Abstract
Members of the formin family of actin filament nucleation factors have been implicated in sarcomere formation, but precisely how these proteins affect sarcomere structure remains poorly understood. Of six formins in the simple nematode Caenorhabditis elegans, only FHOD-1 and CYK-1 contribute to sarcomere assembly in the worm's obliquely striated body-wall muscles. We analyze here the ultrastructure of body-wall muscle sarcomeres in worms with putative null fhod-1 and cyk-1 gene mutations. Contrary to a simple model that formins nucleate actin for thin filament assembly, formin mutant sarcomeres contain thin filaments. Rather, formin mutant sarcomeres are narrower and have deformed thin filament-anchoring Z-line structures. Thus, formins affect multiple aspects of sarcomere structure.
Keywords: Muscle, Sarcomere, Z-line, Thin filament, Formin, C. elegans
Introduction
Contraction of striated muscles depends on the sliding of parallel thin filaments and thick filaments past each other in repeating cytoskeletal units called sarcomeres [1]. Sarcomere ends, called Z-lines, are marked by electron-dense structures that serve as anchor sites for actin-based thin filaments, while the sarcomere center, or M-line, is characterized by a similar anchoring structure for bipolar myosin-based thick filaments (Figure 1). The sarcomere can be further subdivided into a broad central A-band occupied by thick filaments and centered on the M-line, and I-bands that mark thick filament-free zones. Sequentially repeating sarcomeres produce the striations apparent in striated muscle of vertebrates and invertebrates.
Figure 1.
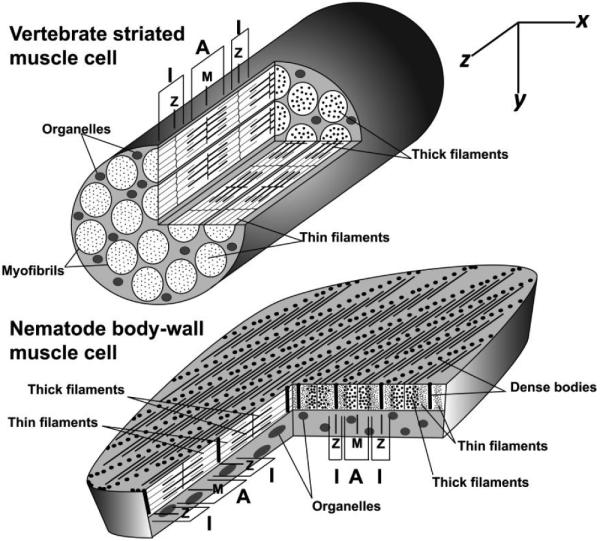
Sarcomere organization in vertebrate and nematode muscle. Sarcomeres in vertebrate striated muscle cells are linked longitudinally in cytoplasmic myofibrils, while sarcomeres in nematode BWM cells are packed in a continuous layer beneath the cell surface that faces the outer body wall. Sarcomeres of both muscles appear similar in longitudinal y/z-sections: Z-lines at sarcomere ends anchor thin filaments; M-lines at sarcomere centers anchor thick filaments; A-bands containing thick filaments are centered on the M-lines; and I-bands lacking thick filaments are centered on the Z-lines. However, the different sarcomere arrangements result in very different appearances in x/y-and x/z-sections. Vertebrate sarcomeres in neighboring myofibrils are in register, resulting in cross striations perpendicular to the thin and thick filaments, visible in y/z - and x/z-sections. In x/y-cross sections of vertebrate striated muscle cells, all sarcomeres are cut at the same position. Thus, the x/y-section at the end of the modeled muscle cuts through I-bands in all sectioned sarcomeres, while the x/y-section near the middle of the muscle cuts A-bands in all sectioned sarcomeres. Conversely, nematode sarcomeres are offset from their neighbors by approximately 1 µm along the z-axis. As a result, striations appear at an oblique angle from thin and thick filaments in x/z-views. In x/y-cross sections of nematode muscle, different sarcomere positions are revealed in neighboring sarcomeres. Thus, the Z-line structure (dense body) for one sarcomere in an x/y-section is bordered by the I-bands of neighboring sarcomeres, which in turn are adjacent to the A-bands of their neighbors.
In each sarcomere, thin filaments emanate from the Z-lines, stretching through the I-bands to overlap with thick filaments in the A-band. The degree of overlap depends on the degree of muscle contraction. In strongly contracted muscle, myosin motor activity pulls the thin filaments toward the sarcomere center, increasing the amount of overlap between the thick and thin filaments. Consequently, the Z-lines are also drawn closer to the A-bands, and I-bands are reduced in width. Conversely, the overlap of thin and thick filaments is minimized and I-bands are widened in stretched muscle.
