Abstract
DNA double-strand breaks are the critical lesions responsible for the majority of ionizing radiation-induced cell killing. Thus, the ability of tumor cells to elicit a DNA damage response following radiation, via activation of DNA repair and cell cycle checkpoints, promotes radiation resistance and tumor cell survival. Consequently, agents which target these DNA damage response pathways are being developed to overcome radiation resistance. Overall, these agents are effective radiosensitizers; however, their mechanisms of tumor cell selectivity are not fully elucidated. In this review, we will focus on the crucial radiation-induced DNA damage responses as well as clinical and translational advances with agents designed to inhibit these responses. Importantly, we describe how synthetic lethality can provide tumor cell selective radiosensitization by these agents and expand the therapeutic window for DNA damage response-targeted agents used in combination with radiation therapy.
Background
There are many factors which influence tumor cell sensitivity or resistance to ionizing radiation including the tumor microenvironment (1, 2), membrane signaling (3), and the immune system (4). However, as the principal target of radiation, DNA damage is the most critical factor governing radiation-induced cell death (5). DNA is subject to an array of different types of damage following exposure to ionizing radiation including base and sugar damage, crosslinks, as well as both single- and double- strand breaks (SSB and DSB, respectively). In particular, DNA DSBs are largely responsible for the cellular lethality of radiation. Hence, the ability of cells to recognize and respond to DSBs is fundamental in determining the sensitivity (or resistance) of cells to radiation. In this review, we will focus on radiation-induced DNA damage and repair as well as the cell cycle checkpoint pathways activated to mitigate this damage, with an emphasis on therapeutic targeting of these pathways to improve radiation therapy outcomes.
Radiation-induced DNA damage
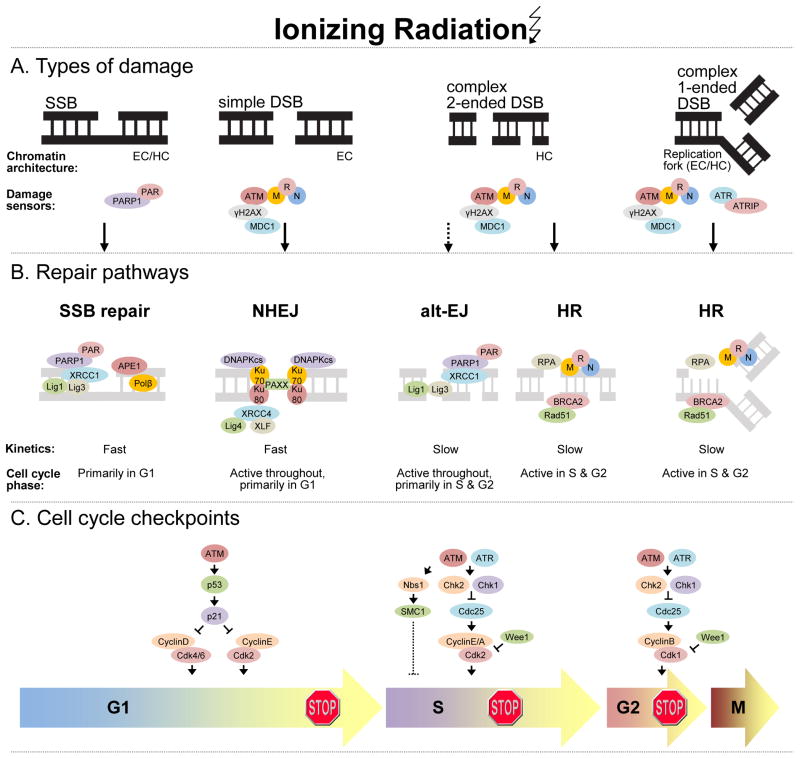
DNA DSBs can be simple or complex depending on several factors including the physical characteristics of the break, the chromatin architecture surrounding the break, and the kinetics with which the break is repaired (6–8). Simple DSBs are often classified as 2-ended DSBs directly induced by radiation, occurring in euchromatic regions that are repaired with fast kinetics (Fig. 1A). On the other hand, complex DSBs are characterized as 2-ended DSBs occurring in close proximity to other types of damage (crosslinks, SSBs, etc.) or within heterochromatic regions. In addition, replication-associated DSBs which occur as a result of a SSB colliding with an active DNA replication fork, referred to herein as 1-ended DSBs, also represent a complex type of DSB. All of these complex types of DSBs are repaired with relatively slow kinetics. Given the involvement of persistent SSBs in the generation of 1-ended DSBs, SSBs are also of relevance to radiation-induced DSBs. SSBs may arise via direct radiation-induced DNA damage or as intermediaries formed during repair of other single strand lesions (e.g. base excision repair of oxidized bases).
Figure 1.
