Abstract
Gene expression changes are linked to air pollutant exposures in in vitro and animal experiments. However, limited data are available on how these outcomes relate to ambient air pollutant exposures in humans. We performed an exploratory analysis testing whether gene expression levels were associated with air pollution exposures in a Los Angeles area cohort of elderly subjects with coronary artery disease. Candidate genes (35) were selected from published studies of gene expression-pollutant associations. Expression levels were measured weekly in 43 subjects (≤12 weeks) using quantitative PCR. Exposures included gaseous pollutants O3, nitrogen oxides (NOx), and CO; particulate matter (PM) pollutants elemental and black carbon (EC, BC); and size-fractionated PM mass. We measured organic compounds from PM filter extracts, including polycyclic aromatic hydrocarbons (PAHs), and determined the in vitro oxidative potential of particle extracts. Associations between exposures and gene expression levels were analyzed using mixed-effects regression models. We found positive associations of traffic-related pollutants (EC, BC, primary organic carbon, PM0.25-2.5 PAH and/or PM0.25 PAH, and NOx) with NFE2L2, Nrf2-mediated genes (HMOX1, NQO1, and SOD2), CYP1B1, IL1B, and SELP. Findings suggest that NFE2L2 gene expression links associations of traffic-related air pollution with phase I and II enzyme genes at the promoter transcription level.
Keywords: longitudinal studies, oxidative stress, transcription, biochemical pathways, particulate matter, air pollution
INTRODUCTION
Air pollution has been linked with adverse cardiovascular outcomes in numerous epidemiological and toxicological studies.1 Traffic-related air pollution is associated with increases in blood pressure, myocardial infarction, atherosclerotic progression, and other outcomes.1 Previously, we showed increases in levels of circulating biomarkers of inflammation and platelet activation in association with exposure to primary (combustion-related) air pollutants in an elderly cohort panel with coronary artery disease (CAD).2,3 That panel study contributed subject data to the present study. In the region of study (Los Angeles, CA), the markers of combustion-related air pollution used (e.g. elemental carbon, EC) are primarily from mobile sources (traffic). We hypothesize that gene expression levels in whole blood along biopathways relevant to these health outcomes will also be associated with traffic-related air pollutant exposures in our cohort.
Air pollutant exposure experiments in vitro show gene expression changes in cells involved in cardiovascular disease progression, including endothelial and epithelial cells, monocytes, and macrophages. Gong et al.4 found diesel exhaust particle exposure increased expression levels in human endothelial cells of antioxidant genes (SOD1, HMOX1, and NQO1), genes linked with vascular inflammation (IL8, CXCL1, and DUSP1) and unfolded protein response (UPR) genes (HSPA8, XBP1, ATF4, and ATF6). In vitro genomic work by Gargalovic et al.5 identified UPR genes of human endothelial cells as mediators of vascular inflammation and atherosclerosis, supporting a possible role for UPR genes in air pollution-related cardiovascular outcomes. Additionally, genes from the nuclear factor erythroid 2-related factor 2 (Nrf2)-mediated oxidative stress response pathway were upregulated in airway epithelial cells after exposure to ambient outdoor particulate matter (PM).6 There is evidence that oxidative stress, an important mechanism of cardiovascular disease progression,7 is increased with elevated air pollution exposure.8 Thus, the Nrf2-mediated oxidative stress response genes may be important in human responses to air pollution exposure.
Using inflammation-prone Sirt1 knock-out mice, Wu et al.9 showed decreased expression of Kruppel-like factor 2 (Klf2) with PM exposure. Klf2, a key transcription factor for coagulation and thrombotic function, downregulates pro-inflammatory genes and has reduced expression levels in circulating monocytes from patients with CAD versus age-matched controls.10 Thus, in a cohort with CAD, air pollution exposure-associated cardiovascular responses may be mediated by KLF2 expression changes.
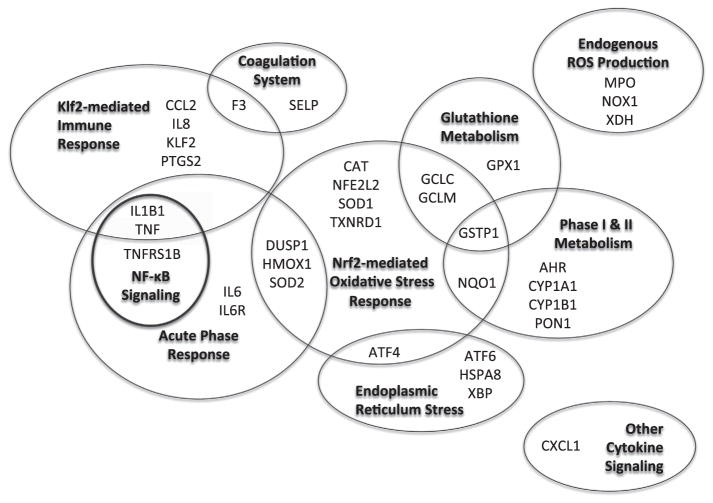
Although human studies have supported potential clinical impacts of traffic-related air pollution,1 the ways that air pollutants bring about such changes on a cellular level are not well understood. Experimental data discussed above are beginning to shed light on this topic, including one study showing upregulation of inflammatory and oxidative stress pathway genes in peripheral blood mononuclear cells of five healthy human subjects exposed to diesel exhaust.11 Studying gene expression in an epidemiologic study can help to support the potential public health relevance of experimental studies. However, to our knowledge, only three exploratory epidemiologic studies have been published on gene expression associations with air pollution from fossil fuel combustion or similar sources12–15 (see McHale et al.16 for review of human genome-wide studies of other inhaled exposures). On the basis of existing human studies, and in vitro and in vivo experiments, we selected 35 genes from 10 biological pathways relevant to air pollution exposure responses and examined changes in candidate gene expression in our cohort panel of elderly subjects with CAD.2,3,17 Biopathways included: coagulation, Klf2-mediated immune response, NF-κB signaling, acute phase response, Nrf2-mediated oxidative stress response, endoplasmic reticulum stress (UPR), glutathione metabolism, phase I and phase II metabolism, endogenous reactive oxygen species (ROS) production, and cytokine signaling (Figure 1, Supplementary Table S1, which include additional references that were informative to the selection of genes). Exploring gene expression changes along these pathways, in an urban cohort panel, can support studies of potential mechanistic pathways through which air pollutants may cause adverse cardiovascular outcomes.
Figure 1.
Candidate genes grouped by biopathway. Gene abbreviations and NCBI identification numbers are listed in Supplementary Table S2.
METHODS
Study Design and Population
This study was a cohort panel design consisting of repeated measures of outcomes and exposures for 43 elderly adults living in three retirement communities in the Los Angeles air basin. Each subject effectively serves as his/her own control. We followed subjects for 12 weeks, 6 weeks each during the warm and cool seasons to provide seasonal differences in air pollutant levels.18 The cohort analyzed in this study represents a subset of three of four retirement communities in the Cardiovascular Health and Air Pollution Study cohort previously described2,3,17 that were closest to the Los Angeles metro area.
