary relationships and possible origins of several venom toxin families (e.g., conopressin, actinoporin) with terebrid homologs are examined through phylogenetic methodologies. Additionally, a preliminary classification of teretoxin gene superfamilies is proposed, based mainly on the molecular evolution and cysteine (Cys) framework of venom peptide genes. The putative teretoxins identified enhance the number of proteins convergently recruited in venom and be can be used to investigate the evolution and possible origins of terebrid venom peptides.
Materials and Methods
Sample Collection
The Tr. anilis and Te. subulata specimens used in this study were collected on a 2011 expedition to Inhaca, an island off of the coast of Mozambique, as described in Castelin et al. (2012). Specimens from this expedition were used to obtain venom ducts for NGS projects and to enhance existing phylogenetic reconstructions of the Terebridae. Specimens are dissected to extract venom ducts that are stored in RNAlater and brought back to The American Museum of Natural History for transcriptome research. Muséum National d'Histoire Naturelle (MNHN) Museum voucher numbers and GenBank accession numbers are listed in supplementary table S3, Supplementary Material online.
RNA Extraction and Sequencing
Total RNA was extracted from four pooled Tr. anilis venom ducts using Qiagen RNeasy Micro kit, with DNase digestion on column, according to manufacturer’s instructions. Due to their extremely small size, four Tr. anilis ducts were required to obtain a workable amount of RNA for transcriptome sequencing. A total of 10 ng of Tr. anilis total RNA was used as template for Clontech’s SMARTer Ultra Low RNA Kit for Illumina Sequencing to perform polyA enriched first strand cDNA synthesis and 12 cycles of polymerase chain reaction amplification according to manufacturer’s instructions. Total RNA and the resulting cDNA library were assessed for quality and concentration with Agilent Bionalyzer, using their high sensitivity DNA chip. cDNA was fragmented using a Covaris S2 Sonicator (range 300–600 bp) and libraries were amplified eight cycles. Library quality was analyzed on the Agilent Bioanalyzer using the DNA High Sensitivity Chip.
The cDNA library was sequenced using Illumina HiSeq 2000 technology at the New York University Center for Genomics and Systems Biology. Library construction was performed with the Kapa Biosystems Kit for sequencing on two paths of an eight-lane Illumina flow cell.
Total RNA was extracted from four pooled Te. subulata venom ducts using TRIzol Reagent and PureLink RNA Mini Kit using standard protocols. RNA quality assessment, library preparation, and Illumina sequencing were performed at The genomics resources core facility at Weill Cornell Medical College. Briefly, generation of cDNA from approximately 0.1 ug of total RNA, adenylation, and adapter ligation was performed with Illumina TruSeq RNA Sample Preparation Kit. Throughout library preparation, cDNA was QC validated for downstream sequencing with the Agilent Bioanalyzer. The Te. subulata cDNA library was sequenced using Illumina HiSeq 2500 technology with a multiplexed sample run in a single lane, using paired end clustering and 101 × 2 cycle sequencing.
Read Processing and De Novo Assembly
Triplostephanus anilis, Illumina HiSeq generated 288,959,674 paired end reads of 100 bp length with 89.43% bases having a quality score ≥ Q30. Raw read quality assessment was performed with FastQC to determine the need for base trimming and adapter removal (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/, last accessed June 10, 2015). Seqtk was used to trim bases from the reads using a default Mott algorithm based on phred score (https://github.com/lh3/seqtk, last accessed June 10, 2015) while Trimmomatic (http://www.usadellab.org/cms/?page=trimmomatic, last accessed June 10, 2015) was used to remove adapter contamination. Ultimately the quality control process yielded a total of 280,143,112 reads. For Te. subulata, paired end reads of 100 bp in length were also generated on the Illumina HiSeq platform, producing 176,799,164 raw reads with 92.42% having a quality score of Q30 or better, with no read trimming deemed necessary.
Given the large number of reads and high depth of coverage, digital normalization was applied to the trimmed reads prior to assembly with Trinity (Grabherr et al. 2011; Haas et al. 2013) and Velvet Oases (Zerbino and Birney 2008; Schulz et al. 2012). Digital normalization reduced the size of sequencing data by eliminating highly redundant reads, while retaining the information found in the full data set. Digital normalization also eased the computational burden by reducing the time and memory requirements of de novo assembly, which is advantageous when performing multiple assemblies (Brown et al. 2012).The de novo assembly programs Trinity (release date April 13, 2014) and Velvet Oases (v. 1.2.10 and v. 0.2.08) were run using default parameters to assemble quality trimmed and normalized Tr. anilis reads. Trinity assemblies were run on both Trinity in silico normalized reads, which reduced the number of reads to 9,204,374, and the full read set prior to normalization. For the Velvet Oases assemblies, two-pass digital normalization reduced the data set to 23,607,286 paired end reads and 102,935 single end or “orphaned” reads. The Velvet Oases assemblies were constructed over a range of odd-numbered kmer values from 25 to 55, and assemblies of kmers 25 and 51 were subsequently chosen for analysis. The Te. subulata de novo assembly was performed only with the Trinity program, on the full set of 176, 799 164 paired end 100 bp Illumina reads.
Basic Local Algorithm Search Tool Annotation
Annotation of the Tr. anilis venom duct transcriptome for putative teretoxins was performed by running BLASTx for each of the four assemblies against a combined database of conotoxins downloaded from Conoserver (www.conoserver.org) and our in house database of teretoxin sequences. The same BLASTx search was performed for the Te. subulata Trinity assembly. After basic local algorithm search tool (BLAST) identification of putative toxins at a value of 1e-3 or lower, teretoxin contigs were subjected to open reading frame (ORF) finding using stand-alone getorf from EMBOSS (http://emboss.sourceforge.net/apps/cvs/emboss/apps/getorf.html). ORFs generated from sequences between two stop codons were translated with the standard genetic code. These translated ORFs were then validated based on the identification of a signal sequence, proregion (when present), and Cys frameworks. All alignments of putative teretoxins to their closest BLAST hit were performed with MUSCLE (Edgar 2004) or ClustalW (Thompson et al. 2002). All the RNA-Seq sequence reads used were submitted to NCBI SRA with BioProject ID 286256.
The Tr. anilis Trinity DN assembly was used for a more general BLAST search against the 1) NCBI nonredundant (nr) protein database, 2) UniProtKB/Swiss-Prot (release 2014_03), and 3) UniProtKB/TrEMBL (release 2014_03). For this assembly, assignment of gene ontology (GO) terms was performed in two parts: phase one of annotation followed a bioinformatics pipeline (SFG) made freely available online by the Palumbi lab (http://sfg.stanford.edu/guide.html; De Wit et al. 2012). After blasting against all three databases, a python script was run on significant BLAST hits in combination with a download of Uniprot flatfiles to extract gene names, general descriptions, and GO categories, which are then combined in a master annotation metatable. Running this pipeline generated 18,390 annotated contigs with an e-value of 1e-6 or lower. The second phase of GO analysis incorporated the use of BLAST2GO (B2G) tools on the SFG results for further mapping, annotation and in particular, the use of GoSlim, which employs a reduced set of GO terms to provide a broad overview of the GO content which were used to identify posttranslational modification enzymes.
The SFG annotation of the Tr. anilis Trinity DN assembly was mined for terms that could potentially relate to posttranslational modifications and Tr. anilis transcripts were extracted from these results. Putative Tr. anilis posttranslational enzymes were then blasted against the Te. subulata assembly to determine if similar transcripts were present in both species. BLAST hits with extremely low e-values (1e-30 or lower) and high query coverage were then aligned with either MUSCLE (Edgar 2004) or ClustalW (Thompson et al. 2002) to compare alignments and verify the integrity of these hits.
Evolutionary Analyses
Phylogeny of the Terebridae
Terebrid sequences of nuclear gene 28S and three mitochondrial genes 16S, 12S, and COI were downloaded from GenBank, concatenated and aligned with MUSCLE (Edgar 2004). The model general time reversible (GTR) with gamma-distributed rate across sites and a proportion of the sites invariable (GTR + G + I) was selected as the best model of sequence evolution for each partition, implementing the AIC with ModelTest 3.7 (Posada and Crandall 1998). The combined molecular data set was analyzed in RAxML 7.4.2 (Stamatakis 2006) using the maximum likelihood (ML) optimality criterion, with GTR + G + I, and a partition scheme allowing individual optimization of parameters for all four genes. Support values were estimated with a rapid bootstrap algorithm with 1,000 pseudoreplicates. Cochlespira pulchella, Iotyrris cingulifera, Conus marmoreus, Conus miles, and Harpa sp. were designated as outgroups.
