Abstract
Pioneering electron microscopy studies defined two primary populations of ribosomes in eukaryotic cells: one freely dispersed through the cytoplasm and the other bound to the surface of the endoplasmic reticulum (ER). Subsequent investigations revealed a specialized function for each population, with secretory and integral membrane protein-encoding mRNAs translated on ER-bound ribosomes, and cytosolic protein synthesis was widely attributed to free ribosomes. Recent findings have challenged this view, and transcriptome-scale studies of mRNA distribution and translation have now demonstrated that ER-bound ribosomes also function in the translation of a large fraction of mRNAs that encode cytosolic proteins. These studies suggest a far more expansive role for the ER in transcriptome expression, where membrane and secretory protein synthesis represents one element of a multifaceted and dynamic contribution to post-transcriptional gene expression.
For decades, the endoplasmic reticulum (ER) has held a special fascination for cell biologists, biochemists and molecular geneticists. Since the pioneering ultrastructural and biochemical studies by Palade and co-workers demonstrating that proteins destined for secretion from the cell are synthesized on ER-bound ribosomes, research into protein biogenesis on the ER has yielded remarkable insights into the mechanisms of protein targeting, translocation and trafficking through the secretory pathway1–7. These findings led to a number of fundamental questions about this system, as proposed by Palade in his Nobel lecture3: how are ribosomes targeted to the ER? What are the functions of ER-bound ribosomes?
In the decades since these initial discoveries, our understanding of ER-localized mRNA translation has coalesced around one model. In this model, the translation of mRNAs encoding secretory or membrane proteins is initiated in the cytosol, and the mRNA–ribosome complexes are selectively recruited to the ER membrane, concurrent with the emergence of a topogenic signal — either a signal sequence at the amino terminus of the nascent polypeptide chain or a transmembrane domain (FIG. 1a). Continued research into the mechanism of selective ER localization of mRNA led to one of the most important discoveries of the era: the identification of the signal recognition particle (SRP)8–10 (BOX 1; FIG. 1). The SRP is a conserved ribonucleoprotein complex that binds early in translation to the hydrophobic core domain that is characteristic of all topogenic signals and directs the co-translational targeting of the ribosome–mRNA–nascent polypeptide chain complex to the ER8,9. This targeting process is mediated by a cognate SRP-interacting ER membrane protein complex, the SRP receptor, and is sequentially coupled to the binding of the translating ribosome to the protein-conducting channel in the ER membrane6,7,11,12. As this pathway operates through a positive selection mechanism, ribosomes engaged in the translation of cytosolic proteins, which lack topogenic signals, are not recruited to the ER and thus remain in the cytosol. Notably, this mechanism is entirely consistent with early models positing that newly exported mRNAs undergo translation on cytosolic ribosomes and, in the case of ribosomes translating signal sequence-encoding mRNAs, then undergo co-translational targeting to the ER13. As a reflection of the extensive and advanced mechanistic elucidation of SRP pathway function, this model has received extensive and rigorous experimental support and has enjoyed widespread acceptance.
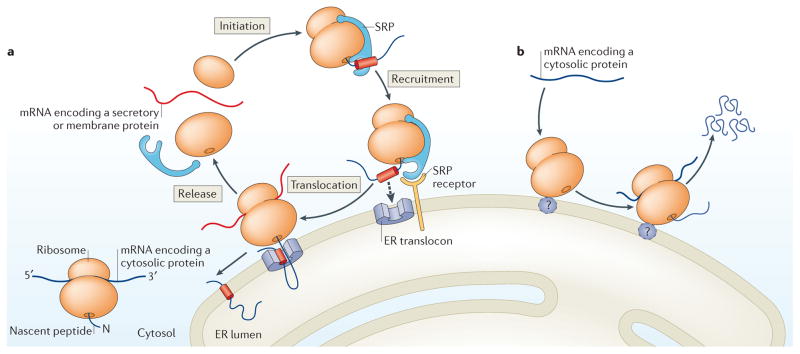
Figure 1. Revisiting translational compartmentalization.
a | The model for translational compartmentalization that has been generally accepted since the discovery of the signal recognition particle (SRP) is presented. Translation initiation begins in the cytosol. Ribosomes translating mRNAs encoding cytosolic proteins that lack a topogenic endoplasmic reticulum (ER)-targeting signal remain in the cytosol. By contrast, ribosomes translating mRNAs that encode a protein containing a signal peptide or a transmembrane domain (such as secretory and integral membrane proteins) are targeted to the ER co-translationally by the SRP. Briefly, following translation initiation in the cytosol, the emerging topogenic signal serves as a targeting signal to the ER. Following docking on the ER translocon, the secretory and membrane proteins are translocated across or into the ER membrane. Upon completion of protein synthesis, ribosomal subunits are recycled into the cytosol. b | A pan-transcriptomic role for the ER in mRNA translation is depicted. Translation initiation can occur directly on the ER. Moreover, mRNAs that encode cytosolic proteins can also be translated by ER-bound ribosomes. Thus, a large fraction of the proteome can be translated by ER-associated ribosomes. Such a diverse and selective translation of mRNAs redefines this ubiquitous organelle as a primary site of proteome synthesis in the cell. Ribosomes translating mRNAs encoding cytosolic proteins associate with the ER via an unknown ribosome receptor (indicated by the question mark).
Box 1. In vitro reconstitution of mRNA localization to the ER.
The development of a robust in vitro reconstitution system of the protein translocation reaction stands as a prominent milestone in the field and one that enabled the discovery of the signal recognition particle (SRP), the SRP receptor and the functional analysis of translocon components, most prominently the SEC61 protein conducting channel complex8–12,71,119,120. An in vitro translation extract, typically derived from wheat germ, is supplemented with an mRNA encoding a secretory protein and rough microsomes in the presence or absence of added SRP (see the figure, part a). Translating free ribosomes are only recruited to the rough microsome when the SRP is present, and the nascent polypeptide chain is translocated across the microsomal (endoplasmic reticulum (ER)) membrane. Thus, this assay system reconstitutes a cytosol-to-ER pathway for the selective localization of signal sequence-encoding mRNAs to the ER, which is consistent with earlier proposals that translation of secretory and membrane proteins is initiated on cytoplasmic ribosomes13,119,120. However, the question of whether the SRP-dependent pathway is the sole mechanism for selective localization of mRNAs to the ER remained unanswered. Using variations of this basic assay system, this question was addressed in subsequent experiments. In one variation, the translation extract was depleted of ribosomes by ultracentrifugation, and the capacity of ER-bound ribosomes to participate in de novo translation was investigated. Under these experimental conditions, native ER- bound ribosomes participated in de novo translation and, notably, translated mRNAs irrespectively of whether they encoded a signal sequence15,17,96. In a further variation (see the figure, part b), rough microsomes were subjected to mild proteolysis to inactivate SRP receptor function. Translation initiation of mRNAs encoding signal sequences still occurred in the absence of the SRP receptor, mediated by ER-bound ribosomes, and their encoded proteins were translocated15,17,96.

Current views regarding mRNA translation on the ER largely reflect our understanding of the SRP pathway and its capacity to selectively localize mRNAs that encode secretory and membrane proteins to microsomal vesicles in vitro5,8,9. In this Review, we return to the very beginning of the field, to questions first posed by Palade and co-workers: are ER-bound ribosomes limited to the translation of secretory and membrane protein-encoding mRNAs? Or is there a broader role for bound ribosomes in mRNA transcriptome expression? The notion that ER-bound ribosomes could have activity beyond the synthesis of secretory and membrane proteins, a possibility suggested by Siekevitz and Palade early in their investigations into the mechanism of protein secretion14, has more recently found support with the advent of new biochemical, cell biological and transcriptomic approaches to the study of mRNA translation in vivo. Considerable data now reveal a diverse role for the ER in mRNA translation, in which ER-bound ribosomes function in the synthesis of a broad fraction of cellular proteins, including both cytosolic and ER-targeted proteins (FIG. 1b). Surprisingly, continuing investigations into the in vivo mechanism of ribosome binding to the ER showed that ribosome traffic to the ER is only weakly coupled to the protein synthesis cycle of initiation, elongation and termination15–17. These findings, together with recent data demonstrating an ER localization of many factors that bind and regulate mRNAs and their translation, suggest a biochemical and regulatory ER translation environment that is distinct from the cytosol. Very recent reports also reveal that the ER-associated translational system is dynamic, with the capacity to rapidly reorganize in response to cellular stimuli or stress18. Taken together, these developments point to a need for re-examining our understanding of how mRNA translation is spatially organized and regulated in eukaryotic cells. In this Review, we discuss a newly emerging model for mRNA translation on the ER, whereby the ER is a primary site of general protein synthesis, as well as a site with exquisite regulatory functions that can selectively influence specific mRNAs by several mechanisms. This new model contributes to the ever-expanding richness of post-transcriptional gene regulation and adds an important new variable of ER localization into the consideration of how the translation of an mRNA may be regulated.
