Abstract
Background
Single Nucleotide Polymorphisms (SNPs) in the CHI3L1 promoter, the gene encoding YKL-40, are associated with circulating YKL-40 levels and asthma prevalence. However, the effects of gene polymorphisms on asthma severity and airway expression of YKL-40 have not been examined.
Objective
Determine the effect of genetic variation in CHI3L1 on asthma severity and YKL-40 expression in individuals from the Yale Center for Asthma and Airways Disease (YCAAD) and the Severe Asthma Research Program (SARP).
Methods
SNPs spanning the CHI3L1 gene were genotyped in 261 YCAAD and 889 SARP individuals, respectively. Association and haplotype analyses were conducted to identify effects on airflow obstruction, YKL-40 levels and asthma severity.
Results
Fifteen SNPs in CHI3L1 were associated with FEV1 and/or serum YKL-40 levels. Rs12141494 (intron 6) was the only SNP in individuals of European ancestry in both cohorts that was associated with serum YKL-40 levels and post-bronchodilator FEV1. Conditional analysis demonstrated that the effect on lung function was independent of the promoter SNP rs4950928 and haplotype analysis demonstrated that G alleles at rs12141494 and rs4950928 are associated with lower YKL-40 levels and higher FEV1 % predicted. In individuals with asthma, the risk allele A at rs12141494 was associated with severe asthma and higher levels of YKL-40 in the airway (P ≤0.05).
Conclusion
In contrast to the promoter SNP, rs4950928, the intronic SNP, rs12141494 in CHI3L1 is associated with asthma severity, lung function and YKL-40 levels in the blood and airway. These data suggest that SNP rs12141494 modulates expression of YKL-40 in the airway and contributes to airway remodeling and asthma severity.
Keywords: Asthma, Asthma severity, Severe asthma, Genetic association studies, Genetic variation, CHI3L1 protein, human, YKL-40 protein, human Airway remodeling
Introduction
YKL-40, a chitinase-like protein, belongs to the chitinase and chitinase-like family of proteins. These evolutionarily conserved molecules interact with chitin, the second most abundant polysaccharide on earth. Human and mechanistic studies have demonstrated that CHI3L1/YKL-40 plays a role in the pathobiology of asthma by modulating innate and adaptive immune and remodeling responses (1). In the first human asthma studies, we demonstrated that elevated serum levels of YKL-40 correlate with asthma severity, airway remodeling, and increased thickness of the sub-epithelial basement membrane in individuals with asthma (2). This work led to mechanistic studies demonstrating that bronchial epithelial cells exposed to YKL-40 generate higher levels of IL-8 and stimulate smooth muscle proliferation in vitro (3). An additional mechanism of YKL-40 increased bronchial smooth muscle proliferation involves the protease activated receptor-2 (4). Taken together, these studies established an important role for YKL-40 as a molecule uniquely juxtaposed between environmental exposure, inflammation, and the development of airway remodeling and severe asthma.
Genetic studies have also demonstrated that variation in CHI3L1 contributes to the pathogenesis of asthma. A Genome-wide Association Study (GWAS) of serum YKL-40 levels in a founder population of European descent by Ober et al. (5), identified an association between the rs4950928 promoter Single Nucleotide Polymorphism (SNP) and circulating YKL-40 levels. This SNP was also associated with the risk of asthma and FEV1 in three populations of European ancestry with mild asthma. Subsequent reports have shown an association with rs4950928 and asthma (6), although the risk allele was opposite of that reported by Ober (5). Additional publications have been conflicting with respect to the effect of genetic variation in the CHI3L1 gene and asthma (7, 8). To date, the effect of genetic variation in CHI3L1 on asthma severity has not been examined.
To determine the effects of genetic variation in CHI3L1 gene on YKL-40 expression in the airway, airway remodeling and asthma severity we examined two cohorts of individuals with asthma from the Yale Center for Asthma and Airways Disease (YCAAD) and the Severe Asthma Research Program (SARP). We hypothesized that genetic variation in CHI3L1 is associated with persistent airflow obstruction, serum YKL-40 levels, asthma severity, and airway expression of YKL-40. To examine this hypothesis, we characterized the effect of SNPs in the CHI3L1 gene on severe asthma traits, determined the interaction between SNPs by haplotype analysis, and correlated identified SNP with airway expression of YKL-40. Ultimately, we identified a novel polymorphism in CHI3L1 that is likely to contribute to airway remodeling and asthma severity through increased production of YKL-40 in the airway.
Methods
Populations
Yale Center for Asthma and Airways Disease
Study participants in the YCAAD cohort based in New Haven, Connecticut, underwent an extensive phenotypic characterization after Institutional Review Board (IRB) approval. Inclusion, exclusion criteria, and study protocol have been described previously (2). Severe asthma was defined as outlined by the SARP clustering algorithm and was modified as follows: Severe asthma included subjects with a baseline FEV1 of less than 68% of predicted (SARP clusters 4 and 5, while non-severe asthma was defined by the presence of a baseline FEV1 equal or greater than 68% predicted (SARP clusters, 1, 2 and 3) (9). For specific details related to definitions and specific study measurements, see online supplement. A total of 259 individuals from this cohort were analyzed.
The Severe Asthma Research Program
Individuals in the SARP cohort completed study visits using established standard operating procedures as previously described (9). The definition for severe asthma outlined above was also used in this cohort. IRB approval at the SARP institutions was obtained for these studies. The characteristics of these subjects have been reported in previous publications (9, 10). A total of 919 subjects from this cohort were analyzed.
Sputum Induction
Individuals in the YCAAD cohort underwent sputum induction with inhaled hypertonic saline. Mucus plugs were removed using a dissecting microscope and washed to remove squamous cell contamination. The cellular and aqueous compartments were separated, cell counts, cell differentials, and viability were determined (Diff-Quik and trypan blue exclusion). Aliquots of supernatants were stored until processing. Samples with >20% squamous cells were considered contaminated and were not processed further.
Measurement of Serum and Sputum YKL-40 Levels
YKL-40 levels were measured in duplicate in serum and sputum specimens using a commercially available enzyme-linked immunosorbent assay (ELISA) kit for YKL-40 (Quidel). The mean value of the two duplicates was used in the statistical analyses. Duplicate samples with coefficients of variation (CV) greater than 20% were re-assayed. The mean CV for all samples was 10%. The limit of detection is 5.4 ng/mL. Values under the limit of detection were defaulted to 0 ng/mL.