Phylogenetic Reconstruction of Putative Venom Toxin Homologs
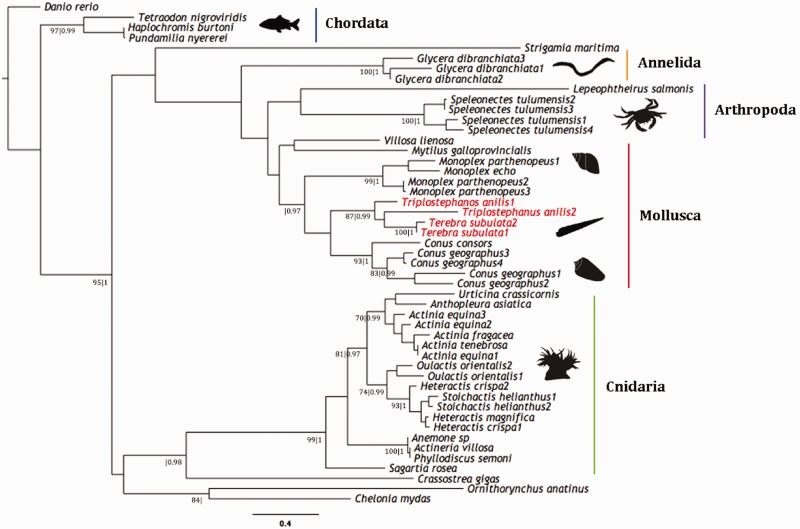
Terebrid transcripts that exhibited homology to toxin families (conopressin/conophysin and actinoporin) and putative orthologs downloaded from GenBank and provided by von Reumont et al. (2014) from selected taxa spanning the Metazoa were aligned with MUSCLE (Edgar 2004). The best fitting model of protein evolution was selected using ProtTest 2.4 (Abascal et al. 2005) following the Akaike Information Criterion (AIC) and used for phylogenetic tree reconstruction under two optimality criteria: ML and Bayesian inference (BI). ProtTest 2.4 identified Whelan and Goldman + I + G as the best model of protein evolution for the actinoporin data set and Jones, Taylor, and Thorton (JTT) + I + G for the conopressin/conophysin data set.
ML analyses were performed with RAxML 7.4.2 (Stamatakis 2006) on both data sets and support values estimated with a rapid bootstrap algorithm with 1,000 pseudoreplicates. BI analyses were also performed on both data sets with MrBayes 3.1.2 (Ronquist et al. 2012). Four Markov chains of 10,000,000 generations each were started from a random tree and run simultaneously, with a sampling frequency of one tree every 100 generations (samplefreq = 100). The consensus trees were calculated after discarding the initial 25% of trees as burnin (burninfrac = 0.25). Different chordate taxa were selected as outgroups for both trees. The topologies of the trees generated for each data set under the two optimality criteria are congruent and thus, for convenience, only the ML tree is shown with both posterior probabilities and bootstrap values indicated above each supported node. GenBank accession numbers provided in supplementary table S3, Supplementary Material online.
Identification of Teretoxin Superfamilies
Like their cone snail counterparts, teretoxin sequences are highly divergent in the mature peptide region but have a conserved signal region. Following conopeptide superfamily classification procedures (Puillandre et al. 2012), only the teretoxin conserved signal sequences were analyzed with phylogenetic methodologies to define new superfamilies. SignalP (Petersen et al. 2011) was used to identify signal sequences of all putative teretoxins recovered from the venom gland transcriptomes of Tr. anilis and Te. subulata and aligned with previously known teretoxins (Imperial et al. 2003, 2007; Kendel et al. 2013; Anand et al. 2014). Although signal sequences are somewhat more conserved, teretoxins are highly divergent, making homology hypothesis doubtful and alignments difficult. Consequently, to make the inference more robust, signal sequences were aligned with two different algorithms: MUSCLE (Edgar 2004) and ClustalW (Thompson et al. 2002). The best fitting model of protein evolution was selected using ProtTest 2.4 (Abascal et al. 2005) following the AIC and used for phylogenetic tree reconstruction under ML and BI optimality criteria. ProtTest 2.4 identified JTT + G as the best model of protein evolution for both data sets.
ML analyses were conducted on both data sets with RAxML 7.4.2 (Stamatakis 2006) and support values estimated with a rapid bootstrap algorithm with 1,000 pseudoreplicates. Additionally, a BI analysis was performed with MrBayes 3.1.2 (Ronquist et al. 2012) in the data set aligned with MUSCLE (Edgar 2004). Four Markov chains of 100,000,000 generations each were started from a random tree and run simultaneously, with a sampling frequency of one tree every 100 generations (samplefreq = 100). The consensus tree was calculated after discarding the initial 25% of trees as burnin (burninfrac = 0.25). To our knowledge, no gene has been identified as an appropriate outgroup for teretoxins and thus, no outgroup was included in any of the analyses.
As only a few teretoxins are known (the vast majority identified in this study) and no superfamilies have been previously defined, we relied on the more extensive knowledge about comparable cone snail toxin superfamilies (Puillandre et al. 2012) and follow similar criteria to define teretoxin superfamilies. The three resulting phylogenetic trees were analyzed and sequence identity levels of the supported clades (bootstrap values ≥ 70 and posterior probabilities ≥ 90) found in two out of three trees were determined with an in house Perl script.
Results and Discussion
Sequencing and De Novo Assembly of Tr. anilis and Te. subulata Venom Duct Transcriptomes
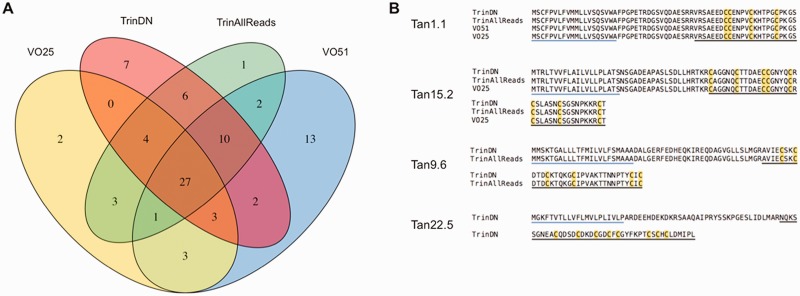
A total of four Tr. anilis assemblies, including Trinity assemblies and Velvet Oases assemblies run at kmers 25 and 51, were used for the identification of novel neuropeptides. The four assemblies were: TrinDN (Trinity assembly from digitally normalized reads), TrinAllReads (Trinity assembly run on the full set of reads), VO25 (Velvet Oases assembly run at kmer = 25), and VO51 (Velvet Oases assembly run at kmer = 51) (fig. 3A). Four assemblies were used to maximize the capture of putative teretoxins, for comparative analyses across assemblies to support the de novo assembly process, and to determine which assembly process should be used in future pipelines. All assemblies identified putative teretoxins, however certain peptides were present only under certain assembly conditions, such as Tan22.5 (fig. 3B). Twenty-seven teretoxins were identified by all four assemblies of Tr. anilis, indicating these were the most valid T. anilis teretoxin transcripts. When directly comparing VO51 and Trinity de novo assembly statistics it would appear as if VO51 was the most effective assembly (supplementary table S1, Supplementary Material online). However, several recent studies have analyzed de novo sequencing assemblers and the validity of measures, such as median contig length, length of contigs, and N50, and found these statistics can be misleading. While generally assembly statistics can identify the continuity of contigs, they cannot attest to contig validity (Kumar and Blaxter 2010; Mundry et al. 2012; Salzberg et al. 2012; Clarke et al. 2013; Lu et al. 2013; O’Neil and Emrich 2013). Conversely, Trinity, which was designed to capture transcript isoforms, has been identified as the best assembler under reference-free conditions, such as with nonmodel systems like the Terebridae (Li et al. 2014). Additionally, when comparing the number of putative teretoxins identified, VO51 and Trinity assemblers found relatively the same number of contigs, 61 for VO51 versus 59 for TrinDN (Fig. 3A). As it is not practical to perform four assemblies for all transcriptomes of interest, the digitally normalized Trinity assembly was chosen as the primary tool for downstream analysis of Tr. anilis and was subsequently chosen as the primary tool for assembly and downstream analysis of Te. subulata. Having well validated high-quality sequence data is an essential first step to studying any transcriptome of interest. As there is no solved terebrid genome, de novo assembly is required. A significant effort was made to utilize multiple assembly programs and bioinformatics approaches for cross validation of findings and to achieve reliable assemblies of Tr. anilis and Te. subulata venom duct transcriptomes to obtain a reliable portrait of Terebridae venom.
Fig. 3.—
Venn diagram showing overlap of putative Tr. anilis teretoxins found in the four different assemblies. (A) Comparative analysis of the four assemblies conducted on Tr. anilis transcriptome sequences: VO25, Velvet Oases assembly with kmer25; TrinDN, Trinity digital normalization assembly; TrinAllReads, Trinity assembly without digital normalization; VO51, Velvet Oases assembly with kmer51. (B) Representative alignments of teretoxin transcripts found in all four, three, two, and only one assembly, respectively. Levels of support range from all four assemblies to single assembly (Tan, Tr. anilis).
