Abstract
The emergence of drug resistant pathogens is often considered a canonical case of evolution by ‘natural’ selection. Here we argue that the strength of selection can be a poor predictor of the rate of resistance emergence. It is possible for a resistant strain to be under negative selection and still emerge in an infection or spread in a population. Measuring the right parameters is a necessary first step towards the development of evidence-based resistance management strategies. We argue that it is the absolute fitness of the resistant strains that matters most, and that a primary determinant of the absolute fitness of a resistant strain when it arises is the ecological context in which it finds itself.
Keywords: Evolutionary rescue, chemotherapy, antibiotics, mutant selection window
Evolutionary Emergence of Resistance
When an infected patient is treated with antimicrobial chemotherapy the population of microbes within the patient begins to decline. During this process of population decline, genotypes resistant to the antimicrobial drug can appear through mutation or horizontal gene transfer. Resistant microbes also might have been present at the start of treatment. If this population of rare resistant genotypes then grows sufficiently in size to cause symptoms or to be transmitted we say that a drug-resistant infection has been established. We refer to this process as the evolutionary emergence of drug resistance.
Different chemotherapeutic protocols (e.g., combination versus mono therapy (1), synergistic versus antagonistic drug combinations (2–4), and high versus low drug concentration (5–9)) result in different likelihoods of resistance emergence. This is because such protocols affect the likelihood of resistant genotypes appearing through mutation, as well as the fitness of resistant and wild type genotypes once they have appeared. An important research objective is therefore to compare the impact of different protocols on the probability and rate of resistance emergence. Such information makes it possible to design protocols that simultaneously maximize treatment efficacy while managing resistance (5). Our goal here is to help progress this enterprise by considering the effect of different treatment protocols on the fitness of resistant and wild type microbes within a patient once they are present.
For the most part, studies of the factors influencing resistance emergence have focused on the selective advantage or disadvantage of drug resistant strains in treated and untreated patients (e.g., (1, 10–13)). Here we suggest that, instead, it is often more appropriate to focus on the absolute fitness of resistant strains in treated and untreated patients rather than their performance relative to sensitive strains (see Gloassary).
We make this argument in two parts. First, we suggest that the selective advantage of resistance is not the most important indicator of resistance emergence within treated patients. This is because, by definition, a focus on selection is a focus on the relative fitness of resistant and wild type microbes. Yet relative fitness tell us little about the extent to which the actual size of the resistant population is changing as a result of treatment. A focus on the absolute fitness of the resistant strain is usually more relevant to resistance emergence because resistance emerges when the absolute abundance of resistant microbes gets sufficiently high. The abundance of resistant microbes relative to that of sensitive microbes is often irrelevant (e.g., when both are very rare).
Second, we ask how different treatment regimens affect absolute fitness. We suggest that different treatment regimens result in different fitnesses of resistant strains by engendering different degrees of competitive release (14), a term borrowed from the ecological literature. Competitive release (defined in the subsection below - also see the Glossary) amplifies the numbers of resistant microbes, thus increasing the probability and rate of resistance emergence. We suggest that a recognition of the distinction between selection and competitive release will better guide future work on resistance management
Absolute versus relative fitness
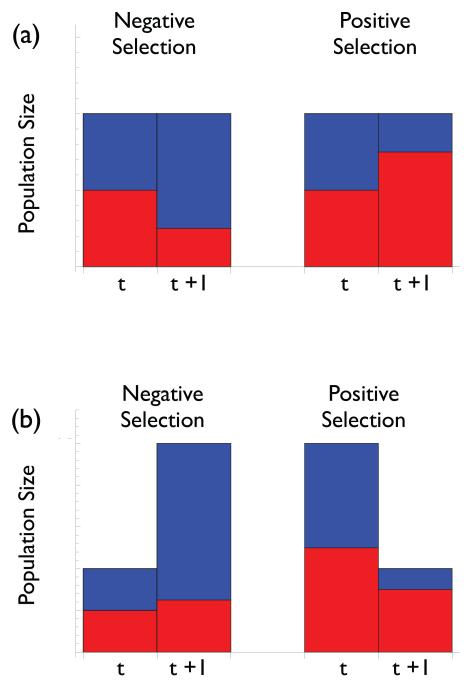
The first part of our argument is the simplest and rests on the important distinction between absolute and relative fitness. Evolution is a change in the genetic composition of a population. From the standpoint of evolution all that matters is the fitness of one type relative to another. The difference in fitness between the resistant and wild type strain is referred to as the selection coefficient (15). If the resistant strain has a higher fitness than the wild type then the selection coefficient will be positive and the resistant strain will come to make up a greater fraction of the population (termed positive selection). Conversely, if the selection coefficient is negative the resistant strain will come to make up a smaller fraction of the population (termed negative selection; Figure 1a).
Figure 1.
The distinction between relative and absolute fitness. Height of bars indicates total population size. Coloring indicates the fraction of the population made up of resistant (red) and wildtype (blue) strains. (a) Between time t and t+1 the left-hand population has undergone negative selection and thus resistant strains make up a smaller fraction of the population. The opposite is true for the right-hand population. (b) Between time t and t+1 the left- and right-hand populations have again undergone negative and positive selection respectively, but the absolute size of the resistant population has nevertheless increased in the case of negative selection and decreased in the case of positive selection.