SNP Genotyping
DNA was extracted from peripheral blood leukocytes using standard protocols. The Sequenom MassARRAY ® system (San Diego, California) genotyping platform was used. Genotyping was performed according to the manufacturer's iPLEX application guide at the Center for Human Genomics at Wake Forest University Health Sciences and the Yale Keck DNA sequencing facility for SARP and YCAAD samples, respectively (11). To identify rare and novel SNPs that could be associated with expression of YKL-40, resequencing of the CHI3L1 gene was conducted on a subset of 6 individuals of European Ancestry in the YCAAD cohort 3 with the CC genotype and 3 with GG genotype at the promoter SNP rs4950928, with high and low YKL-40 levels, respectively; amplicon distribution: 13 kilobases (kb) upstream-5′, 7.8 kb CHI3L1 gene, 1.2 kb downstream-3′) as previously described (12). The identified SNPs were compared to SNPs genotyped in previous publications (5, 6). These SNPs were analyzed with Haploview to identify tagging polymorphisms in the CHI3L1 gene using data from the HapMap project (version 3 release 27) (13). A total of 17 SNPs spanning the CHI3L1 gene, identified through sequencing, previous publications (5, 6) and haplotype tagging were genotyped in YCAAD (discovery). Simultaneously, 14 SNPs identified through haplotype tagging using data from the HapMap project (version 3 release 27) (13) were genotyped in SARP (validation). A total of 9 SNPs present in both datasets tagged all other SNPs reported in this study. Genotyping criteria for SNP quality included the removal of SNPs with low call rates (<80%), SNPs that violated Hardy-Weinberg (P-value <0.05), Minor Allele Frequency (MAF) <4%, samples with low call rates (<98%). Pair-wise marker linkage disequilibrium (LD) was estimated using Lewontin's D' statistic and r2 using Haploview (13).
Statistical Analysis
R software (R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria) was used for data analysis. R stats package version 3.0.1, the SNPassoc version 1.8-5, haplo.stats version 1.6.3 and car version 2.0-21 packages were implemented. Continuous variables were tested using non-parametric tests including Kruskal-Wallis test. Categorical variables were analyzed with chi-square tests. Results are reported as medians and Interquartile Ranges [IQRs] unless otherwise specified. Hardy-Weinberg equilibrium calculation was performed to assess the distribution of the SNPs in each group using Haploview (13). The SNPassoc package applies a generalized linear model to generate the association P-values. Using this statistical package dominant, codominant, recessive and additive models of association were tested for post-bronchodilator FEV1 percent predicted, used as a surrogate marker of airway remodeling, and serum YKL-40 levels on the discovery phase of the study. The additive association model was identified as the best-fit based on the Akaike information criterion (AIC) (14), all reported association values were generated using this model and were adjusted for age and gender, based on the effect these two features have on YKL-40 levels (15), pulmonary function (16), and on the best-fit based on the AIC criterion (14). In the discovery cohort (YCAAD) P values were adjusted for multiple testing with the Benjamini-Hochberg procedure (17)(False Discovery Rate (FDR) < 0.1) to balance power and the potential for false positives. While association values in the validation cohort (SARP) were adjusted for multiple testing with the Bonferroni method (P <0.05) to provide a more stringent and reliable measure of true association. We conducted a conditional analysis on individuals of European ancestry with the rs4950928 CC genotype to evaluate for independent associations on non-promoter SNPs, all P values on the conditional analyses were adjusted with the Bonferroni method. Haplotypes were inferred using the expectation maximization algorithm in Haploview (13). Haplotypes with a frequency greater than 5% were considered in the analysis. A generalized linear model using the Expectation Maximization (EM) algorithm described by Excoffier and Slatkin (18) was implemented in the haplotype analysis with the haplo.stats R package. Individuals with asthma and controls were considered in the discovery, validation, haplotype and conditional analyses. The Levene's test for homogeneity of variance on the two cohorts of individuals with asthma was implemented with the car package. The analysis of asthma severity considered asthma subjects only.
Results
Population characteristics and correlation of circulating YKL-40 levels with asthma severity and airway remodeling
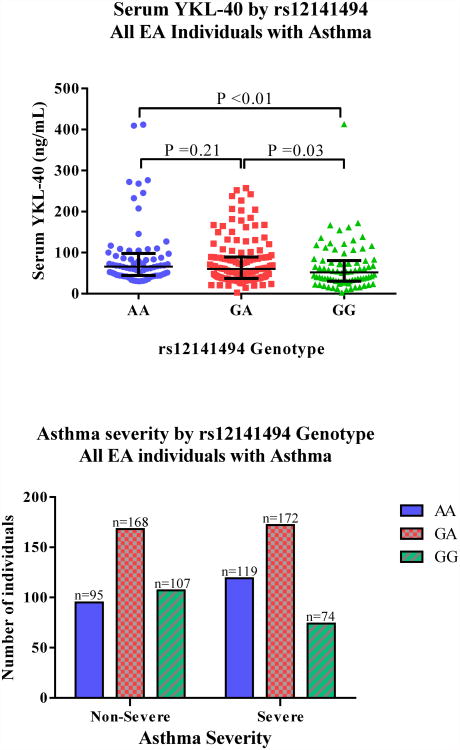
To confirm that serum YKL-40 levels are associated with asthma severity and airway remodeling, two populations of individuals of European ancestry (EA) were evaluated separately: discovery was conducted in YCAAD (n=213; 174 individuals with asthma v. 39 healthy controls) and validation was completed in SARP (n=684; 562 individuals with asthma v. 122 healthy controls). Since the two cohorts are predominantly of EA, individuals of African Ancestry (AA) from both cohorts were pooled together (n=281; 259 individuals with asthma v. 22 healthy controls). Baseline characteristics of all three groups are presented in Table 1. Additional clinical characteristics of the YCAAD cohort are presented in Table E1 (online supplement). Individuals of EA with asthma in YCAAD and SARP had lower FEV1 % predicted compared to healthy controls. These individuals also had higher fractional exhaled nitric oxide (FeNO) and higher serum IgE levels. In individuals of AA, no differences were seen in age, Body Mass Index (BMI), or FeNO between subjects with asthma compared to healthy controls. YKL-40 levels were evaluated in 213 EA YCAAD, 143 EA SARP, and 92 AA and were significantly higher in individuals of EA with severe asthma compared to non-severe, and control subjects (Table 1). YKL-40 was not associated with severe asthma in AA, despite similar median values compared to EA in SARP. This is the result of a lack of statistical power due to the small number of AA individuals with available YKL-40 measurements. Additional characteristics of AA individuals with asthma are presented on Table E2 (online supplement). Post-bronchodilator FEV1 % predicted had a significant correlation with serum YKL-40 levels in individuals of EA in both YCAAD (Spearman rho=-0.19; P <0.01) and SARP (Spearman rho=-0.24; P < 0.01). Concordant with higher serum YKL-40 levels in individuals with severe asthma, the post-bronchodilator FEV1 % predicted was significantly lower in both YCAAD and SARP. These findings indicate that the two EA cohorts have similar characteristics and confirm that serum YKL-40 levels are associated with irreversible airflow obstruction and severe asthma in individuals of European ancestry.
