Abstract
Eukaryotic genomes are colonized by various transposons including short interspersed elements (SINEs). The 5′ region (head) of the majority of SINEs is derived from one of the three types of RNA genes—7SL RNA, transfer RNA (tRNA), or 5S ribosomal RNA (rRNA)—and the internal promoter inside the head promotes the transcription of the entire SINEs. Here I report a new group of SINEs whose heads originate from either the U1 or U2 small nuclear RNA gene. These SINEs, named SINEU, are distributed among crocodilians and classified into three families. The structures of the SINEU-1 subfamilies indicate the recurrent addition of a U1- or U2-derived sequence onto the 5′ end of SINEU-1 elements. SINEU-1 and SINEU-3 are ancient and shared among alligators, crocodiles, and gharials, while SINEU-2 is absent in the alligator genome. SINEU-2 is the only SINE family that was active after the split of crocodiles and gharials. All SINEU families, especially SINEU-3, are preferentially inserted into a family of Mariner DNA transposon, Mariner-N4_AMi. A group of Tx1 non-long terminal repeat retrotransposons designated Tx1-Mar also show target preference for Mariner-N4_AMi, indicating that SINEU was mobilized by Tx1-Mar.
Keywords: SINEU, U1, U2, crocodilians, gharial, alligator, crocodile, transposable elements, Mariner-N4_AMi, Tx1, Tx1-Mar
Introduction
Short interspersed elements (SINEs) are abundant components of eukaryotic genomes. The transposition of SINEs is dependent on the machinery of their counterpart non-long terminal repeat (non-LTR) retrotransposons (also called long interspersed elements, or LINEs) (Kajikawa and Okada 2002; Dewannieux et al. 2003). Based on the origin of their 5′ regions, called heads, most SINEs are classified into three groups. SINE1 has a head derived from 7SL RNA genes and is represented by the primate Alu family (Ullu and Tschudi 1984; Kriegs et al. 2007). SINE1 is distributed only among euarchontoglires (primates, flying lemurs, tree shrews, rodents, and rabbits). SINE2 contains tRNA gene-derived head and is widely distributed among eukaryotes (Sakamoto and Okada 1985; Jurka et al. 2005). The head of SINE3 is derived from 5S rRNA genes (Kapitonov and Jurka 2003). SINE3 is distributed among vertebrates and insects.
Genes of 7SL RNA, tRNA, and 5S rRNA contain internal promoters of RNA polymerase III inside their RNA-encoding regions (Paule and White 2000). In the transposition of SINEs, an RNA transcribed from a SINE locus is reverse transcribed and inserted into a new locus by the mechanism of non-LTR retrotransposons (Kajikawa and Okada 2002; Dewannieux et al. 2003). During the transposition of a non-LTR retrotransposon, a short DNA sequence is often duplicated at both ends of the retrotransposon, creating what are called target site duplications (TSDs). In other cases, several nucleotides are deleted (target site truncation, TST) or the target DNA may be unaltered at all. If RNA sequences containing internal promoters are retrotransposed, they can propagate themselves efficiently. Sequences derived from RNA genes are occasionally added at the 5′ ends of SINEs. For example, a tRNA-derived sequence was added to a SINE with a 7SL RNA-derived head (Nishihara et al. 2002), and a 5S rRNA-derived sequence was added to a SINE with a tRNA-derived head (Nishihara et al. 2006).
Small nuclear RNA (snRNA) is another group of small RNA. U1, U2, U4, U5, and U6 snRNA genes encode the components of the spliceosome, a large ribonucleoprotein complex that catalyzes intron splicing (Valadkhan 2005). U1 snRNA base pairs to the 5′ splice site and U2 pairs with the intron branch point sequence. U1, U2, U4, and U5 snRNAs are transcribed by RNA polymerase II, while U6 snRNA is transcribed by RNA polymerase III (Will and Luhrmann 2001); however, the snRNA genes have similar promoters. All vertebrate snRNA gene promoters contain a distal sequence element, which is located 220 bp upstream of the initiation site and functions as an enhancer, as well as a proximal sequence element, which is a core promoter element located 60 bp upstream of the initiation site. No internal promoter for snRNA genes has been reported.
With some exceptions, SINE and its partner non-LTR retrotransposon that mobilizes the SINE contain short 3′ sequences (tails) similar to each other (Kajikawa and Okada 2002). Non-LTR retrotransposons are classified into more than 30 “clades” (Kapitonov et al. 2009). The Tx1 clade is a group of non-LTR retrotransposons found in animals, and many retrotransposons belonging to the Tx1 clade show target sequence preference (Kojima and Fujiwara 2004). Tx1 from Xenopus laevis is inserted into Tx1D, a nonautonomous piggyBac DNA transposon family (Christensen et al. 2000). Keno is specifically inserted into U2 snRNA genes and has been found in vertebrates, lancelet (Tx1-5_BF), and cnidarians (Tx1-1_HM) (Kojima and Fujiwara 2004; Kapitonov and Jurka 2009). Kibi and Koshi are found in teleost fishes and are specifically inserted into (TC)n and (TTC)n microsatellites, respectively (Kojima and Fujiwara 2004). The target choice for non-LTR retrotransposons is mainly determined by the target specificity of their encoding endonuclease because it cleaves DNA at the initial step of retrotransposition (Takahashi and Fujiwara 2002).
Recently four crocodilian genomes were sequenced (Wan et al. 2013; Green et al. 2014). Among the four sequenced species, Alligator mississippiensis (American alligator) and A. sinensis (Chinese alligator) belong to Alligatoridae, Crocodylus porosus (saltwater crocodile) belongs to Crocodylidae, and Gavialis gangeticus (Indian gharial) belongs to Gavialidae. Crocodylidae and Gavialidae form a clade called “Longirostres” (Green et al. 2014). Crocodilian genomes contain a large amount of old transposon remnants. Among them, CR1 non-LTR retrotransposons and endogenous retroviruses were studied extensively (Chong et al. 2014; Suh et al. 2014). Crocodilian genomes contain a small amount of SINEs with tRNA-derived heads.
