Abstract
Hypothesis
We propose that single-nucleotide polymorphisms (SNPs) in genes of the VEGF-pathway of angiogenesis will associate with survival in non-small cell lung cancer (NSCLC) patients.
Methods
Fifty-three SNPs in VEGF-pathway genes were genotyped in 150 European stage I-III NSCLC patients and tested for associations with patient survival. Replication was performed in an independent cohort of 142 European stage I-III patients. Reporter gene assays were used to assess the effects of SNPs on transcriptional activity.
Results
In the initial cohort, five SNPs associated (q<0.05) with relapse-free survival (RFS). The minor alleles of intronic FLT1 SNPs, rs7996030 and rs9582036, associated with reduced RFS (HR=1.67 [95% CI, 1.22 to 2.29] and HR=1.51 [95% CI, 1.14 to 2.01], respectively) and reduced transcriptional activity. The minor alleles of intronic KRAS SNPs, rs12813551 and rs10505980, associated with increased RFS (HR=0.64 [0.46 to 0.87] and HR=0.64 [0.47 to 0.87], respectively) and the minor allelic variant of rs12813551 also reduced transcriptional activity. Lastly, the minor allele of the intronic KRAS SNP rs10842513 associated with reduced RFS (HR=1.65 [95% CI, 1.16 to 2.37]). Analysis of the functional variants suggests they are located in transcriptional enhancer elements. The negative effect of rs9582036 on RFS was confirmed in the replication cohort (HR=1.69 [0.99 to 2.89], p=0.028) and the association was significant in pooled analysis of both cohorts (HR=1.67 [1.21-2.30], p=0.0001).
Conclusions
The functional FLT1 variant rs9582036 is a prognostic determinant of recurrence in stage I-III NSCLC. Its predictive value should be tested in the adjuvant setting of stage I-III NSCLC.
Keywords: non-small cell lung cancer, SNPs, VEGF-pathway, FLT1, enhancer
INTRODUCTION
Disease stage appears to be the most important prognostic factor in NSCLC patients, but disease recurrence is common even for patients with stage I-III disease1. Molecular biomarkers that identify patients who will develop recurrence and might benefit from adjuvant therapy are sorely needed2. Currently, there are no prognostic molecular markers or expression signatures in clinical use in resectable NSCLC2.
The angiogenic potential of many cancers, including NSCLC, impacts their clinical course. Angiogenesis is an essential event in tumor growth, progression and metastasis formation, and is regulated by several angiogenic cytokines, mainly those of the VEGF-pathway3-5. In NSCLC, tumor vascularization and levels of VEGF-pathway proteins have been found to be associated with patient outcomes6. Trials testing bevacizumab in the adjuvant setting have been negative and an ongoing randomized study (ECOG 1505) might provide the definitive answer to the role of adjuvant bevacizumab.
As angiogenesis is a host-mediated process, germline genetic variation in the VEGF-pathway is likely to affect the angiogenic potential of a tumor7, 8. A small number of single-nucleotide polymorphisms (SNPs) in VEGF-pathway genes have been tested for association with outcomes in NSCLC and other solid tumors (reviewed in9-11), but there is little information about the functional significance of germline variation in genes of the VEGF-pathway. Thus, there is no mechanistic basis to support many of these associations, in part explaining the inconsistent results from these studies11. Without prospective validation of findings already built in at the time of the initial discovery, the demonstration of the clinical validity of biomarkers is a lengthy and difficult process.
The aim of this study was to identify germline variants in the VEGF-pathway genes that associate with NSCLC survival and may act as markers of angiogenic-dependent tumor recurrence. NSCLC patients were genotyped for candidate VEGF-pathway SNPs to test associations with patient survival. To aid the interpretation of these associations, the molecular function of SNPs was characterized by examining their effects on transcriptional activity in reporter gene assays. Finally, the SNPs found to associate with survival were also prospectively tested in a validation cohort of European stage I-III NSCLC patients to provide an independent assessment of our findings.
METHODS
Ethics statement
Tissue banking and related research was approved by institutional review boards at the Medical University of Gdansk, Poland and the General University Hospital of Valencia, Spain. All patients signed an informed consent.
Initial patient cohort
The initial cohort consisted of 150 White European stage I-III NSCLC patients who underwent pulmonary resection. They belonged to a cohort of unselected patients systematically diagnosed with resectable NSCLC with tumor samples collected at the Medical University of Gdansk, Poland. Median follow-up of the study group was 63.4 (range, 13.1 to 82.3) months. Relapse-free survival (RFS) was defined from the date of surgery to the date of local or distant relapse, death of any cause, or last follow-up. Overall survival (OS) was defined from the date of surgery to the date of death of any cause or last follow-up. Relapse of disease was assessed by chest radiograms or CT scans every 3 months for the first two years and every 6-12 months thereafter. The patient characteristics are described in Table 1. Adjuvant therapy was given to only 4.6% of patients12. Postoperative chemotherapy was not routinely administered in the analyzed period per institutional guidelines. Primary tumors were fresh frozen at the time of surgery. All patients signed an informed consent. Further details of the cohort were described previously12.
Table 1.