Compartment-selective mRNA translation
In the landmark study19 that first described the ribosome, Palade noted that these newly discovered particles had a “particular affinity for the endoplasmic reticulum”. Moreover, in cells with extensive ER networks (so-called ‘professional secretory cells’), ribosomes were observed to be predominately associated with the ER. For example, in a pancreatic acinar cell, a cell type that is largely devoted to the secretion of digestive enzymes, ribosomes are mostly ER-bound, with few located in the sparse cytosol19. By contrast, cells with a less demanding secretory role, such as intestinal epithelial cells, generally have a less extensive network of ER and thus a smaller relative fraction of ER-associated ribosomes19. These observations led to the conclusion that the ER was the site of secretory and membrane protein synthesis.
It is well established that the ER is a prominent site of mRNA localization and that secretory and integral membrane proteins are synthesized on ER-bound ribosomes3,14. However, the question of whether the bound ribosomes of the ER are devoted entirely to secretory and integral membrane protein synthesis has received scant attention. Through the lens of history, this is surprising to a degree. In their seminal studies, Siekevitz and Palade noted that ER-bound ribosomes also seemed to be engaged in the synthesis of ‘non-exportable’ proteins14, an observation that was later supported by biochemical studies20. This rather puzzling observation was again raised by Palade in his Nobel lecture3, in which he noted that both cytosolic and ER-bound ribosomes “carry newly synthesized proteins irrespective of the latter’s final destination”. An obvious and appropriate concern, and one that has been discussed and debated early on, is that the presence of mRNAs encoding cytosolic proteins on the ER is due to cross-contamination during cell fractionation. Extensive efforts to address this concern demonstrated that the magnitude of cross-contamination was small and wholly insufficient to account for the magnitude of cytosolic mRNA representation on the ER21–23. This conclusion is in agreement with more recent experiments conducted in Drosophila melanogaster24, in yeast25 and in mammalian tissue culture cells26–28 using multiple, independent experimental approaches29,30. With cross-contamination proving to be an insufficient explanation, it becomes of pressing interest to address an alternative interpretation in which bound ribosomes of the ER have a broader role in transcriptome expression than described in current models.
The ER as a primary compartment for translation
What fraction of total cellular translation is associated with the ER? This simple question is perhaps the most fundamental, and even in very early studies the answer pointed to an unexpectedly large fraction14. Later work demonstrated that in HeLa and HEK293 cells, approximately half of all ribosomes are ER-associated and a similar fraction of the total mRNA is ER-localized31. It is useful to contrast this metric with other well-studied examples of mRNA localization. There are several examples of a particular species of mRNA that localize to discrete areas of the cell; for example, β-actin mRNA localizes to the leading edge of migrating fibroblasts, ASH1 mRNA is found at the yeast bud tip, and oskar and bicoid mRNAs localize to the posterior and anterior poles, respectively, of the D. melanogaster oocyte32,33. These examples illustrate one of the principal functions of mRNA localization: selective targeting of a small number of mRNAs to a discrete cellular locale to enable a specific biological function. Such targeting of mRNAs is often mediated by specific localization motifs (termed ‘zip codes’) that are encoded in the mRNA, most commonly in the 3 untranslated region32,33. By contrast, the ER extends broadly throughout the cytosol in essentially every cell type, is studded with ribosomes over much of that area and associates with a large fraction of mRNAs and by a diverse range of mechanisms34–37 (see below). It is therefore useful to distinguish mRNA localization to the ER from these other modes of mRNA localization; that is, instead of a small number of mRNAs undergoing localization for a specific physiological outcome, localization of mRNAs and translational activity to the ER serves far broader, more general roles in gene expression. Indeed, in a recent study in yeast, ~75% of all cellular translation activity was found to be associated with the ER38.
The composite biological functions of mRNA localization to the ER remain elusive. One aspect, the function of the ER as the site of co-translational protein entry into the secretory pathway, is abundantly clear. However, mRNAs encoding membrane and secretory proteins often constitute a small fraction of transcribed genes39. For example, in HEK293 cells, such mRNAs represent only ~13% of all mRNAs, yet approximately half of all ribosomes are ER-bound28 (FIG. 2a). As noted above, the explanation for this discrepancy is that a large fraction of mRNAs that encode cytosolic proteins are present on the ER and translated by ER-bound ribosomes, as was proposed in the early years of the field21,40,41 and has been revisited more recently25–27,37,42. In further support of this view, alternative methods of fractionating cells into their cytosolic and ER-bound components have enabled a precise definition of the partitioning of ribosomes, mRNAs and other factors between these two compartments29,31. Indeed, these studies have found that mRNAs that encode cytosolic proteins are abundantly present and translated on the ER28,31,42,43. These observations have been reported in studies conducted in diverse organisms, over decades, using different methods of cell fractionation and mRNA compositional analysis.
Figure 2. Distribution of protein synthesis between the cytosol and the ER.
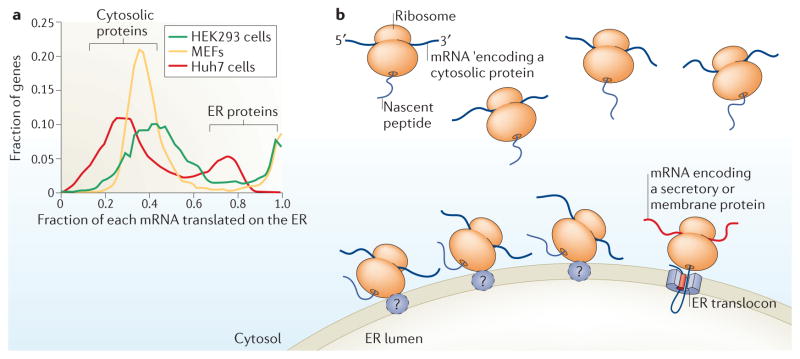
a | The distribution of translation between the cytosol and the endoplasmic reticulum (ER) for each mRNA is displayed as a histogram for HEK293 cells 28, mouse embryonic fibroblasts (MEFs)18 and Huh7 cells (R. Campos, S. Bradrick, D.W.R., M. Garcio-Blanco & C.V.N., unpublished observations). The histogram shows two distinct populations of mRNAs: one that is ER-enriched and one that is less so. The ER-enriched group of mRNAs tends to encode secreted or membrane proteins, whereas the other group generally encodes cytosolic or nuclear proteins. Importantly, essentially all mRNAs, even those encoding cytosolic proteins, are translated to a significant degree on the ER. b | In the cytosol, the translation machinery primarily synthesizes cytosolic proteins, as depicted by the nascent chains entering the cytosol. Data suggest that translation on the ER is also primarily directed towards the synthesis of cytosolic proteins and not only towards the synthesis of membrane and secreted proteins.
A key mechanistic point that cannot be overemphasized is that despite the ER localization of their parent mRNA, the cytosolic protein products are not translocated into the ER but assume their appropriate cytosolic localization37. Thus, these findings decouple the ER localization of an mRNA and its translation from the subcellular destination of its encoded protein. This separation of mRNA localization from protein localization means that mRNAs do not need to associate with the ER or the cytosol in a purely binary manner; a fraction of the population of each mRNA may be represented in each compartment. Indeed, it seems that essentially all mRNAs that encode cytosolic proteins are well represented in both compartments18,25,28,40,41. For an average cytosolic protein-encoding mRNA, approximately half of the mRNA molecules are typically ER-bound, and this number increases in more secretion-oriented, rough ER-enriched cells28,31. These results lead to a surprising conclusion: the majority of translational activity on the ER — 75% in HEK293 cells — is directed towards the synthesis of cytosolic proteins (FIG. 2b). Similarly, mRNAs encoding protein components of other organelles, such as the nucleus and peroxisomes, are well represented on the ER, and those that encode membrane-targeting sequences are highly ER-enriched28. The only broad exceptions to this rule are mitochondria, which have their own, separate translocation machinery45,46, and tail-anchored and small secretory proteins, which associate with the ER post-translationally28,47. The ER should therefore be considered as a site of general cellular protein synthesis, in addition to its established role as a specialized site dedicated to the synthesis of secreted or membrane proteins.
