Abstract
The fluorescent protein Dronpa undergoes reversible photoswitching reactions between the bright ‘on’ and dark ‘off’ states via photoisomerisation and proton transfer reactions. We report the room temperature crystal structure of the fast switching Met159Thr mutant of Dronpa at 2.0 Å resolution in the bright on state. Structural differences with the wild type include shifted backbone positions of strand β8 containing Thr159 as well as an altered A-C dimer interface involving strands β7, β8, β10, and β11. The Met159Thr mutation increases the cavity volume for the p-hydroxybenzylidene-imidazolinone chromophore as a result of both the side chain difference and the backbone positional differences.
Keywords: Dronpa, reversibly photoswitchable fluorescent protein, X-ray structure, room temperature, molecular dynamics
Introduction
Photochromic fluorescent proteins have important applications in super-resolution fluorescence microscopy. Particularly, the class of Reversibly Photoswitchable Fluorescent Proteins (RPFP's) allow repeated cycles of forward and reverse photoconversion 1-3,4,5. The reversibly photoswitchable fluorescent protein Dronpa is a genetically modified form of the ‘22G’ wild type isolated from the coral Pectiniidae 6. The reversible photoswitching reactions of Dronpa involve cis-trans/trans-cis photoisomerisation as well as thermal proton transfer reactions 7,8. While the ‘22G’ wild type form is oligomeric, random and directed mutations created a monomeric form, ‘22Gm3’, designated ‘Dronpa’. Six mutations were introduced : I101N (β5), F114Y (β6), L162S (β8), R194H (β10), N205S (loop β10-β11) and G218E (β11-loop) 9. Subsequent crystallization of the monomeric Dronpa revealed a tetrameric arrangement, seen in P212121, P21212 and P21 crystal forms, which rationalized the effects of the mutations introduced in Dronpa on the dissociation constant and prevalence of monomeric form in dilute solution 10. Stiel et al 11 and Wilmann et al 12 reported similar crystal structures for the on state of Dronpa. Solution NMR measurements identified chemical shift differences between the on and off states for seven residues: Gly36 (β3), Cys62(β3), Met93(central helix), Ala160(β8), Cys171(β9), Asp172(β9), Phe173(β9) from 1H-15N heteronuclear single-quantum coherence (HSQC) spectra 10. In addition, exchange broadening was seen in the off state for a number of residues: 62 (central helix), 65-69 (central helix), 90-91(β4), 132, 140, 142-145(β7), 156-160(β8), 191-195(β10), and 212-214(β11). These results implicate primarily the A-C dimer interface, which undergoes photo-induced changes of both equilibrium geometry as well as changes in the flexibility of the backbone, detected as chemical exchange broadening 10. The on-to-off photoswitching efficiency of Dronpa is relatively low, with an estimated quantum yield of 3.2*10−4 reported for the wild type 6,13. Several fast-switching mutants of Dronpa have been reported. One of these, Dronpa-M159T, was selected in particular for showing an increased quantum yield for on-off photoswitching, but also an improved quantum yield for the off-to-on reaction was reported 11. Specifically, Stiel et al report a 1143-fold increase of the rate of the onoff reaction at room temperature for the M159T mutant, implying an absolute quantum yield of 0.3711, whereas a more recent study of monomeric species in vitro recently reported a ~65 fold increase 14. The off-to-on reaction was found to have increased 2-fold, suggesting a quantum yield of 0.72 relative to the 0.36 value estimated for the wild type 13 whereas a more recent study of monomeric species in vitro recently reported no significant difference 14. Stiel et al argue that the steric repulsion of the Met159 group is the primary reason for the three orders of magnitude acceleration of the M159T mutant. This view appears to be supported when considering the Phe-173-Ser mutant of the tetrameric variant of the EosFP fluorescent protein, named ‘IrisFP’15. This reported mutation indirectly affects the position of the equivalent Met159 side chain, which subsequently allows cis-trans photoisomerisation not seen in the wild type 15. However, oligomerisation also affects the photoswitching efficiencies. The monomeric Dronpa has increased on-to-off rate relative to the ‘22G’ tetrameric parent molecule. In addition, a single point mutation Lys154Asn of Dronpa created a variant which showed a photoconversion rate intermediate between 22G and Dronpa 16. The crystal structure of the Lys154Asn-Dronpa mutant rationalized the effect on the A-C interface in particular 16.