Initial and validation cohorts: patient characteristics, relaspe-free survival (RFS) and overall survival (OS) (log-rank test).
| Initial cohort | Validation cohort | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n (%) | Median OS months (95% CI) | p value | Median RFS months (95% CI) | p value | n (%) | Median OS months (95% CI) | p value | Median RFS months (95% CI) | p value | |
| Sex | 0.08 | 0.14 | 0.26 | 0.26 | ||||||
| Female | 37 (25) | 62 (15.4-NR#) | 49 (13.7-NR) | 20 (14) | NR (NR-NR) | NR (NR-NR) | ||||
| Male | 113 (75) | 23 (17.5-39.8) | 18 (13.1-25.7) | 122 (86) | 67 (30.9-NR) | 44.3 (26.2-NR) | ||||
| Disease stage | <0.0001 | <0.0001 | 0.64 | 0.30 | ||||||
| I | 64 (43) | NR (48.6-NR) | 62 (36.9-NR) | 78 (55) | NR (32.3-NR) | 48.3 (43.5-NR) | ||||
| II | 33 (22) | 37 (20.3-NR) | 22 (16.8-NR) | 35 (25) | 42.9 (29.8-NR) | 26.2 (16.3-NR) | ||||
| III | 53 (35) | 10 (7.8-13.3) | 8 (6.0-11.0) | 29 (20) | NR (17.2-NR) | NR (11.2-NR) | ||||
| Histology | 0.31 | 0.42 | 0.03 | 0.04 | ||||||
| Squamous | 88 (59) | 25 (16.2-41.6) | 17 (11.6-36.9) | 67 (47) | NR (42.6-NR) | 48.3 (26.2-NR) | ||||
| Adenocarcinoma | 42 (28) | 61 (20.9-NR) | 36 (17.7-NR) | 55 (39) | 67 (42.9-NR) | NR (37.9-NR) | ||||
| Others* | 20 (13) | 15 (9.1-NR) | 15 (7.2-NR) | 20 (14) | 17.1 (12.9-NR) | 11.2 (7.3-NR) | ||||
| Ever Smokers | 0.96 | 0.69 | 0.40 | 0.61 | ||||||
| No | 7 (5) | 21 (7.16-NR) | 18 (3.4-NR) | 19 (13) | NR (NR-NR) | NR (17.9-NR) | ||||
| Yes | 143 (95) | 28 (17.8-48.6) | 21(13.5-36.9) | 123 (87) | 67 (32.3-NR) | 45.8(29.2-NR) | ||||
| Age (years) | 0.13 | 0.33 | 0.30 | 0.30 | ||||||
| <65 | 78 (52) | 36 (21.3-NR) | 22 (13.7-62.4) | 74 (52) | NR (42.6-NR) | NR (43.5-NR) | ||||
| >65 Median (range) |
72 (48) | 21 (12.6-48.6) 64 (37-85) |
18 (8.4-38.6) | 68 (48) | 53.3 (27.9-NR) 64 (26-82) |
31.9 (21.1-NR) | ||||
NR, Not reached
Initial cohort = Not otherwise specified, Validation cohort = 6 LCC, 4 ADL-SCC, 2 carcinoid, 8 not otherwise specified.
In the validation cohort, adjuvant therapy was administered to 56 patients and was not associated with either RFS or OS (results not shown).
SNP selection and genotyping in the initial cohort
In the initial patient cohort, 53 SNPs (minor allele frequency (MAF) >5%) in 13 candidate genes (identified from the PharmGKB VEGF signaling pathway web resource: www.pharmgkb.org/pathway/PA2032#tabview=tab1&subtab=) were selected using several approaches: SNPs associated with mRNA expression in lymphoblastoid cell lines (LCL); SNPs identified from our prior study13; SNPs in HIF1A and FLT1 (not expressed in LCLs) predicted to be functional by FastSNP and FuncPred; non-synonymous SNPs predicted to change protein structure according to FastSNP; SNPs identified from previous association studies. Detailed information on the SNPs according to these criteria is provided (see table, Supplemental Digital Content 1, which identifies VEGF-pathway gene SNPs genotyped in the initial cohort).
Genomic DNA prepared from fresh-frozen patient tumor samples (AllPrep DNA/RNA kit, Qiagen, Germantown, MD) was used for genotyping. Five KDR SNPs had been genotyped as described previously14. The remaining SNPs were genotyped by TaqMan® (Applied Biosystems, Foster City, CA) per the manufacturer's instructions using a CFX384 Real-Time System (Bio-Rad, Hercules, CA), and Sanger-based DNA sequencing (Mammalian Genotyping Core at UNC) was used to validate representative samples and determine thresholds for allelic discrimination. SNP allele frequencies were comparable to those previously reported from the HapMap and 1,000 Genomes projects (see table, Supplemental Digital Content 1, which identifies VEGF-pathway gene SNPs genotyped in the initial cohort). For rs9582036 in FLT1 and rs10505980 in KRAS, additional quality control of the genotyping was performed using the genotype calls in the tumor DNA and matching germline DNA in squamous NSCLCs from The Cancer Genome Atlas (see table, Supplemental Digital Content 2, which illustrates FLT1 rs9582036 and KRAS rs10505980 genotype calls from squamous NSCLC genotyped samples from The Cancer Genome Atlas). Linkage disequilibrium (LD) was analyzed using the SHEsis application. No SNPs deviated from Hardy-Weinberg equilibrium (HWE) after controlling for multiple testing using a false discovery rate (FDR) at q<0.05 (see table, Supplemental Digital Content 1, which identifies VEGF-pathway gene SNPs genotyped in the initial cohort).