The ER and cytosol as distinct translation compartments
As the magnitude of compartmentalization of translation to the ER has become clear, evidence has also begun to emerge that the ER and cytosol represent distinct biochemical environments for translation. Depending on their subcellular distribution, mRNAs may be exposed to distinct pools of regulatory factors and translational components, which may affect protein production. Various translational regulators48–51, ribonucleases52–54 and RNA-binding proteins43,55 localize to the ER to promote specialized functions. For example, an emerging body of work demonstrates that small interfering RNA (siRNA)- and microRNA (miRNA)-mediated silencing is highly dependent on the ER; for both transfected siRNAs and endogenous miRNAs, the critical processes of RNA-induced silencing complex (RISC) loading, mRNA binding and Argonaute 2-mediated processing occur in association with the ER membrane51,56. This localization has led to the proposition that although most siRNAs and miRNAs are cytosolic, the ER is the central site for siRNA processing and silencing activity, and cytosolic siRNAs and miRNAs remain inactive. This compartmentalization subjects mRNAs to distinct regulatory environments, where ER-localized mRNAs are targeted for silencing, whereas the same mRNA in the cytosol may escape.
The environments for protein folding also differ between the cytosol and the ER. A recent study found that aggregates of cytosolic proteins are generated by ER-bound ribosomes and are retained in association with the ER membrane38. The mechanism for this preferential aggregation of proteins derived from ER-bound ribosomes is unclear, but it opens the possibility that the folding status of many proteins depends on the localization of its parent mRNA, particularly given that protein structure is highly dependent on co-translational protein folding57,58. These differences could, for example, derive from differences in chaperone concentrations or translation rates between ER-bound and cytosolic ribosomes.
The biochemical environment for translation initiation and elongation on the ER also fundamentally differs from the cytosol. Elongating ribosomes on the ER access a distinct pool of tRNAs, which complete entire cycles of charging and discharging all while maintaining ER association59–61. This finding opens the possibility that cytosolic and ER-bound ribosomes could have divergent elongation rates at particular codons, enabling the optimization of translation of particular mRNAs in a distinct compartment. The localization of the multi-tRNA synthetase complex to the ER could further amplify the differences between compartments62. In addition to elongation-associated factors, the ER has its own abundant pool of eukaryotic initiation factor 2α (eIF2α) and associated regulatory factors that are bound to the ER independently of ribosomes, raising the possibility that initiation could also be subject to local regulation31. The combined result of such compartmentalization is that, on average, mRNAs are more heavily translated by ribosomes and that incorporation of radiolabelled amino acids is enhanced61 on the ER relative to the cytosol.
The emerging picture is that the ER is not only a primary site for translation but a fundamentally distinct compartment for protein synthesis. Accumulating evidence suggests that the ER can perform its translational function with minimal contributions from cytosolic components. Why might evolution select for such compartmentalization? One potential answer lies in the two-dimensional nature of the ER membrane. Collapsing an enzymatic process such as translation into two dimensions probably confers substantial enhancements in enzymatic activity, an idea that is supported by theoretical data63. Compartmentalization may be especially relevant given the numerous tightly regulated stages of translation, many of which would rely on the diffusion of large molecules such as ribosomes and mRNAs. Compartmentalizing a multicomponent process onto a membrane surface also raises several additional and interesting questions. For example, if a ribosome and an mRNA are both tethered to the ER, how do components move relative to one another as the ribosome processes along the mRNA? Are there factors that reduce the energetic barrier that results from having to move two ER-tethered molecules relative to one another? Regardless of the answer, we view the localization of translation to the ER as a prominent example of our emerging understanding that cellular components are intricately organized even when not separated by membranes64,65.
Assembling the translation machinery
What mechanisms confer localization of translation activity to the ER? In answering this question, it is useful to consider that the primary functional unit of translation is the polyribosome, which comprises an mRNA, an array of ribosomes along that mRNA, a series of nascent chains in the process of synthesis and a diverse set of associated RNA-binding proteins (FIG. 3a). Any one of these components may, with sufficient affinity, confer association of an entire polyribosome with the ER, and different classes of polyribosomes may associate with the ER by diverse mechanisms that rely on distinct subsets of these binding interactions.
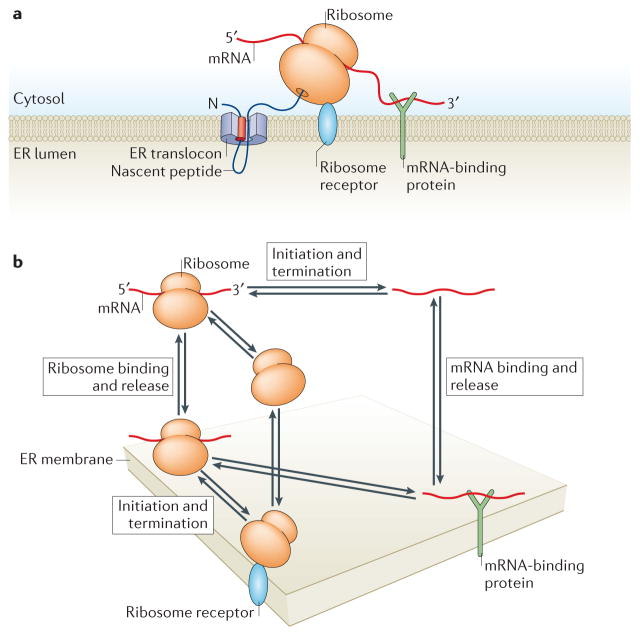
Figure 3. Assembling the translation machinery on the ER.
a | The primary functional unit of translation is the polyribosome, which comprises an mRNA, an array of ribosomes along that mRNA, a series of nascent polypeptide chains in the process of synthesis and a diverse set of associated RNA-binding proteins. A polyribosome may initiate assembly on the endoplasmic reticulum (ER) by any one of its constituent parts. The well-established mechanism for compartmentalizing translation to the ER is the signal recognition particle (SRP) pathway, in which a signal sequence in the nascent polypeptide initiates binding of the ribosome and its elongating polypeptide (FIG. 1a). Several other modes of ER association for ribosomes and mRNAs have also been described. Ribosomes may associate with the ER independently of their encoded nascent chain and with mRNAs subsequent to their association with an ER ribosome receptor. In addition, RNA-binding proteins can confer direct, ribosome-independent mRNA binding. This system requires that only one of these components bind to the ER to enable assembly of an entire polyribosome, and it is also possible that a polyribosome is targeted to the ER via multiple mechanisms in parallel. b | A general framework for ribosome and mRNA movement between the cytosol and the ER is depicted. There are essentially two transitions that occur: first, the association of an mRNA and a ribosome to form an active translational unit and the corresponding dissociation; and second the movement of each of these components between the cytosol and the ER. The rate at which each of these transitions occurs would vary depending on the mechanism of ER recruitment. For example, in the SRP pathway (FIG. 1a), the sequence of events is initiation in the cytosol, movement of the translation unit to the ER and finally dissociation of the ribosomes from the mRNA and release into the cytosol. In an alternative model, ribosomes bind stably to the ER and undergo many successive rounds of mRNA translation without release to the cytosol; that is, rates of ribosome binding and release would be lower than those of translational units that are targeted by the SRP pathway. Measurement of these rates for different classes of mRNAs and ribosomes will be essential for understanding the relative importance of various mechanisms for recruiting translation to the ER.
The SRP pathway is the best-understood mechanism for conferring ribosome and mRNA association with the ER8,9,66,67. During this process, the nascent polypeptide chain serves as a targeting signal for the recruitment of its associated mRNA and ribosomes to the ER, for ribosome binding to the protein conducting channel and for translocation of the nascent polypeptide chain2,7,68,69. However, if the SRP pathway is the sole mechanism for directing ribosome and mRNA traffic to the ER, several criteria must be met. First, only mRNAs that encode substrates for the SRP — that is, those that encode transmembrane and secretory proteins — would undergo localization to the ER. Second, ribosome association with the ER should be strictly dependent on, and mechanistically linked to, translation initiation, elongation and termination. From the available data, neither of these points has been borne out experimentally; a broad range of mRNAs associate with the ER, and ribosomes predominately maintain their association with the ER following termination15,16,28,70. These findings thus offer the opportunity to consider additional, alternative means of polyribosome recruitment and binding to the ER (FIG. 3b).
Ribosome binding to the ER
The decoupling of ribosome binding to the ER from protein synthesis could contribute to polyribosome assembly on the ER70. In this scenario, a ribosome could undergo an entire cycle of initiation, translation and termination while retaining ER association, and confer ER localization to its associated mRNA throughout this process. In addition to the identification of several candidate ER ribosome receptors (see below), there is some evidence to support this model. Ribosomes generally remain associated with the ER even after their dissociation from mRNAs, which indicates that ribosome association with the ER, once established, can be independent of both translation and nascent polypeptide chains. Furthermore, ribosomes bound to the ER are competent for de novo initiation of translation of both cytosolic and membrane proteins17,31. These two observations suggest that mRNA localization to the ER is primarily mediated by the translation activity of bound ribosomes. Considering that most translated mRNAs are found in polyribosomes, it can be readily appreciated that functional association with multiple ER-bound ribosomes would confer a stable state of mRNA binding to the ER. This raises, of course, the question of how ribosomes bind to the ER in vivo.