While sarcomere structure is well studied, sarcomere assembly is still incompletely understood. For example, how thin filaments assemble into parallel arrays is unclear. In vitro, actin filaments assemble from actin monomers through the initial formation of nuclei for filaments, followed by elongation of nuclei by addition of monomers to the filament ends. Some of the mechanisms regulating the elongation of the actin-based thin filaments are understood. Thin filaments elongate at the rapidly polymerizing barbed ends at the Z-lines, as well as the slowly polymerizing pointed ends directed toward the sarcomere center [2]. CapZ negatively regulates barbed end actin dynamics, but thin filament length is ultimately dictated by pointed end elongation, which is regulated negatively by tropomodulin, and positively by WH2-containing proteins such as Drosophila SALS or vertebrate leiomodin-2 [3-6].
Regarding the nucleation of thin filaments, several actin nucleation factors localize to sarcomeres, including N-WASP, leiomodin-2, and formins, and their perturbations disrupt sarcomere organization to varying degrees [7-10]. Using the simple nematode Caenorhabditis elegans, we provided genetic evidence that the contribution of formins may be broadly conserved across the animals [11]. Two C. elegans formins, CYK-1 and FHOD-1, associate with Z-lines in the worm's striated body-wall muscles (BWMs), and in absence of these formins, the assembly of sarcomere arrays is stunted [11]. As formins stimulate the nucleation of actin filaments and also promote elongation by blocking the inhibitory effects of barbed end capping proteins [12], they would seem to be well-suited to initiate the assembly of the long actin filaments that make the core of sarcomere thin filaments. To better understand the effects of these proteins on sarcomere organization, and to determine whether thin filament assembly is formin-dependent, we examine here the ultrastructure of BWM sarcomeres in worms bearing cyk-1 and fhod-1 gene mutations.
Materials and Methods
Worm strains and growth conditions
Worms were maintained on nematode growth medium (NGM) plates with OP50 bacteria as food, and handled using standard laboratory procedures for C. elegans [13], and grown at 20°C. To produce age-synchronized worm populations, NGM/OP50 plates were populated with twenty young adult hermaphrodites. These were allowed to lay eggs for 4.5 hours before their removal, resulting in age-synchronized progeny that reached young adulthood after 3 days. The wild-type strain N2 was obtained from the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN). XA8001 [fhod-1(tm2363) I], DWP8 [cyk-1(ok2300)/(+) III], and DWP9 [fhod-1(tm2363) I; cyk-1(ok2300)/(+) III] were derived as described previously [11] from the strains VC1895 [+/mT1 II; cyk-1(ok2300)/mT1[dpy-10(e128)] III] and FX02363 [fhod-1(tm2363) I, not outcrossed] obtained from the Caenorhabditis Genetics Center and from S. Mitani (National Bioresource Project for the Experimental Animal Nematode C. elegans, Tokyo Women's Medical University School of Medicine, Tokyo, Japan), respectively. For simplicity, we hereafter refer to the mutations fhod-1(tm2363) and cyk-1(ok2300) as fhod-1(−) and cyk-1(−), respectively. Homozygosity of cyk-1(−) results in sterility [14]. Therefore, homozygous cyk-1(−) mutants and doubly-homozygous fhod-1(−);cyk-1(−) mutants had to be selectively picked from the progeny of heterozygous DWP8 and DWP9 parents based on the presence of protruding vulvae on cyk-1(−) animals, and very short and thin bodies of fhod-1(−);cyk-1(−) animals [11].
Transmission electron microscopy (TEM)
Samples were prepared for TEM by modification of the procedure by Hall [15]. Briefly, young adults were anesthetized with 8% ethanol in M9 Buffer, and cut into 2 to 3 pieces in aldehyde fixative (2.5% glutaraldehyde, 1% formaldehyde, 50 mM sodium cacodylate buffer (pH 7.4), 0.2 M sucrose, 1 mM MgCl2) for 2 hours fixation at room temperature, followed by overnight fixation in fresh fixative at 4°C. Fixed samples were washed with 0.2 M sodium cacodylate (pH 7.4), and stained with 1% osmium tetroxide on ice for one hour, and again for 30 min at room temperature. Samples were then washed with 0.1 M sodium acetate buffer (pH 5.2), and stained en-block with 1% uranyl acetate for one hour on ice, and once more for 30 min at room temperature, before washing and embedding in 2% agarose. Stained samples were dehydrated with a graded ethanol series, and infiltrated with EPON embedding resin (EM-grade embedding material from Polysciences, Inc., Warrington, PA), as described [15]. 80-nm thin sections were mounted onto formvar-coated 200-mesh nickel grids stabilized with evaporated carbon film (Electron Microscopy Science, Hatfield, PA), and stained sequentially with 4% uranyl acetate solution in ethanol and lead citrate solution.
