Abstract
In cancer patients, granulocytic myeloid derived suppressor cells (G-MDSCs) expand in number, infiltrating tumor and lymphatic tissues where they suppress an anti-tumor immune response. We report here the development of a liposomal drug delivery system that selectively targets GMDSCs. The liposomes form a disulfide bond with activated complement C3 after intravenous injection and are taken up by G-MDSCs, which express the receptor for activated C3. In vitro experiments utilizing serum from a C3 knockout mouse demonstrate that G-MDSCs take up these liposomes in a C3-dependent manner. After systemic administration to tumor bearing mice, liposomes were incorporated by 22% of G-MDSCs in the blood and were also present in a percentage of G-MDSCs in the tumor (11%), spleen (22%), liver (35%) and lungs (26%). This liposomal system offers a versatile means of targeted drug delivery to G-MDSCs and could be an important tool for restoring anti-tumor immunity in cancer patients.
Keywords: cancer, immunotherapy, myeloid derived suppressor cells, MDSC, liposome, complement C3
Background
Myeloid-derived suppressor cells (MDSCs) infiltrate tumor and lymphoid tissues of cancer patients, inhibiting a T cell response and playing a major role in creating an immunosuppressive environment that allows for unchecked tumor growth and metastasis 1. In human patients, the number of MDSCs increases with cancer stage and poor prognosis. Treatments that repolarize the MDSCs towards an immunostimulatory phenotype have been shown to restore T cell activity and drive the immune system towards an anti-tumor Th1-mediated response 2, 3. Additionally, in tumor bearing mice, depleting, inhibiting or repolarizing MDSCs helps to restore immune function and augments the anti-tumor immune response when combined with immunotherapies 4, 5. Drugs that are somewhat effective in depleting or inhibiting MDSC function include Sunitinib, Axitinib and Gemcitabine, while all-trans-retinoic acid (ATRA) and vitamin D3 have been shown to restore the natural immunostimulatory phenotype of MDSCs 5. Given the growing array of MDSC-effective drugs and the recognition of MDSC as a central player in tumor growth and metastasis, drug delivery techniques including nanoparticle technologies must be developed to aid in the complex targeting of therapeutic agents to MDSC.
MDSCs are composed of monocytic (M-MDSC) and granulocytic (G-MDSC) cell types, both of which suppress T cell function and lead to immune evasion. Both M-MDSCs and G-MDSCs inhibit T cell function by releasing arginase-1 and inducibile nitric oxide synthase which depletes L-arginine necessary for T cell function 6. In addition, MDSCs inhibit T cell function through the release of reactive oxygen species. In mice M-MDSCs are characterized by surface markers CD11b+Ly6C+ while G-MDSCs are characterized by CD11b+Ly6G+Ly6Cmid 7. In humans, who do not have the Ly6C and Ly6G markers, G-MDSCs are typically CD15+CD33+CD11b+HLA-DRlo/neg while M-MDSCs are CD14+CD33+CD11b+HLA-DRlo/neg 8. The difference in surface markers between the two species makes it challenging to develop antibody-conjugated liposomes and nanoparticles targeting MDSCs in a mouse model that will translate directly to a human model. The one surface marker that is common to both M-MDSC and G-MDSC in mice and humans is CD11b, indicative of the complement 3 receptor. To take advantage of this common target, we have recently developed a method that recruits endogenous activated complement C3 of tumor bearing mice to target liposomes for uptake by MDSC cells via the receptor for activated C3.
Liposomes can improve drug delivery efficiency by favorably changing the biodistribution and pharmacokinetics of encapsulated drug compared to unencapsulated drug, but eventually they are cleared from circulation by macrophages of the reticuloendothelial system (RES), primarily in the spleen and liver 9–11. To target a specific cell population other than RES macrophages generally requires conjugating the liposome to a targeting antibody or ligand 12, 13. However, antibodies are expensive, difficult to conjugate and often lose their efficacy when lyophilized for storage, which partly explains why there are no targeted liposome formulations that have advanced to the clinic 14. The method for targeting MDSCs that we have developed differs, because the targeting ligand is the endogenous activated C3 which conjugates to the liposomes subsequent to intravenous injection of the liposomes. The liposomes contain a lipid that is conjugated to PEG and an orthopyridyl disulfide group (OPSS). The OPSS group readily forms disulfide bonds with free sulfhydryl groups and, when liposomes are injected into the blood, activated C3 containing an exposed sulfhydryl group forms a disulfide bond to the liposomes. C3-coated liposomes bind and internalize into cells that have the receptor for activated C3, which in the blood of cancer patients are primarily G-MDSCs. Utilizing this targeting strategy, we show that MDSCs are targeted in vitro by a C3 dependent manner and investigate the biodistribution and MDSC targeting potential of the liposomes in vivo in tumor-bearing mice.
Methods
Reagents
1,2-Dipalmitoyl-sn-glycero-3-phosphocholine (DPPC), 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), 1,2-Dipalmitoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] (DPPG), 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[poly(ethylene glycol)-2000] (DSPEPEG( 2000)) and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[PDP-poly(ethylene glycol)-2000] (DSPE-PEG(2000)-PDP) were purchased from Avanti Polar Lipids (Alabaster, AL). Lissamine rhodamine B 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (RhoPE) was purchased from Life Technologies (Grand Island, NY, USA). Dulbecco’s phosphate buffered saline (DPBS, pH 7.4), fetal bovine serum (FBS), trypsin, and penicillin-streptomycin were purchased from Mediatech, Inc. (Manassas, VA) CL-4B Sepharose gel, used for column purification of liposomes, was obtained from Amersham Biosciences (Uppsala, Sweden). The flow cytometry antibodies, Brilliant Violet 510™ anti-mouse Ly6C antibody and Brilliant Violet 421™ anti-mouse Ly6G were purchased from Biolegend (San Diego, CA). Anti-Mouse CD45 APC, anti-mouse CD11b APC-eFluor® 780, anti-mouse MHC Class II (I-A/I-E) PerCP-eFluor® 710 and anti-mouse CD16/CD32 purified were purchased from Ebioscience (San Diego, CA). Anti CD11c was obtained from BD Biosciences (San Jose, CA) and rat anti-mouse F4/80 FITC was purchased from ABD Serotech (Raleigh, NC). Unless specified otherwise, all other chemicals and reagents were purchased from Thermo Fisher Scientific (Pittsburgh, PA). Goat anti-mouse complement C3 was from MP Biomedicals and secondary donkey anti-goat 800 IgG was purchased from Li-Cor. Isotype IgG control antibody, goat anti-mouse arginase-1, was obtained from MyBioSource (San Diego, CA). C3 deficient serum was obtained from C57BL/6 knockout mice, kindly provided by Dmitri Simberg.