Current views of how ribosomes associate with the ER in vivo are mainly conjecture. This modest understanding of in vivo ribosome–ER interactions is, to a large extent, a consequence of the combined successes in reconstituting protein translocation in vitro and in structural imaging of the ribosome–SEC61 complex71–74. As the protein conducting channel, a ribosome-binding function for the SEC61 complex fulfils a central tenet of the co-translational model of protein translocation across the ER and fits neatly into the accepted model of a dedicated function for ER-bound ribosomes in secretory and membrane protein synthesis75. However, with new insights into the diversity of mRNA translation occurring on ER-bound ribosomes, it is appropriate to ask whether SEC61 functions as the sole ER ribosome receptor, regardless of the compartmental fate of the nascent polypeptide chain.
Early approaches to the identification of ribosome receptors focused on reconstituting ribosome binding in vitro76,77. In these studies, rough microsomes were first stripped of bound ribosomes by washing in high salt and/or EDTA-containing buffers, and the binding or rebinding of isolated, translationally inactive 80S ribosomes were investigated as a proxy assay for in vivo ribosome association with the ER. These approaches demonstrated that 80S ribosomes bind to stripped rough microsomes at low nanomolar affinity and that the large ribosomal subunit bound at higher affinity then the small subunit76,77. From this simple assay system emerged a confusing and contentious body of literature, in which similar experimental approaches were used to identify many ribosome receptors. For example, one study identified p180, an ER membrane protein with an extensive and highly basic decapeptide tandem repeat motif, as the primary ribosome receptor78,79. Subsequent work challenged this conclusion and assigned ribosome-binding function primarily, if not entirely, to the SEC61 translocon component SEC61α80–82. Importantly, when reconstituted, the purified SEC61 complex displayed both protein conducting channel function and the ribosome binding capacity determined in the prior stripped microsome studies, and so gained wide acceptance as the long sought ribosome receptor72,83–85. It should be noted that 80S inactive ribosomes — which were used to both characterize ribosome receptor binding interactions and, in some examples, for reconstitution of the ribosome–SEC61 complexes examined by cryo-electron microscopy — are very poor competitors of SRP-dependent ribosome targeting to native microsomes80,86. This raises the concern that the ER membrane binding activity displayed by inactive 80S ribosomes may be distinct from that used by in vivo targeted, translationally active ribosomes. Alternative approaches to the identification of the ribosome receptor or receptors, in which the binding partners of native, bound ribosomes was determined by chemical crosslinking studies, showed that translocon-associated protein subunit-α (TRAPα), ribophorin I, p180 and the SRP receptor are located in close proximity and/or are bound to the ribosome80,87,88. Furthermore, numerous experimental approaches, including antibody inhibition, have been used to validate a ribosome-binding function for ribophorin I89,90, SEC61β91, SEC62 (REF. 92), ERJ1 (REF. 93) and p180 (REFS 79,94) across yeast and mammals. Combined, these studies paint a confusing picture of ribosome binding to the ER, with SEC61α being widely accepted as the primary (or only) ribosome receptor amid substantial experimental evidence supporting ribosome-binding activity for several other abundant resident ER membrane proteins.
To reconcile the observations of a broad, general role for ER-bound ribosomes in mRNA translation with a conflicting ribosome receptor literature, we first suggest a simple model, whereby many ER proteins separately contribute to the totality of ribosome binding to the ER. In this view, diverse ribosome-binding sites may enable selectivity for structurally distinct subsets of ribosomes, perhaps with different populations of ribosomes being dedicated to the translation of distinct subsets of mRNAs95. A system in which ribosomes have structural heterogeneity or multiple ER-binding sites that enable selective binding of distinct subsets of ribosomes, along with translation of distinct cohorts of mRNAs, would provide a robust basis for the broad compartmentalization of mRNA translation to the ER. Here, we specifically distinguish between ribosome association with the ER and the process of protein translocation. We further speculate that ribosomes that are translating mRNAs with topogenic signals and that are bound to ribosome receptors other than SEC61α could be co-translationally recruited to SEC61 translocons, and that the nascent polypeptide chains could be translocated without a requirement for the translating ribosome to be tightly associated with the protein-conducting channel.
The prominence of the SRP pathway model has carried with it an important implication for ribosome trafficking: that ribosomes exchange rapidly between the ER and cytosol in a manner coupled to the termination of each round of protein synthesis. However, most evidence so far points towards ribosomes associating with the ER across many rounds of protein synthesis. For example, biochemical approaches have pointed towards ribosomes initiating translation while on the ER31,96 and maintaining their ER association following termination of protein synthesis17,96. However, a recent in vivo study using a novel ribosome profiling assay to track ribosome trafficking and translation on the ER found that many ribosomes are recruited to the translocon only upon the emergence of a signal peptide, and depart the ER within, at most, a few cycles of translation97. Although it is unclear whether this observation provides direct evidence for co-translational targeting to the ER or, alternatively, co-translational lateral diffusion of ER-bound ribosomes to the translocon as postulated above, these conclusions highlight the need to better define the in vivo dynamics of ribosome exchange on the ER. It is difficult to reconcile rapid, translation-coupled ribosome exchange with our developing understanding in which the bulk of ER-bound ribosomes are devoted to the translation of cytosolic protein-encoding mRNAs and are rather stably bound to a diversity of ER ribosome receptors. Thus, if most of the mRNA tran-scriptome is translated on the ER, these emerging data indicate that assessments of ribosome exchange dynamics are best performed directly, rather than inferred from the composition of the translated mRNA pool.
mRNA-binding proteins as mediators of ER localization
A solely ribosome-mediated mechanism cannot explain one crucial feature of ER translation: the high enrichment of mRNAs encoding secreted and membrane proteins on the ER25,28. Although approximately half of mRNAs that encode cytosolic proteins are bound to the ER, almost the entirety of the transcriptome encoding secretory and membrane proteins is ER-bound28. As noted above, the activity of the SRP pathway also cannot fully explain this localization, as pharmacological inhibition of translation initiation does not substantially modify mRNA localization27,28. Furthermore, ribosomes and mRNAs diverge in their sensitivity to extraction from the ER, with a subset of mRNAs being efficiently retained even after ribosomes are completely extracted from the ER37,42,43. In addition, neither mutation of the signal sequence nor loss of SRP function markedly compromises the localization of these mRNAs to the ER37,98. These findings point towards a role for the final component of the polyribosome: the mRNA itself and its associated RNA-binding proteins (FIG. 3a).
An mRNA could be tethered to the ER independently of its translation status. Although covalent mRNA modifications that confer ER binding are plausible99,100, no such modifications have yet been identified, and most attention has been focused on RNA-binding proteins that may serve as ER membrane anchors. Such a protein or complex of proteins would require a transmembrane domain and a domain that binds both tightly and stably to RNA. Several proteins that meet these criteria have been identified by proteomic methods, including p180 (REF. 101), ribophorin I (REF. 43), and SEC61α and SEC61β43. Intriguingly, each of these proteins has, at some point, been proposed to act as a ribosome receptor78,79,94,102,103. The newly identified mRNA-binding activity of these proteins leads to an alternative interpretation of prior results, as it is difficult to formally distinguish mRNA-binding activity from ribosome (rRNA) binding. Future work to validate and characterize RNA-binding sites on these proteins, and to identify the subsets of mRNAs that they anchor, is clearly needed. There are also some additional provocative proposals that other factors contribute to ER affinity. For example, collagen-encoding mRNAs were found to rely on the cooperation between the tubulin cytoskeleton and the RNA-binding protein La-related protein 6 (LARP6) to confer stable localization to the ER104. The total impact of the cytoskeleton on mRNA localization to the ER remains to be studied further. Properties of the mRNAs themselves, such as increased uracil content, have also been found to affect ER association105,106. These diverse mechanisms provide a multitude of routes to the ER, enabling the precise regulation of mRNA localization and translation.
