ABSTRACT
Multicellular biofilm formation and surface motility are bacterial behaviors considered mutually exclusive. However, the basic decision to move over or stay attached to a surface is poorly understood. Here, we discover that in Bacillus subtilis, the key root biofilm-controlling transcription factor Spo0A~Pi (phosphorylated Spo0A) governs the flagellum-independent mechanism of social sliding motility. A Spo0A-deficient strain was totally unable to slide and colonize plant roots, evidencing the important role that sliding might play in natural settings. Microarray experiments plus subsequent genetic characterization showed that the machineries of sliding and biofilm formation share the same main components (i.e., surfactin, the hydrophobin BslA, exopolysaccharide, and de novo-formed fatty acids). Sliding proficiency was transduced by the Spo0A-phosphorelay histidine kinases KinB and KinC. We discovered that potassium, a previously known inhibitor of KinC-dependent biofilm formation, is the specific sliding-activating signal through a thus-far-unnoticed cytosolic domain of KinB, which resembles the selectivity filter sequence of potassium channels. The differential expression of the Spo0A~Pi reporter abrB gene and the different levels of the constitutively active form of Spo0A, Sad67, in Δspo0A cells grown in optimized media that simultaneously stimulate motile and sessile behaviors uncover the spatiotemporal response of KinB and KinC to potassium and the gradual increase in Spo0A~Pi that orchestrates the sequential activation of sliding, followed by sessile biofilm formation and finally sporulation in the same population. Overall, these results provide insights into how multicellular behaviors formerly believed to be antagonistic are coordinately activated in benefit of the bacterium and its interaction with the host.
IMPORTANCE
Alternation between motile and sessile behaviors is central to bacterial adaptation, survival, and colonization. However, how is the collective decision to move over or stay attached to a surface controlled? Here, we use the model plant-beneficial bacterium Bacillus subtilis to answer this question. Remarkably, we discover that sessile biofilm formation and social sliding motility share the same structural components and the Spo0A regulatory network via sensor kinases, KinB and KinC. Potassium, an inhibitor of KinC-dependent biofilm formation, triggers sliding via a potassium-perceiving cytosolic domain of KinB that resembles the selectivity filter of potassium channels. The spatiotemporal response of these kinases to variable potassium levels and the gradual increase in Spo0A~Pi levels that orchestrates the activation of sliding before biofilm formation shed light on how multicellular behaviors formerly believed to be antagonistic work together to benefit the population fitness.
INTRODUCTION
How do bacteria move from one location to another in natural niches? Most bacteria are able to swim in aquatic environments powered by rotating flagella, whereas a range of different mechanisms have evolved that facilitate surface spreading (1–4). While swimming is considered to be an individual behavior, cells are able to migrate together and cooperatively during surface translocation (1, 3, 4). Surface movement can depend on the presence of flagella (i.e., swarming), the extension and retraction of type IV pili (i.e., twitching motility), the involvement of focal adhesion complexes (i.e., gliding), or “passive” surface translocation (i.e., sliding). Although the mechanisms of swarming, twitching, and gliding motilities have been extensively studied in most bacteria with appendages, the information about the mechanism of sliding, its regulation, and its importance is sparse. Since its original definition, more than 4 decades ago, the concept of sliding as a passive surface translocation driven by expansive forces in the growing colony has not varied much (2, 3). However, sliding motility represents a heavily exploited mechanism that different pathogens of global importance (i.e., Bacillus anthracis, Salmonella enterica, Staphylococcus aureus, Legionella pneumophila, and mycobacteria) use for spreading (3, 5, 6).
Bacillus subtilis is a Gram-positive endospore-forming bacterium that has been extensively studied due to its diverse differentiation processes (7–11). Different B. subtilis strains swarm on semisolid agar plates (4, 12) or form architecturally complex biofilms with vein-like structures and apical tips (fruiting bodies) that project the formed spores into the air (7, 13–15). In addition, wild and undomesticated B. subtilis isolates (7, 16) have beneficial growth-promoting effects on plants and animals (17, 18) as well as probiotic effects in humans (19–21). If biofilm formation and an active surface motility are antagonistic but important attributes of a bacterium, how then is the collective decision to move over or stay attached to a surface taken and controlled? In this work, we use the model organism B. subtilis to investigate the genetic mechanism and regulatory network of sliding for surface colonization and their relationship with another prominent cooperative surface behavior, i.e., biofilm formation.
RESULTS AND DISCUSSION
The master regulator of sporulation and multicellular biofilm formation, Spo0A, controls sliding motility in B. subtilis.
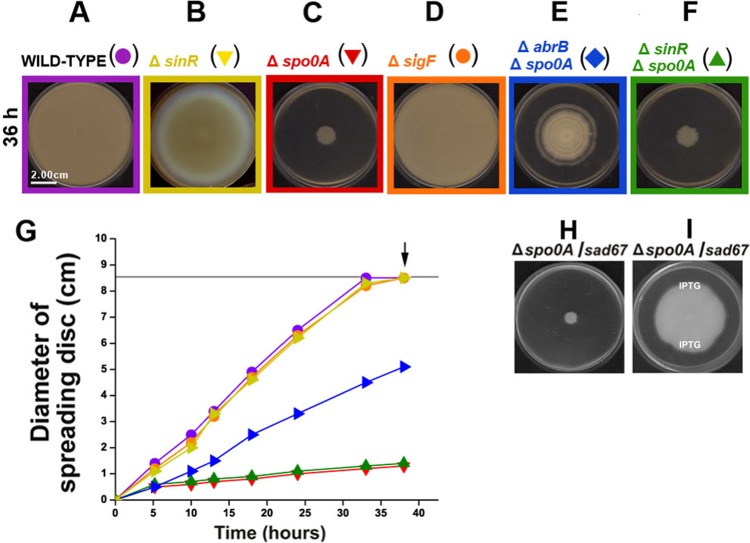
If bacteria use sliding in a cooperative manner to move across surfaces without the necessity for flagella or any other appendages, how does it take place and what are the regulatory networks that induce and control it? To solve this puzzle, we used two wild (undomesticated) B. subtilis strains of different genetic lineages, the Marburg-related strain NCIB3610, able to swarm and slide (7, 12, 22), and the human-probiotic natto-related strain RG4365 (16, 21), which only slides (see Fig. S1 in the supplemental material). It is known that the global transcription factor SinR is essential for the swimming and swarming motilities in B. subtilis (23). While, as expected, the inactivation of sinR in the NCIB3610 strain yields a completely defective swarming phenotype (24), the inactivation of sinR in the RG4365 B. subtilis natto strain yields an almost unaffected sliding-proficient phenotype (Fig. 1A, B, and G). If SinR is not required to slide, is there any other transcription factor that contributes to the regulatory network of sliding? In B. subtilis, other multicellular and developmental programs (biofilm formation, fruiting body formation, and sporulation) are governed by the master transcription factor Spo0A of the phosphorelay signaling pathway (7, 25). Taking into account the dispensability of SinR activity for sliding proficiency, we wondered if this was also the case for Spo0A in sliding. Although swarming of an NCIB3610-isogenic spo0A-deletion strain was not affected (see below), a spo0A-deletion RG4365-derived strain was completely unable to slide on the agar surface (Fig. 1C and G). This result suggested that the master regulator of sporulation and biofilm formation, the protein Spo0A, would also be a key regulator of social sliding. The inactivation of spoIIAC (sigF), the distal gene of the tricistronic spoIIA operon, coding for the first compartment-specific sporulation sigma transcription factor (σF), did not affect the sliding phenotype of wild-type RG4365 cells (Fig. 1D and G) and suggested the independency of sliding from the sporulation program in B. subtilis. It is known that the absence of Spo0A activity results in an increase of the activities of the transcription factors SinR and AbrB (26) that could be responsible for the absence of sliding ability in the spo0A natto strain. If this were the case, the regulatory proteins SinR and/or AbrB could be an inhibitor of sliding motility. As shown in Fig. 1E to G, the inactivation of abrB but not sinR was able to restore (although only partially) the sliding ability of Spo0A-deficient cells and suggested that AbrB was negatively controlling the expression of at least one gene whose product was necessary for sliding proficiency (see below). To confirm the essential role of Spo0A for the proficiency of social sliding in B. subtilis, we constructed an RG4365-derived strain that harbored, in addition to a deletion of the wild-type copy of spo0A, an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible form of Spo0A (Sad67) that is active in the absence of phosphorylation (phosphorelay independent) (27–29). As shown in Fig. 1H and I, the supplementation with IPTG restored the sliding ability of the spo0A-deletion but Sad67-carrying strain and confirmed the essential role of Spo0A for social sliding proficiency in B. subtilis.
FIG 1 .
Revealing the genetic regulation of sliding motility in B. subtilis. (A to G) Sliding phenotype (A to F) and kinetic characterization (G) of different B. subtilis natto strains (see Table S1 in the supplemental material) affected in the expression of key regulators of gene expression. B. subtilis cells were cultured and inoculated on LB-0.7% agar plates as indicated in Materials and Methods. The arrows in panel G indicate the developmental times when the photographs shown in panels A to F were taken. Strain references for the symbols in panel G correspond to the reference colors shown in panels A to F. The horizontal black line at 8.5 cm shows the maximal size of motility related to the size of the agar plate used. Each value is the average from three replicates. (H and I) Active Spo0A (Sad67) triggers sliding motility in the B. subtilis natto strain. In the absence of IPTG supplementation, Spo0A-deficient but Sad67-positive cells are not motile on soft agar plates (H), but in the presence of IPTG, these cells recover full sliding proficiency (I). Solid IPTG, one or two grains, was poured on top of the solidified LB-0.7% agar (at the points indicated in panel I) in order to allow the dissolution of IPTG in the medium and the formation of a continuous gradient of the inducer.
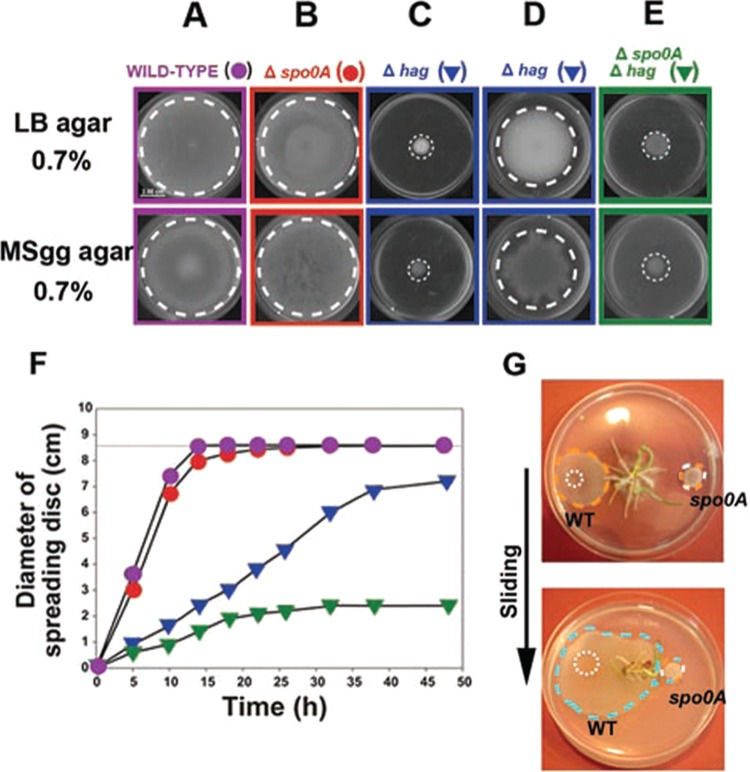
How widespread is sliding motility and how conserved is the role of Spo0A in different B. subtilis isolates? Although flagellum production is essential for swarming motility in the Marburg-related strain NCIB3610 (12), it has been reported that NCIB3610-derived hag strains (unable to make flagella) are able to slide on solid surfaces after longer periods of incubation (24 h or more) (30). Therefore, we wanted to know if the essential role of Spo0A for sliding proficiency in the B. subtilis natto strain is also manifest in Marburg-derived cells. To this end, we analyzed the surface translocation ability of different NCIB3610-derived strains under two experimental conditions: incubation on soft Luria-Bertani (LB) medium as shown in previous work and the conditions previously used by other groups, i.e., soft minimal salts glycerol glutamate (MSgg) agar medium, to investigate the sliding ability of Marburg cells (31). As shown in Fig. 2, the inactivation of spo0A in NCIB3610 cells did not affect their motility behavior (compare Fig. 2A and B). As expected, the interruption of flagellin synthesis in the NCIB3610 hag-derived strain RG4384 (see Table S1 in the supplemental material) blocked the surface translocation in both soft media as monitored at a developmental time of 20 h (Fig. 2C). However, after longer incubation, the NCIB3610 hag cells moved on the agar surface by a sliding mechanism as previously reported (Fig. 2D) (31). Remarkably, the inactivation of spo0A in the Marburg-derived hag strain (Δhag Δspo0A double mutant strain RG4385 [see Table S1]) completely abolished the ability of these flagellum-less and Spo0A-deficient NCIB3610-derived cells to translocate on the agar surface and confirmed the key role of Spo0A as the master regulator of social sliding motility in B. subtilis (Fig. 2E and F).
FIG 2 .