Reporter gene assays
To support the interpretation of the associations between SNPs and survival, we analyzed the molecular effects of five SNPs which passed a FDR threshold (q<0.05) for association with RFS in the initial cohort. Luciferase reporter assays can test the transcriptional effects of genetic variants in potential regulatory genetic regions. When the functional effects of SNPs are unknown (as for the five SNPs associated with RFS in this study), these assays are critical to provide the mechanistic basis of the clinical associations15. The pGL4.26 (Promega, Madison, WI) plasmid with minimal promoter and Firefly luciferase gene was used and the cloning approach is described in Supplemental Digital Content 3. Three DNA clones of each reporter gene construct were prepared for transfection into murine endothelial (SVEC4-10, kind gift from Mark Lingen at the University of Chicago) and human embryonic kidney (HEK-293) cells. Cell culture conditions are described in Supplemental Digital Content 3. Cells were transfected using lipofectamine 2000 (Invitrogen, Carlsbad, CA), the reporter gene construct of interest and Renilla TK plasmid (Promega, Madison, WI). Each construct was transfected in three independent experiments, using triplicate wells. The Dual-Luciferase Reporter Assay kit (Promega, Madison, WI) was used to measure luciferase activity as per the manufacturer's instructions. Luciferase activity was defined as a ratio of Firefly to Renilla luciferase and was normalized to the luciferase activity of the wild-type construct in each experiment.
Validation patient cohort
We tested prospectively whether any of the five SNPs passing the FDR for association with RFS in the initial cohort (Table 2) would associate with RFS in an independent, external validation cohort. Hence, these five SNPs were genotyped in DNA extracted from fresh frozen tumor samples of 142 White European stage I-III NSCLC patients from Spain16. A CONSORT chart is provided (see figure, Supplemental Digital Content 4, which shows the CONSORT chart). Clinical and demographic variables of the patients are provided in Table 1. Patients were systematically diagnosed with operable, histologically confirmed NSCLC at the General University Hospital of Valencia, Spain. Median follow-up of the study group was 37.3 months (95% CI, 29.3 to 43.5). Recurrence of disease was assessed by chest CT scan every 3 months for the first two years and every 6-12 months thereafter, using the same criteria of the initial cohort. Further details of the cohort were described previously16.
Table 2.
Initial and validation cohorts: associations of SNPs with relapse-free survival (RFS) (Cox regression).
| Initial Cohort | Validation Cohort | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP | Alleles | Gene | HR | 95% CI | P value | Q value | HR | 95% CI | P value |
| rs7996030 | G>A | FLT1 | 1.67 | 1.22 to 2.29 | 0.0014 | 0.0490 | 1.37 | 0.78 to 2.42 | 0.1382 |
| rs9582036 | A>C | FLT1 | 1.51 | 1.14 to 2.01 | 0.0044 | 0.0490 | 1.69 | 0.99 to 2.89 | 0.0275 |
| rs10505980 | G>A | KRAS | 0.64 | 0.47 to 0.87 | 0.0049 | 0.0490 | 1.17 | 0.68 to 2.01 | 0.2890 |
| rs12813551 | T>C | KRAS | 0.64 | 0.46 to 0.87 | 0.0052 | 0.0490 | 0.91 | 0.53 to 1.58 | 0.3698 |
| rs10842513 | C>T | KRAS | 1.65 | 1.16 to 2.37 | 0.0060 | 0.0490 | 1.09 | 0.49 to 2.44 | 0.4136 |
In the initial cohort, data are adjusted for stage and pass the false discovery rate (FDR) correction (q<0.05) for multiple testing in additive genetic models. In the validation cohort, data are adjusted for histology, associations are for dominant genetic models (because all five associations in the initial cohort had a dominant model), and the p values are for a one-sided test, as the associations were tested prospectively on the basis of the results obtained from the initial cohort. SNPs are ranked by p value of SNP-RFS associations observed in the initial cohort.
SNP genotyping methods in the validation cohort
Patient tumor specimens were obtained at the time of the surgery and preserved in RNAlater® (Applied Biosystems, Carlsbad, CA) at −80°C until the analysis. Nucleic acid isolation and genotyping using TaqMan SNP assays are described in Supplemental Digital Content 3.
Statistical analyses
Survival was estimated according to the Kaplan-Meier method. Patient characteristics were tested for association with survival using a Mantel-Cox log-rank test to identify potential prognostic factors. The genetic associations with survival in each cohort were then adjusted for these factors, and the independence of the genetic associations was tested using additive Cox proportional hazards models. Since we propose that functional VEGF-pathway SNPs impact tumor angiogenesis and growth, we have chosen RFS as the primary endpoint as it relates to tumor growth or the appearance of new lesions17 and, compared to OS, is less likely to be affected by events (including treatment) occurring after recurrence. In the initial cohort, FDR was applied to correct for multiple testing (a q<0.05 was considered significant), genotypes were coded additively, and associations were adjusted for stage, the only variable associated with RFS in this cohort (Table 1). In the validation cohort, identical statistical methods to those of the initial cohort were used; however, a one-sided test was used because we hypothesized we would observe the same directionality of association in the validation cohort. Moreover, because the mode of heritance for these SNPs in the initial cohort was always dominant, a dominant model was used to test the associations in the validation cohort. Moreover, in the validation cohort, the associations were adjusted for histology, the only variable associated with RFS in this cohort (Table 1). In the combined cohort test, a one-sided test and a dominant model were used, and the associations were not adjusted.