Reassessing SRP function in vivo
With the emergence of alternative models for localizing and tethering translation to the ER, it can be considered that the in vivo role of the SRP may be more specialized than generally thought. It is abundantly clear from in vitro studies that the SRP can confer ER association of ribosomes and mRNAs8, but its precise in vivo function remains unclear. Studies using knockouts or knockdowns of SRP and/or the SRP receptor have provided some insights into this issue. siRNA-mediated silencing of SRP54 subunit expression has no discernible effect on mRNA partitioning — with mRNAs encoding membrane and secretory proteins retaining prominent ER localization37,107 — but does lead to deficiencies in the translocation of a subset of membrane proteins108,109. Furthermore, in yeast, loss of SRP expression is accompanied by the enhanced expression of several cytosolic chaperones108, suggesting that the SRP may function more as a chaperone for membrane and secretory proteins than as a universal mediator of translation localization. Together, the bulk of evidence suggests that the SRP ensures that a subset of membrane and secretory proteins are efficiently targeted to the protein conducting channel as they are synthesized, whereas the localization of the mRNA and ribosome to the ER may be conferred by other mechanisms. This separation of protein translocation and mRNA and ribosome targeting into distinct pathways enables a diverse range of mRNAs that encode proteins targeted to all cellular compartments to be translated on the ER.
mRNA localization during cellular stress
Considering the magnitude of translational compartmentalization to the ER and the unique properties of that compartment compared with the cytosol, the regulation of mRNA localization stands to be an important component of post-transcriptional gene regulation. It is no surprise then that recent studies have begun to link mRNA localization to the ER with the fate of an mRNA during cellular stress response programmes. For example, during oxidative stress, some mRNAs are recruited into stress granules when free in the cytosol, whereas mRNAs that are localized to the ER escape this seqestration44. Not surprisingly, examples of biological processes that take advantage of these properties have begun to emerge.
The most prominent and well-understood instance of mRNA localization contributing to the dynamics of gene expression is during the unfolded protein response (UPR), a translational and transcriptional stress response and homeostasis pathway that is activated in response to a build-up of unfolded proteins in the ER lumen110. Under these stress conditions, the continuing viability of the cell is fundamentally linked to its ability to reduce the influx of new proteins into the ER, as increasing the protein folding load in the ER would further disrupt proteostasis. To meet this need, several processes have evolved that regulate translation specifically on the ER. One of these mechanisms involves the post-transcriptional splicing of the X-box-binding protein 1 (XBP1) mRNA (mammalian cells) or the HAC1 mRNA (yeast) by the ER membrane stress sensor IRE1α54,111,112. The unspliced mRNA, which encodes a transcription factor, is itself localized to the ER113, bringing it into close proximity of IRE1α. Upon activation of the UPR, IRE1α initiates a non-canonical splicing event, yielding the synthesis of a shorter isoform of XBP1, the encoded protein of which in turn activates UPR target gene expression. In this manner, mRNA localization to the ER can serve as a critical conduit through which information regarding the protein folding status of the ER can be transmitted to the nucleus. In addition, the nuclease activity of IRE1α can be directed towards other ER-associated mRNAs, leading to their preferential degradation in a mechanism known as regulated IRE1-dependent decay (RIDD)53,114. Owing to the enrichment of mRNAs encoding ER-resident proteins on this organelle, this activity results in a reduction of protein flux into the ER, ultimately helping to restore proteostasis. These mechanisms demonstrate that the localization of an mRNA can be a determinate variable in its fate during cell stress; ER localization can determine, in the case of XBP1 or HAC1 mRNAs, whether an mRNA is spliced or unspliced or, for all other mRNAs, degraded or left intact.
Furthermore, mRNA localization can be a rapidly changing, dynamic process. In a recent study, it was found that the thousands of mRNAs that encode secretory and membrane proteins were rapidly and almost uniformly released from the ER upon stress induction, whereas mRNAs that encode cytosolic proteins were essentially unaffected18. By removing these mRNAs from the ER and away from the protein-conducting channel, this re-localization suppresses protein flux into the ER lumen and enables the cell time to restore proteostasis. Given the relative stability of ER mRNA localization under most normal, stress-free conditions, this mass re-localization begins to show a surprisingly contrasting picture, whereby some mechanisms of poly-ribosome binding to the ER can be rapidly eliminated while others remain intact. Altogether, the UPR provides a potent example of how mRNA localization is a critical variable in the post-transcriptional regulation of gene expression.
Conclusions and perspectives
The groundbreaking studies of Palade and co-workers identified a fundamental role for the ER in mRNA translation and for both secreted and non-secreted proteins3,14. In the ensuing decades, equally ground-breaking research has revealed fundamental insights into the mechanisms of protein targeting, translocation, trafficking and secretion. With transcriptomic technologies providing key tools for re-investigating the seemingly minor observation that cytosolic proteins were also translated on the ER, a more general role for the ER in transcriptome expression is emerging, whereby the ER has a fundamental and perhaps primary role in protein synthesis. In addition, it is becoming increasingly clear that the biochemical and regulatory ER translation environment differs from the cytosol (FIG. 4). Given that ER localization can confer divergent fates on mRNAs, what cellular processes might modify the localization of an mRNA to the ER? Viruses, many of which replicate via ER-localized viral factories115,116, may specifically compartmentalize the translation machinery to suit their own needs117. Other cellular programmes, such as B cell differentiation, may regulate ER-localized translation to maintain the appropriate composition of the ER proteome118. In these and certainly other instances, the ER is positioned to serve as a critical hub of post-transcriptional gene regulation, and future studies into this regulatory role of the ER will aid our understanding of how cells shape their composition and activity. The coming years will require intense focus on the mechanisms by which the translation machinery is recruited and tethered to the ER so that we may fully understand the role of the ER in gene expression and, more broadly, how such localization shapes the behaviour of the cell.
Figure 4. The ER as a global mRNA translation and regulation hub.
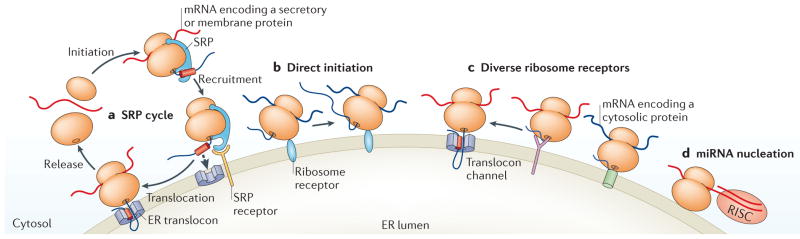
In this schematic, the diverse roles of the endoplasmic reticulum (ER) in global mRNA translation and regulation are depicted. The ER is the entry point of mRNAs encoding secretory and membrane proteins into the secretory pathway. Two modes of localization are shown. a | In one pathway, mRNAs encoding secretory and membrane proteins undergo initiation on free, cytosolic ribosomes and are targeted to the ER via the signal recognition particle (SRP) pathway. b | In the other pathway, many mRNAs, including those encoding secretory and cytosolic proteins undergo direct initiation on stably ER-bound ribosomes. c | Also shown are diverse ribosome receptors that are autonomous to translocation channels. These ribosomes can engage in the synthesis of cytosolic protein-encoding mRNAs, as well as mRNAs encoding topogenic signals. We propose that the receptor-bound, translationally active ribosome encounters an open translocation site by lateral diffusion in the ER membrane, with engagement of the nascent protein at the translocation site bringing the bound ribosome to close physical proximity to the protein-conducting channel. d | MicroRNA (miRNA)-mediated silencing is highly dependent on the ER. For endogenous miRNAs, RNA-induced silencing complex (RISC) loading, mRNA binding and Argonaute 2-mediated processing are associated with the ER membrane, whereas the cytosolic miRNAs remain inactive51,56. Thus, compartmentalization subjects mRNAs to distinct regulatory environments depending on their localization, in which ER-localized mRNAs are targeted for silencing but the same mRNA in the cytosol may escape.
Acknowledgments
The authors thank former members of C.V.N.’s laboratory, in particular M. Potter, R. Seiser, S. Stephens, R. Lerner and S. Jagannathan, for their many contributions to the concepts proposed in this Review, and current laboratory members, in particular J. C.-C. Hsu and A. Hoffman, for their critical feedback and contributions. They also thank S. Shenolikar for his ongoing contribution to the introduction and maturation of these ideas. Work in C.V.N.’s laboratory is supported by a grant from the National Institute of General Medical Sciences of the US National Institutes of Health (GM101533 to C.V.N.). D.W.R. is funded by a Translational Clinical Research Flagship Award entitled ‘National Parkinson’s Disease Translational Clinical Research Programme’ from National Medical Research Council Singapore.