However the probability of resistance emergence is a function of the absolute fitness of resistant microbes, not their fitness relative to that of the wild type. What matters from the standpoint of resistance emergence (in terms of the potential for resistant microbes to cause symptoms or transmit to other hosts) is the abundance of the resistant strain within a patient. The selection coefficient can tell us little about the predicted change in its population size over time. Figure 1b illustrates this point by showing how a resistant population can be under negative selection and nevertheless increase in size, as well as how it can be under positive selection and nevertheless decrease in size. A similar point has recently been made in the context of adaptation to environmental change (16).
The hypothetical scenario illustrated in Figure 1 is extremely simple, but analogous outcomes occur in real disease systems. For example, Box 1 presents data from experimental infections in mice with the malarial parasite Plasmodium chabaudi. It shows clear instances in which the drug resistant clone is under positive selection but is nevertheless decreasing in abundance, as well as instances in which the resistant clone is under negative selection but is nevertheless increasing in abundance, to the point where it has high transmission potential. To summarize then, it is the absolute fitness of the resistant microbes that determines emergence, not their fitness relative to wildtype microbes.
Box 1. Experimental infections with P. chabaudi.
Experimental work with rodent malaria Plasmodium chabaudi in laboratory mice illustrates the important difference between absolute and relative fitness.
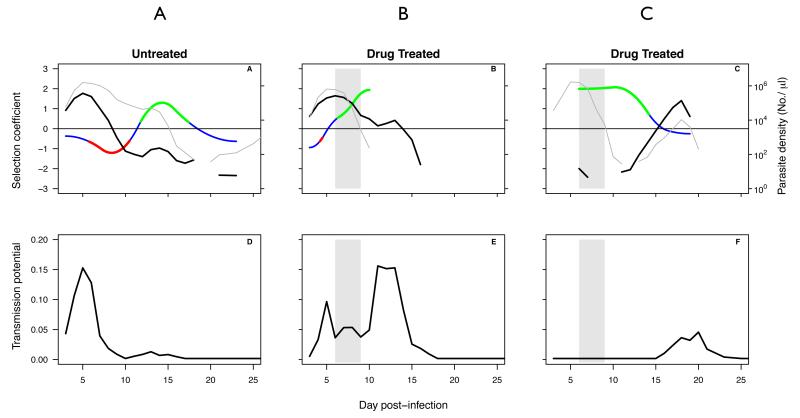
Figure B1-1 shows the complex relationships between selection on resistance measured by the selection coefficient and the absolute fitness of resistant parasites measured either in terms of parasite density (top panels) or transmission to mosquitoes (bottom panels). For example, in the untreated mouse (A), resistance is under very strong positive selection between days 13 and 18 post-infection, even though the abundance of resistant strains is decreasing. This is because the sensitive strain is decreasing in abundance even faster. The same thing occurs following drug treatment in mouse B. In both cases, resistance is not emerging despite strong positive selection. In contrast, following treatment of mouse C, the strong positive selection on resistance declines to zero even though the absolute fitness of the resistant strain is rising following competitive release. That is because the sensitive strain is also relapsing. In that case, resistance is clearly emerging, even though the strength of selection is declining to zero. Thus, the selection coefficient is a poor guide to the rate or probability of resistance emergence.
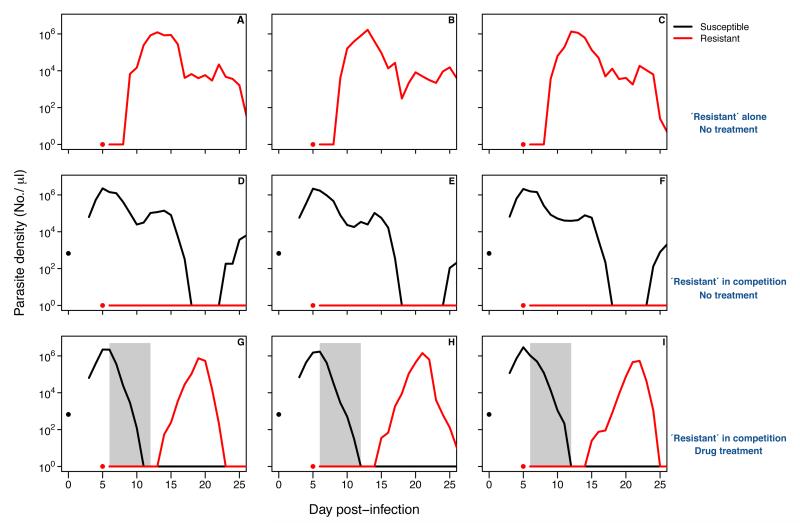
Data from the same experimental system also demonstrate that competitive suppression and competitive release are real biological phenomena. Figure B1-2 shows the kinetics of drug-resistant parasites of P. chabaudi in nine laboratory mice (red line). When they are alone in an infection (top panels), the resistant parasite population rapidly expands to high densities. However, if drug-sensitive parasites have already proliferated to high densities, the resistant parasite population is unable to expand (middle panels). This is competitive suppression. If the sensitive parasites are removed by drug treatment (lower panels), the resistant parasite population is able to expand. This is competitive release. Thus, the probability of resistance emergence is strongly linked to the extent of competitive release. Other experiments have shown that resistance emergence can be constrained by using treatment regimens which less effectively remove sensitive parasites (7).
Competitive release versus selection
Since it is absolute fitness that matters for resistance emergence we must consider how different treatment regimens affect the absolute fitness of resistant microbes. To focus our argument, we consider the contentious question of how the extent of drug pressure affects the probability of resistance emergence (5–9, 17). The term ‘drug pressure’ refers to a variety of factors including the time course of drug concentration during treatment (i.e., pharmacokinetics). However, for simplicity we will refer only to drug concentration. Also, for convenience, in what follows we use the terms ‘fitness’ and (per capita) growth rate interchangeably. We stress however that our arguments hold for any reasonable measure of fitness and any reasonable measure of drug pressure.