Liposome preparation
Liposomes containing DSPE-PEG(2000)-PDP (referred to as OPSS-liposomes) and DSPEPEG( 2000) (referred to as control liposomes) were prepared using the film hydration–extrusion method as described previously 15. Briefly, DPPC/DSPC/DPPG/RhoPE /DSPE-PEG(2000)-PDP in chloroform were mixed at a molar ratio of 80:10:4:1:5 for OPSS-liposomes and for control-liposomes DSPE-PEG(2000) was substituted for DSPE-PEG(2000)-PDP to give a similar ratio. Lipids were dried under a stream of nitrogen for 1 hour and then placed under vacuum for 2 hours to remove any traces of chloroform. The lipid film was rehydrated with 0.5 mL of water at 47 °C, and extruded 9 times through a polycarbonate membrane filter of 400 nm (Avestin, Ottawa, ON, Canada). After extrusion, liposomes were column purified using a CL-4B Sepharose column hydrated in 1x DPBS, pH 7.4 and the collected liposome fraction was diluted to a concentration of 0.875 mg lipid/mL. Liposome size was determined using PL/DNA sizing in a LM20 nanoparticle size analyzer (Nanosight, Amesbury, Wiltshire, United Kingdom)
Liposome binding of activated C3
Dot blot techniques were used to determine if liposomes bind activated C3 when exposed to complete non-heat-inactivated mouse serum. To obtain mouse serum, mouse blood was collected via cardiac puncture. After clotting, the blood was centrifuged at 3000 x g for 3 minutes and the supernatant serum was collected. Ten microliters of OPSS-liposomes or control liposomes were incubated with 10 μL of mouse serum for 1 hour. In addition, a third sample consisted of OPSS-liposomes similarly incubated in mouse serum and subsequently exposed to 10 mM of reducing agent tris(2-carboxyethyl)phosphine) (TCEP) for 30 minutes. Liposomes were isolated from the serum by centrifuging at 70,000 x g for 7 minutes utilizing a Beckman Optima TLX ultracentrifuge centrifuge. Liposomes were centrifuged and rinsed 3 times in PBS before the pellet was rehydrated in 10 μL PBS. A volume of 2 μL of each sample was absorbed onto nitrocellulose paper and then rotated in 1x PBS + 5% milk + 0.1% Tween 20 blocking solution for 1 hour. The nitrocellulose blot was then transferred to a fresh solution of 1x PBS + 5% milk and incubated with 1:1000 dilution of anti-C3 antibody overnight at 4 °C. The next morning, the nitrocellulose blot was rinsed 3x with fresh 1x PBS + 0.1% tween 20 and incubated with 1:10,000 dilution of secondary donkey anti-goat licor (800) antibody for 1 hour. The nitrocellulose blot was rinsed with 1x PBS + 0.1% Tween 20 and imaged using a LiCor imager with Odyssey software to detect the presence of C3 protein associated with the different liposome samples. Additionally, dot blots of OPSS-liposomes incubated in serum were stained with a 1:8000 dilution of anti-C3 antibody or an equivalent concentration of isotype control IgG antibody at a 1:1000 dilution and similarly detected with the LiCor imager.
In vitro uptake of liposomes
An in vitro assay using mouse blood cells was utilized to determine which cells in the blood would take up liposomes after intravenous injection and to test the mechanism of that uptake. Blood was collected from 3 mice that were bearing 4T1 mammary gland tumors. To establish the tumors, 4T1 cells (5 x 104) were maintained in RPMI + 10% FBS+ 1% penicillin–streptomycin and injected into the mammary fat pad of Balb/c mice in a volume of 25 μL. After 9 days, blood from the three mice was collected via cardiac puncture into EDTA-coated tubes. The blood samples were pooled together and spun at 3000 x g for 3 minutes. After removing the supernatant, the blood cells were resuspended in 3 ml of red blood cell lysis buffer (Fisher) and rotated for 5 minutes. The lysis was stopped with 15 ml of 1x DPBS, and cells were again centrifuged at 3000 x g for 3 minutes. Cells were resuspended in RPMI + 1% penicillin–streptomycin and aliquoted into a 96 well plate with 80 μL per well at a concentration to achieve 100,000 cells per well. The cells for this experiment were collected and used the same day without ever having been frozen to avoid any artifacts that might occur from the freezing process. OPSS-liposomes and control-liposomes were mixed with either normal mouse serum, mouse serum from a C3-deficient mouse, or mouse serum that had been heat inactivated (30 minutes at 60 °C). In all cases 10 μL of liposomes were incubated with 10 μL of serum for 1 hour, and liposomes were then added to the 80 uL of cells in each well to bring the final volume in each well up to 100 μL with a final concentration of 10% mouse serum. In addition, one sample utilized OPSS-liposomes that were incubated in mouse serum for 1 hour and subsequently exposed to 10 mM TCEP for 1 hour before being dialyzed for 16 hours against PBS with two buffer changes of 1:50,000 v/v to remove TCEP. Cells were exposed to liposomes for 4 hours before collection and analysis by flow cytometry, to analyze the cells that took up rhodamine-labeled liposomes under the various serum incubation conditions. All animal procedures were approved by the University of Colorado IACUC.