Materials and Methods
1. Protein production and crystallisation
The M159T mutation was introduced into the original expression construct pRESTb-Dronpa, and was expressed in E.coli BL21(DE3), purified by Ni-NTA affinity chromatography and gel filtration chromatography as previously described 8. The extinction coefficient for Dronpa-M159T was taken as 61,732 M−1cm−1 at 489 nm 11. Protein was concentrated to 20 mg/ml and taken up in 50 mM Tris/HCl (pH 7.8) and 120 mM NaCl. Crystals were grown at 22°C in sitting drops with siliconised glass plates (Hampton Research) using 20 % (w/v) PEG (poly[ethylene glycol] monoethyl ether 3350 (Sigma-Aldrich) and 0.14 M Mg(NO3)2. Crystals were mounted in quartz capillaries (Hampton Research).
2. Data collection and processing
Crystals were mounted at room temperature and monochromatic 1° rotation images were collected at the Advanced Photon Source (APS, Argonne, USA), BioCARS, beamline 14 BM-C to 2.0 Å resolution with a 300 mm distance. The wavelength was 0.9787 Å (12.668 keV energy and band pass ΔE/E: 3.1 × 10−4). The detector was ADSC Quantum-315 CCD (315 × 315 mm). The data was indexed and integrated in space group P222 (point group 222) with MOSFLM 17 and scaled and merged in primitive orthorhombic spacegroup P212121 with Scala 18using CCP4 19.
3. Molecular replacement, refinement and validation
The structure was solved by molecular replacement using coordinates from 2Z1O 10, using Molrep 20. The structure was refined with rigid body and restraint refinement using REFMAC5, not applying Non Crystallographic Symmetry restraints 21. Manual refinement and building waters were done using coot 22. The structure was validated using the wwPDB (http://wwpdb-validation.wwpdb.org) server. Structure factors and coordinates with up to 2.04 Å resolution were deposited to the Protein Data Bank (http://www.wwpdb.org) with deposition code 4uts.
4. Molecular dynamics simulations
Molecular Dynamics simulations were performed using Gromacs 23 for the monomer and tetramer structures of the Wild Type Dronpa and the M159T mutant in order to assess the cavity volumes at room temperature. Full Methods are given in the SI.