Identification and Characterization of Putative Teretoxins Using BLAST
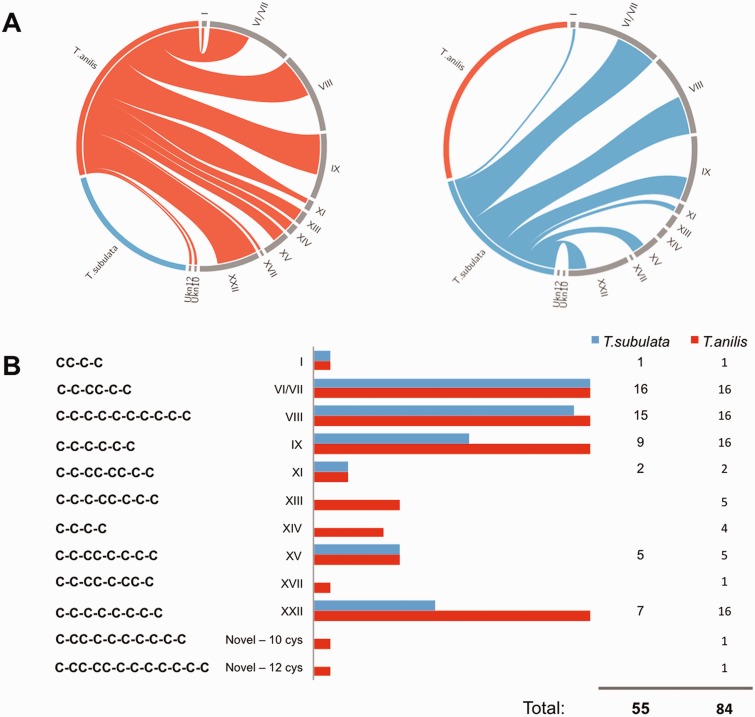
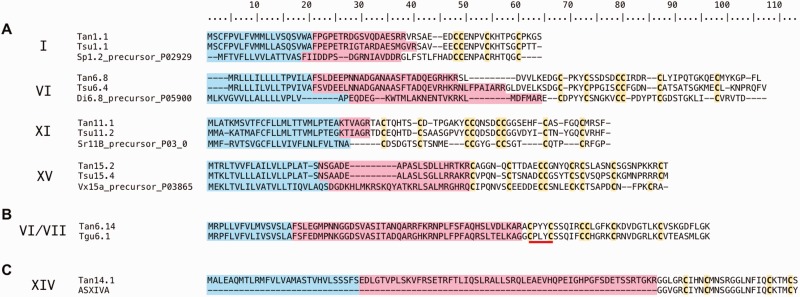
BLAST searches of the venom duct transcriptomes against an in-house local database of cono- and teretoxins created using the Conoserver database and teretoxins identified in the Holford lab yielded 84 putative teretoxins for Tr. anilis, and 55 putative teretoxins for Te. subulata (fig. 4). It should be noted that discrepancies in teretoxin diversity between the Tr. anilis and Te. subulata might be due to the different depth of sequencing of both trasncriptomes. The transcripts were analyzed for the presence of canonical peptide features as identified in conotoxins, namely the presence of an N-terminal signal sequence, an intervening propeptide region ending with several basic residues identifying a cleavage site, and a C-terminal Cys-rich mature peptide (Terlau and Olivera 2004). Of the 139 total putative teretoxin transcripts identified, 105 contain the full signal-pro-mature toxin canonical structure, and 34 have signal-mature sequence without a proregion, which also occasionally occurs in cone snails. All 139 putative teretoxins were organized by Cys framework, that is, Cys pattern of the mature peptide, using the same Roman numeral nomenclature applied to conotoxins (Akondi et al. 2014; fig. 4). Teretoxins found in Tr. anilis and Te. subualta displayed a wide array of Cys frameworks (fig. 4B). This result is promising as it suggests that, similar to cone snails, each terebrid species can produce a unique cocktail of peptides in its venom arsenal (Norton and Olivera 2006; Kaas et al. 2010; Dutertre et al. 2012). Among the 84 Tr. anilis putative teretoxins identified, ten Cys frameworks were previously known from conotoxins, but two are novel frameworks of 10 and 12 Cys respectively, Tan_10Cys, with Cys framework C-CC-C-C-C-C-C-C-C, and Tan_12Cys, with Cys framework C-CC-CC-C-C-C-C-C-C-C (fig. 4B). The diversity of Cys frameworks found suggests that teretoxins may have a wide array of pharmacological targets. Additionally, novel frameworks Tan_10Cys and Tan_12Cys suggest these teretoxins are distinct from conotoxins in structure and function.
Fig. 4.—
Putative teretoxins identified from Tr. anilis and Te. subulata transcriptomes. (A) Circos plot showing the distribution of cysteine frameworks of Tr. anilis and Te. subulata putative teretoxins. The teretoxins identified in Tr. anilis and Te. subulata are not same. Tr. anilis teretoxins are shown in blue and Te. subulata's are shown in red. The diversity of Cys frameworks indicates the two species have different venom arsenals. (B) The Cys frameworks identified among Tr. anilis (red) and Te. subulata (blue) putative teretoxins. Each Cys framework is listed and the number of teretoxins found for each species is identified. Fifty-five Te. subulata and 84 Tr. anilis putative teretoxins were identified in total. Of the 12 Cys frameworks identified, two are novel frameworks found in Tr. anilis venom.
The complex array of Cys patterns found in conoidean venom peptides varies in terms of arrangement and number (Akondi et al. 2014). Conotoxins are classified into gene superfamilies by their conserved signal sequence, which is usually associated with a characteristic Cys framework. Each Cys framework is in turn associated with a different pharmacological activity (Norton and Olivera 2006; Robinson and Norton 2014). A similar approach has been applied here to describe the putative teretoxins identified in Tr. anilis and Te. subulata venom duct transcriptomes.
The most prevalent teretoxin transcripts identified in Tr. anilis and Te. subulata assembled sequences fall into four different Cys frameworks, VI/VII, VIII, IX, and XXII, of varying number and pattern (fig. 4B). Even though the number of Cys frameworks varies between Tr. anilis and Te. subulata, comparative analysis of the types of Cys frameworks highlight strong similarities between the two venoms. Representative alignments of teretoxin transcripts of selected Cys frameworks found in Tr. anilis and Te. subulata transcriptomes along with their closest BLAST hit are illustrated in figure 5. For the alignments shown, Cys frameworks are conserved between teretoxins and conotoxins; however, the intervening residues are largely variable, again supporting the claim that teretoxins may have molecular functions distinct from conotoxins (fig. 5A).
Fig. 5.—
Representative BLAST alignments of T. anilis and Te. subulata teretoxin transcripts. Blue indicates the signal sequence, pink the proregion, and yellow the cysteines. Roman numerals on the left indicate the corresponding cysteine framework assigned to each transcript. (A) Example alignments displaying homology of teretoxins to the top conotoxin analog BLAST hit. There is conservation of Cys patterns in mature peptide toxins between teretoxins and conotoxins. (B) PXY (Pro-X-Tyr), where X can be any intervening residue is a conserved amino acid motif found in a majority of VI/VII teretoxin transcripts. PXY motif underlined in red. (C) Tan14.1 is the only putative teretoxin identified displaying a high degree of sequence identity to a conotoxin (asXIVa).
The VI/VII framework of the O superfamily (C-C-CC-C-C) in cone snails is the most heavily represented with 16 unique sequences in Tr. anilis and 16 unique sequences in Te. subulata (fig. 4B). O superfamily conopeptides, which are active on sodium (Na+), potassium (K+), and calcium (Ca2+) channels, are also a predominant gene family in cone snail venom (Hu et al. 2011). O superfamily peptides produce immobilization and neuromuscular block in the prey by inhibiting ion channel flux (Terlau and Olivera 2004). An intriguing feature of the VI/VII framework putative teretoxins is the repeated appearance of a conserved amino acid motif between the first and second Cys residues, best defined as PXY (Pro-X-Tyr), where X can be any intervening residue. Interestingly, the PXY motif is not prevalent in conotoxins. Only two (P02847 and P04279) of 684 conotoxins with framework VI/VII published on ConoServer present the PXY motif. This stark difference between teretoxins and conotoxins is highly indicative of the functional diversity between these two venom arsenals. An example of PXY motif in teretoxins is illustrated in putative Tr. anilis teretoxin Tan.6.14, which is homologous to a teretoxin (Tgu6.1) previously identified from Terebra guttata (Holford M, unpublished data) (fig. 5B). Similar to the ICK (Inhibitor Cysteine Knot) motif in conopeptides, the PXY motif may determine the structural and functional selectivity of teretoxins to their molecular target. Proline residues are known to affect the secondary structure of proteins (Levitt 1981). Unlike other amino acids, proline is a secondary amine whose cyclic side chain provides rigidity to peptide chains that is usually represented by a “bend” in structure. Tyrosine residues have a reactive phenol (–OH) that can act as an acceptor of phosphate groups. Phosphorylation of phenol groups via receptor tyrosine kinases is a key feature of signal transduction processes (Ullrich and Schlessinger 1990). Proline and Tyrosine together in the PXY motif impart significant conformational and functional properties that may be important to teretoxin peptide function. Although framework VI/VII conotoxins often target Na+, K+, and Ca2+ channels (Lewis et al. 2012), it remains to be seen if a similar pattern will emerge for teretoxins with a PXY motif.
