Abstract
Objectives
To determine the impact of HIV-1 subtype on treatment outcomes and the emergence of drug resistance in the resource limited setting of Kampala, Uganda.
Design
The Joint Clinical Research Centre (JCRC) in Kampala, Uganda has provided over 2000 drug-resistant genotypes (DRGs) over the past 10 years as standard of care for patients failing therapy and 1403 from treatment-naive and experienced patients over the past 10 years have been analyzed for this study.
Method
Viral loads, CD4 cell count, treatment histories and other relevant clinical data was compared with the infecting HIV-1 subtype and DRGs of Ugandan patients failing treatment.
Results
Patients failing HAART with DRGs (n = 937) were more frequently infected with subtype D than expected on the basis of the subtype distribution in the treatment-naive population (n = 655) in Kampala (P < 0.001). Higher proportions of treatment failures among subtype D-infected patients were driven by resistance to nucleoside reverse transcriptase inhibitors (NRTI) (P < 0.0002) more than to non-NRTIs (P > 0.04) or protease inhibitors.
Conclusion
Higher rates of treatment failure among subtype D as compared with subtype A-infected Ugandans was analogous to the faster disease progression in subtype D-infected patients. The mechanism(s) by which drug resistance may emerge faster in subtype D HIV-1 may relate to higher replicative fitness and increased propensity for a CXCR4 tropism.
Keywords: antiretroviral treatment, drug resistance, HIV-1, resource-limited setting, subtypes, Uganda
Introduction
The widespread use of HAART, a combination of at least three antiretroviral drugs in first-line or second-line regimens has substantially improved the prognosis of HIV-infected individuals as well as modified the natural history of the disease in the developing world [1,2]. With the roll out of antiretroviral treatment (ART) to more than 8 million HIV-infected individuals in resource-limited settings, the benefit of therapy has been equally impressive at reducing morbidity and mortality [3] despite initial skepticism about proper implementation [4]. Recommendations on when to start treatment have been controversial especially in Europe and North America [5]. New WHO treatment guidelines of initiating treatment with CD4 cell count less than 350 per microlitre are rarely implemented in resource-limited settings because of feasibility and cost [6].
Treatment failures, defined by rebounds in HIV-1 load [7], are generally caused by intermittent treatment or adherence, poor drug tolerance, and limited treatment monitoring; all of which can lead to the emergence of HIV-1 resistance to antiretroviral drugs. Upon emergence of new drug resistance and cross-resistance to the new ART, second-line treatment regimens require careful monitoring to avoid new treatment failures and resumption of disease [8]. With the limited treatment options available in the developing countries, resistance to nucleoside reverse transcriptase inhibitors (NRTIs), nonnucleoside reverse transcriptase inhibitors (NNRTIs) and protease inhibitors poses a great challenge to planning for various salvage/second-line treatment regimens [9]. Drug resistance testing is often preferred to viral load testing in some resource-limited clinics as circulating virus must be detectable to obtain a drug resistance genotype [10–12].
Drug resistance has been identified for every antiretroviral drug that is currently approved by the US Food and Drug Administration [13]. The rapid emergence of drug resistance has been attributed to the high replication rate of HIV coupled with the low fidelity of its viral reverse transcriptase enzyme [14]. With the NNRTI and protease inhibitor, a mutation may confer cross-resistance to one or more antiretroviral drugs in that drug class [15]. Resistance to NRTIs is the most selective with limited cross-resistance conferred by specific NRTI resistance mutations [15]. The most significant exception may be the drug resistance mutations (e.g. M41L, D67N, K70R, D210W, T215Y/F, and K219E) that confer resistance to the thymidine analogs (stavudine and zidovudine) [16]. Thousands of correlative phenotypic and genotypic drug resistance assays using subtype B HIV-1 isolates were adopted to characterize drug resistance in nonsubtype B infections [17], which dominate the worldwide epidemic. With a few exceptions [18,19], many studies now confirm that subtype B drug resistance mutations also confer drug resistance in HIV-1 of other subtypes. However, relative emergence rates of these drug resistance mutations in nonsubtype B isolates during selective therapy have not been a subject of intense investigation. Most studies conclude that response to the same antiretroviral-treatment regimen is similar in patients regardless of infecting HIV-1 subtype [20–22]. One study does suggest that subtype C-infected patients accumulate resistant mutations at a much slower rate when compared with subtype B [23].
To study the impact of the infecting HIV-1 subtype on antiretroviral-treatment outcomes and drug resistance, we have compiled treatment histories, outcomes and drug resistance results in a cross-sectional study spanning 10 years at the Joint Clinical Research Centre (JCRC) in Kampala, Uganda. Over 2000 samples from patients failing treatment were tested for drug resistance using genotypic assay. From this cohort, more than 900 DRGs were obtained from patients with clinical monitoring and a complete treatment history. We investigated if drug resistance was affected by the infecting HIV-1 subtype, the type of antiretroviral-treatment regimen, the number of successive treatment regimens, and the dates of treatment and/or resistance testing. Our findings suggest that antiretroviral resistance is more common in subtype D than in subtype A or subtype C-infected patients. Emergence of drug resistance is increasing in anti-retroviral-treated, subtype D-infected patients (>55%) over time and with each successive treatment regimen. However, the proportion of subtype D has remained at less than 36% in the untreated population in Kampala, Uganda over the past 10 years. Finally, we compared the frequency of treatment failures by subtype in this cross-sectional cohort to that in a longitudinal cohort of 188 patients (a subset from ∼1600 patients in an ongoing meta-analyses).
Methods
Study population and ethics
JCRC was one of the first institutions in Uganda to offer antiretroviral therapy for HIV-positive patients in accordance with the WHO guidelines at the time. From 1999 to 2010, clinicians attending to antiretroviral-treated patients at the JCRC requested drug resistance testing based on clinical evidence of treatment failure: defined as viral loads more than 2000 copies/ml and/or CD4 cell count less than 250 cells/μl on two successive visits. Patients were exposed to at least two ART drugs starting in early 1990 s whereas all patients received HAART as standard-of-care by 1998. First-line HAART consisted of two NRTIs a thymidine analog (stavudine or zidovudine) and a cytidine analog (emtricitabine or lamivudine). The NNRTI was either nevirapine or efavirenz. Data for these analyses were obtained from the patient care database at the JCRC. IRB approval (EM10–07) was obtained for drug resistance testing and for development for an anonymized database to store the clinical data. Both the clinical and laboratory staff were blinded to this clinical data on ∼2000 patient samples derived from ∼15 000 HIV-infected patients attending the clinic.