Results and Discussion
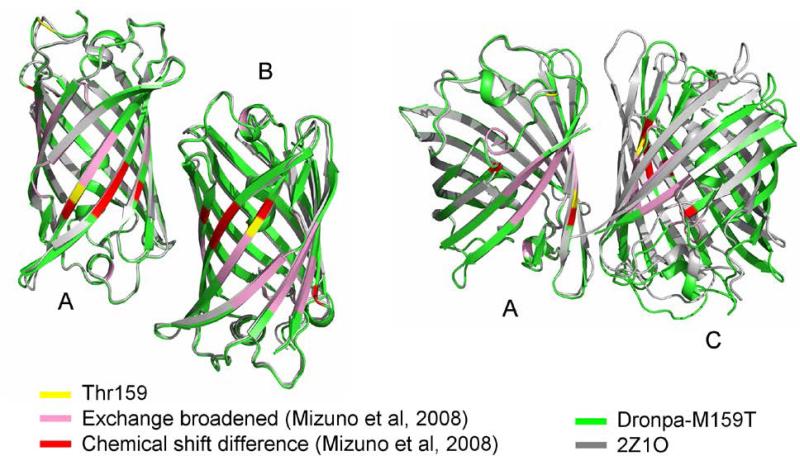
A total number of 195 1° rotation images were collected at room temperature from 4 crystals in 12 wedges and were integrated, merged and scaled together to achieve best completeness and multiplicity (Table 1). Inclusion of selected weaker images which did show radiation damage and had reduced I/σI was acceptable based on R-factor criteria (Table 1). The Laue group probability was 0.958 and a systematic absence probability of 0.893 to give a P212121 Space group with confidence of 0.83. The Dronpa-M159T mutant crystallized under conditions identical to those reported for the wild type Dronpa 11. However, the space group for the reported wild type crystals was orthorhombic body centered I222, and having unit cell dimensions a=75.292 Å, b= 109.627Å, c=275.232Å (2IOV pdb11. The Dronpa-M159T mutant however crystallized in primitive orthorhombic spacegroup P212121 a=75.655 Å, =111.140 Å, c=117.572 Å. Molecular replacement was successful using the coordinates of the on state of the wild type Dronpa including side chains, from coordinates 2Z1O 10. Following rigid body refinement, and cycles of restraint refinement and manual refinement and water building, a tetrameric arrangement of Dronpa-M159T was revealed which does not match previously reported tetrameric arrangements. Table 1 shows the data collection and refinement statistics for the asymmetric unit containing four individually refined chains. It was noted that restraint refinement applying non crystallographic symmetry restraints reduced the Rfree from 21.3% to 20.4% while the Rcryst was almost unaffected at 17.2 % (Table 1). Coordinates submitted to the Protein Data Bank (code 4UTS) were those without NCS restraints, supported by the I/σI value, multiplicity and crystallographic R factors. A comparison with the P212121 wild type structure 2Z1O (unit cell dimensions a=73.492 Å, b=103.516 Å, c=122.866 Å) 10 reveals that the A-B interface is in essence conserved but the A-C interface is significantly altered (Figure 1, Table S1). A structural alignment of the A chains of Dronpa-M159T and Dronpa (2Z1O) resulted in an RMS of 0.251 Å. An analysis of the Debye-Waller factors of the individual chains in the M159T-Dronpa and Dronpa 10 tetramers showed similar distributions for A and D chains, and the B and C chains for both structures (Figure S2). Analysis of the interface surfaces using PISA 24 showed that the buried surface areas are significantly smaller in the M159T mutant (Table S1). Thus, while the M159T has only minimal effects on the structure and fold of the monomer, the interaction between the A and C chains involves fewer interactions and is shifted by ~ 20 Å rotation, or hinging (Figure 1). Figure 1 furthermore locates residues which undergo either chemical shift modification or become exchange broadened in the off state, as reported from HSQC NMR measurements10. This indicates that also in the P212121 Dronpa-M159T structure reported here, the A-C interface concentrates most of the perturbed residues.
Table 1.
Data collection refinement and geometry
| Data Collection | |
|---|---|
| Space Group | P212121 |
| Unit Cell (Å) | a=75.655, b=111.140, c=117.572 |
| Resolution range (Å) | 55.815 - 2.038 |
| Highest resolution shell (Å) | 2.15 – 2.038 |
| Number of observations | 810,104 |
| Number of Unique reflections | 64,823 |
| Completeness (%) | 98.5 (91.0) |
| Mean I/σ(I) | 8.4 (3.9) |
| Multiplicity | 7.4 (7.0) |
| Rmerge (%)1 | 14.9 (43.8) |
| Rmeas (%)2 | 16.1 (47.0) |
| Rp.i.m. (%)3 | 5.9 (16.7) |
| Model and Refinement Statistics | |
| Resolution range (Å) | 55,632 - 2,029 |
| No. of reflections (total) | 64,823 |
| No. of reflections (test) | 3287 |
| Rcryst (%) /NCS4 | 17.2 / 17.4 |
| Rfree (%) /NCS4 | 21.3 / 20.4 |
| Stereochemical parameters | |
| Restraints (RMSD observed) | |
| Bond angle (°) | 2.216 |
| Bond Length (Å) | 0.0194 |
| Av. Isotropic B value (Å2) / Wilson plot B value (Å2) | 29.785 / 26.6 |
| DPI based on Rfree (%) | 15.1 |
| No. Protein residues / atoms | 852 / 6,904 |
| No. Water molecules | 242 |
| Ramachandran plot: residues (%) in favored/allowed | 98.1 / 99.8 |
Values in parentheses are for the highest resolution shell
R-factors without and with applying NCS restraints.