The other heavily represented Cys frameworks in Tr. anilis and Te. subulata transcriptomes include: 16 precursor teretoxin transcripts from T. anilis and 9 from Te. subulata of the six-Cys framework IX peptides (C-C-C-C-C-C), 16 precursor teretoxin transcripts from T. anilis and 7 from Te. subulata of the eight-Cys framework XXII (C-C-C-C-C-C-C-C), and 16 precursor teretoxin transcripts from Tr. anilis and 15 from Te. subulata of the ten-Cys framework VIII (C-C-C-C-C-C-C-C-C-C) (fig. 4B). The eight-Cys framework XXII was only recently observed in conotoxins, but is quite prevalent in T. anilis transcripts and relatively abundant in Te. subulata transcripts, a finding that further supports while there may be commonalities between cone snail and terebrid venoms, the specific cocktail and complexity found in these groups are extremely variable. Frameworks with five or fewer Tr. anilis and Te. subulata teretoxin transcripts, include the eight-Cys frameworks XI (C-C-CC-CC-C-C), XIII (C-C-C-CC-C-C-C) (only in Tr. anilis) and XV (C-C-CC-C-C-C-C), the four-Cys framework XIV (C-C-C-C) (only in Tr. anilis), and a single transcript in both Tr. anilis and Te. subulata for the four-Cys framework I (CC-C-C) (fig. 4B). The low representation of framework I peptides is somewhat surprising, as these peptides are ubiquitous in cone snails, and are known to frequently target nicotinic acetylcholine receptors (nAChRs; Lebbe et al. 2014). In contrast, similar to conotoxins, relatively few teretoxin transcripts are reported for frameworks XI, XIII, and XV all of which contain eight Cys in different conserved patterns (Kaas et al. 2010).
Framework XI (I superfamily), composed of three subfamilies, is one of the best characterized conotoxin families in terms of pharmacology, with some I conotoxins functioning as Na+ channel agonists and K+ channel modulators (Kauferstein et al. 2004). The representative framework XI sequence shown, Tan.11.1, has its closest BLAST hit to a teretoxin identified from Te. guttata, and as such differs significantly in sequence from its conotoxin counterparts (fig. 5B). Framework XIII conotoxins are extremely rare and have only recently been assigned to a novel G superfamily (Aguilar et al. 2013). Five framework XIII transcripts were identified in Tr. anilis transcriptome only. Several framework XIV transcripts are present in the Tr. anilis venom duct, but not in the Te. subulata transcriptome. The T. anilis Tan.14.1 transcript in particular, displays a high degree of sequence identity in the mature peptide region to asXIVa conotoxin (Zugasti-Cruz et al. 2008) (fig. 5C). This is the only example from our assembly of a putative teretoxin transcript displaying a significant degree of sequence identity to a conotoxin. The asXIVa sequence does not have a precursor region as it was identified from venom fractionation; however, the mature peptide almost completely aligns with Tan.14.1. Framework XIV conotoxins are a complex assortment of peptides classified into multiple superfamilies that include A, I2, J, L, M, O1, and O2 (Kaas et al. 2012). Several framework XIV conotoxins have been shown to elicit nAChR and K+ channel inhibition (Imperial et al. 2006; Peng et al. 2006).
It should be noted that even when a potential teretoxin with a well-established conotoxin framework is identified in this study, the overall amino acid composition of the mature peptides is, in most cases, radically different from conotoxins in terms of identity, number, and distribution of intra-Cys residues, Tan14.1 being the exception (fig. 5). As such, it remains to be seen whether teretoxins with the same conserved Cys frameworks found in cone snails will have similar physiological functions. Given their significant differences from conotoxins, teretoxins may have novel molecular targets, thus identifying new mechanisms to modulate cellular function in the discovery and development of biomedical therapeutic agents.
Beyond BLAST Identification of Putative Teretoxins
As terebrids and their peptide toxins are in the preliminary stages of investigation and currently have very low representation in public databases, it is important to have alternatives to BLAST as a means to identify putative peptide toxin transcripts. To this end, an in house software program, termed Pepticomb, was developed and used to analyze the Tr. anilis and Te. subulata transcriptome assemblies. Pepticomb is largely based on source code kindly provided by the developers of the conoprec tool found on the Conoserver website (www.conoserver.org; Kaas et al. 2008, 2012). In stepwise fashion, Pepticomb examines a set of contigs for the presence of a signal sequence, looks for a proregion that terminates in a basic residue cleavage site, and uses regular expressions to mine for permutations of Cys frameworks in the mature peptide with a limited number of intra-Cys residues set by the user. Running this program on the Tr. anilis and Te. subulata assemblies successfully identified all the putative teretoxins already identified via BLAST, as well as additional candidate teretoxins not identified by BLAST (supplementary table S2, Supplementary Material online). These additional candidates include peptides of frameworks I, IX, VI/VII, VIII, XIII, XIV, and XXII, which are Cys frameworks already present in the putative teretoxins identified via BLAST. Pepticomb results also included candidate teretoxins with unknown frameworks (supplementary table S2, Supplementary Material online). Two unknown frameworks with 10- and 12-Cys were found in Tr. anilis, whereas three 8-Cys, three 10-Cys, five 12-Cys, and four 14-Cys unknown frameworks were found in Te. subulata. Interestingly, one of the ten-Cys unknown frameworks (C-CC-C-C-C-C-C-C-C) was found in putative toxins from both Tr. anilis and Te. subulata. The surprising number of novel frameworks for Te. subulata transcriptomes compared with BLAST results suggests there are many more teretoxin Cys frameworks to be discovered.
Given the inherent potential for misassembly or chimeric transcripts in de novo assembly, it is difficult without strong BLAST support to present the additional putative teretoxins identified via Pepticomb as high confidence transcripts. As such, these results are not included in the final count of 139 putative teretoxins found in Tr. anilis and Te. subulata transcriptomes identified via BLAST homology, and are pending further verification by proteomic analysis and other methodologies. Nonetheless, Pepticomb is an important tool for exploring the frontiers not captured by BLAST, and is extremely useful for organizing and validating putative toxin transcripts that are generated from BLAST results. These results were included to demonstrate that there is a significant amount of transcripts not identified by BLAST when working with nonmodel systems. In such instances, alternatives to BLAST techniques, such as Pepticomb and the use of the profile Hidden Markov Model to aid in the identification and classification of terebrid gene superfamilies may yield more robust results, as was recently demonstrated for conotoxins (Laht et al. 2012; Robinson et al. 2014).
Similar to other conoidean venom peptides, the abundance of teretoxins found in our Tr. anilis and Te. subulata transcriptomes is believed to stem in part from high rates of gene duplication, while the hypervariable mature peptide region likely results from strong diversifying selection at specific loci, leading to significant allelic variation (Duda and Palumbi 2000; Chang and Duda 2012). As more terebrid transcriptomes are reconstructed it will be possible to perform comparative evolutionary analyses to identify the drivers of diversification in this family.
Identification of Teretoxin Gene Superfamilies
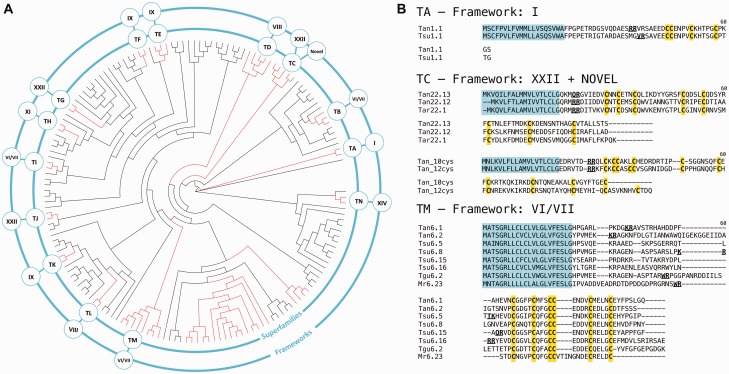
The high sequence similarity of conotoxin precursor regions has led to the grouping of conotoxins into gene superfamilies based on their consensus signal sequence which can provide clues to the evolutionary relationships of conopeptides (Kaas et al. 2010; Robinson and Norton 2014). Using a similar mechanism, we have identified the first, to our knowledge, teretoxin gene superfamilies for the Terebridae (fig. 6).
Fig. 6.—
Teretoxin gene superfamilies. (A) Circular phylogeny of terebrid gene superfamilies (midpoint root) obtained from MUSCLE alignment of teretoxin signal sequences under ML criteria with JTT + G and 1,000 pseudoreplicates. Clades highlighted in red have strong support values but only those that meet additional criteria (see details in text) are designated as teretoxin superfamilies. Inner ring shows superfamily name and outer ring Cys framework of the superfamily. (B) Sequence alignment of representative teretoxin superfamilies and their associated frameworks. Note that superfamily TC includes teretoxins with Cys framework XXII (top) and two novel frameworks (bottom). Also, superfamily TM shares significant sequence identity with the H superfamily of conotoxins (included as an example Mr6.23 from C. marmoreus) Tan, Tr. anilis; Tsu, Te. subulata; Tar, Te. argosyia; Tgu, Te. guttata.