Sequence analysis and genotypic drug resistance
Of the 2000 unique HIV-1 sequences analyzed, 939 had high quality HIV-1 reverse transcriptase (RT) or protease (PR)-RT sequences associated with complete clinical demographics and treatment monitoring information. For approximately 50 samples, low-quality sequence was related to multiple polymorphic sites in the reverse transcriptase or PR-RT sequence preventing the assembly of an average consensus sequence for the patient-derived virus. Over 1000 DRGs were excluded due to limited patient records, viral loads, or CD4 cell count (i.e. less than two viral load and CD4 cell count per year). Finally, we also excluded all DRGs from patients not attending the JCRC clinic for the duration of treatment/care. Approximately, 30% of all DRGs are performed for other HIV/AIDS clinics in Kampala and throughout Uganda. As part of various clinical research studies, genotypes from PR-RT sequencing were also obtained from 269 untreated patients attending the JCRC clinic starting in 1997. To reduce costs, reverse transcriptase sequencing was primarily performed for patients receiving an NNRTI-based regimen while PR-RT sequencing was performed on patients on protease inhibitor-based regimens. Briefly, HIV-1 RNA was extracted from plasma samples using a Qiagen RNA extraction method (Qiagen Inc., Chatsworth, California, USA). Polymerase gene-specific primers were used for PCR as described [10] and in supplementary Table 1, http://links.lww.com/QAD/A334. The PCR products were purified using the QIAquick PCR purification kit (Qiagen, Valencia, California, USA), sequenced using Visible Genetics (Ontario, Canada) (1997–2002 samples), Beckman Coulter sequencing kit and CEQ 8000 instrument (Beckman Coulter Inc., Fullerton, California, USA) (2002–2007 samples) and longer read lengths, and reduced number of repeat testing were obtained from 630 samples (2006– 2010) using the BigDye Terminator cycle sequencing kit and ABI3730xl sequencing platform (Life Technologies, Carlsbad, California, USA). With a few exceptions, all sequencing was performed in the Center for AIDS Research (CFAR) laboratories at the JCRC site. The HIV-1 sequences were edited using BioEdit sequence editor version 7.0.4 and then uploaded into the Stanford University HIV Drug resistance database (http://hivdb. stanford.edu) to obtain the drug resistance profile. Mutations were generally categorized according to the International AIDS Society-USA recommendations [15]. Separate phylogenetic analyses were performed to predict subtype, recombinants, and to exclude any contaminants. Each patient PR-RTor reverse transcriptase sequence was aligned to curated set of subtype A, B, C, D, G, and circulating recombinant forms HIV-1 sequences from the alignment reference sequence database from Los Alamos HIV sequence database. All HIV-1 nucleotide sequences have been submitted to Genbank and are accessible by searching with the citation of this article.
Statistics
Standard t tests, Pearson product moment correlations, and test for proportions were performed for these studies.
Results
Drug resistance genotyping at the Joint Clinical Research Centre over a 10-year span
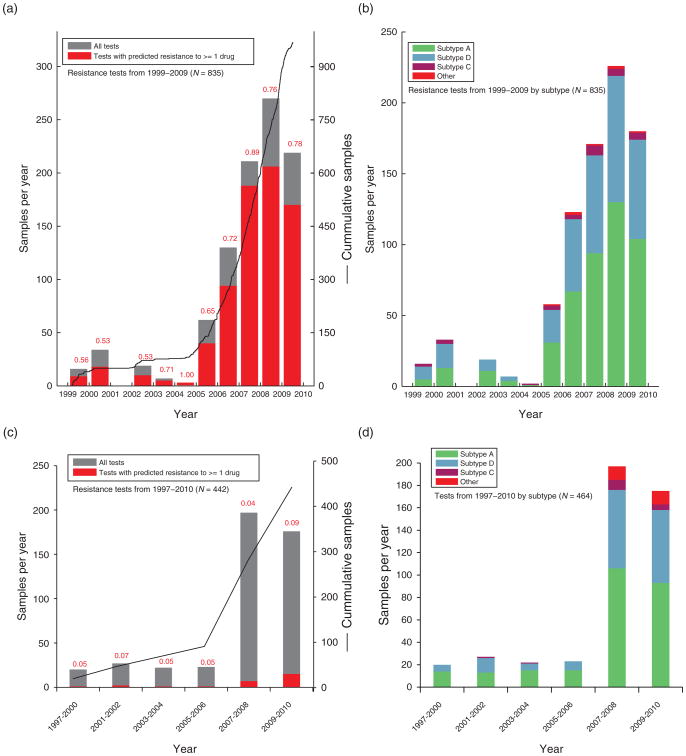
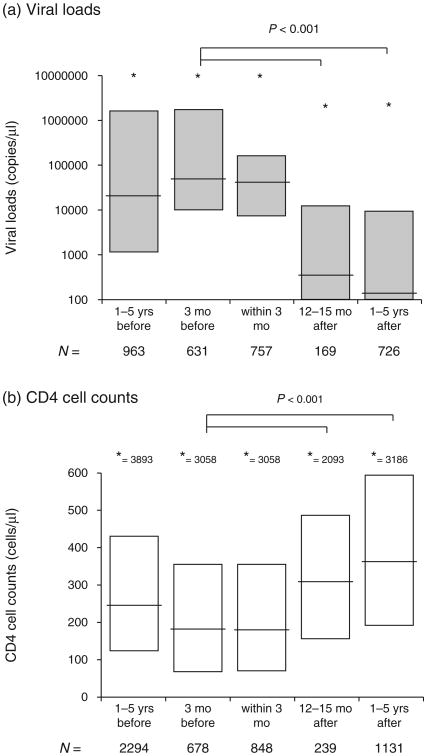
Drug resistance genotyping/testing is requested for those patients receiving antiretroviral treatment and for whom a detectable viral load of more than 2000 copies/ml, CD4 cell count below 250 cells/μl on two consecutive visit, or have decreased more than 200 CD4 cells/μl between visits (Fig. 1). At the time of testing (up to 3 months prior to testing), the median CD4 cell count was 177 cells/μl (n = 678) (25–75% of 67–354 cells/μl) and median viral load was 48 000 copies/ml (n = 678) (10 000–1 750 000) (Fig. 2). The number of drug resistance tests done over a 10-year period is shown in Fig. 1a. Prior to 2004, most of the patients receiving antiretroviral drugs were paying for their medications as well as their treatment monitoring assays. Due to the very high costs of antiretroviral treatment, the cumulative numbers of people receiving treatment was less than 5000 by 2003. Hence, the number of drug resistance tests was much lower prior to 2004. With limited drug supplies and high cost of drugs, poor adherence led to high frequency of treatment failures [10]. With the roll out of antiretroviral treatment by the PEPFAR program in 2004 at the JCRC, the number of patients receiving HAART increased to over 10 000 by 2005 in just Kampala and adherence to treatment improved dramatically with treatment retention rates more than 97%. In the JCRC clinics across Uganda, over 60 000 patients were on HAART by 2007 with an estimated 50% of the HIV-infected Ugandans who required HAART based on the WHO treatment guidelines at the time (i.e., CD4 cell count less than 250 cells/μl).