To begin, it is first helpful to review the main conceptual framework that is used for thinking about the effect of drug concentration on the emergence of resistance. This is the ‘Mutant Selection Window’ (MSW) hypothesis (18–22).
Drug resistance and the ‘Mutant Selection Window’ (MSW) hypothesis
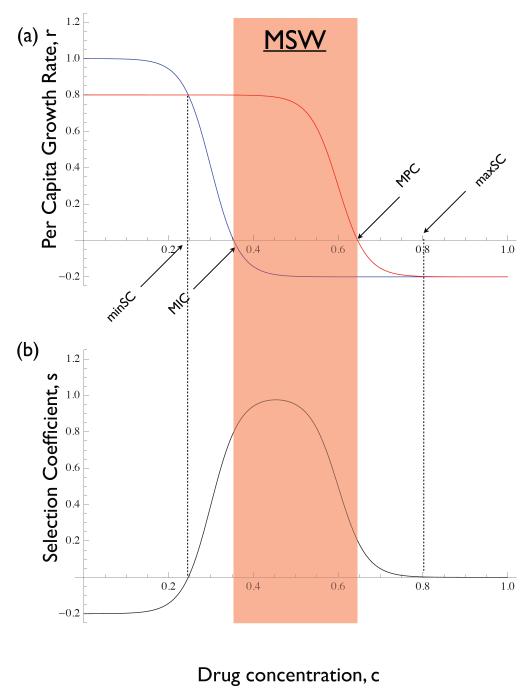
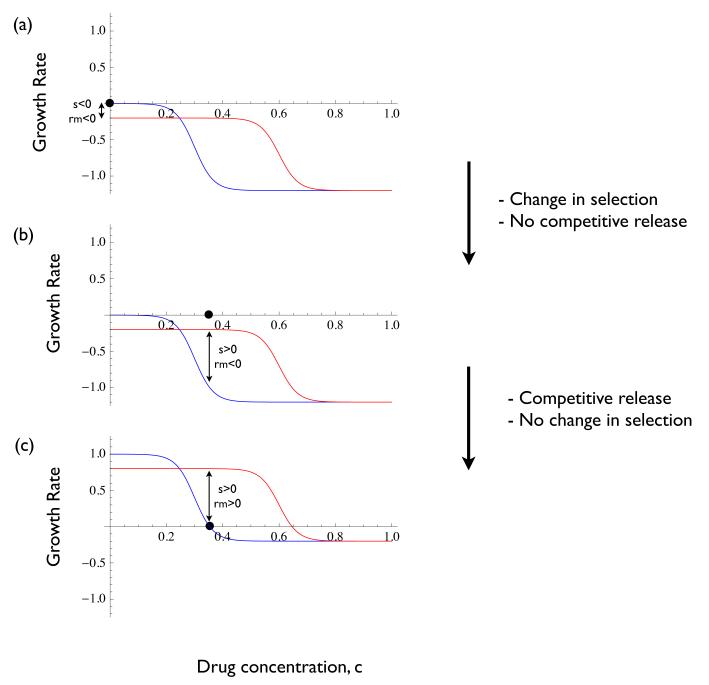
The MSW hypothesis was developed to predict the drug concentrations under which resistance will emerge. It is based on a plot of the per capita growth rate of the wild type and mutant strains as a function of drug concentration, c (20–22). Figure 2a illustrates a hypothetical example displaying several features that are typical of such plots. First, there is a cost to resistance in the absence of the drug, reflected by the resistant strain growth rate being lower than that of the wild type at c = 0. Second, by virtue of the resistant strain being able to withstand higher concentrations of the drug, its growth rate curve will eventually cross that of the wild type at some value of c (labelled ‘minSC’ in Figure 2a). And third, the growth rate curves converge again for very high values of c once the growth rates of both types is negative.
Figure 2.
The mutant selection window hypothesis. (a) per capita growth rates of mutant (red) and wild type (blue) as a function of drug concentration. Many instances of this model in the literature have the growth rate curves asymptoting to zero (as opposed to becoming negative). Strictly speaking they must cross the horizontal axis somewhere and become negative if there exists a drug concentration at which their growth rates are zero. Also labeled are the minimum inhibitory concentration (MIC), the mutant prevention concentration (MPC), the minimum selective concentration (minSC - the smallest concentration for which selection is positive), and the maximum selective concentration (maxCS - the largest concentration for which selection is positive). Shaded red window indicates the ‘mutant selection window’ (MSW) as defined in the literature. (b) The selection coefficient of the mutant is the difference in growth rate between it and the wild type. The mutant will be selectively favoured whenever s > 0. The mutant can be selectively advantageous for drug concentrations lying outside the MSW.
The drug concentration at which the wild type has zero growth rate is called the minimum inhibitory concentration (MIC; Figure 2a). The drug concentration at which the mutant has zero growth rate is called the mutant prevention concentration (MPC; Figure 2a). The mutant selection window (MSW) is then defined to be the range of drug concentrations between the MIC and the MPC. The reasoning is that, within this window of drug concentrations the mutant will be ‘selectively enriched’ whenever it appears (18–20).