Table 1.
Demographic characteristics.
| European Ancestry YCAAD | Controls n=39 | Non-Severe n=137 | Severe n=37 | P value |
|---|---|---|---|---|
| Age (years) | 45 [31-57] | 46 [36-56] | 59 [50-68] | <0.01 |
| Female sex | 27(69%) | 98 (72%) | 23 (62%) | 0.52 |
| Age of Onset | - | 22 [5-40] | 21 [4-45] | 0.46 |
| BMI (Kg/m2) | 25.5 [23.5-27.8] | 26.3 [22.7-31.2] | 29.3 [25.0-31.5] | 0.08 |
| FEV1 Pre-BD PP | 99 [91-105] | 89 [79-98] | 53 [39-59] | <0.01 |
| FEV1 Post-BD PP | 104 [96-110] | 95 [84-106] | 60 [45-70] | <0.01 |
| FeNO (ppb) | 22 [13-29] | 26 [19-60] | 40 [24-54] | 0.02 |
| Total IgE (IU/mL) | 20 [11-72] | 95 [29-225] | 129 [26-289] | 0.61 |
| Serum YKL-40 (ng/mL)* | 43 [31-67] | 65 [37-105] | 83 [37-110] | 0.01 |
| European Ancestry SARP | Controls n=122 | Non-Severe n=234 | Severe n=328 | P value |
| Age (years) | 27 [23-40] | 31 [22-41] | 41 [31-51] | <0.01 |
| Female sex | 68 (66%) | 168 (72%) | 190 (58%) | 0.29 |
| Age of Onset | - | 10 [4-22] | 12 [3-30] | 0.06 |
| BMI (Kg/m2) | 24.4 [21.7-28.4] | 25.1 [22.1-29.3] | 28.7 [24.4-34.0] | <0.01 |
| FEV1 Pre-BD PP | 98 [93-105] | 89 [82-99] | 60 [43-75] | <0.01 |
| FEV1 Post-BD PP | 105 [97-110] | 97 [89-107] | 71 [36-86] | <0.01 |
| FeNO (ppb) | 14 [11-20] | 26 [17-52] | 29 [17-52] | 0.03 |
| Total IgE (IU/mL) | 35 [15-83] | 117 [49-232] | 115 [29-305] | 0.05 |
| Serum YKL-40 (ng/mL)* | 43 [33-53] | 44 [32-60] | 60 [41-101] | <0.01 |
| African Ancestry SARP and YCAAD | Controls n=22 | Non-Severe n=132 | Severe n=127 | P value |
| Age (years) | 35 [29-40] | 29 [22-40] | 36 [22-45] | 0.79 |
| Female sex | 16 (73%) | 88 (66%) | 82 (65%) | 0.48 |
| Age of Onset | - | 8 [2-18] | 4 [1-17] | 0.66 |
| BMI (Kg/m2) | 28.7 [25.8-34.7] | 29.0 [24.7 -36.6] | 32.3 [23.5-37.5] | 0.50 |
| FEV1 Pre-BD PP | 103 [96-110] | 87 [80-96] | 62 [52-77] | <0.01 |
| FEV1 Post-BD PP | 108 [101-117] | 98 [89-107] | 80 [66-93] | <0.01 |
| FeNO (ppb) | 18 [16-28] | 28 [15-55] | 30 [18-44] | 0.63 |
| Total IgE (IU/mL) | 66 [39-162] | 150 [47-376] | 201 [69-569] | 0.02 |
| Serum YKL-40 (ng/mL)* | 41 [29-47] | 39 [23-65] | 60 [32-79] | 0.33 |
Values expressed as n and (percentage) or median and [interquartile range].
Number (Percentage) with available measurements: European Ancestry YCAAD=213 (100%); European Ancestry SARP=143 (22%); African Ancestry SARP and YCAAD=92 (33%)
Definition of Abbreviations: YCAAD = Yale Center for Asthma and Airways Disease; SARP = Severe Asthma Research Program; BMI = Body Mass Index; FEV1 = Forced Expiratory Volume in One Second; Pre-BD = Pre-bronchodilator; PP= Percent of predicted; Post-BD=Post-bronchodilator; FeNO = Fractional exhaled nitric oxide; ppb = parts per billion; IgE = Immunoglobulin E.
CHI3L1 SNPs and Linkage Disequilibrium Structure
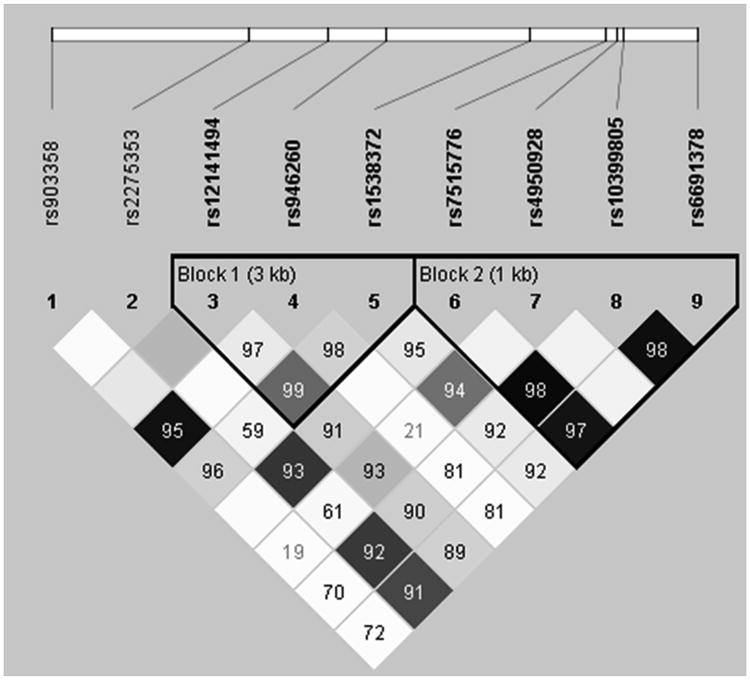
To visualize the LD coefficients in the EA and AA individuals, we generated haplotype maps using Haploview (13). Nine common SNPs in YCAAD and SARP were used to generate a haplotype map for European ancestry, Figure 1 and for African ancestry, Supplemental figure E1A. This analysis revealed two haplotypic blocks, one in the promoter region that includes SNPs in the 5′ region of the gene and the promoter SNP rs4950928, and one including the Intronic/Exonic (I/E) regions of the CHI3L1 gene. These haplotypic blocks show common SNPs and a similar structure to those identified in the HapMap project in the Utah residents with Northern and Western European ancestry from the CEPH collection (CEU group-Figure E1B), and African ancestry in Southwest USA (ASW group- Figure E1C), respectively. This indicates that the SNP distribution and LD patterns seen in our study populations were consistent with HapMap populations of similar ancestry.
