Abstract
β -catenin (encoded by CTNNB1) is a subunit of the cell surface cadherin protein complex that acts as an intracellular signal transducer in the WNT signaling pathway; alterations in its activity have been associated with the development of hepatocellular carcinoma and other liver diseases. Other than WNT, additional signaling pathways also can converge at β-catenin. β-catenin also interacts with transcription factors such as T-cell factor, forkhead box protein O, and hypoxia inducible factor 1α to regulate the expression of target genes. We discuss the role of β-catenin in metabolic zonation of the adult liver. β-catenin also regulates the expression of genes that control metabolism of glucose, nutrients, and xenobiotics; alterations in its activity may contribute to the pathogenesis of nonalcoholic steatohepatitis. Alterations in β-catenin signaling may lead to activation of hepatic stellate cells, which is required for fibrosis. Many hepatic tumors such as hepatocellular adenomas, hepatocellular cancers, and hepatoblastomas have mutations in CTNNB1 that result in constitutive activation of β-catenin, so this molecule could be a therapeutic target. We discuss how alterations in β-catenin activity contribute to liver disease and how these might be used in diagnosis and prognosis, as well as in the development of therapeutics.
Keywords: WNT, β-Catenin, E-Cadherin, Liver Fibrosis, NASH, HCC, Liver Tumors
β-catenin is expressed throughout the adult liver. In hepatocytes, it is observed at the cell surface throughout the hepatic lobule, although it has cytoplasmic and nuclear localization in the hepatocytes that surround the central vein. In the normal adult liver, β-catenin signaling is always active in hepatocytes in the pericentral region, although it forms part of intercellular junctions elsewhere.
Upon injury to the liver (surgical resection, toxic insult, infection, metabolic insult, or tumor growth), β-catenin localization and signaling change. These changes can contribute to reparative responses or to disease development. How is β-catenin signaling regulated in the healthy liver and how does it change during the development of specific diseases? We discuss its roles in WNT signaling and WNT-independent pathways.
Activation of β-Catenin by WNT Signaling
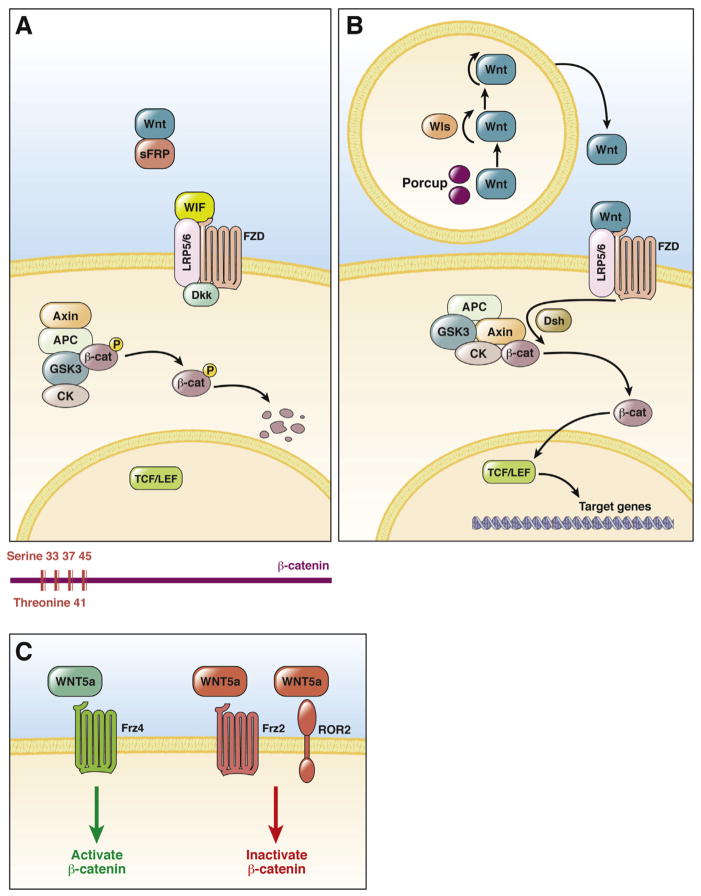
β-catenin is a component of the WNT signaling pathway; it usually is bound to a multiprotein degradation complex comprising casein kinase I, glycogen synthase kinase 3β (GSK3β), adenomatous polyposis coli gene product (APC), diversin, and axin (Figure 1). When WNT signaling is off as a result of the presence of WNT inhibitors such as WNT inhibitory factor, soluble frizzled-related proteins (sFRPs), Dickkopf and cerberus,1 β-catenin is phosphorylated first at serine-45 (S45) by casein kinase I, followed by phosphorylation at S33, S37, and threonine-41 by GSK3β2–4 (Figure 1). Once phosphorylated, β-catenin is recognized by β-transducin repeat-containing protein, which also requires intact lysine-19 and lysine-49 in β-catenin, for ubiquitination and proteosomal degradation of β-catenin5,6 (Figure 2).
Figure 1.
WNT Signaling via β-catenin. (A) In the absence of WNT or in the presence of WNT inhibitors, β-catenin is phosphorylated at specific serine and threonine residues in exon 3 by kinases in the degradation complex, leading to recognition by β-transducin repeat-containing protein for destruction. Mutations affecting these residues can lead to stabilization of the β-catenin protein. (B) WNT protein is palmitoylated and glycosylated by porcupine (Porcup) in the endoplasmic reticulum and transported by wntless (WLS) protein from the Golgi apparatus to the membrane for secretion. Secreted WNT binds to its receptor (FZ) and co-receptors LRP5 or LRP6. The signal is transduced through disheveled to inactivate the β-catenin degradation complex, leading to nuclear translocation of β-catenin. In the nucleus, β-catenin interacts with LEF/TCF transcription factors to regulate the expression of target genes. (C) WNT5A either can activate or inhibit β-catenin signaling, depending on the type of FZD expressed by a cell. WNT5A can activate β-catenin signaling in the presence of frizzled-4 (FZD4), although it inhibited β-catenin in the presence of FZD2 or an alternate WNT receptor called the receptor tyrosine kinase-like orphan receptor 2. β-cat, β-catenin; CK, casein kinase; Dsh, disheveled; P, phosphorylation; WIF, WNT inhibitory factor.
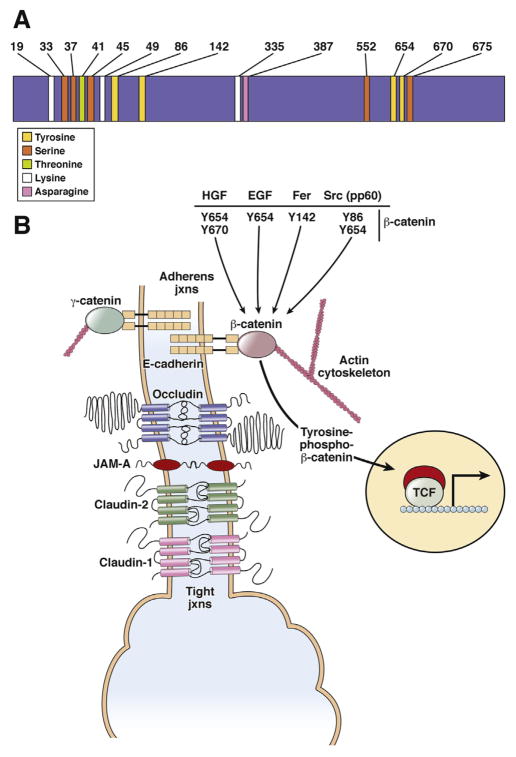
Figure 2.
β-catenin structure and role in adherens junctions. (A) Specific amino acids in β-catenin regulate its stability, activity, and interactions with other proteins. (B) In a normal liver, β-catenin acts as a bridge between the intracytoplasmic tail of E-cadherin and the actin cytoskeleton to regulate cell adhesion. Molecules such as HGF, EGF, FER kinase, and SRC can affect this complex negatively through tyrosine phosphorylation of β-catenin at specific residues. The end result of dissociation of the complex in some cases can result in nuclear translocation and activation of β-catenin. Left: in the absence of β-catenin, γ-catenin is able to maintain AJs by binding to E-cadherin and actin cytoskeleton. γ-Catenin did not compensate for nuclear β-catenin function. EGF, epidermal growth factor; JAM-A, junctional adhesion molecule-A; jxns, junctions.
Binding of active WNT to its cell surface receptor (frizzled) and co-receptor (low-density lipoprotein–related protein 5 or 6 [LRP5/6]) in an autocrine or paracrine manner inactivates the β-catenin degradation complex. For WNT proteins to be bioactive, they must be glycosylated and acylated, which is, mediated by porcupine.7 After acylation, WNT proteins become hydrophobic and require a cargo receptor, Wntless (also called Evenness interrupted), for cellular transport and secretion.8,9 Upon inhibition of degradation complex, β-catenin is released and undergoes nuclear translocation, where it acts as a co-factor for the T-cell factor/lymphoid enhancement factor (TCF/LEF) family of transcription factors to regulate target gene expression (Figure 1). It is important to note that WNT can activate or sometimes inhibit β-catenin as well. An example worth noting is that of WNT5a, which has been considered a prototypical noncanonical WNT. However, its ability to activate β-catenin or inhibit it through activation of the WNT/calcium pathway was shown to be dependent on the form of frizzled available at the time for ligand binding10 (Figure 1C).
Dissociation of β-Catenin From Adherens Junctions
β-catenin is a component of the adherens junctions (AJs). It forms a bridge between the cytoplasmic domain of the cadherins and the actin cytoskeleton11–13 (Figure 2). Specific β-catenin binding sites on the cytoplasmic domain of cadherins have been characterized.14,15 Interactions between β-catenin and E-cadherin are regulated by tyrosine phosphorylation in the carboxy terminal of β-catenin. Phosphorylation of β-catenin destabilizes the cadherin–β-catenin bond and promotes loss of intracellular adhesion.16,17 Conversely, dephosphorylation of β-catenin at tyrosine residues increases the activity of E-cadherin, and β-catenin and α-catenin reassembly.18 After tyrosine phosphorylation of β-catenin, its cytosolic pool is increased, and may increase the transcriptional activity of the β-catenin–TCF complex.19
Nonreceptor kinases such as sarcoma family kinase and Fps/fes (Feline sarcoma) related kinase phosphorylate β-catenin at tyrosine residues, leading to its dissociation from E-cadherin (Figure 2).20,21 The epidermal growth factor receptor and epidermal growth factor receptor 2 are associated with β-catenin.22–24 Epidermal growth factor signaling can induce tyrosine phosphorylation of β-catenin at Y654 to decrease the association between β-catenin and E-cadherin, leading to enhanced β-catenin activity in the nuclei if its degradation by the proteasome is simultaneously inhibited.25
Hepatocyte growth factor (HGF) induces phosphorylation of β-catenin at tyrosine residues 654 (Y654) and Y670 through direct interactions with MET (Figure 2).26–28 In fact, β-catenin is required for liver growth in response to HGF in vivo.29 Hepatoblastomas, which frequently have deletions or missense mutations in exon 3 of the gene that encodes β-catenin (CTNNB1), express high levels of Y654 β-catenin, induced by HGF signaling via MET.30 Fibrolamellar variants of hepatocellular cancers (HCCs) have higher levels of β-catenin phosphorylation at Y654 than nontumor liver tissues.25
Signaling Pathways
TGFβ Pathway
WNT signaling interacts with the transforming growth factor β (TGF-β) pathways. Complexes of β-catenin–TCF and SMAD4 interact with SMAD2 and SMAD3 after TGF-β signaling.32 Subsets of HCC tissues have signs of activation of β-catenin after that of TGF-β.33 Intriguingly, the WNT target BMP and activin membrane-bound inhibitor inhibit the ability of TGF-β to inhibit proliferation, and could be involved in the development of HCC.34
NF-κB Pathway
β-catenin forms a complex with the p65 subunit of the nuclear factor-κB (NF-κB) in hepatocytes to inhibit this transcription factor.35 Loss of β-catenin leads to NF-κB activation of genes that regulate cell survival. Mice with hepatocyte-specific disruption of β-catenin have reduced liver injury in response to tumor necrosis factor-α. Expression of NF-κB target genes increases when β-catenin is inhibited, and decreases when β-catenin is overexpressed.36 Studies are needed to determine whether interactions between NF-κB and β-catenin co-factor could be targeted by therapeutic agents.