Grids with stained sections were viewed using a JEM-1400 Electron Microscope (JEOL USA Inc., Peabody, MA) controlled by TEM Center for JEM-1400 software (Version: 1.4.2.3071; JEOL USA Inc.). TEM images were processed using Gatan Digital Micrograph software (Version: 1.85.1535; Gatan Inc., Pleasanton, CA).
Image analysis and statistical analysis
TEM images were linearly processed using ImageJ software (Image Processing and Analysis in Java, v1.49g; National Institutes of Health, Bethesda, MD) to optimize brightness and contrast. To determine the number of thin filaments surrounding each thick filament, twenty thick filaments were selected from three different samples in regions of the A-band where thin and thick filaments interdigitate. The number of thin filaments surrounding each thick filament was then counted. Lateral boundaries between adjacent sarcomeres are not immediately apparent in cross sections, but we determined relative differences in sarcomere widths based on the distance between adjacent Z-lines in cross sections, measuring this with ImageJ for ten sarcomeres in each strain in each of two independent experiments. Similarly, the number of thick filaments per sarcomere could not be determined from cross sections, but we determined the relative number of thick filaments in adjacent sarcomeres based on counting the number of thick filaments visible between four to eight sets of adjacent Z-lines in each strain analyzed. These results are reported as the mean ± one standard deviation. Dense body morphologies in ten Z-lines for each strain, in each of two independent experiments, were scored as normal singular projections of electron-dense material, as shredded-appearing projections of electron-dense material, or as largely lacking electron-dense material. For all analyses, only cross-sectional views were used. Measured data were exported to Excel for Mac 2011 software (Version 14.4.4; Microsoft, Redmond, WA). One-way analyses of variation were performed using StatPlus:mac software plug-in (Version: 5.8.2.0; AnalystSoft Inc.), and Fisher’s least significant difference post hoc tests were applied to results. Differences in results exceeding the 99% confidence interval were considered to be statistically significant.
Western blot analysis
Young adult wild-type and fhod-1(−) worms were harvested and washed with M9 supplemented with 0.1% Triton X-100, and suspended in a 1:1 worm-to-M9 slurry, Worm slurries were mixed directly with sample buffer in 1.7 mL tubes, boiled for 3 min, ground for 30 sec with a tissue grinder, boiled a further 3 min, and pelleted 15 sec. Samples were subject to SDS-PAGE and probed by western blot for tubulin to normalize concentrations. Normalized samples were then probed by western blot for tubulin, actin, and muscle-specific myosin heavy chain MYO-3. Mouse monoclonal primary antibodies 5-6 anti-MYO-3 (generated by H.F. Epstein, Baylor College of Medicine, Houston, TX; used at 1:2000 dilution) and AA4.3 anti-tubulin (generated by C. Walsh, University of Pittsburgh, Pittsburgh, PA; used at 1:1000 dilution) were obtained through the Developmental Studies Hybridoma Bank (University of Iowa, Iowa City, IA). Mouse monoclonal primary antibody ACTN05(C4) anti-actin (Abcam, Cambridge, MA) was used at 1:500 dilution, and goat anti-mouse HRP secondary antibody (Rockland Immunochemicals, Gilbertsville, PA) was used at 1:3000 dilution. Band intensities were quantified using ImageJ.
Results
fhod-1 and cyk-1 formin mutant sarcomeres are narrow, but have normal banding patterns of thin and thick filaments
C. elegans BWMs are analogous to vertebrate skeletal muscle in that they are striated muscles with well-defined sarcomeres, and they control body movements. However, BWM cells are also distinct from vertebrate striated muscle cells in several ways (Figure 1). Rather than the cylindrical shape typical of vertebrate muscle cells with sarcomeres packed into elongated myofibrils in the cytoplasm, BWM cells are spindle-shaped cells with a flattened, continuous layer of sarcomeres adjacent to the plasma membrane. Moreover, all Z-line and M-line structures of BWM cells are firmly anchored to the plasma membrane and the adjacent basement membrane by integrin-containing complexes, and thus resemble vertebrate costameres that decorate only the most peripheral sarcomeres in striated muscle cells [16,17]. Furthermore, BWM sarcomeres are packed such that their striations are oriented obliquely to the thin and thick filaments, rather than perpendicular as in vertebrates [1,18]. Consequently, while any given cross section of vertebrate muscle viewed by TEM will reveal the same longitudinal position for all sarcomeres in that section, a similar cross section of BWM reveals the entire spectrum of sarcomere positions as it intersects adjacent sarcomeres (Figure 1, x/y-sections).