Here, a new group of SINEs whose heads originate from either the U1 or U2 snRNA gene, designated SINEU, is described. SINEUs are found only from crocodilians. SINEUs are classified into three groups based on their structures. SINEU-1 and SINEU-2 show the recurrent addition of U1- or U2-derived sequences onto their 5′ termini. SINEU-3 resembles an internally deleted U2 snRNA gene and is almost exclusively inserted into a family of Mariner-type DNA transposon, Mariner-N4_AMi, while the other two SINEU families also show weak target preference for this transposon family. This shared target preference suggests that these families’ mobilization is dependent on the transposition machinery of Tx1-type non-LTR retrotransposons. On the basis of these observations, how SINEU elements are originated, transcribed, and transposed are discussed.
Materials and Methods
Characterization of SINEU Families/Subfamilies and Tx1 Families
Genome sequences of three species of crocodilians, A. mississippiensis (American alligator), C. porosus (saltwater crocodile), and G. gangeticus (Indian gharial), were generated by International Crocodilian Genomes Working Group (Green et al. 2014). Transposons including SINEU elements and Tx1 non-LTR retrotransposons were detected by systematic screening of new repetitive sequences, described elsewhere (Green et al. 2014). The classification of crocodilian Tx1 non-LTR retrotransposon families was confirmed with the aid of RTClass1 (Kapitonov et al. 2009). Sequences of all known Tx1 non-LTR retrotransposons were obtained from Repbase (Jurka et al. 2005).
Because of the ancient concurrent activity of closely related SINEU-1 subfamilies, it was difficult to classify subfamilies just based on sequence similarity. Thus, SINEU-1 was classified first by the structural features, such as the presence of a U1- or U2-derived head, and later subclassified based on sequence similarity. BLASTclust in the NCBI BLAST package (http://www.ncbi.nlm.nih.gov/BLAST/, last accessed June 10, 2015) was used to distinguish structurally similar subfamilies.
Target Analysis
Flanking 100-bp or 1,000-bp sequences at both ends of Tx1 non-LTR retrotransposons were analyzed in order to see their target preference or specificity. Target preference for microsatellites was determined manually. Target preference for multicopy genes or transposons was determined by performing Censor (Kohany et al. 2006) against Repbase. If a certain type of sequence is seen in more than 20% of the flanking sequences of a certain non-LTR retrotransposon family, this family is considered target specific.
Copy Number Estimation
SINEU copy numbers are estimated for each species individually based on the results of Censor search against the total set of crocodilian repeat sequences (Green et al. 2014). Hits shorter than 50 bp were excluded. Hits more similar to U1 or U2 snRNA sequences than to the SINEU consensus sequences were also excluded. SINEU-1 subfamilies were not distinguished because they are very old and abundant; instead, the total numbers of all SINEU-1 subfamilies were estimated.
Phylogenetic Analysis
The reverse transcriptase (RT) domain sequences of crocodilian Tx1 non-LTR retrotransposons and Tx1 non-LTR retrotransposons whose target specificity had been characterized were aligned with the aid of MAFFT (Katoh et al. 2005). They were found to be around 530 aa in length and include the motifs 0–7 for RT domains of non-LTR retrotransposons (Malik et al. 1999). The aligned sequences are available on request. Maximum-likelihood trees were constructed by PhyML (Guindon et al. 2010) with bootstrap values (1,000 replicates) using two different amino acid substitution models: RtREV and LG. ProtTest (Abascal et al. 2005) was used to choose the appropriate substitution model and the LG model is the most appropriate based on both the Akaike Information Criteria and the Bayesian Information Criteria. The phylogenetic trees were drawn with the aid of FigTree 1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/, last accessed June 10, 2015).
Results
SINEU with U1 or U2 snRNA Gene-Derived Heads
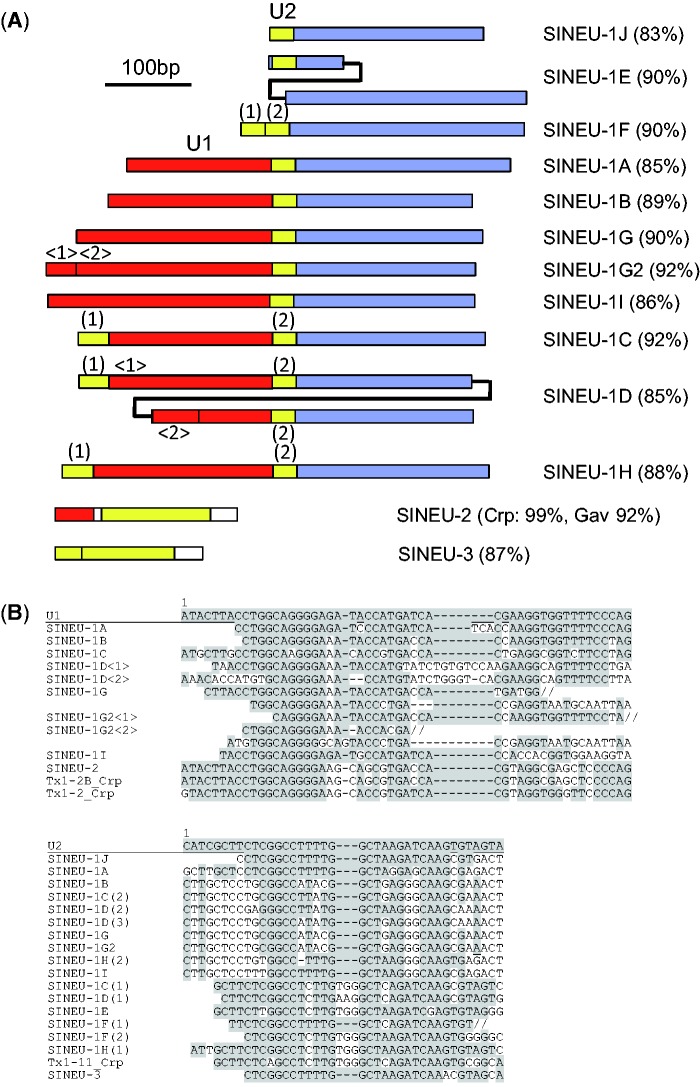
From the characterization of transposable elements from the genomes of three species of crocodilians, A. mississippiensis, C. porosus, and G. gangeticus (Green et al. 2014), SINE families whose 5′ regions originated from either U1 or U2 snRNA genes were characterized. They are designated as SINEU and classified into three families (SINEU-1, SINEU-2, and SINEU-3) based on their structures (fig. 1). No SINEU families were detected outside of crocodilians. SINEU-1 was further classified into 11 subfamilies based on their structures and sequences. The 5′ ends of SINEU elements do not always correspond to the 5′ end of a U1 or U2 snRNA sequence; however, this might be an artifact from constructing consensus sequences from divergent copies. All groups of SINEU have 3′ polyA tail.