Participants were over 65 years of age, with clinically confirmed CAD. This population is expected to have increased susceptibility to adverse associations with air pollution exposure.19 Exclusion criteria included: smoking within previous 12 months; dialysis treatment or uncontrolled hypertension, which alter inflammation and other outcomes; and employment outside their retirement community (where daily exposures were measured). We collected time-invariant subject characteristics (e.g. age, sex, and comorbidities) with questionnaires. This study was approved by the Institutional Review Board of the University of California, Irvine. All subjects provided written informed consent before participation.
Gene Expression
We analyzed 35 candidate genes and 5 reference genes using RNA from whole blood. Blood is a suitable matrix to evaluate systemic impacts, reflecting alterations in both local and distant environments.20 Samples were collected each Friday, at the same time of day for each subject, in PaxGene vacuum tubes (BD Diagnostics, Franklin Lakes, NJ, USA) that inhibit RNA degradation and gene induction before testing.21 Tubes were mixed, immediately frozen, transported to the laboratory and stored at − 80 °C until tested. Total RNA was isolated using a QIAcube™ and PAXgene Blood RNA kits (Qiagen, Valencia, CA, USA). Isolated RNA was reverse-transcribed into first-strand cDNA (ThermoScript RT-PCR kit, Invitrogen, CA, USA) for gene expression analysis.
Candidate genes (Figure 1, Supplementary Table S1) were selected based on either biological function or known pollutant exposure associations. Pathway information is derived from Ingenuity Pathway™ Software Analysis, the PANTHER classification system (http://www.pantherdb.org), and NCBI databases (http://www.ncbi.nlm.nih.gov). Gene expression levels were determined using end-point competitive PCR, with internal standards (the competitors) and sample cDNA co-amplified in the same reaction.22 Products were resolved with linear Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry (MassARRAY™ Quantitative Gene Expression) and concentrations of the target transcripts were calculated from the ratio of the PCR products. All samples from a given subject were assayed on the same plate. Signal acquisition, allele assignment, peak area integration, data processing, and analysis were carried out using the MassARRAY™ platform and software (Sequenom, San Diego, CA, USA). Services were contracted with Immune Sciences Lab (David H. Murdock Research Institute, Kannapolis, NC, USA).
Transcript copy numbers were determined based on the EC50 calculated by non-linear regression analysis of the cDNA and competitor allele frequencies. Copy numbers were normalized using the geNorm algorithm described by Vandesompele et al.23 We calculated the expression stability for a reference (housekeeping) gene as the average pairwise variation with other tested reference genes. We determined the most stable reference genes (ACTB, B2M, and GAPDH) by stepwise exclusion of genes with the highest instability. Gene expression normalization factors for each sample were generated from the geometric mean of reference genes. All calculations were carried out using the qBasePLUS data-analysis and integrated geNORM software (Biogazelle™).
Exposure Measurement
Exposure variables and lags were selected based on a priori hypotheses of their relations to cardiovascular outcomes in a target population with high traffic-related air pollution exposure. The rationale for their measurement is detailed elsewhere.17 The subset tested herein represents pollutants and averaging times with significant findings for cardiovascular and inflammation outcomes tested in our cohort.
We measured the concentrations of air pollutants in the immediate outdoor environment of participating retirement communities in the Los Angeles air basin, an area with some of the highest air pollutant levels in the United States. All exposure data were captured for each week preceding outcome measurements. We used standard federal reference methods to measure gases (nitrogen dioxide/nitrogen oxides, NO2/NOx; ozone, O3; and carbon monoxide, CO). We also measured elemental and organic carbon (EC, OC) (OC_EC Analyzer, Model 3F, Sunset Laboratory, Tigard, OR, USA), and black carbon (BC) (Aethalometer, Magee Scientific, Berkeley, CA, USA) in PM2.5 (PM with aerodynamic diameter <2.5 μm). We estimated primary organic carbon (OCpri) using the “EC tracer” method as we previously described,24 and secondary organic carbon (SOC) as total OC minus OCpri. The above exposures were averaged daily.
We determined daily size-fractionated particle mass for quasi-ultrafine particles (qUFP) <0.25 μm in aerodynamic diameter (PM0.25), accumulation-mode particles 0.25–2.5 μm (PM0.25–2.5) and coarse mode particles 2.5–10μm (PM2.5–10), using collocated Sioutas Personal Cascade Impactors (SKC, Eighty-Four, PA, USA),25 as discussed elsewhere.26 We quantified organic components of PM0.25–2.5 and qUFP from 5-day filter composites using GC/MS, as described elsewhere3,27 (see Supplementary Table S2). Components included polycyclic aromatic hydrocarbons (PAHs), key toxic components of primary organic aerosols that, in Los Angeles, are primarily from fossil fuel combustion. We also measured hopanes, tracers of vehicle exhaust found in vehicle lubricant oils.28 Exposure markers of secondary organic aerosols (SOA) included organic acids, primarily produced by photochemical aging of primary carbons,29 and water-soluble organic carbon (WSOC), a tracer of both photochemically-produced SOA and biomass burning.30,31 We estimated SOA as two times the difference between measured WSOC (from Sievers Total Organic Carbon, TOC; GE) and the fraction of secondary WSOC from wood smoke (estimated as 71% of OC from wood smoke, determined from a chemical mass balance model using levoglucosan as wood smoke tracer32), described in detail elsewhere.27
We measured particle oxidative potential from aqueous extracts of the 5-day filter composites of PM0.25 and PM0.25–2.5 using an in vitro assay of ROS described elsewhere33 and in the Supplementary Material. Briefly, rat alveolar macrophages were exposed to PM extract and 2′7′-dicholorodihy-drofluorescein diacetate. Fluorescence intensity from cellular production of ROS was measured. Un-opsonized Zymosan (a β-1,3-polysachharide of D-glucose) was our positive control and results are reported in μg Zymosan equivalents/m3 air.
Statistical Analysis
The distributions of the gene expression levels were evaluated after applying the natural logarithm of the Biogazelle normalized concentrations. The log-transformed gene expression data for an individual gene was retained if the distribution of all observations for that gene was sufficiently normal. If not, we applied an autoscale standardization method developed for use on the genes that were not log-normally distributed (AHR, CCL2, F3, GCLC, IL6, IL6R, IL8, NQO1, SELP, TNF, and TNFRSF1B).34 This required centering the log-transformed gene expression data relative to the subject-specific mean and scaling the data by the ratio of the individual to the group standard deviations. This transformation normalized the distributions for the purposes of model fitting and parameter estimation. For those genes that were autoscaled, the presented fold-change estimates are interpreted relative to changes in the predictor of interest scaled by the ratio of the individual to the group standard deviation of the outcome.
We analyzed exposure–response relationships using linear mixed-effects models of gene expression predicted by air pollutant level. Each pollutant–gene expression relationship was examined in a separate model. We estimated random intercepts by subject, nested within season and community, to account for correlated within-individual repeated measures. The best fitting covariance structure was autoregressive order 1, as determined by Akaike’s information criterion. Pollutant concentrations were averaged for 1–7 days preceding blood draws and mean-centered by season and community, as in the previous analyses (described in the Supplementary Material).3 We adjusted, a priori, for temperature over the same averaging time as the pollutant. We tested smoothed penalized spline terms adjusting for potential non-linear temperature associations; however, these did not improve the model fit and were not retained. In addition, we performed a post hoc analysis of distributed lag models for NFE2L2 and IL1B to assess lag effects (Supplementary Figures S1 and S2). Much of the published research informing our selection of candidate genes was based on experimental designs, and would not be expected to have the same temporal relationships observed in free-living individuals in an epidemiological investigation like the present. Therefore, in this exploratory study, we evaluated moving averages for 1 through 7 days in linear mixed-effects models and lags 0 through 6 days the distributed lag models, rather than selecting specific lags for specific genes. Because these models concern within-subject associations with variation in air pollution levels, they were not adjusted for individual time-invariant characteristics. Differences in such characteristics between subjects are partly accounted for by the random subject intercepts. Person-weeks when subjects reported any infections were excluded from analysis because infections could markedly alter transcription levels along inflammatory and other pathways (N = 11 person-weeks, 3% of total). To standardize comparisons between pollutants, we expressed results for interquartile range (IQR) increases in pollutant levels. Due to power limitations, we were unable to assess effect modification in this analysis.