The effects of radiation-induced DNA damage. A, Major types of radiation-induced DNA damage with respective DNA damage sensor proteins are illustrated. Radiation induces single-strand breaks (SSB) either directly or indirectly as intermediates of base excision repair. Simple double-strand breaks (DSB) involve two broken DNA ends in close proximity and occur in euchromatin (EC). Complex DSBs involve two broken DNA ends (i.e. 2-ended DSB) in proximity to additional DNA damage (e.g. cross-links, SSBs, etc.) or within heterochromatin (HC), or a DSB within a replication fork (1-ended DSB). B, SSBs and simple DSBs are repaired with fast kinetics by SSB repair and non-homologous end-joining (NHEJ) pathways, respectively. Alternative-end-joining (alt-EJ) is a slow, compensatory repair pathway activated when DNA-PKcs is absent or when NHEJ/HR attempt, but fail to complete repair. Alt-EJ likely contributes to repair of complex 2-ended DSBs. Homologous recombination (HR) operates under slow kinetics and is partly responsible for repair of complex 2-ended DSBs and exclusively responsible for repair of 1-ended DSBs. These repair pathways function in a cell cycle dependent manner, as illustrated. C, Cell cycle checkpoints are activated in response to DNA damage to prevent propagation of cells with damaged DNA and to permit time for DNA repair. The major checkpoints include those occurring in G1, S and G2. While ATM activation is the initial response to radiation-induced DNA DSBs, ATR is subsequently activated and contributes to a sustained cell cycle checkpoint response. Dashed lines represent incompletely understood pathways. Other abbreviations: ATRIP, ATR interacting protein; MRN, Mre11-Rad50-Nbs1
Ataxia telangiectasia mutated (ATM), in cooperation with the trimeric protein complex composed of Mre11-Rad50-Nbs1 (MRN), is the earliest responder to DNA DSBs. Activation and localization of these proteins at sites of DSBs in ‘radiation-induced foci’ initiates phosphorylation of the histone variant, γH2AX and the recruitment of Mediator of DNA damage checkpoint protein 1 (MDC1) as well as additional ATM and MRN molecules, necessary for the orchestration of subsequent DNA DSB repair and cell cycle checkpoints. In addition to ATM, ataxia telangiectasia mutated and RAD3-related (ATR) plays a role in initiating the response to DNA DSBs in S and G2 phases of the cell cycle, especially in the context of replication-associated 1-ended DSBs (9). In contrast to DSBs, direct SSBs are detected by PARP1 which catalyzes the formation of poly-ADP-ribose (PAR) chains on itself and other acceptor proteins to facilitate the recruitment of DNA repair factors such as x-ray cross-complementing protein 1 (XRCC1) and DNA polymerase β (POLβ).
DNA damage repair
DNA DSB repair is comprised of two major and mechanistically distinct processes: non-homologous end-joining (NHEJ) and homologous recombination (HR) (Fig. 1B). NHEJ and HR contribute to the fast and slow components of DSB repair with half-lives in the range of 5–30 minutes and 2–5 hours, respectively (10, 11). Although highly efficient, NHEJ is error prone, given its ability to catalyze simple rejoining reactions between DNA ends with no sequence homology. NHEJ repairs the majority of direct 2-ended radiation-induced DSBs and is the predominant DSB repair mechanism in G1 phase, although activity is present in all phases of the cell cycle. Following recognition of DNA DSBs, NHEJ begins with the stabilization of free DNA ends mediated by binding of the KU70 and KU80 heterodimers and subsequent recruitment of DNA-PKcs. 53BP1 and KU70/80 prevent DNA resection, an initial step of other DSB repair processes. NHEJ proceeds in a DNA-PK dependent manner with recruitment of other core NHEJ proteins including XRCC4 (x-ray cross-complement protein 4), LIG4 (ligase 4), XLF (XRCC4-like factor), and the recently identified PAXX (paralog of XRCC4 and XLF) (12), which together mediate alignment and ligation of DNA ends. NHEJ likely represents the a priori DNA DSB repair mechanism as other DSB repair pathways, discussed below, only occur following initial attempts to repair by NHEJ (13).
Alternative end-joining (alt-EJ; also referred to as alt-NHEJ or B-NHEJ) mechanisms are also being elucidated and represent pathways that may compensate in the absence of core NHEJ proteins or when NHEJ or HR initiate, but fail to complete repair (reviewed in (14)). Alt-EJ is generally distinguished from classical NHEJ based on its lack of requirement of core NHEJ proteins such as DNA-PKcs and KU70/80, its slower kinetics and its dependence on PARP1, XRCC1 and LIG1 or LIG3. In some cases alt-EJ is associated with microhomologous DNA sequences (e.g. MMEJ, microhomology-mediated end-joining) which likely represent a sub-pathway of alt-EJ. The factors involved in alt-EJ (and its probable sub-pathways) are still being elucidated, as it is not yet clear whether other factors found at alt-EJ junctions such as KU70/80, MRN, and CtIP [(carboxy-terminal binding protein) interacting protein] are direct contributors to alt-EJ or are simply remnants from failed repair attempts by NHEJ or HR.
Homologous recombination repair is a relatively slow, although highly accurate repair process owing to its requirement for extensive end resection and homologous DNA sequences. While the contribution of HR to the repair of simple radiation-induced DSBs is likely minor, HR is crucial for the repair of complex DNA DSBs (Fig. 1B). This is due to the fact that as lesions become increasingly complex homologous templates are necessary for faithful repair. Given the requirement for homologous DNA sequences, HR can only operate during the S and G2 phases of the cell cycle when sister chromatids are present. The core components of HR execute the major steps of the pathway beginning with resection of 5′ DNA ends which involves the MRN complex and CtIP, followed by replication protein A (RPA) binding to single stranded 3′ DNA ends. Initiated by BRCA2, RAD51 displaces RPA and mediates strand invasion to a homologous DNA strand forming the characteristic Holliday junction structure, followed by the final steps of HR which include synthesis at the single strand DNA regions, branch migration and junction resolution.
