Abstract
Introducing new reactivity into living organisms is a major challenge in synthetic biology. Despite an increasing interest in both developing aqueous-compatible small molecule catalysts and engineering enzymes to perform new chemistry in vitro, the integration of non-native reactivity into metabolic pathways for small molecule production has been underexplored. Herein we report a biocompatible iron(III) phthalocyanine catalyst capable of efficient olefin cyclopropanation in the presence of a living microorganism. By interfacing this catalyst with E. coli engineered to produce styrene, we synthesize non-natural phenyl cyclopropanes directly from D-glucose in single-vessel fermentations. This process represents the first combination of non-biological carbene-transfer reactivity with cellular metabolism for small molecule production.
Keywords: metabolism, iron, phthalocyanine, synthetic methods, synthetic biology
The field of synthetic biology is changing how important small molecules are manufactured.[1] Using renewable starting materials (e.g. sugar, plant biomass, CO2) metabolic engineers overproduce small molecules in single-vessel fermentations by optimizing both known and de novo biosynthetic pathways in host microorganisms. Despite significant progress in this discipline, a major remaining challenge is engineering organisms to access compounds of non-natural origin. This objective is important because many small molecules of commercial interest are not currently accessible using known enzymatic chemistry. Two strategies have emerged to achieve this goal: engineering non-biological reactivity into enzymes and developing non-enzymatic catalysts that can be interfaced with cellular metabolism.[2,3]
Exploring the reactivity of carbenes has been particularly fruitful for enzyme engineering efforts. Carbene intermediates are accessed and utilized in cells by thiamin diphosphate-dependent enzymes (Figure 1A).[4] Previous studies have introduced ruthenium carbene complexes within artificial metalloenzymes for olefin ring-closing metathesis in water and phosphate buffer.[5] More recent work by the Arnold and Fasan laboratories has extended the types of carbenes involved in enzymatic catalysis to include iron carbenes by engineering hemin-binding proteins to catalyze enantioselective olefin cyclopropanation with ethyl diazoacetate (EDA) both in vitro and in a whole cell format (Figure 1B).[2b,f,g,6] These studies were inspired by the mechanistic similarities between cytochrome P450 oxene-transfer catalysis and transition metal-mediated carbene-transfer reactions. This work provides a biocatalytic route to cyclopropanes, which are found in many bioactive synthetic molecules.[7] Despite the success of these engineering efforts, enzymes that utilize metallocarbene intermediates have not yet been integrated into engineered metabolic pathways.
Figure 1.
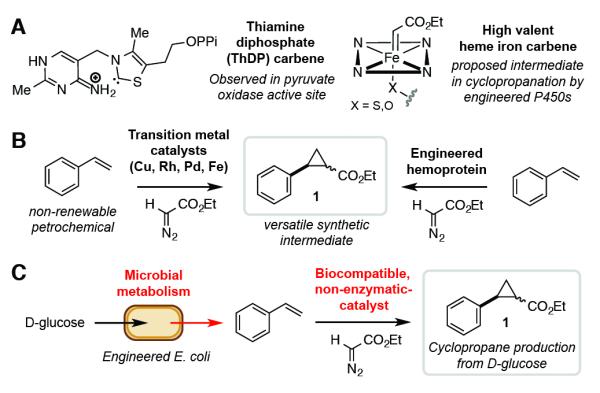
Design of a biocompatible cyclopropanation reaction. A) Carbene intermediates in biological catalysis. B) Current approaches for accessing ethyl-2-phenylcyclopropane-1-carboxylate (1) from styrene using transition metal-mediated carbene chemistry. C) Phenyl cyclopropane production from D-glucose by combining in vivo styrene production with biocompatible chemistry.
We envisioned approaching this challenge using biocompatible chemistry: non-enzymatic reactions capable of modifying metabolites as they are made by living organisms (Figure 1C).[3] Here we describe the identification of an iron phthalocyanine catalyst that cyclopropanates styrene generated by engineered microbial metabolism. This reaction is both a new biocompatible iron-mediated transformation and, to the best of our knowledge, the first example of metallocarbene chemistry being interfaced with cellular metabolism for small molecule production.