Gene expression profiling from whole blood is complicated by the heterogeneity of cell types and their possible differential responses to stressors; changes in the concentration of inflammatory cells may change measured expression levels. Also, expression changes may be diluted if a particular gene is only expressed in a small subset of cells.35,36 However, the aim of this exploratory analysis was not to assess expression levels in particular cell types, but to characterize overall expression levels in peripheral whole blood. Due to IRB limitations in allowed total blood volumes, we were unable to collect repeated measurements of leukocyte differential counts. The use of repeated measures, with subjects effectively serving as their own controls, helps to reduce the effect of inherent biological variability of blood cell distribution between people on model results. Because within-person cell shifts may result in gene expression shifts, we a priori planned to utilize expression of cell surface markers as a means to adjust models for cell distribution as described in the online Supplementary Material. However, these results were not informative, and we present exposure–response models with the limitation that they were not adjusted for gene expression of cell surface markers.
Because we evaluated many models for this analysis, we attempted to quantify the extent of our multiple comparison problem. We used a permutation analysis to simulate the distribution of the Wald statistic for each association estimate under the strong null hypothesis that there is no relationship between gene expression level and pollutant level. We permuted the gene expression level outcomes 26,500 times for each exposure, within each individual subject, and re-ran our mixed-effects model. We then compared our observed Wald statistic with the critical value corresponding to a family-wise level 0.05 test resulting from the simulated distribution after accounting for all comparisons. We used this method rather than the more conservative Bonferroni adjustment because the pollutant exposure levels are correlated, hence the resulting tests are also likely to be correlated. While this approach partially accounts for the observed correlation among tests, the permutation correction is still likely to produce somewhat conservative inference as it bounds the family-wise type I error relative to the maximum observed statistic across all comparisons. This computationally intensive, somewhat conservative adjustment method generated increased P-values that were no longer significant as might be expected. This may be attributable to the limited number of subjects and large number of models. We performed this simulation for a two gene subset (IL1B and NFE2L2) and present one, NFE2L2, as an example in the online Supplementary Material (Supplementary Figure S3, Supplementary Table S4). The observed values presented here are unadjusted with the caveat that these P-values are likely to be non-significant when adjusting the family-wise type I error rate at level 0.05. As such, we view the resulting regression parameter estimates as evidence of potential effects to generate hypotheses for future studies.
RESULTS
Population and Exposures
Table 1 shows cohort characteristics derived from subject questionnaires. Descriptive statistics of air pollutant exposures are listed in Table 2. The pollutant correlations are shown in Supplementary Tables S4 and S5. Briefly, primary combustion-related air pollutants (EC, BC, OCpri, NOX, and CO) were strongly correlated with each other and weakly correlated with secondary photochemically-related pollutants (O3 and SOC). Individual components from a given particle size fraction were most highly correlated with the other components from that size fraction. Quasi-ultrafine PM0.25 hopanes (a tracer of vehicular sources) and PM0.25 PAH had a Spearman correlation coefficient of 0.77, supporting the view that PAH is largely attributable to traffic sources in the study area.
Table 1.
Subject characteristics (N = 43).
| Characteristic | Value |
|---|---|
| Age (years, Mean ±SD) | 84.7 ±5.83 |
| Female | 53.5% |
| Race | |
| Hispanic | 2.3% |
| White | 97.7% |
| Cardiovascular history | |
| Confirmation of CAD | |
| Myocardial Infarction | 44.2% |
| Coronary artery bypass graft or angioplasty | 32.6% |
| Positive angiogram or stress test | 16.3% |
| Clinical diagnosis | 7.0% |
| Congestive heart failure | 27.9% |
| Hypertension (by history) | 76.7% |
| Hypercholesterolemia (by history) | 67.4% |
| Medications | |
| ACE inhibitors and Angiotensin II receptor antagonists | 41.9% |
| HMG-CoA reductase inhibitors (statins) | 51.2% |
Table 2.
Descriptive statistics of outdoor air pollutant exposures in three retirement communities of the Los Angeles air basin.
| Exposurea | Mean (SD) | Median | IQR | Min/Max |
|---|---|---|---|---|
| PM0.25 mass (μg/m3) | 9.89 (3.97) | 9.25 | 6.27 | 3.31/19.3 |
| PM0.25 macrophage ROS (μg Zymosan equivalents/m3) | 41.4 (38.5) | 21.3 | 56.2 | 2.59/147 |
| PM0.25 organic components | ||||
| WSOC (μg/m3)b | 0.50 (0.22) | 0.49 | 0.28 | 0.08/1.01 |
| PAH total (ng/m3) | 1.13 (0.48) | 0.97 | 0.46 | 0.55/2.70 |
| PAH LMW (ng/m3) | 0.41 (0.15) | 0.36 | 0.16 | 0.21/0.74 |
| PAH MMW (ng/m3) | 0.37 (0.18) | 0.34 | 0.18 | 0.11/0.96 |
| PAH HMW (ng/m3) | 0.35 (0.20) | 0.30 | 0.27 | 0.13/1.01 |
| Hopanes (ng/m3) | 0.33 (0.31) | 0.25 | 0.44 | 0.06/1.57 |
| Organic Acids (μg/m3) | 0.26 (0.22) | 0.19 | 0.35 | 0.06/0.96 |
| PM0.25–2.5 Mass (μg/m3) | 12.2 (5.31) | 11 | 7.14 | 3.89/28.1 |
| PM0.25–2.5 Macrophage ROS (μg Zymosan equivalents/m3) | 84.9 (55.8) | 84 | 97.8 | 9.03/203 |
| PM0.25–2.5 organic components | ||||
| WSOC (μg/m3)b | 0.50 (0.29) | 0.48 | 0.36 | 0.16/1.37 |
| PAH total (ng/m3) | 0.53 (0.17) | 0.47 | 0.26 | 0.36/1.01 |
| PAH LMW (ng/m3) | 0.17 (0.03) | 0.15 | 0.03 | 0.14/0.30 |
| PAH MMW (ng/m3) | 0.14 (0.09) | 0.09 | 0.10 | 0.08/0.39 |
| PAH HMW (ng/m3) | 0.22 (0.06) | 0.21 | 0.07 | 0.13/0.41 |
| Hopanes (ng/m3) | 0.49 (0.29) | 0.39 | 0.25 | 0.16/1.45 |
| Organic acids (μg/m3) | 48.8 (38.5) | 40.7 | 41.6 | 9.74/150 |
| PM2.5 components | ||||
| Elemental carbon (μg/m3) | 1.63 (0.60) | 1.58 | 0.82 | 0.36/3.34 |
| Organic carbon (μg/m3) | 6.81 (2.80) | 6.09 | 3.57 | 2.46/13.8 |
| Black carbon (μg/m3) | 1.88 (0.76) | 1.76 | 0.91 | 0.50/4.51 |
| Primary organic carbon (μg/m3) | 4.37 (2.11) | 3.62 | 2.39 | 1.41/10.6 |
| Secondary organic carbon (μg/m3) | 2.76 (1.41) | 2.61 | 1.82 | 0.27/7.65 |
| Particle number (particle no./cm3) | 14,686 (5,910) | 13,331 | 6,729 | 2,019/30,180 |
| Gases | ||||
| NO2 (ppb) | 31.8 (9.58) | 31.3 | 13.2 | 9.91/58.1 |
| NOx (ppb) | 56.5 (30.3) | 50.0 | 35.3 | 11.8/183 |
| CO (ppm) | 0.63 (0.27) | 0.57 | 0.38 | 0.21/1.68 |
| O3 (ppb) | 24.9 (11.4) | 23.5 | 15.2 | 3.83/60.7 |
| Temperature (°C) | 19.6 (5.43) | 19.6 | 8.92 | 8.42/31.4 |
Abbreviations: HMW, high molecular weight (>4 ring); IQR, interquartile range; LMW, low molecular weight (2–3 ring); MMW, medium molecular weight (4 ring); PAH, polycyclic aromatic hydrocarbon; ROS, reactive oxygen species; WSOC, water-soluble organic carbon.