As alluded to above, unrepaired SSBs contribute to the formation of complex DSBs. The SSB repair pathway is a common pathway shared for repair of direct SSBs as well as the SSB intermediates created during base excision repair (BER) by excision of a damaged base and its corresponding phosphodiester bond. Following SSB detection (described above), PARP1 facilitates SSB repair by recruitment of the XRCC1 scaffolding protein to the SSB (Fig. 1B). XRCC1 promotes recruitment and stabilization of SSB repair enzymes, including those which mediate end-processing (e.g. APE1), gap synthesis (e.g. POLβ), and DNA ligation (e.g. LIG1 or LIG3) (reviewed in (15)).
DNA damage-induced cell cycle checkpoints
In response to DNA damage, cell cycle checkpoints are activated to block cell cycle progression and prevent propagation of cells with damaged DNA. The major DNA damage inducible checkpoints occur in G1, S, and G2 and are initiated by the same machinery responsible for DNA DSB repair recognition, ATM and ATR. The G1 checkpoint is mediated mainly by ATM which results in phosphorylation and activation of P53 transcriptional activity, leading to increased transcription of the cyclin-dependent kinase inhibitor, P21, which induces G1 arrest by inhibiting G1 and early S-phase cyclin-CDK complexes (Fig. 1C). Cells in S-phase at the time of radiation exhibit a slowing of DNA synthesis mediated by two distinct pathways, ATM/NBS1/SMC1 and ATM/CHK2/CDC25A/CDK2 (16). While it is not clearly understood how the former pathway regulates DNA synthesis, there is evidence that recruitment of the MRN complex to replication sites by RPA is involved (17). In contrast, the latter pathway negatively regulates DNA replication by preventing loading of the replication factor, Cdc45 onto replication origins (18). Activation of the G2 checkpoint in response to DNA damage prevents entry of cells into mitosis and is initiated by ATM-mediated activation of CHK1 and CHK2. While ATM is the initial activator of this pathway, delayed ATR activation contributes to a sustained checkpoint response and is especially important in the context of replication-associated DNA DSBs (9). Activated CHK1 and CHK2 phosphorylate CDC25C and trigger its cytoplasmic sequestration and inactivation. In the absence of CDC25C phosphatase activity, CDK1 remains bound by inhibitory phosphorylations (catalyzed by the WEE1 kinase) resulting in G2 arrest.
Clinical–Translational Advances
Given the lethality of unrepaired DNA DSBs, the development of agents designed to target proteins involved in the DNA damage response and thus, prevent repair or cell cycle checkpoints in response to DNA damage is an intense area of investigation. While the majority of these agents (Table 1) are effective radiosensitizers, tumor cell selectively remains an outstanding issue in their development. As monotherapy, the main developmental thrust for these agents is based on the principles of synthetic lethality, such that agents targeting a given DNA damage response pathway are strategically being applied to tumors with aberrations in other DNA damage response pathways. This strategy confers tumor cell selectivity since normal tissues have intact DNA damage response pathways and thus are spared from synthetic lethality. For radiotherapy combination studies, however, these synthetic lethal interactions are generally not being utilized, despite their potential to maximize the efficacy of these agents with radiation. Given that modern radiation therapy planning and delivery facilitates the induction of tumor selective DNA damage, radiation should potentiate synthetic lethal interactions between DNA damage response pathways and therefore, may improve the efficacy of some less robust synthetic lethal interactions. In this section we will review synthetic lethal interactions between agents and genes involved in radiation-induced DNA damage responses. We will also begin to address whether extending these synthetic lethal mechanisms to radiation combination studies can improve the therapeutic window for these agents.
Table 1.