Fig. 1. Summary of drug resistance genotype testing performed on treatment-naive and treatment-experienced HIV-infected patients at the Joint Clinical Research Centre (JCRC), Kampala, Uganda over a 10-year period.
The number of drug resistance genotypes (DRGs) performed on samples from treatment failures (a and b) and treatment-naive patients (c and d) over the past 10 years are presented as a percentage with at least one primary drug-resistant mutation (a and c) or based on the infecting HIV-1 subtype in the sample (b and d).
Fig. 2. CD4 cell count and viral loads before and after drug resistance genotyping in Joint Clinical Research Centre (JCRC) patients.
Viral loads (a) and CD4 cell count (b) were measured 1–5 year and 3 months in patients prior to obtaining a drug resistance genotype (DRG). These analyses were also performed within 3 months of the DRG or 12–15 months and 1–5 years following the DRG. Only one CD4 or viral load measurement per patient (with DRG) was factored into the 3 month and 12–15 month analyses. The 1–5 year analyses of CD4 cell count and viral loads before or after the DRG involved several values per patient when available. In (a) *refers to the highest outlying viral load that is scaled by the Y axis. In (b) the highest CD4 cell count is provided as a number, e.g. ‘* = 3893’. yrs, years; mo, months.
The numbers of antiretroviral resistance tests performed by the CFAR laboratory were approximately three-fold higher from 2001 to 2004 and two-fold higher from 2004 to the end of 2009, which again relates to more than 2000 drug resistance tests but only 939 with complete clinical paramaters/demographics. A reduction in PEPFAR funding in 2009 at the JCRC clinics reduced the requests for drug resistance testing. It was difficult to ascertain the impact of DRG on subsequent treatment outcomes because we did not compare with treatment outcomes following failures in which DRG tests were not performed. However, following treatment failure, a DRG test, and a change in treatment regimen, there was significant improvements with a lower median viral load (349 copies/ml) and a higher median CD4 cell count (311 cells/μl) at 12–18 months as compared to the clinical values prior to the DRG test (48 800 copies/ml and 177 cells/μl, respectively) (all P < 0.001; Fig. 2). Virus suppression was maintained even 1–5 years following the DRG test and a change in treatment regimen.
Of the 939 DRGs performed over the past 10 years, 754 or 80% had at least one primary drug resistant mutation. Tests prior to 2005 reflect initiation of treatment with self-paying patients and prior to the PEPFAR rollout of HAART in 2004. During this time, nonstructured treatment interruptions were the norm with many patients. Also, many DRGs prior to 2005 were performed on samples from patients who had stopped a treatment regimen due to cost and were then advised to obtain a DRG prior to starting a new treatment regimen (now available at a lower cost). With these DRGs, primary drug-resistant mutations had reverted to wild type in the intrapatient virus population and was not be detected by our DRG. Thus, less than 60% of these patient samples had one or more primary drug resistance mutations prior to 2005.
Over this 10-year period, we performed DRG and subtyping on 269 treatment-naive patients attending the JCRC Clinic in Kampala as well as an additional 188 patients as part of the PASAR study [24–26] (Fig. 1c and d). In these drug-naive cohorts, 5–9% of these patients harbored a virus with one or more primary drug-resistant mutation(s). There appears to be a slight increase in drug resistance within the treatment-naive population after 2008 but this is subject to further study. Supplementary Fig. 1a and b, http://links.lww.com/QAD/A334 shows the neighbor joining phylogenetic trees from 252 and 456 HIV RT sequences sampled from the treatment-naive and treatment-experienced patients failing therapy, respectively. The pie chart in the middle of Supplementary Fig. 1b, http://links.lww.com/QAD/A334 summarizes the percentages of subtypes from all 939 DRGs performed on patients with treatment failure. Subtype A accounts for ∼55% in both patient groups followed by subtype D and subtype C (<4%). Approximately, 10% are unique intersubtype recombinant forms (URFs) in reverse transcriptase but if the entire genome was sequenced, this may go up to more than 30% [27]. Frequency of subtype D infections decreases and subtype A and C increases from west to east and along the shore of Lake Victoria starting from the Tanzanian to the Kenyan border [28]. The prevalence of subtypes in JCRC Clinics in Kampala has remained stable for over 10 years (Fig. 1). These percentages are consistent with those of other cohorts in Kampala [27].
Preferential emergence of specific drug resistance mutations
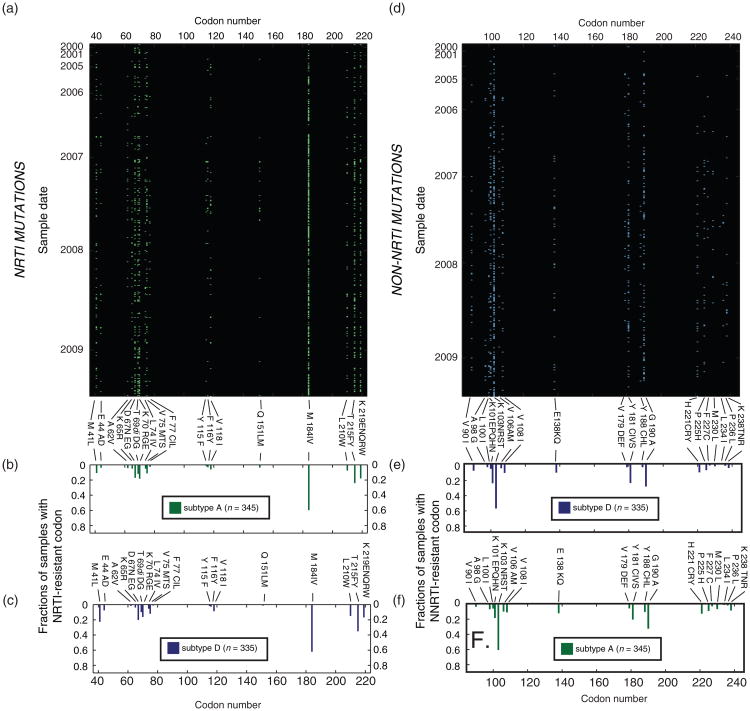
The most common NRTI and NNRTI resistance mutations from the DRGs were M184V and K103N (Fig. 3a and b) due in part to the predominance of lamivudine and nevirapine in treatment regimens. Thymidine analog-associated resistance mutations were also observed at high frequency. Each dot in Fig. 3a and b represents detection of a primary drug-resistant mutation in a DRG. All of the mutations were found at similar frequencies in DRGs performed on subtype A and subtype D-infected patients failing an antiretroviral regimen (upper and lower bar graphs, Fig. 3a and b).