Figure 1.
The Dronpa-M159T tetrameric structure (green) is overlaid with the tetrameric wild type coordinates from 2Z1O (grey) using all atoms in chain A only. The corresponding B chains are aligned whereas the C chains do not. The Thr159 (yellow) is positioned in the A-C interface. In addition, residues 36, 62, 93, 160, 171, 172 and 173 showed chemical shift differences in the off state (red) and residues 62, 65-69, 90, 91, 132, 140, 142-145, 156-160, 191-195 and 212-214 underwent exchange broadening (pink) in HSQC spectroscopy 10 and are also concentrated in the A-C interface.
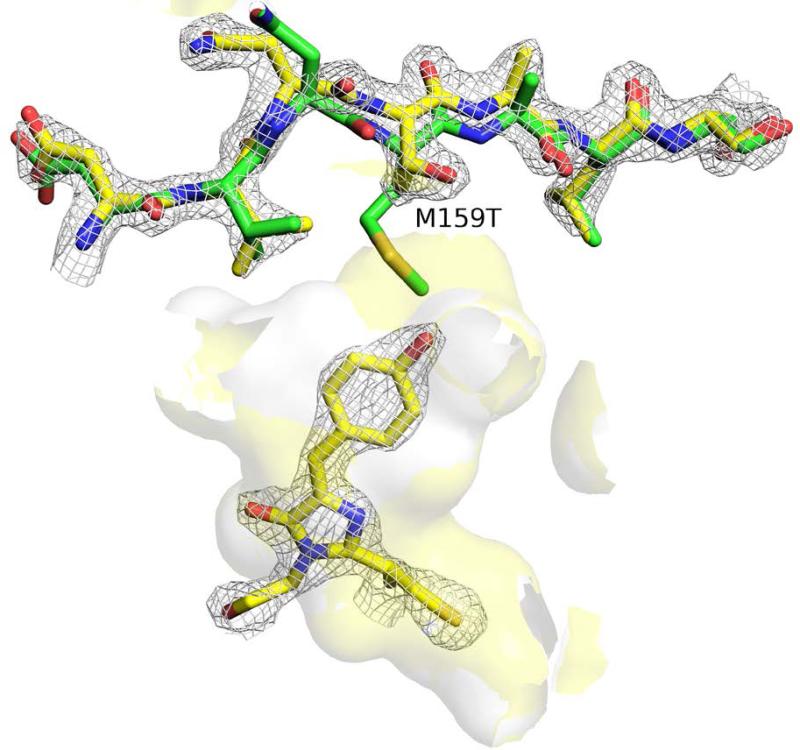
Further comparison between the local structures near the cis- p-hydroxybenzylideneimidazolinone chromophore structures of both Dronpa-M159T and Dronpa shows an enlargement of the chromophore cavity in the case of the Met159T mutant (Figure 2). This is primarily due to the removal of the bulky side chain of Met159, but in addition, a 1.4 Å ‘outward’ shift of the backbone coordinates is clearly observed. Calculation of the cavity volumes using the trj_cavity program 25 of the X-ray structure coordinates resulted in values of 49 and 55 Å3 for Dronpa and the M159T mutant, respectively. We note however that the M159T structure is collected at room temperature, whereas the Dronpa structure was collected at cryogenic temperature. To investigate whether the increase in cavity volume observed in the M159T X-ray structure as compared to Dronpa, is maintained in solution for both the monomer and the tetramer, we determined the volume of the chromophore pocket in our trajectories.