Agents targeting the DNA damage response in clinical and pre-clinical development*
| Target | Agent | Single agent development stage | Combination agent development stage | Reference or clinical trial identifier number(s) |
|---|---|---|---|---|
| ATM | KU55933, KU59403 | Pre-clinical | Pre-clinical (RT, chemo) | 58 |
| AZ32 | - | Pre-clinical (RT) | 22 | |
| ATR | AZD6738 | Phase 1 | Phase 1 (RT, chemo1) | NCT02223923, NCT02264678 |
| VE-821/VE-822, VX-970 | Pre-clinical | Phase 1 (chemo2) | NCT02157792 | |
| Pre-clinical (RT) | 23 | |||
| CHK1 | LY2606368 (Chk1/2) | Phase 2 | Phase 1 (chemo3) | NCT02124148 |
| LY2603618 | Phase 2 | Phase 2 (chemo4) | NCT01139775, NCT00839332 | |
| MK8776 | Phase 1 | Phase 2 (chemo5) | NCT01870596, NCT00779584 | |
| DNA-PK | CC-115 (DNA-PK & mTOR) | Phase 1 | - | NCT01343625 |
| ZSTK474 (PI3 kinase) | Phase 2 | - | NCT01682473 | |
| LIG4 | SCR7 | Pre-clinical | Pre-clinical (RT, chemo) | 24 |
| PARP | Olaparib | Approved | Phase 1 (RT, chemoRT6) | NCT01460888, NCT01562210 |
| Phase 3 (chemo7) | NCT01924533 | |||
| Veliparib | Phase 3 | Phase 1 (RT) | NCT01264432, NCT01589419 | |
| Phase 2 (chemoRT8) | NCT01514201, NCT01386385 | |||
| Phase 3 (chemo9) | NCT02163694, NCT02152982 | |||
| Niraparib | Phase 3 | Phase 1 (chemo10) | NCT01847274, NCT02044120 | |
| RAD51 | RI-1 | Pre-clinical | Pre-clinical (chemo) | 37 |
| B02 | Pre-clinical | Pre-clinical (chemo) | 38 | |
| RPA | Compound 8 | Pre-clinical | - | 45 |
| HAMNO | Pre-clinical | Pre-clinical (chemo) | 46 | |
| SMI MCI13E | Pre-clinical | Pre-clinical (chemo) | 47 | |
| WEE1 | AZD1775 | Phase 1 | Phase 1/2 (RT, chemoRT11) | NCT01922076, NCT02037230 |
| Phase 2 (chemo12) | NCT02272790, NCT01076400 |
Studies representing the most advanced developmental stage to date were selected and do not represent an all-inclusive list.
Carboplatin;
gemcitabine, cisplatin, or etoposide;
cisplatin or cetuximab;
pemetrexed-cisplatin, gemcitabine, or pemetrexed;
cytarabine or gemcitabine;
cisplatin-RT;
paclitaxel;
temozolomide-RT, carboplatin-paclitaxel-RT;
carboplatin-paclitaxel, temozolomide;
Temozolomide;
cisplatin-RT, temozolomide-RT, gemcitabine-RT;
gemcitabine, paclitaxel, carboplatin, or cisplatin
As monotherapy, the prototypical example of synthetic lethality between an agent and a gene involved in the DNA damage response is PARP inhibition in BRCA1/2 mutant cancers. Since this discovery, numerous other synthetic lethal interactions between DNA damage response pathways have been elucidated. For example, ATM defective cancers are particularly susceptible to DNA-PK or ATR inhibition (19, 20). Likewise, ERCC1 defective tumors are sensitive to ATR inhibition (21).
While there is extensive evidence to support the ability of DNA damage response targeted agents to radiosensitize (22–26), there is considerably less data to support their tumor cell selectivity. Perhaps, the best established tumor cell selective mechanism is illustrated by the synthetic lethality occurring between CHK1 or WEE1 inhibitors and mutant TP53. In combination with radiation, CHK1 or WEE1 inhibitors are synthetically lethal in TP53 mutant cancers (26–28). This is due in part to abrogation of the G2 checkpoint by CHK1 or WEE1 inhibitors, which is particularly detrimental in TP53 mutant cancers that lack a G1 checkpoint. In addition, it is plausible that the ability of CHK1/WEE1 inhibitors to inhibit HR (29–31) may also play a role in their selectivity, as TP53 mutant cancer cells are more likely than normal cells to rely on HR for DSB repair due to their inability to arrest in G1 where NHEJ is the dominant DSB repair mechanism. Similarly, TP53 mutant gliomas are preferentially radiosensitized by ATM inhibition, although the mechanism of this selectivity is not fully understood (32).
There are numerous ongoing clinical trials combining PARP1/2 inhibitors with radiation or chemoradiation therapy (Table 1). A suggested model for radiosensitization by PARP inhibition involves inhibition of BER/SSB repair, leading to persistent radiation-induced SSBs that are converted to cytotoxic 1-ended DSBs in replicating cells (33, 34). Given that 1-ended DSBs require HR for repair and that PARP inhibition is highly effective in HR defective (i.e. BRCA mutant) cancers, it is plausible to speculate that BRCA1/2 mutant cancers should be radiosensitized by PARP inhibition. This, however, is likely an oversimplification of the potential mechanisms of interaction between radiation, BRCA mutation and PARP inhibition, underscoring the requirement for thorough investigation in this area. In addition to inhibition of BER/SSB repair, inhibition of alt-EJ is also involved in PARP inhibitor-mediated radiosensitization. This is supported by the findings that radiosensitization is replication-independent in NHEJ deficient cells (33) and that POLβ null cells, which are defective in BER/SSB repair, are maximally radiosensitized by PARP inhibition (35). Taken together, these studies indicate that radiosensitization by PARP inhibitors involves multiple factors including replication, BRCA1/2 mutation, as well as NHEJ and BER/SSB repair status that require careful evaluation in order to maximize the therapeutic index of PARP inhibitors with radiation.
Given the observed synthetic lethal interactions between the BER/SSB and HR pathways, illustrated by the efficacy of PARP inhibitors in BRCA1/2 mutant tumors, the inverse has been investigated to determine whether HR inhibitors are effective in BER defective cancers. Indeed, in BER defective tumors, ATM inhibition or RAD51 deficiency preferentially induces radiosensitization in PP;β deficient relative to POLβ proficient tumor cells (36). These studies underscore the importance of the development of selective HR inhibitors such as those targeting RAD51 (37, 38) and their testing as radiosensitizers, especially in cancers with a high frequency of POLβ mutations (reviewed in (39)).