Differences in reporter assays were analyzed by Student's t-test or ANOVA followed by Dunnett's multiple comparisons test in GraphPad Prism (p<0.05 for significance). All other statistical analyses were carried out using the R Statistical environment along with extension packages.
RESULTS
Five SNPs associate with RFS in the initial NSCLC cohort
Out of the 53 SNPs genotyped in 150 European stage I-III NSCLC patients, five SNPs (two in FLT1 and three in KRAS) associated with RFS and passed the FDR threshold for significance (q<0.05, Table 2). Disease stage was incorporated into these models because it significantly associated with RFS and OS (p<0.0001, Table 1).
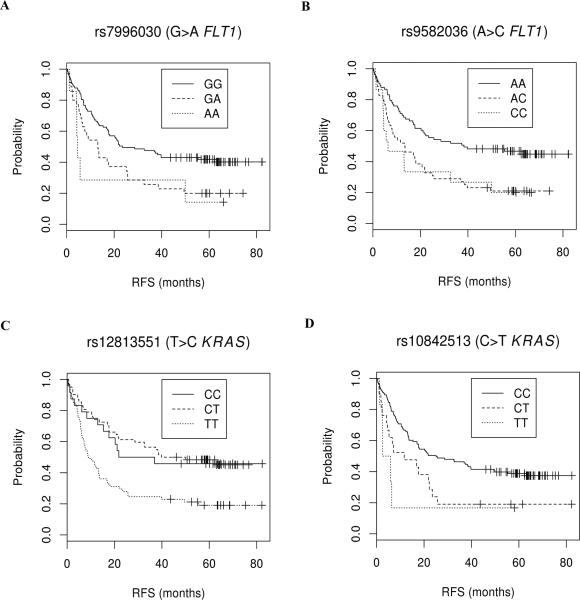
The minor alleles of the intronic FLT1 SNPs rs7996030 and rs9582036 associated with reduced RFS (HR=1.67 [95% CI 1.22 to 2.29] and HR=1.51 [95% CI 1.14 to 2.01], respectively); for rs7996030 in a dominant model, the median RFS of patients with GA+AA genotypes was 2.1 fold shorter than that of GG patients (HR=2.13 [1.38 to 3.28], p=0.0006, Fig. 1A); for rs9582036 in a dominant model (in modest LD with rs79906030, r2=0.44), the median RFS of patients with CA+CC genotypes was 2.9 fold shorter than that of AA patients (HR=1.77 [1.18 to 2.67], p=0.006, Fig. 1B).
Fig. 1. Kaplan-Meier plots of significant SNP-RFS associations after false discovery rate (FDR) correction (q<0.05) in initial cohort.
Symbols denote censored points. (A) rs7996030: median RFS is 11.1 (95% CI, 5.8 to 25.7) and 23.5 (17.7-NR) months for patients with GA+AA and GG genotypes. (B) rs9582036: median RFS is 13.1 (7.2 to 20.7) and 38.3 (21.1 to NR) months for patients with CC+AC and AA genotypes, respectively. (C) rs12813551: this plot is also representative of rs10505980 due to the high LD with rs12813551. Median RFS is 39.2 (21.8 to NR) and 9.1 (7.0 to 16.8) for patients with CC+TC and TT genotypes, respectively. (D) rs10842513: median RFS is 6.6 (4.6 to 22.2) and 25.1 (17.1 to 49.9) months for patients with CC+TC and TT genotypes, respectively. While these results are for associations adjusted for stage, the plots are for unadjusted data.
The minor alleles of the intronic KRAS SNPs rs12813551 and rs10505980 associated with increased RFS (HR=0.64 [0.46 to 0.87] and HR=0.64 [0.47 to 0.87], respectively); for rs12813551 in a dominant model, the median RFS of patients with TC+CC genotypes was 4.3 fold longer than that of TT patients (HR=0.49 [0.32 to 0.74], p=0.0008, Fig. 1C); for rs10505980 in a dominant model (in high LD with rs12813551, r2=0.83), the median RFS of patients with GA+AA genotypes was 3.7 fold longer than that of GG patients (HR=0.49 [0.33 to 0.75], p=0.0008).
The minor allele of the intronic KRAS SNP rs1084251was associated with reduced RFS (HR=1.65 [1.16 to 2.37]), and in a dominant model the median RFS of patients with CT+TT genotypes was 3.8 fold shorter than that of CC patients (HR=1.97 [1.22 to 3.18], p=0.006, Fig. 1D). An additional seven SNPs nominally associated with RFS (p<0.05) but did not pass the FDR threshold (Table 3).
Table 3.
Initial cohort: nominal associations of SNPs with relapse-free survival (RFS, p<0.05) in Cox regression models adjusted for stage.
| SNP | Alleles | Gene | HR | 95% CI | P value | Q value |
|---|---|---|---|---|---|---|
| rs4246229 | A>G | KRAS | 0.71 | 0.54 to 0.94 | 0.0168 | 0.1120 |
| rs34176876 | A>- | KRAS | 0.72 | 0.55 to 0.95 | 0.0191 | 0.1120 |
| rs1570360 | G>A | VEGFA | 1.39 | 1.03 to 1.87 | 0.0290 | 0.1428 |
| rs1951795 | C>A | HIF1A | 1.47 | 1.03 to 2.11 | 0.0341 | 0.1428 |
| rs11549465 | C>T | HIF1A | 1.62 | 1.02 to 2.57 | 0.0392 | 0.1428 |
| rs542403 | A>G | FRS2 | 0.66 | 0.44 to 0.98 | 0.0404 | 0.1428 |
| rs2076139 | C>T | MAPK11 | 0.70 | 0.49 to 0.99 | 0.0417 | 0.1428 |
SNPs are ranked by significance of SNP-RFS associations. The associations with p>0.05 are available upon request.