Fig. 3. Frequency of drug resistance mutations in subtype A and D HIV-1 failing antiretroviral treatment from 1998 to 2009.
The frequency of drug resistance mutations per subtype A (a, b, and f) or subtype D samples (c, d, and e) was examined for this treatment failure cohort. The mutations in reverse transcriptase to nucleoside reverse transcriptase inhibitors (NRTI) in each subtype A-infected patient is shown by single green dot across the reverse transcriptase (a). The frequency of each NRTI mutations within the subtype A population is described in (b) and for subtype D in (c). For the NNRTI resistance mutations, the individual mutations per DRG test for each year is shown in (d) for subtype D infections. The frequency of each NNRTI mutation within the subtype D population is described in (e) and for subtype A in (f).
Impact of HIV-1 subtype on emergence of drug resistance
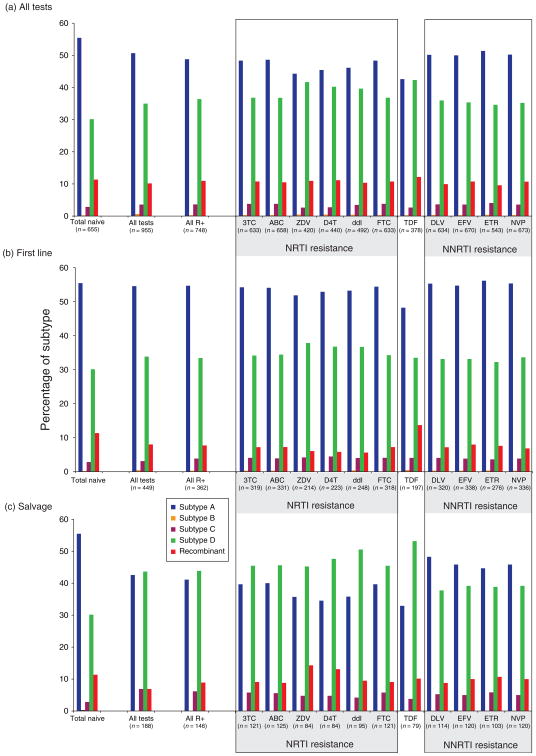
As described above, we combined the subtyping analyses on our treatment-naive cohorts (n = 464) [29–31] with that from other cohorts in Kampala, Uganda [27]. From these analyses (n = 655), the prevalence of sub-types and recombinant forms were quite consistent over 10 years of study with subtype A infections at 55.5%, B – 0.2%, C – 2.8%, D – 30.1%, and URFs – 11.4% (first bar set, Fig. 4). We then analyzed the subtypes of our cross-sectional cohort of patients receiving DRGs and with evidence of treatment failure. The percentage of subtype A dropped with a concomitant increase in subtype D in both patients requiring a DRG (second bar set, Fig. 4a) and in the subset that harbored at least one primary drug resistance mutation (third bar set, Fig. 4a). These findings might suggest that the emergence of anti-retroviral resistance was more common in subtype D than with A infections. A slightly higher proportion of subtype D versus subtype A was observed in antiretroviral-treated patients with antiretroviral resistance than in the treatment-naive population (P = 0.017, Table 1). In addition, increased treatment failures (and drug resistance) with subtype D versus subtype A infections appeared to be associated with NRTI treatment and the appearance of NRTI-resistant mutations (P values = 0.002–0.0147), and not NNRTI treatment/resistance mutations (P > 0.04). We can infer the percentages of infecting subtypes in this antiretroviral-treated population based on those HIV-1 subtypes observed in the treatment-naive JCRC cohort (Figs. 1 and 2).
Fig. 4. Comparing the distribution in HIV-1 subtypes in the treatment-naive population and in patients failing antiretroviral treatment.
The percentage of subtypes A, B, C, D, and unique recombinant forms in the treatment naive population is shown as the first set of bars in panels a, b, and c. Percentage of subtypes in all DRG tests performed on reverse transcriptase coding region are in the second set of bars followed by the subtype percentage in only those samples harboring primary drug resistance mutations. The next set of bars describes the subtype percentages in those samples harboring primary resistance mutations to a specific nucleoside reverse transcriptase inhibitor (NRTI), tenofovir (TDF), and nonnucleoside reverse transcriptase inhibitor (NNRTI). Panel (a) describes the subtype distributions for all patient samples failing any antiretroviral treatment regimen, (b) only those failing first line HAART (one NNRTI + one cytidine analog + one thymidine analog), and (c) only those failing a second line or subsequent salvage regimens. ABC, abacavir; D4T, stavudine; ddI, didanosine; DLV, delaviridine; EFV, efavirenz; ETR, etravirine; FTC, emtricitabine; NVP, nevirapine; TDF, tenofovir; ZDV, zidovudine; 3TC, lamivudine.
