Abstract
Alternative splicing and gene duplication are the two main processes responsible for expanding protein functional diversity. Although gene duplication can generate new genes and alternative splicing can introduce variation through alternative gene products, the interplay between the two processes is complex and poorly understood. Here, we have carried out a study of the evolution of alternatively spliced exons after gene duplication to better understand the interaction between the two processes. We created a manually curated set of 97 human genes with mutually exclusively spliced homologous exons and analyzed the evolution of these exons across five distantly related vertebrates (lamprey, spotted gar, zebrafish, fugu, and coelacanth). Most of these exons had an ancient origin (more than 400 Ma). We found examples supporting two extreme evolutionary models for the behaviour of homologous axons after gene duplication. We observed 11 events in which gene duplication was accompanied by splice isoform separation, that is, each paralog specifically conserved just one distinct ancestral homologous exon. At other extreme, we identified genes in which the homologous exons were always conserved within paralogs, suggesting that the alternative splicing event cannot easily be separated from the function in these genes. That many homologous exons fall in between these two extremes highlights the diversity of biological systems and suggests that the subtle balance between alternative splicing and gene duplication is adjusted to the specific cellular context of each gene.
Keywords: alternative splicing, gene duplication, protein diversity, homologous exons, subfunctionalization
Introduction
Alternative splicing (AS) and gene duplication (GD) are two of the main mechanisms behind the diversification of protein function. Both can increase the numbers of proteins coded within genomes; GD creates initially redundant copies of genes that with time, and following different possible evolutionary paths, can diversify in sequence and function (Conant and Wolfe 2008; Innan and Kondrashov 2010), whereas AS allows genes to code for more than one distinct protein from the same locus (Smith and Valcárcel 2000). The relationship between GD and AS is not well understood, so analyzing the interconnection between the two processes may provide insights into their relative importance in the generation of new protein products.
As GD and AS are both repositories of protein diversity, interplay between the two can be expected. According to the interchangeable model (I-model), or function-sharing model, alternative isoforms that were originally coded within a single gene may separate into different genes after a GD event by means of differential retention of AS patterns in each duplicate gene. The extreme case would be the subfunctionalization of gene duplicates. Here, AS and GD might be regarded as interchangeable repositories of protein diversity. This model has received support from 1) genome-wide analyses reporting a negative correlation between AS and the size of protein families (Kopelman et al. 2005; Su et al. 2006; Talavera et al. 2007), and 2) reports of acceleration of AS divergence after GD (Zhang et al. 2010; Xu et al. 2012), although the validity of some of these results is debated (Talavera et al. 2007; Roux and Robinson-Rechavi 2011; Su and Gu 2012). More recently, Lambert et al. (2015) analyzed exon divergence of zebrafish gene duplicates that are co-orthologs of human genes. Although their analysis does support a general trend of splice isoform separation after GD, their results must be treated with caution as they were based on the comparison of heterogeneous transcriptome annotations that, in the case of zebrafish at least, are far from complete.
In the noninterchangeable (NI-model), the AS-encoded protein diversity is not distributed among gene duplicates. The underlying implication of this model is that it may not be favorable to separate alternative isoforms into different genes, which may indicate that AS in these genes is not just a means to encode protein diversity but also of controlling their expression. The importance of AS in these genes may be related to the balanced production of isoforms or other kinds of regulation linked to the splicing process that may not be attainable with independent genes. In contrast to the I-model or “Function-sharing model,” the NI-model has not been thoroughly investigated. The natural prediction of the NI-model is that alternative exons will be preserved by purifying selection after GD events.
To study the relationship between GD and AS we concentrated on characterizing the evolutionary conservation of mutually exclusive homologous exons (MEHEs), defined here as duplicated exons that are incorporated into alternatively spliced transcripts in a mutually exclusive manner. We chose to focus on MEHEs because they are potentially the most biologically relevant type of AS (Ezkurdia et al. 2012), and because they are particularly adequate for the systematic comparison of isoforms after GD.
Like gene duplicates, MEHEs can evolve new functions or experience a subfunctionalization process within the context of a single gene. As long as these alternative MEHEs have evolved different functions, full subfunctionalization may occur if each of the ancestral MEHEs is retained in a different gene after GD. Indeed, a handful of examples of subfunctionalization driven by splice isoform separation have been reported in the literature (Altschmied et al. 2002; Yu et al. 2003; Pacheco et al. 2004; Cusack and Wolfe 2007; Hultman et al. 2007; Marshall et al. 2013). Unfortunately, a study of true subfunctionalization is not possible in silico, because only experimental evidence can confirm that two different sequences have two different cellular functions. So here we have used the separation of MEHEs among gene duplicates as a proxy for subfunctionalization. As we cannot be sure that the separation of MEHEs is genuine subfunctionalization, the process of separating homologous exons after GD is referred to here as splice isoform separation.
We carefully curated a list of human MEHEs, most of which are predicted to be relevant on the basis of evolutionary conservation, and analyzed the conservation of MEHEs using sequence similarity searches in five distantly related vertebrate species, including lamprey, fugu, zebrafish, spotted gar, and coelacanth. Within this data set we focused on GD events to assess the prevalence of the NI- and I-models. We identified cases of splice isoform separation by looking for differential conservation of MEHEs among gene duplicates. We also identified genes in which MEHEs were preserved after duplication. We discuss the biological implications of each model.
Materials and Methods
We explored the human genome using Ensembl version 75 from February 2014 (Flicek et al. 2013) and compared CCDS annotations to identify genes with MEHEs. CCDS annotations represent high-quality transcript annotations for which the EBI, the NCBI (National Center for Biotechnology Information), the WTSI, and the UCSC (the University of California–Santa Cruz) reached a consensus (Pruitt et al. 2009). Although CCDS annotations are not complete, restricting the set of transcripts to these cases avoids including rare and low frequency human transcript variants. We discarded those CCDS that were part of another CCDS. We found that 5,322 genes contained more than one nonredundant CCDS. We sorted the resulting CCDS by length and defined the longest one as reference. Other transcripts were compared against the reference transcript to identify coding exons that code for at least ten amino acids and are present in one transcript but not in the other, that is, that are MEEs. We then identified whether the resulting pairs of MEEs were homologous (MEHEs) using BLAST v2.2.25 (Altschul et al. 1997) comparisons, with an e value threshold of 0.005. To simplify the analysis and to avoid the inclusion of false positives related to annotation problems, we restricted the set to those genes for which we identified just one pair (or set) of MEHEs in the reference transcript. The final set contained 97 genes.