Protein Kinase A Signaling
Prostaglandin E1 (PGE1) and isoproterenol activate protein kinase A (PKA) and also can activate β-catenin. PKA phosphorylates β-catenin at S675 to help stabilize and activate it.37 Cyclic adenosine monophosphate–dependent activation of PKA leads to phosphorylation of β-catenin at S552 and S675 to promote its ability to regulate transcription38 (Figure 2A). PGE2 activates β-catenin via cyclic adenosine monophosphate and PKA during induction and engraftment of hematopoietic stem cells. PGE2 might cooperate with WNT to induce β-catenin activation during liver regeneration.39 PKA activation was required for triiodothyronine-induced activation of β-catenin, expression of cyclin D1, and proliferation of hepatocytes.40 Recently, phosphorylation of β-catenin at S675 was shown to mediate fibrocystin-defective congenital hepatic fibrosis in mice.41
Nucleus
β-catenin does not bind DNA directly, but it interacts with and regulates the activities of transcription factors. For example, β-catenin activates gene expression by interacting with the TCF/LEF family of transcription factors. β-catenin binds other transcription factors under different conditions. The interactions between β-catenin with various transcription factors allows it to control many different functions of liver cells.
Interactions With TCF4
In the nucleus, β-catenin co-activates the high mobility group box containing the DNA binding protein TCF/LEF family of transcription factors42,43 (see Stadeli et al44 for a full description). Once the TCF-β–catenin complex is formed in the nucleus, expression of target genes is stage- and tissue-specific (Table 1).
Table 1.
Genes Regulated by WNT Signaling to β-Catenin in the Liver
| Target genes | Liver model | Co-factor for TF | Reference |
|---|---|---|---|
| Axin-2 | HCA, hepatoblastoma, and HCC | TCF4 | 142,153 |
| MYC | HB | TCF4 | 142,154 |
| Cyclin-D1 | Normal liver, LD, LR, hepatoblastoma, HCC | TCF4 | 53,142,154–157 |
| Epidermal growth factor receptor | Normal liver and hepatoblastoma | TCF4 | 123 |
| G-protein–coupled receptor 49 | HCC | TCF4 | 158 |
| Glutamate transporter-1 | Normal liver | TCF4 | 159 |
| Glutamine synthetase | Normal liver, HCC, hepatoblastoma, HCA | TCF4 | 51,52,89,96,142,159,160 |
| Ornithine aminotransferase | Normal liver | TCF4 | 159 |
| Survivin | HCC | 161 | |
| T-cell factor-1 | HCC | TCF4 | 162 |
| Lect2 | Normal liver/HCC | TCF4 | 96,163,164 |
| Vascular endothelial growth factor A | HCC and ischemia-reperfusion injury | TCF4/HIF1α | 46,165,166 |
| Regucalcin | Normal liver and HCC | TCF4 | 167 |
| Constitutive androstane receptor | Normal liver | TCF4 | 58 |
| CYP1A2 | Normal liver | TCF4 | 58 |
| Glucose-6-phosphatase | Normal liver | FOXO | 50 |
| Phosphoenol-pyruvate carboxykinase | Normal liver | FOXO | 50 |
| Erythropoietin | Ischemia-reperfusion injury | HIF1α | 46 |
| Claudin-2 | Normal liver | TCF4 | 53 |
LD, liver development; LR, liver regeneration; TF, transcription factor.
Interactions With Hypoxia Inducible Factor 1α
β-catenin interacts with hypoxia inducible factor 1α (HIF1α) during periods of hypoxia. In colorectal cancer cells, during oxygen deprivation, formation of a complex between β-catenin and TCF4 is inhibited, instead β-catenin binds to HIF1α to induce expression of genes that regulate adaptation and survival under hypoxic conditions.45 In ischemia-reperfusion liver injury,46 increased levels of WNT or β-catenin protected cells, whereas inhibition of β-catenin caused tissue damage under conditions of hypoxia. Under conditions of normoxia, β-catenin bound TCF to regulate genes that promote proliferation, whereas under conditions of hypoxia, β-catenin switched from TCF to HIF1α, leading to increased expression of targets such as erythropoietin and vascular endothelial growth factor. This switch could be modulated by N-acetylcysteine, which regulates reactive oxygen species. It therefore was proposed that interactions between β-catenin and HIF1α depend on the redox state of a cell.
Another study showed an opposite role of HIF2α in regulating the activities of β-catenin and TCF4.47 HIF2α assembled with the complex of β-catenin and TCF to facilitate gene transcription and induce tumor cell proliferation. In fact, the effect of HIF2α was opposite to that of HIF1α on β-catenin and proliferation because HIF1α inhibited β-catenin–dependent transcription. A balance of HIF1α vs HIF2α therefore might contribute to the overall activity of β-catenin and determine the response in the form of proliferation vs cellular adaptation and survival when appropriate upstream effectors are present.
Interactions With Forkhead Box Protein O
β-catenin is sensitive to oxidative stress and interacts with forkhead box protein O (FOXO) transcription factors. β-catenin binds directly to FOXO in response to oxidative stress,48 and FOXO and TCF compete for binding to β-catenin. When target genes are activated by β-catenin and FOXO, those regulated by β-catenin and TCF are inactivated.49 FOXO factors respond not only to changes in the redox state of a cell but also to the insulin level, and their interactions with β-catenin have been associated with pathogenesis of nonalcoholic steatohepatitis (NASH).50
Normal and Diseased Liver
β-catenin can be activated via several different pathways, although the WNT signaling pathway has been the most widely studied. Researchers have studied mice with liver-specific knockout of β-catenin or LRP5/6, in which WNT is unable to communicate with β-catenin. These studies have provided evidence for the roles of β-catenin signaling in metabolic zonation, where β-catenin regulates the expression of genes in pericentral hepatocytes.51 Hepatic tissues of mice with liver-specific knockout of β-catenin do not express Glul (encodes glutamine synthetase [GS]), Cyp2e1 (encodes cytochrome p450 2e1), or Cyp1a2.52,53 Furthermore, knockout of LRP5/6 from livers of mice prevents expression of these genes, showing that β-catenin is regulated by WNT signaling in mouse liver.54 Mice with liver-specific knockout of β-catenin have defects in regeneration also observed in LRP5/6 knockout mice, supporting the role of WNT signaling in this process.55
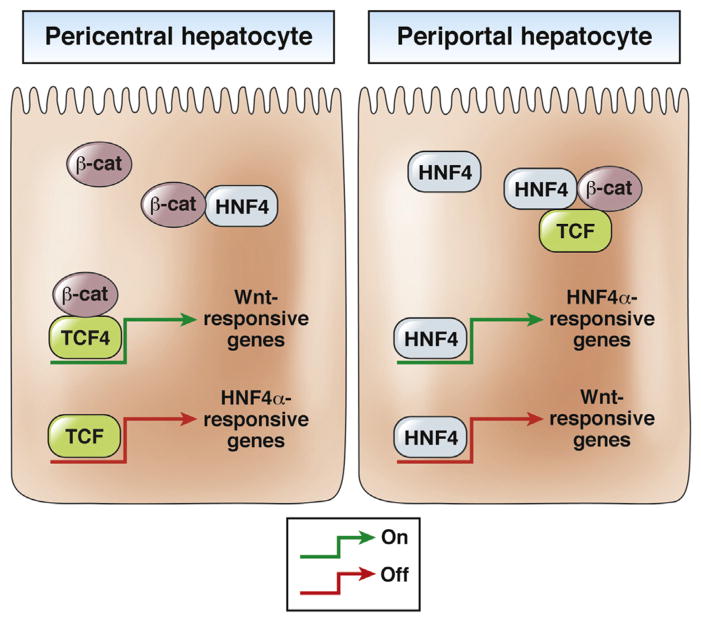
Zonation of hepatic lobule requires expression of specific genes in pericentral vs periportal hepatocytes for optimum hepatic function in regulating metabolism.56 Is WNT signaling alone sufficient to regulate this dynamic process? Although hepatocyte nuclear factor-4α (HNF4α) is considered to be a liver-enriched transcription factor, it also regulates gene expression in periportal hepatocytes.57 Moreover, loss of HNF4α from hepatocytes increases pericentral gene expression by the periportal hepatocytes (similar to mice with liver-specific knockout of APC). There appears to be a mechanism whereby HNF4α suppresses gene activation by β-catenin and TCF4.58
The sequence of the WNT-responsive element in gene promoters is similar to that of the HNF4α-responsive element. Therefore, HNF4α could bind to the TCF4-binding site and vice versa. Further analyses identified interactions between β-catenin, HNF4α, and TCF4 proteins. Although TCF4 and HNF4α might bind to one another’s motifs on target genes, the presence of HNF4α appears to prevent β-catenin–dependent transcription, whereas the presence of β-catenin appears to prevent HNF4α-dependent transcription. This could be the basis of overall metabolic zonation in the liver (Figure 3). Further studies are needed to elucidate these interactions and their regulation.
Figure 3.
Interactions among β-catenin, HNF4α, and TCF4 in periportal and pericentral hepatocytes. (Left panel) β-catenin binds to TCF4 and this complex binds to WNT response elements in promoters of target genes in pericentral hepatocytes. Here, TCF can bind to the HNF4α response elements, making it unavailable for HNF4α to bind and transactivate its target genes. Furthermore, β-catenin can bind HNF4α to prevent its binding to gene promoters. β-catenin signaling via TCF4 therefore is activated and HNF4α transactivation is inactivated in these cells. (Right panel) In periportal hepatocytes, HNF4α binds to HNF4α response elements in promoters of target genes to induce their expression while also occupying WNT response elements, preventing TCF binding. Furthermore, HNF4α can bind β-catenin and TCF to keep them engaged and prevent binding to target gene promoters. HNF4α signaling therefore is activated and β-catenin signaling via TCF4 is inactivated in these cells. β-cat, β-catenin.
NASH
NASH is the most common cause of chronic liver disease in the United States. NASH arises through several metabolic aberrations in tandem, including alterations in glucose and lipid metabolism. These culminate in inflammation, oxidative stress, cell death, and, in some patients, fibrosis and HCC. β-catenin signaling is involved in many of these processes. Variants of the gene that encodes transcription factor 7-like 2, a transcription factor that interacts with β-catenin, have been associated with a risk for type 2 diabetes.59
β-catenin binding to TCF4 activates gene expression, whereas binding to HNF4α inhibits the activity of this transcription factor. The association between TCF4 and HNF4α might be inhibited by β-catenin, especially in the periportal region, which would allow for expression of genes that regulate hepatic glucose production and lipogenesis.58 β-catenin regulates gluconeogenesis at baseline and during starvation, a process that usually is deranged in insulin resistance, via interactions with FOXO1. This interaction regulates the expression of genes encoding glucose-6-phosphatase and phosphoenolpyruvate carboxykinase, which are rate-limiting enzymes in hepatic gluconeogenesis.50 Increased interaction between β-catenin and TCF during hepatic gluconeogenesis had the opposite results—it negatively regulated hepatic gluconeogenesis genes.60 There appears to be a switch that regulates interactions between β-catenin and transcription factors. Further studies are needed to evaluate the mechanism, its regulation and any therapeutic implications. Although it is not clear what factors regulate the function of β-catenin during the development of NASH, WNT signaling appears to be involved.
Although NASH is not always associated with obesity, often body fat, insulin resistance, and type 2 diabetes occur with NASH in patients. WNT signaling might link all these processes with NASH pathogenesis. LRP6+/− mice are protected from hepatic steatosis when fed high-fat diets.61 Reduced WNT signaling in these mice decreased body fat mass and hepatic gluconeogenesis, and increased brown adipose tissue and hepatic sensitivity to insulin. This occurred via decreased mammalian target of rapamycin signaling, increased expression of uncoupling protein 1 and peroxisome proliferator-activated receptor (PPAR)γ coactivator 1-α, increased expression of hepatic leptin, and decreased nuclear β-catenin in hepatocytes. These changes led to lower levels of glucose-6-phosphatase and phosphoenolpyruvate carboxykinase. WNT signaling or nuclear β-catenin therefore could contribute to NASH pathogenesis, although further studies are required.