The MSW model has been extremely influential and has been subjected to numerous empirical tests, particularly in bacteria (23–33). It is worth noting, however, that although this terminology is ingrained in the literature it is technically incorrect. In terms of selection it is not the window between the MIC and the MPC that matters. The mutant has a selective advantage at any drug concentration for which its growth rate lies above that of the wild type (i.e., where the selection coefficient is positive; Figure 2b). Thus the window of selection occurs between the lowest concentration at which the the wild type and mutant have the same growth rate (labeled minSC, for minimum selective concentration in Figure 2a) and the concentration at which the two growth rates again converge as concentration increases (labeled maxSC, for maximum selective concentration in Figure 2a; in practice the value of maxSC might only be reached asymptotically). The distinction with respect to the minSC has been pointed out clearly before (22) but it applies to the maxSC as well.
The misidentification of the MIC and MPC as the boundaries of the window of selective drug concentrations stems from a lack of distinction between relative and absolute fitness. From Figure 2 one can see that the lower boundary of the MSW as classically (and erroneously) defined is the drug concentration at which the wild type’s absolute fitness is zero whereas the upper boundary is the concentration at which the resistant strain’s absolute fitness is zero. But these bounds have nothing to do with selection per se (cf. Figure 2b).
Although the MSW hypothesis gets the terminology incorrect it does correctly identify the MPC as the upper boundary of the window of drug concentrations that allow for the emergence of resistance. But the lower boundary is incorrect because it focuses on the absolute fitness of the wild type rather than that of the resistant strain. And it is for this reason that experiments have been able to show that resistance can emerge at concentrations below the MIC (34–36).
But if we wish to identify the lowest drug concentration at which the resistant strain has a positive growth rate (i.e., positive absolute fitness) Figure 2 reveals a problem. It suggests that the resistant strain has a positive growth rate for all drug concentrations below the MPC. We know from empirical studies, however, that resistance does not emerge for all such concentrations (for instance, it typically does not emerge in the absence of drugs). How can we resolve this apparent contradiction? The answer lies in the fact that graphs like that in Figure 2 portray the relationship between drug concentration and absolute fitness as fixed. In reality these relationships are specific to the conditions under which they are measured.
As a microbial population grows it causes a ‘deterioration’ in its environment that ultimately will halt its own growth (i.e., make its absolute fitness decline to zero). For example, the depletion of resources and/or the stimulation of an immune response produce density dependence that eventually causes the per capita growth rate of the microbe to fall to zero. The data in Box 1 illustrates exactly this point in the case of P. chabaudi, where the growth rate declines to zero (and in fact becomes negative) after two weeks or more. This means that growth rate curves like those in Figure 2 are not fixed, but instead are functions of within-patient variables like resources levels, immune response, etc. Often curves like those in Figure 2 are measured under pristine conditions (i.e., during exponential growth) but it is the change in these curves that arises from changes in the within-patient variables that ultimately halts (and potentially prevents) the growth of wild type and resistant strains (e.g., Figure B1-1). Given that multiple-strain infections are extremely common in most microbes (37, 38), we suggest that quite generally it is the competitive interactions between resistant and wild type strains through such within-patient state variables that ultimately sets the lower boundary on the range of drug concentrations for which resistance can emerge.
Figure B1-1.
Data from three mice infected with pyrimethamine-resistant and sensitive strains of Plasmodium chabaudi at equal densities of 106 : 106 (A,B) or at a ratio of 1 : 105 (C). Mice were either untreated (A), or treated with 8 mg/kg of pyrimethamine for four days (B,C). Top panels show within host dynamics of resistant (thick black lines) and sensitive (thin black lines) parasites, and the selection coefficients (colored lines: green, resistance is under positive selection, red, resistance is under negative selection, blue, the selection coefficient not significantly different from zero). Bottom panels show predicted proportion of mosquitoes infected by resistant parasites for the corresponding mice, based on the densities of transmission stages (data not shown). Gray bars indicate timing and duration of drug treatment. Data from (7); transmission potential estimated as described in (7), selection coefficients as described in (13). Selection coefficients can only be estimated when both clones are present.
Competitive suppression and competitive release
To illustrate our argument, we begin by considering a process we will refer to as competitive suppression. Box 2 provides a specific hypothetical example. Suppose an infection is initiated and no drug treatment is being used. Then the growth of the wild type population will degrade the within-patient environment to a point where its fitness reaches zero. In other words, once density dependence (which acts through the within-patient state variables) has become strong enough, the growth rate curve for the wild type must necessarily cross the horizontal axis at drug concentration c = 0. At this point, because of a cost of resistance, the mutant growth rate curve will lie below that of the wild type and therefore resistance will fail to spread whenever it arises because of competitive suppression (mediated by the within-patient state variable, in this case immunity; Box 2, Figure B2-1a).
Box 2. Competitive Suppression.
Consider a population of wild type and resistant microbes with densities p and pm respectively. Suppose that density dependence acts solely through a shared immune response, as described by the following model:
| (2-1a) |
| (2-1b) |
| (2-1c) |
The λ’s are the per capita birth rates of the two types, as functions of drug concentration, and I is the density of a relevant immune molecule. The parameter a scales the effect of the immune response on the growth rate of the microbes. In this example the growth rates of the wild type and mutant are
| (2-2a) |
| (2-2b) |
illustrating how the growth rate curves (as functions of c) also depend on the within-patient state variable I. The selection coefficient is
| (2-3a) |
| (2-3b) |
illustrating that, in this example, the selection coefficient is independent of the within-patient variables I.