The key regulator of multicellular behavior, Spo0A, controls sliding motility in B. subtilis. (A and B) Spo0A activity is fully dispensable for the swarming proficiency of NCIB3610 cells. (C and D) Inactivation of flagellar synthesis (hag mutation) impairs swarming motility in NCIB3610 cells (C), but after a longer incubation (24 h or more), sliding proficiency is turned on in Hag-deficient cells (D). (E) Spo0A activity is essential for surface translocation ability of Hag-deficient cells. Photos shown in panels A to C and panels D and E correspond to the sliding migration of the indicated strains after 15 h and 40 h of incubation, respectively. (F) Kinetics of swarming and sliding motilities in Spo0A- and Hag-positive or -deficient NCIB3610 cells. (G) Important role of Spo0A and sliding proficiency for plant root colonization. As indicated in the supplemental material, as soon as sanitized wheat seeds germinated on LB-diluted agar plates, 3.0 µl of stationary-phase cultures of wild-type and spo0A mutant cells was inoculated at the points indicated by the white dotted circles in the top panel. After 24 h of incubation, the wild-type (WT) and spo0A cells formed rounded colonies of similar size and appearance that were confined to the point of inoculation (data not shown). After 3 days of bacterial inoculation, wild-type cells, but not the spo0A cells, efficiently slid on the agar surface (the boundaries of the sliding disc are denoted by the orange dashed circle in the top panel). After 5 days (bottom panel), the wild-type cells were able to colonize the root rhizosphere (blue dashed circle) while the spo0A cells remained immobilized. Representative images of several independent experiments are shown.
What might be the importance of bacterial sliding in nature? B. subtilis is a beneficial bacterium that improves plant and animal growth (17, 18) as well as possessing advantageous probiotic properties in humans (19, 21). One desired attribute of a host-colonizing bacterium is the ability to spread over and colonize a particular niche (i.e., the rhizosphere) and establish a long-lasting community (i.e., a biofilm) associated with the host (19, 32). Notably, as shown in Fig. 2G, Spo0A plays a key role in the ability of the plant growth-promoting rhizobacterium B. subtilis (33, 34) to activate social sliding and colonize the root rhizosphere.
Microarray analysis of B. subtilis cells under sliding conditions.
Our initial analysis uncovered the novel roles of the transcription factors AbrB and Spo0A as a repressor and an activator of sliding, respectively. What other genes are important for sliding proficiency in B. subtilis and what is the role of AbrB and Spo0A in their expression? To answer these questions, we performed microarray experiments under different environmental conditions and in various genetic backgrounds. On the one hand, we compared the global gene expression of the spo0A mutant RG4370 with that of the wild-type natto strain RG4365 on LB plates with a 0.7% agar concentration, where, as was shown, the wild-type and the spo0A mutant strains were proficient and impaired in sliding, respectively (see Table S2A in the supplemental material). In a second type of experiment, we examined wild-type cells under sliding-restrictive conditions using LB plates with a 1.5% agar concentration and compared their transcriptome with the pattern of gene expression under sliding-permissive conditions on 0.7% agar plates (see Table S2B). These microarray analyses showed that 310 and 295 genes were significantly (P value, <10−4) up- or downregulated in the spo0A mutant strain compared to the wild-type strain, respectively, while 72 and 100 genes were found to be up- and downregulated, respectively, at an increased agar concentration (see Table S2).
Interestingly, most of the genes belonging to the σD regulon, which is related to flagellum motility and chemotaxis, were activated in the spo0A mutant strain under sliding-permissive conditions. However, the B. subtilis natto strain lacks flagella under this and all tested genetic backgrounds (see Fig. S1E to G in the supplemental material and data not shown). It has been suggested that Marburg-related wild-type cells (i.e., NCIB3610) lack flagellum production for translocation on solid surfaces depending on extracellular surfactin and potassium ion (22). More recently, it was shown that synthesis of the exopolysaccharide (EPS) of the extracellular matrix is genetically coupled to the inhibition of flagellum-mediated motility (23), and as we show below, EPS expression is increased under sliding-permissive conditions.
While mutation in spo0A resulted in differential expression of various genes under sliding-permissive conditions on LB medium, we did not find any sporulation-related gene in the wild-type strain to be differentially expressed under this experimental condition (sliding turned on [see Table S2A in the supplemental material]). One simple explanation for this observation is that under the condition used (i.e., rich LB medium and sliding-permissive conditions), sporulation is not activated in the wild-type strain and, therefore, mutation in spo0A has no effect on these genes in the wild-type strain during sliding. In contrast, we observed elevated expression of sporulation σG-dependent genes in wild-type cells grown under non-sliding-permissive conditions (higher agar concentration [see Table S2B in the supplemental material]). This induction of sporulation genes in samples from 1.5% agar plates is probably due to the fact that under this sliding-restrictive condition, B. subtilis cannot spread, nutrients around cells become limited, and sporulation is started similarly to the conditions during formation of complex colony biofilms (11, 35, 36).
Which other genes are expressed during active sliding? We found that in wild-type cells under sliding-permissive conditions (see Table S2B in the supplemental material) and in comparison to the spo0A mutant strain (see Table S2A), genes related to biofilm matrix production (sipW, tasA, and eps in the case of the spo0A mutant), biofilm surface layer (bslA), fatty acid synthesis (fab), and surfactin synthesis (srfAC in the case of the spo0A mutant) were upregulated.
Sliding but not swarming depends on the bslA and eps genes.
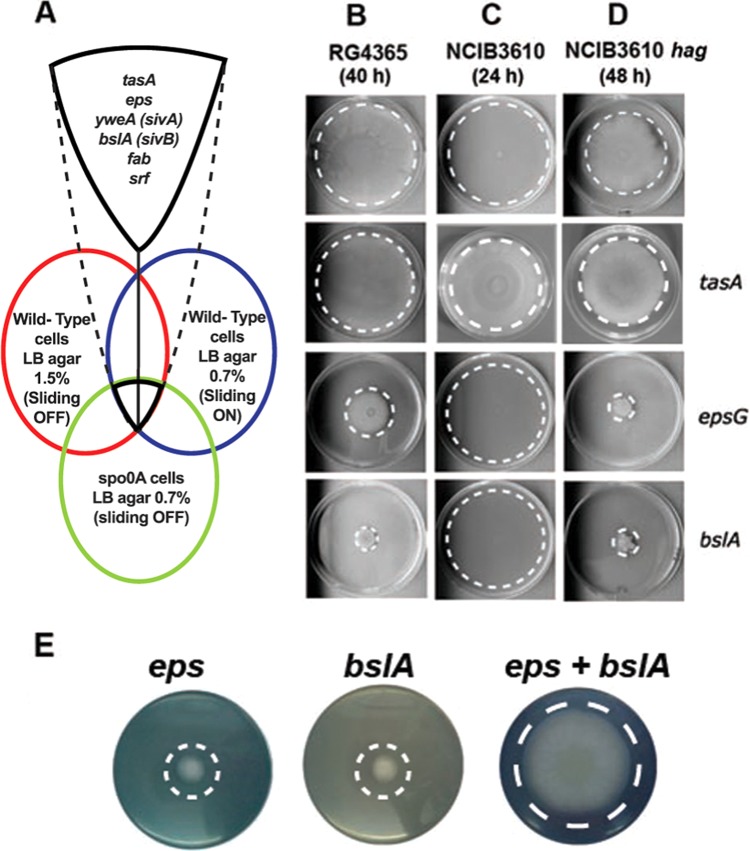
The microarray experiments presented above showed that genes related to biofilm formation and biofilm surface layer are upregulated under the conditions when sliding is feasible and suggest that this gene repertoire could include novel and necessary components of the sliding machinery in B. subtilis (Fig. 3A). Therefore, mutations in bslA, epsG, or tasA genes were introduced into the wild-type B. subtilis natto strain RG4365. Mutations in bslA or epsG abolished sliding of the B. subtilis natto strain (Fig. 3B). On the other hand, mutation of the tasA gene did not alter the sliding properties of the B. subtilis natto strain. Are EPS production and BslA synthesis required for sliding proficiency? We examined the effect of the mutations of bslA and epsG in the B. subtilis NCIB3610 wild-type strain (proficient in swarming and sliding) and its hag derivative (proficient only in sliding). While swarming of bslA and epsG mutants in the NCIB3610 background was not altered (Fig. 3C), sliding properties of hag bslA and hag epsG double mutant strains were decreased similarly to the bslA and epsG single mutants of the B. subtilis natto strain (Fig. 3D). These experiments show that both the BslA protein and the EPS, which are essential components of the biofilm matrix in B. subtilis, are indispensable for sliding. The microarray analysis showed that abrB was downregulated under sliding-permissive conditions (see Table S2A in the supplemental material), which is in agreement with our experimental results that showed the partial restoration of sliding proficiency of the spo0A mutant strain when abrB was also deleted (Fig. 1E). Accordingly, AbrB is a repressor of bslA (37, 38), a gene that here was shown as required for full sliding proficiency (Fig. 3B).
FIG 3 .
Sliding and swarming ability of mutant B. subtilis strains. (A) The cartoon highlights the B. subtilis genes with possible novel roles in sliding motility as suggested by the microarray experiments performed (see text for details). (B) Sliding properties of B. subtilis natto strain derivatives; from top to bottom, wild-type, tasA, epsG, and bslA strains. (C) Swarming of B. subtilis NCIB3610 and its derivatives comparable to those in panel B. (D) Sliding of B. subtilis NCIB3610 hag strain and double mutant hag tasA, hag epsG, and hag bslA strains. (E) Transcomplementation of sliding-deficient strains. Neither bslA nor eps mutant cells are able to slide separately, but when they are poured together, sliding proficiency is restored.
In line with the experiments on the B. subtilis natto strain, swarming and sliding were not decreased after a mutation was introduced into the tasA gene of NCIB3610 or into hag strains, respectively (Fig. 3C and D), suggesting that the other essential component of the biofilm matrix, TasA, has no role in the ability of B. subtilis to slide. It is interesting that based on the microarray analysis, several genes encoding antimicrobial metabolites (i.e., bacilysin, bacillibactin, and plipastatin) were increased under sliding-permissive conditions (see Table S2 in the supplemental material). In this scenario, the known antimicrobial activity of TasA (39) might suggest a role of this protein in protection of sliding cells against predators instead of a crucial role of this protein in motility.
Understanding the role of the cellular components of the sliding machinery in B. subtilis.
The array experiments showed that the operon related to surfactin production (srf) was induced under sliding-permissive conditions (see Table S2 in the supplemental material), a result that is in agreement with the essential role of this surfactant in sliding proficiency (see Fig. S2A). Surfactin is a secreted lipopeptide molecule that in addition to its function in cell-cell communication, as a paracrine signal during multicellularity (10, 40), has been proposed to allow the spreading of multicellular colonies through the production of surfactant waves that decrease the surface tension of the surrounding space (41). In addition, the EPS overproduced during active sliding is also a component of the extracellular matrix that has been identified as a major force driving biofilm growth, due to the osmotic stress generated by its secretion in the extracellular space (42). Therefore, it is feasible that during sliding the secreted surfactin and EPS, by producing waves of surfactant and gradients of osmotic pressure in the intercellular space of the motile community, respectively, constitute two major forces driving the cooperative sliding of the cells sitting on the surface.
In addition, the overexpression of the KinA inhibitor SivA (differential expression of sivA is indicated in Table S2A in the supplemental material) (Fig. 3A) (43) would ensure that sporulation is not triggered during active sliding and suggests that KinA is not the histidine kinase involved in the activation of Spo0A for sliding proficiency (see below).
What would be the role of BslA, the other major molecule overproduced during active sliding? It has been shown that BslA is a hydrophobin-like protein secreted to the extracellular space, where it forms surface layers at both the agar-cell and air-cell interfaces around the biofilm (44, 45). Because of its physiochemical properties, BslA behaves as an elastic and highly hydrophobic layer coating the biofilm (44–46). Here, we also confirm that BslA is a major contributor to the water repellence of sliding cells (see Fig. S2B and C in the supplemental material). Moreover, the deficiency in BslA synthesis allows the aqueous solution to pass through the cells immediately (see Fig. S2C). These results suggest that this hydrophobin-like protein could play a role during active sliding as a protector of sliding cells against surface wetting, as was proposed previously for biofilms (45, 47). In this scenario, we hypothesize that the sliding-deficient phenotype of eps and bslA cells could be circumvented when the two types of cells are present together under sliding-permissive conditions. Supporting this hypothesis, a previous study on B. subtilis biofilm formation also suggests that these components can be shared among strains producing one but not the other component (48). As shown in Fig. 3E, mixing epsG and bslA derivatives of the B. subtilis natto strain restores the sliding ability. Interestingly, during the time that our work was under review, van Gestel et al. showed that sliding of B. subtilis 3610 depends on the division of labor between matrix (EPS) and surfactin producer subpopulations (49). Analyzing a specific set of mutants, they could also show the complete deficiency in sliding of srfA and eps mutants. In contrast to our observation of the dispensability of TasA activity for sliding in B. subtilis natto and Hag-deficient NCIB3610 cells poured on LB soft medium, sliding of wild-type 3610 on MSggN agar medium was only partly impaired in colony expansion (49). This partially different observation of the TasA requirement for sliding may be due to the different media and growth conditions used for the experiments.