Fig. 1.—
Structures of SINEU families/subfamilies. (A) Schematic structures of SINEU families/subfamilies. The regions similar to U1 snRNA gene are colored in red, while those similar to U2 gene are in yellow. The 3′ regions shared among all SINEU-1 subfamilies are in blue. Average identity of each copy to the respective consensus is shown in parentheses. Numbers in parentheses are shown to distinguish multiple U1- or U2-derived sequences and correspond to those shown in (B). (B) Alignment of U1- and U2-derived sequences of SINEU subfamilies and Tx1 families. 5′ sequences of each SINEU subfamily are aligned with U1 and U2 snRNA genes. Nucleotides identical to either U1 or U2 gene are shaded. Double slashes indicate that the following sequences are shown in the next line.
SINEU-1
All 11 SINEU-1 subfamilies share similar 3′ sequences. It is possible that the 3′ half of SINEU-1 originated from a non-LTR retrotransposon that contributed to the mobilization of SINEU-1 copies. However, no non-LTR retrotransposon family on Repbase shows significant similarity to the 3′ half of SINEU-1.
The simplest structure is seen in SINEU-1J. It is the oldest SINEU-1 sequence; the average identity of each copy to the consensus is ∼83%. It contains a 5′-terminal short U2-derived sequence (fig. 1). The subfamilies other than SINEU-1J were generated by sequential additions of U1- or U2-derived sequences to SINEU-1J and/or partial duplications of SINEU-1J. A partial duplication of SINEU-1J could have occurred to generate SINEU-1E. The 5′ addition of a U2-derived sequence could have generated SINEU-1F, while the 5′ addition of a U1-derived sequence could have generated SINEU-1A, B, G, and I. SINEU-1C and SINEU-1H were likely generated by additions of U2-derived sequences onto a SINEU-1A/B/G/I-type ancestor. The 5′ addition of a U1-derived sequence onto SINEU-1G could have generated SINEU-1G2. Finally, SINEU-1D appears to be the fusion of SINEU-1C and SINEU-1B, or it could be the partially duplicated sequence of SINEU-1C. Partial duplication is also seen inside U1- or U2-derived sequences; the clearest example is SINEU-1G (fig. 1B). The U1-derived sequence of SINEU-1G is partially duplicated and TGG trinucleotides likely contributed to the duplication.
SINEU-1C and SINEU-1G2 were the most recently active among SINEU-1 subfamilies; each copy of these subfamilies is ∼92% identical to its respective consensus. In general, a family with more complex structure is newer. The exception is SINEU-1D; the average identity of copies of SINEU-1D to the consensus is ∼85%. This indicates that SINEU-1D had a short life span; it must have become inactive just after its generation. In the history of SINEU-1 subfamilies, at least two events of 5′ addition of a U1 sequence (the birth of SINEU-1A/B/G/I and that of SINEU-1G2) and at least three events of 5′ addition of a U2 sequence (the birth of SINEU-1J, SINEU-1F, and SINEU-1C/H) have occurred.
Orthologous sequences of alligator and crocodile/gharial are ∼93% identical on average and orthologous sequences of crocodile and gharial are 95.7% identical (Green et al. 2014). These values correspond to ∼96.5% and ∼97.8% identity to their ancestral (consensus) sequences, respectively. Therefore even the most recently active subfamilies of SINEU-1 ceased transposing before the split between alligator and crocodile/gharial, which was 80–100 Ma. Because of their relatively old ages, it was difficult to detect TSDs; however ∼10-bp TSDs were occasionally observed (data not shown).
SINEU-2 and SINEU-3
SINEU-2 is composed of 5′ U1 and 3′ U2 sequences (fig. 1). It is found in crocodile and gharial but not in alligator, indicating its recent expansion. The consensus sequences of SINEU-2 from the 2 species, SINEU-2_Crp from crocodile and SINEU-2_Gav from gharial, are ∼94% identical. SINEU-2 remains active or had been active until quite recently in the crocodile lineage, but became inactive at some point in the gharial lineage; many copies of SINEU-2 from crocodile are >99% identical to the SINEU-2_Crp consensus but copies from gharial are only ∼92% identical to the SINEU-2_Gav consensus.
SINEU-2_Crp is the newest SINEU family and thus SINEU-2_Crp insertions were analyzed in detail. SINEU-2_Crp was further classified into four subfamilies (SINEU-2A_Crp to SINEU-2D_Crp) based on sequence variations (supplementary fig. S1, Supplementary Material online). SINEU-2D_Crp shares the structural features with SINEU-2_Gav. SINEU-2A_Crp and SINEU-2B_Crp have the replacement of 53CAGGTG58 by TA, corresponding to the junction between the U1- and U2-derived sequences. SINEU-2B_Crp and SINEU-2C_Crp have the replacement of 20AGCACCGTGACCACGTAGGCGGGCTC45 by GACACCATGATCAATCAGTTGGTTTT inside of the U1-derived head, which is likely due to the recombination of the U1-derived head of SINEU-2 and another U1 sequence at either the DNA or RNA level. These replacements are not seen in SINEU-2_Gav, and thus they were generated after the speciation between crocodiles and gharials.