As a secondary analysis, associations with OS were similarly tested. Nine SNPs nominally associated with OS (p<0.05, see table, Supplemental Digital Content 5, which identifies initial cohort nominal associations of SNPs with OS (p<0.05) in Cox regression models adjusted for stage), but none passed the FDR threshold. However, all five SNPs passing FDR for RFS (Table 2) had an effect on OS concordant with that on RFS (p<0.02 for these SNPs).
Three SNPs demonstrate functional effects on reporter gene expression
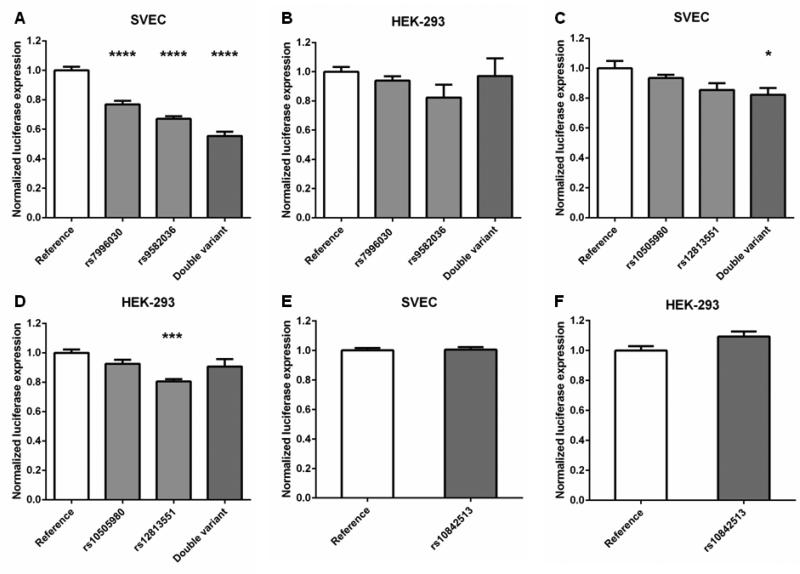
To provide a mechanistic explanation for the genetic associations with RFS, SNPs were assessed for regulatory activity using luciferase reporter gene assays. Genomic regions containing the SNPs were cloned downstream of the luciferase gene in a construct containing a minimal promoter and assays were performed to determine the effects of the SNPs on the transcriptional activity of the minimal promoter. An intronic region containing the minor allele of the FLT1 SNP rs9582036 or rs7996030 (associated with reduced RFS) reduced transcriptional activity in an endothelial cell line (SVEC) by 33% (p<0.0001) and 23% (p<0.0001), respectively, when compared to a reference construct containing the major allelic variants (Fig. 2A). The effect on transcriptional activity was even more pronounced when both minor alleles were tested in the same construct: activity was reduced up to 45% (p<0.0001; Fig. 2A). Neither SNP had a significant effect on transcriptional activity in HEK-293 cells (Fig. 2B), suggesting that the effects of the variants may be cell or tissue type specific, as is characteristic of transcriptional enhancers. Moreover, analysis of ENCODE functional genomic data indicated that the FLT1 region examined contains a putative enhancer element, coincident with rs7996030 and possibly extending to rs9582036 (see Supplemental Digital Content 6A, which illustrates evidence of putative transcriptional enhancer elements at FLT1 loci).
Fig. 2. FLT1 and KRAS SNPs effect transcriptional activity in reporter gene assays.
Relative luciferase activity of FLT1 SNPs in SVEC (A) and HEK-293 (B) cell lines, and KRAS SNPs in SVEC (C, E) and HEK-293 (D, F) cell lines. Values were normalized to those of the reference sequences containing the major allelic variants of the SNPs of interest. Significance was tested by Student's t-test or ANOVA followed by Dunnett's multiple comparisons test in GraphPad Prism (for significance: *p<0.05; ***p<0.001; ****p<0.0001)
For the KRAS SNPs rs12813551 and rs10505980, an intronic region containing the minor alleles of both variants (associated with increased RFS) reduced transcriptional activity in SVEC cells by 18% (p=0.014; Fig. 2C). Neither SNP had a significant effect by itself in SVEC cells, though the minor allele of rs12813551 reduced transcriptional activity by 15% (p=0.051). In HEK-293 cells, the minor allele of rs12813551 reduced transcriptional activity by 19% (p<0.001), but neither the minor allele of rs10505980 nor the construct containing the minor alleles of both SNPs had a significant effect (Fig. 2D). Analysis of ENCODE data suggests that rs12813551 is coincident with a putative transcriptional enhancer while rs10505980 is ~1.5 kb downstream of this element (see Supplemental Digital Content 6B, which illustrates evidence of putative transcriptional enhancer elements at KRAS loci).
An intronic region containing the minor allele of the KRAS SNP rs10842513 (associated with reduced RFS) demonstrated no effect in SVEC cells (Fig. 2E) and only a marginal increase (9%, p=0.054) in transcriptional activity in HEK-293 cells (Fig. 2F). ENCODE data did not provide evidence for substantive transcriptional enhancer activity at this SNP locus (see Supplemental Digital Content 6C, which illustrates lack of evidence of putative transcriptional enhancer elements at KRAS rs10842513 locus).