Gene Expression and Regression Models
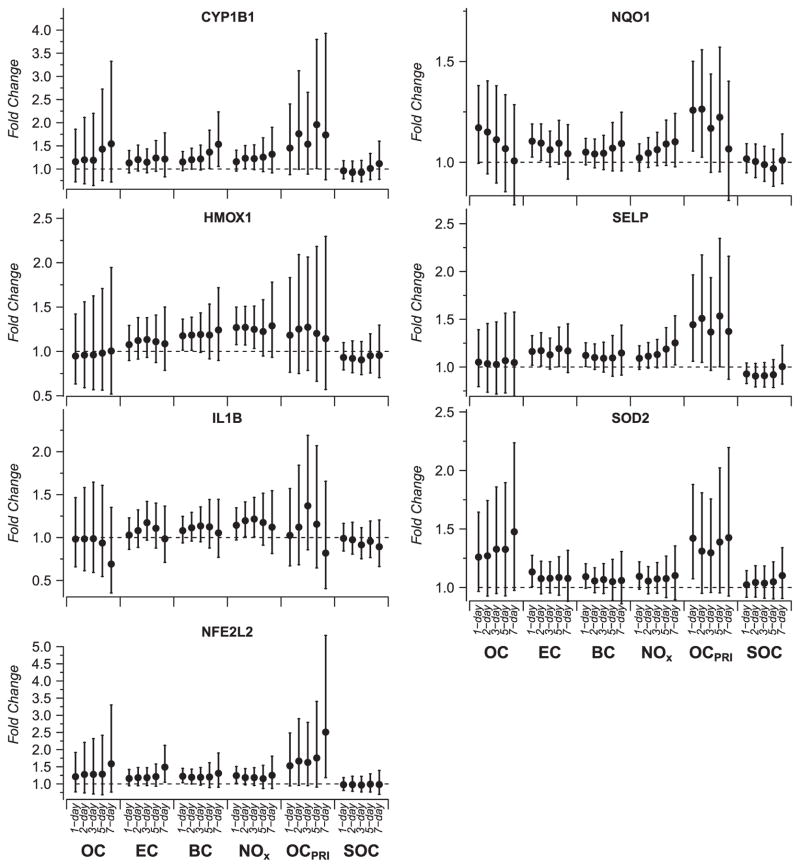
We analyzed repeated quantitative PCR measures of 35 gene expression levels from 43 subjects (360 person-observations per gene on average). Expression levels of five of these candidate genes (CYP1A1, PON1, SOD1, NOX1, and XDH) were too low for quantification, leaving 30 genes for analysis. Using linear mixed-effects models, we found traffic-related air pollutants were associated with expression of genes in multiple pathways including inflammation, platelet activation, and Nrf2-mediated oxidative stress response (Figure 2). We present 95% confidence intervals (CIs) unadjusted for multiple testing because the computationally intense adjustment method, described in Materials and methods, confirmed that our study was underpowered for the number of models we explored. Many of the unadjusted 95% CIs for the fold-change estimates included 1. Figures 2 and 3 show model results for exposures with among the strongest associations. Nevertheless, there was a consistency of associations in terms of their magnitude and direction across each of the genes within the a priori defined pathways. Specifically, we found positive associations of EC, BC, OCpri, and NOX with the Nrf2 gene (NFE2L2), and Nrf2-mediated/linked genes, HMOX1, NQO1, and SOD2. The largest association with NFE2L2 was for the 7-day average of OCpri (2.51 fold-change, unadjusted 95% CI: 1.18–5.53, per IQR increase). Traffic-related air pollutants were positively associated with increased expression of IL1B, SELP, and CYP1B1 whose transcription is not directly Nrf2-mediated. The largest associations found for SELP and CYP1B1 were with OCpri (1.53 fold-change in SELP, unadjusted 95% CI: 1–2.35, and 1.96 fold-change in CYP1B1, unadjusted 95% CI: 1.01–3.80, per IQR increase in 5-day OCpri).
Figure 2.
Fold-change and 95% confidence interval for qPCR expression level of CYP1B1, HMOX1, IL1B, NQO1, NFE2L2, SELP, and SOD2 in association with air pollutants measured hourly, including traffic-related air pollutants (elemental carbon, black carbon, primary organic carbon, and NOx). Time period shown indicates the number of days preceding gene expression measurement over which pollutant concentration was averaged.
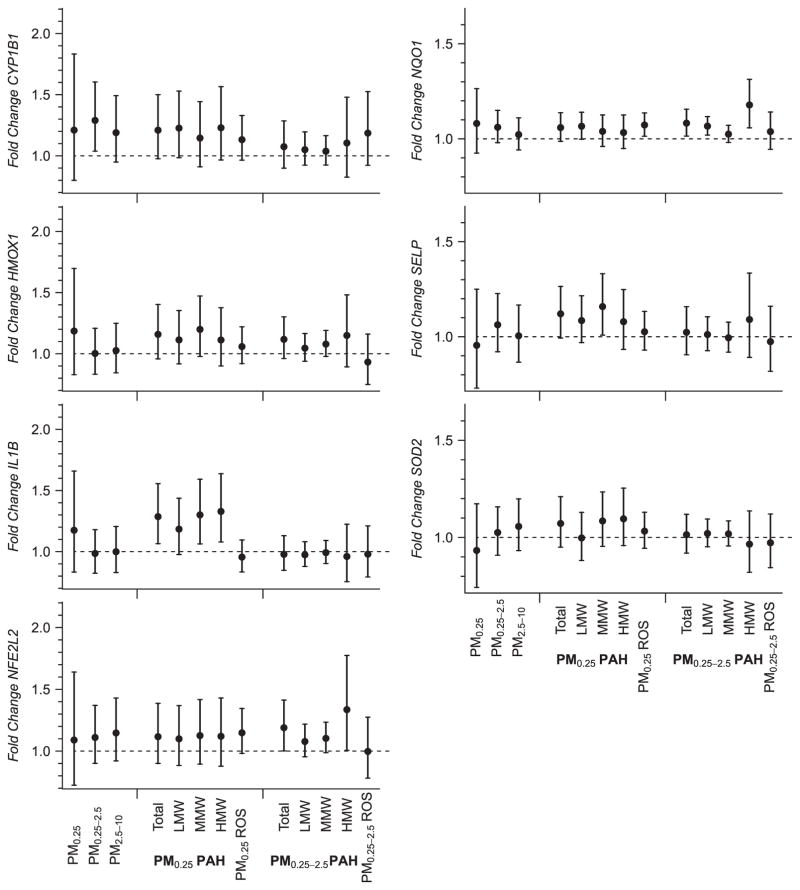
Figure 3.