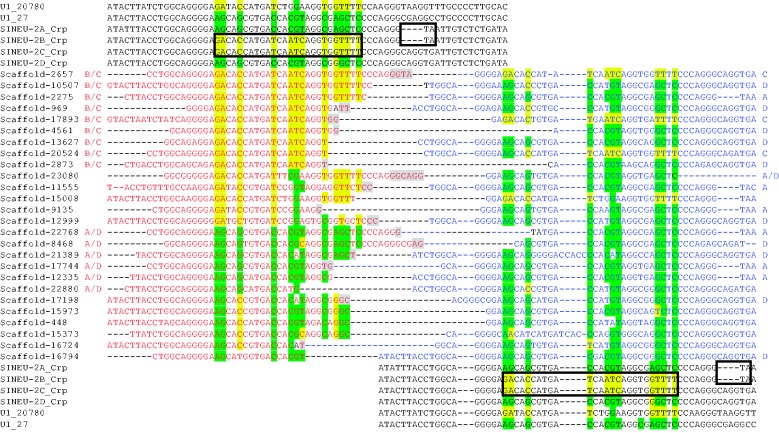
By the comparison of orthologous loci of crocodile and gharial, SINEU-2 insertions were found in the crocodile genome at whose orthologous loci in the gharial genome SINEU-2 are absent (fig. 2). Gharial-specific SINEU-2 insertions were also detected (supplementary fig. S2, Supplementary Material online). Both types of target site alterations (TSD and TST) were observed aside from the cases where no nucleotide was altered upon integration.
Fig. 2.—
Crocodile lineage-specific insertions of SINEU-2 with 5′ extensions. Crp represents the crocodile scaffold sequence and Gav represents the gharial scaffold sequence. SINEU-2 sequences are colored in green with their 5′ extension in blue, and putative TSD sequences characterized based on the corresponding “empty” sites in gharial are in red. (A) SINEU-2A with 5′ extension. Intron sequence for AKHW01109664, which encodes a protein annotated as amisp024556, from alligator is shown. Scaffold-6053 is a crocodile sequence that has the sequence similar to the 5′ extension sequences of SINEU-2A. (B) SINEU-2B with 5′ extension. The 5′ extension sequences show similarity to the consensus of Mariner-N4_AMi.
The SINEU-3 family seems to have originated by internal ∼50 bp deletion of a U2 snRNA gene, although its 3′, ∼40 bp nucleotides are not similar to any part of a U2 gene. The 5′, 32 bp and the middle 83 bp sequences correspond to fragments of U2 gene. This structure is analogous to monomeric Alu (FAM, FLAM, FRAM), which is an internally deleted derivative of 7SL RNA (Quentin 1992; Kriegs et al. 2007).
Copy Numbers of SINEU Families in Crocodilians
Consistent with the ancient activities of SINEU-1 and SINEU-3 families, their copy numbers in the three crocodilian genomes are comparable—around 7,700 and 1,600, respectively. In contrast, the copy number of SINEU-2 is 1,058 in the crocodile genome, while it is only 137 in the gharial genome. Meanwhile, the alligator genome completely lacks SINEU-2 insertions. The sum of SINEU families occupies ∼2 MB of the crocodilian genomes, while the total occupancy of all SINE families is around 15 MB in these genomes (Green et al. 2014). Excluding SINEU, other SINE families are very old in the crocodilian genomes.
Target Preference of SINEU Families
SINEU-3 copies are frequently inserted into Mariner-N4_AMi. Although Mariner-N4_AMi is old and disrupted, the original TSDs for SINEU-3 are expected to have been AGGTTCCCATCAGCAT in many cases. Tandem SINEU-3 insertions are often observed sandwiching a TSD. Although these tandem SINEU-3 copies were reported as SINEU-3B in our previous analysis (Green et al. 2014), they are more likely generated by a sequential insertion of two SINEU-3 copies and should not be considered as a subfamily of SINEU-3. Mariner-N4_AMi is quite abundant in the crocodilian genomes. The copies of Mariner-N4_AMi are generally 86–89% identical to the consensus, but many older insertions are also present. There are 11,801 copies which are >80% identical to the Mariner-N4_AMi consensus in the crocodile genome.
To examine the target specificity of SINEU-3, 347 nearly full-length (>90% in length) SINEU-3 insertions were extracted and their flanking sequences were analyzed. Among them, 313 (90%) are flanked by Mariner-N4_AMi at least on one side. Among 1,548 nearly full-length SINEU-1 insertions, 358 (23%) are flanked by Mariner-N4_AMi. Among 297 nearly full-length SINEU-2 insertions, 39 (13%) are flanked by Mariner-N4_AMi. The target preference of SINEU-1 and SINEU-2 families is much weaker than that of the SINEU-3 family. Given that SINEU copies are often inserted in tandem, which may cause flanking Mariner-N4_AMi sequences to fail to be detected, the presence or absence of Mariner-N4_AMi sequences in the flanking 1,000-bp sequences on both sides of SINEU insertions was examined using Censor (supplementary fig. S3, Supplementary Material online), which confirmed the target preference of SINEU elements.
Because the divergence time of Mariner-N4_AMi (86–89% identity to the consensus) and SINEU-3 is similar (89% identity to the consensus), the possibility that SINEU-3 was amplified hitchhiking Mariner-N4_AMi is raised. To examine this possibility, the 5′-truncated SINEU-3 copies, which are likely to have been inserted independently, were analyzed. Because SINEU-3 is similar to an internally deleted U2 snRNA gene, Censor search with full-length SINEU-3 sequence as a query hits many U2 snRNA genes. To exclude these U2 genes, SINEU-3 copies starting between 11 and 25 and ending between 161 and 169 are used. They expand both regions of the 5′, 32-bp and the middle 83-bp U2-derived sequences, as well as the 3′ sequence unique to SINEU-3. Among 88 copies, 77 copies are flanked with Mariner-N4_AMi at least at one side. Nine of the remaining 11 copies are flanked with copies of other families of SINEU or Tx1 non-LTR retrotransposon that are flanked with Mariner-N4_AMi. The remaining 2 copies are flanked with Tx1-6_AMi and they are not directly associated with Mariner-N4_AMi. In summary, only 2 of the 88 truncated SINEU-3 copies are not associated with Mariner-N4_AMi. It indicates independent insertion of SINEU-3 copies into Mariner-N4_AMi.