Flow cytometry analysis of liposome uptake
Cells were analyzed using flow cytometry to determine the populations that were positive for rhodamine-labeled liposomes. Collected cells were centrifuged at 3000 x g for 3 minutes and resuspended in 1ml of FACS buffer (1x PBS + 1% BSA + 0.01% sodium azide). Cells were counted and aliquoted into flow cytometry tubes to achieve 80,000 cells per tube. The cells were again centrifuged at 3000 x g for 3 minutes and resuspended in 50 μL of FACS buffer that contained 1 ul of CD16/32 blocking antibody to block non-specific binding of IgG to Fc receptors. The cells were incubated in the blocking solution for 10 minutes at room temperature before the addition of 50 μL FACS buffer containing 1 μL each of anti-mouse antibodies against Ly6C, Ly6G, CD45, CD11b, CD11c, MHC II and F4/80 (staining buffer). Cells were incubated in the dark with the staining buffer for 35 minutes at 4 °C. After staining, 2 ml of FACS buffer was added to the cells which were then spun at 3000 x g for 3 minutes, resuspended in 350 μL of FACS solution and analyzed using a Beckman Coulter 4 Laser Gallios with Hypercyt Adaptor and Kaluza 1.3 software.
In vivo liposome biodistribution
An in vivo biodistribution study was performed to determine which cell population takes up liposomes after systemic administration and to observe if those liposome-positive cells are present in the tumor, blood and various organs. 4T1 tumors were established in the mammary gland of Balb/c mice as described for the in vitro experiments. After 21 days, a 200 μL aliquot of OPSS-liposomes or control-liposomes was administered to the mice via tail vein injection. After 4 hours, the mice were euthanized and blood was collected via cardiac puncture. The organs and tumors were removed. Half of the organ or tumor was placed in Hank’s balanced salt solution for analysis by flow cytometry and the other half was placed in OCT embedding compound and frozen on dry ice for histological analysis. To digest the organs and obtain single cell suspensions for flow cytometry analysis, the organs were placed in fresh Hank’s solution containing 1 mg/ml collagenase (for tumor, lungs, liver and spleen) or 0.1 mg/ml elastase and 0.5 mg/ml hyaluronidase (for lungs). Organs were diced with a scalpel and flipped end over end at 37 °C for 1 hour. Homogenates were run through a 70 μm filter and enzymes were quenched with 50 ml of Hanks’s supplemented with 10% FBS. Red blood cells of the liver, spleen and blood were ruptured using red blood cell lysis buffer (BD biosciences), and cells of blood, tumor and organs were spun down and resuspended in 1 ml of FACS solution. Single cell solutions were counted, aliquoted, stained and analysed by flow cytometry for uptake of rhodamine-labeled liposomes as described for the in vitro experiments. For histology studies, 15 μm tissue slices were prepared from frozen tissue blocks and mounted on slides. Fluorescent images were acquired using an Operetta high content imaging system.
Results
OPSS-liposomes disulfide bind activated C3
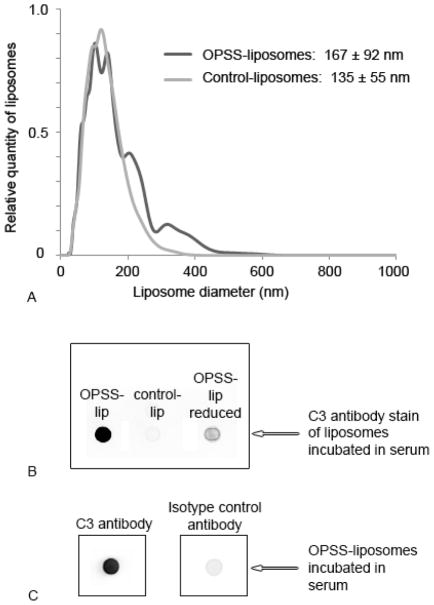
To target MDSCs which are express the receptor for activated C3, liposomes were formulated that contained OPSS groups on the distal end of PEGylated lipid. The OPSS group readily forms disulfide bonds with available sulfhydryl groups 16. Since activated C3 contains an exposed sulfhydryl group, we hypothesized that OPSS-liposomes would form a disulfide bond with C3, allowing for their uptake by MDSCs which express the receptor for activated C3 17. Liposomes with the OPSS group had a diameter of 167 ± 92 nm which was not significantly different from control liposomes with no OPSS group (135 ± 55 nm) (Figure 1A). However there does appear to be minor aggregation of the OPSS-liposomes as evident from the appearance of two additional minor size peaks at approximately 200 and 300 nm (Figure 1A).
Figure 1.
Characterization of OPSS- and control-liposomes. OPSS- and control-liposome size was determined in PBS using a nanoparticle size analyzer (A). Both OPSS- and control-liposomes were incubated in serum and tested for presence of C3 protein using dot blot techniques and anti-C3 antibody. In addition, OPSS-liposomes that had been exposed to serum and then a reducing agent were also assayed to determine if activated C3 was still conjugated to liposomes through a disulfide bond (B). Finally, OPSS-liposomes were exposed to serum and probed with C3 antibody or IgG isotype control antibody (C).
To test the ability of OPSS-liposomes to disulfide bind activated C3, mouse serum was collected from Balb/c mice, liposomes were incubated in the serum, and the liposomes were then isolated by centrifugation and assayed for the presence of C3 by dot blots. OPSS-liposomes which contained the OPSS group showed a strong presence of C3 protein, while control-liposomes that did not contain the OPSS group showed very little association with C3 (Figure 1B). In addition, when the OPSS-liposomes were incubated with serum and then exposed to a reducing agent, the amount of bound C3 greatly decreased, showing that activated C3 was associating with the liposomes through a reducible disulfide bond (Figure 1B). Serum exposed liposomes probed with an isotype IgG control antibody showed very little signal, eliminating the possibility of nonspecific interaction of the primary antibody with the OPSS group on the liposomes (Figure 1C).