While the genes involved in fatty acid (FA) synthesis (i.e., fabF, fabHBA, fabG, etc.) were overexpressed in wild-type cells under sliding-permissive conditions, those genes involved in FA degradation (i.e., fadR, fadA, fadE, etc.) were downregulated at the same time (see Table S2 in the supplemental material). Is an active lipid synthesis, and therefore active membrane formation and remodeling, necessary to slide? In order to confirm the in silico results and test the formulated hypothesis, we proceeded to specifically block de novo FA synthesis in B. subtilis cells grown under swarming- and sliding-supportive conditions. To this end, we treated B. subtilis cells with the antibiotic cerulenin, which is a specific inhibitor of the FabF condensing enzyme (14), at sub-MICs (below 2 µg ⋅ ml−1), which do not affect the vegetative growth of the NCIB3610 and RG4365 strains (50) (see Fig. S3A and B). Our results show that sub-MICs of cerulenin produce a dose-dependent impediment of sliding motility as well as swarming in B. subtilis (Fig. 4A; also see Fig. S3C and D). These results confirm the microarray data and suggest that an active de novo FA synthesis constitutes an overlooked requirement for surface (sliding and swarming) motility.
FIG 4 .
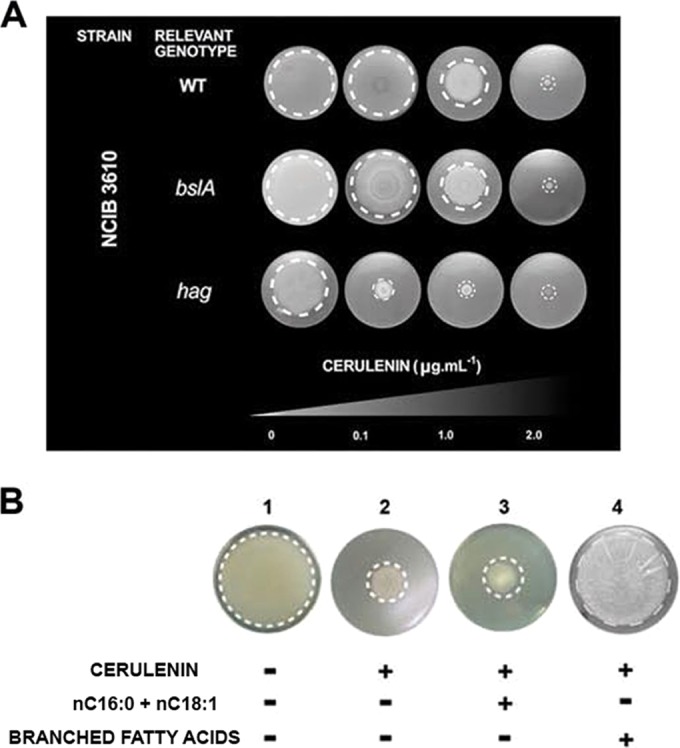
De novo branched fatty acid synthesis is required for swarming and sliding proficiencies in B. subtilis. (A) Dose-dependent inhibitory effect of sub-MICs of cerulenin on swarming and sliding proficiencies of NCIB3610-related wild-type and bslA and hag mutant strains, respectively. Sliding and swarming experiments were performed as indicated in the legend to Fig. 1 but with the inclusion of the indicated cerulenin concentration in the soft agar plates. (B) Exogenous branched FAs but not linear FAs (palmitic [nC16:0] and oleic [nC18:1] acids) restore the sliding proficiency of the B. subtilis natto strain in the absence of de novo FA synthesis.
Surprisingly, the supplementation with exogenous FAs (nC16:0 and nC18:1, palmitic and oleic acids, respectively) of LB soft agar plates containing cerulenin (2 µg ⋅ ml−1) did not bypass the inhibition of surface motility in B. subtilis but allowed the resumption of the planktonic growth of a similar cerulenin-treated culture incubated under shaking conditions (see Fig. S4A in the supplemental material). Why did the addition of nC16:0 and nC18:1 FAs not suppress the negative effect of cerulenin on sliding but allow the resumption of planktonic growth? To answer this question, we analyzed the FA profile in samples of wild-type cells grown under sliding-permissive conditions and liquid shaking culture (see Fig. S4B). B. subtilis, unlike Escherichia coli, synthesizes linear and branched (iso- and anteiso-) saturated fatty acids at 37°C to maintain an adequate membrane fluidity. We found that under active sliding there is a predominance of the synthesis of saturated FAs with lower melting points (anteiso-C15:0 and anteiso-C17:0) and a decrease in the synthesis of the FAs with higher melting points (linear nC15:0 and nC16:0). Overall, during active sliding, the percentage of linear FAs drops from 25.0 to 4.0% while the content of anteiso-FAs rises from 26.0 to 54.0%. Simultaneously, the global content of iso-FAs remains around 50.0%, independently of the growth conditions (see Fig. S4B). The notable increase in the synthesis of low-melting-point anteiso-FAs and the simultaneous decrease in the synthesis of linear FAs would allow the synthesis of membrane lipids with lower melting points and therefore the biogenesis of membranes with higher fluidity. We hypothesize that the synthesis of cellular membranes with a higher fluidity might facilitate the group translocation of B. subtilis cells on solid surfaces in the absence of the propelling force of the flagella (see Fig. S1B). In this scenario, the increase in membrane fluidity that might be required to slide could not be reached with the supply of linear FAs to cerulenin-treated cultures under sliding-permissive conditions. To test this idea, we supplemented B. subtilis cells incubated under sliding-permissive conditions in the presence of cerulenin with branched FAs. As predicted by the hypothesis, the sliding proficiency was fully restored when branched FAs were added as a supplement to the cells with an interrupted de novo FA synthesis (Fig. 4B).
The phosphorelay signaling system coordinates multiple multicellular behaviors in B. subtilis.
Until now, two multicellular behaviors of B. subtilis have been known to be under the control of the phosphorelay signaling system: fruiting body formation (including spore formation) and biofilm development (7, 11, 51). As demonstrated in this work, sliding motility is a type of cooperative behavior under the novel control of Spo0A phosphorylated by inorganic phosphate (Spo0A~Pi) and therefore of the phosphorelay.
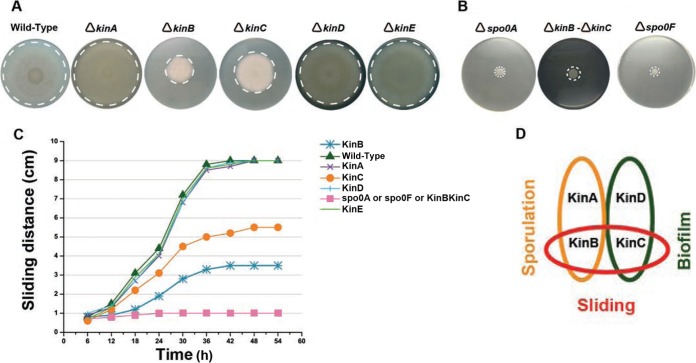
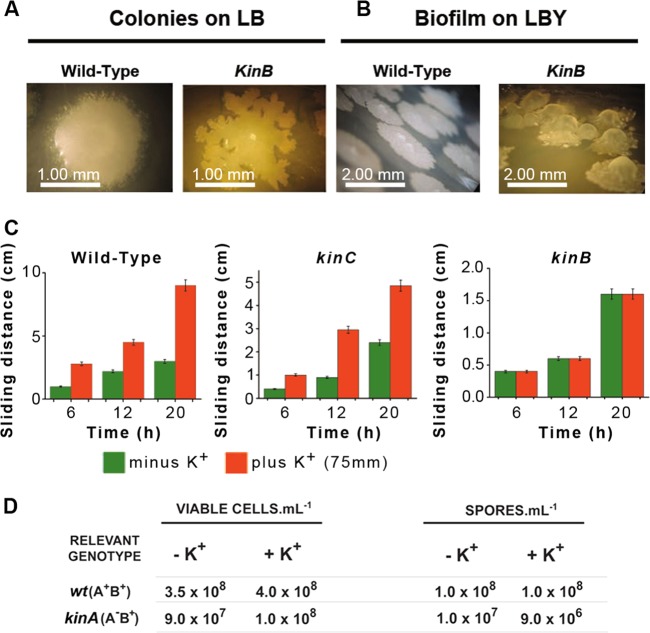
Which phosphorelay histidine kinase governs sliding motility? To solve this question, we constructed RG4365-isogenic phosphorelay-kin mutant strains to analyze their sliding behavior. As shown in Fig. 5A, the wild-type and the kinA, kinD, and kinE single mutant strains show similar and proficient patterns of sliding motility. On the other hand, mainly the kinB and, to a much lesser extent, the kinC single mutant strains display a partial impairment in sliding. To confirm that KinB and KinC are sensor kinases involved in the control of sliding, we constructed a kinB kinC double mutant strain and compared its sliding phenotype with those of the spo0A mutant and the other phosphorelay-defective control strain, the spo0F single mutant, which are completely impaired in surface translocation. As shown in Fig. 5B, the kinB kinC double mutant strain displayed a complete impairment in sliding proficiency that was equivalent in magnitude to the sliding deficiency of the spo0A and spo0F mutant strains. In Fig. 5C, the sliding kinetics of the different phosphorelay mutant strains confirm that KinB and KinC are the two sensor kinases that govern sliding motility in B. subtilis and that KinB activity is more significant than KinC activity for the proficiency in that behavior. As expected, the transcomplementation (into the nonessential amyE locus) of the kinB and kinC mutant strains with a wild-type copy of kinB and kinC, respectively, restored full sliding proficiency (see Fig. S5 in the supplemental material).
FIG 5 .
The phosphorelay sensor kinases KinB and KinC govern social sliding motility in B. subtilis. (A) Sliding phenotype of single kin mutant B. subtilis natto strains (see Table S1 in the supplemental material) after 40 h of incubation on soft LB agar plates at 37°C. (B) Complete sliding-deficient phenotype of spo0A, spo0F, and double kinB kinC mutant strains of the B. subtilis natto strain under conditions of incubation similar to those indicated for panel A. (C) Kinetics of sliding motility of different phosphorelay mutants over time. Note that the line with pink squares is common to the spo0A, spo0F, and kinB kinC mutant strains. (D) Sporulation, biofilm formation (at atmospheric oxygen level), and sliding motility are Spo0A-dependent developmental programs that B. subtilis preferentially regulates by duos of phosphorelay sensor kinases.
Interestingly, the sliding-controlling KinC kinase (this work) has been proposed (along with KinD) to govern the onset of biofilm formation (10, 33–35, 52). Further, KinA and KinB have been suggested to alter biofilm development on certain media and at reduced oxygen levels (53). We confirm (data not shown) that in the RG4365 natto strain, as well as in the NCIB3610 strain (33, 34), both sensor kinases, KinC and KinD, govern the onset of biofilm development and extracellular matrix production in response to plant-derived polysaccharides that constitute one of the signals able to induce both kinases (33).
In toto, B. subtilis employs duos of phosphorelay histidine kinases to control different multicellular behaviors. The histidine kinase duos KinA/KinB, KinC/KinD, and KinB/KinC govern the onset of sporulation and fruiting body formation (7, 8, 28, 54, 55), biofilm development under atmospheric oxygen pressure (10, 33–35, 51, 52), and social sliding (this work), respectively (Fig. 5D).
The sliding signal.
What is the nature of the signal, acting on KinB and/or KinC, which is responsible for triggering sliding motility in B. subtilis? To answer this fundamental question, we had two premises. First, the sliding-inducing signal, responsible for the autophosphorylation of KinB and/or KinC, should not be strong enough to trigger KinB~Pi-/KinC~Pi-dependent activation (phosphorylation) of Spo0A to the high levels of the regulator (Spo0A~Pi) needed for spore formation, because we did not observe induction of sporulation genes under conditions of active sliding (see Table S2 in the supplemental material). Second, the same signal that activates the autophosphorylation of KinB and/or KinC sensor kinases to make Spo0A~Pi and induce sliding motility would prevent initial KinB~Pi-dependent and/or KinC~Pi-dependent biofilm formation.
A recent report suggested that KinB is controlled by the respiratory apparatus via its second transmembrane segment (53). It is proposed that under conditions of reduced electron transport, KinB becomes active (formation of KinB~Pi) via a redox switch involving its second transmembrane segment with one or more cytochromes to induce biofilm formation and sporulation (53). We envision that under active sliding, in rich soft medium, the physiological conditions of the sliding cells would be different from the conditions of sessile cells forming a biofilm. Under conditions of biofilm formation, a crowded population of cells exists encased in the biofilm matrix with nutrients that become rapidly exhausted (11). Furthermore, if a reduced electron transport triggers KinB~Pi-dependent biofilm formation and sporulation, surface translocation (sliding) would not be activated at the same time since the two are antagonist responses (53). Therefore, we consider it unlikely that the status of the respiratory apparatus, sensed by the second transmembrane domain of KinB, could be the physiological condition triggering sliding.