Five vertebrate species, all distantly related to human, were selected to explore the evolutionary conservation of MEHEs. Selected taxa included lamprey (Smith et al. 2013), spotted gar (Amores et al. 2011), zebrafish (Howe et al. 2013), fugu (Aparicio et al. 2002), and coelacanth (Amemiya et al. 2013), and were retrieved from Ensembl v75. The genomes of these target species were scanned using TBLASTN without low complexity filtering (−F F) and with an e-value threshold of 0.1 to find similarity matches to query human MEHEs. We merged overlapping similarity hits using bedtools v2.17.0 (Quinlan and Hall 2010) and determined whether they overlapped (or were close enough to) annotated genes. To set a distance threshold for assigning nonoverlapping hits to neighbor genes, we calculated the 95 percentile of gene lengths for each species and required a hit to be closer than that threshold (this threshold is highly variable between target species: 27,992, 41,583, 96,255, 109,349, and 151,632 bp for fugu, lamprey, spotted gar, zebrafish, and coelacanth, respectively). We carefully reviewed those cases in which similar hits were ambiguously assigned to multiple neighbor genes. Finally, we identified those cases in which multiple nonoverlapping hits belonged to the same gene, as these cases are candidates for having conserved MEHEs.
For genes with multiple similar hits (usually 2) to the query MEHEs, we determined whether each hit was most similar to each of the query MEHEs. In addition, we determined whether the genes to which hits were assigned were orthologs of the query human gene according to EnsemblCompara (Vilella et al. 2009). We also annotated whether query and target genes were part of the same phylogenetic tree in EnsemblCompara and whether, if not orthologous, the alternative paralogous relationship was set as confident according to the EnsemblCompara pipeline (we determined whether the gene is considered an ortholog of a different human gene that the query gene with confidence). Uncertain cases were carefully reviewed.
To date the origin of the MEHEs, we assumed that when two species share a pair of MEHEs these have not been acquired independently. This is equivalent to inferring ancestral character states with Dollo parsimony (Farris 1977). Phylogenetic analyses (particular cases) and the degree of similarity (in general) support this assumption. In certain cases, the presence of human paralogs with the same MEHEs allowed dating the evolutionary origin of MEHEs at the corresponding GD event, which may be an older age than that inferred looking at the presence of MEHEs in the five target species.
We conducted detailed evolutionary analyses to validate and characterize potential splice isoform separation cases, that is, cases in which gene duplicates retained or lost different MEHEs. Multiple sequence alignments were built with MAFFT v7.123b (Katoh and Standley 2013), handled and visualized with Jalview (Waterhouse et al. 2009). Phylogenetic trees were reconstructed for exons and/or genes under maximum likelihood with Phyml v3.1 (Gouy et al. 2010; Guindon et al. 2010), using 1,000 replicates of nonparametric bootstrapping and choosing the best-fit model of evolution with ProtTest v2.4 (Abascal et al. 2005). The selection of taxa varied depending on each particular case (alignments and trees are available from the author upon request). Tree figures were prepared with FigTree (http://tree.bio.ed.ac.uk/software/figtree/, March 2015).
Results
AS of MEHEs Is Highly Conserved in Vertebrates
We identified 97 genes with a single set of MEHEs from among the set of human CCDS (consensus coding sequences) transcripts (see Materials and Methods). To assess the evolutionary conservation of the corresponding AS events, we relied on direct sequence searches against target genomes with TBLASTN rather than comparing annotations of the isoforms in the corresponding species because the gene annotations of all species apart from human are still not close to being complete. For each BLAST hit we determined whether it corresponded to annotated or new genes and/or exons, and whether the corresponding genes were considered orthologs or paralogs of the query human gene in the EnsemblCompara database. We carefully analyzed each of the cases.
We assessed the validity of relying on sequence similarities rather than on comparison of transcript annotations to trace the evolution of MEHEs across species. Careful curation revealed that in a few cases (4) the MEHEs were conserved even though TBLASTN was not able to detect them, mainly because these exons were too short or highly divergent. Despite this, our assessment of transcript annotation qualities in target species showed that sequence-based approaches are still much better. We found that transcript annotations are usually incomplete and, importantly, of very heterogeneous quality across species. We estimated the number of nonannotated genes and exons (see supplementary material, Supplementary Material online) and found that although 93.7% of the MEHEs identified with TBLASTN were annotated in fugu, only 53.6% of the TBLASTN-identified MEHEs were annotated in lamprey (supplementary table S1, Supplementary Material online).
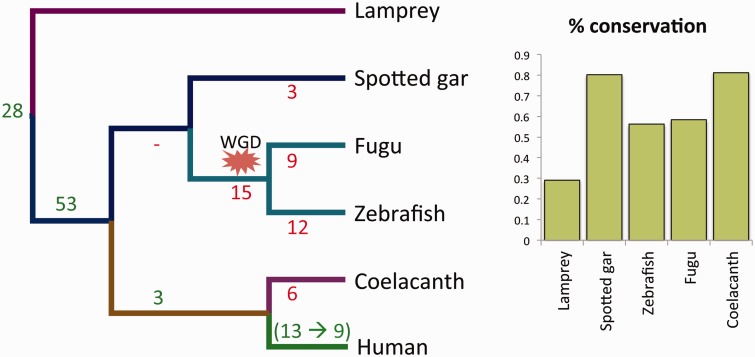
For 84 of 97 genes we found that both MEHEs are present in at least one of the five target species (fig. 1 and supplementary table S1, Supplementary Material online). We determined that for almost all of the cases orthologous relationships could be established between each of the target MEHEs and each of the query MEHEs, implying that the large majority of MEHEs have not duplicated independently in different lineages (see supplementary material, Supplementary Material online). Hence, we can infer that these 84 MEHEs (or the majority of them) originated at least 400 Ma.
Fig. 1.—
(A) The date of origin and loss of the 97 human MEHE AS patterns shown against the phylogeny of human and five distant vertebrate species. Gain of AS event is shown in green, and the inferred number of AS losses in red. (B) The percentage of conservation of the 97 human AS events in each species.
In 41 of the 84 cases, the MEHEs were conserved in all four species of jawed vertebrates. MEHE conservation reached lamprey in 28 genes, despite the distant relationship between lamprey and human (∼500 Ma; Kumar and Hedges, 2011). Up to 80 cases have been conserved in at least one bony fish, more frequently in spotted gar (77 cases) than in fugu and zebrafish (56 and 54, respectively; fig. 1B). The larger number of losses in teleosts is probably the result of the whole-genome duplication experienced in their ancestor.
We carefully revised the 13 cases for which no conservation was detected in any of the target species to check whether the MEHEs appeared later in the human lineage or whether the lack of significant sequence similarity was due to low sequence conservation and/or exons that were too short. We found that 4 of these 13 cases were indeed present in at least one of the target species. Consequently, we ended with a total of 88 of 97 cases of human MEHEs of ancient origin (90.7%). We did not include these four cases as part of the comparative analysis because we have no objective way to establish their conservation across the five target species.