Efforts are underway to extend these synthetic lethal interactions beyond genes directly involved in the DNA damage response to common oncogenes such as KRAS and MYC that, when deregulated, lead to replication stress, genomic instability, endogenous DNA damage, and ultimately an increased reliance on DNA damage response pathways such as those mediated by ATR/CHK1 (40–43). Underscoring the potential efficacy of targeting ATR/CHK1 in KRAS/MYC-driven cancers, initial studies have suggested that CHK1 inhibition (alone or in combination with a PARP inhibitor) confers greater radiosensitization in isogenic KRAS mutant vs. KRAS wild-type cancer cells (reviewed in (44)). Strategies for directly targeting oncogene-induced replication stress through the use of small molecules that disrupt RPA-mediated protein-protein or protein-single-stranded DNA interactions are also underway (45–47). Taken together, these types of studies will determine whether oncogene-induced replication stress is a targetable phenomenon and may provide a rationale for extending the use of other agents targeting the DNA damage response to cancers with oncogene-induced replication stress. Future studies are necessary to determine whether radiation enhances these interactions.
In order to pair a targeted agent with a tumor cell defect in a specific DNA damage response pathway, reproducible assays which capture the activity of these pathways in human tissues are necessary. While numerous mutations in genes involved in individual repair pathways have been found to influence sensitivity to DNA damage response-targeted agents, a complete understanding of the genes, their specific mutations, and their mutational landscape that lead to overall functional defects in DNA damage response pathways is still lacking (48). Therefore, functional assays provide valuable insights into the global activity of a given DNA damage response pathway. For example, HR status has been determined in breast tumor biopsies by assessing RAD51 focus formation following ex vivo-irradiation (49). The utility of this functional HR assay is illustrated by its ability to identify HR defective cancers beyond those with characteristic BRCA1/2 mutations (50). Furthermore, functional assays have been used to assess BER in tumor biopsies and may prove useful for predicting sensitivity to DNA damage response-targeted agents (51). Although functional assays for NHEJ are more difficult since core NHEJ proteins do not form readily visible foci at DSBs due to their low quantities, some success in this area was demonstrated by detection of phopsphorylated-KU70 foci (52).
In the absence of obvious defects in DNA damage response pathways, an additional strategy for achieving synthetic lethality is to use pairs of agents strategically selected to target epistatic DNA damage response pathways. In an effort to extend the synthetic lethality of PARP inhibitors beyond BRCA1/2 mutant tumors, agents that inhibit HR are an intense area of investigation. To date, many agents which indirectly inhibit HR have been identified and are being combined with PARP inhibitors, such as inhibitors of EGFR (53), PI3K (54), HSP90 (55), CHK1 (56) and WEE1 (30). Some of these agent pairs such as those targeting WEE1-PARP or HSP90-PARP appear to be especially effective when given in combination with radiation (reviewed in (44)), supporting the notion that radiation-induced DNA damage can potentiate these interactions. Finally, based on the consistent observation that PARP inhibitors in combination with radiation lead to a more robust G2 checkpoint (likely due to increased DNA damage), agents which inhibit the G2 checkpoint such as Chk1 and Wee1 inhibitors lead to improved radiosensitization, especially in TP53 mutant cancers which lack a G1 checkpoint (30, 56, 57). Given that pairs of agents do not afford the same tumor cell selectivity that an agent paired with a tumor specific mutation does, the tumor cell selectivity of agent pairs needs to be carefully investigated.
In recent years, the number of known synthetic lethal interactions between DNA damage response pathways has expanded considerably and in some instances these interactions are being exploited in pre-clinical radiation studies. Despite these advancements however, none of the current clinical radiation studies (Table 1) are incorporating this information. In order to improve the therapeutic window for these DNA damage response-targeted agents in combination with radiotherapy, mechanisms of synthetic lethality in the context of radiosensitization need to be further investigated. In addition, both functional and genomic assays which permit identification of DNA damage response pathway defects in tumor cells need to continue to be developed. The large number of DNA damage-targeted agents in development and the frequency of tumor defects in DNA damage response pathways offer great potential to improve the efficacy of radiation therapy.
Acknowledgments
The authors thank Dr. Thomas Wilson for helpful discussions and Dr. Qiang Zhang for manuscript review and editing.
Grant Support
M.A. Morgan was supported by the NIH under award number R01CA163895. T.S. Lawrence was supported by the NIH under award numbers R01CA138723, P3CA130810, and P50CA130810 and an A. Alfred Taubman Scholarship.
Footnotes
Disclosure of Potential Conflicts of Interest
M.A. Morgan reports receiving a commercial research grant from AstraZeneca. No potential conflicts of interest were disclosed by the other author.