In order to establish a classification that is phylogenetically relevant and reflects the evolution of teretoxins, we performed a phylogenetic analysis including the signal sequences of all putative teretoxins identified here and previously published (Imperial et al. 2007; Kendel et al. 2013). Criteria similar to those proposed by Puillandre et al. (2012) for conotoxins were used to define teretoxin superfamilies. Namely: 1) A new teretoxin superfamily should represent an independent lineage, not be nested within another clade, and have strong support values. 2) Sequence identity within the potential superfamily should be at least 60%. And 3) The Cys pattern should be different from the one found in the sister clade. Following this scheme, we have identified 14 new teretoxin gene superfamilies (fig. 6). The putative teretoxin superfamilies are named using a two-letter nomenclature system, in which the first letter is always a T (to distinguish teretoxin from conotoxin superfamilies) and a second letter is designated in alphabetical order, starting with A. Consequently, the first teretoxin superfamily identified here has been named TA superfamily, and this scheme was followed to assign names to all 14 teretoxins superfamilies, T[A-N], identified in our data sets. Most teretoxin gene superfamilies identified include representatives from different terebrid species, suggesting that the superfamilies are credible and will be further validated as additional terebrid transcriptomes become available.
Each superfamily, by definition, is characterized by a conserved signal sequence, and generally by a unique conserved Cys framework (fig. 6B). Thus, superfamily TA is characterized by Cys framework I; superfamilies TB, TI, and TM include teretoxins with framework VI/VII; framework VIII is found in superfamilies TD and TL; framework IX characterizes superfamilies TE, TF, and TK; superfamily TH presents framework XI; superfamily TN is characterized by framework XIV; framework XXII is present in superfamilies TC, TG, and TJ; and two novel 10- and 12-Cys frameworks are also found in superfamily TC. Therefore, TC superfamily represents an exception to the superfamily unique Cys framework rule, because it includes teretoxins characterized by framework XXII and also two novel 10- and 12-Cys frameworks (fig. 6B). As recently reported for conotoxins, it is at times possible for a superfamily with a conserved signal sequence to be assigned to more than one Cys framework due to gene duplication and other gene expression mechanisms (Robinson and Norton 2014).
All teretoxin signal sequences were compared with the consensus sequences of conotoxin superfamilies to identify potential similarities that might provide evidence of a common origin. Analysis of teretoxin signal peptides largely displayed low sequence identity with cone snails signal peptides, with the exception of TM superfamily transcripts with signal sequence MATSGRLLCLCLVLGLVF and six-Cys framework C-C-CC-C-C. This conserved signal sequence has strong identity (>80%) with the recently described conotoxin H gene superfamily found in C. marmoreus and Conus victoriae (Dutertre et al. 2012; Robinson et al. 2014) (fig. 6B). However, the general lack of similarity between teretoxins signal sequences and the consensus sequences of the 26 conotoxin superfamilies described to date, supports prior hypotheses regarding the potential to identify undescribed venom peptide toxin superfamilies and functions in the Terebridae (Puillandre and Holford 2010).
Identification of Terebrid Venom Proteins with Posttranslational Functions
Posttranslational modifications are prominent in conotoxins (Craig et al. 1999; Buczek et al. 2005; Wang et al. 2007; Safavi-Hemami et al. 2010); however preliminary research, based on limited molecular and proteomic data, suggested that teretoxins were not posttranslationally modified (Imperial et al. 2003; Imperial et al. 2007). In the interest of gaining an overview of posttranslational enzymes present in the terebrid venom arsenal, GO analyses were performed on Tr. anilis and Te. subulata assembled transcriptomes. Annotation of Tr. anilis and Te. subulata transcriptomes for the presence of posttranslational enzymes yielded a number of candidate proteins similar to those found in conotoxins (fig. 7 and supplementary fig. S1, Supplementary Material online). These proteins are described below.
Fig. 7.—
Posttranslational modification enzymes found in Terebrid venom-duct transcriptomes. (A) Posttranslational modification enzymes found in Tr. anilis and Te. subulata transcriptomes, with respective amino acid length and accession number. (B) Representative BLAST alignment to tyrosyl sulfotransferase from Littorina sitkana, which is responsible for sulfation of tyrosine in conotoxins.
γ-Glutamyl Carboxylase
Vitamin K dependent γ-glutamyl carboxylase is a posttranslational enzyme found in the cone snail venom duct that catalyzes the addition of a carboxyl group to specific glutamate residues (Czerwiec et al. 2002). Transcripts identified from the Tr. anilis and Te. subulata transcriptomes display a near perfect match to Conus textile vitamin K-dependent γ-glutamyl carboxylase (supplementary fig. S1A, Supplementary Material online). Although carboxylation of glutamate residues is an established posttranslational modification in conotoxins, it should be noted that this carboxylase is highly conserved in a wide variety of organisms, including humans and Drosophila. In mammalian systems, it plays an important regulatory role in the blood clotting cascade through the carboxylation of blood-clotting proteins such as prothrombin (Suttie 1988).
Peptidyl-Glycine α-Amidating Monooxygenase
Another important enzyme of interest in conotoxin posttranslational modification is peptidyl-glycine α-amidating monooxygenase (PAM), which modifies peptides containing a C-terminal glycine residue through cleavage of the glycine followed by amidation of the preceding residue (Ul-Hasan et al. 2013). Annotation of Tr. anilis and Te. subulata assemblies yielded transcripts with homology to PAM A, found in Conus bullatus (Hu et al. 2011) and in the oyster Crassotrea gigas (Zhang et al. 2012; supplementary fig. S1B, Supplementary Material online). C-terminal amidation is a very common posttranslational modification in venom peptides and therefore it is not surprising to find the PAM enzyme in terebrid venom.
Prolyl 4-Hydroxylase
Hydroxylation of conotoxin mature peptide prolines happens in as many as one out of two residues, presumably through the activity of PH4, although this specific enzyme has not been characterized for any cone snail species (Lopez-Vera et al. 2008). Preliminary studies have indicated that hydroxyprolination may be important for both conotoxin bioactivity and oxidative folding (Lopez-Vera et al. 2008). Proline hydroxylation is also widely present in humans as the result of PH4 activity, with diverse protein substrates, however, the best-known role is the stabilization of the collagen triple helix (Gorres and Raines 2010). Several PH4 transcripts are present in Tr. anilis and Te. subulata assemblies, with the most predominant being PH4-1, a catalytic subunit of the larger protein, showing strong sequence identity to the PH4-1 sequence of Cr. gigas (supplementary fig. S1C, Supplementary Material online). Although not characterized for cone snails, the presence of PH4-1 in terebrid venom suggests a homolog of this enzyme may also be found in cone snail venom.
Tyrosyl Sulfotransferase and Glutaminyl-Peptide Cyclotransferase
Other identified Tr. anilis and Te. subulata protein transcripts with potential posttranslational function include tyrosyl sulfotransferase and glutaminyl-peptide cyclotransferase, which are responsible for sulfation of tyrosine and N-terminal cyclization of glutamine to pyroglutamate in conotoxins, respectively (Buczek et al. 2005; Craig et al. 1999; fig. 7B and supplementary fig. S1D, Supplementary Material online). The terebrid tyrosyl sulfotransferase transcripts identified displayed homology to that of the sea snail Littorina sitkana, whereas the terebrid glutaminyl-peptide cyclotransferase identified was most similar to that of Cr. gigas. Homology of these enzymes to other mollusks suggests conserved distribution of these genes in gastropods.
Although the identification of protein transcripts potentially implicated in posttranslational modification of teretoxins is of great interest, it remains to be seen whether these modifications will be identified on the proteomic level. It is possible that the posttranslational enzymes identified are involved in common housekeeping processes and not used to modify teretoxins in the venom duct. The recognition sequences for posttranslational modification enzymes within the conotoxin precursor structure have still to be defined, and it is not yet elucidated how, for example, some prolines are selected for hydroxylation whereas others remain unchanged. It has been proposed that glutamate carboxylation in conotoxins depends on a specific sequence found in the proregion of the peptide, but this is one of the few cases where the process is characterized (Buczek et al. 2005; Wang et al. 2007; Safavi-Hemami et al. 2010). The identification of similar putative posttranslational modification enzymes and folding proteins in terebrids and cone snails indicates that certain venom compounds may be conserved throughout the Conoidea superfamily.
Identification and Evolution of Putative Venom Toxin Homologs
Phylogenetic hypotheses of aligned amino acid sequences were constructed to evaluate orthology predictions and evolutionary relationships of selected venom toxins (conopressin/conophysin and actinoporins). Identification of putative toxin homologs in the Terebridae expands the range of protein types convergently recruited into venom from predatory organisms (Casewell et al. 2013).
Conopressin/Conophysin
Putative Terebridae homologs to conopressin/conophysin peptides are referred to as terepressin/terephysin peptides, respectively. Phylogenetic analysis of these transcripts indicates that the two Terebridae conopressin/conophysin-like transcripts cluster together in a well-supported clade that is sister group to another strongly supported clade, which includes C. geographus and Conus radiatus sequences (fig. 8). Moreover, the two terepressin/terephysin transcripts from Tr. anilis (anilis_comp44965_c5_seq1_1) and Te. subulata (subulata_comp99487_c0_seq1_1), group together with all other mollusk and annelid sequences incorporated in the analysis, in a well-supported Lophotrochozoan clade that is sister to another strongly supported clade including all the Ecdysozoan sequences (fig. 8). The phylogenetic position of the putative terepressin/terephysins indicates that both transcripts are conopressin/conophysin orthologs.