Splice Isoform Separation by Retention of Different Homologous Exons after GD
We identified cases in which alternative isoforms ancestrally coded by a single gene became separated into different genes by means of GD coupled to differential loss and conservation of MEHEs in each paralog. In such cases, protein diversity initially encoded through AS becomes distributed in different genes, supporting the I-model. We found a total of ten cases of this kind (table 1), nine of which are new. We also identified a case (CUX1) of differential conservation of nonhomologous mutually exclusive exons (MEEs). Among the 11 cases, the following 7 experienced complete splice isoform separation: CALU, CUX1, MARVELD3, PGM1, PDLIM3, RNF128, and U2AF1. In the remaining four cases (CACNA1C/1D, CDC42, FYN, and SLC8A3) splice isoform separation was detected between two paralogs but at the same time other paralogs conserved the ancestral pattern of AS of MEHEs. These 11 cases represent a very significant increase with respect to those reported in the literature. The cases of PGM1 and CUX1 are described in the supplementary material, Supplementary Material online (supplementary figs. S1 and S2, Supplementary Material online), whereas CALU, MARVELD3, and CACNA1C/CACNA1D are described below. Splice isoform separation of PDLIM3 occurred in platypus and splice isoform separation (and subfunctionalization) of U2AF1 has been already described in the literature (Pacheco et al. 2004).
Table 1.
The 11 Cases in Which Each Duplicated Gene Lost or Retained One of the Ancestral MEHEs in a Concerted Manner (Splice Isoform Separation) Are Shown, Indicating Which Genes and Lineages Are Affected
| Human Gene | Origin of MEHEs | Human Exons (GRCh38) | Differential Conservation of Ancestral MEHEs in Lineage (Genes) |
|---|---|---|---|
| CACNA1C, CACNA1D | Vertebrates | 12:2504435–2504539, 12:2504841–2504945; 12:2633628–2633712, 12:2634296–2634374 (CACNA1C) | Vertebrates (CACNA1S and CACNA1F) |
| CALU | Jawed vertebrates | 7:128754261–128754455, 7:128754528–128754722 | Teleosts (CALUA and CALUB) |
| CDC42 | Jawed vertebrates | 1:22091427–22091517, 1:22089942–22090032 | Zebrafish (CDC42L and CDC42L2) |
| CUX1 | Bilaterians | 7:101816011–102258233, 7:101816031–102249042; 7:101815904–102283957, 7:101816031–102283090a | Zebrafish (CUX1A and CUX1B) |
| FYN | Chordates | 6:111700103–111700268, 6:111699514–111699670 | Vertebrates (many genes, e.g., FRK vs. SRC, YES1 . . .) |
| MARVELD3 | Vertebrates | 16:71640389–71641027, 16:71634192–71634803 | Lamprey, spotted gar, zebrafish, fugu (also other vertebrates) |
| PDLIM3 | Chordates (?) | 4:185508298–185508562, 4:185514702–185514890 | Platypus (ENSOANG00000006867, ENSOANG00000013438) |
| PGM1 | Vertebrates (?) | 1:63623460–63623760, 1:63593488–63593734 | Teleosts (PGM1 and PGM5) |
| RNF128 | Jawed vertebrates | X:106726913–106727397, X:106694002–106694408 | Zebrafish and cave fish (Otophysa) (RNF128A and ENSDARG00000029890) |
| SLC8A3 | Jawed vertebrates | 14:70060835–70060939, 14:70063822–70063929 | Spotted gar, fugu, coelacanth … (SLC8A4b, SLC8A2a) |
| U2AF1 | Jawed vertebrates | 21:6493043–6493110, 21:6492130–6492197 | Fugu, tilapia and stickleback (Percomorphaceae?) (U2AF1 and ENSTRUG00000013815) |
Note.—Genes in bold indicate cases undergoing complete splice isoform separation.
aCUX1 is not a case of homologous but of nonhomologous MEEs.
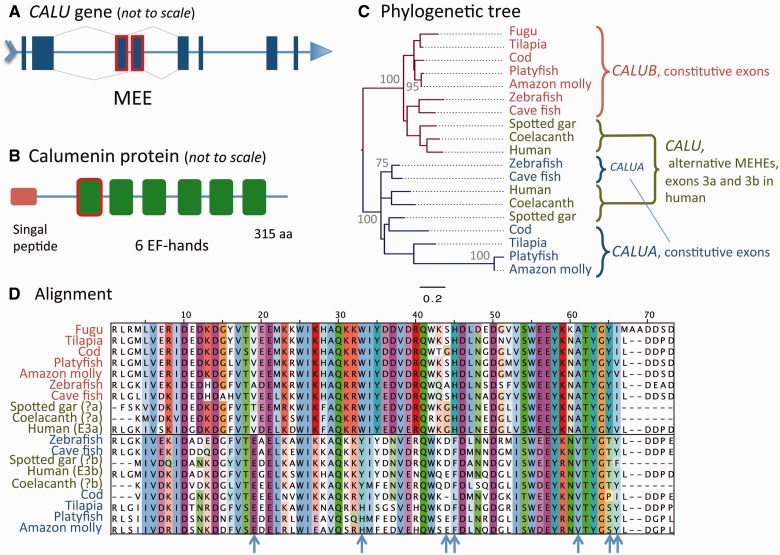
The CALU gene is ubiquitously expressed and encodes a protein (calumenin) distributed throughout the secretory pathway (Vorum et al. 1999), known to inhibit vitamin-K-dependent protein carboxylation (Wajih et al. 2004) and involved in protein sorting and folding (Tsukumo et al. 2009; Wang et al. 2012). CALU contains six calcium-binding EF-hand domains, the first of which is coded by one of two MEHEs (fig. 2A and B). Little is known about the functional role of the splicing of MEHEs exons in CALU. Calumenin MEHEs may be differentially expressed in human primary tumors (Dutertre et al. 2010). This, together with the observation that CALU is a phosphorylation substrate of v-Src (Shah and Shokat 2002), suggests that it may participate in signal transduction pathways related to transformation (Honoré 2009).
Fig. 2.—
Splice isoform separation of CALU in teleosts by differential retention of ancestral MEHEs (A) that code for the first EF-hand domain (B) is strongly supported by the position in the ML exon tree of two distinct teleost genes, CALUA and CALUB, each within the group of monophyly defined by each ancestral MEHE (C; with the best-fit evolutionary model LG+I+G). Numbers close to nodes indicate cases with more than 70% of bootstrap support based on 1,000 replicates. The multiple sequence alignment reveals some positions (blue arrows) with specific conservation patterns between MEHEs of human, spotted gar and coelacanth, and between duplicated genes in zebrafish and other teleosts (D).
Genomic BLAST revealed similarities specific to each of the human alternative exons within the corresponding CALU loci of spotted gar and coelacanth, allowing us to date the origin of the MEHEs of CALU to the ancestor of jawed vertebrates. We found that the pattern of AS was lost in fugu and zebrafish. There are two orthologs of human CALU in zebrafish (CALUA and CALUB), which originated from a duplication event in the ancestor of teleosts (one of these duplicates was later lost in fugu; fig. 2C). Interestingly, each zebrafish ortholog specifically retained one of the ancestral alternative exons while losing the other. By exploring other species that present multiple orthologs to human CALU we also found differential exon losses in all other teleosts but tetraodon and stickleback, which, as fugu, lost one of the duplicated genes (fig. 2C). Hence, the process of splice isoform separation took place in the ancestor of teleosts right after the duplication of CALU.