Figure 2.
The cavity available to the cis anionic chromophore in the M159T structure (yellow surface) is enlarged relative to Dronpa (white surface) as a result of the reduced side chain as well as a 1.4 Å ‘outward retraction’ of the side chain (Dronpa-M159T: yellow scheme / Dronpa: green scheme 2Z1O 10). The cavity volume shown does not include crystal waters. 2Fo-Fc electron density is contoured at the 2.0 sigma level for chromophore and residues 156-171 for the Dronpa-M159T structure.
Figure S4 shows the volumes of the cavity for monomeric Dronpa, monomeric M159T and tetrameric M159T as a function of simulation time. Focusing on the 100-point running average, we see that the volume of the cavity in M159T monomer (55 (+/− 19) Å3) is indeed larger in solution than Dronpa (37 (+/− 10) Å3). Furthermore, we also notice a small, but consistent difference in the cavity volume between the M159T monomer and tetramer, with the cavities in the tetramer on average being slightly smaller (47 (+/− 14) Å3) than the cavity in the monomer (55 (+/− 2.3) Å3). This result suggests that without the protein-protein contacts, the cavity can relax and increase its volume. We speculate furthermore that this volume increase, albeit very small, may also help reducing the steric hindrance for the isomerization of the chromophore and increase the kinetics of this process. It is stressed that a 65-fold enhancement of the on-off kinetics at room temperature corresponds to a small ~10 kJ/mol activation energy difference. The small but reproducible cavity volume differences found in this study may therefore be in line with the significant rate enhancement. By comparing the isomerization rates in the monomer and the oligomeric states, we indeed found that the reactions occurs more efficiently in the monomer 14.
Experimentally it was observed from time resolved UV-VIS spectroscopy measurements that in dilute solution, the relative on-to-off photoswitching rate constant with continuous 473 nm illumination increased 65-fold relative to the unmodified Dronpa samples, at pH 7.8 and 3 × 10−6 M concentration, favoring monomeric forms 14. We further noted distinctly reduced on-off phototransformation kinetics with increased protein concentration for both the wild type and mutant samples, in approximately the same concentration ranges. While the oligomerisation constant is not yet well determined for Dronpa-M159T, Mizuno et al found a value of 1.3 × 10−4 M for Dronpa 26. In general agreement with similar conclusion by Nguyen Bich, et al (2012)16 and Mizuno, et al (2010) 26, self-association strongly affects the on-to-off photoswitching kinetics of the Dronpa-M159T mutant as well. Mizuno et al (2008) already noted that Gly218Glu mutation of 22G, affecting the A-C interface, also yielded photochromic behavior 26. A Lys145Asn mutation of Dronpa was shown to have a self-association constant which was lower than Dronpa in the on state 16. Zhou et al subsequently reported Lys145Asn-Dronpa was tetrameric at concentrations from 10 to 100 μM in the on state, but converted to monomeric species in the off state after illumination with 500 nm light 27. The importance of mutation in the β7 strand was also in agreement with the observation of exchange broadening seen for 142-145 region of strand β7 in the off state of Dronpa 10. The light-induced reversible association/dissociation of Lys145Asn-Dronpa was exploited for optogenetics control in a series of chimeric constructs in cells27. Furthermore, Kao et al28 found that increased viscosity of the medium decreases the on-to-off photoswitching kinetics of Dronpa and even more pronounced in double mutant Met159Ala/Val157Ile-Dronpa. It was proposed that structural flexibility, increased in either monomeric form or low viscosity media, aid the cis-trans photoisomerisation efficiency, from a kinetic model that assumes parallel, competing pathways for radiative decay, photoswitching and nonradiative decay28.