Interestingly, it has been proved that intracellular potassium represents a negative signal for KinC (10, 52). Potassium is a major intracellular ion that impairs KinC activation through interaction with the cytoplasmic PAS-PAC sensor domain of the kinase (10). The intracellular potassium concentration decreases as B. subtilis cells reach the late logarithmic phase when newly synthesized surfactin, through its membrane pore formation activity, and the putative potassium channel YugO secrete the ion to the outside of the cell (10, 52). In this model, the surfactin/YugO-mediated intracellular drop in potassium concentration activates KinC. Curiously, in contrast to the round colonies formed on LB and LBY (LB medium supplemented with 4.0% yeast extract; see also reference 14) agar plates by the wild-type RG4365 strain and its isogenic kinC derivative, the kinB mutant strain forms colonies and biofilms with a tendril-shaped morphology that are very similar to the morphology of wild-type B. subtilis colonies grown on CM (casein digest-mannitol medium) plates, a solid medium with potassium deficiency (22) (Fig. 6A and B). Basically, low (micromolar) and high (millimolar) levels of potassium ions favor tendril-like and rounded colony formation, respectively (22). Due to the similar colony phenotypes of the kinB mutant and the wild-type strain grown on solid medium with low levels of potassium, we were motivated to investigate if potassium is involved in the regulation of KinB. Remarkably, as shown in Fig. 6C, we discovered a dose-dependent positive effect of potassium ions on sliding motility of the wild-type and kinC strains, which are proficient in kinB expression. This sliding stimulation was observed at potassium levels between 50 mM and 100 mM (data not shown) (with an optimal sliding-stimulatory concentration of 75 mM) that are comparable to the potassium concentrations that inhibited KinC from triggering biofilm formation (10, 52). In contrast, there was no effect of potassium supplementation on the sliding proficiency of the kinB mutant (Fig. 6C). These results strongly suggest that potassium represents a positive signal for KinB activation. A closer examination of the colony and biofilm phenotypes of the kinB mutant strain (Fig. 6A and B) seems to indicate that KinB might inhibit KinC from stimulating biofilm formation, and we are currently investigating this phenomenon.
FIG 6 .
Potassium is the physiological signal that regulates sliding motility in B. subtilis. (A and B) Tendril-like morphology of RG4365-isogenic kinB colonies and biofilms (complex colonies) formed on LB (A) or LBY (B) medium. (C) Potassium stimulates sliding of KinB-positive (wild-type and kinC mutant strains) but not KinB-deficient (kinB strain) cells. (D) Potassium does not represent a signal for sporulation proficiency. Sporulation proficiencies of wild-type (wt) (kinA+ kinB+) and kinA mutant (kinA kinB+) strains in the presence and absence of added potassium ions (75 mM) are shown. Viable cells and spores were determined after 30 h of growth in SM as previously described (27). Results presented in panels C and D are representative of three experiments performed separately.
It is known that the sensor histidine kinase KinB is, in addition to KinA, the main sporulation kinase of B. subtilis (54). Although a kinB mutant strain is proficient in sporulation (Spo+ phenotype), a double kinA kinB mutant strain is almost unable to sporulate (Spo0 phenotype) (54). Therefore, we were interested to investigate if the positive effect of potassium on KinB-dependent sliding proficiency was also valid for spore formation. As shown in Fig. 6D, potassium did not stimulate sporulation in either of the two KinB-proficient strains, i.e., wild-type (KinA+ KinB+) and kinA mutant (KinA− KinB+), that were analyzed. Consequently, potassium constitutes a specific signal for sliding motility that precisely fulfills the two hypothesized premises to be ineffective in triggering sporulation (first premise, Fig. 6D) but, at the same time, strong enough to trigger sliding motility (second premise, Fig. 6C) (10, 52). In addition, the dual role of potassium ions (present at high intracellular levels at early times of growth) as activators and inhibitors of KinB and KinC activities, respectively, points to KinB as the phosphorelay kinase responsible for the start of the cooperative sliding movement (see below).
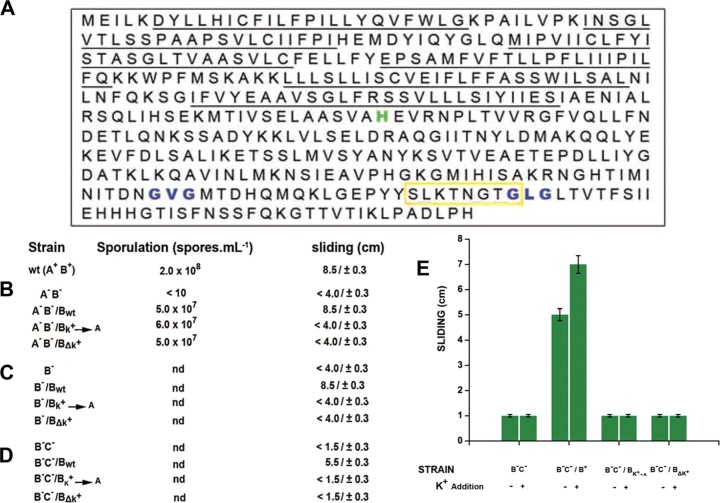
Does potassium represent a direct or an indirect signal to activate KinB? Searching for conserved domains and sequence motifs present in KinB that might be involved in the potassium response, we discovered a disregarded sequence (SLKTNGTG) residing on the ATP-binding region of KinB that is absent in the sequences of the other four phosphorelay sensor kinases (56) (Fig. 7A). This sequence possesses a significant homology to the highly conserved K+-filter (selectivity) sequence of the pore loop domain (P-domain) of potassium channels (T/S-x-x-T-x-G-x-G consensus sequence) (57, 58). Despite the many protein motifs and domains present in different types of potassium channels (58), the KinB K+-selectivity-like sequence (here called K* for simplicity) is the only common element related to K+ channels. While active KinB is a dimer (59), typical potassium channels are tetramers made up of predominantly identical subunits clustered to form the ion permeation pathway across the membrane (60–62). In addition to the absence of the pore motifs that surround the selectivity filter (Fig. 7B), KinB also lacks the different domains that have been described in different potassium channels (58, 60, 61). Furthermore, the K* resides in the cytosolic region of the kinase, while in all known (eukaryotic and prokaryotic) potassium channels the K+-filter resides in transmembrane domains. While these topological features exclude KinB as a potassium channel, they open the possibility that the kinase might sense the intracellular concentration of the ion throughout its cytosolic K*. Therefore, we tested if the K* plays a role or not in sliding motility. To this end, we constructed two types of kinB mutant strains harboring specific mutations in the K* (see Fig. S6 in the supplemental material). In one case, three out of the four conserved amino acids of the consensus K* were replaced by alanines (the fourth conserved amino acid, G, of the consensus sequence was not altered as it overlaps with a predicted ATPase motif of the kinase [Fig. 7A]) to give rise to the mutant KinBK*→A (see Fig. S6). In the second constructed kinB mutant strain, 7 out of the 8 amino acids of the K* were deleted (mutant strain KinBΔK* [see Fig. S6]). One important consideration for both mutant strains before analysis of their roles in sliding is that they must be functional (i.e., promote spore formation). In this sense, the sliding-promoting activity of KinB should be separable from its biofilm/sporulation-promoting activities, a scenario that would explain why KinB-dependent sliding proficiency and KinB-dependent biofilm/spore formation are not simultaneously activated (see below).
FIG 7 .
KinB harbors a cytosolic selectivity filter motif responsive to potassium ions that specifically allows sliding proficiency. (A) Amino acid sequence of KinB. The six continuous underlines indicate the six transmembrane domains of KinB. The histidine highlighted in green corresponds to the residue of autophosphorylation; the blue amino acid triplets represent the top and bottom sites of the ATP-binding domain of the kinase. The yellow box highlights the cytosolic sequence in KinB with homology to the potassium selectivity filter sequence present in potassium channels. (B to D) Sporulation and sliding proficiencies are separable KinB functions. KinB mutant strains affected in the integrity of the selective filter sequence were able to restore full sporulation proficiency of a Spo0 kinA kinB double mutant strain (A−B− in panel B) but did not restore KinB-dependent sliding activity in that A−B− background (B), either in kinB (B−) (C)- or in kinB kinC (B−C−) (D)-deficient mutant strains. Bk+→A and BΔk+ indicate KinB proteins with Ala-exchanged and Ala-deletion K+-filter domains, respectively. (E) Mutation of the potassium selectivity sequence in KinB abolished the ability of B. subtilis to slide in response to potassium addition. Sliding and sporulation proficiencies were measured as indicated in Materials and Methods. Results presented in panels B to E are representative of four experiments performed separately after 40 h of incubation.
Remarkably, while the complementation of a kinA kinB double mutant strain (originally Spo0 and deficient in sliding) with a wild-type copy of kinB restored sporulation and sliding proficiencies, both types of alterations in KinB (KinBK*→A and KinBΔK*) were able to complement full sporulation proficiency but did not restore the KinB-driven sliding proficiency in that genetic (kinA kinB) background (Fig. 7B). Simultaneously, both constructed KinB mutants failed to restore sliding proficiency of kinB (Fig. 7C) and kinB kinC (Fig. 7D) mutant strains and were also insensitive to the stimulation of sliding after potassium supplementation (Fig. 7E). These results confirm that the K+-selectivity-like sequence present in KinB (K*) plays an essential role in potassium sensing, leading to sliding proficiency that is located on a different part of the KinB protein than the electron transport-sensing transmembrane segment 2 responsible for triggering biofilm and spore formation (54).
Potassium constitutes the signal for the fine-tuned interconnection of social sliding and biofilm development.
Potassium represents the most abundant ion in the cytoplasm (~200 mM in E. coli versus 7 mM content in LB medium) (58). Unlike most other intracellular cations, the high intracellular concentrations of potassium do not interfere significantly with vegetative growth. However, apart from the stimulatory effect of potassium on KinB (this work), it is known that potassium is a strong inhibitor of KinC activity (10, 52), but enigmatically, the activity of both histidine kinases (KinB and KinC) is required for full sliding proficiency (Fig. 5A and B). The simplest explanation for this apparent paradox is that KinB and KinC should work at different times of sliding development accompanying the drop in the intracellular concentration of potassium that happens during the transition from the log phase to the early stationary phase. We favor a scenario (i.e., in LB soft agar plates) in which, at the onset of sliding and at the edge of an active sliding colony (where the youngest cells would be present), the intracellular potassium concentration would be high because cells have plenty of nutrients and are in log phase. During this time, KinB (which is the first phosphorelay kinase to be expressed and therefore is present early on in the cell) (63) should be active (due to the potassium stimulus) in driving the synthesis of the sliding machinery while KinC activity would remain low because of its reduced expression at early times of growth and because of the presence of high levels of intracellular potassium (10, 52) (Fig. 8A, left image). As soon as sliding cells approach the late log phase on LB soft agar plates (probably at the inner, older portion of the sliding disc), surfactin and the potassium exporter YugO are expressed on the cellular membrane (10, 52). Therefore, the intracellular potassium concentration decreases, and KinB activity is downregulated and replaced by active KinC to maintain the appropriate levels of Spo0A~Pi required for the continued expression of genes needed for full sliding proficiency (i.e., bslA, srf, eps, and fab) and biofilm formation (see below) at the inner part of the sliding disc (Fig. 8A, right image). Supporting the view of this spatiotemporal regulation of the kinases, when wild-type (KinA- and KinB-positive) B. subtilis cells are loaded on an optimized soft agar medium that in addition to sliding motility also allows biofilm formation (i.e., soft LBY agar) (14), it is possible to observe the formation of structures typical of a biofilm in the inner part of the sliding disc, while the outer borders remain flat (Fig. 8B, left image). This result strongly suggests that (under sliding-permissive conditions) KinC and KinB are active at the interior and at the edge of the sliding disc, respectively (Fig. 8A, right image). In agreement with this result, when kinC (KinB-positive and KinC-deficient) cells were incubated under similar conditions, the sliding disc, as expected, was smaller than the sliding disc formed by wild-type cells, and more importantly, no structure that resembles a biofilm was formed (Fig. 8B, middle image). Concordantly, a kinB strain (KinC-positive and KinB-deficient) formed typical biofilm wrinkle-like structures at the inside and outside regions of the slowly sliding cells (Fig. 8B, bottom right images). Overall, the former results (Fig. 6 to 8) suggest that potassium ions and KinB~Pi act earlier than KinC~Pi to activate the onset of sliding and confirm, once the potassium concentrations start to be different in distinct regions of the sliding disc, the spatiotemporal regulation of KinB and KinC (Fig. 8A). In addition, the KinC~Pi-dependent biofilm formation observed at the interior (older) part of the sliding disc after incubation in a medium that allows biofilm and sliding (LBY soft agar plates) (left and middle images in Fig. 8B) suggests that the potassium-mediated activation of KinB to make Spo0A~Pi and start sliding motility (Fig. 6C and 7) is not only insufficient to induce KinB~Pi-dependent spore formation (Fig. 6D) but also insufficient to trigger the onset of KinB~Pi-dependent biofilm formation (middle image in Fig. 8B).
FIG 8 .
Spatiotemporal regulation of the sliding-inducing kinases. (A) At the onset of sliding, as soon as cells were poured on LBY-0.7% agar plates, KinB was the first-acting kinase while KinC remained inactive. This differential activity of KinB and KinC is due to the intracellular potassium input that activates and inhibits each kinase, respectively (left panel). As progression of sliding continues, there is a drop in the intracellular potassium concentration in the cells at the inner part of the sliding disc. Under this physiological condition, KinB and KinC become inactive and active inside the sliding disc, respectively, while KinB remains active at the newest part (border) of the sliding community. (B) Wild-type B. subtilis RG4365 and its isogenic derivatives mutated in kinC or kinB were loaded on soft agar plates of LBY medium and incubated for 20 h at 37°C. Under these conditions of simultaneous stimulation of sliding and biofilm formation proficiencies, the formation of a structured biofilm in the inner part of the wild-type sliding disc is observed. In contrast, in the sliding disc of the kinC strain no biofilm structure is formed, suggesting that the biofilm observed in the inner part of the sliding disc of wild-type cells is a product of the KinC activity. In both cases (wild-type and kinC strains), the borders of the sliding discs are flat and unstructured. In the case of the kinB cells (right panel), KinC activity drives the formation of typical wrinkled structures, representative of a mature biofilm, inside and at the borders of the colony. (C) Two models for the temporal progression of multicellularity in B. subtilis as described in the text. (D) β-Galactosidase production from PabrB-lacZ in B. subtilis cells grown on petri dishes filled with media that favor the expression of different social behaviors under the control of Spo0A~Pi: sliding motility (LB-0.7% agar; green line), biofilm formation (LBY-1.5% agar; red line), sporulation (SM-1.5% agar; black line), or none (LB-1.5% agar; blue line) (see text for details). Cells were taken from the petri dishes at the times indicated in the figure and assayed as described in the supplemental material. The results shown are representative of three independent experiments made in duplicate. M.U., Miller units. (E to H) Five microliters of an overnight culture of the natto strain RG4382 (Δspo0A::Ery/Pspac-spo0A-sad67Cat) was inoculated on the middle of solidified LBY-0.7% agar medium prepared with different concentrations of IPTG as shown in the figure. After 20 h of incubation at 37°C, photographs were taken and the sporulation frequency (after elution of the cells from the petri dishes) was determined as described in Materials and Methods. The results are representative of seven independent experiments performed in triplicate.