We began our studies by investigating the efficiency of olefin cyclopropanation under conditions compatible with the growth of E. coli. We initially examined the tetraphenylporphyrin iron(III) chloride (FeTPPCl) catalyzed cyclopropanation of 4-vinyl anisole and EDA. This reaction has been previously reported to occur under aqueous alkaline conditions using diazomethane, and also in aqueous phosphate buffer (pH 7.0) under anaerobic conditions using the hemin cofactor as a catalyst (29% conversion).[2g,8] We performed the FeTPPCl-mediated reaction in water, phosphate buffer (pH 7.0) and growth media of increasing complexity. We found that while the reaction was moderately efficient in water, phosphate buffer and growth media provided significantly higher conversions and diastereoselectivities (Table 1). Unlike other biocompatible transformations, cyclopropanation proceeded efficiently in the complex medium LB.[3a] The reasons for the increased conversion in media relative to water are unclear, however similar effects observed for other reactions run in highly ionic solvents are hypothesized to arise from rate acceleration due to an increased hydrophobic effect.[9] Finally, the addition of bacterial cells (E. coli BL21(DE3), OD600=0.5) had no detrimental effect on product conversion or selectivity (Entries 5 and 7).
Table 1.
FeTPPCl-catalyzed cyclopropanations in aqueous media and in the presence of E. coli.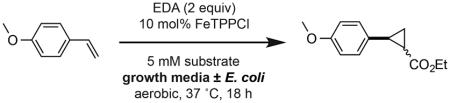
| Entry | Growth Medium | E.coli cells added? | Yield (%) | trans:cis |
|---|---|---|---|---|
| 1 | H2O | no | 46 | 3:1 |
| 2 | 0.1 M K2HPO4 | no | 77 | 3.7:1 |
| 3 | M9-glucose | no | 71 | 3.7:1 |
| 4 | M9CA-glucose | no | 72 | 4:1 |
| 5 | M9CA-glucose | yes | 75 | 3.9:1 |
| 6 | LB | no | 71 | 4:1 |
| 7 | LB | yes | 70 | 4:1 |
Reactions were performed using 4-vinylanisole (5 mM), EDA (10 mM) and FeTPPCl (0.5 mM) in sealed Hungate tubes under an atmosphere of air. All cultures were grown in the presence of kanamycin (50 mg/L) and isopropyl β-D-1-thiogalactopyranoside (IPTG; 0.2 mM). E. coli BL21 cells transformed with an empty pET-29b(+) expression plasmid (OD600= 0.5) were used. Product concentrations in crude culture extracts were determined by 1H-NMR relative to an internal standard of 1,3,5-trimethoxybenzene. All data is shown as an average of three experiments to one standard deviation.
We next conducted a catalyst screen in M9CA-glucose media containing E. coli (Table 2 and Table S1). Placing electron-withdrawing and electron-donating substituents on the porphyrin ring system influenced product diastereoselectivities but diminished conversion relative to FeTPPCl (Entries 2 and 3). Replacement of the four aromatic rings of the TPP ring system with aliphatic substituents abolished catalytic activity (Table S1). Hemin was unreactive under the reaction conditions (Entry 4), despite being previously reported to catalyze this transformation in vitro.[2g] Ferric phthalocyanine (FePcCl), proved to be an exceptional cyclopropanation catalyst, affording product in 95% yield (Entry 5). Catalyst loading of FePcCl could be reduced to as low as 1 mol% with only a moderate loss in conversion. No product was detected using FeCl3 or in the absence of added catalyst, ruling out the possibility that either endogenously biosynthesized metal ion complexes or ferric ligands produced by E. coli contribute to catalysis in vivo.
Table 2.
Catalyst screen in the presence of E. coli.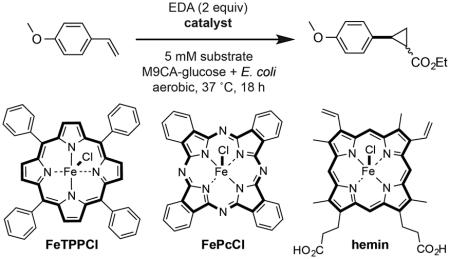
| Entry | Catalyst (mol%) | Yield (%) 75 | trans:cis |
|---|---|---|---|
| 1 | FeTPPCl (10) | 3.9:1 | |
| 2 | Fe(F20TPP)Cl (10) | 55 | 5.1:1 |
| 3 | Fe(OMe)4TPPCl (10) | 7 | 2.9:1 |
| 4 | hemin (10) | 0 | – |
| 5 | FePcCl (10) | 95 | 3.4:1 |
| 6 | FePcCl (5) | 93[a] | 3.4:1 |
| 7 | FePcCl (2.5) | 90[b] | 3.7:1 |
| 8 | FePcCl (1) | 80[b] | 4.0:1 |
Reactions were performed as described in Table 1. All data is shown as an average of three experiments.