Target Preference of Tx1 Families from Crocodilians
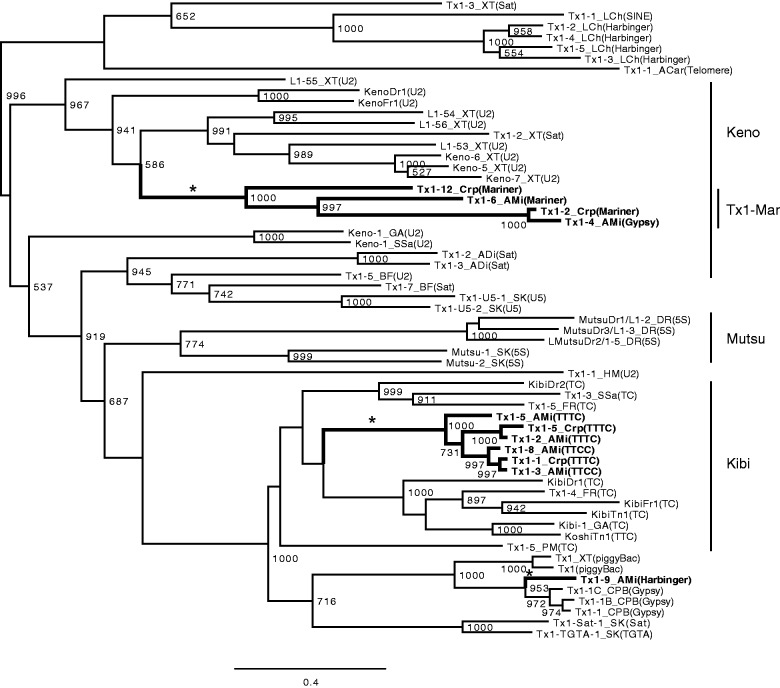
Some families of Tx1 non-LTR retrotransposons from crocodilians are also preferentially inserted into Mariner-N4_AMi. On the basis of the phylogeny of RT sequences, crocodilian Tx1 families are classified into three groups (fig. 3, asterisks). One group is coclustered with Keno. Most crocodilian Tx1 families in this group are preferentially inserted into Mariner-N4_AMi and its related transposons (supplementary figs. S3 and S4, Supplementary Material online). This Tx1 group was designated “Tx1-Mar.” Exceptions are Tx1-8_Crp and Tx1-4_AMi, preferentially inserted into specific groups of LTR retrotransposons.
Fig. 3.—
Phylogeny of Tx1 non-LTR retrotransposons. Only Tx1 families whose targets are known were analyzed with crocodilian Tx1 families. Bootstrap values of 1,000 replicates are shown at nodes if they are over 500. Names of crocodilian families are in bold. Three lineages of crocodilian Tx1 families are indicated by asterisks. Target sequences are shown in parentheses after names. The transposon superfamilies are shown when the targets are transposon, while the repeat units are shown when the targets are microsatellites. U2, U2 snRNA; Sat, satellites; U5, U5 snRNA; 5S, 5S rRNA.
The second group is composed of Tx1-1_Crp, Tx1-5_Crp, Tx1-6_Crp, Tx1-1_Gav, Tx1-2_AMi, Tx1-3_AMi, Tx1-5_AMi, and Tx1-8_AMi. They are preferentially inserted into (TTCC)n or (TTTC)n microsatellites (supplementary fig. S5, Supplementary Material online). It is consistent with their phylogenetic positions close to the Kibi and Koshi families that are specifically inserted into (TC)n and (TTC)n microsatellites, respectively (Kojima and Fujiwara 2004).
The last group includes only Tx1-9_AMi. Tx1-9_AMi is preferentially inserted at ∼300 bp downstream from the 5′ ends of three families of Harbinger DNA transposons (supplementary fig. S4, Supplementary Material online). The 5′, ∼400-bp sequences of these three transposons are ∼90% identical to one another.
Tx1-Mar Families Likely trans-mobilize SINEU Families
The shared target preference of Tx1-Mar and SINEU strongly suggest that SINEU families are mobilized by the machinery of Tx1-Mar non-LTR retrotransposons. The average identity of copies of Tx1-Mar families to the consensus is 85–87%. The average identity of copies of SINEU-1 and SINEU-3 is 85–91%. Although most SINEU-2 copies in the crocodile genome are > 90% identical to the consensus, a minority (9%) are 85–90% identical. This implies concurrent transposition activities of Tx1-Mar families and SINEU families.
Another indication of the contribution of Tx1-Mar families to the mobilization of SINEU families is that some Tx1-Mar families have U1- or U2-originated sequences at their 5′ ends. Tx1-2_Crp, Tx1-2B_Crp and Tx1-4_AMi have a U1-originated head, which is quite similar to that of SINEU-2, while Tx1-11_Crp has a U2 head almost identical to those of SINEU-1C, SINEU-1E, and SINEU-1H (fig. 1B).
Inside Mariner-N4_AMi, there are two regions frequently inserted by both Tx1-Mar and SINEU copies. One is around position 240 of Mariner-N4_AMi, and here Tx1-Mar and SINEU copies are inserted in the opposite direction to Mariner-N4_AMi. The other frequently inserted region is around position 830, and here Tx1-Mar and SINEU copies are inserted in the same direction as Mariner-N4_AMi. The sequences around these two regions do not have recognizable similarity. Tx1-10_AMi, Tx1-10_Crp, and Tx1-8_Crp prefer to be inserted around position 830 rather than around position 240, while other families show preference to be inserted around position 240 (supplementary fig. S6, Supplementary Material online). Interestingly, SINEU-3 insertions are never found around position 830, while SINEU-1 and SINEU-2 insertions are often seen there (supplementary fig. S6, Supplementary Material online). This indicates that different Tx1-Mar families contributed to the mobilization of SINEU-1/2 and SINEU-3.
5′ Extension of SINEU-2 Copies Indicates their Birth and Transcription Mechanism
As the nature of sequences transposed by the non-LTR retrotransposon machinery (Kojima 2010), many SINEU-2 copies are 5′-truncated. The frequent 5′-truncation may cause the difference in the 5′ ends among consensus sequences of SINEU-1 subfamilies. In the case of SINEU-2, some copies include the 5′ end of U1 snRNA (supplementary fig. S1, Supplementary Material online). The 5′ flanking sequences of some apparent full-length SINEU-2 copies are similar to one another, and they can be classified into several groups. Although the possibility that these flanking sequences represent the true 5′ termini of SINEU-2 cannot be excluded at present, these shared flanking sequences are called “5′ extension” hereafter. These 5′ extension sequences may have been generated by different mechanisms such as template jump (Bibillo and Eickbush 2004) or 5′-transduction (Damert et al. 2009).