Fibrosis
The first studies showing increased expression of WNT pathway components in hepatic fibrosis used genomic analysis from primary biliary cirrhosis livers.62,63 These studies identified increased expression of WNT13, WNT5a, β-catenin, and others, although a cause-and-effect relationship was not addressed. In the meantime, the role of WNT signaling was and continues to be reported in pulmonary and renal fibrosis.64–67 Hepatic fibrosis is a wound healing response and represents a common end point of many chronic liver diseases, which, owing to lack of treatment, remains a substantive unmet clinical need. Hepatic fibrosis is mostly a function of hepatic stellate cells (HSCs), which are a source of extracellular matrix deposition within injured livers. The role of WNT signaling in hepatic stellate cell biology has begun to unravel.
A study directly investigating gene expression differences in quiescent vs activated HSCs identified, among others, an up-regulated expression of WNT5a and Fz2.68 Subsequent studies further have identified aberrant WNT5a expression in fibrotic livers, and its suppression led to reduced HSC activation.69,70 As has been discussed earlier, WNT5A can have opposing roles on β-catenin signaling based on receptors expressed in a cell type (Figure 1C). Whether the role of WNT5A in promoting fibrosis is caused by inhibition, activation, or independent of β-catenin needs further clarification. Unfortunately, there is evidence for all 3 in the literature at the present time. As mentioned previously, one study showed no change in nuclear translocation of β-catenin after a WNT5A increase that was evident during stellate cell activation concomitant with up-regulation of FZ2.68
Another study showed that activation of WNT/β-catenin signaling in primary rat HSC by an inhibitor of GSK3β decreased synthesis of α-smooth muscle actin and WNT5A, and induced the expression of glial fibrillary acidic protein.71 This study further showed that activation of WNT signaling decreased DNA synthesis and prevented HSCs from entering the cell cycle to eventually show the role of WNT signaling to β-catenin in maintaining their quiescence. However, a growing body of literature supports the activation of β-catenin by WNT signaling during the process of HSC activation and fibrosis.72–74 In fact, different molecules have been used to block WNT signaling, directly or indirectly, to show an overall antifibrotic effect both in vitro and in vivo.
Activating pregnane X receptor by rifampicin, among other things, also inhibited WNT signaling and reduced HSC proliferation and transdifferentiation to active myofibroblasts.75 Another study showed that necdin, a melanoma antigen family protein that promotes neuronal and myogenic differentiation while inhibiting adipogenesis, is expressed in HSCs. Necdin is induced during HSC activation and its silencing reversed them to quiescence through PPARγ and suppression of WNT/β-catenin signaling.76 Another study showed that Septin 4 a subunit of the septin cytoskeleton specifically expressed in quiescent HSCs, is down-regulated through transdifferentiation to activated myofibroblasts. Loss of Septin 4 in HSCs coincided with decreased expression of WNT inhibitor Dickkopf protein 2, increased WNT signaling via β-catenin, and increased fibrosis.77
Finally, another study supported the concept that the canonical WNT pathway promotes fibrogenesis, based on an analysis of mesoderm-specific transcript homologue, a strong negative regulator of WNT/β-catenin signaling.78 The investigators showed that mesoderm-specific transcript homologue expression in HSCs alleviated carbon tetrachloride–induced collagen deposition in liver tissue by decreasing the expression of β-catenin, α-smooth muscle actin, and Smad3 both in vivo and in vitro.
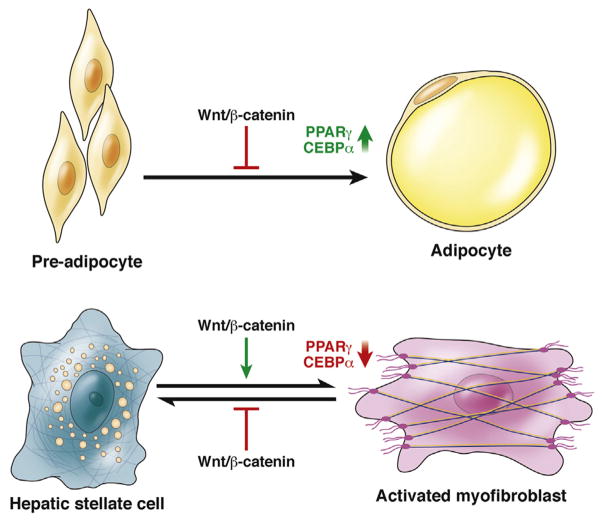
WNT–β-catenin signaling might activate HSCs via negative regulation of adipogenesis.79 Expression of WNT1 or β-catenin with the S33Y mutation in preadipocytes prevented their differentiation to adipocytes at least in part through inhibition of expression of CCAAT enhancer binding protein-α and PPARγ (Figure 4). Conversely, blockade of β-catenin signaling, either through dominant-negative TCF4 or axin, facilitated adipogenic differentiation of the preadipocytes. Quiescent HSCs contain lipid droplets and their activation to myofibroblasts involves loss of adipogenic genes including PPARγ and CCAAT enhancer binding protein-α, therefore this event is analogous to differentiation of adipocytes to preadipocytes. Gain-of-function mutations in adipogenic transcription factors such as PPARγ and sterol regulatory element binding protein-1c can reverse a culture-induced myofibroblast into a quiescent HSC phenotype. Taken together, these observations indicate that activation of WNT signaling via β-catenin could be involved in HSC activation by inhibiting adipogenic programs; inhibition of this signaling pathway could contribute to the adipogenic gene profile of a quiescent HSC. Inhibiting WNT signaling to β-catenin therefore might block hepatic fibrosis. Further studies are needed to determine the identity and cell sources of the factors that activate β-catenin HSCs.
Figure 4.
WNT signaling to β-catenin during adipogenesis and activation of hepatic stellate cells. Inhibition of WNT signaling to β-catenin is required for differentiation of pre-adipocytes into adipocytes, regulating expression of genes that control adipogenesis such as PPARγ and CCAAT enhancer binding protein-α (CEBPα). HSC differentiation into active myofibroblasts is analogous to de-differentiation of adipocytes to preadipocytes (losing adipogenic properties). WNT activation of β-catenin promotes this process by negatively regulating the expression of PPARα and CEBPα. WNT signaling via β-catenin therefore might be inhibited to treat patients with fibrosis: it would activate the expression of genes involved in adipogenesis-induced quiescence of activated stellate cells.
Liver Cancer
Focal Nodular Hyperplasia
Focal nodular hyperplasia (FNH) is a benign hepatic tumor characterized by the presence of almost-normal-looking hepatocytes that are arranged in plates, which are usually 1- to 2-cells thick. These structures are separated by fibrotic bands and surround a central fibrous scar that contains a dystrophic arterial blood vessel. It has been speculated that FNH may originate from a hyperplastic lesion composed of reactive polyclonal proliferating hepatocytes, which could be the result of increased blood flow caused by local vascular malformation.80 There is also a well-known association of FNH with vascular disorders such as Rendu–Osler–Weber syndrome or hereditary telangiectasia. The molecular basis of FNH remains largely obscure. However, it is clear that FNH is polyclonal in nature in a significant subset of cases. Recently, transcriptomic analysis of FNH identified activation of WNT to the β-catenin pathway without any mutations in the gene encoding β-catenin, consistent with its polyclonal origin.81 Interestingly, immunostaining for glutathione synthetase (GS), whose expression is regulated by WNT–β-catenin signaling, shows a map-like staining of FNH in the liver that now is used in diagnosis.82 Although the significance of these findings is unclear, these observations might result from an alternate mechanism of β-catenin activation such as growth factor–dependent activation in the face of altered blood flow in the lesion.26,83 Alternatively, especially because these tumors are thought to be a result of vascular disturbances, there may be areas of hypoxia within the tumors.80 Hypoxia has been shown to activate WNT signaling, therefore changes in β-catenin signaling, indicated by increased levels of GS, may be secondary to areas of relative hypoxia within FNH, which might promote growth of this benign tumor.84
Hepatocellular Adenomas
Hepatocellular adenomas (HCAs) are characterized by monoclonal proliferation of well-differentiated hepatocytes that usually are arranged in sheets and cords. There is a classic absence of a portal triad and interlobular bile ducts in HCAs. Although initially considered a well-defined homogeneous entity, it now is known that several molecular classes of this tumor type exist that dictate tumor origin, behavior, and eventually determines prognosis and helps to determine treatment options.82 At least 3 classes of HCAs with a known molecular basis have been defined and at least 2 of these have WNT signaling via β-catenin activation and implications in disease behavior.
Biallelic inactivating mutations in HNF1A or TCF1 genes have been identified in approximately 30%–40% of HCAs.85 These tumors have marked steatosis and excess glycogen accumulation. HCAs with HNF1A inactivation have an extremely low risk of malignant transformation; which is almost always observed in tumors with activation of β-catenin and interleukin 6 (inflammatory).
Another subset of HCAs (approximately 10%–15%) have WNT activation of β-catenin activation owing to mutations in exon-3 of CTNNB1.86 HCAs with these mutations in β-catenin develop in combination with other mutations in inflammatory HCAs.87 In fact, approximately half of HCAs with active β-catenin are of the inflammatory type. Some HCAs were found to have mutations outside of exon 3 of CTNNB1.88 These mutations affected codons 335 (exon 7) and 387 (exon 8). These HCAs initially were categorized as either unclassified or inflammatory. The mutations led to β-catenin activation, although to a lesser extent than classic exon 3 mutations, in vitro and in vivo.
Overall, mutations in β-catenin occurred more frequently in HCAs that developed in men. These HCAs were found to have cholestasis and cell dysplasia in histologic analyses. A target of β-catenin, GS, is up-regulated in β-catenin–mutated HCA at the RNA and protein level. Because detection of nuclear β-catenin by staining often is challenging, samples also can be analyzed by a GS analysis to determine if β-catenin is mutated in HCAs with greater sensitivity.89 HCAs with mutations in β-catenin have a greater propensity for malignant transformation.86
The third group of HCAs that account for approximately half of all adenomas are of the inflammatory type. The characteristic feature of inflammatory HCA is the activation of the janus kinase (JAK)–signal transducer and activator of transcription (STAT) pathway. These tumors are characterized histologically by polymorphic inflammatory in-filtrates. This type of HCA has mutations in FYN-related kinase, interleukin 6 ST (encodes gp130), STAT3, and GNAS complex locus.87,88 All of these mechanisms lead to activation of STAT3 and expression of genes, including those encoding cytokines and chemokines, which promote tumor infiltration by lymphocytes. Importantly, a subset of this tumor type also has mutations in CTNNB1, irrespective of the molecular driver, and poses an increased risk of malignant transformation.87
HCC
Patients with HCC have poor prognoses—HCC has the highest mortality and morbidity rates of any cancer worldwide. It usually develops in patients with chronic liver injury or cirrhosis, caused by factors such as infection, metabolic disease, or biliary disease.90 There are limited therapeutic options for advanced HCC. Sorafenib has been approved by the US Food and Drug Administration for treatment of stage 4 unresectable HCC. It only modestly increases survival time; therefore, new specific treatments are needed.91
HCC cells from animal models and patients have constitutive activation of WNT–β-catenin signaling. Liver cancer cells were shown by immunohistochemical analyses to have abnormal localization of cadherins and catenins.92 In mouse models, mutations in CTNNB1 and altered expression were detected in approximately 25% of all HCCs and in up to 50% of all hepatic tumors.93 Several subsequent studies corroborated these observations in patients and currently approximately 8%–44% of all HCCs show mutations in CTNNB1, the most well-understood mechanism of β-catenin activation (Table 2). These mutations, similar to HCA, affect exon 3 at serine/threonine sites or adjacent amino acids that hinder phosphorylation and eventually degradation of β-catenin protein, leading to its stabilization and nuclear translocation. Mutations also have been reported in other components of the degradation complex of β-catenin including AXIN1 in approximately 3%–16%94–96 and AXIN2 in approximately 3% of all HCC cases95 (Table 2). Additional mechanisms also have been described and include overexpression of FRZ7,97,98 WNT3 up-regulation,99 inactivation of GSK3β,100 methylation of sFRP1,101 epigenetic inactivation of several sFRPs,102 TGF-β–dependent activation of β-catenin,33 and β-catenin activation by receptor tyrosine kinases, especially in the fibrolamellar subset of HCCs.25
Table 2.