We used TEM to examine cross sections of wild-type young adult worms, and young adults bearing mutations of the fhod-1 and cyk-1 formin genes. Notably, both formin gene mutations eliminate sequence that codes for the formin homology-2 domain critical for formin-mediated actin nucleation, and thus are predicted to be null alleles with regard to actin filament assembly [11]. For all worms examined, including fhod-1(−);cyk-1(−) double mutants, complete BWM sarcomeres were apparent (Figure 2). Because preparation of the animals for TEM induced BWM contraction, the thick filament-deficient I-bands were reduced to small strips and thick filament-containing A-bands occupied most of each sarcomere. Electron-dense material usually, but not always (see below), occupied the Z-lines at the center of each I-band.
Figure 2.
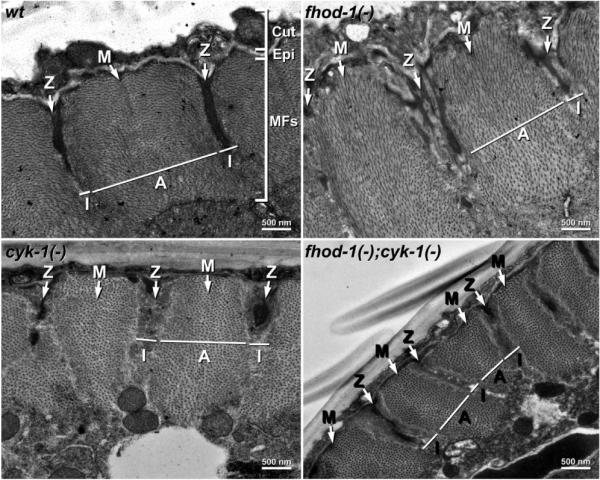
TEM of wild-type and formin mutant sarcomeres. Cross section (x/y) views through the body walls of young adult wild-type and indicated formin mutant worms show the outer cuticle (Cut), the thin epidermis (Epi), and the thick layer of BWM myofilaments (MFs) organized into sarcomeres. The positions of example Z-lines, M-lines, A-bands, and I-bands are indicated.
Considering that formins can stimulate actin filament assembly in vitro and in vivo, we had expected the formin mutants would be specifically deficient in thin filaments. Surprisingly, thin filaments were abundant for all strains near the Z-lines, and were also present through much of the A-bands where they interdigitated with thick filaments (Figure 3). Aside from a small central region of the A-bands, we observed no large regions containing thick filaments but lacking thin filaments, as might have been expected if thin filament assembly were dramatically reduced relative to thick filament formation. However, where thin and thick filaments interdigitated, the packing ratio of thin filaments to thick filaments was moderately less in the fhod-1(−);cyk-1(−) double mutant sarcomeres, with an average of 8.9 thin filaments surrounding each thick filament, compared to an average of 12 in wild-type BWMs [19].
Figure 3.
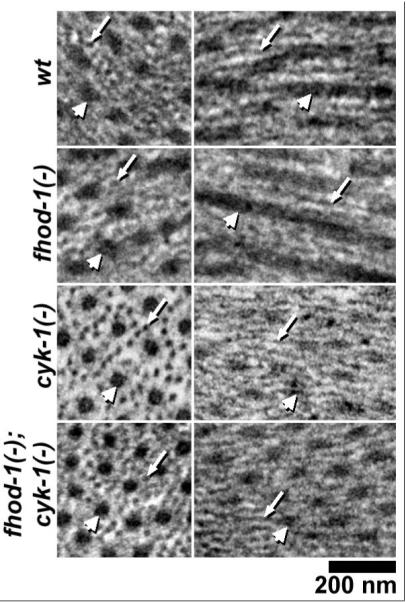
Formin mutant sarcomeres contain thin filaments. Thin filaments (small arrows) and thick filaments (large arrows) are seen in cross- or nearly cross-section (left) and in longitudinal- or nearly longitudinal-section (right) in the indicated worm strains.
fhod-1(−) and cyk-1(−) mutant sarcomeres have reduced amounts of thin and thick filaments
Using the distance from Z-line to Z-line in cross sections as an indication of relative sarcomere width, we observed narrower widths in fhod-1(−) and cyk-1(−) mutants than in wild-type, and even narrower widths in double mutants (Figure 4A). This was mirrored by a significant reduction in the number of thick filaments visible between adjacent Z-lines in cross sections, with only 660 ± 52 thick filaments between Z-lines in fhod-1(−);cyk-1(−) double mutants, compared to 1210 ± 176 thick filaments in wild-type animals (p < 0.001). Because the packing ratio of thin to thick filaments is also modestly reduced in the double mutant, we can also reason that the number of thin filaments in each sarcomere is also reduced. These results, combined with the reduced number of sarcomeres in the formin mutants [11], point to a great loss in the number of thick and thin filaments present in formin mutant BWMs.