References
- 1.Wilson WR, Hay MP. Targeting hypoxia in cancer therapy. Nat Rev Cancer. 2011;11:393–410. doi: 10.1038/nrc3064. [DOI] [PubMed] [Google Scholar]
- 2.Baumann M, Krause M, Hill R. Exploring the role of cancer stem cells in radioresistance. Nat Rev Cancer. 2008;8:545–54. doi: 10.1038/nrc2419. [DOI] [PubMed] [Google Scholar]
- 3.Toulany M, Rodemann HP. Membrane receptor signaling and control of DNA repair after exposure to ionizing radiation. Nuklearmedizin. 2010;49 (Suppl 1):S26–30. [PubMed] [Google Scholar]
- 4.Formenti SC, Demaria S. Radiation therapy to convert the tumor into an in situ vaccine. Int J Radiat Oncol Biol Phys. 2012;84:879–80. doi: 10.1016/j.ijrobp.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mladenov E, Magin S, Soni A, Iliakis G. DNA double-strand break repair as determinant of cellular radiosensitivity to killing and target in radiation therapy. Front Oncol. 2013;3:113. doi: 10.3389/fonc.2013.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Groth P, Orta ML, Elvers I, Majumder MM, Lagerqvist A, Helleday T. Homologous recombination repairs secondary replication induced DNA double-strand breaks after ionizing radiation. Nucleic Acids Res. 2012;40:6585–94. doi: 10.1093/nar/gks315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodarzi AA, Jeggo P, Lobrich M. The influence of heterochromatin on DNA double strand break repair: Getting the strong, silent type to relax. DNA Repair (Amst) 2010;9:1273–82. doi: 10.1016/j.dnarep.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 8.Shibata A, Conrad S, Birraux J, Geuting V, Barton O, Ismail A, et al. Factors determining DNA double-strand break repair pathway choice in G2 phase. EMBO J. 2011;30:1079–92. doi: 10.1038/emboj.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flynn RL, Zou L. ATR: a master conductor of cellular responses to DNA replication stress. Trends Biochem Sci. 2011;36:133–40. doi: 10.1016/j.tibs.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iliakis G, Dahm-Daphi J, Dikomey E. DNA repair and cell cycle regulation after ionizing irradiation. In: Molls M, Vaupel P, Nieder C, Anscher MS, editors. The impact of tumor biology on cancer treatment and multidisciplinary strategies. Berlin, Heidelberg: Springer-Verlag; 2009. pp. 251–71. [Google Scholar]
- 11.Jeggo PA, Geuting V, Lobrich M. The role of homologous recombination in radiation-induced double-strand break repair. Radiother Oncol. 2011;101:7–12. doi: 10.1016/j.radonc.2011.06.019. [DOI] [PubMed] [Google Scholar]
- 12.Ochi T, Blackford AN, Coates J, Jhujh S, Mehmood S, Tamura N, et al. DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science. 2015;347:185–8. doi: 10.1126/science.1261971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kakarougkas A, Jeggo PA. DNA DSB repair pathway choice: an orchestrated handover mechanism. Br J Radiol. 2014;87:20130685. doi: 10.1259/bjr.20130685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dueva R, Iliakis G. Alternative pathways of non-homologous end joining (NHEJ) in genomic instability and cancer. Transl Cancer Res. 2013;2:163–77. [Google Scholar]
- 15.Caldecott KW. DNA single-strand break repair. Exp Cell Res. 2014;329:2–8. doi: 10.1016/j.yexcr.2014.08.027. [DOI] [PubMed] [Google Scholar]
- 16.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–23. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 17.Olson E, Nievera CJ, Liu E, Lee AY, Chen L, Wu X. The Mre11 complex mediates the S-phase checkpoint through an interaction with replication protein A. Mol Cell Biol. 2007;27:6053–67. doi: 10.1128/MCB.00532-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falck J, Petrini JH, Williams BR, Lukas J, Bartek J. The DNA damage-dependent intra-S phase checkpoint is regulated by parallel pathways. Nat Genet. 2002;30:290–4. doi: 10.1038/ng845. [DOI] [PubMed] [Google Scholar]
- 19.Riabinska A, Daheim M, Herter-Sprie GS, Winkler J, Fritz C, Hallek M, et al. Therapeutic targeting of a robust non-oncogene addiction to PRKDC in ATM-defective tumors. Science Transl Med. 2013;5:189ra78. doi: 10.1126/scitranslmed.3005814. [DOI] [PubMed] [Google Scholar]
- 20.Reaper PM, Griffiths MR, Long JM, Charrier JD, Maccormick S, Charlton PA, et al. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol. 2011;7:428–30. doi: 10.1038/nchembio.573. [DOI] [PubMed] [Google Scholar]
- 21.Mohni KN, Kavanaugh GM, Cortez D. ATR pathway inhibition is synthetically lethal in cancer cells with ERCC1 deficiency. Cancer Res. 2014;74:2835–45. doi: 10.1158/0008-5472.CAN-13-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karlin JD, Tokarz M, Beckta J, Farhan A, Pike K, Barlaam B, et al. A novel ATM kinase inhibitor effectively radiosensitizes glioblastoma in mice. Int J Radiat Oncol Biol Phys. 90:S35. [Google Scholar]