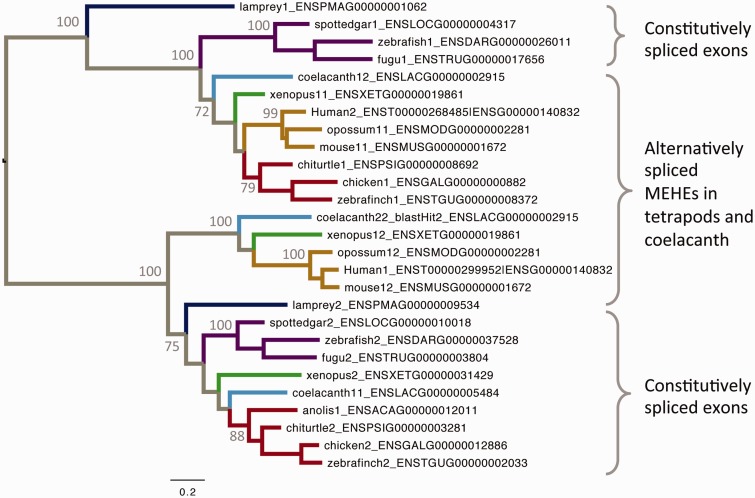
MARVELD3 belongs to the occludin family, whose members are components of tight junctions (Steed et al. 2009) and share a MARVEL domain that contains four transmembrane helices and is typically involved in membrane apposition events (Sánchez-Pulido et al. 2002). MARVELD3 acts by coupling tight junctions to the MEKK1–JNK (c-Jun-N-terminal kinase) pathway, so determining cell behavior and survival (Steed et al. 2009). Indeed, MARVELD3 is downregulated during epithelial–mesenchymal transition in human pancreatic cancer cells (Kojima et al. 2011) and loss of MARVELD3 expression increases cell migration and proliferation, whereas re-expression reverts the metastatic phenotype (Steed et al. 2009). The human MARVELD3 gene contains two MEHEs (E3a and E3b) that code for the C-terminal half of the protein that contains the MARVEL domain. Both isoforms are widely expressed in epithelial and endothelial cells (Steed et al. 2009) and share a less-conserved and highly acidic N-terminal region that is predicted to be disordered and responsible for the interaction with the MEKK1–JNK signaling pathway (Steed et al. 2009). No functional differences have been described yet between the two isoforms.
Our analysis revealed a complex evolutionary history for MARVELD3, and we had to consider other vertebrates to clarify it. The pattern of AS, previously reported as specific to mammals (Steed et al. 2009), is also observed in coelacanth and Xenopus. In all vertebrates but mammals, that is, in lamprey, ray-finned fishes, coelacanth, Xenopus and reptiles (including birds), there are two MARVELD3 genes instead of one. With the exception of coelacanth and Xenopus, species with duplicated MARVELD3 show no AS. Interestingly, the phylogenetic reconstruction of these exons reveals two clearly defined lineages (groups of orthology), each covering the whole set of analyzed vertebrates. In species with duplicated genes, each of the two separated exons maps to a different group of orthology. In species with AS, each alternative exon maps to each group of orthology. In coelacanth and Xenopus both things happen, as one of their duplicated genes conserved the AS pattern. Although other alternative hypotheses could be proposed, we believe that the most parsimonious interpretation for this complex scenario is that originally, in the ancestor of vertebrates, MARVELD3 acquired the pattern of AS. Then, this ancestral gene duplicated and one of the paralogs lost the pattern of AS. Later, after the split of the major vertebrate lineages, some lineages lost the paralog that had no AS (mammals) whereas other lineages lost one of the AS isoforms from the paralog that did have AS (fig. 3). According to the phylogenetic tree, splice isoform separation occurred at least three times (at the ancestors of lamprey, ray-finned fishes, and reptiles) whereas a single gene loss event took place in the ancestor of mammals. Remarkably, despite several gene and exon losses, both original splice isoforms have been always kept, either within the same or different genes, which might be taken as an indication of their biological relevance and functional independence.
Fig. 3.—
The ML phylogenetic tree of MARVELD3 exons (LG+I+G+F evolutionary model), which shows the evolutionary relationship between equivalent homologous exons in different species. The exons exist either in the form of alternatively spliced exons or as constitutively spliced exons in separate genes. The numbers at each internal node indicate bootstrap support.
AS of MEHEs Is Conserved in Human Paralogs
The cases of differential conservation of MEHEs in duplicated genes reveal that the protein diversity encoded with AS can be distributed between independent genes. To explore the validity of the alternative NI-model, we looked for cases in which MEHEs were conserved between paralogs after GD.
We identified 21 clusters of human paralogs, comprising 54 genes, with the same pattern of MEHEs (table 2). The great majority of these paralogs duplicated a long time ago, in the ancestor of jawed vertebrates or earlier. These MEHEs are of special interest because they have ancient origins and have been conserved along different gene lineages. Hence, these are genes for which AS may be resilient to GD and support the NI-model. The following examples illustrate how relevant the AS of MEHEs of these genes might be.
Table 2.