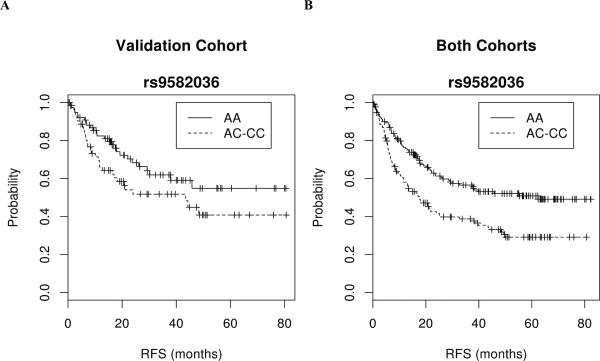
The association of rs9582036 and RFS replicates in validation cohort
We prospectively tested whether any of the five SNPs passing the FDR for the association with RFS in the initial cohort (Table 2) associated with RFS in an independent, external validation cohort of 142 European stage I-III NSCLC patients (Table 1)16. Out of the five SNPs, only rs9582036 in FLT1 had a significant and concordant effect (Table 2): the RFS of patients with the AC+CC genotypes was shorter than that of AA patients (HR=1.57 [0.93 to 2.66], p=0.045, Fig. 3A). When this association was adjusted for histology (the only parameter associated with RFS, Table 1), the HR was 1.69 [0.99 to 2.89], p=0.028. The adjustment for stage (as in the initial cohort) did not improve this association (HR=1.57 [95% CI 0.92 to 2.66], p=0.049).
Fig 3. Kaplan-Meier plots of the association between the rs9582036 SNP in FLT1 and RFS in the validation cohort (A) and in the combined initial and validation cohorts (B).
Symbols denote censored points. The plots are for unadjusted data.
In a pooled analysis of both cohorts (n=292), the HR of rs9582036 for RFS (adjusted for stage, using a dominant model) was 1.67 (95%CI 1.21 to 2.30), with a marked separation between the two survival curves (p=0.0001, Fig. 3B); without adjusting for stage, the HR was 1.81 [1.32 to 2.49], p=0.0001).
DISCUSSION
The VEGF-pathway genetic studies of NSCLC outcome have focused so far on a small number of genes and genetic variants, in particular VEGFA9. We rationally selected 53 SNPs from 13 VEGF-pathway genes and identified five SNPs in FLT1 and KRAS that associated with RFS after correction for multiple testing. We subsequently validated the association of the FLT1 SNP rs9582036 with RFS in an independent cohort of stage I-III European NSCLC patients. Molecular studies of germline SNPs associated with patient outcome are essential to guide the assessment of their clinical utility15 and we found functional evidence to support the clinical associations of the two FLT1 SNPs (rs7996030 and rs9582036) and one of the KRAS SNPs (rs12813551). Furthermore, all three functional variants were found to be located in or adjacent to putative transcriptional enhancer elements. The finding that the FLT1 SNPs had an additive effect on gene expression is consistent with a recent proposal that multiple enhancer variants cooperatively act to modestly alter gene expression and account for genetic associations with disease18.
The most important result of this study is that the variant allele (C) of the FLT1 SNP rs9582036 is associated with shorter RFS in both the initial and the validation cohorts (Table 2, Fig. 1 and Fig. 3). rs9582036 is a common germline variant (frequency of about 30% in Caucasians) and this association is supported by the finding that this variant is functional. A putative transcriptional enhancer element is located immediately adjacent to rs9582036 and possibly extends into the locus of this SNP in lung tissue (see Supplemental Digital Content 6A, which illustrates evidence of putative transcriptional enhancer elements at FLT1 loci). The minor allele of rs9582036 reduces transcriptional activity and, thus, suggests that this variant may act through this element to reduce FLT1 expression. Intriguingly, the effect of rs9582036 was only observed in the SVEC cell line, indicating that the effect may be specific to endothelial cells and that this variant may have effects on angiogenesis in vivo.
There is evidence to suggest that the clinical association and the molecular effect of rs9582036 reconcile with the biology of FLT1. FLT1 encodes the VEGF receptor 1 (VEGFR-1), a mediator of tumor endothelial function19. Its level of phosphorylation in response to VEGFA is low20, 21, and its soluble form is expressed by endothelial tumor cells, sequestering VEGFA through the formation of a complex22. Soluble VEGFR-1 also inhibits VEGFA in a dominant manner by heterodimerizing with the ligand binding region of VEGFR-223, the most potent receptor of VEGFA which mediates most of the pro-angiogenic effects of VEGFA. Enhanced in vivo expression of soluble VEGFR-1 by tumor cells inhibits solid tumor growth, impedes metastatic nodule development, and extends host survival19. In addition to its soluble form, upregulation of membrane-anchored VEGFR-1 in endothelial cells contributes to the readjustment of the tumor endothelial phenotype in response to increased oxygen supply and vessel normalization24. These lines of evidence are strongly suggestive of FLT1 being a negative regulator of neovascularization (see review19). Hence, collectively, these findings fit a mechanistic model where reduced endothelial expression of VEGFR-1, mediated by rs9582036, could accelerate NSCLC recurrence through increased angiogenesis.