A significant reduction of the on-to-off photoswitching kinetics were seen in crystals of Dronpa-M159T relative to solutions, although optical measurements of crystals involved conversion of larger optical density. Estimates to extrapolate to reduced optical density still yielded reduced switching kinetics by approximately order of magnitude. Specifically, time resolved micro-spectroscopy showed full conversion in less than 5 seconds of optically thick crystalline samples, having an optical density in excess of 6 at the maximum wavelength 489nm, with continuous illumination at 473nm and 7.6 mW power in a 500 μm 1/e2 width spot while maintaining the temperature at 18° C with a cryo-stream. These conditions correspond to full conversion approximately 50 μm optical depth of the P212121 crystal form of Dronpa-M159T.
In conclusion, the altered A-C interface contact of the Met159Thr mutant compared to Dronpa can be reconciled with its increased photoswitching efficiency in the framework of photo-induced structural flexibility.
Supplementary Material
Acknowledgements
This work was supported by the Engineering and Physical Sciences Research Council (EPSRC) via award EP/I003304/1.
Footnotes
The work was performed at Advanced Photon Source Argonne National Laboratory Argonne, IL 60439 USA
References
- 1.Chudakov DM, Matz MV, Lukyanov S, Lukyanov KA. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol Rev. 2010;90(3):1103–1163. doi: 10.1152/physrev.00038.2009. [DOI] [PubMed] [Google Scholar]
- 2.Chudakov DM, Lukyanov S, Lukyanov KA. Tracking intracellular protein movements using photoswitchable fluorescent proteins PS-CFP2 and Dendra2. Nat Protoc. 2007;2(8):2024–2032. doi: 10.1038/nprot.2007.291. [DOI] [PubMed] [Google Scholar]
- 3.Lippincott-Schwartz J, Manley S. Putting super-resolution fluorescence microscopy to work. Nat Methods. 2009;6(1):21–23. doi: 10.1038/nmeth.f.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiedenmann J, Oswald F, Nienhaus GU. Fluorescent proteins for live cell imaging: opportunities, limitations, and challenges. IUBMB Life. 2009;61(11):1029–1042. doi: 10.1002/iub.256. [DOI] [PubMed] [Google Scholar]
- 5.Nienhaus K, Ulrich Nienhaus G. Fluorescent proteins for live-cell imaging with super-resolution. Chem Soc Rev. 2014;43(4):1088–1106. doi: 10.1039/c3cs60171d. [DOI] [PubMed] [Google Scholar]
- 6.Ando R, Mizuno H, Miyawaki A. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science. 2004;306(5700):1370–1373. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- 7.Andresen M, Stiel AC, Trowitzsch S, Weber G, Eggeling C, Wahl MC, Hell SW, Jakobs S. Structural basis for reversible photoswitching in Dronpa. Proc Natl Acad Sci U S A. 2007;104(32):13005–13009. doi: 10.1073/pnas.0700629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warren MM, Kaucikas M, Fitzpatrick A, Champion P, Sage JT, van Thor JJ. Ground-state proton transfer in the photoswitching reactions of the fluorescent protein Dronpa. Nat Commun. 2013;4:1461. doi: 10.1038/ncomms2460. [DOI] [PubMed] [Google Scholar]
- 9.Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc Natl Acad Sci U S A. 2002;99(20):12651–12656. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizuno H, Mal TK, Walchli M, Kikuchi A, Fukano T, Ando R, Jeyakanthan J, Taka J, Shiro Y, Ikura M, Miyawaki A. Light-dependent regulation of structural flexibility in a photochromic fluorescent protein. Proc Natl Acad Sci U S A. 2008;105(27):9227–9232. doi: 10.1073/pnas.0709599105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stiel AC, Trowitzsch S, Weber G, Andresen M, Eggeling C, Hell SW, Jakobs S, Wahl MC. 1.8 A bright-state structure of the reversibly switchable fluorescent protein Dronpa guides the generation of fast switching variants. Biochem J. 2007;402(1):35–42. doi: 10.1042/BJ20061401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilmann PG, Turcic K, Battad JM, Wilce MC, Devenish RJ, Prescott M, Rossjohn J. The 1.7 A crystal structure of Dronpa: a photoswitchable green fluorescent protein. J Mol Biol. 2006;364(2):213–224. doi: 10.1016/j.jmb.2006.08.089. [DOI] [PubMed] [Google Scholar]