Twenty-six SINEU-2 copies have 5′ extensions similar to U1 snRNA genes (fig. 4). The 5′ extension sequences show some nucleotide variations. The variations of SINEU-2 head sequences and CpG decay can explain most variations of 5′ extension sequences, but it does not exclude the possibility that they are derived from variations of U2 snRNA genes. The downstream SINEU-2 sequences also show variations and belong to different subfamilies. Even when the 5′ extension sequence corresponds to one subfamily of SINEU-2, the subfamilies of the 5′ extension and of the downstream SINEU-2 are often different; for example, a SINEU-2A copy in the scaffold-2275 has a 5′ extension identical to the heads of SINEU-2B and C. Thus, template slippage and partial duplication can be excluded as the main mechanism of this extension. The positions of 3′ truncation of the 5′ extension sequences are different by at most 22 nucleotides. Short sequence homology between the extension sequences and SINEU-2 sequences is occasionally observed at the junctions. These observations clearly show that SINEU-2 heads were generated through multiple events of 5′ sequence addition onto SINEU-2 copies, not by the retrotransposition of a single master copy.
Fig. 4.—
Alignment of SINEU-2_Crp copies with the 5′ extension sequences similar to U1 snRNA genes. The consensus sequences of SINEU-2_Crp subfamilies and also two representative U1 snRNA gene sequences (in scaffold-20780 and scaffold-27) are shown. The regions distinguishing different SINEU-2_Crp subfamilies are boxed. The 5′ extension sequences are colored in red and the downstream SINEU-2_Crp sequences are in blue. Nucleotides different among SINEU-2_Crp subfamilies are highlighted by either green or yellow. Nucleotides that can be aligned as either the 5′ extension sequences or the downstream SINEU-2_Crp sequences are shaded. If the sequence corresponds to a specific SINEU-2_Crp subfamily or subfamilies, it is indicated.
Some 5′ extension sequences were dissimilar from U1 genes and could be classified into two groups (fig. 2). They belong to the SINEU-2A_Crp and SINEU-2B_Crp subfamilies, and have 31 and 42 nucleotide-long extensions, respectively. The 5′ extension sequence of SINEU-2A_Crp is similar to the introns of a multicopy gene family encoding a RING zinc finger and a B-Box zinc finger from various amniotes. Three copies of this gene family from the alligator genome include a copy of SINEU-1B, just downstream from a sequence similar to the 5′ extension sequence of SINEU-2A_Crp. A similar sequence was also found in scaffold-6053 from the crocodile genome. SINEU-1B shares a U1-originated sequence with SINEU-2, and so the 5′ extension of SINEU-2A_Crp was likely derived from either template switch during reverse transcription or recombination between SINEU-2 and a copy of SINEU-1B inserted into a copy of the multicopy genes. The 5′ extension of SINEU-2B_Crp shows weak similarity to nucleotides 275-231 of Mariner-N4_AMi, indicating that the master copy for these SINEU-2B_Crp insertions is a SINEU-2B_Crp copy inserted into a Mariner-N4_AMi copy.
Discussion
Retrotransposition of snRNAs and the Birth of SINEU Families
Retrotransposed copies of U1 and U2 snRNA genes were first reported in Van Arsdell et al. (1981). The retrotransposition of snRNAs in three ways has been previously reported in the human genome. Tailless retropseudogenes of snRNA are 3′-truncated at different positions and flanked by TSDs, indicating that they are trans-mobilized by the machinery of L1 (Van Arsdell et al. 1981; Schmitz et al. 2004; Kojima 2010). Buzdin et al. (2002) and Garcia-Perez et al. (2007) reported retrotransposed copies of snRNA-derived sequences followed by fragments of L1, Alu, or processed pseudogenes; they are transposed by template switch during L1 retrotransposition. One and two copies for U1 and U2 snRNA retrotransposition, respectively, were reported in the combination of Alu fragments. Additionally, U1 or U2 snRNA sequences followed by polyA tracts and flanked by TSDs were reported (Garcia-Perez et al. 2007).
Even though the 5′ addition of U1- or U2-derived sequences is quite common for SINEU elements, as observed in the case of SINEU-2_Crp (fig. 4), SINEU families are not the sum of independently generated chimeric retrocopies/processed pseudogenes. First, no non-LTR retrotransposon family that shares the 3′ terminus with SINEU-1 subfamilies could be detected despite the systematic survey of crocodilian repetitive sequences (Green et al. 2014). If SINEU-1 copies had been generated independently by template switch, many non-LTR retrotransposon copies that share the 3′ terminus with SINEU-1 families should be found, because only a minority of non-LTR retrotransposon copies experience template switch. Second, two subfamilies of SINEU-2 having 5′ extension (fig. 2) were found, indicating the presence of at least two master copies for SINEU-2. SINEU-3, which is composed of just two U2 snRNA gene fragments, is unlikely to be a sum of retrocopies because the internal deletion is shared among copies.
Both trans-mobilization and template switch may have contributed to the evolution of SINEU families. Template switch likely contributed to the addition of U1- or U2-originated sequences onto the 5′ ends of SINEU families. It is also observed in the case of 5′ extension for SINEU-2 copies. The common ancestor of SINEU-1 subfamilies, which was likely similar to SINEU-1J, might have originated by a single event of template switch to a U2 snRNA. SINEU-2 may have been generated by template switch to a U1 snRNA from a trans-mobilized U2 snRNA. SINEU-3 may have originated from a trans-mobilized U2 snRNA, followed by an internal deletion, similarly to monomeric Alu elements like FAM and FLAM (Quentin 1992; Kriegs et al. 2007).