Fold-change and 95% confidence interval for qPCR expression level of CYP1B1, HMOX1, IL1B, NQO1, NFE2L2, SELP, and SOD2 in association with particulate matter size fractions, particulate matter polycyclic aromatic hydrocarbons, and particulate matter oxidative potential (measured as macrophage ROS production in vitro). Time period shown indicates the number of days preceding gene expression measurement over which pollutant concentration was averaged.
Figure 3 shows gene expression fold-change estimates for mass concentrations of PM size fractions, size-fractionated particle components, and oxidative potential of particle extracts. Findings for PAH from particle extracts in Figure 3 are consistent with those for other PM markers of primary combustion (EC, BC, and primary OC) (Figure 2). Overall, positive associations were seen for CYP1B1, HMOX1, IL1B, NQO1, NFE2L2, SELP, and SOD2 in relation to PAHs, but did not differ substantially by PAH molecular weight class. Among the strongest of these associations was qUFP PAH and IL1B; we show a 1.33 fold-change in IL1B (unadjusted 95% CI: 1.08–1.64) per IQR increase in qUFP high molecular weight PAH. Accumulation mode PAH of high molecular weight had positive associations (unadjusted P<0.05) with NQO1 (1.18 fold-change, unadjusted 95% CI: 1.06–1.31, per IQR increase) and NFE2L2 (1.34 fold-change increase, unadjusted 95% CI: 1.01–1.77, per IQR increase), as well as positive but non-significant associations with CYP1B1, HMOX1, and SELP.
PM0.25 in vitro ROS was associated with increases in NFE2L2 gene expression (1.15 fold-change per IQR increase in PM0.25 ROS, unadjusted 95% CI: 0.98–1.34). PM0.25 in vitro ROS was also positively associated with NQO1 and CYP1B1, while PM0.25–2.5 ROS was only associated with CYP1B1. PM from biomass burning was positively associated (unadjusted P<0.05) with HMOX1 (not shown), and was positively, but not significantly, associated with expression of several others (ATF4, ATF6, GCLM, IL1B, KLF2, MPO, and XBP1). Photochemically-related secondary pollutant exposures were not associated with gene expression, including size-fractionated particle components (SOA and organic acids), and PM2.5 SOC and O3 (see Supplementary Table S6). Other air pollutants measured daily (total OC and CO) were not associated with the expression of genes (not shown). We also found no trends of association of any pollutants with downregulation of genes.
Air pollutant exposures were not associated with expression of any of the following genes: AHR, CCL2, CXCL1, DUSP1, F3, GCLC, GPX-1, GSTP1, HSPA8, IL6, IL6R, IL8, PTGS2, TNF, and TNFRS1B.
The online Supplementary Material (Supplementary Figures S1 and S2) shows results of a sensitivity analysis to assess lag effects using a polynomial distributed lag model. Results for a few models showed that the strongest effects could be from exposure within 24 h, but most models were largely uninformative suggesting that cumulative average exposures were a better reflection of the underlying exposure–response relationships.
To assess the significance of our inference that genes mediated by Nrf2 were differentially expressed relative to those that are not, we performed a post hoc pathway analysis based on the methods described for Gene Set Analysis.37 The details of this analysis can be found in the online Supplementary Material. Using this method, the P-value for the differential effect of the Nrf2 pathway was non-significant at 0.28.
DISCUSSION
We have shown that candidate gene expression from whole blood is associated with traffic-related air pollution exposures among elderly subjects with CAD living in the Los Angeles air basin. We found many positive associations within the Nrf2-mediated oxidative stress response pathway, providing preliminary support for our hypothesis that gene expression in pathways relevant to pro-oxidant exposures would be associated with traffic-related air pollutant exposures. This hypothesis-driven exploratory test of probable pathways for air pollutant responses generated results that could help bridge the gap between population-level epidemiological studies and both in vivo and in vitro toxicological experiments. To our knowledge, this is the first cohort panel study to show such gene expression changes in relation to urban air pollution exposures in the outdoor home environment.
Among the genes with positive associations, the Nrf2 gene is of particular interest. This transcription factor regulates Phase II and other antioxidant genes by binding antioxidant response elements (AREs) of their promoter regions. In a study of cardiac fibroblasts, Nrf2 expression was shown to be critical for total SOD and NQO1 enzymatic activity.38 In vitro evidence showed that diesel exhaust particles increased Nrf2 protein expression,39 while urban fine and ultrafine PM induced expression of both Nrf2-mediated oxidative stress response pathway genes and xenobiotic metabolism signaling pathway genes.6 Mouse models showed that UFP exposure increased NFE2L2, SOD2, CAT, NQO1, and ATF4 gene expression.40 Our results in human subjects, together with these and other studies, support the probable role of Nrf2 in air pollution exposure–response relationships.
Further supporting the importance of Nrf2, we found positive associations of NFE2L2 expression with PAHs. PAHs are markers of air pollution from fossil fuel combustion sources known to activate the aryl hydrocarbon receptor (AHR). Once bound, the AHR interacts with xenobiotic response elements (XREs) in the promoter regions of xenobiotic metabolism genes, initiating PAH metabolism to pro-oxidant compounds. Thus, PAH metabolites could then trigger Nrf2-ARE. Conversely, Nrf2 has been shown to bind to a functional ARE in the promoter of AHR in mouse embryonic fibroblasts.41 Thus, NFE2L2 expression can lead to AHR-XRE activation. Formerly considered separate response pathways, this recent evidence identifies crosstalk between the xenobiotic and antioxidant response pathways. We found increased NFE2L2 expression along with increased expression in downstream genes in both AHR-XRE and Nrf2-ARE pathways. Much of the existing literature describes Nrf2 as a master regulator of antioxidant response,38 and our novel findings here are in agreement with this. Our previous observations of this cohort panel showed that traffic-related air pollutants were associated with increased blood pressure,42 ambulatory electrocardiographic evidence of cardiac ischemia (ST segment depression),43 and ventricular arrhythmia.44 Nrf2 pathway involvement in gene expression–air pollution exposure associations suggests oxidative stress as a potential mode of action by which these exposures could cause the adverse cardiovascular outcomes observed in many air pollution studies.1
In our analysis of genes mediated by Nrf2, including AHR and CYP1B1, we found a non-significant maxmean statistic, though the trend among these genes suggests that they may be differentially expressed in association with the exposures we measured. This was partly driven by several Nrf2-linked genes that were not significantly associated with air pollutants (AHR, ATF4, CAT, DUSP1, GCLC, GCLM, GSTP1, and TXNRD1). We also considered the phase I and II metabolism and Nrf2-mediated oxidative stress response pathways linked, but expression levels of three candidate genes from the AHR-XRE pathway (SOD1, CYP1A1, and PON1) could not be included as they were below the limit of detection. Nevertheless, because of the established role of Nrf2 in regulating oxidative stress responses,38 our findings highlight the possible importance of the Nrf2 pathway in humans with ambient traffic-related air pollution exposures.