Groups of Human Paralogs with Homologous Patterns of AS along with the Date of the Corresponding Duplication Events and the Relative Position of MEHEs within the Gene
| Human Paralogs | Description | Duplication Ancestor | Region Affected and AS Role |
|---|---|---|---|
| ACSL1, ACSL6 | Acyl-CoA synthetase long-chain | Jawed vertebrates | Internal |
| ACTN1, ACTN2, ACTN4 | Alpha-actinin | Vertebrates. One AS conserved in fruitfly ACTN | Two pairs of internal MEHEs. Actin-binding domain (fig. 4). Tissue specificity (Waites et al. 1992) |
| ASIC1, ASIC2 | Acid-sensing ion channel | Vertebrates | 5 prime. N-terminus and first transmembrane helix of the channel |
| CACNA1A, CACNA1B, CACNA1E | Voltage-dependent L-type calcium channel subunit alpha-1 | Vertebrates | Internal. Cytoplasmic C-terminal region. Fine tuning of channel properties (Lipscombe et al. 2013) |
| CACNA1C, CACNA1D | Voltage-dependent L-type calcium channel subunit alpha-1 | Vertebrates | Two pairs of internal MEHEs. End of first ion transport domain, beginning of last ion transport domain |
| CLDN10, CLDN18 | Claudin | Vertebrates. MEHEs also found in C. savygnii | 5 prime. PMP22_Claudin domain. Permeability for anions or cations (Günzel et al. 2009) |
| CYP4F2, CYP4F3 | Cytochrome P450, family 4, subfamily F | Catarrhini | Internal. Beginning of p450 domain |
| DEFB110, DEFB119 | Beta-defensin | Amniotes | 3 prime. A signal peptide is shared between isoforms, while the extracellular domain, with many conserved Cys, is alternatively spliced |
| DNM1, DNM2 | Dynamin | Vertebrates | Internal. Dynamin_M domain |
| FGFR1, FGFR2, FGFR3 | Fibroblast growth factor receptor | Vertebrates, jawed vertebrates. MEHEs also found in tunicates | Internal. C-terminal half of the third Ig-like domain. Interaction with FGF and heparan sulfate proteoglycans (Olsen et al. 2004) |
| GNAL, GNAS | Guanine nucleotide-binding protein G(olf/s) subunit alpha | Jawed vertebrates | 5 prime. N-terminal region predicted disordered and beginning of G-alpha domain |
| GRIA1, GRIA2, GRIA3, GRIA4 | AMPA glutamate receptor | Vertebrates | Internal. Ligand-gated ion channel domain. Channel-gating kinetics (Partin et al. 1996) |
| ITGA3, ITGA6 | Integrin alpha | Vertebrates | 3 prime. Cytoplasmic C-termini. Interaction with HPS5 (Fukushi et al. 2004). Tissue specificity (De Melker et al. 1997) |
| MAPK8, MAPK9, MAPK10 | Mitogen-activated protein kinase/JNK. | Vertebrates | Internal. Kinase domain. Different affinities for ATF-2, Elk-i and Jun transcription factors (Gupta et al. 1996) |
| MEF2A, MEF2C, MEF2D | Myocyte-specific enhancer factor. | Jawed vertebrates | Internal. Holliday junction regulator protein family C-terminal repeat |
| NRG1, NRG2 | Pro-neuregulin | Jawed vertebrates | Internal. Tissue specificity, cell localization, etc. (Liu et al. 2011) |
| PDLIM3, LDB3 | PDZ and LIM domain protein 3 (ALP), LIM domain-binding protein 3 (Enigma) | Chordates? (not in the same Ensembl tree) | Tissue specific AS affecting the small ZM domain responsible for alpha-actinin-2 binding (Faulkner et al. 1999) |
| SCN2A, SCN3A, SCN5A, SCN8A, SCN9A | Sodium channel protein subunit alpha | Amniotes, vertebrates | Internal. Beginning/middle of first ion transport domain. Developmental and tissue specificities (Gazina et al. 2010) |
| SLC44A2, SLC44A5 | Choline transporter-like protein | Vertebrates | 3 prime. Cytoplasmic C-terminal tail |
| SLC8A1, SLC8A3 | Sodium/calcium exchanger | Vertebrates | Internal in calx-beta motif. May modulate the dynamic properties of Ca2+ sensing (Khananshvili 2013) |
| TPM1, TPM2, TPM3, TPM4 | Tropomyosin alpha chain | Vertebrates | Several: 5 prime, internal, 3 prime. Developmental and tissue specificities (reviewed in Gunning et al. 2005) |
Note.—Groups in bold indicate cases in which all the paralogs descending from the last GD event conserved the ancestral MEHEs.
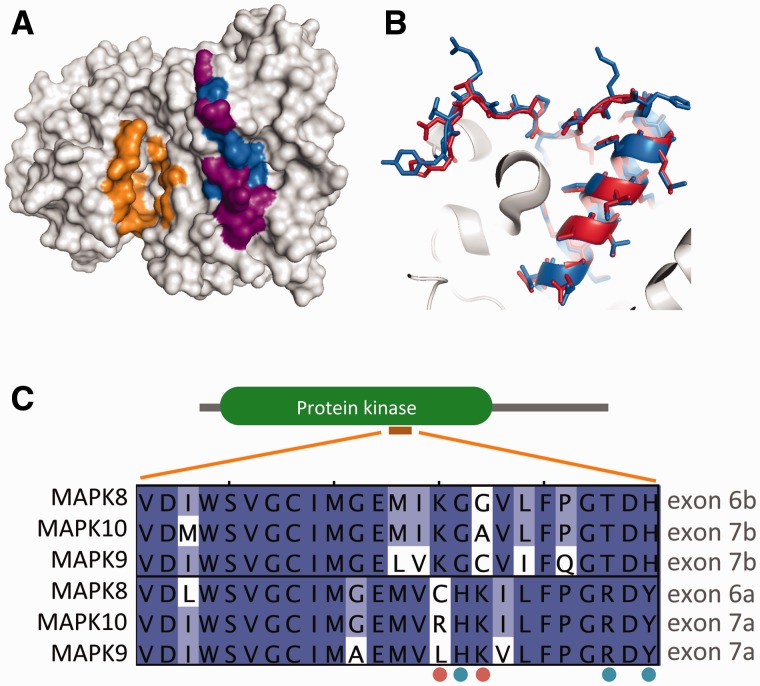
The strongest support for the NI-model comes from the JNKs, AMPA glutamate receptors, and myocyte-specific enhancer factors (MEF2s). In these cases, MEHEs were conserved in all the members of their families despite ancient GD events. In the case of JNKs (MAPK8, MAPK9, and MAPK10), the MEHEs code for part of the kinase domain (fig. 4). The biological significance of these MEHEs may relate to different ligand-binding specificities (Gupta et al. 1996), but remains unclear (Seki et al. 2012). In the case of AMPA glutamate receptors (GRIA1, GRIA2, GRIA3, and GRIA4), MEHEs already existed in the ancestor of vertebrates (already reported in Chen et al. 2006) and code for the flip and flop exons (supplementary fig. S5, Supplementary Material online). Although these exons have almost identical amino acid sequences, their use yields important functional variations (Partin et al. 1996).
Fig. 4.—
The 3D-structure of human MAPK8 (pdb code 3O17) is shown in (A) emphasizing the region corresponding to the MEHEs (blue), which of the residues coded by the MEHEs differ between alternative MAPK8 isoforms (purple) and the location of the active ATP-binding site (orange). (B) Direct comparison between the two alternative human MAPK8 isoforms (3O17 in blue, 1UKH in red), showing that most differences are found within the loop. (C) Multiple sequence alignment of MEHEs of JNKs (E6a and E6b in MAPK8), highlighting residues that are specifically conserved within each ancestral exon (blue dots) or that are conserved in one but variable in the other (orange dots).