Table 1. Statistical analyses of proportions of subtype A and D in the treatment-naive and antiretroviral-resistant populations in Kampala, Uganda.
| Population of tested individuals | Conditions | Subtype A | Subtype D | P-value |
|---|---|---|---|---|
| Treatment naive (2000–2010) | 254 | 138 | ||
| All treatments (2000–2010) | All tests | 484 | 334 | 0.0604 |
| All test with resistance | 365 | 272 | 0.0171 | |
| All tests with resistance to: | ||||
| __NRTIs/NtRTIs | ||||
| ____3TC | 306 | 233 | 0.0136 | |
| ____ABC | 320 | 242 | 0.0147 | |
| ____ZDV | 186 | 175 | 0.0002 | |
| ____D4t | 200 | 177 | 0.0009 | |
| ____ddI | 227 | 195 | 0.0014 | |
| ____FTC | 306 | 233 | 0.0136 | |
| ____TDF | 161 | 160 | 0.0001 | |
| __NNRTIs | ||||
| ____DLV | 318 | 228 | 0.0424 | |
| ____EFV | 335 | 237 | 0.0513 | |
| ____ETR | 279 | 188 | 0.1285 | |
| ____NVP | 338 | 237 | 0.0595 | |
| First-line treatment (2000–2010) | All tests | 245 | 152 | 0.3691 |
| All test with resistance | 198 | 121 | 0.4523 | |
| All tests with resistance to: | ||||
| __NRTIs/NtRTIs | ||||
| ____3TC | 173 | 109 | 0.3594 | |
| ____ABC | 179 | 114 | 0.3200 | |
| ____ZDV | 111 | 81 | 0.1015 | |
| ____D4t | 118 | 82 | 0.1675 | |
| ____ddI | 132 | 91 | 0.1670 | |
| ____FTC | 173 | 109 | 0.3594 | |
| ____TDF | 95 | 66 | 0.1999 | |
| __NNRTIs | ||||
| ____DLV | 177 | 106 | 0.5479 | |
| ____EFV | 185 | 112 | 0.4980 | |
| ____ETR | 155 | 89 | 0.7449 | |
| ____NVP | 186 | 113 | 0.4833 | |
| Salvage therapy (2000–2010) | All tests | 80 | 82 | 0.0007 |
| All test with resistance | 60 | 64 | 0.0011 | |
| All tests with resistance to: | ||||
| __NRTIs/NtRTIs | ||||
| ____3TC | 48 | 55 | 0.0008 | |
| ____ABC | 50 | 57 | 0.0007 | |
| ____ZDV | 30 | 38 | 0.0012 | |
| ____D4t | 29 | 40 | 0.0003 | |
| ____ddI | 34 | 48 | 0.0001 | |
| ____FTC | 48 | 55 | 0.0008 | |
| ____TDF | 26 | 42 | 0.0000 | |
| __NNRTIs | ||||
| ____DLV | 55 | 43 | 0.1116 | |
| ____EFV | 55 | 47 | 0.0432 | |
| ____ETR | 46 | 40 | 0.0495 | |
| ____NVP | 55 | 47 | 0.0432 |
ABC, abacavir; D4T, stavudine; ddI, didanosine; DLV, delaviridine; EFV, efavirenz; ETR, etravirine; FTC, emtricitabine; NNRTI, nonnucleoside reverse transcriptase inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; NVP, nevirapine; TDF, tenofovir; ZDV, zidovudine; 3TC, lamivudine.
Interestingly, there is only a slight trend for increased treatment failures in subtype D versus subtype A infections during first line HAART using a cytidine analog + thymidine analog + NNRTI (Fig. 4b). The most significant skewing of subtypes was observed in those patients failing a second line or subsequent salvage HAART (Fig. 4c). With salvage treatment, increased emergence of drug resistance with subtype D was highly significant (P = 0.0007) but this was largely attributable to NRTI (P < 0.0012) rather than NNRTI selection pressure (P > 0.04). Finally, we examined the impact of protease inhibitor selective pressure on emergence of drug resistance among sub-type D and A patients but the results were inconclusive as only 14 out of 150 patients developed protease inhibitor resistance.
Discussion
Since 1999, DRG testing has been provided as a standard-of-care assay for patients receiving HAART at the JCRC. Due to the high costs of the antiretroviral drugs, demands for this assay were minimal until the rollout of HAART with PEPFAR in 2004. Treatment failures were obviously more prevalent in late 1990 s with a two NNRTI regimen than with HAART [10]. The majority of treatment failure samples harbored antiretroviral-resistant virus (>70%). The slightly lower level of drug resistance prior to 2005 relates to the number of samples collected from patients that initiated sub-optimal treatment, had interrupted or stopped therapy (due primarily to cost), and/or were re-initiating treatment. Thus, virus in these samples may have reverted back to wild type. Since the vast majority of patients were treated with an NNRTI, a cytidine analog, and a thymidine analog, it was not surprising to observe a high prevalence of the ‘classical’ primary drug resistance mutations such as M184V, K103N, Y181C, and so on. The prevalence of these mutations was not affected by subtype.
Treatment failure and drug resistance were more frequent in patients infected with subtype D than those infected withsubtypeA,C,orrecombinantforms.Toclarify,itis important to place these observations into context. We determined HIV-1 subtypes of 464 treatment-naive patients prior to initiating a HAART regimen [29–31] with an additional 191 samples derived from other studies [10,27]. Subtype A was 55% while subtype D was 30%. In the 939 treatment failures compared with 655 treatment-naive patients, there was a significant shift in the frequency of subtype D infections (50 versus 55%) and of subtype A infections (34 versus 30%) but no significant difference for subtype B, C, or recombinants. This shift to a higher frequency of failures among subtype D-infected patients was only a trend with first-line HAART and was highly significant with salvage treatment. With the failures to second-line or subsequent salvage treatment, subtype frequency shift was quite evident with subtype D (42% salvage failure versus 55% naive) and subtype A infections (43 versus 34%), this subtype shift was also observed with primary resistance in the naive cohorts.
It is unclear why treatment failures and resistance may be more frequent in subtype D versus subtype A-infected individuals in Uganda. Previous studies have reported faster disease progression in subtype D versus subtype A-infected patients in natural history cohorts [32,33]. Using dual virus competitions in primary human T cells and macrophages, subtype D has slightly greater replicative fitness than subtype A HIV-1 when competed against isolates of the same coreceptor usage [34]. By extension, we and others have also shown that replicative fitness is tightly associated with disease progression [35–38]. The faster replicating CXCR4-using viruses were more common in subtype D as compared to subtype A infections [39]. Faster disease progression, higher prevalence of CXCR4-tropism, and higher replication fitness of HIV-1 subtype D over A are not necessarily direct factors affecting treatment success and the emergence of resistance but this association with pathogenesis and response to treatment should be explored. Any treatment interruption, adverse event, or even poor drug penetration into specific tissues may promote residual virus replication. Ultimately, the virus with faster replication kinetics and higher turnover (e.g. subtype D) would lead to accelerated evolution and may result in a higher probability to achieve drug resistance.