- 23.Fokas E, Prevo R, Pollard JR, Reaper PM, Charlton PA, Cornelissen B, et al. Targeting ATR in vivo using the novel inhibitor VE-822 results in selective sensitization of pancreatic tumors to radiation. Cell Death Dis. 2012;3:e441. doi: 10.1038/cddis.2012.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srivastava M, Nambiar M, Sharma S, Karki SS, Goldsmith G, Hegde M, et al. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell. 2012;151:1474–87. doi: 10.1016/j.cell.2012.11.054. [DOI] [PubMed] [Google Scholar]
- 25.Senra JM, Telfer BA, Cherry KE, McCrudden CM, Hirst DG, O’Connor MJ, et al. Inhibition of PARP-1 by olaparib (AZD2281) increases the radiosensitivity of a lung tumor xenograft. Mol Cancer Ther. 2011;10:1949–58. doi: 10.1158/1535-7163.MCT-11-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bridges KA, Hirai H, Buser CA, Brooks C, Liu H, Buchholz TA, et al. MK-1775, a novel Wee1 kinase inhibitor, radiosensitizes p53-defective human tumor cells. Clin Cancer Res. 2011;17:5638–48. doi: 10.1158/1078-0432.CCR-11-0650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell JB, Choudhuri R, Fabre K, Sowers AL, Citrin D, Zabludoff SD, et al. In vitro and in vivo radiation sensitization of human tumor cells by a novel checkpoint kinase inhibitor, AZD7762. Clin Cancer Res. 2010;16:2076–84. doi: 10.1158/1078-0432.CCR-09-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vance S, Parsels LA, Parsels JD, Maybaum J, Lawrence T, Morgan MA. Radiosensitization of pancreatic cancer cells by combined Chk1 and PARP1 inhibition using AZD7762 and olaparib [abstract]. Proceedings of the 102nd Annual Meeting of the American Association for Cancer Research; 2011 Apr 2–6; Orlando, Florida. Philadelphia (PA): AACR; 2011. p. Abstract nr 2657. [Google Scholar]
- 29.Morgan MA, Parsels LA, Zhao L, Parsels JD, Davis MA, Hassan MC, et al. Mechanism of radiosensitization by the Chk1/2 inhibitor AZD7762 involves abrogation of the G2 checkpoint and inhibition of homologous recombinational DNA repair. Cancer Res. 2010;70:4972–81. doi: 10.1158/0008-5472.CAN-09-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karnak D, Engelke CG, Parsels LA, Kausar T, Wei D, Robertson JR, et al. Combined inhibition of Wee1 and PARP1/2 for radiosensitization in pancreatic cancer. Clin Cancer Res. 2014;20:5085–96. doi: 10.1158/1078-0432.CCR-14-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krajewska M, Heijink AM, Bisselink YJ, Seinstra RI, Sillje HH, de Vries EG, et al. Forced activation of Cdk1 via wee1 inhibition impairs homologous recombination. Oncogene. 2013;32:3001–8. doi: 10.1038/onc.2012.296. [DOI] [PubMed] [Google Scholar]
- 32.Biddlestone-Thorpe L, Sajjad M, Rosenberg E, Beckta JM, Valerie NC, Tokarz M, et al. ATM kinase inhibition preferentially sensitizes p53-mutant glioma to ionizing radiation. Clin Cancer Res. 2013;19:3189–200. doi: 10.1158/1078-0432.CCR-12-3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loser DA, Shibata A, Shibata AK, Woodbine LJ, Jeggo PA, Chalmers AJ. Sensitization to radiation and alkylating agents by inhibitors of poly(ADP-ribose) polymerase is enhanced in cells deficient in DNA double-strand break repair. Mol Cancer Ther. 2010;9:1775–87. doi: 10.1158/1535-7163.MCT-09-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bridges KA, Toniatti C, Buser CA, Liu H, Buchholz TA, Meyn RE. Niraparib (MK-4827), a novel poly(ADP-Ribose) polymerase inhibitor, radiosensitizes human lung and breast cancer cells. Oncotarget. 2014;5:5076–86. doi: 10.18632/oncotarget.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chalmers AJ, Lakshman M, Chan N, Bristow RG. Poly(ADP-ribose) polymerase inhibition as a model for synthetic lethality in developing radiation oncology targets. Semin Radiat Oncol. 2010;20:274–81. doi: 10.1016/j.semradonc.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 36.Neijenhuis S, Verwijs-Janssen M, van den Broek LJ, Begg AC, Vens C. Targeted radiosensitization of cells expressing truncated DNA polymerase {beta} Cancer Res. 2010;70:8706–14. doi: 10.1158/0008-5472.CAN-09-3901. [DOI] [PubMed] [Google Scholar]
- 37.Budke B, Logan HL, Kalin JH, Zelivianskaia AS, Cameron McGuire W, Miller LL, et al. RI-1: a chemical inhibitor of RAD51 that disrupts homologous recombination in human cells. Nucleic Acids Res. 2012;40:7347–57. doi: 10.1093/nar/gks353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang F, Mazin AV. A small molecule inhibitor of human RAD51 potentiates breast cancer cell killing by therapeutic agents in mouse xenografts. PLoS One. 2014;9:e100993. doi: 10.1371/journal.pone.0100993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Starcevic D, Dalal S, Sweasy JB. Is there a link between DNA polymerase beta and cancer? Cell Cycle. 2004;3:998–1001. [PubMed] [Google Scholar]