Figure 1. Haplotype map for individuals of European ancestry in the CHI3L1 gene in YCAAD and SARP.
Identification of CHI3L1 polymorphisms associated with serum YKL-40 and irreversible airflow obstruction
To identify polymorphisms in CHI3L1 that are associated with serum YKL-40 or post-bronchodilator FEV1 % predicted, individuals of EA in the YCAAD cohort (discovery) were analyzed using an additive model and adjusted for multiple comparisons (FDR < 0.10) as described in methods (Table 2 summarizes the significant associations in the EA and AA groups). Consistent with previous reports (5), multiple SNPs were associated with serum YKL-40 in the discovery cohort including rs4950928. In individuals of EA with the rs4950928 CC genotype serum YKL-40 levels were higher compared to those with the GG genotype (72 vs. 34 ng/mL, P <0.01) (Figure E2A). In addition, four CHI3L1 SNPs (rs12141494, rs880633, rs884209 and rs6698204) were associated with post-bronchodilator FEV1 % predicted (Table 2).
Table 2.
Significant associations of genetic variation in CHI3L1 with lung function and YKL-40 protein levels.
| Cohort | SNP | Position relative to TSS | Genomic coordinates | Location | Minor Allele | MAF | HW P-value | Risk Allele | FEV1 post-BD PP P-value | Serum YKL-40 P-value |
|---|---|---|---|---|---|---|---|---|---|---|
| Discovery EA YCAAD | ||||||||||
| rs884209 | 8462 | 201413912 | 3′ UTR | A | 0.44 | 0.57 | A | 0.085 | 0.160 | |
| rs12141494 | 4326 | 201418048 | Intron 6 | A | 0.45 | 0.44 | A | 0.085 | 0.170 | |
| rs880633 | 2950 | 201419424 | Exon5 | C | 0.45 | 0.53 | C | 0.085 | 0.164 | |
| rs1538372 | 1219 | 201421155 | Intron 2 | A | 0.37 | 0.08 | G | 0.213 | 0.031 | |
| rs4950928 | -131 | 201422505 | Promoter | G | 0.22 | 0.85 | C | 0.213 | 8.1 × 10-5 | |
| rs10399931 | -329 | 201422703 | 5′ UTR | C | 0.26 | 0.52 | C | 0.136 | 1.1 × 10-4 | |
| rs10920579 | -3221 | 201425595 | 5′ UTR | A | 0.21 | 0.96 | G | 0.221 | 6.7 × 10-5 | |
| rs6698204 | -3432 | 201425806 | 5′ UTR | G | 0.44 | 0.58 | A | 0.085 | 0.039 | |
| rs4950929 | -4375 | 201426749 | 5′ UTR | G | 0.21 | 0.96 | T | 0.221 | 6.7 × 10-5 | |
| rs946263 | -9630 | 201432004 | 5′ UTR | G | 0.19 | 0.95 | A | 0.198 | 6.7 × 10-5 | |
| rs2486064 | -13013 | 201435387 | 5′ UTR | A | 0.41 | 0.38 | G | 0.660 | 0.030 | |
| Validation EA SARP | ||||||||||
| rs2364572 | 9165 | 201413209 | 3′ UTR | T | 0.04 | 0.42 | T | 0.006 | NA | |
| rs2275353 | 5678 | 201416696 | Intron 7 | A | 0.18 | 1.00 | G | 0.017 | 1 | |
| rs12141494 | 4326 | 201418048 | Intron 6 | G | 0.46 | 0.75 | A | 0.005 | 0.001 | |
| rs1538372 | 1219 | 201421155 | Intron 2 | A | 0.31 | 1 | G | 1 | 0.006 | |
| rs4950928 | -131 | 201422505 | Promoter | G | 0.20 | 0.82 | C | 1 | 0.011 | |
| rs10399805 | -247 | 201422621 | 5′ UTR | A | 0.14 | 0.65 | G | 0.035 | 1 | |
| rs946261 | -2122 | 201424496 | 5′ UTR | G | 0.38 | 0.27 | A | 0.011 | 0.053 | |
| African Ancestry | ||||||||||
| rs6691378 | -1371 | 201423745 | 5′ UTR | A | 0.27 | 0.72 | A | 1 | 0.006 |
For all the genotyped SNPs and phenotypic associations, see online supplement, Table E2.
Genomic coordinates in Chromosome 1 using the Human Genome NCBI Build 36.1 (March, 2006)
All P values are adjusted for age and gender. Followed by correction for multiple comparison. FDR threshold for the discovery-EA YCAAD cohort < 0.1. Bonferroni threshold for the Validation EA SARP and AA cohorts < 0.05
Definition of Abbreviations: SNP = Single Nucleotide Polymorphism; TSS: Transcription start site; MAF = Minor Allele Frequency; HW = Hardy-Weinberg; FEV1 = Forced Expiratory Volume in One Second; PP= Percent of predicted; Post-BD=Post-bronchodilator; YCAAD = Yale Center for Asthma and Airways Disease; SARP = Severe Asthma Research Program; UTR = Untranslated Region.