Based on stable subtype distributions in the treatment naive population and the slightly higher frequency of treatment failure with subtype D infections, we would require a cohort size of 2000 treatment failures in the first year from approximately 30 000 treatment naive individuals starting HAART to observe significant shift in subtype distribution in a first-line treatment study. Thus, we have initiated meta-analyses with approximately 3000 DRGs available from various Ugandan treatment studies. However, it is important to note that some of these studies are based in southwest Uganda (e.g. Rakai district) where subtype D prevalence in the treatment-naive population is higher than in Kampala [28]. We have attempted to examine the rate of treatment failure in different subtype by enrolling 1600 treatment naive JCRC patients into several clinical studies sponsored by different funding agencies but with similar end-point analyses [10,24,40–42]. Treatment failure was typically less than 15% during the first 2 years of follow-up during first-line HAARTand of these failures, ∼70% harbor drug-resistant viruses. For example, one of these studies enrolled 188 treatment-naive patients at the JCRC [24]. The proportion of subtypes was similar as in other JCRC cohorts over the past 10 years (Figs 1 and 2; Supplementary Fig. 1, http://links.lww.com/QAD/A334). Of 25 treatment failures in the first year, only 18 harbored DRGs and there was no obvious or statistically significant difference in subtype distribution [24]. Statistical significance for increased treatment failures in subtype D over subtype A may be achieved with extending the study follow-up period. With our meta-analyses of our longitudinal cohort with ∼1600 patients, follow-up is generally 2–3 years with first-line treatment failures reaching more than 20% at the JCRC.
Based on the shift in subtype D and subtype A frequencies in the treatment failure versus treatment-naive populations, we estimate a hazard ratio of 1.28 for treatment failure comparing subtype D versus a subtype A-infected individual. The risk of treatment failure in subtype D versus subtype A-infected patients appears different based on the type of treatment regimen. In general, higher levels of NRTI resistance over NNRTI resistance in treatment failure appeared necessary to shift proportions of subtype D over subtype A. For unknown reasons, resistance to tenofovir, stavudine, and didanosine appear to be more prominent in subtype D than in subtype A during treatment failure. These findings appear complex but may be related to the residual antiviral activities and/or replicative fitness of specific DRGs. The higher intrinsic fitness of subtype D over subtype A [34] may help compensate for loss in fitness associated with drug resistance mutations. K103N and Y181C in HIV-1 confer high-level primary resistance to efavirenz, and nevirapine, respectively, but have minimal cost on replicative fitness [43]. Resistance to other NRTI are known to emerge more slowly, confer more moderate levels of drug resistance, and are associated with higher fitness costs. The exception would be the lamivudine/emtricitabine treatment and emergence of M184V in which M184V confers a high level of resistance (>1000-fold) but also high replicative fitness costs [44]. Of all the NRTIs, resistance to lamivudine and emtricitabine in treatment failures is associated with the lowest shift in subtype distribution from the treatment-naive cohort.
Conclusion
These findings would suggest that replication kinetics and relative compensation for drug resistance mutations within specific HIV-1 backbone sequence could impact treatment outcome. Of great significance for HIV treatment in Africa, specific HIV-1 subtypes such as subtype D may have higher intrinsic fitness, lead to faster disease progression, may have higher rates of treatment failures, thus, infections with subtype D may require more stringent treatment monitoring.
Acknowledgments
F.K., I.N., H.N., E.N., and L.B. performed all the DRGs and experimentation in this article. S.M., J.A. and D.T. performed all the data analyses. T.I. and D.T. performed all the phylogenetic analyses. B.R., F.S., M.S., A.B., C.K., and P.M. recruited all of the patients for this study and analyzed the clinical data. C.K., P.M., R.S., and E.A. had the concept, procured the funding, and directed the overall aspects of this research study. E.A. was the principal investigator of the entire study, directed the clinical laboratory in Uganda, and supervised the overall direction of this research.
JCRC Drug Resistance Working group includes: Leonard Bagenda (Center for AIDS Research Uganda Laboratories, Joint Clinical Research Centre), Hannah Nanyonjo (Center for AIDS Research Uganda Laboratories, Joint Clinical Research Centre), Taina Immonen (Department of Mathematics, Case Western Reserve University, Cleveland, Ohio, USA), Francis Ssali (TREAT, Joint Clinical Research Centre, Kampala, Uganda), Michael Semanda (TREAT, Joint Clinical Research Centre, Kampala, Uganda), and Aggrey Bukuru (TREAT, Joint Clinical Research Centre, Kampala, Uganda).
We would like to thank the patients attending the JCRC clinics for this anonymized dataset as well as the clinical staff at the JCRC.
The work was supported by National Institutes of Health National Institute of AIDS and Infectious Diseases grant AI025879, AI49170, and AI36219. All experiments and drug resistance genotyping was performed in the Uganda Laboratory Core at the Joint Clinical Research Centre (JCRC), Kampala, Uganda and supported by the CWRU/UH Center for AIDS Research (AI36219).
Footnotes
Conflicts of interest: None of the authors have a conflict of interest with this study.