Mutations Detected in CTNNB1
| Cases with mutations in exon 3 of CTNNB1 (%) | Information |
|---|---|
| 41/125 (33)168 | An additional 15.2% had mutations in AXIN1 |
| 9/32 (28)25 | Tyrosine-654 was phosphorylated β-catenin in the fibrolamellar variety of HCC |
| 20/45 (44)96 | An additional 7 patients had AXIN1 mutations |
| 15/45 (33)169 | No GSK3B mutations |
| 16/38 (42)158 | Multiple mutations in 2 patients |
| 14/73 (19)95 | 1 insertion between S33 and G34 |
| 5/62 (8)110 | Aflatoxin study |
| 7/60 (12)168 | 62% of tumors had cytoplasmic β-catenin staining |
| 57/434 (13)108 | 34 had mutations at GSK3B; 17 had mutations at codons 32 and 34 |
| 9/22 (41)107 | Multiple mutations in 1 patient |
| 12/35 (34)115 | Multiple mutations in 2 patients |
| 21/119 (18)106 | Multiple mutations in a patient |
| 9/38 (24)169 | Aberrant accumulation of β-catenin in the nucleus, cytoplasm and membrane was seen in 39% of cases |
| 8/31 (26)93 | 2 patients had mutations at D32 |
Mutations in CTNNB1 and AXIN1/2 lead to WNT autonomous activation of β-catenin, whereas other mechanisms such as WNT signaling via overexpressed frizzled or decreased expression of WNT inhibitors would activate β-catenin in a WNT-dependent manner. Is the extent of activation of WNT signaling owing to these diverse mechanisms comparable? Also, is the target gene expression caused by disparate mechanisms of β-catenin activation similar? A study showed significant correlation between CTNNB1 mutations and overexpression of the target genes GS, G-protein–coupled receptor 49, and glutamate transporter-1 (P = .0001), but not for other target genes such as ornithine aminotransferase, LECT2, c-MYC, or cyclin D1.96 This study showed GS to be a good immunohistochemical marker of β-catenin activation in HCC, which also was validated independently.25 However, no increase in the expression of GS, G-protein coupled receptor 49, or glutamate transporter-1 was evident in HCC with AXIN1 mutations. A mouse model with conditional loss of AXIN1 in hepatocytes led to HCC that showed only some β-catenin targets to be up-regulated and did not show an increase in nuclear β-catenin or GS.103 A tissue-array–based study also showed distinct patterns of β-catenin and GS staining in HCC.104 Thus, it is likely that various mechanisms of β-catenin activation in HCC may lead to differing qualitative and quantitative WNT signaling with a distinct impact on tumor phenotype. Indeed, β-catenin active HCC owing to mutations in CTNNB1, AXIN1, or additional modes of β-catenin activation, all have been shown to have a distinct phenotype in transcriptome classifications of HCC.33,96,105
Is there a predilection of β-catenin activation in HCC based on disease etiology? Could variations in mutation frequencies be reflective of differences in geographic, dietary, and other etiologic factors influencing the molecular pathogenesis of HCC? One study detected an inverse correlation between β-catenin mutations and loss of heterozygosity in the genome suggesting chromosomal instability (involving tumor-suppressor genes) and mutations in CTNNB1 representing alternative modes of tumor progression.106 Higher frequency of CTNNB1 mutations in HCC associated with hepatitis C virus (HCV) infection has been reported with more than 40% of tumors showing mutations (mostly S45).107 HBV-related HCC had less-frequent CTNNB1 mutations.108 A recent study, however, showed that in patients with a low number of HBV DNA copies per hepatocyte, additional risk factors such as alcohol, HCV infection, or NASH might cooperate to influence disease progression to HCC.109 Although CTNNB1 mutations were clustered into the G5 and G6 groups (in which G4, G5, and G6 mostly included patients without HBV infection),105 tumors in the G4–G6 group contained a small subset of HBV-positive tumors, co-infected with HCV or additional disease modifiers, and CTNNB1 mutations.109 Few aflatoxin-associated HCCs had mutations in CTNNB1, although 45% of these tumors showed increased accumulation of β-catenin, compared with healthy liver tissue.110
Distinct molecular signatures of HCC have been identified in cirrhotic vs noncirrhotic livers. Although these findings are preliminary, they indicate that unique pathogenetic events separate the subsets. HCCs from non-cirrhotic livers more frequently have activation of WNT signaling to β-catenin than HCCs from cirrhotic livers, which frequently have altered P53 activity.90 Along similar lines, another study reported more frequent alterations in WNT signaling via β-catenin in HCV-associated HCCs, compared with those associated with alcoholism, which have alterations in RB1- and P53-regulated pathways.111
Researchers found that 59% of HCCs have somatic mutations in the promoter region of telomerase reverse-transcriptase (TERT) that increased its expression. This mutation also was observed in 25% of cirrhotic preneoplastic nodules and in 44% of HCAs that evolved into HCC.112 What was intriguing was the fact that in HCCs, there was a significant association between CTNNB1 and TERT promoter mutations, therefore these appear to synergize to promote neoplastic transformation—especially in a subset of cirrhotic preneoplastic nodules.112
There is debate about the overall effects of β-catenin mutations and activation in patients with HCC, and how these affect their prognosis.113 CTNNB1 mutations have been associated with both better prognosis and a more differentiated tumor type,105,114 and more proliferating and poorly differentiated HCC.33,115,116 Overall, prognosis is a complex attribute and in HCC it is dependent on not only multiple tumor characteristics, such as vascular invasion, metastasis, and nodularity, but also the state of nontumor tissues, including fibrosis, cirrhosis, hepatic dysfunction, and extrahepatic factors. Additional studies are required to address if alterations in gene expression with CTNNB1 mutations or other changes in β-catenin activation affect tumor phenotype and patient prognosis.96,117
Decreased hepatic fibrosis in CTNNB1-mutated tumors has been reported in a few studies.25,118 Whether β-catenin mutations and cirrhosis are mutually exclusive, non-cooperating risk factors for the development of HCC or their concurrence decreases the threshold of neoplastic transformation. Although not tested directly for CTNNB1 mutations, a small but significant subset of HCV patients (which are known to favor β-catenin gene mutations for the development of HCC) developed HCC without evidence of advanced fibrosis.119 Similarly, small subsets of HCAs that progress to HCC in patients often show CTNNB1 mutations and no fibrosis.86 To experimentally address any cooperation of advanced fibrosis and CTNNB1 mutations in the development of HCC, we induced fibrosis in S45-mutant–β-catenin transgenic and control mice with thioacetamide and showed no difference in HCC development.104 Thus, CTNNB1 mutations and cirrhosis may not cooperate and so may be independent contributors to tumorigenesis.
The WNT–β-catenin pathway in HCC in experimental models deserves mention. The first question that has been answered is if β-catenin mutation, activation, or over-expression by itself is sufficient for HCC initiation. None of the transgenic mice overexpressing either wild-type or stable mutants of β-catenin thus far have shown spontaneous HCC.120–123 However, several studies now suggest that β-catenin collaborates with other signaling pathways to contribute to hepatocarcinogenesis. β-catenin was shown to cooperate with an activated renin-angiotensin system (RAS) in HCC.124 Mice with heterozygous disruption of LKB1 that express transgenic β-catenin have accelerated progression of HCC compared with mice with LKB1 disruption.125 The chemical carcinogen diethylnitrosamine, which normally induces HCC by activating RAS,126 induces earlier formation of larger HCCs in mice with the S45 mutation in β-catenin.122 These findings indicate that alterations in β-catenin activity are involved in the development of HCC, although additional factors are required for tumorigenesis.
In clinical studies, somatic mutations in TERT promoter that led to its overexpression frequently were observed with activation of WNT–β-catenin signaling, especially those caused by CTNNB1 mutation.112 The exception to this rule was the liver-specific APC deletion mutants, which develop spontaneous HCC secondary to β-catenin activation.127,128 However, because APC also inhibits proliferation by binding directly to DNA, its β-catenin–independent effects may cooperate with β-catenin to lead to HCC in this model.129
Specific chemical carcinogenesis models can be used to induce HCC with preferential activation of β-catenin via its mutations. In the C3H strain, a single intraperitoneal injection of DEN at 75 μg/g body weight to 6-week-old male mice will lead to HCC with predominant RAS mutations.126 Interestingly, when these mice were fed phenobarbitol (0.05% in diet) 3 weeks after DEN injection, 80% of the liver tumors that developed had β-catenin mutations whereas RAS mutations were undetectable. These findings indicate that phenobarbital selects for cells that contain β-catenin mutations.130 Similar to the observation in patients, tumors with β-catenin mutations in mice also show tumor-wide GA staining and can serve as a good biomarker. New models of liver cancer, such as transgenic lines, and those that involve administration of chemical carcinogens (such as 2-amino-3, 4-dimethylimidazo [4,5-f] quinoline, which selectively activates β-catenin) could improve our understanding of the role and regulation of this pathway and to test new therapeutic strategies.131
Hepatoblastoma
Hepatoblastoma is the most common malignant hepatic tumor of early childhood. Up to 90% of these tumors are composed of cells with abundant nuclear and cytoplasmic localization of β-catenin caused by in-frame mutations in CTNNB1.132–135
Mutations in AXIN1 have been detected in less than 10% of HBs.95 APC mutations have been identified in familial cases136 and in sporadic cases.137 A comprehensive study in 85 HB patients showed 65 cases with missense mutations and interstitial deletions affecting exon 3 of CTNNB1.138 The same study identified loss-of-function mutations in APC and AXIN1 genes. Therefore, 82% of hepatoblastomas in this group contained an activating mutation in β-catenin. Hepatoblastoma is a component of syndromes such as Beckwith–Wiedemann syndrome.134,139 Therefore, β-catenin appears to be involved in the pathogenesis of hepatoblastoma.
However, the exact role of β-catenin in hepatoblastoma is unclear. During normal liver development, β-catenin signaling is activated during hepatic induction, hepatoblast expansion, hepatocyte maturation, and biliary differentiation.140,141 The major difference between β-catenin signaling in normal hepatic development vs hepatoblastoma is the extent and molecular basis of its activation. Although during normal development β-catenin likely is activated spatiotemporally by proteins such as WNT, fibroblast growth factor, and HGF, during the development of hepatoblastoma, β-catenin activation occurs autonomously from ligand because of deletions or mutations. Mutations that activate β-catenin do so uncontrollably and constitutively. Genes regulated by WNT signaling, such as those encoding c-MYC, cyclin-D1, GS, epidermal growth factor receptor, axin-2, and others have been reported to be up-regulated in different histologic subtypes of hepatoblastoma.142
The 2 major histologic subtypes of hepatoblastoma are the embryonal and fetal variety, which, as the name suggests, are composed of embryonal (or immature) or fetal (or more differentiated) hepatoblasts, although these tumors can have components of both. In fact, more nuclear β-catenin was evident in embryonal hepatoblastoma (or in embryonal areas of a hepatoblastoma) and coincided with a lack of GS (a β-catenin target in mature hepatocytes), whereas fetal hepatoblastoma showed membranous, cytoplasmic, and nuclear β-catenin and coincided with GS expression in the tumor (or fetal component of a hepatoblastoma).143 Because we reported full-length β-catenin protein during early liver development and a calpain-cleaved truncated form missing the amino terminal in late liver development in mouse, we used β-catenin antibodies directed against its amino- or carboxy-terminal to distinguish the 2 histologic subtypes. We showed that irrespective of the monoallelic mutations in CTNNB1, and based on the normal wild-type allele that always is present in the tumors, we were able to distinguish embryonal hepatoblastoma, which are positive for amino- and carboxy-terminal β-catenin, from fetal hepatoblastoma, which was positive only for carboxy-terminal β-catenin, based on immunohistochemical analyses.
Interstitial deletions in CTNNB1 are more frequent than missense mutations in hepatoblastoma.143 Whether there are any functional differences between point vs deletion mutants of the β-catenin gene needs to be studied further. Intriguingly, when an amino-terminal deletion mutant of β-catenin is overexpressed in the liver using an adenoviral approach or by generation of a transgenic mouse over-expressing β-catenin under liver-specific promoter, the mice never show hepatoblastoma.120,121 In addition, transgenic mice expressing a point mutant form of β-catenin do not show hepatoblastoma.122 It is likely that oncogenic β-catenin is by itself insufficient to induce hepatoblastoma and may be cooperating with another pathway.