It is unclear whether the associations of estimated biomass burning PM with HMOX1 and MPO are meaningful and further work is needed. However, those results support the involvement of oxidative stress in the known associations of biomass burning with adverse respiratory outcomes.45
We also showed IL1B and SELP were positively associated with traffic-related air pollutant exposure. In our previous work, we found associations between levels of circulating sP-selectin (a protein marker of platelet activation encoded by SELP), and air pollution exposure, indicating that subjects at higher exposures had increased coagulation potential.2 P-selectin has an established role in vascular disease progression.46 IL-1β contributes to atherosclerosis progression by mediating vascular injury responses.47 Together, the increases we found in IL1B and SELP may signal endothelial damage or dysfunction that could trigger heightened coagulation. Because these genes do not rely on the Nrf2-mediated oxidative stress response, and neither shows a strong association with PM oxidative potential, we speculate that induction of these genes may occur via non-oxidative stress mechanisms.
Though our study is limited in the number of subjects (N = 43), the use of many repeated measures for each individual (≤12) allowed us to detect trends in expression at low fold-change levels. While many unadjusted 95% CIs for the fold change estimates included 1, there was overall consistency in the associations. Using false discovery rate logic, we would expect approximately 140 significant associations (30 genes over 19 exposures with 5 lags yields 2850 models, at alpha = 0.05). However, this approach neglects correlations between our variables of interest and does not have an intrinsic known null distribution. Using a simulated null distribution for our variables, our results did not reach significance with adjustment for multiple comparisons, as our study is likely underpowered for the number of models we analyzed. Considering models for different averaging times, multiple positive associations of similar magnitude, even if some are non-significant, suggest a possible overall relationship between a given gene and air pollutant. In addition, if our findings resulted from chance, we would not expect to see trends within pollutant classes or grouping of results by hypothesized biological pathways. Specifically, primary combustion-related air pollutants (EC, BC, OCpri, NOX, and CO) were strongly correlated with each other and likely were similar surrogates for the underlying causal pollutants components, which could have included PAH (a specific group of primary combustion-related air pollutant chemicals). Furthermore, we developed a priori hypotheses regarding specific candidate gene expression changes using published data; thus, our interpretations of these exploratory results are supported by biological plausibility and experimental evidence.
We are aware that some of our associations could have resulted from unmeasured confounding or high variability between individuals.20 Many of the factors affecting variability identified by Dumeaux et al.20 were either not relevant or controlled in our study. Of these, we could not control for blood cell subtype variation, but did exclude observations when subjects experienced infections to limit the effect infections would have had on cell type distribution and inflammation. There is work describing the use of methylation profiles to identify variations in cell type in whole blood.48,49 However, we did not collect repeated DNA samples in this panel study; therefore, we were unable to utilize methods like these to further characterize the gene expression relations with specific cell subsets. While we modeled random intercepts in mixed models to account for time-invariant subject differences, unmeasured time-variant factors (e.g. diet), could have affected the study results. Furthermore, the dose–response relationships between pollutants and outcomes, and potential compensatory mechanisms and feedback pathways complicate these reported associations in ways we cannot capture.
We selected genes for their possible associations with cardiovascular outcomes, thus, in our population with pre-existing cardiovascular disease, changes in these genes may occur due to underlying cardiovascular disease changes, rather than air pollution exposure. Disease progression is an unmeasured time-variant factor, but we do not expect substantial, rapid progression for our entire cohort during or across these short 6-week study periods, which took place across a 5½ month period for each subject. In addition, these genes were also selected for their relevance to toxicant metabolism and detoxification pathways. While we cannot definitively state that our associations are robust, the results are of substantial interest in understanding our previously reported findings in the same cohort, namely, significant associations of protein biomarkers and acute cardiovascular outcomes with fossil fuel combustion-linked air pollutant exposures.2,3,17,42,43,50 Additionally, we cannot generalize our results to younger, healthier, and more diverse populations, or other geographical areas because pollutant composition varies in urban areas other Los Angeles. We modeled associations using outdoor air pollutant levels. Our recent results show that, in our study locations, indoor levels of markers of traffic pollution, including EC and PAH, were strongly affected by outdoor levels.51 In addition, we determined from daily diaries that the percentage of time spent outdoors while at home to be low (3.7%), and subjects spent an additional 7.4% of their time away from the community at both indoor and outdoor locations. Therefore, our exposure assessments based on measurements at outdoor community environments are likely to accurately reflect the exposures of our study participants. While similar combustion-related air pollutants could show comparable associations in other regions, future studies are needed to examine these gene expression associations in other cohorts.
CONCLUSIONS
We report novel associations of gene expression changes with traffic-related air pollution exposures in a Los Angeles cohort of elderly subjects with CAD in this exploratory panel study analysis. We infer that the observed associations are coherent with experimental mechanistic data and with published associations between air pollution exposure from fossil fuel combustion sources and adverse cardiovascular outcomes, including hospital admissions and mortality.1 Results suggest that further work is warranted to test whether NFE2L2 gene expression may link air pollution from fossil fuel combustion with phase I and phase II enzyme genes via pathway crosstalk at the promoter transcription level. Additional research is needed to confirm these associations in other populations and to more thoroughly characterize the role of the Nrf2-mediated oxidative stress response pathway in cardiovascular responses to traffic-related and other air pollutant exposures.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Environmental Health Sciences at the National Institutes of Health (NIH, R01-ES12243 and R21-ES016420), the California Air Resources Board (contracts 03-329 and 09-341), and the National Center for Research Resources at the National Institutes of Health (NIH, MO1-RR00827). SW is supported by the National Institute of Environmental Health Sciences at the National Institutes of Health (NIH, F30 ES21107) and the Stanley Behrens UCI Graduate Division Public Impact Fellowship. ND is supported by the University of Southern California Provost’s PhD fellowship. The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the funding agencies. We thank the Department of Epidemiology and General Clinical Research Center, University of California Irvine; Department of Civil and Environmental Engineering, University of Southern California (USC); the Wisconsin State Laboratory of Hygiene; the California Air Resources Board; and the South Coast Air Quality Management District.
Footnotes
CONFLICT OF INTEREST
CS receives royalties from SKC for the air sampling device (Sioutas Personal Cascade Impactor Sampler) used in this research.