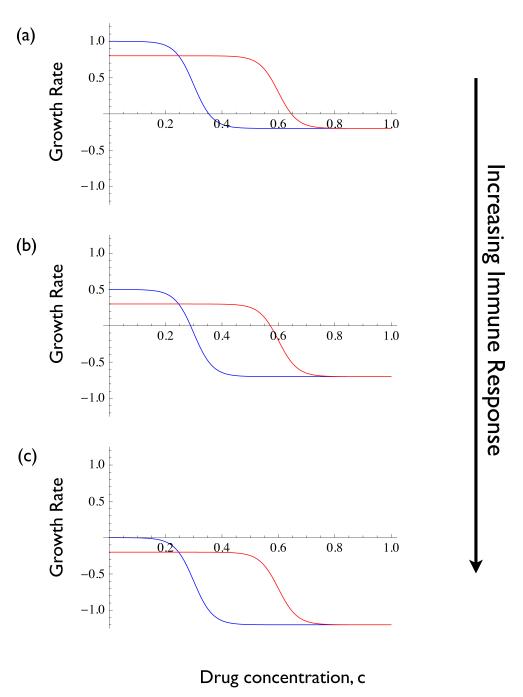
Figure B2-1 illustrates what this means in terms of the growth rate functions, like those of Figure 2. We assume that the drug concentration is held at c = 0 and the infection starts with only wild type individuals. Panel (a) is the ‘pristine’ environment, before any immune response has developed. Panels (b) and (c) show the growth curves for increasing immune responses, I. As the wild type grows, its stimulates an increasing immune response. This is the deterioration of the within-patient environment (from the standpoint of microbial growth). The mutant curve shows the growth rate that a mutant would have if it appeared under the different conditions. The growth curves are eventually pushed downwards until the growth rate of the wild type at c = 0 is zero. At this point, any mutant that appears will have a negative growth rate because of the cost of resistance. In other words, it will be competitively suppressed through the within-patient variable I and thus will not spread even though it would have a positive growth rate if it caused an infection on its own.
Figure B2-1.
Theoretical growth rate curves for wild type (blue) and resistant (red) genotypes. (a) Curves at the beginning of an infection, (b) Curves as the infection develops, and (c) Curves once density dependence through the immune response is strong enough to halt wild type growth.
From this hypothetical example we can see that ultimately what matters from the standpoint of the emergence of resistance is the resistant genotype’s absolute fitness rm when it appears. If rm > 0 then it can emerge. And a key observation is that we can have positive selection but nevertheless rm < 0 and therefore emergence is impossible. Likewise, we can have negative selection but nevertheless rm > 0 and therefore emergence will occur. The latter can happen if, for example, the drug concentration is zero and if both the resistant and wild type strains were present at the start of an infection. In this case the wild type will not yet have caused a change in the within-patient environment (i.e., no immune response or resource depletion will have occurred), and therefore both strains will grow despite the fact that the resistant strains suffers a cost of resistance (e.g., see Box 2, Figure B2-1a). Only once density-dependence (competitive suppression) sets in can the cost of resistance reduce the absolute fitness of resistant strains to zero.
With this idea of competitive suppression in hand, we can now define competitive release. Competitive release is the increase in the absolute mutant fitness rm that comes from removing the wild type with chemotherapy. Competitive release therefore necessarily arises through an ecological interaction between the wild type and the mutant, as mediated through some element of the within-patient environment. Box 3 illustrates the phenomenon of competitive release in the context of the hypothetical example from Box 2, and highlights how it is distinct from selection.
Box 3. Competitive Release versus Selection.
We continue with the example from Box 2. Suppose that the infection initially contains only the wild type, and the drug concentration is c = 0. The wildtype microbe then grows to the point where a large enough immune response is stimulated to stop wild type growth. In this case the wild type growth curve passes through the horizontal axis at c = 0, the growth rate of any mutant that appears is negative, and the selection coefficient is negative (Figure B3-1a; this is also Figure B2-1c, which illustrates the case where the resistant strain is competitively suppressed).
Now suppose that the drug is administered in way that achieves a constant concentration of c = 0.35 (Figure B3-1b). Immediately the selective coefficient becomes positive, but any mutant that appears will still have a negative growth rate and therefore will not spread. The wild type also now has a negative growth rate, however, and therefore its population will decline. As it does so, the within-patient environmental state will rebound (in this example, I from Box 2 decays), eventually lifting the competitive suppression of the mutant and allowing it to have a positive growth rate (i.e., it experiences competitive release; Figure B3-1c). Thus, in this example, the selection coefficient changes from panel (a) to (b) but not from panel (b) to (c). However, the mutant growth rate changes from panel (b) to (c) through competitive release, and it is this release that allows the mutant to spread.
To make this idea more precise it is helpful to introduce some notation. Suppose x is the within-patient state variable (e.g., density of resources, immune cells, etc), and suppose that x0 is the value of this variable in the absence of infection (i.e., the ‘pristine’ environment). We will use r(c, x) and rm(c, x) for the growth rates of the wild type and mutant, to indicate that they are functions of both the drug concentration and the within-patient state variable x.