G-MDSCs take up OPSS liposomes in vitro
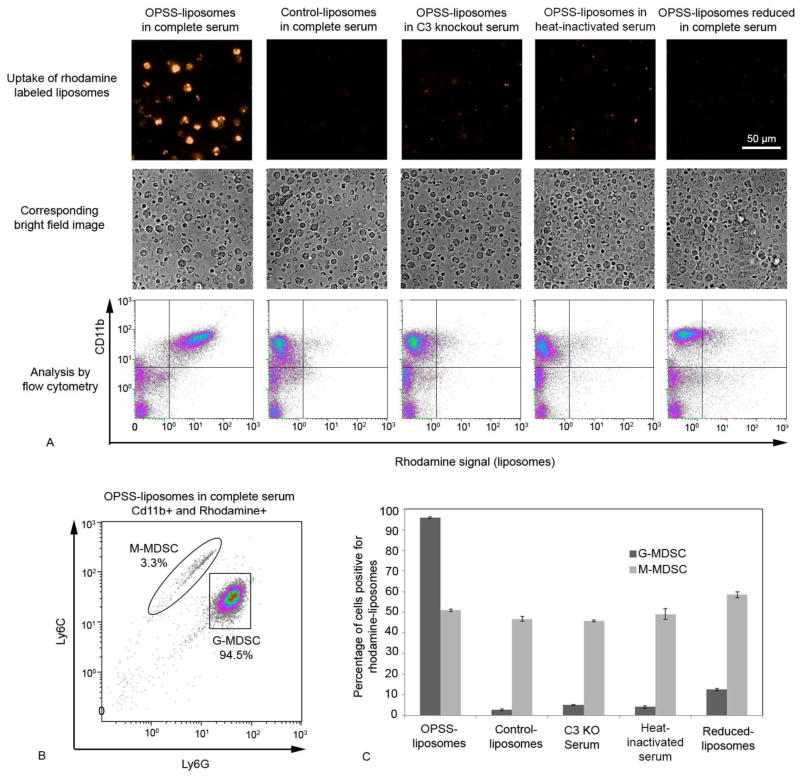
The ability of OPSS-liposomes to target MDSCs through a C3-dependent mechanism was tested using blood cells that were isolated from mice and subsequently exposed to liposomes that had been incubated in an array of different serum conditions (Figure 2A). OPSS-liposomes incubated in complete serum were taken up specifically by cells that expressed the receptor for activated C3. FACS analysis of blood cells with respect to CD11b and rhodamine indicated that 85.2 ± 0.4% of the CD11b+ cells had taken up rhodamine-labeled OPSS-liposomes (upper right quadrant of first panel, Fig 2A) compared to 3.2 ± 0.4% of CD11b− cells (lower right quadrant of first panel, Fig 2A). Control-liposomes that lack the OPSS group were also incubated in complete serum, but were only taken up by 5.9 ± 0.3% of the CD11b+ cells (upper right quadrant of second panel, Fig 2A).
Figure 2.
In vitro analysis of OPSS-liposome uptake by MDSCs. Rhodamine labeled OPSS-and control-liposomes were incubated in serum and then exposed to blood cells from mice with 4T1 mammary tumors to determine the cell populations that took up liposomes. Controls included OPSS-liposomes incubated in C3 knockout serum, OPSS-liposomes incubated in heat-inactivated serum and OPSS-liposomes that were exposed to serum and then reduced. Representative fluorescent and brightfield images with corresponding flow cytometry data illustrate the level of rhodamine uptake by Cd11b positive cells under different experimental conditions. Quadrant lines were used to estimate the percentage of cells that were positive for both CD11b and rhodamine signal (upper right quadrant). Experiments were done in triplicate to yield the data reported in the text (mean +/− s.e.) (A). Cells in each experiment that were positive for both CD11b and rhodamine were re-plotted according to their Ly6C and Ly6G levels, thereby differentiating G-MDSC and M-MDSC populations. Enclosure lines were used to estimate the percentages of G-MDSC and M-MDSCs (B). Bars represent the mean percentages of G-MDSC versus M-MDSC populations positive for rhodamine liposomes from triplicate analyses of data under different experimental conditions (C). Data are expressed as mean ± SE (n=3).
To study the degree of liposome uptake by M-MDSC versus G-MDSCs, the cells that were both CD11b+ and rhodamine positive (upper right quadrant, Fig. 2A) were re-analyzed with respect to Ly6C and Ly6G surface markers (Figure 2B). In each of triplicate analyses, more than 90% of these cells were G-MDSCs and fewer than 5% were M-MDSCs (Figure 2B). The M-MDSC and G-MDSC populations were then individually analyzed in triplicate to determine the percentage of each population that had engulfed liposomes (Figure 2C). The OPSS-liposomes incubated in complete mouse serum were taken up readily by G-MDSCs in vitro with 95.8 ± 0.4% of GMDSCs positive for the presence of rhodamine liposomes (Figure 2C). Control-liposomes incubated in the same serum conditions showed limited uptake, with only 2.6 ± 0.3% of GMDSCs testing positive for rhodamine. Other control conditions included OPSS-liposomes incubated in C3 deficient serum and OPSS-liposomes incubated in heat-inactivated serum which denatures the complement components of serum. Again, there was limited uptake with less than 5% of G-MDSCs positive for rhodamine OPSS-liposomes under both of these conditions with impaired C3 activity. Finally, OPSS-liposomes were incubated in serum to allow the binding of activated C3 and then exposed to reducing conditions to cleave any disulfide bonds. The exposure to a reducing agent greatly reduced the uptake with only 12.5 ± 0.6% of G-MDSC testing positive for liposomes. The limited uptake of control-liposomes shows the necessity of the OPSS group for G-MDSC targeting, while the other controls using various serum conditions indicate the dependence of this uptake on activated C3 disulfide binding to the OPSS-liposomes.