Eighty percent of hepatoblastomas have simultaneous nuclear β-catenin and Yes-associated protein, a component of the Hippo signaling pathway.144 In hepatoblastoma cells, but not in HCC cells, suppression of one led to inhibition of the other, suggesting synergism between the 2 pathways. Overexpression of a mutant form of β-catenin and active Yap using the sleeping beauty transposon/transposase led to extensive development of hepatoblastoma and death of mice at about 12 weeks.144 Interactions among Yap, β-catenin, TCF4, and TEA domain are involved in the pathogenesis of hepatoblastoma. This model will be invaluable in understanding the biology of the disease and provide an opportunity to test therapies.
Future Directions
Alterations in the expression and activity of β-catenin affect hepatic pathophysiology. These effects require extensive characterization. We do not completely understand how β-catenin is regulated and what determines its interaction with various transcription factors, under different conditions, or in different cell types. Although β-catenin controls regeneration via cyclin-D1 expression and metabolic zonation by regulating the expression of pericentral genes, the identity of specific WNT proteins that modulate β-catenin activity in these events remains unknown.
Reagents that alter β-catenin activity could be developed as therapeutics. In patients with end-stage liver disease of any etiology or patients receiving liver transplants, activation of β-catenin could induce liver regeneration. It is important to identify factors that regulate β-catenin activity. We have shown that triiodothyronine can activate β-catenin in cells40,145 and studies in animals are planned.
As protocols are optimized to induce differentiation of stem cells into hepatocytes, these should include strategies to modify β-catenin activity, which could increase the efficiency of the process. Similarly, based on its role in regulating target genes in the pericentral hepatocytes, spatiotemporal activation of β-catenin activation might be used to generate more physiologic, functional hepatic tissue.
The roles of β-catenin activation in hepatic disorders such as NASH and hepatic fibrosis requires further study; current evidence is preliminary or controversial. However, we have much knowledge about the role and regulation of β-catenin in hepatic tumor development. Subsets of HCAs, HCCs, and hepatoblastomas show an unquestionable activation of β-catenin owing to mutations in key components of the WNT pathway or other mechanisms. Although it is not clear whether all mechanisms leading to β-catenin activation in tumors are equally bad, strategies to inhibit this protein are expected to be of notable benefit when β-catenin activation is secondary to mutations in CTNNB1.
Studies have shown the therapeutic benefits of inhibiting β-catenin. Although it is not clear how β-catenin inhibition would affect HCC growth and progression, it would down-regulate target genes that have important roles in cancer cell proliferation (such as those encoding cyclin-D1 and c-MYC) viability (such as survivin) or metabolism (such as GS), angiogenesis (such as vascular endothelial growth factor A), or stem cell expansion (such as epithelial cell adhesion molecule). Interesting β-catenin inhibitors that could have clinical applications include ICG-001 or its derivative PRI-724, PMED-1, and others.146–148 Anti-sense and small inhibitor RNAs also might be used to reduce levels of β-catenin in HCCs or other tumor types. In a recent study, β-catenin reduction in CTNNB1-mutated HCCs in a murine model led to complete tumor response, showing a clear benefit of therapeutic targeting of this molecule.149 This lays the groundwork of precision medicine for HCC treatment.150
Could β-catenin be a safe therapeutic target owing to its role at AJs in addition to being the WNT signaling component? Although it would be helpful to inhibit nuclear β-catenin in HCC cells, effects on β-catenin at the membrane actually could increase tumor cell motility and migration by disrupting the AJ. Interestingly, β-catenin inhibition increased levels of γ-catenin at the cell membrane to maintain the AJ.151,152 γ-catenin, however, could not compensate for the loss of β-catenin in the WNT signaling pathway in the liver, in vivo, or in hepatoma cells. It will be pertinent to continue to define the exact molecular basis of γ-catenin stabilization after β-catenin suppression to spare it when γ-catenin is targeted therapeutically for a subset of HCCs.
Acknowledgments
Funding
This study was funded by National Institutes of Health grants 1R01DK62277, 1R01DK100287, and 1R01DK095498; and an Endowed Chair for Experimental Pathology.
Abbreviations used in this paper
- AJ
adherens junction
- APC
adenomatous polyposis coli gene product
- FNH
focal nodular hyperplasia
- FOXO
forkhead box protein O
- GS
glutathione synthetase
- GSK3β
glycogen synthase kinase 3β
- HBV
hepatitis B virus
- HCA
hepatocellular adenoma
- HCC
hepatocellular cancer
- HCV
hepatitis C virus
- HGF
hepatocyte growth factor
- HIF1α
hypoxia inducible factor 1α
- HNF4α
hepatocyte nuclear factor-4α
- HSC
hepatic stellate cell
- LEF
lymphoid enhancement factor
- LRP5/6
low-density lipoprotein–related protein 5 or 6
- NASH
nonalcoholic steatohepatitis
- NF-κB
nuclear factor-κB
- PGE
prostaglandin E
- PKA
_________________________
- PPAR
peroxisome proliferator-activated receptor
- RAS
renin-angiotensin system
- S
serine
- sFRP
soluble frizzled-related protein
- STAT
signal transducer and activator of transcription
- TCF
T-cell factor
- TERT
telomerase reverse-transcriptase
- TGF
transforming growth factor
Footnotes
Conflicts of interest
Satdarshan Pal Monga is a paid consultant for the AbbVie Hepatic Disease Steering Committee.
References
- 1.Cruciat CM, Niehrs C. Secreted and transmembrane wnt inhibitors and activators. Cold Spring Harb Perspect Biol. 2013;5:a015081. doi: 10.1101/cshperspect.a015081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behrens J, Jerchow BA, Wurtele M, et al. Functional interaction of an axin homolog, conductin, with beta-catenin, APC, and GSK3beta. Science. 1998;280:596–599. doi: 10.1126/science.280.5363.596. [DOI] [PubMed] [Google Scholar]
- 3.Liu C, Li Y, Semenov M, et al. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- 4.Amit S, Hatzubai A, Birman Y, et al. Axin-mediated CKI phosphorylation of beta-catenin at Ser 45: a molecular switch for the Wnt pathway. Genes Dev. 2002;16:1066–1076. doi: 10.1101/gad.230302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aberle H, Bauer A, Stappert J, et al. Beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winer IS, Bommer GT, Gonik N, et al. Lysine residues Lys-19 and Lys-49 of beta-catenin regulate its levels and function in T cell factor transcriptional activation and neoplastic transformation. J Biol Chem. 2006;281:26181–26187. doi: 10.1074/jbc.M604217200. [DOI] [PubMed] [Google Scholar]
- 7.Barrott JJ, Cash GM, Smith AP, et al. Deletion of mouse Porcn blocks Wnt ligand secretion and reveals an ectodermal etiology of human focal dermal hypoplasia/Goltz syndrome. Proc Natl Acad Sci U S A. 2011;108:12752–12757. doi: 10.1073/pnas.1006437108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banziger C, Soldini D, Schutt C, et al. Wntless, a conserved membrane protein dedicated to the secretion of Wnt proteins from signaling cells. Cell. 2006;125:509–522. doi: 10.1016/j.cell.2006.02.049. [DOI] [PubMed] [Google Scholar]
- 9.Bartscherer K, Pelte N, Ingelfinger D, et al. Secretion of Wnt ligands requires Evi, a conserved transmembrane protein. Cell. 2006;125:523–533. doi: 10.1016/j.cell.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 10.Mikels AJ, Nusse R. Purified Wnt5a protein activates or inhibits beta-catenin-TCF signaling depending on receptor context. PLoS Biol. 2006;4:e115. doi: 10.1371/journal.pbio.0040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knudsen KA, Soler AP, Johnson KR, et al. Interaction of alpha-actinin with the cadherin/catenin cell-cell adhesion complex via alpha-catenin. J Cell Biol. 1995;130:67–77. doi: 10.1083/jcb.130.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nieset JE, Redfield AR, Jin F, et al. Characterization of the interactions of alpha-catenin with alpha-actinin and beta-catenin/plakoglobin. J Cell Sci. 1997;110:1013–1022. doi: 10.1242/jcs.110.8.1013. [DOI] [PubMed] [Google Scholar]
- 13.Wheelock MJ, Knudsen KA. Cadherins and associated proteins. In Vivo. 1991;5:505–513. [PubMed] [Google Scholar]
- 14.Huber AH, Weis WI. The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin. Cell. 2001;105:391–402. doi: 10.1016/s0092-8674(01)00330-0. [DOI] [PubMed] [Google Scholar]
- 15.Jou TS, Stewart DB, Stappert J, et al. Genetic and biochemical dissection of protein linkages in the cadherin-catenin complex. Proc Natl Acad Sci U S A. 1995;92:5067–5071. doi: 10.1073/pnas.92.11.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ozawa M, Kemler R. Altered cell adhesion activity by pervanadate due to the dissociation of alpha-catenin from the E-cadherin catenin complex. J Biol Chem. 1998;273:6166–6170. doi: 10.1074/jbc.273.11.6166. [DOI] [PubMed] [Google Scholar]
- 17.Roura S, Miravet S, Piedra J, et al. Regulation of E-cadherin/catenin association by tyrosine phosphorylation. J Biol Chem. 1999;274:36734–36740. doi: 10.1074/jbc.274.51.36734. [DOI] [PubMed] [Google Scholar]
- 18.Hu P, O’Keefe EJ, Rubenstein DS. Tyrosine phosphorylation of human keratinocyte beta-catenin and plakoglobin reversibly regulates their binding to E-cadherin and alpha-catenin. J Invest Dermatol. 2001;117:1059–1067. doi: 10.1046/j.0022-202x.2001.01523.x. [DOI] [PubMed] [Google Scholar]
- 19.Piedra J, Martinez D, Castano J, et al. Regulation of beta-catenin structure and activity by tyrosine phosphorylation. J Biol Chem. 2001;276:20436–20443. doi: 10.1074/jbc.M100194200. [DOI] [PubMed] [Google Scholar]
- 20.Rosato R, Veltmaat JM, Groffen J, et al. Involvement of the tyrosine kinase fer in cell adhesion. Mol Cell Biol. 1998;18:5762–5770. doi: 10.1128/mcb.18.10.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Behrens J, Vakaet L, Friis R, et al. Loss of epithelial differentiation and gain of invasiveness correlates with tyrosine phosphorylation of the E-cadherin/beta-catenin complex in cells transformed with a temperature-sensitive v-SRC gene. J Cell Biol. 1993;120:757–766. doi: 10.1083/jcb.120.3.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoschuetzky H, Aberle H, Kemler R. Beta-catenin mediates the interaction of the cadherin-catenin complex with epidermal growth factor receptor. J Cell Biol. 1994;127:1375–1380. doi: 10.1083/jcb.127.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanai Y, Ochiai A, Shibata T, et al. c-erbB-2 gene product directly associates with beta-catenin and plakoglobin. Biochem Biophys Res Commun. 1995;208:1067–1072. doi: 10.1006/bbrc.1995.1443. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi K, Suzuki K, Tsukatani Y. Induction of tyrosine phosphorylation and association of beta-catenin with EGF receptor upon tryptic digestion of quiescent cells at confluence. Oncogene. 1997;15:71–78. doi: 10.1038/sj.onc.1201160. [DOI] [PubMed] [Google Scholar]