Glossary
- Topogenic signal
A hydrophobic region of a protein that is targeted to the endoplasmic reticulum
- Signal recognition particle (SRP)
A ribonucleoprotein complex that targets nascent secretory and membrane proteins to the endoplasmic reticulum as they emerge from the ribosomes
- Microsomal vesicles
Vesicles that are derived from the endoplasmic reticulum and that are commonly used for in vitro studies of translation and protein translocation
- Multi-tRNA synthetase
An assembly of many tRNA synthetases that serves to concentrate the charging of tRNAs with amino acids
- Rough microsomes
Highly purified, translation-competent endoplasmic reticulum, generally obtained from canine pancreas
- Proteostasis
The status and health of protein folding in the cell
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Caro LG, Palade GE. Protein synthesis, storage, and discharge in the pancreatic exocrine cell. An Autoradiogr Study. J Cell Biol. 1964;20:473–495. doi: 10.1083/jcb.20.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson AE, van Waes MA. The translocon: a dynamic gateway at the ER membrane. Annu Rev Cell Dev Biol. 1999;15:799–842. doi: 10.1146/annurev.cellbio.15.1.799. [DOI] [PubMed] [Google Scholar]
- 3.Palade G. Intracellular aspects of the process of protein synthesis. Science. 1975;189:347–358. doi: 10.1126/science.1096303. This paper provides an important overview of the early understanding of the field and a summary of outstanding questions. [DOI] [PubMed] [Google Scholar]
- 4.Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 5.Blobel G. Protein targeting (Nobel lecture) Chembiochem. 2000;1:86–102. doi: 10.1002/1439-7633(20000818)1:2<86::AID-CBIC86>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 6.Walter P, Johnson AE. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- 7.Cross BC, Sinning I, Luirink J, High S. Delivering proteins for export from the cytosol. Nature Rev Mol Cell Biol. 2009;10:255–264. doi: 10.1038/nrm2657. [DOI] [PubMed] [Google Scholar]
- 8.Walter P, Blobel G. Translocation of proteins across the endoplasmic reticulum. II. Signal recognition protein (SRP) mediates the selective binding to microsomal membranes of in-vitro-assembled polysomes synthesizing secretory protein. J Cell Biol. 1981;91:551–556. doi: 10.1083/jcb.91.2.551. This landmark study reports the discovery of the SRP mechanism of cytosol-to-ER mRNA localization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walter P, Ibrahimi I, Blobel G. Translocation of proteins across the endoplasmic reticulum. I Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory protein. J Cell Biol. 1981;91:545–550. doi: 10.1083/jcb.91.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noriega TR, Chen J, Walter P, Puglisi JD. Real-time observation of signal recognition particle binding to actively translating ribosomes. Elife. 2014;3:e04418. doi: 10.7554/eLife.04418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilmore R, Blobel G, Walter P. Protein translocation across the endoplasmic reticulum. I Detection in the microsomal membrane of a receptor for the signal recognition particle. J Cell Biol. 1982;95:461–469. doi: 10.1083/jcb.95.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer DI, Krause E, Dobberstein B. Secretory protein translocation across membranes-the role of the ‘docking protein’. Nature. 1982;297:647–650. doi: 10.1038/297647a0. [DOI] [PubMed] [Google Scholar]
- 13.Blobel G, Dobberstein B. Transfer of proteins across membranes. I Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J Cell Biol. 1975;67:835–851. doi: 10.1083/jcb.67.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siekevitz P, Palade GE. A cytochemical study on the pancreas of the guinea pig. 5 In vivo incorporation of leucine-1-C14 into the chymotrypsinogen of various cell fractions. J Biophys Biochem Cytol. 1960;7:619–630. doi: 10.1083/jcb.7.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Potter MD, Seiser RM, Nicchitta CV. Ribosome exchange revisited: a mechanism for translation-coupled ribosome detachment from the ER membrane. Trends Cell Biol. 2001;11:112–115. doi: 10.1016/s0962-8924(00)01905-x. [DOI] [PubMed] [Google Scholar]
- 16.Seiser RM, Nicchitta CV. The fate of membrane-bound ribosomes following the termination of protein synthesis. J Biol Chem. 2000;275:33820–33827. doi: 10.1074/jbc.M004462200. [DOI] [PubMed] [Google Scholar]
- 17.Potter MD, Nicchitta CV. Regulation of ribosome detachment from the mammalian endoplasmic reticulum membrane. J Biol Chem. 2000;275:33828–33835. doi: 10.1074/jbc.M005294200. [DOI] [PubMed] [Google Scholar]
- 18.Reid DW, Chen Q, Tay AS, Shenolikar S, Nicchitta CV. The unfolded protein response triggers selective mRNA release from the endoplasmic reticulum. Cell. 2014;158:1362–1374. doi: 10.1016/j.cell.2014.08.012. This study identifies a role for dynamic ER mRNA localization in cellular stress responses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palade GE. A small particulate component of the cytoplasm. J Biophys Biochem Cytol. 1955;1:59–68. doi: 10.1083/jcb.1.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hicks SJ, Drysdale JW, Munro HN. Preferential synthesis of ferritin and albumin by different populations of liver polysomes. Science. 1969;164:584–585. doi: 10.1126/science.164.3879.584. [DOI] [PubMed] [Google Scholar]
- 21.Mueckler MM, Pitot HC. Structure and function of rat liver polysome populations. I Complexity, frequency distribution, and degree of uniqueness of free and membrane-bound polysomal polyadenylate-containing RNA populations. J Cell Biol. 1981;90:495–506. doi: 10.1083/jcb.90.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mechler B, Vassalli P. Membrane-bound ribosomes of myeloma cells. III The role of the messenger RNA and the nascent polypeptide chain in the binding of ribosomes to membranes. J Cell Biol. 1975;67:25–37. doi: 10.1083/jcb.67.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mechler B, Vassalli P. Membrane-bound ribosomes of myeloma cells. II Kinetic studies on the entry of newly made ribosomal subunits into the free and the membrane-bound ribosomal particles. J Cell Biol. 1975;67:16–24. doi: 10.1083/jcb.67.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kopczynski CC, et al. A high throughput screen to identify secreted and transmembrane proteins involved in Drosophila embryogenesis. Proc Natl Acad Sci USA. 1998;95:9973–9978. doi: 10.1073/pnas.95.17.9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diehn M, Eisen MB, Botstein D, Brown PO. Large-scale identification of secreted and membrane-associated gene products using DNA microarrays. Nature Genet. 2000;25:58–62. doi: 10.1038/75603. [DOI] [PubMed] [Google Scholar]
- 26.Diehn M, Bhattacharya R, Botstein D, Brown PO. Genome-scale identification of membrane-associated human mRNAs. PLoS Genet. 2006;2:e11. doi: 10.1371/journal.pgen.0020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lerner RS, et al. Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA. 2003;9:1123–1137. doi: 10.1261/rna.5610403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reid DW, Nicchitta CV. Primary role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling. J Biol Chem. 2012;287:5518–5527. doi: 10.1074/jbc.M111.312280. This report defines the translation that occurs on cytosolic and ER-bound ribosomes during homeostasis and cell stress. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stephens SB, Dodd RD, Lerner RS, Pyhtila BM, Nicchitta CV. Analysis of mRNA partitioning between the cytosol and endoplasmic reticulum compartments of mammalian cells. Methods Mol Biol. 2008;419:197–214. doi: 10.1007/978-1-59745-033-1_14. [DOI] [PubMed] [Google Scholar]
- 30.Jagannathan S, Nwosu C, Nicchitta CV. Analyzing mRNA localization to the endoplasmic reticulum via cell fractionation. Methods Mol Biol. 2011;714:301–321. doi: 10.1007/978-1-61779-005-8_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jagannathan S, Reid DW, Cox AH, Nicchitta CV. De novo translation initiation on membrane-bound ribosomes as a mechanism for localization of cytosolic protein mRNAs to the endoplasmic reticulum. RNA. 2014;120:489–498. doi: 10.1261/rna.045526.114. This paper presents findings that translation initiation occurs on the ER. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palacios IM, Johnston DS. Getting the message across: the intracellular localization of mRNAs in higher eukaryotes. Annu Rev Cell Dev Biol. 2001;17:569–614. doi: 10.1146/annurev.cellbio.17.1.569. [DOI] [PubMed] [Google Scholar]
- 34.Cui XA, Zhang Y, Hong SJ, Palazzo AF. Identification of a region within the placental alkaline phosphatase mRNA that mediates p180-dependent targeting to the endoplasmic reticulum. J Biol Chem. 2013;288:29633–29641. doi: 10.1074/jbc.M113.482505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loya A, et al. The 3′-UTR mediates the cellular localization of an mRNA encoding a short plasma membrane protein. RNA. 2008;14:1352–1365. doi: 10.1261/rna.867208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicchitta CV, Lerner RS, Stephens SB, Dodd RD, Pyhtila B. Pathways for compartmentalizing protein synthesis in eukaryotic cells: the template-partitioning model. Biochem Cell Biol. 2005;83:687–695. doi: 10.1139/o05-147. [DOI] [PubMed] [Google Scholar]