While liposomes in G-MDSCs were clearly taken up by a C3 dependent mechanism, this was not the case for M-MDSCs. In all conditions tested, between 45–60% of M-MDSC cells took up rhodamine labeled liposomes indicating that complement is not necessary for liposome internalization by these cells (Figure 2C). However, because the M-MDSCs are a minority population, the total uptake of liposomes by these cells was small (< 5%) compared to the uptake by the G-MDSCs (Figure 2B).
G-MDSC uptake of OPSS-liposomes in vivo
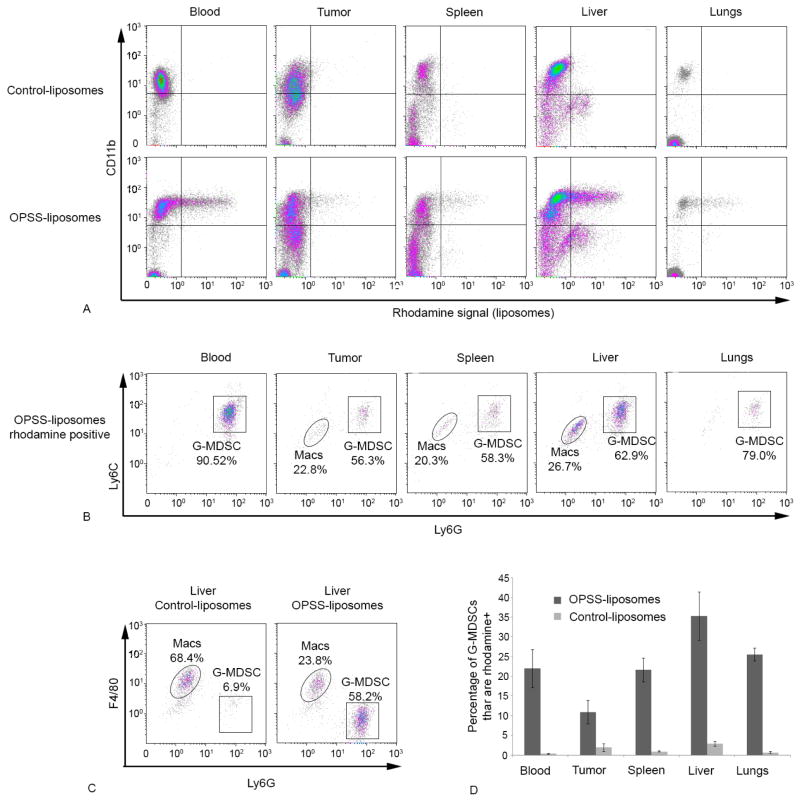
After establishing that G-MDSCs take up OPSS-liposomes in vitro by a C3 dependent mechanism, a biodistribution study was performed in vivo to determine which cells took up liposomes administered systemically and how those cells were distributed among the blood, tumor and various organs. Balb/c mice bearing 4T1 mammary tumors were injected with either OPSS-liposomes or control-liposomes and, at 4 hours post-injection, the various tissues and tumors were collected and analyzed by flow cytometry. In mice injected with OPSS-liposomes, a significant portion of the CD11b+ cells in the blood and various tissues were also positive for rhodamine stained liposomes (Figure 3A). As with the in vitro data, the majority of the cells that took up rhodamine-labeled OPSS-liposomes were G-MDSCs. FACS analysis of rhodamine positive cells with respect to Ly6G and Ly6C markers indicated that 91.3 ± 3.1% of the cells that took up liposomes in the blood were G-MDSC and that very few other cells, including MMDSCs, tested positive for the rhodamine-labeled liposomes. Figure 3B shows a representative analysis of this experiment that was run in triplicate. At this 4 hour time point, when the total GMDSC population in the blood was analyzed, it was found that OPSS-liposomes had been taken up by 22.0 ± 4.9 of the G-MDSCs while only 0.3 ± 0.1% of the G-MDSCs took up liposomes in mice that were injected with control-liposomes (Figure 3D & Table 1).
Figure 3.
In vivo uptake of OPSS- and control-liposomes by G-MDSCs. OPSS- and control-liposomes were administered intravenously and the blood, tumor, spleen, liver and lung cells were analyzed by flow cytometry after 4 hours to determine which cells were positive for liposome uptake. Representative flow cytometry graphs (panel A) show CD11b signal plotted against rhodamine signal. Analysis of the rhodamine positive cells (both upper and lower right quadrants of graphs in panel A) using the Ly6G and Ly6C markers showed that G-MDSCs are the predominant cell population that took up OPSS-liposomes (B). However, triplicate analysis of liver cells by F480 and Ly6G revealed that a significant percentage of the rhodamine positive cells were macrophages (Macs) (C). The bar graph (D) shows the percentage of G-MDSCs that have taken up liposomes in animals that were injected with either OPSS- or control-liposomes. Data are expressed as mean ± SE (n=3), and actual numbers are given in Table I.
Table 1.
Percentage of G-MDSCs and macrophages that were positive for OPSS- or control-liposomes in different tissues. Macrophages were not detected in high enough numbers to analyze in the blood samples. Data are expressed as mean ± SE of triplicate samples. G-MDSC were chosen based on the positive expression of Ly6C, Ly6G and CD11b, while selected macrophages were positive for CD11b and F4/80.
| G-MDSCs | Macrophages | |||
|---|---|---|---|---|
| OPSS | Control | OPSS | Control | |
| Blood | 22.0 ± 4.9 | 0.3 ± 0.1 | NA | NA |
| Tumor | 10.8 ± 3.0 | 1.9 ± 1 | 3.7 ± 1.3 | 2.4 ± 0.6 |
| Spleen | 21.6 ± 3.0 | 0.9 ± 0.2 | 33.9 ± 2.9 | 27.5 ± 2.0 |
| Liver | 35.3 ± 6.2 | 2.9 ± 0.6 | 79.2 ± 3.8 | 70.4 ± 1.4 |
| Lungs | 25.5 ± 1.6 | 0.7 ± 0.3 | 56.0 ± 15.9 | 42.4 ± 12.9 |
Analysis of the tumor, spleen, liver and lungs showed that 10.8 ± 3.0, 21.6 ± 3.0, 35.3 ± 6.2 and 25.5 ± 1.6% of the G-MDSC had taken up OPSS-liposomes respectively while mice injected with control-liposomes had minimal liposome uptake by G-MDSC in all of these tissues (Figure 3D & Table 1).