Figure 4.
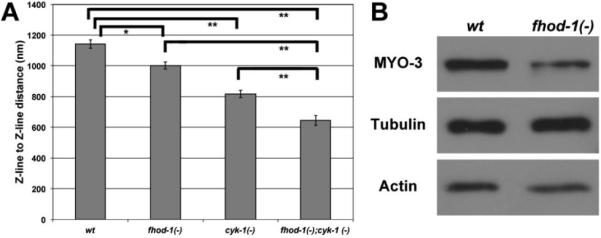
Formin mutants have small sarcomeres, and fhod-1 mutants have reduced levels of some muscle proteins. (A) The distance between Z-lines in x/y cross-sections was measured as a proxy for sarcomere width in the indicated strains. Shown are mean values of two independent experiments (n=10 for each experiment), with standard error of the mean indicated by error bars. Significant differences are indicated by (*) for p<0.01 or (**) for p<0.001. (B) Whole worm extracts of wild-type and fhod-1(−) worms were probed for muscle-specific MYO-3 and muscle-enriched actin, with tubulin used as a loading control. MYO-3 was reduced ~50% and actin is reduced ~10% in the mutant worms relative to wild-type.
To determine whether this reduction in thick and thin filaments reflected a reduced presence of filament components, we probed whole worm extracts of wild-type and fhod-1(−) mutants by western blotting for actin, which is enriched in BWMs, and for a BWM-specific myosin heavy chain, MYO-3, using tubulin as a loading control not specifically expressed in muscle. Importantly, loss of FHOD-1 results in BWM and egg-laying muscle defects, but does not appear to affect non-muscle tissues, validating this comparison [11]. Compared to wild-type animals, fhod-1(−) mutants express approximately 10% less actin, and 50% less MYO-3 (Figure 4B). We did not similarly analyze animals defective for cyk-1, as disruption of cyk-1 by mutation or RNAi impairs development of a variety of non-muscle tissues and results in very poor growth [11], and thus would likely impair expression of any putative loading control, including tubulin.
fhod-1 and cyk-1 mutant sarcomeres have defective Z-line structures
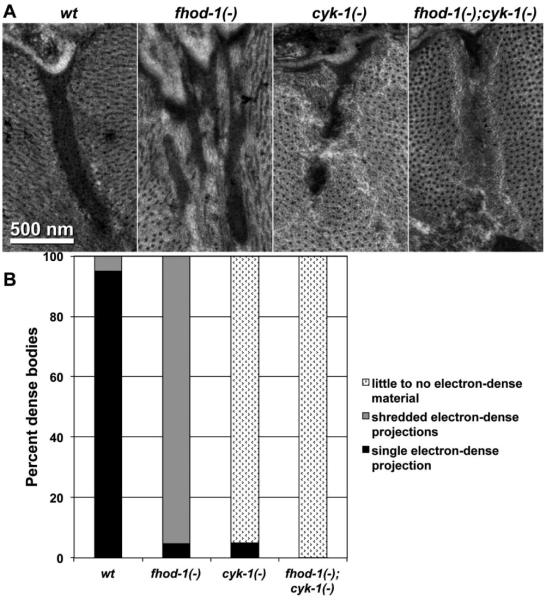
The majority of thin filament-anchoring Z-line structures in BWM cells are dense bodies. These appear as electron-dense finger-like projections that extend from the plasma membrane into the muscle cytoplasm (Figure 5A, wt). Absences of FHOD-1 and CYK-1 affected dense bodies in different ways. The Z-line positions of fhod-1(−) sarcomeres contained electron-dense material, but usually with a shredded appearance rather than singular projections (Figure 5). Conversely, the Z-lines of cyk-1(−) and fhod-1(−);cyk-1(−) sarcomeres often contained very little to no electron-dense material, but were often occupied by an amorphous mass of unidentified material surrounded by the thin filaments (Figure 5).
Figure 5.
(A) Z-lines of worms with the indicated genotypes are shown. A singular electron-dense body is seen in wild-type, while the fhod-1(−) Z-line contains multiple, thinner bodies, and the cyk-1(−) and fhod-1(−);cyk-1(−) Z-lines contain reduced amounts of electron-dense material. (B) Z-lines in worms with the indicated genotypes were visually categorized as containing: singular electron-dense bodies, multiple shredded electron-dense bodies, or very little to no electron-dense material. Results shown are representative of two independent experiments that scored twenty Z-lines of each strain.