References
- 1.Mermin J, Were W, Ekwaru JP, Moore D, Downing R, Behumbiize P, et al. Mortality in HIV-infected Ugandan adults receiving antiretroviral treatment and survival of their HIV-uninfected children: a prospective cohort study. Lancet. 2008;371:752–759. doi: 10.1016/S0140-6736(08)60345-1. [DOI] [PubMed] [Google Scholar]
- 2.Campbell TB, Smeaton LM, Kumarasamy N, Flanigan T, Klingman KL, Firnhaber C, et al. Efficacy and safety of three antiretroviral regimens for initial treatment of HIV-1: a randomized clinical trial in diverse multinational settings. PLoS Med. 2012;9:e1001290. doi: 10.1371/journal.pmed.1001290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ojikutu B, Makadzange AT, Gaolathe T. Scaling up ART treatment capacity: lessons learned from South Africa, Zimbabwe, and Botswana. Curr Infect Dis Rep. 2008;10:69–73. doi: 10.1007/s11908-008-0012-0. [DOI] [PubMed] [Google Scholar]
- 4.Taegtmeyer M, Chebet K. Overcoming challenges to the implementation of antiretroviral therapy in Kenya. Lancet Infect Dis. 2002;2:51–53. doi: 10.1016/s1473-3099(01)00173-6. [DOI] [PubMed] [Google Scholar]
- 5.Wood R, Lawn SD. Should the CD4 threshold for starting ART be raised? Lancet. 2009;373:1314–1316. doi: 10.1016/S0140-6736(09)60654-1. [DOI] [PubMed] [Google Scholar]
- 6.Crowley S, Rollins N, Shaffer N, Guerma T, Vitoria M, Lo YR. New WHO HIV treatment and prevention guidelines. Lancet. 2009;375:874–875. doi: 10.1016/S0140-6736(09)62064-X. [DOI] [PubMed] [Google Scholar]
- 7.Gupta RK, Hill A, Sawyer AW, Cozzi-Lepri A, von Wyl V, Yerly S, et al. Virological monitoring and resistance to first-line highly active antiretroviral therapy in adults infected with HIV-1 treated under WHO guidelines: a systematic review and meta-analysis. Lancet Infect Dis. 2009;9:409–417. doi: 10.1016/S1473-3099(09)70136-7. [DOI] [PubMed] [Google Scholar]
- 8.Simcock M, Sendi P, Ledergerber B, Keller T, Schupbach J, Battegay M, et al. A longitudinal analysis of healthcare costs after treatment optimization following genotypic antiretroviral resistance testing: does resistance testing pay off? Antivir Ther. 2006;11:305–314. [PubMed] [Google Scholar]
- 9.Hamers RL, Derdelinckx I, van VM, Stevens W, Rinke de Wit TF, Schuurman R. The status of HIV-1 resistance to antiretroviral drugs in sub-Saharan Africa. Antivir Ther. 2008;13:625–639. [PubMed] [Google Scholar]
- 10.Richard N, Juntilla M, Abraha A, Demers K, Paxinos E, Galovich J, et al. High prevalence of antiretroviral resistance in treated Ugandans infected with nonsubtype B human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 2004;20:355–364. doi: 10.1089/088922204323048104. [DOI] [PubMed] [Google Scholar]
- 11.Diaz RS, Sucupira MC, Vergara TR, Brites C, Bianco RD, Bonasser FF, et al. HIV-1 resistance testing influences treatment decision-making. Braz J Infect Dis. 2010;14:489–494. [PubMed] [Google Scholar]
- 12.Badolato R, Schumacher RF, Rodella E, Gargiulo F, Torti C, Notarangelo LD, et al. Genotyping for guiding drug choice in human immunodeficiency virus-infected children failing multiple antiretroviral treatment regimens. Pediatr Infect Dis J. 2005;24:747–749. doi: 10.1097/01.inf.0000172910.79381.64. [DOI] [PubMed] [Google Scholar]
- 13.Arts EJ, Hazuda DJ. HIV-1 antiretroviral drug therapy. Cold Spring Harb Perspect Med. 2012;2:a007161. doi: 10.1101/cshperspect.a007161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mansky LM. HIV mutagenesis and the evolution of antiretroviral drug resistance. Drug Resist Updat. 2002;5:219–223. doi: 10.1016/s1368-7646(02)00118-8. [DOI] [PubMed] [Google Scholar]
- 15.Johnson VA, Brun-Vezinet F, Clotet B, Gunthard HF, Kuritzkes DR, Pillay D, et al. Update of the drug resistance mutations in HIV-1: December 2010. Top HIV Med. 2010;18:156–163. [PubMed] [Google Scholar]
- 16.Larder BA, Kemp SD. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT) Science. 1989;246:1155–1158. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- 17.Kantor R, Katzenstein DA, Efron B, Carvalho AP, Wynhoven B, Cane P, et al. Impact of HIV-1 subtype and antiretroviral therapy on protease and reverse transcriptase genotype: results of a global collaboration. PLoS Med. 2005;2:e112. doi: 10.1371/journal.pmed.0020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao Y, Paxinos E, Galovich J, Troyer R, Baird H, Abreha M, et al. Characterization of a subtype D human immunodeficiency virus type 1 isolate that was obtained from an untreated individual and that is highly resistant to nonnucleoside reverse transcriptase inhibitors. J Virol. 2004;78:5390–5401. doi: 10.1128/JVI.78.10.5390-5401.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wainberg MA, Brenner BG. The impact of HIV genetic polymorphisms and subtype differences on the occurrence of resistance to antiretroviral drugs. Mol Biol Int. 2012;2012:256982. doi: 10.1155/2012/256982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bocket L, Cheret A, Deuffic-Burban S, Choisy P, Gerard Y, de lT X, et al. Impact of human immunodeficiency virus type 1 subtype on first-line antiretroviral therapy effectiveness. Antivir Ther. 2005;10:247–254. [PubMed] [Google Scholar]
- 21.Geretti AM, Harrison L, Green H, Sabin C, Hill T, Fearnhill E, et al. Effect of HIV-1 subtype on virologic and immunologic response to starting highly active antiretroviral therapy. Clin Infect Dis. 2009;48:1296–1305. doi: 10.1086/598502. [DOI] [PubMed] [Google Scholar]
- 22.Easterbrook PJ, Smith M, Mullen J, O'Shea S, Chrystie I, de Ruiter RA, et al. Impact of HIV-1 viral subtype on disease progression and response to antiretroviral therapy. J Int AIDS Soc. 2010;13:4. doi: 10.1186/1758-2652-13-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soares EA, Santos AF, Sousa TM, Sprinz E, Martinez AM, Silveira J, et al. Differential drug resistance acquisition in HIV-1 of subtypes B and C. PLoS One. 2007;2:e730. doi: 10.1371/journal.pone.0000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamers RL, Sigaloff KC, Wensing AM, Wallis CL, Kityo C, Siwale M, et al. Patterns of HIV-1 drug resistance after first-line antiretroviral therapy (ART) failure in 6 sub-Saharan African countries: implications for second-line ART strategies. Clin Infect Dis. 2012;54:1660–1669. doi: 10.1093/cid/cis254. [DOI] [PubMed] [Google Scholar]