Flow cytometry analysis of samples from tumor, liver and spleen revealed a population of rhodamine-positive cells with low Ly6G and Ly6C signal (Figure 3B). Further analysis of these samples with respect to F4/80, a macrophage marker, indicated that these cells were macrophages. Non-targeted liposomes are known to be cleared rapidly by macrophages of the spleen and liver 9. In the livers of mice injected with control-liposomes, 70.4 ± 1.4 % of the macrophages took up liposomes (Table 1) and this represented 63.5 ± 7.2% of the total cell population in the liver that was positive for rhodamine liposomes. Figure 3C shows a representative analysis of the experiment that was run in triplicate. OPSS-liposomes were also taken up readily by macrophages, with 79.2 ± 3.8% (Table 1) of all macrophages testing positive for rhodamine. However, this represented only 22.2 ± 1.8% of the total cell population that was positive for rhodamine-labeled cells in the liver (Figure 3C). The dominant liver cell population that took up OPSS-liposomes, was G-MDSCs which made up 62.2 ± 1.8% of the liposome-positive cells (Figure 3C). In addition, analysis of flow cytometry data reveals that the average rhodamine signal from the positive G-MDSC fraction was 2.4-fold greater than the signal from the positive macrophage population, showing that individual G-MDSCs took up liposomes to a greater extent than the macrophage cells. These results show that the OPSS-group changes the biodistribution of injected liposomes and significantly targets the G-MDSC cell population in vivo.
Our conclusions from FACS analysis were confirmed by histology of tissue samples from mice injected with OPSS- or control-liposomes. Mice injected with control-liposomes showed very little evidence of liposomes within the cells, while mice injected with OPSS-liposomes had liposome-containing cells distributed in the blood, tumor, spleen, liver and lungs (Figure 4).
Figure 4.
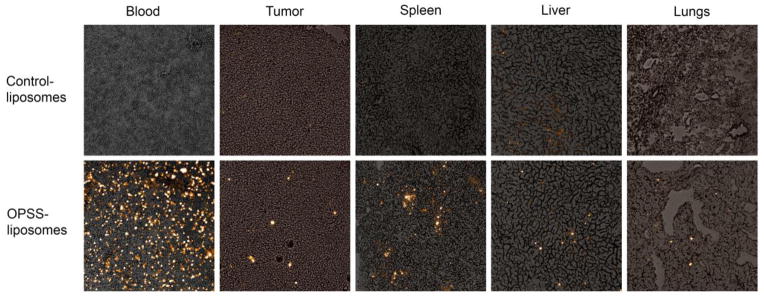
Histology images of OPSS- and control-liposomes in the blood and various tissues. Images show rhodamine labeled liposomes taken up by cells in the blood, tumor, spleen, liver and lungs. The top panel are tissues from mice injected with control-liposomes and the bottom panel are tissues from mice injected with OPSS-liposomes.
Discussion
Current techniques for targeting myeloid-derived suppressor cells involve systemic delivery of drugs with the aim of either depleting, inhibiting or differentiating the immunosuppressive MDSC population 4, 5, 18. These techniques, which primarily use chemotherapeutic agents (e.g. Sunitinib) and hydrophobic molecules (e.g. ATRA), suffer from non-specific toxicity and low drug solubility, ultimately limiting the dose delivered to MDSCs. This work describes a liposomal system which is designed to overcome these limitations by targeting G-MDSCs in the blood via their receptors for activated C3. The system offers the possibility of versatile drug delivery since hydrophilic drugs can be encapsulated in the aqueous core of the liposomes or hydrophobic molecules can be incorporated into the lipid bilayer of the liposomes. With the increased specificity incurred by targeting the receptor for activated C3 on G-MDSCs, it should be possible to achieve greater concentrations of drug in the G-MDSC population while reducing toxic side effects.
G-MDSCs offer an attractive target for liposomal drug delivery because of their prominent role in tumor immune evasion and recent evidence showing that they promote metastasis 19, 20. In addition, since MDSCs circulate in the blood, they are much more accessible to intravenously administered liposomes than cells within the extravascular space of tumor tissue 21, 22. MDSCs that take up liposomes in the blood would subsequently infiltrate the tumor and other tissues in the body. To target the MDSC population, the liposomes in our delivery system have an OPSS group attached to them which readily binds activated complement C3 circulating in the blood. In vitro experiments showed that C3 binds to OPSS-liposomes through a disulfide dependent mechanism and that these C3-coated liposomes are taken up by G-MDSCs in culture. The liposomes are also taken up by M-MDSCs, but control experiments showed that this is not a C3 dependent mechanism. In contrast to G-MDSCs, the M-MDSCs readily internalize controlliposomes that do not have the OPSS group attached in vitro. These experiments show the possible merit of using endogenous C3 for G-MDSC targeting in vivo. If successful, our system has the added practical benefits of lowering liposome production cost, increasing storage stability, and likely lowering the immunogenic effects that protein ligands might provoke.