Supplementary Information accompanies the paper on the Journal of Exposure Science and Environmental Epidemiology website (http://www.nature.com/jes)
References
- 1.Brook RD, Rajagopalan S, Pope CA, 3rd, Brook JR, Bhatnagar A, Diez-Roux AV, et al. Particulate matter air pollution and cardiovascular disease: an update to the scientific statement from the American Heart Association. Circulation. 2010;121:2331–2378. doi: 10.1161/CIR.0b013e3181dbece1. [DOI] [PubMed] [Google Scholar]
- 2.Delfino RJ, Staimer N, Tjoa T, Gillen DL, Polidori A, Arhami M, et al. Air pollution exposures and circulating biomarkers of effect in a susceptible population: clues to potential causal component mixtures and mechanisms. Environ Health Perspect. 2009;117:1232–1238. doi: 10.1289/ehp.0800194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delfino RJ, Staimer N, Tjoa T, Arhami M, Polidori A, Gillen DL, et al. Association of biomarkers of systemic inflammation with organic components and source tracers in quasi-ultrafine particles. Environ Health Perspect. 2010;118:756–762. doi: 10.1289/ehp.0901407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gong KW, Zhao W, Li N, Barajas B, Kleinman M, Sioutas C, et al. Air-pollutant chemicals and oxidized lipids exhibit genome-wide synergistic effects on endothelial cells. Genome Biol. 2007;8:R149. doi: 10.1186/gb-2007-8-7-r149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gargalovic PS, Gharavi NM, Clark MJ, Pagnon J, Yang WP, He A, et al. The unfolded protein response is an important regulator of inflammatory genes in endothelial cells. Arterioscler Thromb Vasc Biol. 2006;26:2490–2496. doi: 10.1161/01.ATV.0000242903.41158.a1. [DOI] [PubMed] [Google Scholar]
- 6.Huang YC, Karoly ED, Dailey LA, Schmitt MT, Silbajoris R, Graff DW, et al. Comparison of gene expression profiles induced by coarse, fine, and ultrafine particulate matter. J Toxicol Environ Health A. 2011;74:296–312. doi: 10.1080/15287394.2010.516238. [DOI] [PubMed] [Google Scholar]
- 7.Lee R, Margaritis M, Channon KM, Antoniades C. Evaluating oxidative stress in human cardiovascular disease: methodological aspects and considerations. Curr Med Chem. 2012;19:2504–2520. doi: 10.2174/092986712800493057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delfino RJ, Staimer N, Vaziri ND. Air pollution and circulating biomarkers of oxidative stress. Air Qual Atmos Health. 2011;4:37–52. doi: 10.1007/s11869-010-0095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Z, Liu MC, Liang M, Fu J. Sirt1 protects against thrombomodulin down-regulation and lung coagulation after particulate matter exposure. Blood. 2012;119:2422–2429. doi: 10.1182/blood-2011-04-350413. [DOI] [PubMed] [Google Scholar]
- 10.Das H, Kumar A, Lin Z, Patino WD, Hwang PM, Feinberg MW, et al. Kruppel-like factor 2 (KLF2) regulates proinflammatory activation of monocytes. Proc Natl Acad Sci USA. 2006;103:6653–6658. doi: 10.1073/pnas.0508235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peretz A, Peck EC, Bammler TK, Beyer RP, Sullivan JH, Trenga CA, et al. Diesel exhaust inhalation and assessment of peripheral blood mononuclear cell gene transcription effects: an exploratory study of healthy human volunteers. Inhal Toxicol. 2007;19:1107–1119. doi: 10.1080/08958370701665384. [DOI] [PubMed] [Google Scholar]
- 12.Pettit AP, Brooks A, Laumbach R, Fiedler N, Wang Q, Strickland PO, et al. Alteration of peripheral blood monocyte gene expression in humans following diesel exhaust inhalation. Inhal Toxicol. 2012;24:172–181. doi: 10.3109/08958378.2012.654856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu MT, Lee TC, Wu IC, Su HJ, Huang JL, Peng CY, et al. Whole genome expression in peripheral-blood samples of workers professionally exposed to polycyclic aromatic hydrocarbons. Chem Res Toxicol. 2011;24:1636–1643. doi: 10.1021/tx200181q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Leeuwen DM, Pedersen M, Hendriksen PJ, Boorsma A, van Herwijnen MH, Gottschalk RW, et al. Genomic analysis suggests higher susceptibility of children to air pollution. Carcinogenesis. 2008;29:977–983. doi: 10.1093/carcin/bgn065. [DOI] [PubMed] [Google Scholar]
- 15.van Leeuwen DM, van Herwijnen MH, Pedersen M, Knudsen LE, Kirsch-Volders M, Sram RJ, et al. Genome-wide differential gene expression in children exposed to air pollution in the Czech Republic. Mutat Res. 2006;600:12–22. doi: 10.1016/j.mrfmmm.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 16.McHale CM, Zhang L, Hubbard AE, Smith MT. Toxicogenomic profiling of chemically exposed humans in risk assessment. Mutat Res. 2010;705:172–183. doi: 10.1016/j.mrrev.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delfino RJ, Staimer N, Tjoa T, Polidori A, Arhami M, Gillen DL, et al. Circulating biomarkers of inflammation, antioxidant activity, and platelet activation are associated with primary combustion aerosols in subjects with coronary artery disease. Environ Health Perspect. 2008;116:898–906. doi: 10.1289/ehp.11189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sioutas C, Delfino RJ, Singh M. Exposure assessment for atmospheric ultrafine particles (UFPs) and implications in epidemiologic research. Environ Health Perspect. 2005;113:947–955. doi: 10.1289/ehp.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Klot S, Peters A, Aalto P, Bellander T, Berglind N, D’Ippoliti D, et al. Ambient air pollution is associated with increased risk of hospital cardiac readmissions of myocardial infarction survivors in five European cities. Circulation. 2005;112:3073–3079. doi: 10.1161/CIRCULATIONAHA.105.548743. [DOI] [PubMed] [Google Scholar]
- 20.Dumeaux V, Olsen KS, Nuel G, Paulssen RH, Borresen-Dale AL, Lund E, et al. Deciphering normal blood gene expression variation--The NOWAC postgenome study. PLoS Genet. 2010;6:e1000873. doi: 10.1371/journal.pgen.1000873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rainen L, Oelmueller U, Jurgensen S, Wyrich R, Ballas C, Schram J, et al. Stabilization of mRNA expression in whole blood samples. Clin Chem. 2002;48:1883–1890. [PubMed] [Google Scholar]
- 22.Elvidge GP, Price TS, Glenny L, Ragoussis J. Development and evaluation of real competitive PCR for high-throughput quantitative applications. Anal Biochem. 2005;339:231–241. doi: 10.1016/j.ab.2005.01.040. [DOI] [PubMed] [Google Scholar]
- 23.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polidori A, Arhami M, Sioutas C, Delfino RJ, Allen R. Indoor/Outdoor relationships, trends, and carbonaceous content of fine particulate matter in retirement homes of the Los Angeles Basin. J Air Waste Manag Assoc. 2007;57:366–379. doi: 10.1080/10473289.2007.10465339. [DOI] [PubMed] [Google Scholar]
- 25.Singh M, Misra C, Sioutas C. Field evaluation of a personal cascade impactor sampler (PCIS) Atmos Environ. 2003;37:4781–4793. [Google Scholar]