Based upon the results of this study, we hypothesize that carriers of the minor allele of rs9582036 FLT1 variant may be less responsive to angiogenesis inhibition, as a result of a potentiated angiogenic tumor phenotype. Indeed, rs9582036 is predictive of shorter survival in bevacizumab-treated patients with metastatic pancreatic and renal-cell carcinoma25. The negative predictive effect is also observed in other nonrandomized studies of anti-VEGF therapies: bevacizumab (colorectal26) and sunitinib (renal cell27). Because results from different tumor types might not be necessarily extended to the setting of stage I-III NSCLC, more molecular and cellular studies in ex vivo angiogenesis models are needed to dissect the resulting effect of FLT1 genetic variation on both the basal NSCLC endothelial phenotypes and response to VEGF blockade.
A major question is whether there is now sufficient evidence to regard rs9582036 as a validated prognostic marker of recurrence in stage I-III NSCLC. RFS was the primary endpoint in our study, and for a genetic association that relates to the biology of the tumor, RFS should be robust to confounders which could not be accounted for. We acknowledge that the demonstration of a prognostic role of rs9582036 in FLT1 for cancer-related OS is probably warranted before this novel marker could be used to inform treatment decisions in stage I-III NSCLC patients. In this setting, interventions after recurrence should be taken into account to avoid the confounding related to imbalances in active therapies. The lack of recommendations on the use of adjuvant chemotherapy in higher-risk NSCLC following surgery in the cancer institute treating study patients may also represent a limitation of the present study.
Our study did not replicate the effect of three KRAS SNPs on RFS (Table 2). These findings may have been false positives or the effects of the SNPs could have been confounded by hidden heterogeneity between the two cohorts. The patients in the validation cohort were not enrolled to match the characteristics of the initial cohort; therefore intrinsic patient differences across the two cohorts might hamper the detection of true positive associations. Furthermore, only rs12813551 demonstrated functionality and its effects were modest in the reporter gene assays. It may be that these SNPs are in linkage disequilibrium with the causal variants underlying the clinical associations and, thus, fine mapping studies may help clarify these associations. Nonetheless, the KRAS SNP rs12813551 should still be tested in other studies as its molecular effect is consistent with the clinical association and the oncogenic biology of KRAS in NSCLC. Moreover, a negative prognostic impact of increased KRAS expression in operable NSCLC has been previously demonstrated28 and the minor allele of rs12813551, associated with increased RFS, had a negative effect on transcriptional activity.
In summary, we identified several associations between VEGF-pathway SNPs and NSCLC outcome, and provided biological interpretations through molecular studies. A prospective validation study has selected rs9582036 in FLT1 as a germline variant associated with poor prognosis in stage I-III NSCLC. These results provide the foundation for testing the prognostic and predictive value of functional VEGF-pathway SNPs in NSCLC patients administered chemotherapy and anti-VEGF therapies.
Supplementary Material
Acknowledgments
We would like to acknowledge Jessie Bishop and Anna Sorin for their help in editing and formatting this paper.
Funding:
This work was supported by NIH/NCI K07CA140390-01, Cancer Research Foundation Young Investigator Award, American Cancer Society (Illinois Division)/LUNGevity Foundation, The Joan Scarangello Foundation to Conquer Lung Cancer, Wendy Will Case Cancer Fund, Grant ST-23 from Medical University of Gdansk, Poland, grant from PN I+D+I 2008-2011, Instituto de Salud Carlos III, (PS09/01149 and PI12/02838), Subdirección General de Redes y Centros de Investigación Cooperativa, Red Temática de Investigación Cooperativa en Cáncer (RD06/0020/1024 and RD12/0036/0025), Spain.
Footnotes
Conflicts of Interest: The authors do not have any conflicts of interst to disclose.
REFERENCES
- 1.Chansky K, Sculier JP, Crowley JJ, et al. The International Association for the Study of Lung Cancer Staging Project: prognostic factors and pathologic TNM stage in surgically managed non-small cell lung cancer. J Thorac Oncol. 2009;4(7):792–801. doi: 10.1097/JTO.0b013e3181a7716e. [DOI] [PubMed] [Google Scholar]
- 2.Subramanian J, Simon R. Gene expression-based prognostic signatures in lung cancer: ready for clinical use? J Natl Cancer Inst. 2010;102(7):464–74. doi: 10.1093/jnci/djq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Folkman J. Angiogenesis in cancer, vascular, rheumatoid and other disease. Nat Med. 1995;1(1):27–31. doi: 10.1038/nm0195-27. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Hoff P, Machado K. Role of angiogenesis in the pathogenesis of cancer. Cancer Treat Rev. 2012;38(7):825–33. doi: 10.1016/j.ctrv.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 6.Salgia R. Prognostic significance of angiogenesis and angiogenic growth factors in nonsmall cell lung cancer. Cancer. 2011;117(17):3889–99. doi: 10.1002/cncr.25935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koukourakis MI, Papazoglou D, Giatromanolaki A, et al. VEGF gene sequence variation defines VEGF gene expression status and angiogenic activity in non-small cell lung cancer. Lung Cancer. 2004;46(3):293–8. doi: 10.1016/j.lungcan.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 8.Azam F, Mehta S, Harris AL. Mechanisms of resistance to antiangiogenesis therapy. Eur J Cancer. 2010;46(8):1323–32. doi: 10.1016/j.ejca.2010.02.020. [DOI] [PubMed] [Google Scholar]