If an infection starts with the wild type and the drug concentration is zero (i.e., c = 0), then its initial growth rate will be positive; that is r(0, x0) > 0. As the wild type population grows, x changes, eventually reaching a value (denoted by x*) at which r(0, x*) = 0. This corresponds to Figure B3-1a in Box 3. Now suppose we introduce drug treatment at a concentration c. The wild type growth rate will be r(c, x*), and this will be negative, meaning that the wild type will now decrease in abundance. This corresponds to Figures B3-1b in Box 3. As the wild type decreases, the within-patient variable x will rebound towards its ‘pristine’ value. Competitive release is defined as the difference rm(c, x) – rm(c, x*). This is the change in mutant growth rate that results from the within-patient environment rebounding from x* to x, when the drug concentration is at c (see Figures B3-1c in Box 3). On the other hand, the selection coefficient is rm(c, x) – r(c, x), which is the difference between mutant and wild type growth rates when the environment is at x and the drug concentration is c.
Figure B3-1.
Theoretical growth rate curves for wild type (blue) and resistant (red) genotypes. (a) Curves once density dependence through the immune response is strong enough to halt wild type growth but before treatment begins, (b) Curves immediately after treatment begins, and (c) Curves once treatment has caused competitive release.
Although we have discussed the concepts of competitive suppression and competitive release in abstract terms, they are biologically very real phenomena (Box 1). Resistant strains grow well on their own (Fig B1-2 top panels). When the susceptible strain is already present and has degraded the environment within a mouse, the resistant strain can no longer grow (competitive suppression, Fig B1-2 middle panels). Removing sensitive parasites with drugs allows the environment within a mouse to support growth of resistant parasites (competitive release, Fig B1-2, bottom panels).
Figure B1-2.
The kinetics of infection in nine mice infected with pyrimethamine-resistant (red) and sensitive (black) P. chabaudi. All mice were infected with approx. 25 resistant parasites on day 5 (red dots). Mice D-I were also infected with 106 sensitive parasites five days earlier (black dots), mice G-I were treated with 8 mg/kg of pyrimethamine for seven days to eliminate sensitive parasites. Gray bars show period of drug treatment. Note, formally, the flat red lines denote times below PCR detection, and not necessarily zero densities. Data are from (7).
Concluding remarks
It is now commonplace to view the spread of drug resistance through the lens of evolutionary biology, with the goal of using advances in this area of fundamental science to help address the important applied problems that resistance poses. Here we have, in essence, argued that there is a critical ecological process that underlies the emergence of resistance; namely, competitive release. Understanding, and potentially controlling, the initial emergence of resistance therefore requires that we understand how competition works, and how contrasting treatment strategies affect this process of competitive release. While we have focused attention on the problem of drug resistance and infectious diseases, it is also worth noting that similar issues arise in other instances of adaptation to novel environments. These range from the emergence of resistance in cancer chemotherapy, to invasive species biology, to adaptation to climate change (16).
Outstanding Questions.
What drug treatment regimens best reduce the absolute fitness of resistant microbes?
How common is competitive release?
What are the mechanisms of competitive suppression and competitive release?
How do host responses contribute to competitive suppression (e.g., through strain-transcending immunity)?
Is resistance emergence more or less likely in acute self-resolving infections?
Acknowledgements
For discussion, we thank W. Nelson and members of the Research and Policy in Infectious Disease Dynamics program of the Science and Technology Directorate, the Department of Homeland Security, the Fogarty International Center, the National Institutes of Health, and particularly those at the Princeton RAPIDD workshop organized by J. Metcalf and R. Kouyos. We also thank G. Teitzel, D. Kennedy, B. Levin, and two anonymous referees for helpful comments on the manuscript. This research was supported by a grant from the Natural Sciences and Engineering Research Council of Canada (TD), a Society in Science - Branco Weiss Fellowship and Marie Curie IIF Fellowship (SH), and by the National Institute of General Medical Sciences (R01GM089932) (AFR). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Glossary
- Drug resistance
a heritable reduction in the drug sensitivity of a microbe.
- Resistance emergence
when a population of rare resistant microbes within a patient increases sufficiently in size to cause symptoms or to be transmitted.
- Absolute Abundance
number of pathogens at some point in time.
- Relative Abundance
a synonym for frequency.
- Growth rate (per capita)
rate of change of abundance per individual microbe.
- Fitness
a term that refers to reproductive success of pathogen and involves both reproduction and survival. It is measured in terms of genetic representation in the next generation.
- Absolute fitness
the fitness of a pathogen clone, independent of the fitness of any other clone; it usually involves some measure of change in absolute abundance like per capita growth rate.
- Relative fitness
the fitness of a pathogen clone relative to the fitness of another clone(s); it usually involves some measure of change in relative abundance (e.g., frequency).
- Selection coefficient
a measure of relative fitness; often absolute fitness of the resistant strain minus the absolute fitness of the wildtype.
- Negative selection
when the selection coefficient is negative; in this case the resistant clone will decrease in frequency.
- Positive selection
when the selection coefficient is positive; in this case the resistant clone will increase in frequency.
- Natural selection
any process by which the forms (variants) of organisms in a population that are best adapted to a particular environment increase in relative frequency as compared with less well-adapted forms over a number of generations (39).
- Competitive suppression
the decrease in absolute fitness of a resistant clone as a result of the wildtype consuming shared resources and/or stimulating a cross-reactive immune response.
- Competitive release
the increase in absolute fitness of a resistant clone that occurs when the wildtype is removed by chemotherapy; this increase in absolute fitness arises through the increased resource abundance and/or decreased immune response that occurs upon the removal of the wildtype.