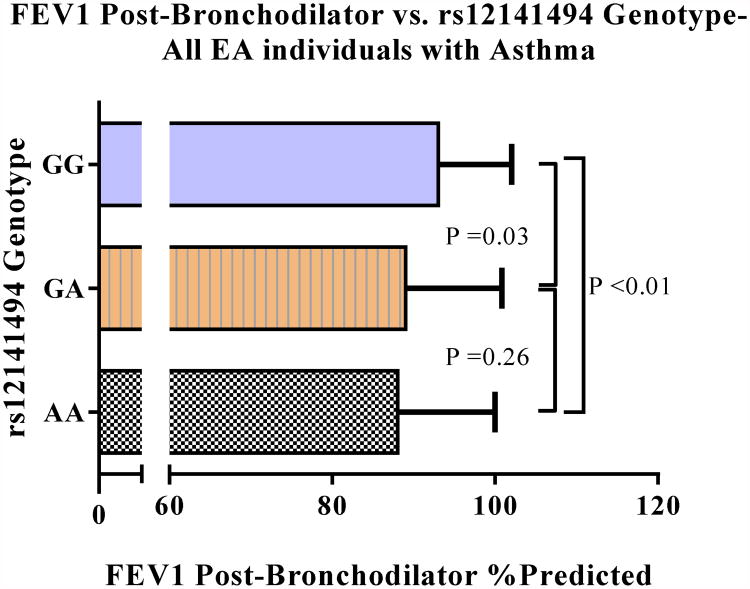
Validation of SNPs with an effect on serum YKL-40 levels and post-bronchodilator FEV1 % predicted that were identified in the discovery cohort was conducted in individuals of EA in the SARP cohort. These association values were adjusted for multiple comparisons (Bonferroni < 0.05) as described in methods (Table 2). This analysis identified multiple SNPs associated with serum YKL-40 or post-bronchodilator FEV1. Consistent with the findings in the discovery cohort, individuals of EA with the rs4950928 CC genotype have higher serum YKL-40 levels compared to those with the GG genotype (53 vs. 16 ng/mL, P <0.01) (Figure E2B). Importantly, of the SNPs identified in the discovery cohort, only rs12141494 was significantly associated with both post-bronchodilator FEV1 % predicted (P < 0.01) and serum YKL-40 levels (P < 0.01) in the SARP cohort. Specifically the rs12141494 A allele was associated with decreased post-bronchodilator FEV1 percent predicted (Effect size) (95% Confidence Interval (CI)): -3.6 (-5.6/-1.6)). No associations were seen between the promoter SNP rs4950928 and pulmonary function in any of the cohorts (Table 2). In individuals of African ancestry only one SNP was associated with the quantitative traits evaluated, therefore, we restricted further analyses to individuals of European ancestry. Taken together, these analyses indicate that the non-promoter SNP, rs12141494 in intron 6, is strongly associated with persistent airflow obstruction and serum YKL-40 levels in individuals of European ancestry.
Effect of CHI3L1 Haplotypes on serum YKL-40 and lung function
To define potential interactions of CHI3L1 SNPs validated above on lung function and YKL-40 levels, a haplotype model was tested on all individuals of EA with asthma and healthy controls from the YCAAD and SARP cohorts. Of the nine SNPs genotyped in both cohorts, five SNPs were selected for this analysis based on their significant association with FEV1 % predicted or serum YKL-40 levels in both the discovery and validation steps (Table 2). Haplotype A present in 18% of individuals of EA, involving the SNP interaction between the minor allele in the promoter SNP rs4950928 (G) and the (G) allele in rs12141494 was associated with improved post-bronchodilator FEV1 and lower YKL-40 (Table 3). While haplotype B in 14 % of the population was associated with improved post-bronchodilator FEV1, involving the (G) allele in rs12141494. This analysis demonstrates that G alleles at both the promoter and the rs12141494 intronic SNP have a protective effect characterized by lower YKL-40 levels and higher post-bronchodilator FEV1 % predicted, demonstrating an interaction between promoter and non-promoter SNPs in the CHI3L1 gene.
Table 3. Haplotype analysis in individuals of European ancestry in YCAAD and SARP.
| Haplotype | Frequency | Phenotype | Effect size and direction | P Value | rs2275353 | rs12141494 | rs1538372 | rs495928 | rs1399805 |
|---|---|---|---|---|---|---|---|---|---|
| A | 18% | FEV1 PP Post-BD | 4 | <0.01 | G | G | A | G | G |
| A | 20% | Serum YKL-40 | -24 | <0.01 | G | G | A | G | G |
| B | 14% | FEV1 PP Post-BD | 6 | <0.01 | A | G | G | C | A |
| B | 15% | Serum YKL-40 | -0.6 | 0.93 | A | G | G | C | A |
| C | 12% | FEV1 PP Post-BD | 2 | 0.19 | G | G | A | C | G |
| C | 13% | Serum YKL-40 | -0.1 | 0.99 | G | G | A | C | G |
| Base | 51% | G | A | G | C | G |
Identification of CHI3L1 SNPs associated with asthma severity
To determine the effect CHI3L1 SNPs on asthma severity, individuals of EA with asthma from YCAAD and SARP were combined (n=736). The Levene's test for homogeneity of variance was used before combining individuals from the two cohorts. Asthma severity and serum YKL-40 levels were included as independent variables. No differences in the variability of the two groups were seen. A linear regression analysis using an additive model of association was conducted in the combined group to identify their relationship with asthma severity (Table 4). The Hardy-Weinberg values for these SNPs were evaluated before implementing the association test to maintain the fundamental assumptions of equilibrium seen in the discovery and validation groups (Table 4). Of the SNPs evaluated, only rs12141494 was associated with asthma severity in individuals of EA (Bonferroni-adjusted P=0.04). The G allele had a protective effect on severity (OR (95% CI): 0.7 (0.6/0.9)), and was associated with lower serum YKL-40 levels (Bonferroni-adjusted P 0.04; Effect size (95% CI): -14.3 (-24.1/-4.4) ng/mL) and higher post-bronchodilator FEV1 % predicted (Bonferroni-adjusted P <0.01; Effect size (95% CI): 3.8 (1.9/5.7) % predicted). This observation indicates that rs12141494 is strongly associated with the severe asthma phenotype, including SARP defined severity, serum YKL-40 levels and persistent airflow obstruction.
Table 4.