The Transcription of SINEU
An open question about SINEU elements is how they are transcribed. A function of SINE heads is to promote transcription. The 7SL RNA, tRNA, and 5S rRNA genes include an internal promoter for RNA polymerase III (Paule and White 2000). U1 and U2 snRNA genes, however, are transcribed by RNA polymerase II, and their promoters are located upstream of transcribed regions (Egloff et al. 2008). OligoT stretch, which works as a pol III terminator (Bogenhagen and Brown 1981), is observed inside of most SINEU families, eliminating the pol III transcription of SINEU. No sequence similar to RNA polymerase I, II, or III promoter was found in any SINEU families. Censor search with SINEU consensus sequences as queries could not detect any sequence that originated from either SINEU elements in the alligator or the gharial cDNA libraries sequenced in the crocodilian genome sequencing project. There are no direct data showing the transcription start site of SINEU. The 5′ ends of SINEU copies may correspond to the transcription start site although the possibility that all copies are truncated cannot be excluded.
The analysis of 5′ extensions of SINEU-2 indicated that some copies of SINEU-2 were transcribed from the 5′ flanking regions. A subfamily of SVA, a composite nonautonomous retrotransposon family found in hominids, is transcribed from the promoter of the MAST2 gene and this subfamily of SVA contains a part of the MAST2 gene sequence (Damert et al. 2009; Hancks et al. 2009). It is possible that SINEU is transcribed from upstream promoters. The master copy of SINEU-2B_Crp was likely inserted in a copy of Mariner-N4_AMi in the opposite direction. The 5′ ends of the extension for SINEU-2B_Crp correspond to position 275 in the consensus sequence of MarinerN-4_AMi. Considering the fact that many SINEU copies are inserted near position 240 of Mariner-N4_AMi in the opposite direction, it is possible that they were also transcribed from the flanking MarinerN-4_AMi sequence. The dependence of transcription on the flanking sequence is observed in the case of R2, a non-LTR retrotransposon family specifically inserted into 28S rRNA genes (Eickbush DG and Eickbush TH 2010). If the target is transcribed, there is no need to include its own promoter. Besides, the cotranscription with target RNA genes can strengthen their target-specific integration (Eickbush et al. 2000; Fujimoto et al. 2004). The dependence of transcription on the flanking sequence may be another strategy to verify the transcription of SINEs.
Is SINEU a New Group of SINEs or a New Group of Retroposons?
When the terms SINEs and LINEs were proposed, little was known about retrotransposons and they were distinguished merely by length. Currently, SINEs are generally considered as retrotransposons (or retroposons) transcribed by RNA polymerase III (Kramerov and Vassetzky 2011). Many short retroposons first annotated as SINE are recognized as nonautonomous LINEs. Bov-A was originally described as a SINE (Jobse et al. 1995), but it was revealed to be a nonautonomous LINE whose 5′ and 3′ parts were originated from the 5′ end and 3′ end of Bov-B LINE. Bov-tA, which was generated by a fusion of tRNA-derived head and Bov-A, on the other hand, is considered as a SINE (Shimamura et al. 1999). Nonautonomous LINE originated by an internal deletion is observed for other LINE groups such as L1 (Bao and Jurka 2010) and Vingi (Kojima et al. 2011). Other SINEs generated by a fusion of nonautonomous LINE and tRNA-derived sequence are also reported, such as SINE2-1_ACar, which was generated by a fusion of a tRNA plus 5′ and 3′ regions of Vingi-2_ACar (Jurka 2010; Kojima et al. 2011).
There are some nonautonomous retroposons mobilized by non-LTR retrotransposon (LINE) but not classified to either nonautonomous LINE or SINE. SVA is a primate composite retroposon ∼2-kb long, transcribed by RNA polymerase II, and mobilized by L1 (Raiz et al. 2012). Sadhu is a group of retroposons found in Arabidopsis (Rangwala et al. 2006). They are ∼900-bp long. They do not encode any protein. They do not have recognizable pol II or pol III promoter either.
If SINEU is truly transcribed by RNA polymerase II, it would not satisfy the criteria for SINE. SINEU may be classified to a new group of retroposons, neither LINE nor SINE. Still I prefer to call SINEU as SINE based on its structural similarity to SINE. SINEU is a short retroposon shorter than 500 bp, except for SINEU-1D, which is a dimeric SINEU-1 family. SINEU contains 5′ sequences derived from small RNA. The structure of SINEU-3 resembles monomeric Alu. One may revise the classification of SINEU when more new short retroposon families that does not satisfy the present SINE criteria are characterized.
Supplementary Material
Supplementary figures S1–S6 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The author thank the International Crocodilian Genomes Working Group for allowing access to the genome sequences. Research reported in this publication was supported by the National Library of Medicine of the National Institute of Health under Award Number P41LM006252. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Institute of Health.