References
- [1].zur Wiesch PA, Kouyos R, Engelstader J, Regoes R, Bonhoeffer S. Population biological principles of drug-resistance evolution in infectious diseases. The Lancet, Infectious Diseases. 2011;11:236–247. doi: 10.1016/S1473-3099(10)70264-4. [DOI] [PubMed] [Google Scholar]
- [2].Chait R, Craney A, Kishony R. Antiobiotic interactions that select against resistance. Nature. 2007;446:668–671. doi: 10.1038/nature05685. [DOI] [PubMed] [Google Scholar]
- [3].Hegreness M, Shoresh N, Damian D, Hartl D, Kishony R. Accelerated evolution of resistance in multi drug environments. Proceedings of the National Academy of Science. 2008;105:13977–13981. doi: 10.1073/pnas.0805965105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Pena-Miller R, Laehnemann D, Jansen G, Fuentes-Hernandez A, Rosenthiel P, Schulemburg H, Beardmore R. When the most potent combination of antibiotics selects for the greatest bacterial load: the smile-frown transition. PLoS Biology. 2013;11:e1001540. doi: 10.1371/journal.pbio.1001540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Read AF, Day T, Huijben S. The evolution of drug resistance and the curious orthodoxy of aggressive chemotherapy. Proceedings of the National Academy of Science. 2012;108:10871–10877. doi: 10.1073/pnas.1100299108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Geli P, Laxminarayan R, Dunne M, Smith DL. One-size-fits-all? optimizing treatment duration for bacterial infections. PLoS One. 2012;7:e29838. doi: 10.1371/journal.pone.0029838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Huijben S, Bell AS, Sim DG, Salathe R, Tomasello D, Mideo N, Day T, Read AF. Aggressive chemotherapy and the selection of drug resistant pathogens. PLoS Pathogens. 2013;9:e1003578. doi: 10.1371/journal.ppat.1003578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ankomah P, Levin BR. Exploring the collaboration between antibiotics and the immune response in the treatment of acute, self-limiting infections. Proceedings of the National Academy of Science. 2014;111:8331–8338. doi: 10.1073/pnas.1400352111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kouyos RD, Metcalf CJE, R., et al. Biger. The path of least resistance: aggressive or moderate treatment? Proceedings of the Royal Society, B. 2014;281:20140566. doi: 10.1098/rspb.2014.0566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Olofsson SK, Cars O. Optimizing drug exposure to minimize selection of antibiotic resistance. Clinical Infectious Diseases. 2007;45:S129–S136. doi: 10.1086/519256. [DOI] [PubMed] [Google Scholar]
- [11].Palmer AC, Kishony R. Understanding, predicting and manipulating the genotypic evolution of antibiotic resistance. Nature Reviews Genetics. 2013;14:243–248. doi: 10.1038/nrg3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Huijben S, Nelson WA, Wargo AR, Sim DG, Drew DR, Read AF. Chemotherapy, within-host ecology and the fitness of drug resistant malaria parasites. Evolution. 2010;64:2952–2968. doi: 10.1111/j.1558-5646.2010.01068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Huijben S, Sim D, Nelson WA, Read AF. The fitness of drug resistant malaria parasites in a rodent model: multiplicity of infection. Journal of Evolutionary Biology. 2011;24:2410–2422. doi: 10.1111/j.1420-9101.2011.02369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].de Roode JC, Culleton R, Bell AS, Read AF. Competitive release of drug resistance following drug treatment of mixed Plasmodium chabaudi infections. Malaria Journal. 2004;3:33. doi: 10.1186/1475-2875-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Otto SP, Day T. A biologist’s guide to mathematical modeling in ecology and evolution. Princeton University Press; 2007. [Google Scholar]
- [16].Bell G. Evolutionary rescue and the limits of adaptation. Philosophical Transactions of the Royal Society, B. 2013;368:20120080. doi: 10.1098/rstb.2012.0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].van den Bosch F, Paveley N, Shaw M, Hobbelen P, Oliver R. The dose rate debate: does the risk of fungicide resistance increase or decrease with dose? Plant Pathology. 2011;60:597606. [Google Scholar]
- [18].Zhao X, Drlica K. Restricting the selection of antibiotic-resistant mutants: a general strategy derived from fluoroquinolone studies. Clinical Infectious Diseases. 2001;33:S147–S156. doi: 10.1086/321841. [DOI] [PubMed] [Google Scholar]
- [19].Zhao X, Drlica K. Restricting the selection of antibiotic-resistant mutant bacteria: measurement and potential use of the mutant selection window. The Journal of Infectious Diseases. 2002;185:561–565. doi: 10.1086/338571. [DOI] [PubMed] [Google Scholar]
- [20].Drlica K, Zhao X. Mutant selection window hypothesis updated. Clinical Infectious Diseases. 2007;44:681688. doi: 10.1086/511642. [DOI] [PubMed] [Google Scholar]
- [21].Canton R, Morosini M-I. Emergence and spread of antibiotic resistance following exposure to antibiotics. FEMS Microbiology Reviews. 2011;35:977–991. doi: 10.1111/j.1574-6976.2011.00295.x. [DOI] [PubMed] [Google Scholar]
- [22].Andersson DI, Hughes D. Evolution of antibiotic resistance at non-lethal drug concentrations. Drug Resistance Updates. 2012;15:162–172. doi: 10.1016/j.drup.2012.03.005. [DOI] [PubMed] [Google Scholar]