How can sliding and the sessile lifestyle of biofilm formation, which are social behaviors believed to be antagonistic to each other, be positively controlled by the same regulatory pathway (Spo0A~Pi), and why are the biofilm formation and sporulation pathways not activated upon increased KinB autophosphorylation during promotion of sliding motility? Fujita and Losick reported that a gradual increase in the levels and activity of Spo0A results in the expression of different sets of genes in B. subtilis (28). For instance, high levels of Spo0A~Pi stimulate sporulation (fruiting body formation) and low levels of Spo0A~Pi stimulate biofilm formation (64). Therefore, we hypothesize that sliding and biofilm formation developments require, as suggested by Chai et al. (64) for the cases of sporulation and biofilm formation, different levels of Spo0A~Pi to become active. Until now, it was clearly demonstrated that the sliding-permissive conditions used in this work (i.e., soft LB agar) and the potassium input that activates KinB (which at the same time inhibits KinC activity) (10) are adequate to support sliding (Fig. 1 and 6C) but insufficient to trigger sporulation (Fig. 6D) and biofilm formation (Fig. 8B, middle image, KinB+ KinC− strain). Therefore, sliding motility in B. subtilis seems to be activated before biofilm formation and sporulation. But how is sliding and not the other developmental pathways triggered when KinB become active? In other words, which behavior needs lower levels of Spo0A~Pi to be triggered? We hypothesize two alternative progressions. In one scenario (Fig. 8C, model I), the surface-committed cells first slide and later on form the biofilm structures at the center. In the other situation (Fig. 8C, model II), the surface-attached cells first produce a sessile biofilm and cells at the edge of the biofilm engage in sliding later on. In both scenarios, the social behavior that is triggered first (sliding or biofilm formation) is the one that requires the smaller amount of Spo0A~Pi (28, 64). One approach to monitor the in vivo levels of Spo0A~Pi is to measure the expression of abrB, the most sensitive reporter of the Spo0A~Pi levels present in the cell (27, 28). Spo0A~Pi is a strong repressor of abrB, and very low levels of Spo0A~Pi (insufficient to trigger biofilm and sporulation) are sufficient to downregulate abrB (27, 28). Therefore, to obtain more insight into the levels of Spo0A~Pi present during sliding and biofilm, we measured the levels of β-galactosidase (β-Gal) activity driven by the expression of an abrB-lacZ transcriptional fusion under conditions that favor sporulation (growth on sporulation medium [SM]-1.5% agar plates), sliding (growth on LB-0.7% agar plates), biofilm (growth on LBY-1.5% agar plates), or none of the abovementioned behaviors (growth on LB-1.5% agar plates). At different times, cells were removed from the petri dishes and the abrB-driven β-galactosidase activity was measured (Fig. 8D). As expected, the abrB expression was the lowest and highest (indicating the highest and smallest amounts of Spo0A~Pi, respectively) when the cells were grown on SM- and LB-1.5% agar plates, respectively (Fig. 8D). Interestingly, the levels of abrB expression under conditions of active sliding (growth on LB-0.7% agar plates) were significantly higher than the AbrB levels observed under conditions of active biofilm formation (growth on LBY-1.5% agar plates). These results (Fig. 8D) suggest that the levels of Spo0A~Pi required to trigger sliding motility are lower than the levels of Spo0A~Pi needed to trigger biofilm formation, and therefore, the former behavior (sliding motility) would be triggered before biofilm formation when B. subtilis is attached and committed to a sessile differentiation (model I in Fig. 8C).
To confirm this interpretation, the Δspo0A sad67 strain (see Table S1 in the supplemental material) (27–29), where the synthesized active Spo0A (Sad67) level depends on the supplemental IPTG, was cultivated on LBY-0.7% agar plates supplemented with different amounts of IPTG. In this experiment, we hypothesized that the behavior (biofilm formation, sliding, or fruiting body formation-sporulation) expressed at the lowest IPTG concentration would reflect the multicellular B. subtilis response that requires the smallest amount of active Spo0A (Sad67) to be produced. As expected, in the absence of IPTG addition (Fig. 8E), sad67 is not expressed and B. subtilis is unable to display (because Spo0A activity is completely absent in Δspo0A cells) any of its different multicellular behaviors. As soon as the LBY-0.7% agar medium is supplemented with a small amount of IPTG (0.01 µM), B. subtilis cells start to slide (Fig. 8F). When the IPTG concentration is increased to 10 µM (and therefore more active Spo0A is produced in the surface-committed cells), B. subtilis triggers complex colony biofilm formation (Fig. 8G). At the largest amount of supplemental IPTG (1,000 µM), the growth of B. subtilis is restricted (because high levels of active Spo0A inhibit vegetative division) (27–29) and B. subtilis directly induces the formation of fruiting bodies filled with spores (Fig. 8H). Overall, these results strongly suggest that the increase in the levels of active Spo0A (Spo0A~Pi) (28, 64) allows the expression of the different behaviors of B. subtilis following a temporal sequence of social sliding motility, multicellular biofilm formation, and finally fruiting body formation (sporulation) (see Fig. S7).
Conclusions.
In toto, we demonstrated how the model organism B. subtilis can cope with the basic, although fundamental, decision that it must take when it is attached and committed to a surface: to move or remain in place. We present a novel mechanistic model, containing a Spo0A command that explains the coordinated expression of the different behaviors of B. subtilis. In this model, the spatiotemporal regulation of KinB and KinC by potassium determines the Spo0A~Pi amount, which in turn orchestrates the onset and sequential progression of sliding motility, biofilm formation, and finally sporulation/fruiting body formation (Fig. 8; see also Fig. S7 in the supplemental material). Recent publications demonstrate the necessity of biofilm formation for the efficient colonization of the root surface and biocontrol properties of B. subtilis (33, 34). The common signal (potassium) and the common regulatory network under Spo0A control (the phosphorelay) (25) for sliding (this work), biofilm formation (this work and references 10 and 32, to ,34), and sporulation-fruiting body formation (54) and our in vitro experiments (Fig. 2G) suggest that sliding might also contribute to the ability of the plant growth-promoting and biocontrol bacterium B. subtilis to reach the root surface and efficiently colonize the rhizosphere. Once again, B. subtilis offers an example of simplicity in how distinct prokaryotic social behaviors previously believed to be antagonistic and independent from each other, i.e., surface motility, biofilm formation, and sporulation, might work together to benefit the bacterium and the host.
MATERIALS AND METHODS
Strains and growth media.
The three wild-type B. subtilis strains used in this study were the domesticated, laboratory reference strain JH642 and the two undomesticated and wild B. subtilis strains NCIB3610 and natto RG4365. These strains and their isogenic derivatives (see Table S1 in the supplemental material) were grown in Luria-Bertani (LB) and Schaeffer’s sporulation medium (SM) as indicated previously (27). For experiments in biofilm formation, the biofilm-enhancer medium LBY was used (14). Cloning of kinB and kinC genes for complementation and site-directed mutagenesis is described below and in Table S3 in the supplemental material.
Spreading (swarming and sliding) experiments.
For surface motility (swarming and sliding), LB plates fortified with 0.7% agar and dried for 1 h were inoculated with 1 µl of 8 × 107 cells ⋅ ml−1 grown to mid-log phase at 37°C in LB broth. The inoculated petri dishes were then incubated at 37°C for 40 h. Each data point shown in the figures represents an average from 6 independent experiments. Data from one representative experiment are shown. Flagellum staining was performed as described previously (14).
DNA transformation and complementation experiments.
Transformation of B. subtilis, to obtain isogenic derivatives of the parental strains, was carried out as previously described (27, 50). When appropriate, antibiotics were included at the following final concentrations: 1 µg ⋅ ml−1 erythromycin (Ery), 5 µg ⋅ ml−1 kanamycin (Kan), 5 µg ⋅ ml−1 chloramphenicol (Cat), 75 µg ⋅ ml−1 spectinomycin (Spc), and 2.5 µg ⋅ ml−1 phleomycin (Pheo). Surfactin and cerulenin were obtained from Sigma-Aldrich. For those plates supplemented with IPTG in Fig. 1I, one or two grains of IPTG was carefully added on top of the soft LB agar after solidification in order to allow IPTG diffusion and gradient formation.
β-Gal assays.
The β-galactosidase (β-Gal) activity from liquid cultures was assayed as previously described (14, 27). In the case of the assays of β-Gal activity driven from colonies, cells were resuspended at a final concentration of 3 × 109 CFU ⋅ ml−1 before measurement of the β-galactosidase activity (14, 27).
Transcriptome analysis.
Cultures of the B. subtilis natto strain (RG4365) and its spo0A derivative (RG4370) were inoculated onto the middle of LB plates containing 0.7% or 1.5% agar. The bacterial biomass was removed from the plates with a spatula after 24 h of growth, and samples were stored at −80°C. At least three independent biological replicates were included. The pellets were immediately frozen in liquid nitrogen and stored at −80°C. RNA extraction was performed with the Macaloid/Roche protocol (37, 65) with two additional steps of phenol-chloroform washing. RNA concentration and purity were assessed using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific). RNA samples were reverse transcribed into cDNA using the Superscript III reverse transcriptase kit (Invitrogen, Carlsbad, CA, USA) and labeled with Cy3 or Cy5 monoreactive dye (GE Healthcare Amersham, The Netherlands). Labeled and purified cDNA samples (NucleoSpin extract II; Biokè, Leiden, The Netherlands) were hybridized in Ambion Slidehyb #1 buffer (Ambion Europe Ltd.) at 48°C for 16 h. The arrays were constructed as described elsewhere (66). Briefly, specific oligonucleotides for all 4,107 open reading frames of B. subtilis 168 were spotted in duplicate onto aldehyde-coated slides (Cell Associates) and further handled using standard protocols for aldehyde slides. Slide spotting, slide treatment after spotting, and slide quality control determination were done as described previously (67). After hybridization, slides were washed for 5 min in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) with 0.5% SDS, washed 2 times for 5 min each in 1× SSC with 0.25% SDS, washed for 5 min in 1× SSC-0.1% SDS, dried by centrifugation (2 min, 2,000 rpm), and scanned in a GenePix 4200AL microarray scanner (Axon Instruments, CA, USA). Fluorescent signals were quantified using ArrayPro 4.5 (Media Cybernetics Inc., Silver Spring, MD) and further processed and normalized with MicroPrep (68). CyberT (69) was used to perform statistical analysis. Genes with a Bayes P value of ≤1.0 × 10−4 were considered significantly affected.
Cloning of kinB and kinC genes and site-directed mutagenesis.
The coding regions of kinB and kinC were amplified with oTB56-oTB57 and oTB61-oTB62 oligonucleotide pairs, respectively (oligonucleotide sequences are indicated in Table S3 in the supplemental material). The PCR products were digested with BamHI and EcoRI enzymes and cloned into the corresponding sites of pTB16. pTB16 is an amyE integration vector with a kanamycin resistance gene. pTB16 was created by amplifying the kanamycin resistance gene from pDG782 (70) with primers oDG1 and oDG2, restricting it with StuI and BamHI, and ligating it to the corresponding sites of PCR-amplified vector that was obtained with oX1 and oX2 on pX (71) plasmid as the template (see Table S3). Site-directed mutants of the kinB gene were obtained using an overlapping fragment PCR method (72), using polymerase X enzyme (Roboklon GmbH, Berlin, Germany). The 5′ region of kinB was amplified with oTB56-oTB58 oligonucleotides, while the 3′ region, containing mutations for 3 amino acid exchanges (S383A, T386A, and G388A) or a 7-amino-acid deletion (S383 to T389), was obtained with oTB59-oTB57 or oTB60-oTB57, respectively (see Table S3). The PCR fragments were used in a second round of PCR as a template accompanied with oTB56-oTB57 oligonucleotides, and fusion fragments were cloned in the same way as the wild-type kinB gene. The plasmid constructs obtained were sequenced before transformation into B. subtilis integrating into the amyE locus.
Fatty acid analysis.