Literature Cited
- Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105. [DOI] [PubMed] [Google Scholar]
- Bao W, Jurka J. 2010. Origin and evolution of LINE-1 derived “half-L1” retrotransposons (HAL1). Gene 465:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibillo A, Eickbush TH. 2004. End-to-end template jumping by the reverse transcriptase encoded by the R2 retrotransposon. J Biol Chem. 279:14945–14953. [DOI] [PubMed] [Google Scholar]
- Bogenhagen DF, Brown DD. 1981. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell 24:261–270. [DOI] [PubMed] [Google Scholar]
- Buzdin A, et al. 2002. A new family of chimeric retrotranscripts formed by a full copy of U6 small nuclear RNA fused to the 3′ terminus of l1. Genomics 80:402–406. [DOI] [PubMed] [Google Scholar]
- Chong AY, et al. 2014. Evolution and gene capture in ancient endogenous retroviruses—insights from the crocodilian genomes. Retrovirology 11:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S, Pont-Kingdon G, Carroll D. 2000. Target specificity of the endonuclease from the Xenopus laevis non-long terminal repeat retrotransposon, Tx1L. Mol Cell Biol. 20:1219–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damert A, et al. 2009. 5′-Transducing SVA retrotransposon groups spread efficiently throughout the human genome. Genome Res. 19:1992–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T. 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 35:41–48. [DOI] [PubMed] [Google Scholar]
- Egloff S, O'Reilly D, Murphy S. 2008. Expression of human snRNA genes from beginning to end. Biochem Soc Trans. 36:590–594. [DOI] [PubMed] [Google Scholar]
- Eickbush DG, Eickbush TH. 2010. R2 retrotransposons encode a self-cleaving ribozyme for processing from an rRNA cotranscript. Mol Cell Biol. 30:3142–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush DG, Luan DD, Eickbush TH. 2000. Integration of Bombyx mori R2 sequences into the 28S ribosomal RNA genes of Drosophila melanogaster. Mol Cell Biol. 20:213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto H, et al. 2004. Integration of the 5′ end of the retrotransposon, R2Bm, can be complemented by homologous recombination. Nucleic Acids Res. 32:1555–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Perez JL, Doucet AJ, Bucheton A, Moran JV, Gilbert N. 2007. Distinct mechanisms for trans-mediated mobilization of cellular RNAs by the LINE-1 reverse transcriptase. Genome Res. 17:602–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, et al. 2014. Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science 346:1254449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321. [DOI] [PubMed] [Google Scholar]
- Hancks DC, Ewing AD, Chen JE, Tokunaga K, Kazazian HH., Jr 2009. Exon-trapping mediated by the human retrotransposon SVA. Genome Res. 19:1983–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobse C, et al. 1995. Evolution and recombination of bovine DNA repeats. J Mol Evol. 41:277–283. [PubMed] [Google Scholar]
- Jurka J. 2010. SINE elements from tetrapods. Repbase Reports 10:635–637. [Google Scholar]
- Jurka J, et al. 2005. Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 110:462–467. [DOI] [PubMed] [Google Scholar]
- Kajikawa M, Okada N. 2002. LINEs mobilize SINEs in the eel through a shared 3′ sequence. Cell 111:433–444. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. 2003. A novel class of SINE elements derived from 5S rRNA. Mol Biol Evol. 20:694–702. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. 2009. Young families of Tx1 non-LTR retrotransposons from the amphioxus genome. Repbase Reports 9:838–854. [Google Scholar]
- Kapitonov VV, Tempel S, Jurka J. 2009. Simple and fast classification of non-LTR retrotransposons based on phylogeny of their RT domain protein sequences. Gene 448:207–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohany O, Gentles AJ, Hankus L, Jurka J. 2006. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics 7:474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima KK. 2010. Different integration site structures between L1 protein-mediated retrotransposition in cis and retrotransposition in trans. Mobile DNA 1:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima KK, Fujiwara H. 2004. Cross-genome screening of novel sequence-specific non-LTR retrotransposons: various multicopy RNA genes and microsatellites are selected as targets. Mol Biol Evol. 21:207–217. [DOI] [PubMed] [Google Scholar]
- Kojima KK, Kapitonov VV, Jurka J. 2011. Recent expansion of a new Ingi-related clade of Vingi non-LTR retrotransposons in hedgehogs. Mol Biol Evol. 28:17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramerov DA, Vassetzky NS. 2011. Origin and evolution of SINEs in eukaryotic genomes. Heredity (Edinb) 107:487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegs JO, Churakov G, Jurka J, Brosius J, Schmitz J. 2007. Evolutionary history of 7SL RNA-derived SINEs in Supraprimates. Trends Genet. 23:158–161. [DOI] [PubMed] [Google Scholar]
- Malik HS, Burke WD, Eickbush TH. 1999. The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol. 16:793–805. [DOI] [PubMed] [Google Scholar]
- Nishihara H, Smit AF, Okada N. 2006. Functional noncoding sequences derived from SINEs in the mammalian genome. Genome Res. 16:864–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara H, Terai Y, Okada N. 2002. Characterization of novel Alu- and tRNA-related SINEs from the tree shrew and evolutionary implications of their origins. Mol Biol Evol. 19:1964–1972. [DOI] [PubMed] [Google Scholar]
- Paule MR, White RJ. 2000. Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res. 28:1283–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentin Y. 1992. Fusion of a free left Alu monomer and a free right Alu monomer at the origin of the Alu family in the primate genomes. Nucleic Acids Res. 20:487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiz J, et al. 2012. The non-autonomous retrotransposon SVA is trans-mobilized by the human LINE-1 protein machinery. Nucleic Acids Res. 40:1666–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangwala SH, et al. 2006. Meiotically stable natural epialleles of Sadhu, a novel Arabidopsis retroposon. PLoS Genet. 2:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Okada N. 1985. Rodent type 2 Alu family, rat identifier sequence, rabbit C family, and bovine or goat 73-bp repeat may have evolved from tRNA genes. J Mol Evol. 22:134–140. [DOI] [PubMed] [Google Scholar]
- Schmitz J, Churakov G, Zischler H, Brosius J. 2004. A novel class of mammalian-specific tailless retropseudogenes. Genome Res. 14:1911–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamura M, Abe H, Nikaido M, Ohshima K, Okada N. 1999. Genealogy of families of SINEs in cetaceans and artiodactyls: the presence of a huge superfamily of tRNA(Glu)-derived families of SINEs. Mol Biol Evol. 16:1046–1060. [DOI] [PubMed] [Google Scholar]
- Suh A, et al. 2014. Multiple lineages of ancient CR1 retroposons shaped the early genome evolution of amniotes. Genome Biol Evol. 7:205–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Fujiwara H. 2002. Transplantation of target site specificity by swapping the endonuclease domains of two LINEs. Embo J. 21:408–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu E, Tschudi C. 1984. Alu sequences are processed 7SL RNA genes. Nature 312:171–172. [DOI] [PubMed] [Google Scholar]
- Valadkhan S. 2005. snRNAs as the catalysts of pre-mRNA splicing. Curr Opin Chem Biol. 9:603–608. [DOI] [PubMed] [Google Scholar]
- Van Arsdell SW, et al. 1981. Direct repeats flank three small nuclear RNA pseudogenes in the human genome. Cell 26:11–17. [DOI] [PubMed] [Google Scholar]
- Wan QH, et al. 2013. Genome analysis and signature discovery for diving and sensory properties of the endangered Chinese alligator. Cell Res. 23:1091–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol. 13:290–301 [DOI] [PubMed] [Google Scholar]