- 40.Gilad O, Nabet BY, Ragland RL, Schoppy DW, Smith KD, Durham AC, et al. Combining ATR suppression with oncogenic Ras synergistically increases genomic instability, causing synthetic lethality or tumorigenesis in a dosage-dependent manner. Cancer Res. 2010;70:9693–702. doi: 10.1158/0008-5472.CAN-10-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grabocka E, Pylayeva-Gupta Y, Jones MJ, Lubkov V, Yemanaberhan E, Taylor L, et al. Wild-type H- and N-Ras promote mutant K-Ras-driven tumorigenesis by modulating the DNA damage response. Cancer Cell. 2014;25:243–56. doi: 10.1016/j.ccr.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rohban S, Campaner S. Myc induced replicative stress response: how to cope with it and exploit it. Biochim Biophys Acta. 2014 Apr 13; doi: 10.1016/j.bbagrm.2014.04.008. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 43.Murga M, Campaner S, Lopez-Contreras AJ, Toledo LI, Soria R, Montana MF, et al. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nat Struct Mol Biol. 2011;18:1331–5. doi: 10.1038/nsmb.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morgan MA, Parsels LA, Maybaum J, Lawrence TS. Improving the efficacy of chemoradiation with targeted agents. Cancer Discov. 2014;4:280–91. doi: 10.1158/2159-8290.CD-13-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frank AO, Feldkamp MD, Kennedy JP, Waterson AG, Pelz NF, Patrone JD, et al. Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach. J Med Chem. 2013;56:9242–50. doi: 10.1021/jm401333u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glanzer JG, Liu S, Wang L, Mosel A, Peng A, Oakley GG. RPA inhibition increases replication stress and suppresses tumor growth. Cancer Res. 2014;74:5165–72. doi: 10.1158/0008-5472.CAN-14-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neher TM, Bodenmiller D, Fitch RW, Jalal SI, Turchi JJ. Novel irreversible small molecule inhibitors of replication protein A display single-agent activity and synergize with cisplatin. Mol Cancer Ther. 2011;10:1796–806. doi: 10.1158/1535-7163.MCT-11-0303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hemann MT. From breaking bad to worse: exploiting homologous DNA repair deficiency in cancer. Cancer Discov. 2014;4:516–8. doi: 10.1158/2159-8290.CD-14-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Willers H, Taghian AG, Luo CM, Treszezamsky A, Sgroi DC, Powell SN. Utility of DNA repair protein foci for the detection of putative BRCA1 pathway defects in breast cancer biopsies. Mol Cancer Res. 2009;7:1304–9. doi: 10.1158/1541-7786.MCR-09-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naipal KA, Verkaik NS, Ameziane N, van Deurzen CH, Ter Brugge P, Meijers M, et al. Functional ex vivo assay to select homologous recombination-deficient breast tumors for PARP inhibitor treatment. Clin Cancer Res. 2014;20:4816–26. doi: 10.1158/1078-0432.CCR-14-0571. [DOI] [PubMed] [Google Scholar]
- 51.Slyskova J, Korenkova V, Collins AR, Prochazka P, Vodickova L, Svec J, et al. Functional, genetic, and epigenetic aspects of base and nucleotide excision repair in colorectal carcinomas. Clin Cancer Res. 2012;18:5878–87. doi: 10.1158/1078-0432.CCR-12-1380. [DOI] [PubMed] [Google Scholar]
- 52.Schuler N, Palm J, Kaiser M, Betten D, Furtwangler R, Rube C, et al. DNA-damage foci to detect and characterize DNA repair alterations in children treated for pediatric malignancies. PLoS One. 2014;9:e91319. doi: 10.1371/journal.pone.0091319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nowsheen S, Cooper T, Stanley JA, Yang ES. Synthetic lethal interactions between EGFR and PARP inhibition in human triple negative breast cancer cells. PLoS One. 2012;7:e46614. doi: 10.1371/journal.pone.0046614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ibrahim YH, Garcia-Garcia C, Serra V, He L, Torres-Lockhart K, Prat A, et al. PI3K inhibition impairs BRCA1/2 expression and sensitizes BRCA-proficient triple-negative breast cancer to PARP inhibition. Cancer Discov. 2012;2:1036–47. doi: 10.1158/2159-8290.CD-11-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dungey FA, Caldecott KW, Chalmers AJ. Enhanced radiosensitization of human glioma cells by combining inhibition of poly(ADP-ribose) polymerase with inhibition of heat shock protein 90. Mol Cancer Ther. 2009;8:2243–54. doi: 10.1158/1535-7163.MCT-09-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vance S, Liu E, Zhao L, Parsels JD, Parsels LA, Brown JL, et al. Selective radiosensitization of p53 mutant pancreatic cancer cells by combined inhibition of Chk1 and PARP1. Cell Cycle. 2011;10:4321–9. doi: 10.4161/cc.10.24.18661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu HR, Wang X, Wang Y. A stronger DNA damage-induced G2 checkpoint due to over-activated CHK1 in the absence of PARP-1. Cell Cycle. 2006;5:2364–70. doi: 10.4161/cc.5.20.3355. [DOI] [PubMed] [Google Scholar]
- 58.Cremona CA, Behrens A. ATM signalling and cancer. Oncogene. 2014;33:3351–60. doi: 10.1038/onc.2013.275. [DOI] [PubMed] [Google Scholar]