- 25.Cieply B, Zeng G, Proverbs-Singh T, et al. Unique phenotype of hepatocellular cancers with exon-3 mutations in beta-catenin gene. Hepatology. 2009;49:821–831. doi: 10.1002/hep.22695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonvini P, An WG, Rosolen A, et al. Geldanamycin abrogates ErbB2 association with proteasome-resistant beta-catenin in melanoma cells, increases beta-catenin-E-cadherin association, and decreases beta-catenin-sensitive transcription. Cancer Res. 2001;61:1671–1677. [PubMed] [Google Scholar]
- 27.Monga SP, Mars WM, Pediaditakis P, et al. Hepatocyte growth factor induces Wnt-independent nuclear translocation of beta-catenin after Met-beta-catenin dissociation in hepatocytes. Cancer Res. 2002;62:2064–2071. [PubMed] [Google Scholar]
- 28.Shibamoto S, Hayakawa M, Takeuchi K, et al. Tyrosine phosphorylation of beta-catenin and plakoglobin enhanced by hepatocyte growth factor and epidermal growth factor in human carcinoma cells. Cell Adhes Commun. 1994;1:295–305. doi: 10.3109/15419069409097261. [DOI] [PubMed] [Google Scholar]
- 29.Zeng G, Apte U, Micsenyi A, et al. Tyrosine residues 654 and 670 in beta-catenin are crucial in regulation of Met-beta-catenin interactions. Exp Cell Res. 2006;312:3620–3630. doi: 10.1016/j.yexcr.2006.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Apte U, Zeng G, Muller P, et al. Activation of Wnt/beta-catenin pathway during hepatocyte growth factor-induced hepatomegaly in mice. Hepatology. 2006;44:992–1002. doi: 10.1002/hep.21317. [DOI] [PubMed] [Google Scholar]
- 31.Purcell R, Childs M, Maibach R, et al. HGF/c-Met related activation of beta-catenin in hepatoblastoma. J Exp Clin Cancer Res. 2011;30:96. doi: 10.1186/1756-9966-30-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishita M, Hashimoto MK, Ogata S, et al. Interaction between Wnt and TGF-beta signalling pathways during formation of Spemann’s organizer. Nature. 2000;403:781–785. doi: 10.1038/35001602. [DOI] [PubMed] [Google Scholar]
- 33.Hoshida Y, Nijman SM, Kobayashi M, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385–7392. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sekiya T, Adachi S, Kohu K, et al. Identification of BMP and activin membrane-bound inhibitor (BAMBI), an inhibitor of transforming growth factor-beta signaling, as a target of the beta-catenin pathway in colorectal tumor cells. J Biol Chem. 2004;279:6840–6846. doi: 10.1074/jbc.M310876200. [DOI] [PubMed] [Google Scholar]
- 35.Nejak-Bowen K, Kikuchi A, Monga SP. Beta-catenin-NF-kappaB interactions in murine hepatocytes: a complex to die for. Hepatology. 2013;57:763–774. doi: 10.1002/hep.26042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du Q, Zhang X, Cardinal J, et al. Wnt/beta-catenin signaling regulates cytokine-induced human inducible nitric oxide synthase expression by inhibiting nuclear factor-kappaB activation in cancer cells. Cancer Res. 2009;69:3764–3771. doi: 10.1158/0008-5472.CAN-09-0014. [DOI] [PubMed] [Google Scholar]
- 37.Hino S, Tanji C, Nakayama KI, et al. Phosphorylation of beta-catenin by cyclic AMP-dependent protein kinase stabilizes beta-catenin through inhibition of its ubiquitination. Mol Cell Biol. 2005;25:9063–9072. doi: 10.1128/MCB.25.20.9063-9072.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taurin S, Sandbo N, Qin Y, et al. Phosphorylation of beta-catenin by cyclic AMP-dependent protein kinase. J Biol Chem. 2006;281:9971–9976. doi: 10.1074/jbc.M508778200. [DOI] [PubMed] [Google Scholar]
- 39.Goessling W, North TE, Loewer S, et al. Genetic interaction of PGE2 and Wnt signaling regulates developmental specification of stem cells and regeneration. Cell. 2009;136:1136–1147. doi: 10.1016/j.cell.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fanti M, Singh S, Ledda-Columbano GM, et al. Triiodothyronine induces hepatocyte proliferation by protein kinase A-dependent beta-catenin activation in rodents. Hepatology. 2014;59:2309–2320. doi: 10.1002/hep.26775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spirli C, Locatelli L, Morell CM, et al. Protein kinase A-dependent pSer(675) -beta-catenin, a novel signaling defect in a mouse model of congenital hepatic fibrosis. Hepatology. 2013;58:1713–1723. doi: 10.1002/hep.26554. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Riese J, Yu X, Munnerlyn A, et al. LEF-1, a nuclear factor coordinating signaling inputs from wingless and decapentaplegic. Cell. 1997;88:777–787. doi: 10.1016/s0092-8674(00)81924-8. [DOI] [PubMed] [Google Scholar]
- 43.Brannon M, Gomperts M, Sumoy L, et al. A beta-catenin/XTcf-3 complex binds to the siamois promoter to regulate dorsal axis specification in Xenopus. Genes Dev. 1997;11:2359–2370. doi: 10.1101/gad.11.18.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stadeli R, Hoffmans R, Basler K. Transcription under the control of nuclear Arm/beta-catenin. Curr Biol. 2006;16:R378–R385. doi: 10.1016/j.cub.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 45.Kaidi A, Williams AC, Paraskeva C. Interaction between beta-catenin and HIF-1 promotes cellular adaptation to hypoxia. Nat Cell Biol. 2007;9:210–217. doi: 10.1038/ncb1534. [DOI] [PubMed] [Google Scholar]
- 46.Lehwald N, Tao GZ, Jang KY, et al. Wnt-beta-catenin signaling protects against hepatic ischemia and reperfusion injury in mice. Gastroenterology. 2011;141:707–718. 718e1–5. doi: 10.1053/j.gastro.2011.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi H, Chun YS, Kim TY, et al. HIF-2alpha enhances beta-catenin/TCF-driven transcription by interacting with beta-catenin. Cancer Res. 2010;70:10101–10111. doi: 10.1158/0008-5472.CAN-10-0505. [DOI] [PubMed] [Google Scholar]
- 48.Essers MA, de Vries-Smits LM, Barker N, et al. Functional interaction between beta-catenin and FOXO in oxidative stress signaling. Science. 2005;308:1181–1184. doi: 10.1126/science.1109083. [DOI] [PubMed] [Google Scholar]
- 49.Hoogeboom D, Essers MA, Polderman PE, et al. Interaction of FOXO with beta-catenin inhibits beta-catenin/T cell factor activity. J Biol Chem. 2008;283:9224–9230. doi: 10.1074/jbc.M706638200. [DOI] [PubMed] [Google Scholar]
- 50.Liu H, Fergusson MM, Wu JJ, et al. Wnt signaling regulates hepatic metabolism. Sci Signal. 2011;4:ra6. doi: 10.1126/scisignal.2001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benhamouche S, Decaens T, Godard C, et al. Apc tumor suppressor gene is the “zonation-keeper” of mouse liver. Dev Cell. 2006;10:759–770. doi: 10.1016/j.devcel.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 52.Sekine S, Lan BY, Bedolli M, et al. Liver-specific loss of beta-catenin blocks glutamine synthesis pathway activity and cytochrome p450 expression in mice. Hepatology. 2006;43:817–825. doi: 10.1002/hep.21131. [DOI] [PubMed] [Google Scholar]
- 53.Tan X, Behari J, Cieply B, et al. Conditional deletion of beta-catenin reveals its role in liver growth and regeneration. Gastroenterology. 2006;131:1561–1572. doi: 10.1053/j.gastro.2006.08.042. [DOI] [PubMed] [Google Scholar]
- 54.Yang J, Mowry LE, Nejak-Bowen KN, et al. Beta-catenin signaling in murine liver zonation and regeneration: a Wnt-Wnt situation! Hepatology. 2014;60:964–976. doi: 10.1002/hep.27082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Monga SP. Role and regulation of beta-catenin signaling during physiological liver growth. Gene Expr. 2014;16:51–62. doi: 10.3727/105221614X13919976902138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gebhardt R, Hovhannisyan A. Organ patterning in the adult stage: the role of Wnt/beta-catenin signaling in liver zonation and beyond. Dev Dyn. 2010;239:45–55. doi: 10.1002/dvdy.22041. [DOI] [PubMed] [Google Scholar]
- 57.Colletti M, Cicchini C, Conigliaro A, et al. Convergence of Wnt signaling on the HNF4alpha-driven transcription in controlling liver zonation. Gastroenterology. 2009;137:660–672. doi: 10.1053/j.gastro.2009.05.038. [DOI] [PubMed] [Google Scholar]
- 58.Gougelet A, Torre C, Veber P, et al. T-cell factor 4 and beta-catenin chromatin occupancies pattern zonal liver metabolism in mice. Hepatology. 2014;59:2344–2357. doi: 10.1002/hep.26924. [DOI] [PubMed] [Google Scholar]
- 59.Grant SF, Thorleifsson G, Reynisdottir I, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 60.Ip W, Shao W, Chiang YT, et al. The Wnt signaling pathway effector TCF7L2 is upregulated by insulin and represses hepatic gluconeogenesis. Am J Physiol Endocrinol Metab. 2012;303:E1166–E1176. doi: 10.1152/ajpendo.00249.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu W, Singh R, Choi CS, et al. Low density lipoprotein (LDL) receptor-related protein 6 (LRP6) regulates body fat and glucose homeostasis by modulating nutrient sensing pathways and mitochondrial energy expenditure. J Biol Chem. 2012;287:7213–7223. doi: 10.1074/jbc.M111.286724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shackel NA, McGuinness PH, Abbott CA, et al. Identification of novel molecules and pathogenic pathways in primary biliary cirrhosis: cDNA array analysis of intra-hepatic differential gene expression. Gut. 2001;49:565–576. doi: 10.1136/gut.49.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tanaka A, Leung PS, Kenny TP, et al. Genomic analysis of differentially expressed genes in liver and biliary epithelial cells of patients with primary biliary cirrhosis. J Autoimmun. 2001;17:89–98. doi: 10.1006/jaut.2001.0522. [DOI] [PubMed] [Google Scholar]
- 64.Chilosi M, Poletti V, Zamo A, et al. Aberrant Wnt/beta-catenin pathway activation in idiopathic pulmonary fibrosis. Am J Pathol. 2003;162:1495–1502. doi: 10.1016/s0002-9440(10)64282-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.He W, Dai C, Li Y, et al. Wnt/beta-catenin signaling promotes renal interstitial fibrosis. J Am Soc Nephrol. 2009;20:765–776. doi: 10.1681/ASN.2008060566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morrisey EE. Wnt signaling and pulmonary fibrosis. Am J Pathol. 2003;162:1393–1397. doi: 10.1016/S0002-9440(10)64271-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Surendran K, McCaul SP, Simon TC. A role for Wnt-4 in renal fibrosis. Am J Physiol Renal Physiol. 2002;282:F431–F441. doi: 10.1152/ajprenal.0009.2001. [DOI] [PubMed] [Google Scholar]
- 68.Jiang F, Parsons CJ, Stefanovic B. Gene expression profile of quiescent and activated rat hepatic stellate cells implicates Wnt signaling pathway in activation. J Hepatol. 2006;45:401–409. doi: 10.1016/j.jhep.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 69.Rashid ST, Humphries JD, Byron A, et al. Proteomic analysis of extracellular matrix from the hepatic stellate cell line LX-2 identifies CYR61 and Wnt-5a as novel constituents of fibrotic liver. J Proteome Res. 2012;11:4052–4064. doi: 10.1021/pr3000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiong WJ, Hu LJ, Jian YC, et al. Wnt5a participates in hepatic stellate cell activation observed by gene expression profile and functional assays. World J Gastroenterol. 2012;18:1745–1752. doi: 10.3748/wjg.v18.i15.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kordes C, Sawitza I, Haussinger D. Canonical Wnt signaling maintains the quiescent stage of hepatic stellate cells. Biochem Biophys Res Commun. 2008;367:116–123. doi: 10.1016/j.bbrc.2007.12.085. [DOI] [PubMed] [Google Scholar]
- 72.Ge WS, Wang YJ, Wu JX, et al. Beta-catenin is overexpressed in hepatic fibrosis and blockage of Wnt/beta-catenin signaling inhibits hepatic stellate cell activation. Mol Med Rep. 2014;9:2145–2151. doi: 10.3892/mmr.2014.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cheng JH, She H, Han YP, et al. Wnt antagonism inhibits hepatic stellate cell activation and liver fibrosis. Am J Physiol Gastrointest Liver Physiol. 2008;294:G39–G49. doi: 10.1152/ajpgi.00263.2007. [DOI] [PubMed] [Google Scholar]