While our system appears to target the CR3 (CD11b) positive G-MDSCs, further study is required to fully describe the cellular specificity of OPSS-liposomes and the C3 breakdown products responsible for uptake into MDSCs. In vivo, C3 complement is activated to C3b, exposing a thioester which can interact with hydroxyl group-bearing nucleophiles and also with the OPSS group on our liposomes 23. C3b is taken up by the CR1 receptor and also recruits factor B, forming the complex C3bB which activates additional complement C3. C3b is further metabolized to iC3b and C3dg, breakdown products which can target receptors including CR2 (iC3b, C3dg), CR3 (iC3b), CRIg (iC3b), and CR4 (iC3b) expressed at various levels by macrophages, dendritic cells and leukocytes. Additional flow cytometry experiments will help to elucidate the mechanism of liposome uptake and all the cell types being targeted.
Liposomes administered systemically through intravenous injection are taken up primarily by the macrophages of the liver and spleen 9–11. We find that control non-OPSS liposomes show this same biodistribution, localizing to macrophages that reside in the spleen and liver, with little presence in any other cell population. However, adding an OPSS group to the liposomes results in a very different biodistribution. Although macrophages still engulf the OPSS-liposomes, GMDSCs are the dominant cell population taking up the liposomes. G-MDSCs in the blood readily take up rhodamine labeled liposomes and G-MDSCs in the tumor, spleen, liver and lungs also contain labeled liposomes. Liposomes do not penetrate well into these tissues whereas MDSCs are known to rapidly infiltrate extravascular tissues. It seems likely, therefore, that the G-MDSCs in our system are internalizing the liposomes as they circulate through the blood and are carrying those liposomes into the tumor, spleen, liver and lungs 24. Given recent evidence that immunosuppressive MDSCs are involved in facilitating metastasis, it is important to note that our liposomal-targeted MDSCs infiltrate into those tissues which are possible metastatic sites 19, 20. An effective strategy for reducing metastatic tumor burden might be to target GMDSCs in the blood with a drug responsible for phenotype reversal of these cells from immunosuppressive to immunostimulatory. Subsequent infiltration of these cells into tissue would not promote metastasis, but instead activate an immune response against invading cancer cells.
The most promising therapeutic candidate for our targeted drug delivery system is all-trans-retinoic acid (ATRA), an agent which has been shown to differentiate MDSCs in vivo and significantly enhance the anti-tumor effect of immunotherapies 3, 4, 25, 26. In human cancer patients, treatment with ATRA decreased the number of circulating MDSCs and increased the antigen response of CD4+ and CD8+ T cells 3, 5. However, oral administration of ATRA resulted in heterogeneous drug levels which limited the effectiveness of treatment. In comparison, greater stability and plasma concentration of ATRA can be achieved by incorporating it into liposomes27, 28. Our results indicate that targeted delivery to G-MDSCs in vivo can be achieved by attaching an OPSS group to the liposome surface, which then in turn binds endogenous activated C3. This targeting system may be applicable to liposomal formulations of drugs like ATRA for eliciting differentiation of G-MDSCs in vivo and therapeutic treatment of cancers.
Acknowledgments
We are very grateful for funding and support from the Susan G. Komen Foundation (#KG111128) and NIH/NIGMS (#1 RO1GM093287) that made this work possible.
We would like to thank Dr. Richard Kullberg and Dr. Anthony Elias for their advice throughout the research and with preparation of this manuscript. We also acknowledge Brian Reid and the University of Colorado High Throughput and High Content Screening Facility.
Abbreviations
- MDSC
myeloid derived suppressor cell
- M-MDSC
monocytic myeloid derived suppressor cell
- G-MDSC
granulocytic myeloid derived suppressor cell
- ATRA
all-trans-retinoic acid
- RES
reticuloendothelial system
- OPSS
orthopyridyl disulfide
Footnotes
The authors declare no competing financial interest. In addition, this is an original work that has not been submitted to any journal other than Nanomedicine. The work of others that is described in the paper has been appropriately referenced.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Khaled YS, Ammori BJ, Elkord E. Myeloid-derived suppressor cells in cancer: Recent progress and prospects. Immunol Cell Biol. 2013;91:493–502. doi: 10.1038/icb.2013.29. [DOI] [PubMed] [Google Scholar]
- 2.Diaz-Montero CM, Salem ML, Nishimura MI, Garrett-Mayer E, Cole DJ, Montero AJ. Increased circulating myeloid-derived suppressor cells correlate with clinical cancer stage, metastatic tumor burden, and doxorubicin-cyclophosphamide chemotherapy. Cancer Immunol Immunother. 2009;58:49–59. doi: 10.1007/s00262-008-0523-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mirza N, Fishman M, Fricke I, Dunn M, Neuger AM, Frost TJ, et al. All-trans-retinoic acid improves differentiation of myeloid cells and immune response in cancer patients. Cancer Res. 2006;66:9299–307. doi: 10.1158/0008-5472.CAN-06-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ugel S, Delpozzo F, Desantis G, Papalini F, Simonato F, Sonda N, et al. Therapeutic targeting of myeloid-derived suppressor cells. Curr Opin Pharmacol. 2009;9:470–81. doi: 10.1016/j.coph.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 5.Najjar YG, Finke JH. Clinical perspectives on targeting of myeloid derived suppressor cells in the treatment of cancer. Front Oncol. 2013;3:49. doi: 10.3389/fonc.2013.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raber PL, Thevenot P, Sierra R, Wyczechowska D, Halle D, Ramirez ME, et al. Subpopulations of myeloid-derived suppressor cells impair t cell responses through independent nitric oxide-related pathways. Int J Cancer. 2014;134:2853–64. doi: 10.1002/ijc.28622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ribechini E, Greifenberg V, Sandwick S, Lutz MB. Subsets, expansion and activation of myeloid-derived suppressor cells. Med Microbiol Immunol. 2010;199:273–81. doi: 10.1007/s00430-010-0151-4. [DOI] [PubMed] [Google Scholar]