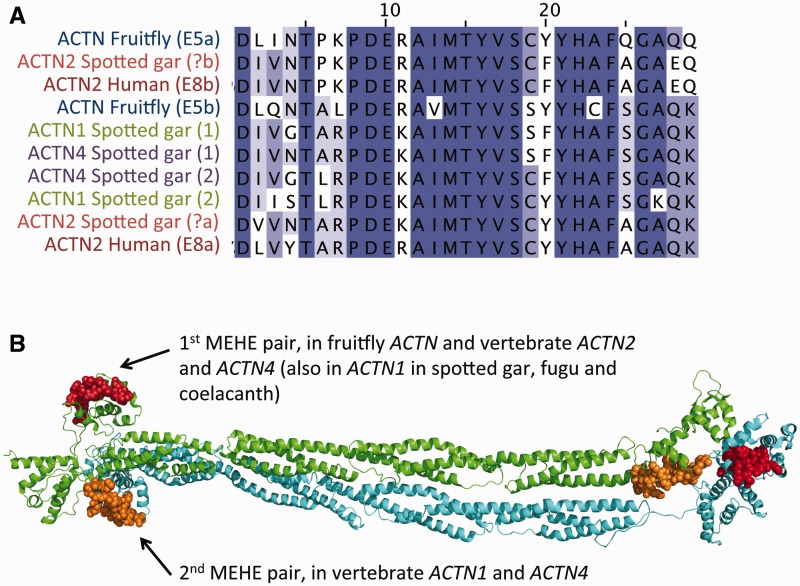
The importance of AS is particularly clear in the case of human ACTN2 and ACTN4 genes, which code for alpha-actinin 2 and 4, respectively. Alpha-actinins are important cytoskeletal proteins with multiple roles and many interacting partners. Interestingly, some of these partners also have MEHEs, for example PDLIM3, with MEHEs that affect a region involved in actinin-binding. The actinin family has two pairs of MEHEs that are distant in sequence, but close in the dimeric structure (fig. 5B). In human, ACTN4 has both pairs of MEHEs, ACTN1 and ACTN2 each share a different pair of MEHEs with ACTN4, and ACTN3 has no MEHEs.
Fig. 5.—
The multiple sequence alignment (A) of a pair of MEHEs from different alpha actinins (corresponding to exons 8a and 8b in human ACTN2) reveals the ancient ancestry of this AS event (it first appeared in the ancestor of bilaterians) and how the original pattern has been conserved in multiple gene lineages despite several GD events. Alternatively spliced MEHEs are highlighted by using same colors. Human ACTN4 has two MEHE events, one conserved in ACTN2 (see above) and another that is found in ACTN1, which are spatially close in the 3D dimeric structure of alpha actinin, within the actin-binding regions shown in (B). The structure corresponds to the cryoEM model of chicken ACTN1 (pdb:1SJJ; Liu et al. 2004).
Importantly, the MEHEs in ACTN2 and ACTN4 (and also in ACTN1 in fugu, spotted gar and coelacanth, but not in human or mouse ACTN1) have particularly ancient ancestry. Fruitfly and Caenorhabditis elegans have the same pattern of AS, which allows dating the origin of these MEHEs back to the ancestor of bilaterians (Barstead et al. 1991). This clearly points toward a key functional role of AS for alpha-actinins.
We identified other interesting examples, like the paralogs of the SCN2A gene or the fibroblast growth factor receptors, which are described in the supplementary material, Supplementary Material online (supplementary figs. S3–S5, Supplementary Material online).
The Complex Case of the CACNA1 Family MEHEs
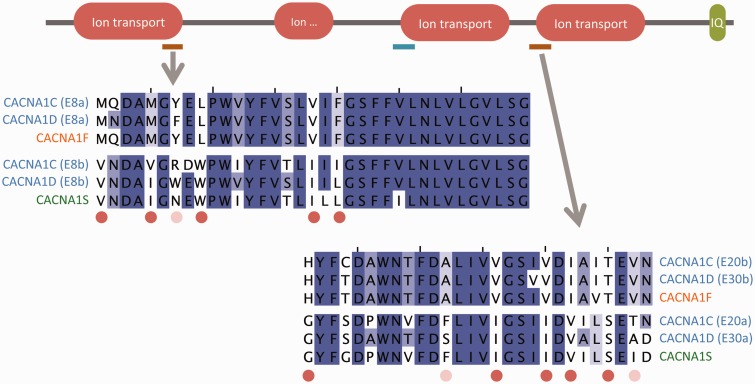
The genes CACNA1C, CACNA1D, CACNA1S, and CACNA1F code for alpha subunits of voltage-gated calcium channels. These four paralogs form a monophyletic group that originated from GDs in the ancestor of jawed vertebrates. There are two pairs of MEHEs whose origin also dates back to the ancestor of vertebrates, but predating the GD events. The genes CACNA1C and CACNA1D both conserved the two pairs of ancestral MEHEs (fig. 6), whereas CACNA1F and CACNA1S experienced a process of loss/retention of different ancestral exons that, interestingly, affected the two pairs of MEHEs (fig. 6). CACNA1C presents an additional pair or MEHEs that may have evolved later in the ancestor of sarcopterygians, as it is conserved in coelacanth, Xenopus, and mammals.
Fig. 6.—
Multiple sequence alignments of two sets of homologous exons from human genes CACNA1C and CACNA1D, along with the equivalent exons from the CACNA1F and CACNA1S paralogs. After duplication CACNA1F retained one homologous exon from each pair of ancestral MEHEs and CACNA1S the other. CACNA1C also has a third pair of MEHEs at the beginning of the third ion transport domain (blue). Exon numbering is distinct in CACNA1C and CACNA1D.
This case is particularly interesting because although conservation of AS within CACNA1C and CACNA1D supports the NI-model, the pattern of exon losses in CACNA1F and CACNA1S, which may be seen as a case of splice isoform separation, supports the I-model. A similar pattern occurs with the MEHEs of genes SLC8A3 and SLC8A1. These examples illustrate the complexity of the interaction between AS and GD. The evolutionary fate of MEHEs after GD may depend on subtle characteristics of each gene and extreme models may not be realistic.
Discussion
We have determined that the AS of mutually exclusively spliced homologous exons is highly conserved in vertebrates. We found evidence of ancient ancestry (>400 Ma) for about 91% of the human MEHEs. Other studies that have compared AS across mammals have provided substantially lower estimates of evolutionary conservation of splicing events (Modrek and Lee 2002; Boue et al. 2003; Thanaraj et al. 2003; Pan et al. 2005; Yeo et al. 2005; Mudge et al. 2011). Modrek and Lee estimated that only 25% of all “minor” alternative exons (regardless of splice type) were conserved between mouse and human. This is in sharp contrast to the conservation of MEHEs, in particular since our taxa selection comprised species that are much more distantly related than human and mouse. The difference in conservation suggests that MEHEs are likely to be much more functionally relevant than other types of alternative exons. The relevance of MEHEs is also supported by strong evidence indicating that the corresponding alternative isoforms reach the protein level much more frequently than would be expected based on the background frequencies of annotated AS events in the transcriptome (Ezkurdia et al. 2012).
For the reasons stated above, MEHEs are particularly amenable to study the relationship between GD and AS and explore the validity of two extreme models. Although we found support for both the I- and NI-models, many cases fell between these two extremes (e.g., one of the duplicated genes conserved the two MEHEs, whereas the other lost one MEHE), reflecting subtle differences in the relative importance of AS, as might be expected from the diversity of biological systems and the large divergence times. Indeed, there were genes and splicing events that provided evidence for both extreme models. For example, within the family of CACNA1C, CACNA1D, CACNA1F, and CACNA1S, two of the paralogs (CACNA1C and CACNA1D) support the NI-model based on conservation of ancestral MEHEs, whereas the other two paralogs (CACNA1F and CACNA1S) support the I-model based on the process of complementary loss/retention of alternative exons.