- 26.Arhami M, Sillanpää M, Hu S, Olson MR, Schauer JJ, Sioutas C, et al. Size-segregated inorganic and organic components of PM in the communities of the Los Angeles Harbor. Aerosol Sci Technol. 2009;43:145–160. [Google Scholar]
- 27.Arhami M, Minguillon MC, Polidori A, Schauer JJ, Delfino RJ, Sioutas C, et al. Organic compound characterization and source apportionment of indoor and outdoor quasi-ultrafine particulate matter in retirement homes of the Los Angeles Basin. Indoor Air. 2010;20:17–30. doi: 10.1111/j.1600-0668.2009.00620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schauer JJ, Kleeman MJ, Cass GR, Simoneit BRT. Measurement of emissions from air pollution sources. 5.C1 – C32 organic compounds from gasoline-powered motor vehicles. Environ Sci Technol. 2002;36:1169–1180. doi: 10.1021/es0108077. [DOI] [PubMed] [Google Scholar]
- 29.Stone EA, Snyder DC, Sheesley RJ, Sullivan AP, Weber RJ, Schauer JJ, et al. Source apportionment of fine organic aerosol in Mexico City during the MILAGRO experiment 2006. Atmos Chem Phys. 2008;8:1249–1259. [Google Scholar]
- 30.Weber RJ, Sullivan AP, Peltier RE, Russell A, Yan B, Zheng M, et al. A study of secondary organic aerosol formation in the anthropogenic-influenced southeastern United States. J Geophys Res. 2007;112:D13302. [Google Scholar]
- 31.Docherty KS, Stone EA, Ulbrich IM, DeCarlo PF, Snyder DC, Schauer JJ, et al. Apportionment of primary and secondary organic aerosols in Southern California during the 2005 study of organic aerosols in Riverside (SOAR-1) Environ Sci Technol. 2008;42:7655–7662. doi: 10.1021/es8008166. [DOI] [PubMed] [Google Scholar]
- 32.Sannigrahi P, Sullivan AP, Weber RJ, Ingall ED. Characterization of water-soluble organic carbon in urban atmospheric aerosols using solid-state 13C NMR spectroscopy. Environ Sci Technol. 2005;40:666–672. doi: 10.1021/es051150i. [DOI] [PubMed] [Google Scholar]
- 33.Landreman AP, Shafer MM, Hannigan MP, Schauer JJ. A macrophage-based method for the assessment of the reactive oxygen species (ROS) activity of atmospheric particulate matter (PM) and application to routine (daily-24 h) aerosol monitoring studies. Aerosol Sci Technol. 2008;42:946–957. [Google Scholar]
- 34.Willems E, Leyns L, Vandesompele J. Standardization of real-time PCR gene expression data from independent biological replicates. Anal Biochem. 2008;379:127–129. doi: 10.1016/j.ab.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 35.Wurmbach E, Gonzalez-Maeso J, Yuen T, Ebersole BJ, Mastaitis JW, Mobbs CV, et al. Validated genomic approach to study differentially expressed genes in complex tissues. Neurochem Res. 2002;27:1027–1033. doi: 10.1023/a:1020900720328. [DOI] [PubMed] [Google Scholar]
- 36.Fan H, Hegde PS. The transcriptome in blood: challenges and solutions for robust expression profiling. Curr Mol Med. 2005;5:3–10. doi: 10.2174/1566524053152861. [DOI] [PubMed] [Google Scholar]
- 37.Efron B, Tibshirani R. On testing the significance of sets of genes. Ann Appl Stat. 2007;1:107–129. [Google Scholar]
- 38.Zhu H, Itoh K, Yamamoto M, Zweier JL, Li Y. Role of Nrf2 signaling in regulation of antioxidants and phase 2 enzymes in cardiac fibroblasts: protection against reactive oxygen and nitrogen species-induced cell injury. FEBS Lett. 2005;579:3029–3036. doi: 10.1016/j.febslet.2005.04.058. [DOI] [PubMed] [Google Scholar]
- 39.Li N, Alam J, Venkatesan MI, Eiguren-Fernandez A, Schmitz D, Di Stefano E, et al. Nrf2 is a key transcription factor that regulates antioxidant defense in macrophages and epithelial cells: protecting against the proinflammatory and oxidizing effects of diesel exhaust chemicals. J Immunol. 2004;173:3467–3481. doi: 10.4049/jimmunol.173.5.3467. [DOI] [PubMed] [Google Scholar]
- 40.Araujo JA, Barajas B, Kleinman M, Wang X, Bennett BJ, Gong KW, et al. Ambient particulate pollutants in the ultrafine range promote early atherosclerosis and systemic oxidative stress. Circ Res. 2008;102:589–596. doi: 10.1161/CIRCRESAHA.107.164970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin S, Wakabayashi N, Misra V, Biswal S, Lee GH, Agoston ES, et al. NRF2 modulates aryl hydrocarbon receptor signaling: influence on adipogenesis. Mol Cell Biol. 2007;27:7188–7197. doi: 10.1128/MCB.00915-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delfino RJ, Tjoa T, Gillen DL, Staimer N, Polidori A, Arhami M, et al. Traffic-related air pollution and blood pressure in elderly subjects with coronary artery disease. Epidemiology. 2010;21:396–404. doi: 10.1097/EDE.0b013e3181d5e19b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delfino RJ, Gillen DL, Tjoa T, Staimer N, Polidori A, Arhami M, et al. Electrocardiographic ST-segment depression and exposure to traffic-related aerosols in elderly subjects with coronary artery disease. Environ Health Perspect. 2011;119:196–202. doi: 10.1289/ehp.1002372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bartell S, Tjoa T, Longhurst J, Sioutas C, Delfino RJ. Particulate air pollution, ambulatory heart rate variability and arrhythmia in elderly subjects with coronary artery disease. Environ Health Perspect. 2013;121:1135–1141. doi: 10.1289/ehp.1205914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naeher LP, Brauer M, Lipsett M, Zelikoff JT, Simpson CD, Koenig JQ, et al. Woodsmoke health effects: a review. Inhal Toxicol. 2007;19:67–106. doi: 10.1080/08958370600985875. [DOI] [PubMed] [Google Scholar]
- 46.Woollard KJ, Chin-Dusting J. Therapeutic targeting of p-selectin in atherosclerosis. Inflamm Allergy Drug Targets. 2007;6:69–74. doi: 10.2174/187152807780077345. [DOI] [PubMed] [Google Scholar]
- 47.Galea J, Armstrong J, Gadsdon P, Holden H, Francis SE, Holt CM, et al. Interleukin-1 beta in coronary arteries of patients with ischemic heart disease. Arterioscler Thromb Vasc Biol. 1996;16:1000–1006. doi: 10.1161/01.atv.16.8.1000. [DOI] [PubMed] [Google Scholar]
- 48.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Accomando WP, Wiencke JK, Houseman EA, Nelson HH, Kelsey KT. Quantitative reconstruction of leukocyte subsets using DNA methylation. Genome Biol. 2014;15:R50. doi: 10.1186/gb-2014-15-3-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bartell SM, Longhurst J, Tjoa T, Sioutas C, Delfino RJ. Particulate air pollution, ambulatory heart rate variability, and cardiac arrhythmia in retirement community residents with coronary artery disease. Environ Health Perspect. 2013;121:1135–1141. doi: 10.1289/ehp.1205914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hasheminassab S, Daher N, Shafer MM, Schauer JJ, Delfino RJ, Sioutas C, et al. Chemical characterization and source apportionment of indoor and outdoor fine particulate matter (PM2.5) in retirement communities of the Los Angeles Basin. Sci Total Environ. 2014;490:528–537. doi: 10.1016/j.scitotenv.2014.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.