Approximately 20 mg of the sample was weighed in a screw-cap 4-ml glass vessel. For the transesterification, a methanol-hydrochloric acid solution was freshly prepared by adding 1 ml of acetyl chloride to 20 ml of methanol (73). Butylated hydroxytoluene (BHT; 3 µg ⋅ ml−1) was added to prevent autoxidation of polyunsaturated fatty acids. Afterward, 1.5 ml of this solution was added to the sample. The solution was overlaid with nitrogen, and the vessel was tightly closed. After vortexing, the vessel was heated at 90°C for 1 h. Once cooled to room temperature, 1 ml of water and 1.5 ml of hexane were added for extraction of fatty acid methyl esters (FAMEs). The tubes were vortexed and centrifuged (6 min at 720 relative centrifugal force [RCF]). The hexane phase was isolated. The extraction procedure with hexane was repeated, and the combined organic solutions were dried over anhydrous Na2SO4 and concentrated to dryness under a gentle stream of nitrogen. The residue was redissolved in 0.2 ml (or 0.1 ml) of hexane (depending on the final concentration).
The Supelco 37-component FAME mix, the Supelco bacterial acid methyl ester mix, and the Supelco triglyceride mix (Supelco, Bellefonte, PA, USA) were used as reference standards to identify the FAMEs.
An aliquot of the sample (1 µl) was analyzed on a Finnigan trace instrument (Thermo Fisher Scientific, Dreieich, Germany) equipped with a ZB5 column (15 m by 0.25 mm by 0.25 µm) with a 10-m Guardian end (Phenomenex, Aschaffenburg, Germany). Mass spectra were measured in electron impact (EI) mode at 70 eV. Helium at 1.5 ml ⋅ min−1 served as the carrier gas. The gas chromatography (GC) injector (split ratio, 1:15), transfer line, and ion source were set at 250°C, 280°C, and 200°C, respectively. FAMEs were eluted under programmed conditions from 50°C (2 min) followed by 10°C ⋅ min−1 to 168°C, 1°C ⋅ min−1 to 177°C, and 10°C ⋅ min−1 to 320°C.
Preparation of branched fatty acid extract from B. subtilis.
One hundred milliliters of a B. subtilis RG3465 culture was grown in LB broth at 37°C until the end of the vegetative phase (~6 h of growth); only saturated fatty acids, mainly branched fatty acids, are made at this temperature (74). Total membrane lipids (phospholipids [PLs] and glycolipids [GLs]) were extracted as previously described (74). Then, the PLs and GLs were hydrolyzed with cold diazomethane to obtain free fatty acids (FAs), which were completely dried and resuspended in 1 ml of ethanol (74). For the complementation experiments on cerulenin-treated B. subtilis cells, the free FAs were used at a 1:100 dilution rate.
Light microscopy and photography.
The developed swarm and slide plates were visualized with a Stemi 2000 (Zeiss) stereomicroscope using a KL1500LCD (Zeiss) illumination system. A Power-Shot A80 (Canon) system was used to capture the photographs for swarm and slide images.
Plant root colonization experiments.
Two sanitized wheat seeds were deposited on top of previously solidified 1/10-diluted LB in 0.7% agar plates and incubated in moisture chambers at 25°C with exposure to periods of 12 h of illumination. After 3 days of incubation, when wheat seeds started to germinate, 3.0 µl of stationary-phase cultures of wild-type and spo0A mutant cells was inoculated at the points indicated by the white dashed circles in Fig. 2. After 24 h of incubation and during the subsequent days, top-to-bottom pictures were taken to show the sliding and plant root colonization abilities of wild-type and spo0A B. subtilis cells.
Microarray data accession number.
Microarray data have been deposited in the Gene Expression Omnibus database under accession no. GSE43840.
SUPPLEMENTAL MATERIAL
Spreading behavior of the undomesticated B. subtilis natto strain. Surface motility of undomesticated B. subtilis Marburg-related strain NCIB3610 (A), human-probiotic natto-related strain RG4365 (B), and domesticated laboratory strain JH642 (C) 10 h, 18 h, and 36 h after inoculation. Briefly, petri dishes containing LB medium fortified with 0.7% agar were centrally inoculated with any of the three B. subtilis strains and incubated at 37°C (see Materials and Methods for details). (A) In these pictures, it is possible to appreciate the rapid ability of the NCIB3610 strain to swarm on the agar surface. It was previously reported that robust swarming motility, a type of social motility that relies on groups of cells (rafts) that express numerous flagella and differentiate to migrate coordinately along a solid substrate, was present in the NCIB3610 strain (4, 12). (B) The strain RG4365 needed a longer incubation to start to move (as would be expected from a strain that is able only to slide), but after the first 5 h of incubation, it was possible to observe a uniform and highly hydrophobic monolayer of cells that cooperatively and uniformly advanced in large filaments made of close cell-to-cell contacts, predominantly along the longitudinal axis, across the agar plate; see amplified views (phase-contrast and fluorescence images) of RG4365 cells on the right. (C) As expected, the domesticated B. subtilis strain JH642 was unable to move because it harbors a mutation in a gene (sfp) needed for surfactin synthesis and a frameshift mutation in the regulatory gene swrA (24). In contrast, the undomesticated strains NCIB3610 and RG4365 do not harbor those mutations and therefore are able to spread on solid surfaces (12, 75). (D) Kinetics of motility diameter of the various strains: NCIB3610 (red diamonds), RG4365 (green circles), and JH642 (blue squares). Arrows in panel D indicate time points with representative pictures above, while a line at 8.5 cm shows the maximal size of motility related to the size of the plate used. The rate of surface motility was maximal (0.75 cm ⋅ h−1) for the strain NCIB3610, followed by the natto strain RG4365 (0.30 cm ⋅ h−1). (E and F) One essential factor for the swimming and swarming ability of the strain NCIB3610 is the flagellum, the proposed motor for those motilities (12). In contrast, the RG4365 strain grown in shaking liquid culture was completely nonmotile (data not shown). Cell samples for flagellum visualization were prepared as indicated previously (12) after 10 h (for NCIB3610 strain [E]) and 20 h (for RG4365 strain [F]) of inoculation on motility plates. These time points represent the stages of maximal surface motility for each wild-type strain (see panel D). Samples were taken from the borders of the surface motility discs, and cells were carefully separated and diluted (from left to right in panels E and F) in order to improve the visualization of flagella. Pictures on the right side of panels E and F are representative photographs of wild-type NCIB3610 cells (E) and RG4365 cells (F) taken from liquid cultures grown until late exponential phase (optical density at 525 nm [OD525], 0.9) in LB medium with aeration (200 rpm). Under all the examined conditions (swarming and swimming), B. subtilis NCIB3610 cells harbored flagella and were motile (E). In contrast, wild-type B. subtilis RG4365 cells were motile only on soft agar plates but without harboring flagella (F). In liquid culture, the RG4365 cells also were devoid of flagella and completely nonmotile (right image in panel F). Cells were stained for flagella as described in Materials and Methods. Arrows indicate the positions of some flagella shown as examples. (G) To confirm the dispensability of flagellum production for the surface-associated motility (sliding) in RG4365, we proceeded to delete the hag gene (75), which is involved in the synthesis of the flagellin subunit of the flagellum (76). As shown (left photograph), the isogenic hag mutant strain RG4387 (see Table S1) was able to slide on the agar surface at the same rate as the wild-type parental strain RG4365 after 24 h of incubation at 37°C. Similarly, the hag inactivation in the NCIB3610 strain abolished its ability to swarm under similar incubation conditions (right photograph). Alignment of the Hag protein sequences based on the available genomes (75) showed that the sequence of the Hag protein in the B. subtilis natto strain contains a 6-amino-acid insertion and a 5-amino-acid deletion at different positions; however, the total sizes of the proteins are similar in various Bacilli (304 and 305 amino acids in strain 168 and the natto strain, respectively). The promoter regions of the hag genes on the genomes of strain 168 and the natto strain are highly similar (98% identical), and the σD binding sites are identical in the two strains. Download
Surfactin production and the hydrophobin BslA are essential for sliding motility in B. subtilis. (A) After addition of synthetic surfactin to surfactin-deficient RG4365 and NCIB3610 cells, surface motilities were reestablished. The supplementation of the petri dishes with synthetic surfactin (Sigma-Aldrich; see Materials and Methods) restored the sliding and swarming ability of B. subtilis natto strain and Marburg-derived sfrAA cells and confirmed that surfactin is the essential biosurfactant that allows the surface-associated translocation (sliding and swarming) of B. subtilis. (B) BslA contributes to the hydrophobicity of B. subtilis under surface motility conditions. Cellular hydrophobicity was calculated using the BATH assay (77) between a hydrocarbon (octane or heptane) and cultures of NCIB3610, RG4365, and bslA natto cells. (C) Surface water repellence of wild-type and bslA natto cells. After overnight incubation on soft LB agar plates at 37°C, 5.0 µl of a hydrophilic staining solution was carefully poured on the patch of motile cells and the colony of the bslA mutant. The swarming- and sliding-proficient cells (NCIB3610 and RG4365, respectively) prevented the water penetration into the cell matrix. In contrast, the bslA-deficient cells were unable to repel the water, which almost immediately and uniformly penetrated throughout them. Download
Sub-MICs of cerulenin do not affect planktonic growth of B. subtilis cells but impair social surface translocation. NCIB3610 (A) and RG4365 (B) wild-type cells were grown in LB broth at 200 rpm and 37°C in the absence or presence of different concentrations of cerulenin. The averages from three independent experiments are shown. Cerulenin concentrations were as follows: 5.0 µg ⋅ ml−1 (red triangles), 2.0 µg ⋅ ml−1 (blue triangles), 1.0 µg ⋅ ml−1 (green triangles), 0.1 µg ⋅ ml−1 (yellow circles), and 0 (orange squares). (C and D) Dose-dependent inhibitory effect of different cerulenin concentrations on swarming (C) and sliding (D) proficiencies of B. subtilis cells. Download
(A) Supplementation with straight fatty acids (palmitic [nC16:0] and oleic [nC18:1] fatty acids) does not restore the sliding proficiency of wild-type RG4365 cells treated with a sub-MIC of cerulenin (left) but allows restart of the growth of RG4365 cells in the presence of the antibiotic under planktonic conditions (right). The graphs are the results of one representative experiment repeated five times. Symbols are as follows: red circles, 2.0 µg ⋅ ml−1 cerulenin (left graph) and 5.0 µg ⋅ ml−1 cerulenin (right graph); blue triangles, 2.0 µg ⋅ ml−1 cerulenin plus nC16:0 and nC18:1 and 5.0 µg ⋅ ml−1 cerulenin plus nC16:0 and nC18:1 (left and right graphs, respectively); green circles, no cerulenin. (B) Profile of fatty acids (FAs) synthesized de novo by wild-type B. subtilis RG4365 cells under conditions of planktonic growth in LB broth (right circle) and active sliding on soft agar LB plates (left circle). The averages from eight biological replicates are presented. Download
Transcomplementation of kinB-deficient and kinC-deficient B. subtilis strains. RG4365-derived kinB and kinC mutants were transcomplemented with wild-type kinB and kinC alleles (see Table S1), respectively, which were ectopically integrated into the nonessential locus amyE. Download
KinB harbors a potassium-sensing domain with homology to the selectivity filter of potassium channels. (A) Representative cartoon of the membrane topology of a typical prokaryotic potassium channel harboring six transmembrane domains. The region of the potassium pore (P) surrounded by two alpha helices that dips into the membrane is highlighted to indicate the components of the pore region: turret, pore helix, selectivity filter, and the loop that precedes the sixth transmembrane domain (TM6). The membrane-embedded selectivity filter for potassium is emphasized in green. (B) Representative drawing of the membrane topology of KinB. The known cytoplasmic domains (HK and ATPase) of the kinase are indicated. The position of the putative potassium-sensing domain harbored by the kinase is also highlighted in green (see text for details). (C) The 8-amino-acid sequence of KinB (S-L-K-T-N-G-T-G) with homology to the selectivity filter present in potassium channels (S/T-X-X-T-X-G-X-G) (56, 60, 78). Out of the 8 amino acids of the motif, position seven of potassium channels is generally assigned to an aromatic amino acid (mainly tyrosine [Y] but also phenylalanine can be present) (56, 57, 60, 79). In KinB, this position is occupied by a threonine (T). However, it has been shown that nonconservative substitutions of the Y residue in position 7 of the selectivity filter leave selectivity for potassium intact, and data show that an aromatic group at position 7 is not essential in determining potassium selectivity (56). More recently, it was found that other amino acids (i.e., leucine, aspartic acid, etc.) can be present in position 7 of the selectivity filter of functional potassium channels (57, 79). Download
Working model for the spatiotemporal regulation of the different social behaviors in B. subtilis. (A) When B. subtilis is attached and committed to a surface (i.e., in soft LB agar plates), its social behaviors fall under the control of the master regulator Spo0A. Initially, KinB becomes autophosphorylated under the positive input of a high intracellular concentration of potassium ions. At the same time, the high level of potassium inhibits KinC activation (autophosphorylation). Not indicated in the cartoon is the possibility that KinB might inhibit KinC in its proficiency to trigger biofilm formation (see text for details). At this early time of development, the levels of formed KinB~Pi are enough to produce low levels of active Spo0A (Spo0A~Pi) that trigger expression of the sliding machinery. Because the two phenomena, sliding and biofilm formation, share essentially similar structural components, there is the possibility that the expression levels of these components under the initial input of KinB~Pi might be different during sliding and biofilm formation in order to allow one or the other behavior and/or that other essential components of the sliding and biofilm formation machineries are differently regulated and have not yet been identified (9, 31–33). (B) At middle times of sliding development, there should be a drop in the intracellular concentration of potassium ions in cells located at the inner part of the sliding disc that leads to the downregulation of KinB and activation of KinC, respectively. KinC~Pi feeds the phosphorelay to produce higher levels of Spo0A~Pi to maintain active sliding in soft LB agar plates. Under special conditions of cultivation (i.e., in soft LBY agar plates), KinC~Pi would also trigger biofilm formation at the inner part of the sliding disc. At the border of both sliding discs (in LB and LBY agar plates), KinB and KinC remain active and inactive, respectively, due to the high intracellular concentration of potassium ions in the cells (not shown in the cartoon). (C) At later times of development, KinC remains active and KinB and KinA become activated by nutrient deficiency as previously described (53). The three activated kinases (KinA~Pi, KinB~Pi, and KinC~Pi) produce high levels of Spo0A~Pi to allow fruiting body formation (filled with spores) at the tips of the biofilm. Download
List of strains used in this study.