Support for the NI-model, in which protein diversity encoded by AS is not distributed among gene duplicates, came from 21 groups of human paralogs in which some or all duplicated genes conserved ancestral patterns of AS for long evolutionary periods. The best examples may be those of the JNKs and AMPA glutamate receptors. In both cases, all the multiple paralogs are related by ancient GD events but have conserved the ancestral MEHEs. The NI-model suggests that the control of expression by AS has a role that is tightly linked with the biological function of the gene.
Support for the I-model came from 11 examples in which each gene duplicate specifically retained one of the two ancestral MEHEs, that is, of splice isoform separation. As a result of the process of concerted loss and retention of ancestral MEHEs, the net protein diversity is conserved but distributed among different genes. For some genes, there may even be advantages to separating the alternative isoforms. At the very extreme of this model we found MARVELD3, for which the splice isoform separation process may have taken place independently in at least three different lineages.
Although we conducted no experimental confirmation, long-standing conservation of MEHEs very likely reflects the existence of functional differences between the homologous exons. If true, the identified cases of splice isoform separation would support a process of subfunctionalization in which ancestral functions have been partitioned between paralogs. The reported cases would add to the handful of cases of this kind reported in the literature (Altschmied et al. 2002; Yu et al. 2003; Pacheco et al. 2004; Cusack and Wolfe 2007; Hultman et al. 2007; Marshall et al. 2013). Evolutionary analysis may explore how these gene duplicates evolved once ancestral isoforms were uncoupled, and whether this uncoupling affected the evolution of accompanying constitutive exons and/or eventually had an adaptive value. In practical terms, having each human isoform represented by a distinct gene in a target species may facilitate the experimental characterization of each isoform function by, for instance, specific gene knockout experiments or gene expression analysis.
The curated analysis presented here throws light on the general aspects of the complex interplay between GD and AS as repositories of protein diversity, and also represents a guide for bettering our understanding of the role of AS for each specific gene.
Supplementary Material
Supplementary material, appendix, tables S1 and S2, and figures S1–S5 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors acknowledge two anonymous referees for their valuable suggestions and corrections. This work was supported by the National Institute of Health (grant number U41 HG007234) and the Spanish Ministry of Economics and Competiveness (grant number BIO2012-40205).
Literature Cited
- Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105. [DOI] [PubMed] [Google Scholar]
- Altschmied J, et al. 2002. Subfunctionalization of duplicate mitf genes associated with differential degeneration of alternative exons in fish. Genetics 161:259–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemiya CT, et al. 2013. The African coelacanth genome provides insights into tetrapod evolution. Nature 496:311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amores A, Catchen J, Ferrara A, Fontenot Q, Postlethwait JH. 2011. Genome evolution and meiotic maps by massively parallel DNA sequencing: spotted gar, an outgroup for the teleost genome duplication. Genetics 188:799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio S, et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297:1301–1310. [DOI] [PubMed] [Google Scholar]
- Barstead RJ, Kleiman L, Waterston RH. 1991. Cloning, sequencing, and mapping of an alpha-actinin gene from the nematode Caenorhabditis elegans. Cell Motil Cytoskeleton. 20:69–78. [DOI] [PubMed] [Google Scholar]
- Boue S, Letunic I, Bork P. 2003. Alternative splicing and evolution. Bioessays 25:1031–1034. [DOI] [PubMed] [Google Scholar]
- Chen Y-C, Lin W-H, Tzeng D-W, Chow W-Y. 2006. The mutually exclusive flip and flop exons of AMPA receptor genes were derived from an intragenic duplication in the vertebrate lineage. J Mol Evol. 62:121–131. [DOI] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. 2008. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet. 9:938–950. [DOI] [PubMed] [Google Scholar]
- Cusack BP, Wolfe KH. 2007. When gene marriages don’t work out: divorce by subfunctionalization. Trends Genet. 23:270–272. [DOI] [PubMed] [Google Scholar]
- De Melker AA, et al. 1997. The A and B variants of the alpha 3 integrin subunit: tissue distribution and functional characterization. Lab Invest. 76:547-563. [PubMed] [Google Scholar]
- Dutertre M, et al. 2010. Exon-based clustering of murine breast tumor transcriptomes reveals alternative exons whose expression is associated with metastasis. Cancer Res. 70:896–905. [DOI] [PubMed] [Google Scholar]
- Ezkurdia I, et al. 2012. Comparative proteomics reveals a significant bias toward alternative protein isoforms with conserved structure and function. Mol Biol Evol. 29:2265–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris JS. 1977. Phylogenetic analysis under Dollo’s Law. Syst Biol. 26:77-88. [Google Scholar]
- Faulkner G, et al. (1999). ZASP: a new Z-band alternatively spliced PDZ-motif protein. J Cell Biol. 146:465-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, et al. 2013. Ensembl 2013. Nucleic Acids Res. 41:D48–D55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushi JI, Makagiansar IT, Stallcup WB. 2004. NG2 proteoglycan promotes endothelial cell motility and angiogenesis via engagement of galectin-3 and α3β1 integrin. Mol Cell Biol. 15:3580-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazina EV, et al. 2010. Differential expression of exon 5 splice variants of sodium channel alpha subunit mRNAs in the developing mouse brain. Neuroscience 166:195–200. [DOI] [PubMed] [Google Scholar]
- Gloria-Bottini F, et al. 2007. Phosphoglucomutase genetic polymorphism and body mass. Am J Med Sci. 334:421–425. [DOI] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 27:221–224. [DOI] [PubMed] [Google Scholar]
- Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321. [DOI] [PubMed] [Google Scholar]
- Gunning PW, Schevzov G, Kee AJ, Hardeman EC. 2005. Tropomyosin isoforms: divining rods for actin cytoskeleton function. Trends Cell Biol. 15:333-341. [DOI] [PubMed] [Google Scholar]
- Günzel D, et al. 2009. Claudin-10 exists in six alternatively spliced isoforms that exhibit distinct localization and function. J Cell Sci. 122:1507-1517. [DOI] [PubMed] [Google Scholar]
- Gupta S, et al. 1996. Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J. 15:2760–2770. [PMC free article] [PubMed] [Google Scholar]
- Honoré B. 2009. The rapidly expanding CREC protein family: members, localization, function, and role in disease. Bioessays 31:262–277. [DOI] [PubMed] [Google Scholar]
- Howe K, et al. 2013. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496:498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultman KA, Bahary N, Zon LI, Johnson SL. 2007. Gene duplication of the zebrafish kit ligand and partitioning of melanocyte development functions to kit ligand a. PLoS Genet. 3:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H, Kondrashov F. 2010. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 11:97–108. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khananshvili D. 2013. The SLC8 gene family of sodium–calcium exchangers (NCX)–structure, function, and regulation in health and disease. Mol Aspects Med. 34:220-235. [DOI] [PubMed] [Google Scholar]