Fig. 8.—
Conopressin/conophysin phylogeny. Phylogenetic reconstruction of conopressin/conophysin using sequences from representatives across the Metazoa and putative terepressin/terephysins from Tr. anilis and Te. subulata. Tree generated under ML optimality criteria with JTT + I + G and 1,000 pseudoreplicates. Bayesian analysis with four Markov chains of 10,000,000 generations each, yielded same topology. Bootstrap values (≥70) and posterior probabilities (≥0.90) are shown below each supported node (bs|pp). Terebridae sequences are highlighted in red and clades representing major phyla are indicated by colored vertical bars and closed circles.
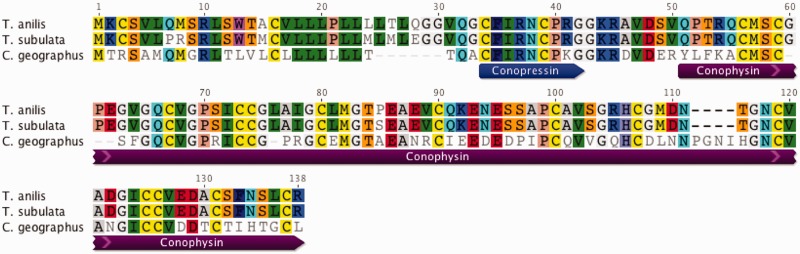
Conopressins, short, nine residue peptides with two Cys, such as CFIRNCPK, were originally discovered in the venoms of C. geographus and Conus striatus, and have been characterized as homologs of vasopressin/oxytocin hormonal neurotransmitters based on their strong sequence similarity (Cruz et al. 1987). Despite the strong similarity to vasopressin, conopressin-T, isolated from the venom of Conus tulipa, has been shown to act as a V1-vasopressin receptor antagonist and a partial oxytocin receptor agonist, generating considerable interest in having conopressins serve as templates for drug design (Dutertre et al. 2008). The identified terepressins (CFIRNCPR), share strong sequence identity with conopressins, possibly sharing similar functions and thus, also represent interesting targets for drug development.
Conophysins, which belong to the neurophysin peptide family, are one of the longest peptides ever identified in cone snail venom, with 14 Cys residues and 7 disulfide bridges. Conophysins were first characterized from the venom of C. radiatus, and their physiological role is currently unknown. Although the terepressin portion of the transcript shows homology to conopressin-G, the signal peptide and conophysin-like sequence, identified here as terephysin, are much more variable compared with the signal and conophysin sequences recently identified in C. geographus (Dutertre et al. 2014). The two Terebridae full precursor transcripts characterized provide evidence for terepressin and terephysin being expressed together, similar to vasopressin/neurophysin homologs from other organisms (fig. 9). Vasopressin/neurophysin hormonal neurotransmitters are emerging as viable targets for novel treatments for mental disorders, such as autism, social anxiety disorder, and schizophrenia (Meyer-Lindenberg et al. 2011). Although it is not clear what role terepressin/terephysin plays in predation, their similarity to vasopressin/neurophysin suggests they may be used as hormonal neurotransmitters to manipulate mental disorders.
Fig. 9.—
Putative terepressin/terephysins. Alignment of conopressin/conophysin-like sequences identified from Tr. anilis and Te. subulata venom duct transcriptomes with conopressin/conophysin sequence from C. geographus. Similar to vasopressin/neurophysin homologs from other organisms, the two Terebridae full precursor transcripts characterized are expressed together as one gene.
Actinoporin
Four actinoporin-like transcripts, referred to as tereporins were also identified from the transcriptomes of Tr. anilis and Te. subulata (subulata_comp82089_c0_seq1, subulata_comp86348_c1_seq1, subulata_comp30976_c0_seq2, subulata_comp30976_c0_seq1; fig. 10). Actinoporins are highly conserved pore-forming cytolytic toxins that lack Cys residues and are ubiquitous within sea anemones (Macek 1992; Anderluh and Maček 2002; García-Ortega et al. 2011; von Reumont et al. 2014), but have also been isolated from the venoms of other organisms such as mollusks (Shiomi et al. 2002), annelids (von Reumont et al. 2014) and chordates (Warren et al. 2008). It has been suggested that they have active roles in predation, defense and digestion and are lethal to mollusks, crustaceans, fish, and small mammals (Giese et al. 1996; García et al. 2009; García-Ortega et al. 2011; von Reumont et al. 2014). The toxic activity of actinoporin involves the formation of pores within biological membranes that result in a colloid-osmotic shock leading to cell death (García-Ortega et al. 2011). A phylogenetic analysis was conducted including actinoporin sequences from representatives across the Metazoa, with a special focus on taxa closely related to the Terebridae, such as members of the Lophotrochozoa (Bouchet et al. 2011; fig. 10). The four Terebridae actinoporin-like transcripts cluster together in a strongly supported clade and are the sister group of another well-supported clade that includes actinoporin-like sequences from C. geographus and C. radiatus. The phylogenetic reconstruction of actinoporin sequences including the new tereporin transcripts supports the orthology predictions for these putative teretoxins.
Fig. 10.—
Actinoporin phylogeny. Phylogenetic reconstruction of actinoporin using sequences from representatives across the Metazoa and actinoporin-like sequences from Tr. anilis and Te. subulata. Tree generated under ML optimality criteria with Whelan and Goldman + I + G and 1,000 pseudoreplicates. Bayesian analysis with four Markov chains of 10,000,000 generations each, yielded same topology. Bootstrap values (≥70) and posterior probabilities (≥0.90) are shown below each supported node (bs|pp). Terebridae sequences are highlighted in red and colored vertical bars indicate clades representing major phyla.
Conclusion
This study applied NGS and state of the art bioinformatics tools to present the first comprehensive analysis of terebrid venom duct transcriptomes, with a particular focus on identification of disulfide rich peptides that can be used as probes for investigating venom evolution in the Terebridae and as potential bioactive compounds to develop novel therapeutics. One hundred and thirty-nine putative teretoxins were identified from Tr. anilis and Te. subulata venoms. Apart from the significant number of potential new teretoxins for investigating cellular processes, comparative analyses of the assembled terebrid venom duct transcriptomes enabled the: 1) Identification of two novel Cys frameworks, Tan_10Cys and Tan_12Cys, with conserved signal sequences, but varying Cys frameworks and intra-Cys amino acid residues, a feature that is also being reported in conotoxins. 2) Identification of a PXY motif, which, similar to the ICK motif in conotoxins, may have structural and function implications on teretoxin bioactivity. 3) Identification of teretoxin gene superfamilies and associated Cys frameworks. 4) Characterization of enzymes that could be linked to posttranslational modification of teretoxins, pending validation by proteomic analyses of terebrid venom duct components. 5) The characterization of convergent evolution of venom proteins conopressin/conophysin and actinoporins. These results lay the foundation for understanding the complex evolutionary relationships among teretoxins and pave the way for further comparative analyses of teretoxins to conoidean venom peptide toxins and to other venomous organisms.
De novo assembly of the transcriptomes of nonmodel organisms presents an ongoing challenge. To alleviate this problem, a working bioinformatics pipeline was established with this work that can be applied to subsequent transcriptome efforts for other species of terebrids. To ensure the quality of the putative teretoxins identified, two different assembly programs, Trinity and Velvet Oases, were run at different parameters, and cross-correlated to validate identified teretoxin transcripts. Cross validation, especially in cases where all four assemblies identified an exact match, provides a high degree of confidence in the integrity of the putative teretoxin transcripts identified. The expansive number of terebrid venom duct transcripts assembled allowed for a high depth of coverage that is reflected not only in the number of teretoxins captured but also in the analysis of other components of the transcriptome, such as identification of toxin-related genes, and of transcripts that can be assigned GO terms to form a global portrait of venom expression and venom convergence. The presence of different transcripts coding for enzymes involved in posttranslational modifications, the same modifications found extensively in conotoxins, raises interesting questions regarding previous research suggesting that these modifications are low or absent in terebrid toxins (Imperial et al. 2003, 2007). All terebrid transcripts identified require validation with other methodologies such as proteomic characterization, which will be part of the future directions of this research.
Analyses of Tr. anilis and Te. subulata venom duct transcriptomes implies that while teretoxins share organizational features with conotoxins, they differ substantially in terms of distribution of Cys frameworks, amino acid composition, and average length. For example, the paucity of Cys framework XI (I superfamily) transcripts, directly contrasts cone snail transcriptomes in which I superfamily conotoxins are largely abundant and characterized as Na+ channel agonists and K+ channel modulators. Such structural differences between teretoxins and conotoxins indicate teretoxins are novel compounds with molecular targets that may be distinct from conotoxins, making terebrid marine snails an attractive resource for investigating cellular physiologies. As a result, customary model organisms such as Drosophila melanogaster, Caenorhabditis elegans, and Mus musculus, while still gold standards, no longer corner the market, and it is possible with ever advancing sequencing technologies to literally scrape the ocean floor for organisms such as the Terebridae, which produce novel genes and gene products that can be used to investigate fundamental questions pertaining to gene evolution and adaptive change.