- 74.Myung SJ, Yoon JH, Gwak GY, et al. Wnt signaling enhances the activation and survival of human hepatic stellate cells. FEBS Lett. 2007;581:2954–2958. doi: 10.1016/j.febslet.2007.05.050. [DOI] [PubMed] [Google Scholar]
- 75.Haughton EL, Tucker SJ, Marek CJ, et al. Pregnane X receptor activators inhibit human hepatic stellate cell transdifferentiation in vitro. Gastroenterology. 2006;131:194–209. doi: 10.1053/j.gastro.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 76.Zhu NL, Wang J, Tsukamoto H. The Necdin-Wnt pathway causes epigenetic peroxisome proliferator-activated receptor gamma repression in hepatic stellate cells. J Biol Chem. 2010;285:30463–30471. doi: 10.1074/jbc.M110.156703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yanagida A, Iwaisako K, Hatano E, et al. Downregulation of the Wnt antagonist Dkk2 links the loss of Sept4 and myofibroblastic transformation of hepatic stellate cells. Biochim Biophys Acta. 2011;1812:1403–1411. doi: 10.1016/j.bbadis.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 78.Li W, Zhu C, Li Y, et al. Mest attenuates CCl4-induced liver fibrosis in rats by inhibiting the Wnt/beta-catenin signaling pathway. Gut Liver. 2014;8:282–291. doi: 10.5009/gnl.2014.8.3.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ross SE, Hemati N, Longo KA, et al. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289:950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- 80.Wanless IR, Albrecht S, Bilbao J, et al. Multiple focal nodular hyperplasia of the liver associated with vascular malformations of various organs and neoplasia of the brain: a new syndrome. Mod Pathol. 1989;2:456–462. [PubMed] [Google Scholar]
- 81.Rebouissou S, Couchy G, Libbrecht L, et al. The beta-catenin pathway is activated in focal nodular hyperplasia but not in cirrhotic FNH-like nodules. J Hepatol. 2008;49:61–71. doi: 10.1016/j.jhep.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 82.Nault JC, Bioulac-Sage P, Zucman-Rossi J. Hepatocellular benign tumors-from molecular classification to personalized clinical care. Gastroenterology. 2013;144:888–902. doi: 10.1053/j.gastro.2013.02.032. [DOI] [PubMed] [Google Scholar]
- 83.Chen YW, Jeng YM, Yeh SH, et al. P53 gene and Wnt signaling in benign neoplasms: beta-catenin mutations in hepatic adenoma but not in focal nodular hyperplasia. Hepatology. 2002;36:927–935. doi: 10.1053/jhep.2002.36126. [DOI] [PubMed] [Google Scholar]
- 84.Mazumdar J, O’Brien WT, Johnson RS, et al. O2 regulates stem cells through Wnt/beta-catenin signalling. Nat Cell Biol. 2010;12:1007–1013. doi: 10.1038/ncb2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bluteau O, Jeannot E, Bioulac-Sage P, et al. Biallelic inactivation of TCF1 in hepatic adenomas. Nat Genet. 2002;32:312–315. doi: 10.1038/ng1001. [DOI] [PubMed] [Google Scholar]
- 86.Zucman-Rossi J, Jeannot E, Nhieu JT, et al. Genotype-phenotype correlation in hepatocellular adenoma: new classification and relationship with HCC. Hepatology. 2006;43:515–524. doi: 10.1002/hep.21068. [DOI] [PubMed] [Google Scholar]
- 87.Rebouissou S, Amessou M, Couchy G, et al. Frequent in-frame somatic deletions activate gp130 in inflammatory hepatocellular tumours. Nature. 2009;457:200–204. doi: 10.1038/nature07475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pilati C, Letouze E, Nault JC, et al. Genomic profiling of hepatocellular adenomas reveals recurrent FRK-activating mutations and the mechanisms of malignant transformation. Cancer Cell. 2014;25:428–441. doi: 10.1016/j.ccr.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 89.Bioulac-Sage P, Rebouissou S, Thomas C, et al. Hepatocellular adenoma subtype classification using molecular markers and immunohistochemistry. Hepatology. 2007;46:740–748. doi: 10.1002/hep.21743. [DOI] [PubMed] [Google Scholar]
- 90.Tannapfel A, Wittekind C. Genes involved in hepatocellular carcinoma: deregulation in cell cycling and apoptosis. Virchows Arch. 2002;440:345–352. doi: 10.1007/s00428-002-0617-x. [DOI] [PubMed] [Google Scholar]
- 91.Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 92.Ihara A, Koizumi H, Hashizume R, et al. Expression of epithelial cadherin and alpha- and beta-catenins in nontumoral livers and hepatocellular carcinomas. Hepatology. 1996;23:1441–1447. doi: 10.1053/jhep.1996.v23.pm0008675162. [DOI] [PubMed] [Google Scholar]
- 93.de La Coste A, Romagnolo B, Billuart P, et al. Somatic mutations of the beta-catenin gene are frequent in mouse and human hepatocellular carcinomas. Proc Natl Acad Sci U S A. 1998;95:8847–8851. doi: 10.1073/pnas.95.15.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Satoh S, Daigo Y, Furukawa Y, et al. AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1. Nat Genet. 2000;24:245–250. doi: 10.1038/73448. [DOI] [PubMed] [Google Scholar]
- 95.Taniguchi K, Roberts LR, Aderca IN, et al. Mutational spectrum of beta-catenin, AXIN1, and AXIN2 in hepatocellular carcinomas and hepatoblastomas. Oncogene. 2002;21:4863–4871. doi: 10.1038/sj.onc.1205591. [DOI] [PubMed] [Google Scholar]
- 96.Zucman-Rossi J, Benhamouche S, Godard C, et al. Differential effects of inactivated Axin1 and activated beta-catenin mutations in human hepatocellular carcinomas. Oncogene. 2007;26:774–780. doi: 10.1038/sj.onc.1209824. [DOI] [PubMed] [Google Scholar]
- 97.Merle P, de la Monte S, Kim M, et al. Functional consequences of frizzled-7 receptor overexpression in human hepatocellular carcinoma. Gastroenterology. 2004;127:1110–1122. doi: 10.1053/j.gastro.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 98.Nambotin SB, Lefrancois L, Sainsily X, et al. Pharmacological inhibition of Frizzled-7 displays anti-tumor properties in hepatocellular carcinoma. J Hepatol. 2011;54:288–299. doi: 10.1016/j.jhep.2010.06.033. [DOI] [PubMed] [Google Scholar]
- 99.Kim M, Lee HC, Tsedensodnom O, et al. Functional interaction between Wnt3 and Frizzled-7 leads to activation of the Wnt/beta-catenin signaling pathway in hepatocellular carcinoma cells. J Hepatol. 2008;48:780–791. doi: 10.1016/j.jhep.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ban KC, Singh H, Krishnan R, et al. GSK-3beta phosphorylation and alteration of beta-catenin in hepatocellular carcinoma. Cancer Lett. 2003;199:201–208. doi: 10.1016/s0304-3835(03)00421-x. [DOI] [PubMed] [Google Scholar]
- 101.Shih YL, Shyu RY, Hsieh CB, et al. Promoter methylation of the secreted frizzled-related protein 1 gene SFRP1 is frequent in hepatocellular carcinoma. Cancer. 2006;107:579–590. doi: 10.1002/cncr.22023. [DOI] [PubMed] [Google Scholar]
- 102.Takagi H, Sasaki S, Suzuki H, et al. Frequent epigenetic inactivation of SFRP genes in hepatocellular carcinoma. J Gastroenterol. 2008;43:378–389. doi: 10.1007/s00535-008-2170-0. [DOI] [PubMed] [Google Scholar]
- 103.Feng GJ, Cotta W, Wei XQ, et al. Conditional disruption of Axin1 leads to development of liver tumors in mice. Gastroenterology. 2012;143:1650–1659. doi: 10.1053/j.gastro.2012.08.047. [DOI] [PubMed] [Google Scholar]
- 104.Lee JM, Yang J, Newell P, et al. Beta-catenin signaling in hepatocellular cancer: implications in inflammation, fibrosis, and proliferation. Cancer Lett. 2014;343:90–97. doi: 10.1016/j.canlet.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Boyault S, Rickman DS, de Reynies A, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 106.Legoix P, Bluteau O, Bayer J, et al. Beta-catenin mutations in hepatocellular carcinoma correlate with a low rate of loss of heterozygosity. Oncogene. 1999;18:4044–4046. doi: 10.1038/sj.onc.1202800. [DOI] [PubMed] [Google Scholar]
- 107.Huang H, Fujii H, Sankila A, et al. Beta-catenin mutations are frequent in human hepatocellular carcinomas associated with hepatitis C virus infection. Am J Pathol. 1999;155:1795–1801. doi: 10.1016/s0002-9440(10)65496-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hsu HC, Jeng YM, Mao TL, et al. Beta-catenin mutations are associated with a subset of low-stage hepatocellular carcinoma negative for hepatitis B virus and with favorable prognosis. Am J Pathol. 2000;157:763–770. doi: 10.1016/s0002-9440(10)64590-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Amaddeo G, Cao Q, Ladeiro Y, et al. Integration of tumour and viral genomic characterisations in HBV-related hepatocellular carcinomas. Gut. 2015;64:820–829. doi: 10.1136/gutjnl-2013-306228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Devereux TR, Stern MC, Flake GP, et al. CTNNB1 mutations and beta-catenin protein accumulation in human hepatocellular carcinomas associated with high exposure to aflatoxin B1. Mol Carcinog. 2001;31:68–73. doi: 10.1002/mc.1041. [DOI] [PubMed] [Google Scholar]
- 111.Edamoto Y, Hara A, Biernat W, et al. Alterations of RB1, p53 and Wnt pathways in hepatocellular carcinomas associated with hepatitis C, hepatitis B and alcoholic liver cirrhosis. Int J Cancer. 2003;106:334–341. doi: 10.1002/ijc.11254. [DOI] [PubMed] [Google Scholar]
- 112.Nault JC, Mallet M, Pilati C, et al. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat Commun. 2013;4:2218. doi: 10.1038/ncomms3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nejak-Bowen KN, Monga SP. Beta-catenin signaling, liver regeneration and hepatocellular cancer: sorting the good from the bad. Semin Cancer Biol. 2011;21:44–58. doi: 10.1016/j.semcancer.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yuan RH, Jeng YM, Hu RH, et al. Role of p53 and beta-catenin mutations in conjunction with CK19 expression on early tumor recurrence and prognosis of hepatocellular carcinoma. J Gastrointest Surg. 2011;15:321–329. doi: 10.1007/s11605-010-1373-x. [DOI] [PubMed] [Google Scholar]
- 115.Nhieu JT, Renard CA, Wei Y, et al. Nuclear accumulation of mutated beta-catenin in hepatocellular carcinoma is associated with increased cell proliferation. Am J Pathol. 1999;155:703–710. doi: 10.1016/s0002-9440(10)65168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Suzuki T, Yano H, Nakashima Y, et al. Beta-catenin expression in hepatocellular carcinoma: a possible participation of beta-catenin in the dedifferentiation process. J Gastroenterol Hepatol. 2002;17:994–1000. doi: 10.1046/j.1440-1746.2002.02774.x. [DOI] [PubMed] [Google Scholar]
- 117.Austinat M, Dunsch R, Wittekind C, et al. Correlation between beta-catenin mutations and expression of Wnt-signaling target genes in hepatocellular carcinoma. Mol Cancer. 2008;7:21. doi: 10.1186/1476-4598-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dal Bello B, Rosa L, Campanini N, et al. Glutamine synthetase immunostaining correlates with pathologic features of hepatocellular carcinoma and better survival after radiofrequency thermal ablation. Clin Cancer Res. 2010;16:2157–2166. doi: 10.1158/1078-0432.CCR-09-1978. [DOI] [PubMed] [Google Scholar]
- 119.Lok AS, Seeff LB, Morgan TR, et al. Incidence of hepatocellular carcinoma and associated risk factors in hepatitis C-related advanced liver disease. Gastroenterology. 2009;136:138–148. doi: 10.1053/j.gastro.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cadoret A, Ovejero C, Saadi-Kheddouci S, et al. Hepatomegaly in transgenic mice expressing an oncogenic form of beta-catenin. Cancer Res. 2001;61:3245–3249. [PubMed] [Google Scholar]
- 121.Harada N, Miyoshi H, Murai N, et al. Lack of tumorigenesis in the mouse liver after adenovirus-mediated expression of a dominant stable mutant of beta-catenin. Cancer Res. 2002;62:1971–1977. [PubMed] [Google Scholar]