- 37.Pyhtila B, et al. Signal sequence- and translation-independent mRNA localization to the endoplasmic reticulum. RNA. 2008;14:445–453. doi: 10.1261/rna.721108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou C, et al. Organelle-based aggregation and retention of damaged proteins in asymmetrically dividing cells. Cell. 2014;159:530–542. doi: 10.1016/j.cell.2014.09.026. This study identifies a distinct protein folding and aggregation environment on the ER. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 40.Mueckler MM, Pitot HC. Structure and function of rat liver polysome populations. II Characterization of polyadenylate-containing mRNA associated with subpopulations of membrane-bound particles. J Cell Biol. 1982;94:297–307. doi: 10.1083/jcb.94.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mechler B, Rabbitts TH. Membrane-bound ribosomes of myeloma cells. IV mRNA complexity of free and membrane-bound polysomes. J Cell Biol. 1981;88:29–36. doi: 10.1083/jcb.88.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Q, Jagannathan S, Reid DW, Zheng T, Nicchitta CV. Hierarchical regulation of mRNA partitioning between the cytoplasm and the endoplasmic reticulum of mammalian cells. Mol Biol Cell. 2011;22:2646–2658. doi: 10.1091/mbc.E11-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jagannathan S, et al. Multifunctional roles for the protein translocation machinery in RNA anchoring to the endoplasmic reticulum. J Biol Chem. 2014;289:25907–25924. doi: 10.1074/jbc.M114.580688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Unsworth H, Raguz S, Edwards HJ, Higgins CF, Yague E. mRNA escape from stress granule sequestration is dictated by localization to the endoplasmic reticulum. FASEB J. 2010;24:3370–3380. doi: 10.1096/fj.09-151142. [DOI] [PubMed] [Google Scholar]
- 45.Williams CC, Jan CH, Weissman JS. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science. 2014;346:748–751. doi: 10.1126/science.1257522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dudek J, Rehling P, van der Laan M. Mitochondrial protein import: common principles and physiological networks. Biochim Biophys Acta. 2013;1833:274–285. doi: 10.1016/j.bbamcr.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 47.Johnson N, Powis K, High S. Post-translational translocation into the endoplasmic reticulum. Biochim Biophys Acta. 2013;1833:2403–2409. doi: 10.1016/j.bbamcr.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 48.Zhou W, Brush MH, Choy MS, Shenolikar S. Association with endoplasmic reticulum promotes proteasomal degradation of GADD34 protein. J Biol Chem. 2011;286:21687–21696. doi: 10.1074/jbc.M110.212787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brush MH, Weiser DC, Shenolikar S. Growth arrest and DNA damage-inducible protein GADD34 targets protein phosphatase 1α to the endoplasmic reticulum and promotes dephosphorylation of the α subunit of eukaryotic translation initiation factor 2. Mol Cell Biol. 2003;23:1292–1303. doi: 10.1128/MCB.23.4.1292-1303.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jousse C, et al. Inhibition of a constitutive translation initiation factor 2α phosphatase, CReP, promotes survival of stressed cells. J Cell Biol. 2003;163:767–775. doi: 10.1083/jcb.200308075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li S, et al. MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell. 2013;153:562–574. doi: 10.1016/j.cell.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwarz DS, Blower MD. The calcium-dependent ribonuclease XendoU promotes ER network formation through local RNA degradation. J Cell Biol. 2014;207:41–57. doi: 10.1083/jcb.201406037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- 54.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 55.Batlle M, Marsellach FX, Huertas D, Azorin F. Drosophila vigilin, DDP1, localises to the cytoplasm and associates to the rough endoplasmic reticulum. Biochim Biophys Acta. 2011;1809:46–55. doi: 10.1016/j.bbagrm.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 56.Stalder L, et al. The rough endoplasmatic reticulum is a central nucleation site of siRNA-mediated RNA silencing. EMBO J. 2013;32:1115–1127. doi: 10.1038/emboj.2013.52. This report defines the ER as essential for the execution of siRNA function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O’Brien EP, Vendruscolo M, Dobson CM. Kinetic modelling indicates that fast-translating codons can coordinate cotranslational protein folding by avoiding misfolded intermediates. Nature Commun. 2014;5:2988. doi: 10.1038/ncomms3988. [DOI] [PubMed] [Google Scholar]
- 58.Fedorov AN, Baldwin TO. Contribution of cotranslational folding to the rate of formation of native protein structure. Proc Natl Acad Sci USA. 1995;92:1227–1231. doi: 10.1073/pnas.92.4.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Negrutskii BS, Deutscher MP. Channeling of aminoacyl-tRNA for protein synthesis in vivo. Proc Natl Acad Sci USA. 1991;88:4991–4995. doi: 10.1073/pnas.88.11.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Negrutskii BS, Deutscher MP. A sequestered pool of aminoacyl-tRNA in mammalian cells. Proc Natl Acad Sci USA. 1992;89:3601–3604. doi: 10.1073/pnas.89.8.3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stephens SB, Nicchitta CV. Divergent regulation of protein synthesis in the cytosol and endoplasmic reticulum compartments of mammalian cells. Mol Biol Cell. 2008;19:623–632. doi: 10.1091/mbc.E07-07-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.David A, et al. RNA binding targets aminoacyl-tRNA synthetases to translating ribosomes. J Biol Chem. 2011;286:20688–20700. doi: 10.1074/jbc.M110.209452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCloskey MA, Poo MM. Rates of membrane-associated reactions: reduction of dimensionality revisited. J Cell Biol. 1986;102:88–96. doi: 10.1083/jcb.102.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saks V, Beraud N, Wallimann T. Metabolic compartmentation — a system level property of muscle cells: real problems of diffusion in living cells. Int J Mol Sci. 2008;9:751–767. doi: 10.3390/ijms9050751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hudder A, Nathanson L, Deutscher MP. Organization of mammalian cytoplasm. Mol Cell Biol. 2003;23:9318–9326. doi: 10.1128/MCB.23.24.9318-9326.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walter P, Blobel G. Translocation of proteins across the endoplasmic reticulum III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J Cell Biol. 1981;91:557–561. doi: 10.1083/jcb.91.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Walter P, Blobel G. Translocation of proteins across the endoplasmic reticulum. Oncodev Biol Med. 1982;4:137–146. [PubMed] [Google Scholar]
- 68.Rapoport TA. Protein transport across the endoplasmic reticulum membrane. FEBS J. 2008;275:4471–4478. doi: 10.1111/j.1742-4658.2008.06588.x. [DOI] [PubMed] [Google Scholar]
- 69.Zimmermann R, Eyrisch S, Ahmad M, Helms V. Protein translocation across the ER membrane. Biochim Biophys Acta. 2011;1808:912–924. doi: 10.1016/j.bbamem.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 70.Stephens SB, et al. Stable ribosome binding to the endoplasmic reticulum enables compartment-specific regulation of mRNA translation. Mol Biol Cell. 2005;16:5819–5831. doi: 10.1091/mbc.E05-07-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beckmann R, et al. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell. 2001;107:361–372. doi: 10.1016/s0092-8674(01)00541-4. [DOI] [PubMed] [Google Scholar]
- 72.Gorlich D, Rapoport TA. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- 73.Menetret JF, et al. Single copies of Sec61 and TRAP associate with a nontranslating mammalian ribosome. Structure. 2008;16:1126–1137. doi: 10.1016/j.str.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Voorhees RM, Fernandez IS, Scheres SH, Hegde RS. Structure of the mammalian ribosome-Sec61 complex to 3.4 Å resolution. Cell. 2014;157:1632–1643. doi: 10.1016/j.cell.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Simon SM, Blobel G. A protein-conducting channel in the endoplasmic reticulum. Cell. 1991;65:371–380. doi: 10.1016/0092-8674(91)90455-8. [DOI] [PubMed] [Google Scholar]
- 76.Borgese D, Blobel G, Sabatini DD. In vitro exchange of ribosomal subunits between free and membrane-bound ribosomes. J Mol Biol. 1973;74:415–438. doi: 10.1016/0022-2836(73)90037-5. [DOI] [PubMed] [Google Scholar]
- 77.Borgese N, Mok W, Kreibich G, Sabatini DD. Ribosomal-membrane interaction: in vitro binding of ribosomes to microsomal membranes. J Mol Biol. 1974;88:559–580. doi: 10.1016/0022-2836(74)90408-2. [DOI] [PubMed] [Google Scholar]
- 78.Savitz AJ, Meyer DI. Identification of a ribosome receptor in the rough endoplasmic reticulum. Nature. 1990;346:540–544. doi: 10.1038/346540a0. [DOI] [PubMed] [Google Scholar]
- 79.Savitz AJ, Meyer DI. 180 kD ribosome receptor is essential for both ribosome binding and protein translocation. J Cell Biol. 1993;120:853–863. doi: 10.1083/jcb.120.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Collins PG, Gilmore R. Ribosome binding to the endoplasmic reticulum — a 180 kD protein identified by crosslinking to membrane-bound ribosomes is not required for ribosome binding activity. J Cell Biol. 1991;114:639–649. doi: 10.1083/jcb.114.4.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nunnari JM, Zimmerman DL, Ogg SC, Walter P. Characterization of the rough endoplasmic reticulum ribosome-binding activity. Nature. 1991;352:638–640. doi: 10.1038/352638a0. [DOI] [PubMed] [Google Scholar]