24 h reaction.
48 h reaction.
The major byproduct of the reaction at 10 mol% catalyst loading is diethyl maleate, which arises from EDA dimerization. Unexpectedly, at lower catalyst loadings we observed a new byproduct, diethyl succinate (Figure 2 and Table S2). We hypothesized this product arose from E. coli using diethyl maleate as a terminal electron acceptor for anaerobic respiration under the progressively oxygen-limiting conditions of the reaction set-up. We also verified that living cells were required for diethyl succinate production. The formation of this byproduct at 2.5 mol% catalyst loading therefore confirms that the E. coli are alive under our optimal reaction conditions.
Figure 2.
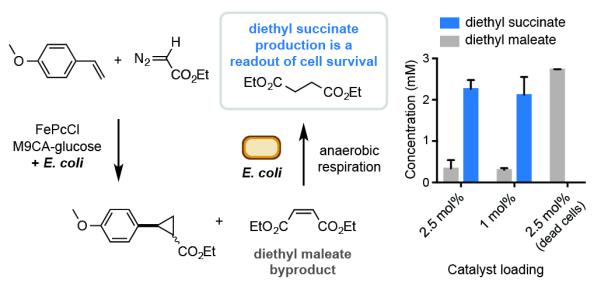
A reaction byproduct reports on E. coli survival under the cyclopropantion reaction conditions. Reactions were performed as described in Table 1.
Having shown that the FePcCl-catalyzed cyclopropanation was compatible with E. coli, we next attempted the reaction with styrene generated via bacterial metabolism. Styrene production from D-glucose has been achieved in the L-phenylalanine overproducing strain E. coli NST74 by introducing two enzymes: L-phenylalanine ammonia lyase from Arabidopsis thaliana (PAL2), which converts L-phenylalanine to trans-cinnamic acid, and a decarboxylase from Saccharomyces cerevisiae (FDC1) that generates styrene from trans-cinnamic acid (Figure 3A).[10]
Figure 3.
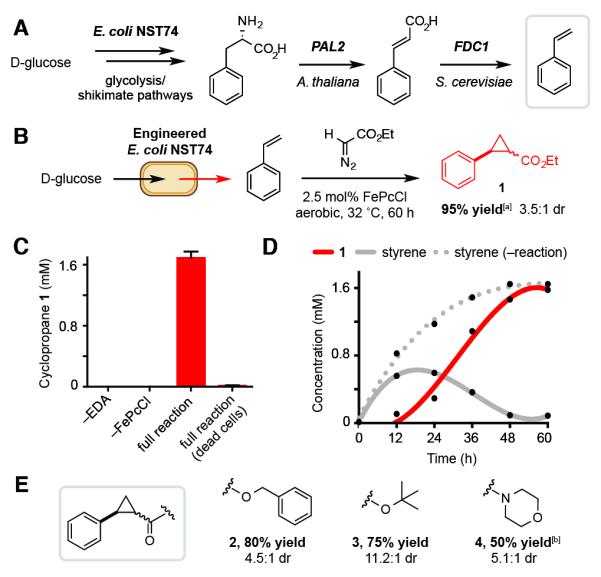
The biocompatible cyclopropanation reaction can be interfaced with microbial styrene production. A) Engineered pathway for styrene production in the L-phenylalanine overproducer E. coli NST74. B) Cyclopropanation using metabolically generated styrene. C) Cyclopropane production requires all reaction components and living E. coli. D) Metabolite production during fermentations. E) Additional cyclopropanes accessed via this approach. Metabolite concentrations were determined by GC relative to an internal standard of 1,3,5-trimethoxybenzene. Yields in Section E are of isolated material from 800 mL cultures. All data is shown as an average of three independent experiments to one standard deviation. [a] 93% isolated yield. [b] 72 h reaction.