- 122.Nejak-Bowen KN, Thompson MD, Singh S, et al. Accelerated liver regeneration and hepatocarcinogenesis in mice overexpressing serine-45 mutant beta-catenin. Hepatology. 2010;51:1603–1613. doi: 10.1002/hep.23538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tan X, Apte U, Micsenyi A, et al. Epidermal growth factor receptor: a novel target of the Wnt/beta-catenin pathway in liver. Gastroenterology. 2005;129:285–302. doi: 10.1053/j.gastro.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Harada N, Oshima H, Katoh M, et al. Hepatocarcinogenesis in mice with beta-catenin and Haras gene mutations. Cancer Res. 2004;64:48–54. doi: 10.1158/0008-5472.can-03-2123. [DOI] [PubMed] [Google Scholar]
- 125.Miyoshi H, Deguchi A, Nakau M, et al. Hepatocellular carcinoma development induced by conditional beta-catenin activation in Lkb1+/− mice. Cancer Sci. 2009;100:2046–2053. doi: 10.1111/j.1349-7006.2009.01284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bauer-Hofmann R, Klimek F, Buchmann A, et al. Role of mutations at codon 61 of the c-Ha-ras gene during diethylnitrosamine-induced hepatocarcinogenesis in C3H/He mice. Mol Carcinog. 1992;6:60–67. doi: 10.1002/mc.2940060110. [DOI] [PubMed] [Google Scholar]
- 127.Colnot S, Decaens T, Niwa-Kawakita M, et al. Liver-targeted disruption of Apc in mice activates beta-catenin signaling and leads to hepatocellular carcinomas. Proc Natl Acad Sci U S A. 2004;101:17216–17221. doi: 10.1073/pnas.0404761101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Haramis AP, Hurlstone A, van der Velden Y, et al. Adenomatous polyposis coli-deficient zebrafish are susceptible to digestive tract neoplasia. EMBO Rep. 2006;7:444–449. doi: 10.1038/sj.embor.7400638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Qian J, Sarnaik AA, Bonney TM, et al. The APC tumor suppressor inhibits DNA replication by directly binding to DNA via its carboxyl terminus. Gastroenterology. 2008;135:152–162. doi: 10.1053/j.gastro.2008.03.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Aydinlik H, Nguyen TD, Moennikes O, et al. Selective pressure during tumor promotion by phenobarbital leads to clonal outgrowth of beta-catenin-mutated mouse liver tumors. Oncogene. 2001;20:7812–7816. doi: 10.1038/sj.onc.1204982. [DOI] [PubMed] [Google Scholar]
- 131.Huang H, Ushijima T, Nagao M, et al. Beta-catenin mutations in liver tumors induced by 2-amino-3,4-dimethylimidazo[4,5-f]quinoline in CDF1 mice. Cancer Lett. 2003;198:29–35. doi: 10.1016/s0304-3835(03)00273-8. [DOI] [PubMed] [Google Scholar]
- 132.Koch A, Denkhaus D, Albrecht S, et al. Childhood hepatoblastomas frequently carry a mutated degradation targeting box of the beta-catenin gene. Cancer Res. 1999;59:269–273. [PubMed] [Google Scholar]
- 133.Udatsu Y, Kusafuka T, Kuroda S, et al. High frequency of beta-catenin mutations in hepatoblastoma. Pediatr Surg Int. 2001;17:508–512. doi: 10.1007/s003830000576. [DOI] [PubMed] [Google Scholar]
- 134.Wei Y, Fabre M, Branchereau S, et al. Activation of beta-catenin in epithelial and mesenchymal hepatoblastomas. Oncogene. 2000;19:498–504. doi: 10.1038/sj.onc.1203356. [DOI] [PubMed] [Google Scholar]
- 135.Jeng YM, Wu MZ, Mao TL, et al. Somatic mutations of beta-catenin play a crucial role in the tumorigenesis of sporadic hepatoblastoma. Cancer Lett. 2000;152:45–51. doi: 10.1016/s0304-3835(99)00433-4. [DOI] [PubMed] [Google Scholar]
- 136.Kurahashi H, Takami K, Oue T, et al. Biallelic inactivation of the APC gene in hepatoblastoma. Cancer Res. 1995;55:5007–5011. [PubMed] [Google Scholar]
- 137.Oda H, Imai Y, Nakatsuru Y, et al. Somatic mutations of the APC gene in sporadic hepatoblastomas. Cancer Res. 1996;56:3320–3323. [PubMed] [Google Scholar]
- 138.Cairo S, Armengol C, De Reynies A, et al. Hepatic stem-like phenotype and interplay of Wnt/beta-catenin and Myc signaling in aggressive childhood liver cancer. Cancer Cell. 2008;14:471–484. doi: 10.1016/j.ccr.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 139.Fukuzawa R, Hata J, Hayashi Y, et al. Beckwith-Wiedemann syndrome-associated hepatoblastoma: wnt signal activation occurs later in tumorigenesis in patients with 11p15. 5 uniparental disomy. Pediatr Dev Pathol. 2003;6:299–306. doi: 10.1007/s10024-003-1009-1. [DOI] [PubMed] [Google Scholar]
- 140.Lade AG, Monga SP. Beta-catenin signaling in hepatic development and progenitors: which way does the WNT blow? Dev Dyn. 2011;240:486–500. doi: 10.1002/dvdy.22522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shin D, Monga SP. Cellular and molecular basis of liver development. Compr Physiol. 2013;3:799–815. doi: 10.1002/cphy.c120022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lopez-Terrada D, Gunaratne PH, Adesina AM, et al. Histologic subtypes of hepatoblastoma are characterized by differential canonical Wnt and Notch pathway activation in DLK+ precursors. Hum Pathol. 2009;40:783–794. doi: 10.1016/j.humpath.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 143.Armengol C, Cairo S, Fabre M, et al. Wnt signaling and hepatocarcinogenesis: the hepatoblastoma model. Int J Biochem Cell Biol. 2011;43:265–270. doi: 10.1016/j.biocel.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 144.Tao J, Calvisi DF, Ranganathan S, et al. Activation of beta-catenin and Yap1 in human hepatoblastoma and induction of hepatocarcinogenesis in mice. Gastroenterology. 2014;147:690–701. doi: 10.1053/j.gastro.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gebhardt R. Speeding up hepatocyte proliferation: how triiodothyronine and beta-catenin join forces. Hepatology. 2014;59:2074–2076. doi: 10.1002/hep.26984. [DOI] [PubMed] [Google Scholar]
- 146.Delgado ER, Yang J, So J, et al. Identification and characterization of a novel small-molecule inhibitor of beta-catenin signaling. Am J Pathol. 2014;184:2111–2122. doi: 10.1016/j.ajpath.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Emami KH, Nguyen C, Ma H, et al. A small molecule inhibitor of beta-catenin/CREB-binding protein transcription [corrected] Proc Natl Acad Sci U S A. 2004;101:12682–12687. doi: 10.1073/pnas.0404875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lenz HJ, Kahn M. Safely targeting cancer stem cells via selective catenin coactivator antagonism. Cancer Sci. 2014;105:1087–1092. doi: 10.1111/cas.12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Delgado E, Okabe H, Preziosi M, et al. Complete response of Ctnnb1-mutated tumours to beta-catenin suppression by locked nucleic acid antisense in a mouse hepatocarcinogenesis model. J Hepatol. 2015;62:380–387. doi: 10.1016/j.jhep.2014.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Calvisi DF. Molecularly targeted therapies for human hepatocellular carcinoma: should we start from beta-catenin inhibition? J Hepatol. 2015;62:257–259. doi: 10.1016/j.jhep.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 151.Wickline ED, Awuah PK, Behari J, et al. Hepatocyte gamma-catenin compensates for conditionally deleted beta-catenin at adherens junctions. J Hepatol. 2011;55:1256–1262. doi: 10.1016/j.jhep.2011.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wickline ED, Du Y, Stolz DB, et al. γ-Catenin at adherens junctions: mechanism and biologic implications in hepatocellular cancer after beta-catenin knockdown. Neoplasia. 2013;15:421–434. doi: 10.1593/neo.122098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yan D, Wiesmann M, Rohan M, et al. Elevated expression of axin2 and hnkd mRNA provides evidence that Wnt/beta-catenin signaling is activated in human colon tumors. Proc Natl Acad Sci U S A. 2001;98:14973–14978. doi: 10.1073/pnas.261574498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ranganathan S, Tan X, Monga SP. beta-Catenin and met deregulation in childhood hepatoblastomas. Pediatr Dev Pathol. 2005;8:435–447. doi: 10.1007/s10024-005-0028-5. [DOI] [PubMed] [Google Scholar]
- 155.Gotoh J, Obata M, Yoshie M, et al. Cyclin D1 over-expression correlates with beta-catenin activation, but not with H-ras mutations, and phosphorylation of Akt, GSK3 beta and ERK1/2 in mouse hepatic carcinogenesis. Carcinogenesis. 2003;24:435–442. doi: 10.1093/carcin/24.3.435. [DOI] [PubMed] [Google Scholar]
- 156.Sekine S, Gutierrez PJ, Lan BY, et al. Liver-specific loss of beta-catenin results in delayed hepatocyte proliferation after partial hepatectomy. Hepatology. 2007;45:361–368. doi: 10.1002/hep.21523. [DOI] [PubMed] [Google Scholar]
- 157.Tan X, Yuan Y, Zeng G, et al. Beta-catenin deletion in hepatoblasts disrupts hepatic morphogenesis and survival during mouse development. Hepatology. 2008;47:1667–1679. doi: 10.1002/hep.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yamamoto Y, Sakamoto M, Fujii G, et al. Overexpression of orphan G-protein-coupled receptor, Gpr49, in human hepatocellular carcinomas with beta-catenin mutations. Hepatology. 2003;37:528–533. doi: 10.1053/jhep.2003.50029. [DOI] [PubMed] [Google Scholar]
- 159.Cadoret A, Ovejero C, Terris B, et al. New targets of beta-catenin signaling in the liver are involved in the glutamine metabolism. Oncogene. 2002;21:8293–8301. doi: 10.1038/sj.onc.1206118. [DOI] [PubMed] [Google Scholar]
- 160.Loeppen S, Schneider D, Gaunitz F, et al. Over-expression of glutamine synthetase is associated with beta-catenin-mutations in mouse liver tumors during promotion of hepatocarcinogenesis by phenobarbital. Cancer Res. 2002;62:5685–5688. [PubMed] [Google Scholar]
- 161.Zhang T, Otevrel T, Gao Z, et al. Evidence that APC regulates survivin expression: a possible mechanism contributing to the stem cell origin of colon cancer. Cancer Res. 2001;61:8664–8667. [PubMed] [Google Scholar]
- 162.Roose J, Huls G, van Beest M, et al. Synergy between tumor suppressor APC and the beta-catenin-Tcf4 target Tcf1. Science. 1999;285:1923–1926. doi: 10.1126/science.285.5435.1923. [DOI] [PubMed] [Google Scholar]
- 163.Okabe H, Delgado E, Lee JM, et al. Role of leukocyte cell-derived chemotaxin 2 as a biomarker in hepatocellular carcinoma. PLoS One. 2014;9:e98817. doi: 10.1371/journal.pone.0098817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ovejero C, Cavard C, Perianin A, et al. Identification of the leukocyte cell-derived chemotaxin 2 as a direct target gene of beta-catenin in the liver. Hepatology. 2004;40:167–176. doi: 10.1002/hep.20286. [DOI] [PubMed] [Google Scholar]
- 165.Delgado E, Bahal R, Yang J, et al. β-Catenin knockdown in liver tumor cells by a cell permeable gamma guanidine-based peptide nucleic acid. Curr Cancer Drug Targets. 2013;13:867–878. doi: 10.2174/15680096113139990081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhang X, Gaspard JP, Chung DC. Regulation of vascular endothelial growth factor by the Wnt and K-ras pathways in colonic neoplasia. Cancer Res. 2001;61:6050–6054. [PubMed] [Google Scholar]
- 167.Nejak-Bowen KN, Zeng G, Tan X, et al. Beta-catenin regulates vitamin C biosynthesis and cell survival in murine liver. J Biol Chem. 2009;284:28115–28127. doi: 10.1074/jbc.M109.047258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wong CM, Fan ST, Ng IO. beta-Catenin mutation and overexpression in hepatocellular carcinoma: clinicopathologic and prognostic significance. Cancer. 2001;92:136–145. doi: 10.1002/1097-0142(20010701)92:1<136::aid-cncr1301>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 169.Kondo Y, Kanai Y, Sakamoto M, et al. Beta-catenin accumulation and mutation of exon 3 of the beta-catenin gene in hepatocellular carcinoma. Jpn J Cancer Res. 1999;90:1301–1309. doi: 10.1111/j.1349-7006.1999.tb00712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]