- 82.Prinz A, Behrens C, Rapoport TA, Hartmann E, Kalies KU. Evolutionarily conserved binding of ribosomes to the translocation channel via the large ribosomal RNA. EMBO J. 2000;19:1900–1906. doi: 10.1093/emboj/19.8.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Görlich D, Prehn S, Hartmann E, Kalies KU, Rapoport TA. A mammalian homolog of Sec61p and SecYp is associated with ribosomes and nascent polypeptides during translocation. Cell. 1992;71:489–503. doi: 10.1016/0092-8674(92)90517-g. [DOI] [PubMed] [Google Scholar]
- 84.Kalies KU, Gorlich D, Rapoport TA. Binding of ribosomes to the rough endoplasmic reticulum mediated by the Sec61p-complex. J Cell Biol. 1994;126:925–934. doi: 10.1083/jcb.126.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schaletzky J, Rapoport TA. Ribosome binding to and dissociation from translocation sites of the endoplasmic reticulum membrane. Mol Biol Cell. 2006;17:3860–3869. doi: 10.1091/mbc.E06-05-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Murphy ECI, Zheng T, Nicchitta CV. Identification of a novel stage of ribosome/nascent chain association with the endoplasmic reticulum membrane. J Cell Biol. 1997;136:1213–1226. doi: 10.1083/jcb.136.6.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kreibich G, Freienstein CM, Pereyra BN, Ulrich BL, Sabatini DD. Proteins of rough microsomal membranes related to ribosome binding. II Cross-linking of bound ribosomes to specific membrane proteins exposed at the binding sites. J Cell Biol. 1978;77:488–506. doi: 10.1083/jcb.77.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kreibich G, Ulrich BL, Sabatini DD. Proteins of rough microsomal membranes related to ribosome binding. I Identification of ribophorins I and II, membrane proteins characteristics of rough microsomes. J Cell Biol. 1978;77:464–487. doi: 10.1083/jcb.77.2.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Harada Y, Li H, Li H, Lennarz WJ. Oligosaccharyltransferase directly binds to ribosome at a location near the translocon-binding site. Proc Natl Acad Sci USA. 2009;106:6945–6949. doi: 10.1073/pnas.0812489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yu YH, Sabatini DD, Kreibich G. Antiribophorin antibodies inhibit the targeting to the ER membrane of ribosomes containing nascent secretory polypeptides. J Cell Biol. 1990;111:1335–1342. doi: 10.1083/jcb.111.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Levy R, Wiedmann M, Kreibich G. In vitro binding of ribosomes to the β subunit of the Sec61p protein translocation complex. J Biol Chem. 2001;276:2340–2346. doi: 10.1074/jbc.M004867200. [DOI] [PubMed] [Google Scholar]
- 92.Muller L, et al. Evolutionary gain of function for the ER membrane protein Sec62 from yeast to humans. Mol Biol Cell. 2010;21:691–703. doi: 10.1091/mbc.E09-08-0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Blau M, et al. ERj1p uses a universal ribosomal adaptor site to coordinate the 80S ribosome at the membrane. Nature Struct Mol Biol. 2005;12:1015–1016. doi: 10.1038/nsmb998. [DOI] [PubMed] [Google Scholar]
- 94.Ueno T, Kaneko K, Sata T, Hattori S, Ogawa-Goto K. Regulation of polysome assembly on the endoplasmic reticulum by a coiled-coil protein, 180. Nucleic Acids Res. 2012;40:3006–3017. doi: 10.1093/nar/gkr1197. This paper provides a thorough analysis of the potential impact of p180 in regulating ribosome binding to the ER. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mauro VP, Edelman GM. The ribosome filter hypothesis. Proc Natl Acad Sci USA. 2002;99:12031–12036. doi: 10.1073/pnas.192442499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Potter MD, Nicchitta CV. Endoplasmic reticulum ribosomes reside in stable association with the translocon following termination of protein synthesis. J Biol Chem. 2002;277:23314–23320. doi: 10.1074/jbc.M202559200. [DOI] [PubMed] [Google Scholar]
- 97.Jan CH, Williams CC, Weissman JS. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science. 2014;346:1257521. doi: 10.1126/science.1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kraut-Cohen J, et al. Translation- and SRP-independent mRNA targeting to the endoplasmic reticulum in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2013;24:3069–3084. doi: 10.1091/mbc.E13-01-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 100.Ovodov SYu, Alakhov YuB. mRNA acetylated at 2′-OH-groups of ribose residues is functionally active in the cell-free translation system from wheat embryos. FEBS Lett. 1990;270:111–114. doi: 10.1016/0014-5793(90)81246-k. [DOI] [PubMed] [Google Scholar]
- 101.Cui XA, Zhang H, Palazzo AF. p180 promotes the ribosome-independent localization of a subset of mRNA to the endoplasmic reticulum. PLoS Biol. 2012;10:e1001336. doi: 10.1371/journal.pbio.1001336. This report identifies a role for p180 in promoting selective mRNA localization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hortsch M, Avossa D, Meyer DI. Characterization of secretory protein translocation: ribosome-membrane interaction in endoplasmic reticulum. J Cell Biol. 1986;103:241–253. doi: 10.1083/jcb.103.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ueno T, et al. Enhancement of procollagen biosynthesis by p180 through augmented ribosome association on the endoplasmic reticulum in response to stimulated secretion. J Biol Chem. 2010;285:29941–29950. doi: 10.1074/jbc.M109.094607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang H, Stefanovic B. Role of LARP6 and nonmuscle myosin in partitioning of collagen mRNAs to the ER membrane. PLoS ONE. 2014;9:e108870. doi: 10.1371/journal.pone.0108870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Polyansky AA, Hlevnjak M, Zagrovic B. Analogue encoding of physicochemical properties of proteins in their cognate messenger RNAs. Nature Commun. 2013;4:2784. doi: 10.1038/ncomms3784. This paper provides a statistical approach that links the chemical properties of mRNAs and their encoded proteins to ER localization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Prilusky J, Bibi E. Studying membrane proteins through the eyes of the genetic code revealed a strong uracil bias in their coding mRNAs. Proc Natl Acad Sci USA. 2009;106:6662–6666. doi: 10.1073/pnas.0902029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.del Alamo M, et al. Defining the specificity of cotranslationally acting chaperones by systematic analysis of mRNAs associated with ribosome-nascent chain complexes. PLoS Biol. 2011;9:e1001100. doi: 10.1371/journal.pbio.1001100. This report demonstrates that loss of the SRP does not compromise mRNA localization at a transcriptome scale. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mutka SC, Walter P. Multifaceted physiological response allows yeast to adapt to the loss of the signal recognition particle-dependent protein-targeting pathway. Mol Biol Cell. 2001;12:577–588. doi: 10.1091/mbc.12.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ren YG, et al. Differential regulation of the TRAIL death receptors DR4 and DR5 by the signal recognition particle. Mol Biol Cell. 2004;15:5064–5074. doi: 10.1091/mbc.E04-03-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 111.Lee K, et al. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16:452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Aragon T, et al. Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature. 2009;457:736–740. doi: 10.1038/nature07641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yanagitani K, et al. Cotranslational targeting of XBP1 protein to the membrane promotes cytoplasmic splicing of its own mRNA. Mol Cell. 2009;34:191–200. doi: 10.1016/j.molcel.2009.02.033. [DOI] [PubMed] [Google Scholar]
- 114.Gaddam D, Stevens N, Hollien J. Comparison of mRNA localization and regulation during endoplasmic reticulum stress in Drosophila cells. Mol Biol Cell. 2013;24:14–20. doi: 10.1091/mbc.E12-06-0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Risco C, et al. Endoplasmic reticulum–Golgi intermediate compartment membranes and vimentin filaments participate in vaccinia virus assembly. J Virol. 2002;76:1839–1855. doi: 10.1128/JVI.76.4.1839-1855.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sodeik B, et al. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J Cell Biol. 1993;121:521–541. doi: 10.1083/jcb.121.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Desmet EA, Anguish LJ, Parker JS. Virus-mediated compartmentalization of the host translational machinery. mBio. 2014;5:e01463–14. doi: 10.1128/mBio.01463-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wiest DL, et al. Membrane biogenesis during B cell differentiation: most endoplasmic reticulum proteins are expressed coordinately. J Cell Biol. 1990;110:1501–1511. doi: 10.1083/jcb.110.5.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Blobel G, et al. Translocation of proteins across membranes: the signal hypothesis and beyond. Symp Soc Exp Biol. 1979;33:9–36. [PubMed] [Google Scholar]
- 120.Walter P, Blobel G. Purification of a membrane-associated protein complex required for protein translocation across the endoplasmic reticulum. Proc Natl Acad Sci USA. 1980;77:7112–7116. doi: 10.1073/pnas.77.12.7112. [DOI] [PMC free article] [PubMed] [Google Scholar]