(A) Array analysis of spo0A and wild-type cells under sliding-permissive conditions; (B) array analysis of wild-type cells under sliding-permissive and sliding-restrictive conditions.
Oligonucleotides used in this study.
ACKNOWLEDGMENTS
We thank N. R. Stanley Wall for the kind gift of strains NRS1502 and NRS2097 and J. Stülke for strain GP901. We thank Tamara Spalding, Adrian Rovetto, and Walter Ramirez for their assistance during the microarray experiments and genetic constructions. We also thank Rafael Carlucci for figure and caption design.
Work in the laboratory of R.R.G. is supported by CONICET (National Research Council of Argentina), ANCyT (National Agency of Science and Technology of Argentina), and the aid of the Pew Latin-American Program in Biological Sciences (Philadelphia, USA), the Fulbright Committee (Washington, DC, USA), and former Fundación Antorchas (Buenos Aires, Argentina). P.D.O./D.V. and C. L./V.D. are PhD fellows from ANCyT and CONICET, respectively. Work in the laboratory of A.T.K. is supported by a Marie Curie Career Integration Grant (PheHetBacBiofilm), Grant KO4741/2-1 from the Deutsche Forschungsgemeinschaft (DFG) within the framework of the DFG Priority Programme SPP1617, a JSMC (Jena School for Microbial Communication) startup fund, and BacFoodNet COST Action FA1202. R.G.-M., E.M., and T.H. are funded by CONACyT-DAAD, JSMC, and IMPRS, respectively.
Footnotes
Citation Grau RR, de Oña P, Kunert M, Leñini C, Gallegos-Monterrosa R, Mhatre E, Vileta D, Donato V, Hölscher T, Boland W, Kuipers OP, Kovács ÁT. 2015. A duo of potassium-responsive histidine kinases govern the multicellular destiny of Bacillus subtilis. mBio 6(4):e00581-15. doi:10.1128/mBio.00581-15.
REFERENCES
- 1.Harshey RM. 2003. Bacterial motility on a surface: many ways to a common goal. Annu Rev Microbiol 57:249–273. doi: 10.1146/annurev.micro.57.030502.091014. [DOI] [PubMed] [Google Scholar]
- 2.Henrichsen J. 1972. Bacterial surface translocation: a survey and a classification. Bacteriol Rev 36:478–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jarrell KF, McBride MJ. 2008. The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol 6:466–476. doi: 10.1038/nrmicro1900. [DOI] [PubMed] [Google Scholar]
- 4.Kearns DB. 2010. A field guide to bacterial swarming motility. Nat Rev Microbiol 8:634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martínez A, Torello S, Kolter R. 1999. Sliding motility in mycobacteria. J Bacteriol 181:7331–7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park SY, Pontes MH, Groisman EA. 2015. Flagella-independent surface motility in Salmonella enterica serovar Typhimurium. Proc Natl Acad Sci U S A 112:1850–1855. doi: 10.1073/pnas.1422938112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Branda SS, González-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. 2001. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A 98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Higgins D, Dworkin J. 2012. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol Rev 36:131–148. doi: 10.1111/j.1574-6976.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kovács AT, Smits WK, Mirończuk AM, Kuipers OP. 2009. Ubiquitous late competence genes in Bacillus species indicate the presence of functional DNA uptake machineries. Environ Microbiol 11:1911–1922. doi: 10.1111/j.1462-2920.2009.01937.x. [DOI] [PubMed] [Google Scholar]
- 10.López D, Fischbach MA, Chu F, Losick R, Kolter R. 2009. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc Natl Acad Sci U S A 106:280–285. doi: 10.1073/pnas.0810940106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. 2013. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol 11:157–168. doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kearns DB, Losick R. 2003. Swarming motility in undomesticated Bacillus subtilis. Mol Microbiol 49:581–590. doi: 10.1046/j.1365-2958.2003.03584.x. [DOI] [PubMed] [Google Scholar]
- 13.Abee T, Kovács AT, Kuipers OP, van der Veen S. 2011. Biofilm formation and dispersal in Gram-positive bacteria. Curr Opin Biotechnol 22:172–179. doi: 10.1016/j.copbio.2010.10.016. [DOI] [PubMed] [Google Scholar]
- 14.Pedrido ME, Pedrido ME, de Oña P, Ramirez W, Leñini C, Goñi A, Grau R. 2013. Spo0A links de novo fatty acid synthesis to sporulation and biofilm development in Bacillus subtilis. Mol Microbiol 87:348–367. doi: 10.1111/mmi.12102. [DOI] [PubMed] [Google Scholar]
- 15.Trejo M, Douarche C, Bailleux V, Poulard C, Mariot S, Regeard C, Raspaud E. 2013. Elasticity and wrinkled morphology of Bacillus subtilis pellicles. Proc Natl Acad Sci U S A 110:2011–2016. doi: 10.1073/pnas.1217178110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lombardía E, Rovetto AJ, Arabolaza AL, Grau RR. 2006. A LuxS-dependent cell-to-cell language regulates social behavior and development in Bacillus subtilis. J Bacteriol 188:4442–4452. doi: 10.1128/JB.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bais HP, Fall R, Vivanco JM. 2004. Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol 134:307–319. doi: 10.1104/pp.103.028712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emmert EA, Handelsman J. 1999. Biocontrol of plant disease: a (Gram-) positive perspective. FEMS Microbiol Lett 171:1–9. doi: 10.1111/j.1574-6968.1999.tb13405.x. [DOI] [PubMed] [Google Scholar]
- 19.Cutting SM. 2011. Bacillus probiotics. Food Microbiol 28:214–220. doi: 10.1016/j.fm.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Senesi S, Ricca E, Henriques A, Cutting S. 2004. Bacillus spores as probiotic products for human use, p 131–141. In Ricca E, Henriques A, Cutting S (ed), Bacterial spore formers: probiotics and emerging applications. Taylor & Francis Group Ltd, New York, NY. [Google Scholar]
- 21.VidyaLaxme B, Rovetto A, Grau R, Agrawal R. 2014. Synergistic effects of probiotic Leuconostoc mesenteroides and Bacillus subtilis in malted ragi (Eleucine corocana) food for antagonistic activity against V. cholerae and other beneficial properties. J Food Sci Technol 51:3072–3082. doi: 10.1007/s13197-012-0834-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kinsinger RF, Shirk MC, Fall R. 2003. Rapid surface motility in Bacillus subtilis is dependent on extracellular surfactin and potassium ion. J Bacteriol 185:5627–5631. doi: 10.1128/JB.185.18.5627-5631.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blair KM, Turner L, Winkelman JT, Berg HC, Kearns DB. 2008. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science 320:1636–1638. doi: 10.1126/science.1157877. [DOI] [PubMed] [Google Scholar]
- 24.Kearns DB, Chu F, Branda SS, Kolter R, Losick R. 2005. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol 55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- 25.Burbulys D, Trach KA, Hoch JA. 1991. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell 64:545–552. doi: 10.1016/0092-8674(91)90238-T. [DOI] [PubMed] [Google Scholar]
- 26.Molle V, Fujita M, Jensen ST, Eichenberger P, González-Pastor JE, Liu JS, Losick R. 2003. The Spo0A regulon of Bacillus subtilis. Mol Microbiol 50:1683–1701. doi: 10.1046/j.1365-2958.2003.03818.x. [DOI] [PubMed] [Google Scholar]
- 27.Arabolaza AL, Nakamura A, Pedrido ME, Martelotto L, Orsaria L, Grau RR. 2003. Characterization of a novel inhibitory feedback of the anti-anti-sigma SpoIIAA on Spo0A activation during development in Bacillus subtilis. Mol Microbiol 47:1251–1263. [DOI] [PubMed] [Google Scholar]
- 28.Fujita M, Losick R. 2005. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev 19:2236–2244. doi: 10.1101/gad.1335705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ireton K, Rudner DZ, Siranosian KJ, Grossman AD. 1993. Integration of multiple developmental signals in Bacillus subtilis through the Spo0A transcription factor. Genes Dev 7:283–294. doi: 10.1101/gad.7.2.283. [DOI] [PubMed] [Google Scholar]
- 30.Kinsinger RF, Kearns DB, Hale M, Fall R. 2005. Genetic requirements for potassium ion-dependent colony spreading in Bacillus subtilis. J Bacteriol 187:8462–8469. doi: 10.1128/JB.187.24.8462-8469.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fall R, Kearns DB, Nguyen T. 2006. A defined medium to investigate sliding motility in a Bacillus subtilis flagella-less mutant. BMC Microbiol 6:31. doi: 10.1186/1471-2180-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rudrappa T, Bais HP. 2007. Arabidopsis thaliana root surface chemistry regulates in planta biofilm formation of Bacillus subtilis. Plant Signal Behav 2:349–350. doi: 10.4161/psb.2.5.4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beauregard PB, Chai Y, Vlamakis H, Losick R, Kolter R. 2013. Bacillus subtilis biofilm induction by plant polysaccharides. Proc Natl Acad Sci U S A 110:E1621–E1630. doi: 10.1073/pnas.1218984110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y, Cao S, Chai Y, Clardy J, Kolter R, Guo JH, Losick R. 2012. A Bacillus subtilis sensor kinase involved in triggering biofilm formation on the roots of tomato plants. Mol Microbiol 85:418–430. doi: 10.1111/j.1365-2958.2012.08109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aguilar C, Vlamakis H, Guzman A, Losick R, Kolter R. 2010. KinD is a checkpoint protein linking spore formation to extracellular-matrix production in Bacillus subtilis biofilms. mBio 1(1):e00035-10. doi: 10.1128/mBio.00035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vlamakis H, Aguilar C, Losick R, Kolter R. 2008. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev 22:945–953. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kovács AT, Kuipers OP. 2011. Rok regulates yuaB expression during architecturally complex colony development of Bacillus subtilis 168. J Bacteriol 193:998–1002. doi: 10.1128/JB.01170-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verhamme DT, Murray EJ, Stanley-Wall NR. 2009. DegU and Spo0A jointly control transcription of two loci required for complex colony development by Bacillus subtilis. J Bacteriol 191:100–108. doi: 10.1128/JB.01236-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stöver AG, Driks A. 1999. Secretion, localization, and antibacterial activity of TasA, a Bacillus subtilis spore-associated protein. J Bacteriol 181:1664–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.López D, Vlamakis H, Losick R, Kolter R. 2009. Paracrine signaling in a bacterium. Genes Dev 23:1631–1638. doi: 10.1101/gad.1813709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Angelini TE, Roper M, Kolter R, Weitz DA, Brenner MP. 2009. Bacillus subtilis spreads by surfing on waves of surfactant. Proc Natl Acad Sci U S A 106:18109–18113. doi: 10.1073/pnas.0905890106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seminara A, Angelini TE, Wilking JN, Vlamakis H, Ebrahim S, Kolter R, Weitz DA, Brenner MP. 2012. Osmotic spreading of Bacillus subtilis biofilms driven by an extracellular matrix. Proc Natl Acad Sci U S A 109:1116–1121. doi: 10.1073/pnas.1109261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garti-Levi S, Eswara A, Smith Y, Fujita M, Ben-Yehuda S. 2013. Novel modulators controlling entry into sporulation in Bacillus subtilis. J Bacteriol 195:1475–1483. doi: 10.1128/JB.02160-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hobley L, Ostrowski A, Rao FV, Bromley KM, Porter M, Prescott AR, MacPhee CE, Van Aalten DM, Stanley-Wall NR. 2013. BslA is a self-assembling bacterial hydrophobin that coats the Bacillus subtilis biofilm. Proc Natl Acad Sci U S A 110:13600–13605. doi: 10.1073/pnas.1306390110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kobayashi K, Iwano M. 2012. BslA (YuaB) forms a hydrophobic layer on the surface of Bacillus subtilis biofilms. Mol Microbiol 85:51–66. doi: 10.1111/j.1365-2958.2012.08094.x. [DOI] [PubMed] [Google Scholar]
- 46.Kovács AT, van Gestel J, Kuipers OP. 2012. The protective layer of biofilm: a repellent function for a new class of amphiphilic proteins. Mol Microbiol 85:8–11. doi: 10.1111/j.1365-2958.2012.08101.x. [DOI] [PubMed] [Google Scholar]
- 47.Epstein AK, Pokroy B, Seminara A, Aizenberg J. 2011. Bacterial biofilm shows persistent resistance to liquid wetting and gas penetration. Proc Natl Acad Sci U S A 108:995–1000. doi: 10.1073/pnas.1011033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Gestel J, Weissing FJ, Kuipers OP, Kovács AT. 2014. Density of founder cells affects spatial pattern formation and cooperation in Bacillus subtilis biofilms. ISME J 8:2069–2079. doi: 10.1038/ismej.2014.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Gestel J, Vlamakis H, Kolter R. 2015. From cell differentiation to cell collectives: Bacillus subtilis uses division of labor to migrate. PLoS Biol 13:e1002141. doi: 10.1371/journal.pbio.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gottig N, Pedrido ME, Méndez M, Lombardía E, Rovetto A, Philippe V, Orsaria L, Grau R. 2005. The Bacillus subtilis SinR and RapA developmental regulators are responsible for inhibition of spore development by alcohol. J Bacteriol 187:2662–2672. doi: 10.1128/JB.187.8.2662-2672.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mhatre E, Monterrosa RG, Kovács AT. 2014. From environmental signals to regulators: modulation of biofilm development in Gram-positive bacteria. J Basic Microbiol 54:616–632. doi: 10.1002/jobm.201400175. [DOI] [PubMed] [Google Scholar]