Discussion
Genetic studies and in vitro studies with cultured cells have implicated formin family proteins in promoting sarcomere organization. Pointing to their clinical relevance, one allele of the human formin gene fhod3 has been associated with increased incidence of hypertrophic cardiomyopathy, while another allele was present in one case of adult onset dilated cardiomyopathy [20,21]. Moreover, expression of FHOD3 and the related FHOD1 formin change differentially during human cardiomyopathies [10,22], suggesting they may play a role in disease progression, or perhaps compensation.
However, determining the functions of formins in sarcomere formation has been difficult. One complication is the multiplicity of formin isoforms present in mammalian systems. This was recently highlighted by an siRNA-based study that showed 6 mouse formins contribute non-redundantly to cardiomyocyte sarcomere formation or maintenance [23]. Using C. elegans as a simpler animal model, we have shown that only two formins, FHOD-1 and CYK-1, contribute to sarcomere formation and maintenance in the worm's BWMs. Mutations of fhod-1 and cyk-1, but no other formin gene, result in BWM development that is highly stunted, with many fewer sarcomeres assembling per muscle cell [11].
To better understand the contribution of formins to sarcomere organization in this simpler system, we used TEM to examine BWMs in worms mutated singly or doubly for fhod-1 and cyk-1. Considering that many formins stimulate the nucleation and elongation of long, unbranched actin filaments in vitro, the most straightforward hypothesis is that formins initiate the assembly of actin-based thin filaments. However, sarcomeres in formin mutant BWMs, including those of fhod-1(−);cyk-1(−) double mutants, contain thin filaments, and their sarcomeric organization is mostly normal (Figures 2 and 3). Thus, thin filament assembly does not appear to be strictly formin-dependent, but there are two caveats to this conclusion. First, the ratio of thin filaments to myosin-based thick filaments is reduced in the fhod-1(−);cyk-1(−) double mutant, suggesting formins have some preferential stimulatory effect on thin filaments. Second, we derive cyk-1(−) single mutants and fhod-1(−);cyk-1(−) double mutants from heterozygous parents due to the requirement for maternally-provided CYK-1 for embryonic cell division [14,24], and cannot rule out the possibility that maternal CYK-1 drives residual thin filament assembly in these formin mutant BWMs.
Formin mutant sarcomeres exhibit defects beyond thin filament density. They are narrower than wild-type sarcomeres, with each formin loss having an additive effect (Figure 2 and 4A). Also, formin mutant Z-lines are aberrant, with loss of FHOD-1 and CYK-1 having distinct effects. In wild-type sarcomeres, the primary Z-line structures of BWM sarcomeres are finger-shaped projections called dense bodies. These structures anchor thin filaments, and connect the contractile lattice to the extracellular matrix [16]. We had shown that FHOD-1 localizes in a diffuse distribution that envelops and links dense bodies, and CYK-1 colocalizes with dense bodies [11]. By TEM, we see that the absence of FHOD-1 results in thinner, branching strands of electron-dense material at Z-lines, while loss of CYK-1 leaves Z-lines very deficient in electron-dense material, but containing variable amounts of amorphous material surrounded by thin filaments (Figure 5). Thus, CYK-1 may contribute to the formation or maintenance of the electron-dense material at the Z-line, while FHOD-1 may be important for maintaining the coherence of the electron-dense material as a singular dense body. The formin mutant sarcomeres also appear to have wider I-bands than wild-type animals (Figure 2). The reason for this difference is not clear, but might reflect poor contractility of the muscles, particularly for cyk-1(−) mutants that have motility defects [11], or as a secondary consequence of the increased space occupied by the frayed dense bodies of fhod-1(−) mutants.
Our results suggest formins play a general role in sarcomere enlargement, and in the formation of Z-line structures. This resembles a growing body of evidence from other genetic systems. In mice, the FHOD-1-related formin encoded by fhod3 is expressed in heart muscle, and a fhod3 null mutation results in sarcomeres that contain thin filaments, but are sparse and narrow, and have aberrantly small Z-line bodies in the place of normal Z-discs [25]. Similarly, a hypomorphic allele of the mouse formin gene daam1 also results in narrow cardiomyocyte sarcomeres with aberrant Z-discs [26], and disruption of the related Drosophila daam gene has similar effects in somatic and cardiac fly muscles [27]. In all these cases, the mutant sarcomeres are deficient in multiple components, including thin filaments, thick filaments, and Z-line material.