Significant SNP associations in individuals of European ancestry with asthma.
| SNP | Location | Minor Allele | MAF | HW P-value | Risk Allele | FEV1 post-BD PP | Serum YKL-40 | Asthma Severity |
|---|---|---|---|---|---|---|---|---|
| rs12141494 | Intron 6 | G | 0.47 | 0.11 | A | 9.2 × 10-4 | 0.043 | 0.040 |
| rs1538372 | Intron 2 | A | 0.31 | 0.07 | G | 0.348 | 0.012 | 1 |
| rs4950928 | Promoter | G | 0.20 | 0.16 | C | 0.633 | 1.2 × 10-5 | 1 |
Effect of rs12141494 on the expression of YKL-40 in the airway
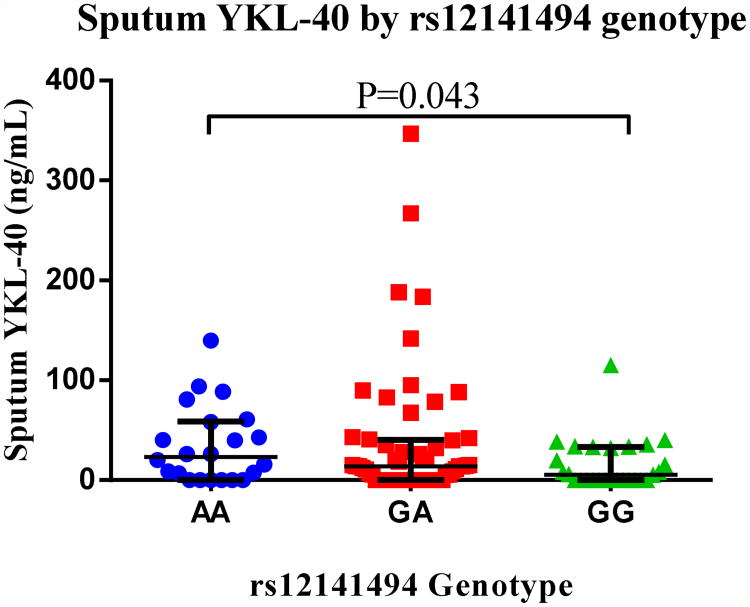
To determine the contribution of rs12141494 to the airway expression of YKL-40, we quantified the levels of YKL-40 in the sputum supernatants of individuals with asthma in the YCAAD cohort that underwent sputum induction as described in methods. Individuals of EA homozygous for the risk allele A had higher YKL-40 sputum levels compared to individuals homozygous for the G allele (23 [0-59] vs. 6 [0-33], ng/mL, P =0.043) (Figure 3). The promoter SNP rs4950928 was not associated with sputum levels (data not shown). This observation, shows that rs12141494 has an effect on the expression of YKL-40 in the airway of individuals with asthma and is consistent with the findings that the rs12141494 G allele is associated with lower levels of YKL-40, milder asthma and better lung function compared to individuals carrying the A allele.
Figure 3.
The rs12141494 AA genotype in individuals of European ancestry with asthma is associated with higher YKL-40 sputum levels.
In silico analysis of functional genomic impact of intronic CHI3L1 SNPs
An in silico analysis for the functional impact of rs12141494 was conducted using the Human Splicing Finder Version 2.4.1 (http://www.umd.be/HSF/HSF.html). The exon 6 CHI3L1 sequence was analyzed by selecting 1000 bp on the 5′ and 3′ flanks of exon 6 to include rs12141494. Mutation analysis of the position 1344 from C to T (complementary to G and A), identified enhancement in the splice site strength according to the Max Entropy algorithm (19), suggesting a potential effect of rs12141494 on pre-messenger RNA processing. Analysis of CHI3L1 mRNA expression in the circulation of a subset of individuals in YCAAD demonstrates a significant genotype effect concordant with that seen at the protein level in the airway (data not shown).
Discussion
These studies are the first to examine the effect of genetic variation in CHI3L1 on asthma severity and demonstrate that the intronic SNP, rs12141494, is associated with disease severity, airflow obstruction, and YKL-40 levels in the serum. This effect is independent of the promoter SNP rs4950928 and rs12141494 was the only SNP found to be associated with airway expression of the CHI3L1 gene product, YKL-40. Taken together, these findings suggest that rs12141494 contributes to asthma severity by altering airway expression of YKL-40 and contributes to airway remodeling, persistent airflow obstruction, and asthma severity.
Previous population studies of genetic variation in the CHI3L1 gene and asthma have shown an association with the promoter SNP rs4950928, asthma prevalence, and YKL-40 expression, however, the risk allele reported in the studies has differed. A European population study (6), identified the G allele in rs4950928 as the risk allele for atopic asthma in never smokers, while the study by Ober et al (5) reported that the C allele was associated with an increased risk of asthma and higher YKL-40 levels in the circulation. An additional publication has suggested that CHI3L1 polymorphisms are not associated with asthma (7), and in Korean children, other promoter polymorphisms are associated with atopy but not with asthma (8). In the current study, we confirmed that the C allele at rs4950928 is associated with higher serum levels of YKL-40 in two cohorts of individuals of European ancestry (Figure E2). Our study was underpowered to detect associations in individuals of African ancestry, therefore additional studies are needed to evaluate these associations in these populations. Interestingly, we did not find that the rs4950928 polymorphism was associated with airflow obstruction or airway levels of YKL-40 in either of the two populations of asthmatics that were examined which suggests that this polymorphism does not directly contribute to asthma severity.
Since serum YKL-40 levels were associated with severe asthma and lung function impairment, we suspected that other promoter or non-promoter polymorphisms could contribute to severe asthma. This led us to discover that a novel intronic SNP, rs12141494, is associated with airway expression of YKL-40 and persistent airflow obstruction in severe asthma. This SNP was not examined in previous genetic studies of CHI3L1 in asthma and was identified by overlapping direct DNA sequencing data of the CHI3L1 gene and HapMap data. Analysis of the CHI3L1 LD structure shows that rs12141494 resides in intron 6 within one of two haplotypic blocks separate from the promoter SNP rs4950928 (Figure 1). This SNP was associated with post-bronchodilator FEV1 % predicted and serum YKL-40 levels, and unlike rs4950928, was associated with the levels of YKL-40 in the sputum. The observation that the additive model had the best fit in conjunction with the quantitative differences seen in these phenotypes, suggests that there is a gene-dosage effect of the risk allele in these associations. The effect of this SNP on lung function persisted after controlling for the risk genotype (CC) at rs4950928 (Table E4) and evaluation of haplotypes, with the A allele associated with higher serum YKL-40 levels, lower lung function, and more severe asthma (Table 3). Individuals with asthma that are homozygous for the A allele of rs12141494 have significantly higher YKL-40 sputum levels, compared to homozygotes for the G allele, further supporting our finding that the rs12141494 genotype has a functional effect on the airway expression of YKL-40 (Figure 3). This suggests that rs12141494 is linked to severe asthma traits, whereas rs4950928 is not. Rs12141494 may contribute to circulating YKL-40 levels through production of YKL-40 in the airway and this effect may be enhanced by the presence of the rs4950938 C allele.