- 25.Hamers RL, Sigaloff KC, Kityo C, Mugyenyi P, de Wit TF. HIV-1 drug resistance in antiretroviral-naive patients in sub-Saharan Africa. Lancet Infect Dis. 2013;13:196–197. doi: 10.1016/S1473-3099(13)70012-4. [DOI] [PubMed] [Google Scholar]
- 26.Sigaloff KC, Hamers RL, Wallis CL, Kityo C, Siwale M, Ive P, et al. Second-line antiretroviral treatment successfully resup-presses drug-resistant HIV-1 after first-line failure: prospective cohort in Sub-Saharan Africa. J Infect Dis. 2012;205:1739–1744. doi: 10.1093/infdis/jis261. [DOI] [PubMed] [Google Scholar]
- 27.Ssemwanga D, Ndembi N, Lyagoba F, Bukenya J, Seeley J, Vandepitte J, et al. HIV type 1 subtype distribution, multiple infections, sexual networks, and partnership histories in female sex workers in Kampala, Uganda. AIDS Res Hum Retroviruses. 2012;28:357–365. doi: 10.1089/aid.2011.0024. [DOI] [PubMed] [Google Scholar]
- 28.Kaleebu P, Ross A, Morgan D, Yirrell D, Oram J, Rutebemberwa A, et al. Relationship between HIV-1 Env subtypes A and D and disease progression in a rural Ugandan cohort. AIDS. 2001;15:293–299. doi: 10.1097/00002030-200102160-00001. [DOI] [PubMed] [Google Scholar]
- 29.Richard N, Juntilla M, Abraha A, Demers K, Paxinos E, Galovich J, et al. High prevalence of antiretroviral resistance in treated Ugandans infected with nonsubtype B human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 2004;20:355–364. doi: 10.1089/088922204323048104. [DOI] [PubMed] [Google Scholar]
- 30.Troyer RM, Lalonde MS, Fraundorf E, Demers KR, Kyeyune F, Mugyenyi P, et al. A radiolabeled oligonucleotide ligation assay demonstrates the high frequency of nevirapine resistance mutations in HIV type 1 quasispecies of NVP-treated and untreated mother-infant pairs from Uganda. AIDS Res Hum Retroviruses. 2008;24:235–250. doi: 10.1089/aid.2007.0138. [DOI] [PubMed] [Google Scholar]
- 31.Hamers RL, Wallis CL, Kityo C, Siwale M, Mandaliya K, Conradie F, et al. HIV-1 drug resistance in antiretroviral-naive individuals in sub-Saharan Africa after rollout of antiretroviral therapy: a multicentre observational study. Lancet Infect Dis. 2011;11:750–759. doi: 10.1016/S1473-3099(11)70149-9. [DOI] [PubMed] [Google Scholar]
- 32.Kaleebu P, French N, Mahe C, Yirrell D, Watera C, Lyagoba F, et al. Effect of human immunodeficiency virus (HIV) type 1 envelope subtypes A and D on disease progression in a large cohort of HIV-1-positive persons in Uganda. J Infect Dis. 2002;185:1244–1250. doi: 10.1086/340130. [DOI] [PubMed] [Google Scholar]
- 33.Baeten JM, Chohan B, Lavreys L, Chohan V, McClelland RS, Certain L, et al. HIV-1 subtype D infection is associated with faster disease progression than subtype A in spite of similar plasma HIV-1 loads. J Infect Dis. 2007;195:1177–1180. doi: 10.1086/512682. [DOI] [PubMed] [Google Scholar]
- 34.Abraha A, Nankya IL, Gibson R, Demers K, Tebit DM, Johnston E, et al. CCR5- and CXCR4-tropic subtype C human immunodeficiency virus type 1 isolates have a lower level of pathogenic fitness than other dominant group M sub-types: implications for the epidemic. J Virol. 2009;83:5592–5605. doi: 10.1128/JVI.02051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quinones-Mateu ME, Ball SC, Marozsan AJ, Torre VS, Albright JL, Vanham G, et al. A dual infection/competition assay shows a correlation between ex vivo human immunodeficiency virus type 1 fitness and disease progression. J Virol. 2000;74:9222–9233. doi: 10.1128/jvi.74.19.9222-9233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Troyer RM, Collins KR, Abraha A, Fraundorf E, Moore DM, Krizan RW, et al. Changes in human immunodeficiency virus type 1 fitness and genetic diversity during disease progression. J Virol. 2005;79:9006–9018. doi: 10.1128/JVI.79.14.9006-9018.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blaak H, van't Wout AB, Brouwer M, Hooibrink B, Hovenkamp E, Schuitemaker H. In vivo HIV-1 infection of CD45RA(R) CD4(R) T cells is established primarily by syncytium-inducing variants and correlates with the rate of CD4(R) T cell decline. Proc Natl Acad Sci U S A. 2000;97:1269–1274. doi: 10.1073/pnas.97.3.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lassen KG, Lobritz MA, Bailey JR, Johnston S, Nguyen S, Lee B, et al. Elite suppressor-derived HIV-1 envelope glycoproteins exhibit reduced entry efficiency and kinetics. PLoS Pathog. 2009;5:e1000377. doi: 10.1371/journal.ppat.1000377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaleebu P, Nankya IL, Yirrell DL, Shafer LA, Kyosiimire-Lugemwa J, Lule DB, et al. Relation between chemokine receptor use, disease stage, and HIV-1 subtypes A and D: results from a rural Ugandan cohort. J Acquir Immune Defic Syndr. 2007;45:28–33. doi: 10.1097/QAI.0b013e3180385aa0. [DOI] [PubMed] [Google Scholar]
- 40.Reynolds SJ, Sendagire H, Newell K, Castelnuovo B, Nankya I, Kamya M, et al. Virologic versus immunologic monitoring and the rate of accumulated genotypic resistance to first-line anti-retroviral drugs in Uganda. BMC Infect Dis. 2012;12:381. doi: 10.1186/1471-2334-12-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mugyenyi P, Walker AS, Hakim J, Munderi P, Gibb DM, Kityo C, et al. Routine versus clinically driven laboratory monitoring of HIV antiretroviral therapy in Africa (DART): a randomised noninferiority trial. Lancet. 2010;375:123–131. doi: 10.1016/S0140-6736(09)62067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lyagoba F, Dunn DT, Pillay D, Kityo C, Robertson V, Tugume S, et al. Evolution of drug resistance during 48 weeks of zidovudine/lamivudine/tenofovir in the absence of real-time viral load monitoring. J Acquir Immune Defic Syndr. 2010;55:277–283. doi: 10.1097/QAI.0b013e3181ea0df8. [DOI] [PubMed] [Google Scholar]
- 43.Iglesias-Ussel MD, Casado C, Yuste E, Olivares I, Lopez-Galindez C. In vitro analysis of human immunodeficiency virus type 1 resistance to nevirapine and fitness determination of resistant variants. J Gen Virol. 2002;83(Pt 1):93–101. doi: 10.1099/0022-1317-83-1-93. [DOI] [PubMed] [Google Scholar]
- 44.Wainberg MA, Drosopoulos WC, Salomon H, Hsu M, Borkow G, Parniak MA, et al. Enhanced fidelity of 3TC selected mutant HIV-1 reverse transcriptase. Science. 1996;271:1282–1285. doi: 10.1126/science.271.5253.1282. [DOI] [PubMed] [Google Scholar]