Supplementary Material
Supplementary figure S1 and tables S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The expedition in Mozambique was carried out at Estação Marítima de Biologia da Inhaca under a Memorandum of Cooperation between Muséum National d'Histoire Naturelle (MNHN, Paris) and the Faculty of Science of University Eduardo Mondlane (Maputo), and the authors thank Prof. Amalia Uamusse for generously hosting our research project. Authors wish to specially thank Philippe Maestrati and José Rosado, and the rest of participants in the Inhaca, Mozambique expedition: Magalie Castelin, Virginie Héros, Pierre Lozouet, Ellen Strong, Alexander Fedosov, Laurent Charles, Emmanuel Vassard, Gabriel Albano, Sergio Mapanga, Arlindo Fernando Machel, Daniela de Abreu, Mizeque Julio Mafambissa, and Mito Nhaca. The authors are also thankful to Mgavi Brathwaite (Program Manager and Academic Adviser, MS in Bioinformatics, NYU Polytechnic School of Engineering) and Ekta Sharma (graduate student, MS in Bioinformatics, NYU Polytechnic School of Engineering) for their help and to Quentin Kaas for sharing conoprec source code. This work was supported by the National Science Foundation (1247550), the Camille and Henry Dreyfus Foundation, National Institutes of Health (MD007599), and PSC-CUNY Enhanced Collaborative Grant (CIRG2064) to M.H. W.G.Q. support provided by The National Institutes of Health - National Institute of Allergy and Infectious Diseases (AI107955). Y.K. funding provided by Russian Foundation for Basic Research grant (14-04-00481) and the MNHN as part of its program of visiting scientists. A.V., J.G., and M.E.W. support provided by The Graduate Center of the City University of New York Science Scholarship. A.V. acknowledges additional funding from The Weissman School of Arts and Sciences of Baruch College, City University of New York. G.R. was supported by an HHMI Undergraduate Science Education Award to Hunter College (52007535).
Literature Cited
- Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105. [DOI] [PubMed] [Google Scholar]
- Aguilar MB, et al. 2013. Precursor De13.1 from Conus delessertii defines the novel G gene superfamily. Peptides 41:17–20. [DOI] [PubMed] [Google Scholar]
- Akondi KB, et al. 2014. Discovery, synthesis, and structure-activity relationships of conotoxins. Chem Rev. 114:5815–5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand P, et al. 2014. Sample limited characterization of a novel disulfide-rich venom peptide toxin from terebrid marine snail Terebra variegata. PLoS One 9:e94122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderluh G, Maček P. 2002. Cytolytic peptide and protein toxins from sea anemones (Anthozoa: Actiniaria). Toxicon 40:111–124. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay PK, Stevenson BJ, Cady MT, Olivera BM, Wolstenholme DR. 2006. Complete mitochondrial DNA sequence of a Conoidean gastropod, Lophiotoma (Xenuroturris) cerithiformis: gene order and gastropod phylogeny. Toxicon 48:29–43. [DOI] [PubMed] [Google Scholar]
- Bouchet P, Kantor YI, Sysoev A, Puillandre N. 2011. A new operational classification of the Conoidea (Gastropoda). J Molluscan Stud. 77:273–308. [Google Scholar]
- Brown CT, Howe AC, Zhang AQ, Pyrkosz AB, Brom TH. 2012 A reference-free algorithm for computational normalization of shotgun sequencing data. arXiv:1203.4802 [q-bio.GN] http://arxiv.org/abs/1203.4802. [Google Scholar]
- Buczek O, Bulaj G, Olivera BM. 2005. Conotoxins and the posttranslational modification of secreted gene products. Cell Mol Life Sci. 62: 3067–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casewell NR, Wüster W, Vonk FJ, Harrison RA, Fry BG. 2013. Complex cocktails: the evolutionary novelty of venoms. Trends Ecol Evol. 28:219–229. [DOI] [PubMed] [Google Scholar]
- Castelin M, et al. 2012. Macroevolution of venom apparatus innovations in auger snails (Gastropoda; Conoidea; Terebridae). Mol Phylogenet Evol. 64:21–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D, Duda TF. 2012. Extensive and continuous duplication facilitates rapid evolution and diversification of gene families. Mol Biol Evol. 29:2019–2029. [DOI] [PubMed] [Google Scholar]
- Clarke K, Yang Y, Marsh R, Xie LL, Zhang KK. 2013. Comparative analysis of de novo transcriptome assembly. Sci China Life Sci. 56:156–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AG, Bandyopadhyay P, Olivera BM. 1999. Post-translationally modified neuropeptides from Conus venoms. Eur J Biochem. 264: 271–275. [DOI] [PubMed] [Google Scholar]
- Cruz LJ, et al. 1987. Invertebrate vasopressin/oxytocin homologs. characterization of peptides from Conus geographus and Conus straitus venoms . J Biol Chem. 262:15821–15824. [PubMed] [Google Scholar]
- Czerwiec E, et al. 2002. Expression and characterization of recombinant vitamin K-dependent gamma-glutamyl carboxylase from an invertebrate, Conus textile. Eur J Biochem. 269:6162–6172. [DOI] [PubMed] [Google Scholar]
- De Wit P, et al. 2012. The simple fool’s guide to population genomics via RNA-Seq: an introduction to high-throughput sequencing data analysis. Mol Ecol Resour. 12:1058–1067. [DOI] [PubMed] [Google Scholar]
- Duda TF, Palumbi SR. 2000. Evolutionary diversification of multigene families: allelic selection of toxins in predatory cone snails. Mol Biol Evol. 17:1286–1293. [DOI] [PubMed] [Google Scholar]
- Duda TF, Jr, Palumbi SR. 1999. Molecular genetics of ecological diversification: duplication and rapid evolution of toxin genes of the venomous gastropod Conus. Proc Natl Acad Sci U S A. 96:6820–6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutertre S, et al. 2008. Conopressin-T from Conus tulipa reveals an antagonist switch in vasopressin-like peptides. J Biol Chem. 283:7100–1708. [DOI] [PubMed] [Google Scholar]
- Dutertre S, et al. 2012. Deep venomics reveals the mechanism for expanded peptide diversity in cone snail venom. Mol Cell Proteomics. 1–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutertre S, et al. 2014. Evolution of separate predation- and defence-evoked venoms in carnivorous cone snails. Nat Commun. 5:3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutertre S, Lewis RJ. 2010. Use of venom peptides to probe ion channel structure and function. J Biol Chem. 285:13315–13320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escoubas P, King GF. 2009. Venomics as a drug discovery platform. Expert Rev Proteomics. 6:221–224. [DOI] [PubMed] [Google Scholar]
- Fry BG, et al. 2009. The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annu Rev Genomics Hum Genet. 10:483–511. [DOI] [PubMed] [Google Scholar]
- Fry BG, Wüster W. 2004. Assembling an arsenal: origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences. Mol Biol Evol. 21:870–883. [DOI] [PubMed] [Google Scholar]
- García T, et al. 2009. Pharmacological effects of two cytolysins isolated from the sea anemone Stichodactyla helianthus. J Biosci. 34:891–898. [DOI] [PubMed] [Google Scholar]
- García-Ortega L, et al. 2011. The behavior of sea anemone actinoporins at the water-membrane interface. Biochim Biophys Acta Biomembr. 1808:2275–2288. [DOI] [PubMed] [Google Scholar]
- Giese C, Mebs D, Werding B. 1996. Resistance and vulnerability of crustaceans to cytolytic sea anemone toxins. Toxicon 34:955–958. [DOI] [PubMed] [Google Scholar]
- Gorres KL, Raines RT. 2010. Prolyl 4-hydroxylase. Crit Rev Biochem Mol Biol. 45:106–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 29(7):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 8(8):1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holford M, Puillandre N, Modica MV, et al. 2009. Correlating molecular phylogeny with venom apparatus occurrence in panamic auger snails (Terebridae). PLoS One 4:e7667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holford M, Puillandre N, Terryn Y, et al. 2009. Evolution of the toxoglossa venom apparatus as inferred by molecular phylogeny of the Terebridae. Mol Biol Evol. 26:15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Bandyopadhyay PK, Olivera BM, Yandell M. 2011. Characterization of the Conus bullatus genome and its venom-duct transcriptome characterization of the Conus bullatus genome and its venom-duct transcriptome. BMC Genomics 60:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imperial JS, et al. 2003. The augertoxins: biochemical characterization of venom components from the toxoglossate gastropod Terebra subulata. Toxicon 42:391–398. [DOI] [PubMed] [Google Scholar]
- Imperial JS, et al. 2006. A novel conotoxin inhibitor of Kv1.6 channel and nAChR subtypes defines a new superfamily of conotoxins. Biochemistry 45:8331–8340. [DOI] [PubMed] [Google Scholar]