- 9.Eng L, Azad AK, Habbous S, et al. Vascular Endothelial Growth Factor Pathway Polymorphisms as Prognostic and Pharmacogenetic Factors in Cancer: A Systematic Review and Meta-analysis. Clin Cancer Res. 2012;18:4526–37. doi: 10.1158/1078-0432.CCR-12-1315. [DOI] [PubMed] [Google Scholar]
- 10.Lambrechts D, Lenz HJ, de Haas S, et al. Markers of response for the antiangiogenic agent bevacizumab. J Clin Oncol. 2013;31(9):1219–30. doi: 10.1200/JCO.2012.46.2762. [DOI] [PubMed] [Google Scholar]
- 11.Schneider BP, Shen F, Miller KD. Pharmacogenetic biomarkers for the prediction of response to antiangiogenic treatment. Lancet Oncol. 2012;13(10):e427–36. doi: 10.1016/S1470-2045(12)70275-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dziadziuszko R, Merrick DT, Witta SE, et al. Insulin-like growth factor receptor 1 (IGF1R) gene copy number is associated with survival in operable non-small-cell lung cancer: a comparison between IGF1R fluorescent in situ hybridization, protein expression, and mRNA expression. J Clin Oncol. 2010;28(13):2174–80. doi: 10.1200/JCO.2009.24.6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pare-Brunet L, Glubb D, Evans P, et al. Discovery and functional assessment of gene variants in the vascular endothelial growth factor pathway. Hum Mutat. 2014;35(2):227–35. doi: 10.1002/humu.22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glubb DM, Cerri E, Giese A, et al. Novel functional germline variants in the VEGF receptor 2 gene and their effect on gene expression and microvessel density in lung cancer. Clin Cancer Res. 2011;17(16):5257–67. doi: 10.1158/1078-0432.CCR-11-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glubb DM, Innocenti F. The architecture of pharmacogenomic associations: structures with functional foundations or castles made of sand? Pharmacogenomics. 2013;14(1):1–4. doi: 10.2217/pgs.12.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanmartin E, Sirera R, Uso M, et al. A gene signature combining the tissue expression of three angiogenic factors is a prognostic marker in early-stage non-small cell lung cancer. Ann Surg Oncol. 2014;21(2):612–20. doi: 10.1245/s10434-013-3330-x. [DOI] [PubMed] [Google Scholar]
- 17.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92(3):205–16. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 18.Corradin O, Saiakhova A, Akhtar-Zaidi B, et al. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 2014;24(1):1–13. doi: 10.1101/gr.164079.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olsson AK, Dimberg A, Kreuger J, et al. VEGF receptor signalling -in control of vascular function. Nat Rev Mol Cell Biol. 2006;7(5):359–71. doi: 10.1038/nrm1911. [DOI] [PubMed] [Google Scholar]
- 20.Waltenberger J, Claesson-Welsh L, Siegbahn A, et al. Different signal transduction properties of KDR and Flt1, two receptors for vascular endothelial growth factor. J Biol Chem. 1994;269(43):26988–95. [PubMed] [Google Scholar]
- 21.Seetharam L, Gotoh N, Maru Y, et al. A unique signal transduction from FLT tyrosine kinase, a receptor for vascular endothelial growth factor VEGF. Oncogene. 1995;10(1):135–47. [PubMed] [Google Scholar]
- 22.Kendall RL, Thomas KA. Inhibition of vascular endothelial cell growth factor activity by an endogenously encoded soluble receptor. Proc Natl Acad Sci U S A. 1993;90(22):10705–9. doi: 10.1073/pnas.90.22.10705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kendall RL, Wang G, Thomas KA. Identification of a natural soluble form of the vascular endothelial growth factor receptor, FLT-1, and its heterodimerization with KDR. Biochem Biophys Res Commun. 1996;226(2):324–8. doi: 10.1006/bbrc.1996.1355. [DOI] [PubMed] [Google Scholar]
- 24.Mazzone M, Dettori D, Leite de Oliveira R, et al. Heterozygous deficiency of PHD2 restores tumor oxygenation and inhibits metastasis via endothelial normalization. Cell. 2009;136(5):839–51. doi: 10.1016/j.cell.2009.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lambrechts D, Claes B, Delmar P, et al. VEGF pathway genetic variants as biomarkers of treatment outcome with bevacizumab: an analysis of data from the AViTA and AVOREN randomised trials. Lancet Oncol. 2012;13(7):724–33. doi: 10.1016/S1470-2045(12)70231-0. [DOI] [PubMed] [Google Scholar]
- 26.Hansen TF, Christensen R, Andersen RF, et al. The predictive value of single nucleotide polymorphisms in the VEGF system to the efficacy of first-line treatment with bevacizumab plus chemotherapy in patients with metastatic colorectal cancer: results from the Nordic ACT trial. Int J Colorectal Dis. 2012;27(6):715–20. doi: 10.1007/s00384-011-1382-6. [DOI] [PubMed] [Google Scholar]
- 27.Beuselinck B, Karadimou A, Lambrechts D, et al. VEGFR1 single nucleotide polymorphisms associated with outcome in patients with metastatic renal cell carcinoma treated with sunitinib -a multicentric retrospective analysis. Acta Oncol. 2014;53:103–12. doi: 10.3109/0284186X.2013.770600. [DOI] [PubMed] [Google Scholar]
- 28.Han N, Dol Z, Vasieva O, Hyde R, Liloglou T, Raji O, et al. Progressive lung cancer determined by expression profiling and transcriptional regulation. Int J Oncol. 2012;41(1):242–52. doi: 10.3892/ijo.2012.1421. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.