The similarity of the effects of formin loss across these organisms suggests these formins may function in a conserved manner. One possible mechanism for such global effects is that formins might influence production of muscle-specific proteins, as occurs with FHOD-1-related mammalian FHOD1 and CYK-1-related mammalian Dia1 and Dia2 formins in smooth muscle cells [28,29]. Actin polymerization driven by these formins can indirectly trigger myocardin-related transcription factor-dependent expression of smooth muscle cell genes, such as those encoding smooth muscle α-actin or SM22. Consistent with a similar mechanism occurring in C. elegans, worms lacking FHOD-1 have significantly reduced levels of a BWM myosin heavy chain isoform, and even a moderately reduced amount of actin, which is more broadly expressed (Figure 4B). While our results do not rule out direct stimulation of thin filament assembly by formins in striated muscle, they do suggest formins have broader effects on sarcomere formation, possibly through a mechanism similar to what occurs in vertebrate smooth muscle.
Acknowledgements
We would like to thank Nakatsugawa Masako of the Electron Microscopy Core at Upstate Medical University for her help in developing a working EM protocol and with thin-sectioning, Dr. David Mitchell for training on sectioning and staining TEM samples and JEM-1400 Electron Microscope operation and image acquisition, Sharon Chase and Dr. Mira Krendel for general help and advice, and the Caenorhabditis Genetics Center (University of Minnesota) and the National Bioresource Project for the Experimental Animal Nematode C. elegans (Tokyo Women's Medical University School of Medicine) for worm strains. The monoclonal antibodies 5-6 developed by Dr. Henry F. Epstein and AA4.3 developed by Dr. Charles Walsh were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242. The National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number R01AR064760 supported this work.
Abbreviations
- BWM
Body-Wall Muscle
- FH2
Formin Homology-2
- TEM
Transmission Electron Microscopy
References
- 1.Huxley H, Hanson J. Changes in the cross-striations of muscle during contraction and stretch and their structural interpretation. Nature. 1954;173:973–976. doi: 10.1038/173973a0. [DOI] [PubMed] [Google Scholar]
- 2.Ono S. Dynamic regulation of sarcomeric actin filaments in striated muscle. Cytoskeleton. 2010;67:677–692. doi: 10.1002/cm.20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Littlefield R, Almenar-Queralt A, Fowler VM. Actin dynamics at pointed ends regulates thin filament length in striated muscles. Nat Cell Biol. 2001;3:544–551. doi: 10.1038/35078517. [DOI] [PubMed] [Google Scholar]
- 4.Mardahl-Dumesnil M, Fowler VM. Thin filaments elongate from their pointed ends during myofibril assembly in Drosophila indirect flight muscles. J Cell Biol. 2001;155:1043–1053. doi: 10.1083/jcb.200108026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai J, Hartwig JH, Perrimon N. SALS, a WH2-domain-containing protein, promotes sarcomeric actin filament elongation from pointed ends during Drosophila muscle growth. Dev Cell. 2007;13:828–842. doi: 10.1016/j.devcel.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Tsukada T, Pappas CT, Moroz N, Antin PB, Kostyukova AS, et al. Leiomodin-2 is an antagonist of tropomodulin-1 at the pointed end of the thin filaments in cardiac muscle. J Cell Sci. 2010;123:3136–3145. doi: 10.1242/jcs.071837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chereau D, Boczkowska M, Skwarek-Maruszewska A, Fujiwara I, Hayes DB, et al. Leiomodin is an actin filament nucleator in muscle cells. Science. 2008;320:239–243. doi: 10.1126/science.1155313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taniguchi K, Takeya R, Suetsugu S, Kan-O M, Narusawa M, et al. Mammalian formin fhod3 regulates actin assembly and sarcomere organization in striated muscles. J Biol Chem. 2009;284:29873–29881. doi: 10.1074/jbc.M109.059303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takano K, Watanabe-Takano H, Suetsugu S, Kurita S, Tsujita K, et al. Nebulin and N-WASP cooperate to cause IFG-1-induced sarcomeric actin filament formation. Science. 2010;330:1536–1540. doi: 10.1126/science.1197767. [DOI] [PubMed] [Google Scholar]