- Kojima T, et al. 2011. Downregulation of tight junction-associated MARVEL protein marvelD3 during epithelial-mesenchymal transition in human pancreatic cancer cells. Exp Cell Res. 317:2288–2298. [DOI] [PubMed] [Google Scholar]
- Kopelman NM, Lancet D, Yanai I. 2005. Alternative splicing and gene duplication are inversely correlated evolutionary mechanisms. Nat Genet. 37:588–589. [DOI] [PubMed] [Google Scholar]
- Kumar S, Hedges SB. 2011. TimeTree2: species divergence times on the iPhone. Bioinformatics 27:2023–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert MJ, Cochran WO, Wilde BM, Olsen KG, Cooper CD. 2015. Evidence for widespread subfunctionalization of splice forms in vertebrate genomes. Genome Res. 25:624–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscombe D, Andrade A, Allen SE. 2013. Alternative splicing: functional diversity among voltage-gated calcium channels and behavioral consequences. Biochim Biophys Acta. 1828:1522-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Taylor DW, Taylor KA. 2004. A 3-D reconstruction of smooth muscle alpha-actinin by CryoEm reveals two different conformations at the actin-binding region. J Mol Biol. 338:115–125. [DOI] [PubMed] [Google Scholar]
- Liu X, et al. 2011. Specific regulation of NRG1 isoform expression by neuronal activity. J Neurosci. 31:8491-8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall AN, Montealegre MC, Jiménez-López C, Lorenz MC, van Hoof A. 2013. Alternative splicing and subfunctionalization generates functional diversity in fungal proteomes. PLoS Genet. 9:e1003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek B, Lee C. 2002. A genomic view of alternative splicing. Nat Genet. 30:13–19. [DOI] [PubMed] [Google Scholar]
- Mudge JM, et al. 2011. The origins, evolution, and functional potential of alternative splicing in vertebrates. Mol Biol Evol. 28:2949–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen SK, et al. 2004. Insights into the molecular basis for fibroblast growth factor receptor autoinhibition and ligand-binding promiscuity. Proc Natl Acad Sci U S A. 101:935–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco TR, et al. 2004. Diversity of vertebrate splicing factor U2AF35: identification of alternatively spliced U2AF1 mRNAS. J Biol Chem. 279:27039–27049. [DOI] [PubMed] [Google Scholar]
- Pan Q, et al. 2005. Alternative splicing of conserved exons is frequently species-specific in human and mouse. Trends Genet. 21:73–77. [DOI] [PubMed] [Google Scholar]
- Partin KM, Fleck MW, Mayer ML. 1996. AMPA receptor flip/flop mutants affecting deactivation, desensitization, and modulation by cyclothiazide, aniracetam, and thiocyanate. J Neurosci. 16:6634–6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, et al. 2009. The consensus coding sequence (CCDS) project: identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 19:1316–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux J, Robinson-Rechavi M. 2011. Age-dependent gain of alternative splice forms and biased duplication explain the relation between splicing and duplication. Genome Res. 21:357–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Pulido L, Martín-Belmonte F, Valencia A, Alonso MA. 2002. MARVEL: a conserved domain involved in membrane apposition events. Trends Biochem Sci. 27:599–601. [DOI] [PubMed] [Google Scholar]
- Seki E, Brenner DA, Karin M. 2012. A liver full of JNK: signaling in regulation of cell function and disease pathogenesis, and clinical approaches. Gastroenterology 143:307–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah K, Shokat KM. 2002. A chemical genetic screen for direct v-Src substrates reveals ordered assembly of a retrograde signaling pathway. Chem Biol. 9:35–47. [DOI] [PubMed] [Google Scholar]
- Smith CW, Valcárcel J. 2000. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci. 25:381–388. [DOI] [PubMed] [Google Scholar]
- Smith JJ, et al. 2013. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet. 45:415–421, 421e1–e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steed E, Rodrigues NTL, Balda MS, Matter K. 2009. Identification of MarvelD3 as a tight junction-associated transmembrane protein of the occludin family. 14:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Gu X. 2012. Revisit on the evolutionary relationship between alternative splicing and gene duplication. Gene 504:102–106. [DOI] [PubMed] [Google Scholar]
- Su Z, Wang J, Yu J, Huang X, Gu X. 2006. Evolution of alternative splicing after gene duplication. Genome Res. 16:182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera D, Vogel C, Orozco M, Teichmann SA, de la Cruz X. 2007. The (in)dependence of alternative splicing and gene duplication. PLoS Comput Biol. 3:e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegtmeyer LC, et al. 2014. Multiple phenotypes in phosphoglucomutase 1 deficiency. N Engl J Med. 370:533–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanaraj TA, Clark F, Muilu J. 2003. Conservation of human alternative splice events in mouse. 31:2544–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukumo Y, Tsukahara S, Saito S, Tsuruo T, Tomida A. 2009. A novel endoplasmic reticulum export signal: proline at the +2-position from the signal peptide cleavage site. J Biol Chem. 284:27500–27510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilella AJ, et al. 2009. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 19:327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorum H, Hager H, Christensen BM, Nielsen S, Honoré B. 1999. Human calumenin localizes to the secretory pathway and is secreted to the medium. Exp Cell Res. 248:473–481. [DOI] [PubMed] [Google Scholar]
- Waites GT, et al. 1992. Mutually exclusive splicing of calcium-binding domain exons in chick alpha-actinin. J Biol Chem. 267:6263-6271. [PubMed] [Google Scholar]
- Wajih N, Sane DC, Hutson SM, Wallin R. 2004. The inhibitory effect of calumenin on the vitamin K-dependent gamma-carboxylation system. Characterization of the system in normal and warfarin-resistant rats. J Biol Chem. 279:25276–25283. [DOI] [PubMed] [Google Scholar]
- Wang Q, et al. 2012. The intracellular transport and secretion of calumenin-1/2 in living cells. PLoS One 7:e35344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Guo C, Shan H, Kong H. 2012. Divergence of duplicate genes in exon-intron structure. Proc Natl Acad Sci U S A. 109:1187–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Van Nostrand E, Holste D, Poggio T, Burge CB. 2005. Identification and analysis of alternative splicing events conserved in human and mouse. Proc Natl Acad Sci U S A. 102:2850–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W-P, Brenner S, Venkatesh B. 2003. Duplication, degeneration and subfunctionalization of the nested synapsin-Timp genes in Fugu. Trends Genet. 19:180–183. [DOI] [PubMed] [Google Scholar]
- Zhang PG, Huang SZ, Pin A, Adams KL. 2010. Extensive divergence in alternative splicing patterns after gene and genome duplication during the evolutionary history of Arabidopsis . Mol Biol Evol. 27:1686–1697 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.