- [23].Sindelar G, Zhao X, Liew A, Dong Y, Lu T, Zhou J, Domagala J, Drlica K. Mutant prevention concentration as a measure of fluoroquinolone potency against mycobacteria. Antimicrobial Agents and Chemotherapy. 2000;44:3337–3343. doi: 10.1128/aac.44.12.3337-3343.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Firsov AA, Vostrov SN, Lubenko IY, Drlica K, Portnoy YA, Zinner SH. In vitro pharmacodynamic evaluation of the mutant selection window hypothesis using four fluoroquinolones against Staphylococcus aureus. Antimicrobial Agents and Chemother apy. 2003;47:1604–1613. doi: 10.1128/AAC.47.5.1604-1613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zinner SH, Lubenko IY, Gilbert D, Simmons K, Zhao X, Drlica K, Firsov AA. Emergence of resistant Streptococcus pneumoniae in an in vitro dynamic model the simulates moxifloxacin concentrations inside and outside the mutant selection window: related changes in susceptibility, resistance frequency and bacterial killing. Journal of Antimicrobial Chemotherapy. 2003;52:616–622. doi: 10.1093/jac/dkg401. [DOI] [PubMed] [Google Scholar]
- [26].Firsov AA, Vostrov SN, Lubenko IY, Arzamastsev AP, Portnoy YA, Zinner SH. Abt492 and levofloxacin: comparison of their pharmacodynamics and their abilities to prevent the selection of resistant Staphylococcus aureus in an in vitro model. Journal of Antimicrobial Chemotherapy. 2004;54:178–186. doi: 10.1093/jac/dkh242. [DOI] [PubMed] [Google Scholar]
- [27].Tam VH, Louie A, Deziel MR, Liu W, Leary R, Drusano GL. Bacterial-population responses to drug-selective pressure: examination of garenoxacin’s effect on Pseudomonas aeruginosa. Journal of Infectious Diseases. 2005;192:420–428. doi: 10.1086/430611. [DOI] [PubMed] [Google Scholar]
- [28].Firsov AA, Smirnova MV, Lubenko IY, Vostrov SN, Portnoy YA, Zinner SH. Testing the mutant selection window hypothesis with Staphylococcus aureus exposed to daptomycin and vancomycin in an in vitro model. Journal of Antimicrobial Chemotherapy. 2006;58:1185–1192. doi: 10.1093/jac/dkl387. [DOI] [PubMed] [Google Scholar]
- [29].Olofsson SK, Marcusson LL, Lindgren PK, Hughes D, Cars O. Selection of ciprofloxacin resistance in Escherichia coli in an in vitro kinetic model: relation between drug exposure and mutant prevention concentration. Journal of Antimicrobial Chemotherapy. 2006;57:1116–1121. doi: 10.1093/jac/dkl135. [DOI] [PubMed] [Google Scholar]
- [30].Tam VH, Louie A, Deziel MR, Liu W, Drusano GL. The relationship between quinolone exposures and resistance amplification is characterized by an inverted u: a new paradigm for optimizing pharmacodynamics to counterselect resistance. Antimicrobial Agents and Chemotherapy. 2007;51:744–747. doi: 10.1128/AAC.00334-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Firsov AA, Lubenko IY, Smirnova MV, Strukova EN, Zinner SH. Enrichment of fluoroquinolone-resistant Staphylococcus aureus: oscillating ciprofloxacin concentrations simulated at the upper and lower portions of the mutant selection window. Antimicrobial Agents and Chemotherapy. 2008;52:1924–1928. doi: 10.1128/AAC.01371-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhu Y-L, Mei Q, Cheng J, Liu Y-Y, Ye Y, Li J-B. Testing the mutant selection window in rabbits infected with methicillin-resistant Staphylococcus aureus exposed to vancomycin. Journal of Antimicrobial Chemotherapy. 2012;67:2700–2706. doi: 10.1093/jac/dks280. [DOI] [PubMed] [Google Scholar]
- [33].Zinner SH, Gilbert D, Greer K, Portnoy YA, Firsov A. Concentration-resistance relationships with Pseudomonas aeruginosa exposed to doripenem and ciprofloxacin in an in vitro model. Journal of Antimicrobial Chemotherapy. 2013;68:881–887. doi: 10.1093/jac/dks463. [DOI] [PubMed] [Google Scholar]
- [34].Negri M-C, Lipsitch M, Blazquez J, Levin BR, Baquero F. Concentration-dependent selection of small phenotypic differences in tem /beta-lactamase-mediated antibiotic resistance. Antimicrobial Agents and Chemotherapy. 2000;44:2485–2491. doi: 10.1128/aac.44.9.2485-2491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Gullberg E, Cao S, Berg OG, Ilback C, Sandergren L, Hughes D, An dersson DI. Selection of resistant bacteria a very low antibiotic concentrations. PLOS Pathogens. 2011;7:e1002158. doi: 10.1371/journal.ppat.1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hughes D, Andersson DI. Selection of resistance at lethal and non-lethal antibiotic concentrations. Current Opinion in Microbiology. 2012;15:555–560. doi: 10.1016/j.mib.2012.07.005. [DOI] [PubMed] [Google Scholar]
- [37].Read AF, Taylor LH. The ecology of genetically diverse infections. Science. 2001;292:1099–1102. doi: 10.1126/science.1059410. [DOI] [PubMed] [Google Scholar]
- [38].Balmer O, Tanner M. Prevalence and implications of multiple-strain infections. Lancet, infection. 2011;11:868–878. doi: 10.1016/S1473-3099(11)70241-9. [DOI] [PubMed] [Google Scholar]
- [39].Baquero F, Nombela C, Cassell G, Gutiérrez-Fuentes J. Evolutionary Biology of Bacterial and Fungal Pathogens. American Society of Microbiology; 2007. [Google Scholar]