What are the possible functional effects of rs12141494 on the architectural changes characteristic of airway remodeling? The presence of higher YKL-40 levels in the airway of individuals with asthma and the rs12141494 A allele suggests that increased expression of YKL-40 is the primary effect of this polymorphism. Bronchial epithelial cells exposed to YKL-40 have been shown to increase expression of IL-8, which, in turn, leads to proliferation and migration of bronchial smooth muscle cells in vitro (3). In addition, epithelial expression of YKL-40 correlates with bronchial smooth muscle mass and activation of the protease activated receptor-2, AKT, ERK and p38 pathways (4). Therefore, higher local production of YKL-40 in the bronchial epithelium in individuals with the AA rs12141494 genotype may contribute to changes in airway structure, persistent airflow obstruction, and higher levels of YKL-40 in the circulation. This is supported by the results of the haplotype analysis that reveal that the G alleles at both SNPs are associated with lower levels of YKL-40 (Table 3). In addition, in silico analysis suggests that the effect of the rs12141494 A allele on pre-messenger RNA processing enhances the strength of the intron 6 splice site, resulting in increased CHI3L1 messenger RNA and YKL-40 protein levels. While analysis of mRNA expression in the circulation of a subset of individuals in YCAAD demonstrates a genotype effect concordant with that seen at the protein level in the airway.
In summary, these studies identify a novel SNP in CHI3L1, rs12141494, that is associated with persistent airflow obstruction, YKL-40 levels in the airway and blood, and severe asthma. The functional effect of the risk allele A may increase YKL-40 in the airway and circulation and may have synergistic effects in conjunction with the CHI3L1 SNP rs4950928. We used a rigorous definition of asthma, two cohorts of individuals with asthma that are representative of centers across the United States, and a single genotyping methodology to reduce bias to conduct these studies. While future studies will determine the effect of rs12141494 on other ethnic groups and the cellular control of YKL-40 synthesis and expression, these observations add to the body of evidence demonstrating that genetic variation in CHI3L1 and YKL-40 play important roles in the development of airway remodeling and severe asthma.
Supplementary Material
Figure E1. Haplotype maps for the CHI3L1 gene in individuals of African ancestry in YCAAD and SARP (A), CEU HapMap (B) and ASW HapMap (C).
Figure E2. Variation in the promoter SNP rs4950928 in the CHI3L1 gene and effect in YKL-40 protein levels in individuals of European ancestry in YCAAD (A) and SARP (B).
Figure E3. rs12141494 and airflow obstruction in individuals of European Ancestry in YCAAD (A) and SARP (B)
Table E1. Asthma Related Healthcare Utilization, Symptom Control and Comorbidities, YCAAD
Values expressed as median and [interquartile range] or percentages.
Definition of Abbreviations: ACT = Asthma Control Test; OCS = Oral corticosteroid; LABA = Long acting beta-agonist;
*Includes individuals using tiotropium or ipratropium
Table E2. Distribution of individuals with asthma of African ancestry per cohort.
*Number (Percentage) of individuals with available measurements: African Ancestry YCAAD=46 (100%); African Ancestry SARP=46 (20%).
Table E3. Association of Genetic variation in CHI3L1 with airflow obstruction phenotypes and YKL-40 protein levels
Definition of Abbreviations: EA: European Ancestry; TSS: Transcription Start Site; YCAAD = Yale Center for Asthma and Airways Disease; SARP = Severe Asthma Research Program; MAF: Minor Allele Frequency; HW: Hardy-Weinberg; FEV1 = Forced Expiratory Volume in One Second; Post-BD=Post-bronchodilator; UTR: Untranslated Region.
Table E4. Significant associations in individuals of European ancestry restricted to individuals with the CC genotype at rs4950928.
Definition of Abbreviations: FEV1 = Forced Expiratory Volume in One Second; Post-BD=Postbronchodilator
Table E5. SNP associations in individuals of European ancestry with asthma.
Definition of Abbreviations: MAF: Minor Allele Frequency; HW: Hardy-Weinberg; FEV1 = Forced Expiratory Volume in One Second; Post-BD=Post-bronchodilator; UTR: Untranslated Region.
Table E6. Unadjusted SNP associations in individuals of European ancestry with asthma per cohort
Figure 2.
The A allele of rs12141494 in individuals of European ancestry with asthma is associated with worse airflow obstruction (A), higher serum YKL-40 levels (B) and increased asthma severity (C).
Key Messages.
Intronic variation, rs12141494, in the CHI3L1 gene encoding YKL-40 is associated with persistent airflow obstruction, airway remodeling and severe asthma.
Individuals with the AA risk genotype at rs12141494 have higher sputum levels of YKL-40, suggesting that its effect may be mediated through increased protein levels in the airway.
Acknowledgments
Sources of support - NIH Training grants T32HL007778-18 and T15LM007056-26.
R01HL095390-03. R01HL069116, and GCRC RR03186.