- Imperial JS, et al. 2007. Venomous auger snail Hastula (Impages) hectica (Linnaeus, 1758): molecular phylogeny, foregut anatomy and comparative toxinology. J Exp Zool B Mol Dev Evol. 308:744–756. [DOI] [PubMed] [Google Scholar]
- Kaas Q, et al. 2008. ConoServer, a database for conopeptide sequences and structures. Bioinformatics 24:445–446. [DOI] [PubMed] [Google Scholar]
- Kaas Q, Westermann J-C, Craik DJ. 2010. Conopeptide characterization and classifications: an analysis using ConoServer. Toxicon 55: 1491–1509. [DOI] [PubMed] [Google Scholar]
- Kaas Q, Yu R, Jin A-H, Dutertre S, Craik DJ. 2012. ConoServer: updated content, knowledge, and discovery tools in the conopeptide database. Nucleic Acids Res. 40:D325–D330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor YI, Taylor JD. 2000. Formation of marginal radular teeth in Conoidea (Neogastropoda) and the evolution of the hypodermic envenomation mechanism. J Zool Lond. 252:251–262. [Google Scholar]
- Kauferstein S, et al. 2004. Novel conopeptides of the I-superfamily occur in several clades of cone snails. Toxicon 44:539–548. [DOI] [PubMed] [Google Scholar]
- Kendel Y, et al. 2013. Venomous secretions from marine snails of the Terebridae family target acetylcholine receptors. Toxins 5:1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh CY, Kini RM. 2012. From snake venom toxins to therapeutics—cardiovascular examples. Toxicon 59:497–506. [DOI] [PubMed] [Google Scholar]
- Kumar S, Blaxter ML. 2010. Comparing de novo assemblers for 454 transcriptome data. BMC Genomics 11:571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laht S, et al. 2012. Identification and classification of conopeptides using profile Hidden Markov Models. Biochim Biophys Acta. 1824:488–492. [DOI] [PubMed] [Google Scholar]
- Lebbe EKM, Peigneur S, Wijesekara I, Tytgat J. 2014. Conotoxins targeting nicotinic acetylcholine receptors: an overview. Mar Drugs. 12: 2970–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt M. 1981. Effect of proline residues on protein folding. J Mol Biol. 145:251–263. [DOI] [PubMed] [Google Scholar]
- Lewis RJ, Dutertre S, Vetter I, Christie MJ. 2012. Conus venom peptide pharmacology. Pharmacol Rev. 64:259–298. [DOI] [PubMed] [Google Scholar]
- Li B, et al. 2014. Evaluation of de novo transcriptome assemblies from RNA-Seq data. Genome Biol. 15:553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Vera E, Walewska A, Skalicky JJ, Olivera BM, Bulaj G. 2008. Role of hydroxyprolines in the in vitro oxidative folding and biological activity of conotoxins. Biochemistry 47:1741–1751. [DOI] [PubMed] [Google Scholar]
- Lu BX, Zeng ZB, Shi TL. 2013. Comparative study of de novo assembly and genome-guided assembly strategies for transcriptome reconstruction based on RNA-Seq. Sci China Life Sci. 56:143–155. [DOI] [PubMed] [Google Scholar]
- Macek P. 1992. Polypeptide cytolytic toxins from sea anemones (Actiniaria). FEMS Microbiol Immunol. 5:121–129. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Domes G, Kirsch P, Heinrichs M. 2011. Oxytocin and vasopressin in the human brain: social neuropeptides for translational medicine. Nat Rev Neurosci. 12:524–538. [DOI] [PubMed] [Google Scholar]
- Miljanich GP. 1997. Venom peptides as human pharmaceuticals. Sci Med. 4(5):6–15. [Google Scholar]
- Miljanich GP. 2004. Ziconotide: neuronal calcium channel blocker for treating severe chronic pain. Curr Med Chem. 11:3029–3040. [DOI] [PubMed] [Google Scholar]
- Miller B. 1970. Feeding mechanisms of the family Terebridae. Annu Rep Am Malacol Union. 971:72–74. [Google Scholar]
- Mundry M, Bornberg-Bauer E, Sammeth M, Feulner PGD. 2012. Evaluating characteristics of de novo assembly software on 454 transcriptome data: a simulation approach. PLoS One 7:e31410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton RS, Olivera BM. 2006. Conotoxins down under. Toxicon 48: 780–798. [DOI] [PubMed] [Google Scholar]
- O’Neil ST, Emrich SJ. 2013. Assessing de novo transcriptome assembly metrics for consistency and utility. BMC Genomics 14:465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivera BM. 2000. w-Conotoxin MVIIA: from marine snail venom to analgesic drug. In: Drugs from the Sea. Fusetani N, editor. Karger: Basel; p. 75–85. [Google Scholar]
- Peng C, et al. 2006. Discovery of a novel class of conotoxin from Conus litteratus, lt14a, with a unique cysteine pattern. Peptides 27:2174–2181. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 8:785–786. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818. [DOI] [PubMed] [Google Scholar]
- Puillandre N, et al. 2008. Starting to unravel the toxoglossan knot: molecular phylogeny of the ‘turrids’ (Neogastropoda: Conoidea). Mol Phylogenet Evol. 47:1122–1134. [DOI] [PubMed] [Google Scholar]
- Puillandre N, Holford M. 2010. The Terebridae and teretoxins: combining phylogeny and anatomy for concerted discovery of bioactive compounds. BMC Chem Biol. 10:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puillandre N, Koua D, Favreau P, Olivera BM, Stocklin R. 2012. Molecular phylogeny, classification and evolution of conopeptides. J Mol Evol. 74:297–309. [DOI] [PubMed] [Google Scholar]
- Robinson SD, et al. 2014. Diversity of conotoxin gene superfamilies in the venomous snail, Conus victoriae. PLoS One 9:e87648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SD, Norton RS. 2014. Conotoxin gene superfamilies. Mar Drugs. 12:6058–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safavi-Hemami H, Bulaj G, Olivera BM, Williamson NA, Purcell AW. 2010. Identification of Conus peptidylprolyl cis-trans isomerases (PPIases) and assessment of their role in the oxidative folding of conotoxins. J Biol Chem. 285:12735–12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg SL, et al. 2012. GAGE: a critical evaluation of genome assemblies and assembly algorithms. Genome Res. 22:557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz MH, Zerbino DR, Vingron M, Birney E. 2012 Oases: Robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi K, Kawashima Y, Mizukami M, Nagashima Y. 2002. Properties of proteinaceous toxins in the salivary gland of the marine gastropod (Monoplex echo). Toxicon 40:563–571. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- Suttie JW. 1988. Vitamin K-dependent carboxylation of glutamyl residues in proteins. Biofactors 1:55–60. [PubMed] [Google Scholar]
- Taylor JD, Kantor YI, Sysoev AV. 1993. Foregut anatomy, feeding mechanisms, relationships and classification of the Conoidea (=Toxoglossa) (Gastropoda). Bull Nat Hist Mus Lond. 59:125–170. [Google Scholar]
- Terlau H, Olivera BM. 2004. Conus venoms: a rich source of novel ion channel-targeted peptides. Physiol Rev. 84:41–68. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Higgins DG. 2002. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. Chapter 2:Unit 2.3. [DOI] [PubMed] [Google Scholar]
- Tsetlin V. 1999. Snake venom a-neurotoxins and other ‘three finger’ proteins. Eur J Biochem. 264:281–286. [DOI] [PubMed] [Google Scholar]
- Twede VD, Miljanich G, Olivera BM, Bulaj G. 2009. Neuroprotective and cardioprotective conopeptides: an emerging class of drug leads. Curr Opin Drug Discov Devel. 12:231–239. [PMC free article] [PubMed] [Google Scholar]
- Ul-Hasan S, et al. 2013. Characterization of the peptidylglycine α-amidating monooxygenase (PAM) from the venom ducts of neogastropods, Conus bullatus and Conus geographus. Toxicon 74:215–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich A, Schlessinger J. 1990. Signal transduction by receptors with tyrosine kinase activity. Cell 61:203–212. [DOI] [PubMed] [Google Scholar]
- von Reumont BM, et al. 2014. A Polychaete’s powerful punch: venom gland transcriptomics of glycera reveals a complex cocktail of toxin homologs. Genome Biol Evol. 6:2406–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonk FJ, et al. 2013. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc Natl Acad Sci U S A. 110:20651–20656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-Q, Han Y-H, Shao X-X, Chi C-W, Guo Z-Y. 2007. Molecular cloning, expression and characterization of protein disulfide isomerase from Conus marmoreus. FEBS J. 274:4778–4787. [DOI] [PubMed] [Google Scholar]
- Warren WC, et al. 2008. Genome analysis of the platypus reveals unique signatures of evolution. Nature 453:175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, et al. 2012. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490:49–54. [DOI] [PubMed] [Google Scholar]
- Zugasti-Cruz A, Aguilar MB, Falcón A, Olivera BM, Heimer de la Cotera EP. 2008. Two new 4-Cys conotoxins (framework 14) of the vermivorous snail Conus austini from the Gulf of Mexico with activity in the central nervous system of mice. Peptides 29:179–185 [DOI] [PMC free article] [PubMed] [Google Scholar]