- 10.Iskratsch T, Lange S, Dwyer J, Kho AL, dos Remedios C, et al. Formin follows function: a muscle-specific isoform of FHOD3 is regulated by CK2 phosphorylation and promotes myofibril maintenance. J Cell Biol. 2010;191:1159–1172. doi: 10.1083/jcb.201005060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mi-Mi L, Votra S, Kemphues K, Bretscher A, Pruyne D. Z-line associated formins promote contractile lattice growth and maintenance in striated muscles of C. elegans. J Cell Biol. 2012;198:87–102. doi: 10.1083/jcb.201202053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol. 2010;11:62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- 13.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swan KA, Severson AF, Carter JC, Martin PR, Schnabel H, et al. cyk-1: a C. elegans FH gene required for a late step in embryonic cytokinesis. J Cell Sci. 1998;111:2017–2027. doi: 10.1242/jcs.111.14.2017. [DOI] [PubMed] [Google Scholar]
- 15.Hall DH. Electron microscopy and three-dimensional image reconstruction. Methods Cell Biol. 1995;48:395–436. doi: 10.1016/s0091-679x(08)61397-7. [DOI] [PubMed] [Google Scholar]
- 16.Moerman DG, Williams BD. Sarcomere assembly in C. elegans muscle. Wormbook. 2006;16:1–16. doi: 10.1895/wormbook.1.81.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sparrow JC, Schöck F. The initial steps of myofibril assembly: integrins pave the way. Nat Rev Mol Cell Biol. 2009;10:293–298. doi: 10.1038/nrm2634. [DOI] [PubMed] [Google Scholar]
- 18.Francis GR, Waterston RH. Muscle organization in Caenorhabditis elegans: localization of proteins implicated in thin filament attachment and I-band organization. J Cell Biol. 1985;101:1532–1549. doi: 10.1083/jcb.101.4.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altun Z, Hall D. In: Muscle system, overview and somatic muscle. Herdon LA, editor. WormAtlas; 2009. [Google Scholar]
- 20.Wooten EC, Hebl VB, Wolf MJ, Greytak SR, Orr NM, et al. Formin Homology 2 Domain Containing 3 variants associated with hypertrophic cardiomyopathy. Circ Cardiovasc Genet. 2013;6:10–18. doi: 10.1161/CIRCGENETICS.112.965277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arimura T, Takeya R, Ishikawa T, Yamano T, Matsuo A, et al. Dilated cardiomyopathy-associated FHOD3 variant impairs the ability to induce activation of transcription factor serum response factor. Circ J. 2013;77:2290–2296. doi: 10.1253/circj.cj-13-0255. [DOI] [PubMed] [Google Scholar]
- 22.Dwyer J, Pluess M, Iskratsch T, DosRemedios CG, Ehler E. The formin FHOD1 in cardiomyocytes. Anat Rec (Hoboken) 2014;297:1560–1570. doi: 10.1002/ar.22984. [DOI] [PubMed] [Google Scholar]
- 23.Rosado M, Barber CF, Berciu C, Feldman S, Birren SJ, et al. Critical roles for multiple formins during cardiac myofibril development and repair. Mol Biol Cell. 2014;25:811–827. doi: 10.1091/mbc.E13-08-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Severson AF, Baillie DL, Bowerman B. A Formin Homology protein and a profilin are required for cytokinesis and Arp2/3-independent assembly of cortical microfilaments in C. elegans. Curr Biol. 2002;12:2066–2075. doi: 10.1016/s0960-9822(02)01355-6. [DOI] [PubMed] [Google Scholar]
- 25.Kan-O M, Takeya R, Abe T, Kitajima N, Nishida M, et al. Mammalian formin Fhod3 plays an essential role in cardiogenesis by organizing myofibrillogenesis. Biol Open. 2012;1:889–896. doi: 10.1242/bio.20121370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li D, Hallett MA, Zhu W, Rubart M, Liu Y, et al. Dishevelled-associated activator of morphogenesis 1 (Daam1) is required for heart morphogenesis. Development. 2011;138:303–315. doi: 10.1242/dev.055566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molnár I, Migh E, Szikora S, Kalmár T, Végh AG, et al. DAAM is required for thin filament formation and sarcomerogenesis during muscle development in Drosophila. PLoS Genet. 2014;10:e1004166. doi: 10.1371/journal.pgen.1004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Staus DP, Blaker AL, Taylor JM, Mack CP. Diaphanous 1 and 2 regulate smooth muscle cell differentiation by activating the myocardin-related transcription factors. Arterioscler Thromb Vasc Biol. 2007;27:478–486. doi: 10.1161/01.ATV.0000255559.77687.c1. [DOI] [PubMed] [Google Scholar]
- 29.Staus DP, Blaker AL, Medlin MD, Taylor JM, Mack CP. Formin homology domain-containing protein 1 regulates smooth muscle cell phenotype. Arterioscler Thromb Vasc Biol. 2011;31:360–367. doi: 10.1161/ATVBAHA.110.212993. [DOI] [PMC free article] [PubMed] [Google Scholar]