-FAMRI Young Clinical Scientist Award 113393
Abbreviations
- SNP
Single Nucleotide Polymorphism
- CHI3L1
Chitinase 3-like 1
- YCAAD
Yale Center for Asthma and Airways Disease
- SARP
Severe Asthma Research Program
- FEV1
Forced Expiratory Volume 1 second
- GWAS
Genome-wide Association Study
- IRB
Institutional Review Board
- ELISA
Enzyme-linked immunosorbent assay
- CV
Coefficient of variation
- kb
kilobase
- MAF
Minor Allele Frequency
- LD
Linkage Disequilibrium
- AIC
Akaike Information Criterion
- FDR
False Discovery Rate
- IQR
Interquartile Range
- EM
Expectation Maximization
- EA
European Ancestry
- AA
African Ancestry
- FeNO
Fractional exhaled nitric oxide
- IgE
Immunoglobulin E
- BMI
Body Mass Index
- CEU
Utah residents with Northern and Western European Ancestry from the CEPH collection
- ASW
African ancestry in Southwest USA
- DNA
Deoxyribonucleic acid
- RNA
Ribonucleic acid
- OR
Odds Ratio
- CI
Confidence Interval
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lee CG, Dela Cruz CS, Herzog E, Rosenberg SM, Ahangari F, Elias JA. YKL-40, a chitinase-like protein at the intersection of inflammation and remodeling. Am J Respir Crit Care Med. 2012;185(7):692–4. doi: 10.1164/rccm.201202-0203ED. [DOI] [PubMed] [Google Scholar]
- 2.Chupp GL, Lee CG, Jarjour N, Shim YM, Holm CT, He S, et al. A chitinase-like protein in the lung and circulation of patients with severe asthma. N Engl J Med. 2007;357(20):2016–27. doi: 10.1056/NEJMoa073600. [DOI] [PubMed] [Google Scholar]
- 3.Tang H, Sun Y, Shi Z, Huang H, Fang Z, Chen J, et al. YKL-40 induces IL-8 expression from bronchial epithelium via MAPK (JNK and ERK) and NF-κB pathways, causing bronchial smooth muscle proliferation and migration. J Immunol. 2013;190(1):438–46. doi: 10.4049/jimmunol.1201827. [DOI] [PubMed] [Google Scholar]
- 4.Bara I, Ozier A, Girodet PO, Carvalho G, Cattiaux J, Begueret H, et al. Role of YKL-40 in bronchial smooth muscle remodeling in asthma. Am J Respir Crit Care Med. 2012;185(7):715–22. doi: 10.1164/rccm.201105-0915OC. [DOI] [PubMed] [Google Scholar]
- 5.Ober C, Tan Z, Sun Y, Possick JD, Pan L, Nicolae R, et al. Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N Engl J Med. 2008;358(16):1682–91. doi: 10.1056/NEJMoa0708801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rathcke CN, Holmkvist J, Husmoen LL, Hansen T, Pedersen O, Vestergaard H, et al. Association of polymorphisms of the CHI3L1 gene with asthma and atopy: a populations-based study of 6514 Danish adults. PLoS One. 2009;4(7):e6106. doi: 10.1371/journal.pone.0006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu AC, Lasky-Su J, Rogers CA, Klanderman BJ, Litonjua A. Polymorphisms of chitinases are not associated with asthma. J Allergy Clin Immunol. 2010;125(3):754–7. 7.e1–7.e2. doi: 10.1016/j.jaci.2009.12.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sohn MH, Lee JH, Kim KW, Kim SW, Lee SH, Kim KE, et al. Genetic variation in the promoter region of chitinase 3-like 1 is associated with atopy. Am J Respir Crit Care Med. 2009;179(6):449–56. doi: 10.1164/rccm.200809-1422OC. [DOI] [PubMed] [Google Scholar]
- 9.Moore WC, Meyers DA, Wenzel SE, Teague WG, Li H, Li X, et al. Identification of asthma phenotypes using cluster analysis in the Severe Asthma Research Program. Am J Respir Crit Care Med. 2010;181(4):315–23. doi: 10.1164/rccm.200906-0896OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore WC, Bleecker ER, Curran-Everett D, Erzurum SC, Ameredes BT, Bacharier L, et al. Characterization of the severe asthma phenotype by the National Heart, Lung, and Blood Institute's Severe Asthma Research Program. J Allergy Clin Immunol. 2007;119(2):405–13. doi: 10.1016/j.jaci.2006.11.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet. 2009 doi: 10.1002/0471142905.hg0212s60. Chapter 2:Unit 2.12. [DOI] [PubMed] [Google Scholar]
- 12.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74(12):5463–7. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 14.AKAIKE H. A New Look at the Statistical Model Identification. IEEE Transactions on Automatic Control. 1974:716–23. [Google Scholar]
- 15.Bojesen SE, Johansen JS, Nordestgaard BG. Plasma YKL-40 levels in healthy subjects from the general population. Clin Chim Acta. 2011;412(9-10):709–12. doi: 10.1016/j.cca.2011.01.022. [DOI] [PubMed] [Google Scholar]
- 16.Crapo RO, Morris AH, Gardner RM. Reference values for pulmonary tissue volume, membrane diffusing capacity, and pulmonary capillary blood volume. Bull Eur Physiopathol Respir. 1982;18(6):893–9. [PubMed] [Google Scholar]
- 17.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995;57:12. [Google Scholar]
- 18.Excoffier L, Slatkin M. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol. 1995;12(5):921–7. doi: 10.1093/oxfordjournals.molbev.a040269. [DOI] [PubMed] [Google Scholar]
- 19.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11(2-3):377–94. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure E1. Haplotype maps for the CHI3L1 gene in individuals of African ancestry in YCAAD and SARP (A), CEU HapMap (B) and ASW HapMap (C).
Figure E2. Variation in the promoter SNP rs4950928 in the CHI3L1 gene and effect in YKL-40 protein levels in individuals of European ancestry in YCAAD (A) and SARP (B).
Figure E3. rs12141494 and airflow obstruction in individuals of European Ancestry in YCAAD (A) and SARP (B)
Table E1. Asthma Related Healthcare Utilization, Symptom Control and Comorbidities, YCAAD
Values expressed as median and [interquartile range] or percentages.
Definition of Abbreviations: ACT = Asthma Control Test; OCS = Oral corticosteroid; LABA = Long acting beta-agonist;
*Includes individuals using tiotropium or ipratropium
Table E2. Distribution of individuals with asthma of African ancestry per cohort.
*Number (Percentage) of individuals with available measurements: African Ancestry YCAAD=46 (100%); African Ancestry SARP=46 (20%).
Table E3. Association of Genetic variation in CHI3L1 with airflow obstruction phenotypes and YKL-40 protein levels
Definition of Abbreviations: EA: European Ancestry; TSS: Transcription Start Site; YCAAD = Yale Center for Asthma and Airways Disease; SARP = Severe Asthma Research Program; MAF: Minor Allele Frequency; HW: Hardy-Weinberg; FEV1 = Forced Expiratory Volume in One Second; Post-BD=Post-bronchodilator; UTR: Untranslated Region.
Table E4. Significant associations in individuals of European ancestry restricted to individuals with the CC genotype at rs4950928.
Definition of Abbreviations: FEV1 = Forced Expiratory Volume in One Second; Post-BD=Postbronchodilator
Table E5. SNP associations in individuals of European ancestry with asthma.
Definition of Abbreviations: MAF: Minor Allele Frequency; HW: Hardy-Weinberg; FEV1 = Forced Expiratory Volume in One Second; Post-BD=Post-bronchodilator; UTR: Untranslated Region.
Table E6. Unadjusted SNP associations in individuals of European ancestry with asthma per cohort