We confirmed that FePcCl was effective under the conditions required for styrene production by performing the cyclopropanation reaction with 1.5 mM styrene in phosphate-limited minimal media (MM1) containing E. coli BL21(DE3) and D-glucose (82% yield, 1.7:1 dr). Serial dilutions and plate counts directly from cultures with and without reaction components showed no difference in survival (Figure S1). Together these results suggested that our reaction would be compatible with in vivo styrene production. Accordingly, the PAL2 and FDC1 genes were introduced into E. coli NST74 on a single expression plasmid (pTrc99A_PAL2-FDC1). Under optimized conditions this strain accumulated 1.65 mM styrene in the culture medium over 48 h (Figure S5). We next attempted the cyclopropanation reaction by adding FePcCl (2.5 mol%) and EDA (2 equiv) to cultures at the point of induction of the styrene-producing pathway (OD600=0.5–0.6, t=0 h). After 48 h we observed cyclopropane 1 (44% conversion, 3.0:1 dr). 1H-NMR analysis of the reaction extract showed no unreacted EDA and significant levels of EDA byproducts, indicating that competing carbene dimerization was likely limiting reaction conversion. This issue was circumvented by adding EDA portion-wise over the course of the fermentation, which increased the yield of 1 to 81%. By adding an additional equivalent of EDA and extending the reaction time to 60 h we obtained 1 in 95% yield by GC (3.5:1 dr, Figure 3B).
We performed a series of control experiments to confirm that cyclopropanation required the presence of catalyst, EDA, and living E. coli (Figure 3C). To obtain information about the timing and rate of cyclopropanation relative to in vivo styrene production, we analyzed product distributions in fermentations with and without the reaction components (Figure 3D, Figure S4). In the presence of FePcCl and EDA, styrene reaches a maximum concentration of 0.6 mM after 18 h and then steadily depletes as 1 accumulates. Ultimately the concentration of 1 equals the concentration of styrene produced in the absence of the reaction components. This observation confirms that the biocompatible cyclopropanation is interfaced with styrene output from E. coli and that after 18 h the rate of styrene consumption via cyclopropanation likely exceeds that of styrene generation via metabolism. This analysis also demonstrates that the reaction components have a minimal effect on the overall levels of styrene production. Interestingly, in the absence of FePcCl accumulating EDA significantly inhibits styrene production after 12 h (P<0.05), indicating that this reagent is toxic to E. coli and that the activity of the catalyst prevents this adverse effect in the full reaction (Figure S5). A preliminary investigation of the cyclopropanation using a three-phase test suggested that catalysis by FePcCl occurs at a solid-liquid interface as no product was detected using polymer-supported styrene (Figure S6). This result indicates that FePcCl is likely functioning as a heterogeneous catalyst in this transformation.
We evaluated the scope of the in vivo cyclopropanation by examining other diazoacetate derivatives. Accessing additional electron poor (acceptor) carbenes provided cyclopropanes 2–4 in good isolated yields (Figure 3E). We also re-examined hemin as a catalyst to test the extent to which the results of our initial screening procedure predicted catalyst performance with metabolically generated styrene. Hemin remained a less efficient cyclopropanation catalyst (27% conversion after 60 h, Table S4), confirming the utility of our catalyst identification strategy. The low reactivity of hemin also resulted in EDA accumulation, which dramatically reduced overall styrene production levels to 0.23 mM. This finding shows that the identity of the non-enzymatic catalyst can influence multiple variables of in vivo reactions. Overall, these studies not only represent a new route to cyclopropanes but also a potential roadmap for the discovery of other biocompatible reactions.
The combined use of enzymatic and non-enzymatic catalysis for chemical synthesis can provide unique benefits over the use of chemo- or biocatalysis alone.[3c,11] In the case of this biocompatible cyclopropanation, the generation of styrene from D-glucose is attractive as it avoids the use of non-renewable petroleum streams. By intercepting biologically-produced styrene in the fermentation vessel we also circumvent the challenges associated with its isolation, including volatility and reactivity (polymerization during gas stripping).[12] Additionally, the use of bench- and air-stable phthalocyanines circumvents the need for strictly anaerobic conditions during cyclopropanation (as is required by many engineered biocatalysts). This feature is critical for overall cyclopropane production as an aerobic growth environment is needed for maximum styrene production by engineered E. coli.[10] Notably, this requirement has likely made incorporating cyclopropanating enzymes into engineered metabolic pathways challenging.