- 13.Habuchi S, Ando R, Dedecker P, Verheijen W, Mizuno H, Miyawaki A, Hofkens J. Reversible single-molecule photoswitching in the GFP-like fluorescent protein Dronpa. Proc Natl Acad Sci U S A. 2005;102(27):9511–9516. doi: 10.1073/pnas.0500489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaucikas M, Tros M, van Thor JJ. Photoisomerisation and proton transfer in the forward and reverse photoswitching of the fast-switching M159T mutant of the Dronpa fluorescent protein. J Phys Chem B. 2014 doi: 10.1021/jp506640q. doi: 10.1021/jp506640q. [DOI] [PubMed] [Google Scholar]
- 15.Adam V, Lelimousin M, Boehme S, Desfonds G, Nienhaus K, Field MJ, Wiedenmann J, McSweeney S, Nienhaus GU, Bourgeois D. Structural characterization of IrisFP, an optical highlighter undergoing multiple photo-induced transformations. Proc Natl Acad Sci U S A. 2008;105(47):18343–18348. doi: 10.1073/pnas.0805949105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen Bich N, Moeyaert B, Van Hecke K, Dedecker P, Mizuno H, Hofkens J, Van Meervelt L. Structural basis for the influence of a single mutation K145N on the oligomerization and photoswitching rate of Dronpa. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 12):1653–1659. doi: 10.1107/S0907444912039686. [DOI] [PubMed] [Google Scholar]
- 17.Leslie AGW, Powell HR. Processing Diffraction Data with Mosflm. In: Randy J, Read JLS, editors. Evolving Methods for Macromolecular Crystallography. Vol. 245. Springer; 2007. pp. 41–51. [Google Scholar]
- 18.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 1):72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 19.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vagin AA, Isupov MN. Spherically averaged phased translation function and its application to the search for molecules and fragments in electron-density maps. Acta Crystallogr D Biol Crystallogr. 2001;57(Pt 10):1451–1456. doi: 10.1107/s0907444901012409. [DOI] [PubMed] [Google Scholar]
- 21.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hess B, Kutzner C, Van der Spoel D, E L. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J Chem Theo Comp. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 24.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 25.Paramo T, East A, Garzon D, Ulmschneider MB, Bond P. Efficient Characterization of Protein Cavities within Molecular Simulation Trajectories: trj_cavity. J Chem Theo Comput. 2014;10:2151–2164. doi: 10.1021/ct401098b. [DOI] [PubMed] [Google Scholar]
- 26.Mizuno H, Dedecker P, Ando R, Fukano T, Hofkens J, Miyawaki A. Higher resolution in localization microscopy by slower switching of a photochromic protein. Photochem Photobiol Sci. 2010;9(2):239–248. doi: 10.1039/b9pp00124g. [DOI] [PubMed] [Google Scholar]
- 27.Zhou XX, Chung HK, Lam AJ, Lin MZ. Optical control of protein activity by fluorescent protein domains. Science. 2012;338(6108):810–814. doi: 10.1126/science.1226854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kao YT, Zhu X, Min W. Protein-flexibility mediated coupling between photoswitching kinetics and surrounding viscosity of a photochromic fluorescent protein. Proc Natl Acad Sci U S A. 2012;109(9):3220–3225. doi: 10.1073/pnas.1115311109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.