- 8.Favaloro J, Liyadipitiya T, Brown R, Yang S, Suen H, Woodland N, et al. Myeloid derived suppressor cells are numerically, functionally and phenotypically different in patients with multiple myeloma. Leuk Lymphoma. 2014:1–8. doi: 10.3109/10428194.2014.904511. [DOI] [PubMed] [Google Scholar]
- 9.Moghimi SM, Szebeni J. Stealth liposomes and long circulating nanoparticles: Critical issues in pharmacokinetics, opsonization and protein-binding properties. Prog Lipid Res. 2003;42:463–78. doi: 10.1016/s0163-7827(03)00033-x. [DOI] [PubMed] [Google Scholar]
- 10.Medina OP, Zhu Y, Kairemo K. Targeted liposomal drug delivery in cancer. Curr Pharm Des. 2004;10:2981–9. doi: 10.2174/1381612043383467. [DOI] [PubMed] [Google Scholar]
- 11.Ohara Y, Oda T, Yamada K, Hashimoto S, Akashi Y, Miyamoto R, et al. Effective delivery of chemotherapeutic nanoparticles by depleting host kupffer cells. Int J Cancer. 2012;131:2402–10. doi: 10.1002/ijc.27502. [DOI] [PubMed] [Google Scholar]
- 12.Gosk S, Moos T, Gottstein C, Bendas G. Vcam-1 directed immunoliposomes selectively target tumor vasculature in vivo. Biochim Biophys Acta. 2008;1778:854–63. doi: 10.1016/j.bbamem.2007.12.021. [DOI] [PubMed] [Google Scholar]
- 13.Cheng WW, Allen TM. Targeted delivery of anti-cd19 liposomal doxorubicin in b-cell lymphoma: A comparison of whole monoclonal antibody, fab’ fragments and single chain fv. J Control Release. 2008;126:50–8. doi: 10.1016/j.jconrel.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 14.van der Meel R, Vehmeijer LJ, Kok RJ, Storm G, van Gaal EV. Ligand-targeted particulate nanomedicines undergoing clinical evaluation: Current status. Adv Drug Deliv Rev. 2013;65:1284–98. doi: 10.1016/j.addr.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 15.Kullberg M, Mann K, Anchordoquy TJ. Targeting her-2+ breast cancer cells with bleomycin immunoliposomes linked to llo. Mol Pharm. 2012;9:2000–8. doi: 10.1021/mp300049n. [DOI] [PubMed] [Google Scholar]
- 16.Sun X, Zhang G, Patel D, Stephens D, Gobin AM. Targeted cancer therapy by immunoconjugated gold-gold sulfide nanoparticles using protein g as a cofactor. Ann Biomed Eng. 2012;40:2131–9. doi: 10.1007/s10439-012-0575-7. [DOI] [PubMed] [Google Scholar]
- 17.Mitchell DA, Ilyas R, Dodds AW, Sim RB. Enzyme-independent, orientation-selective conjugation of whole human complement c3 to protein surfaces. J Immunol Methods. 2008;337:49–54. doi: 10.1016/j.jim.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 18.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–74. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao Y, Slaney CY, Bidwell BN, Parker BS, Johnstone CN, Rautela J, et al. Bmp4 inhibits breast cancer metastasis by blocking myeloid-derived suppressor cell activity. Cancer Res. 2014;74:5091–102. doi: 10.1158/0008-5472.CAN-13-3171. [DOI] [PubMed] [Google Scholar]
- 20.Keskinov AA, Shurin MR. Myeloid regulatory cells in tumor spreading and metastasis. Immunobiology. 2014 doi: 10.1016/j.imbio.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 21.Duncan R, Sat-Klopsch YN, Burger AM, Bibby MC, Fiebig HH, Sausville EA. Validation of tumour models for use in anticancer nanomedicine evaluation: The epr effect and cathepsin b-mediated drug release rate. Cancer Chemother Pharmacol. 2013;72:417–27. doi: 10.1007/s00280-013-2209-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lammers T, Kiessling F, Hennink WE, Storm G. Drug targeting to tumors: Principles, pitfalls and (pre-) clinical progress. J Control Release. 2012;161:175–87. doi: 10.1016/j.jconrel.2011.09.063. [DOI] [PubMed] [Google Scholar]
- 23.Alcorlo M, Martinez-Barricarte R, Fernandez FJ, Rodriguez-Gallego C, Round A, Vega MC, et al. Unique structure of ic3b resolved at a resolution of 24 a by 3d-electron microscopy. Proc Natl Acad Sci U S A. 2011;108:13236–40. doi: 10.1073/pnas.1106746108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ueha S, Shand FH, Matsushima K. Myeloid cell population dynamics in healthy and tumor-bearing mice. Int Immunopharmacol. 2011;11:783–8. doi: 10.1016/j.intimp.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 25.Gabrilovich DI, Velders MP, Sotomayor EM, Kast WM. Mechanism of immune dysfunction in cancer mediated by immature gr-1+ myeloid cells. J Immunol. 2001;166:5398–406. doi: 10.4049/jimmunol.166.9.5398. [DOI] [PubMed] [Google Scholar]
- 26.Kusmartsev S, Cheng FD, Yu B, Nefedova Y, Sotomayor E, Lush R, et al. All-trans-retinoic acid eliminates immature myeloid cells from tumor-bearing mice and improves the effect of vaccination. Cancer Research. 2003;63:4441–9. [PubMed] [Google Scholar]
- 27.Mehta K, Sadeghi T, McQueen T, Lopez-Berestein G. Liposome encapsulation circumvents the hepatic clearance mechanisms of all-trans-retinoic acid. Leuk Res. 1994;18:587–96. doi: 10.1016/0145-2126(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 28.Ozpolat B, Lopez-Berestein G, Adamson P, Fu CJ, Williams AH. Pharmacokinetics of intravenously administered liposomal all-trans-retinoic acid (atra) and orally administered atra in healthy volunteers. J Pharm Pharm Sci. 2003;6:292–301. [PubMed] [Google Scholar]