The use of an inexpensive and highly abundant early transition metal catalyst adds to the appeal of this process. Indeed, interactions between microorganisms and iron are widespread in natural habitats, and evolutionary pressure to accommodate the reactivity of iron-containing minerals could account for the biocompatibility of our catalyst. The success of this cyclopropanation suggests that iron-mediated reactions are a promising source of new biocompatible reactions. Natural microbe-mineral interactions may also be good future starting-points for designing additional biocompatible non-enzymatic transformations.
In summary, we have shown that biocompatible metallocarbene-transfer catalysis and engineered metabolism can be combined for small molecule production. Integrating metallocarbene chemistry with the metabolism of living organisms represents a new approach to constructing non-natural molecules. Important future challenges include developing biocompatible, chiral catalysts for enantioselective cyclopropanation and engineering pathways for the production of substituted styrene substrates. Finally, other non-enzymatic reactions that use styrene may also be good targets for biocompatible reaction development and allow access to further structural diversity from this single, engineered metabolic pathway.
Supplementary Material
Acknowledgements
The authors acknowledge Professor David Nielsen (Arizona State University) for generously providing plasmids, and Professor Kristala Jones Prather (MIT) for helpful discussions during the preparation of this manuscript.
Footnotes
This work was supported by the National Institutes of Health (DP2 GM105434), the Searle Scholars Program, and the European Commission (Marie Curie IOF fellowship to S. W).
Supporting information for this article is given via a link at the end of the document.
References
- [1].a) Keasling J. Science. 2010;330:1355–1358. doi: 10.1126/science.1193990. [DOI] [PubMed] [Google Scholar]; b) Lee JW, Na D, Park JM, Lee J, Choi S, Lee SY. Nat. Chem. Biol. 2012;8:536–546. doi: 10.1038/nchembio.970. [DOI] [PubMed] [Google Scholar]; c) Becker J, Wittermann C. Angew. Chem. Int. Ed. 2015;54:3328–3350. doi: 10.1002/anie.201409033. Angew. Chem.2015, 127, 3383–3407. [DOI] [PubMed] [Google Scholar]; d) Coelho PS, Arnold FH, Lewis JC. In: Comprehensive Organic Synthesis. Molander GA, Knochel P, editors. Vol. 9. Elsevier; Oxford, UK: 2014. pp. 390–420. [Google Scholar]
- [2].a) Renata H, Wang ZJ, Arnold FH. Angew. Chem. Int. Ed. 2015;54:3351–3367. doi: 10.1002/anie.201409470. Angew. Chem.2015, 127, 3408–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Coelho PS, Brustard EM, Kannan A, Arnold FH. Science. 2013;339:307–309. doi: 10.1126/science.1231434. [DOI] [PubMed] [Google Scholar]; c) McIntosh JA, Coelho PS, Farwell CC, Wang ZJ, Lewis JC, Brown TR, Arnold FH. Angew. Chem. Int. Ed. 2013;52:9309–9312. doi: 10.1002/anie.201304401. Angew. Chem.2013, 125, 9479–9482. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Hyster TK, Farwell CC, Buller AR, McIntosh JA, Arnold FH. J. Am. Chem. Soc. 2014;136:15505–15508. doi: 10.1021/ja509308v. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Farwell CC, McIntosh JA, Hyster TK, Wang ZJ, Arnold FH. J. Am. Chem. Soc. 2014;136:8766–8771. doi: 10.1021/ja503593n. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Heel T, McIntosh JA, Dodani SC, Meyerowitz JT, Arnold FH. ChemBioChem. 2014;15:2556–2562. doi: 10.1002/cbic.201402286. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Bordeaux M, Tyagi V, Fasan R. Angew. Chem. Int. Ed. 2015;54:1744–1748. doi: 10.1002/anie.201409928. Angew. Chem.2015, 127, 1764–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Singh R, Kolev JN, Sutera PA, Fasan R. ACS Catal. 2015;5:1685–1691. doi: 10.1021/cs5018612. [DOI] [PMC free article] [PubMed] [Google Scholar]; i) Tyagi V, Bonn RB, Fasan R. Chem. Sci. 2015;6:2488–2494. doi: 10.1039/c5sc00080g. [DOI] [PMC free article] [PubMed] [Google Scholar]; j) Sreenilayam G, Fasan R. Chem. Commun. 2015;51:1532–1534. doi: 10.1039/c4cc08753d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].a) Sirasani G, Tong L, Balskus EP. Angew. Chem. Int. Ed. 2014;53:7785–7788. doi: 10.1002/anie.201403148. Angew. Chem.2014, 126, 7919–7922. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lee Y, Umeano A, Balskus EP. Angew. Chem. Int. Ed. 2013;52:11800–11803. doi: 10.1002/anie.201307033. Angew. Chem.2013, 125, 12016–12019. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wallace S, Balskus EP. Curr. Opin. Biotechnol. 2014;30:1–8. doi: 10.1016/j.copbio.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Wallace S, Schultz EE, Balskus EP. Curr. Opin. Chem. Biol. 2015;25:71–79. doi: 10.1016/j.cbpa.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Meyer D, Neumann P, Ficner R, Tittmann K. Nat. Chem. Biol. 2013;9:488–490. doi: 10.1038/nchembio.1275. [DOI] [PubMed] [Google Scholar]
- [5].a) Lo C, Ringenberg MR, Gnandt D, Wilson Y, Ward TR. Chem. Commun. 2011;47:12065–12067. doi: 10.1039/c1cc15004a. [DOI] [PubMed] [Google Scholar]; b) Mayer C, Gillingham DG, Ward TR, Hilvert D. Chem. Commun. 2011;47:12068–12070. doi: 10.1039/c1cc15005g. [DOI] [PubMed] [Google Scholar]; c) Matsuo T, Imai C, Yoshida T, Saito T, Hayashi T, Hirota S. Chem. Commun. 2012;48:1662–1664. doi: 10.1039/c2cc16898g. [DOI] [PubMed] [Google Scholar]
- [6].a) Coelha PS, Wang ZJ, Ener ME, Baril SA, Kannan A, Arnold FH. Nat. Chem. Biol. 2013;9:485–487. doi: 10.1038/nchembio.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wang ZJ, Renata H, Peck NE, Farwell CC, Coelho PS, Arnold FH. Angew. Chem. Int. Ed. 2014;53:6810. doi: 10.1002/anie.201402809. Angew. Chem.2014, 126, 6928–6931. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Renata H, Wang ZJ, Kitto RZ, Arnold FH. Catal. Sci. Technol. 2014;4:3640–3643. doi: 10.1039/C4CY00633J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lebel H, Marcoux J-F, Molinaro C, Charette AB. Chem. Rev. 2003;103:977–1050. doi: 10.1021/cr010007e. [DOI] [PubMed] [Google Scholar]
- [8].Morandi B, Carreira EM. Science. 2012;335:1471–1474. doi: 10.1126/science.1218781. [DOI] [PubMed] [Google Scholar]
- [9].Breslow R, Rizzo CJ. J. Am. Chem. Soc. 1991;113:4340–4341. [Google Scholar]
- [10].McKenna R, Nielsen DR. Metab. Eng. 2011;13:544–554. doi: 10.1016/j.ymben.2011.06.005. [DOI] [PubMed] [Google Scholar]
- [11].a) Köhler V, Turner NJ. Chem. Commun. 2015;51:450–464. doi: 10.1039/c4cc07277d. [DOI] [PubMed] [Google Scholar]; b) Denard CA, Huang H, Bartlett MJ, Lu L, Tan Y, Zhao H, Hartwig JF. Angew. Chem. Int. Ed. 2014;53:465–469. doi: 10.1002/anie.201305778. Angew. Chem.2014, 126, 475–479. [DOI] [PubMed] [Google Scholar]; c) Wang ZJ, Clary KN, Bergman RG, Raymond KN, Toste FD. Nat. Chem. 2013;5:100–103. doi: 10.1038/nchem.1531. [DOI] [PubMed] [Google Scholar]
- [12].McKenna R, Moya L, McDaniel M, Nielsen DR. Bioprocess. Biosyst. Eng. 2015;38:165–174. doi: 10.1007/s00449-014-1255-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