- 52.Lundberg ME, Becker EC, Choe S. 2013. MstX and a putative potassium channel facilitate biofilm formation in Bacillus subtilis. PLoS One 8:e60993. doi: 10.1371/journal.pone.0060993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kolodkin-Gal I, Elsholz AK, Muth C, Girguis PR, Kolter R, Losick R. 2013. Respiration control of multicellularity in Bacillus subtilis by a complex of the cytochrome chain with a membrane-embedded histidine kinase. Genes Dev 27:887–899. doi: 10.1101/gad.215244.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trach KA, Hoch JA. 1993. Multisensory activation of the phosphorelay initiating sporulation in Bacillus subtilis: identification and sequence of the protein kinase of the alternate pathway. Mol Microbiol 8:69–79. doi: 10.1111/j.1365-2958.1993.tb01204.x. [DOI] [PubMed] [Google Scholar]
- 55.Wang L, Grau R, Perego M, Hoch JA. 1997. A novel histidine kinase inhibitor regulating development in Bacillus subtilis. Genes Dev 11:2569–2579. doi: 10.1101/gad.11.19.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Szurmant H, Hoch JA. 2013. Statistical analyses of protein sequence alignments identify structures and mechanisms in signal activation of sensor histidine kinases. Mol Microbiol 87:707–712. doi: 10.1111/mmi.12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heginbotham L, Lu Z, Abramson T, MacKinnon R. 1994. Mutations in the K+ channel signature sequence. Biophys J 66:1061–1067. doi: 10.1016/S0006-3495(94)80887-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuo MM, Haynes WJ, Loukin SH, Kung C, Saimi Y. 2005. Prokaryotic K+ channels: from crystal structures to diversity. FEMS Microbiol Rev 29:961–985. doi: 10.1016/j.femsre.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 59.Fabret C, Feher VA, Hoch JA. 1999. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J Bacteriol 181:1975–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choe S. 2002. Potassium channel structures. Nat Rev Neurosci 3:115–121. doi: 10.1038/nrn727. [DOI] [PubMed] [Google Scholar]
- 61.Derst C, Karschin A. 1998. Evolutionary link between prokaryotic and eukaryotic K+ channels. J Exp Biol 201:2791–2799. [PubMed] [Google Scholar]
- 62.Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 63.Jiang M, Shao W, Perego M, Hoch JA. 2000. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol Microbiol 38:535–542. doi: 10.1046/j.1365-2958.2000.02148.x. [DOI] [PubMed] [Google Scholar]
- 64.Chai Y, Chu F, Kolter R, Losick R. 2008. Bistability and biofilm formation in Bacillus subtilis. Mol Microbiol 67:254–263. doi: 10.1111/j.1365-2958.2007.06040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Hijum SA, de Jong A, Baerends RJ, Karsens HA, Kramer NE, Larsen R, den Hengst CD, Albers CJ, Kok J, Kuipers OP. 2005. A generally applicable validation scheme for the assessment of factors involved in reproducibility and quality of DNA-microarray data. BMC Genomics 6:77. doi: 10.1186/1471-2164-6-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Hijum SA, de Jong A, Buist G, Kok J, Kuipers OP. 2003. UniFrag and GenomePrimer: selection of primers for genome-wide production of unique amplicons. Bioinformatics 19:1580–1582. doi: 10.1093/bioinformatics/btg203. [DOI] [PubMed] [Google Scholar]
- 67.Kuipers OP, de Jong A, Baerends RJ, van Hijum SA, Zomer AL, Karsens HA, den Hengst CD, Kramer NE, Buist G, Kok J. 2002. Transcriptome analysis and related databases of Lactococcus lactis. Antonie Van Leeuwenhoek 82:113–122. doi: 10.1023/A:1020691801251. [DOI] [PubMed] [Google Scholar]
- 68.van Hijum SA, Garcia de la Nava J, Trelles O, Kok J, Kuipers OP. 2003. MicroPreP: a cDNA microarray data pre-processing framework. Appl Bioinformatics 2:241–244. [PubMed] [Google Scholar]
- 69.Baldi P, Long AD. 2001. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 70.Guérout-Fleury AM, Shazand K, Frandsen N, Stragier P. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335–336. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- 71.Kim L, Mogk A, Schumann W. 1996. A xylose-inducible Bacillus subtilis integration vector and its application. Gene 181:71–76. doi: 10.1016/S0378-1119(96)00466-0. [DOI] [PubMed] [Google Scholar]
- 72.Yang L, Ukil L, Osmani A, Nahm F, Davies J, De Souza CP, Dou X, Perez-Balaguer A, Osmani SA. 2004. Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot Cell 3:1359–1362. doi: 10.1128/EC.3.5.1359-1362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rodríguez-Ruiz J, Belarbi E, Sánchez JLG, Alonso DL. 1998. Rapid simultaneous lipid extraction and transesterification for fatty acid analyses. Biotechnol Tech 12:689–691. doi: 10.1023/A:1008812904017. [DOI] [Google Scholar]
- 74.Grau R, de Mendoza D. 1993. Regulation of the synthesis of unsaturated fatty acids by growth temperature in Bacillus subtilis. Mol Microbiol 8:535–542. doi: 10.1111/j.1365-2958.1993.tb01598.x. [DOI] [PubMed] [Google Scholar]
- 75.Nishito Y, Osana Y, Hachiya T, Popendorf K, Toyoda A, Fujiyama A, Itaya M, Sakakibara Y. 2010. Whole genome assembly of a natto production strain Bacillus subtilis natto from very short read data. BMC Genomics 11:243. doi: 10.1186/1471-2164-11-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mirel DB, Chamberlin MJ. 1989. The Bacillus subtilis flagellin gene (hag) is transcribed by the sigma 28 form of RNA polymerase. J Bacteriol 171:3095–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Y, Miller RM. 1992. Enhanced octadecane dispersion and biodegradation by a Pseudomonas rhamnolipid surfactant (biosurfactant). Appl Environ Microbiol 58:3276–3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goldstein SA, Colatsky TJ. 1996. Ion channels: too complex for rational drug design? Neuron 16:913–919. doi: 10.1016/S0896-6273(00)80114-2. [DOI] [PubMed] [Google Scholar]
- 79.Johansson I, Blatt MR. 2006. Interactive domains between pore loops of the yeast K+ channel TOK1 associate with extracellular K+ sensitivity. Biochem J 393:645–655. doi: 10.1042/BJ20051380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Spreading behavior of the undomesticated B. subtilis natto strain. Surface motility of undomesticated B. subtilis Marburg-related strain NCIB3610 (A), human-probiotic natto-related strain RG4365 (B), and domesticated laboratory strain JH642 (C) 10 h, 18 h, and 36 h after inoculation. Briefly, petri dishes containing LB medium fortified with 0.7% agar were centrally inoculated with any of the three B. subtilis strains and incubated at 37°C (see Materials and Methods for details). (A) In these pictures, it is possible to appreciate the rapid ability of the NCIB3610 strain to swarm on the agar surface. It was previously reported that robust swarming motility, a type of social motility that relies on groups of cells (rafts) that express numerous flagella and differentiate to migrate coordinately along a solid substrate, was present in the NCIB3610 strain (4, 12). (B) The strain RG4365 needed a longer incubation to start to move (as would be expected from a strain that is able only to slide), but after the first 5 h of incubation, it was possible to observe a uniform and highly hydrophobic monolayer of cells that cooperatively and uniformly advanced in large filaments made of close cell-to-cell contacts, predominantly along the longitudinal axis, across the agar plate; see amplified views (phase-contrast and fluorescence images) of RG4365 cells on the right. (C) As expected, the domesticated B. subtilis strain JH642 was unable to move because it harbors a mutation in a gene (sfp) needed for surfactin synthesis and a frameshift mutation in the regulatory gene swrA (24). In contrast, the undomesticated strains NCIB3610 and RG4365 do not harbor those mutations and therefore are able to spread on solid surfaces (12, 75). (D) Kinetics of motility diameter of the various strains: NCIB3610 (red diamonds), RG4365 (green circles), and JH642 (blue squares). Arrows in panel D indicate time points with representative pictures above, while a line at 8.5 cm shows the maximal size of motility related to the size of the plate used. The rate of surface motility was maximal (0.75 cm ⋅ h−1) for the strain NCIB3610, followed by the natto strain RG4365 (0.30 cm ⋅ h−1). (E and F) One essential factor for the swimming and swarming ability of the strain NCIB3610 is the flagellum, the proposed motor for those motilities (12). In contrast, the RG4365 strain grown in shaking liquid culture was completely nonmotile (data not shown). Cell samples for flagellum visualization were prepared as indicated previously (12) after 10 h (for NCIB3610 strain [E]) and 20 h (for RG4365 strain [F]) of inoculation on motility plates. These time points represent the stages of maximal surface motility for each wild-type strain (see panel D). Samples were taken from the borders of the surface motility discs, and cells were carefully separated and diluted (from left to right in panels E and F) in order to improve the visualization of flagella. Pictures on the right side of panels E and F are representative photographs of wild-type NCIB3610 cells (E) and RG4365 cells (F) taken from liquid cultures grown until late exponential phase (optical density at 525 nm [OD525], 0.9) in LB medium with aeration (200 rpm). Under all the examined conditions (swarming and swimming), B. subtilis NCIB3610 cells harbored flagella and were motile (E). In contrast, wild-type B. subtilis RG4365 cells were motile only on soft agar plates but without harboring flagella (F). In liquid culture, the RG4365 cells also were devoid of flagella and completely nonmotile (right image in panel F). Cells were stained for flagella as described in Materials and Methods. Arrows indicate the positions of some flagella shown as examples. (G) To confirm the dispensability of flagellum production for the surface-associated motility (sliding) in RG4365, we proceeded to delete the hag gene (75), which is involved in the synthesis of the flagellin subunit of the flagellum (76). As shown (left photograph), the isogenic hag mutant strain RG4387 (see Table S1) was able to slide on the agar surface at the same rate as the wild-type parental strain RG4365 after 24 h of incubation at 37°C. Similarly, the hag inactivation in the NCIB3610 strain abolished its ability to swarm under similar incubation conditions (right photograph). Alignment of the Hag protein sequences based on the available genomes (75) showed that the sequence of the Hag protein in the B. subtilis natto strain contains a 6-amino-acid insertion and a 5-amino-acid deletion at different positions; however, the total sizes of the proteins are similar in various Bacilli (304 and 305 amino acids in strain 168 and the natto strain, respectively). The promoter regions of the hag genes on the genomes of strain 168 and the natto strain are highly similar (98% identical), and the σD binding sites are identical in the two strains. Download
Surfactin production and the hydrophobin BslA are essential for sliding motility in B. subtilis. (A) After addition of synthetic surfactin to surfactin-deficient RG4365 and NCIB3610 cells, surface motilities were reestablished. The supplementation of the petri dishes with synthetic surfactin (Sigma-Aldrich; see Materials and Methods) restored the sliding and swarming ability of B. subtilis natto strain and Marburg-derived sfrAA cells and confirmed that surfactin is the essential biosurfactant that allows the surface-associated translocation (sliding and swarming) of B. subtilis. (B) BslA contributes to the hydrophobicity of B. subtilis under surface motility conditions. Cellular hydrophobicity was calculated using the BATH assay (77) between a hydrocarbon (octane or heptane) and cultures of NCIB3610, RG4365, and bslA natto cells. (C) Surface water repellence of wild-type and bslA natto cells. After overnight incubation on soft LB agar plates at 37°C, 5.0 µl of a hydrophilic staining solution was carefully poured on the patch of motile cells and the colony of the bslA mutant. The swarming- and sliding-proficient cells (NCIB3610 and RG4365, respectively) prevented the water penetration into the cell matrix. In contrast, the bslA-deficient cells were unable to repel the water, which almost immediately and uniformly penetrated throughout them. Download
Sub-MICs of cerulenin do not affect planktonic growth of B. subtilis cells but impair social surface translocation. NCIB3610 (A) and RG4365 (B) wild-type cells were grown in LB broth at 200 rpm and 37°C in the absence or presence of different concentrations of cerulenin. The averages from three independent experiments are shown. Cerulenin concentrations were as follows: 5.0 µg ⋅ ml−1 (red triangles), 2.0 µg ⋅ ml−1 (blue triangles), 1.0 µg ⋅ ml−1 (green triangles), 0.1 µg ⋅ ml−1 (yellow circles), and 0 (orange squares). (C and D) Dose-dependent inhibitory effect of different cerulenin concentrations on swarming (C) and sliding (D) proficiencies of B. subtilis cells. Download
(A) Supplementation with straight fatty acids (palmitic [nC16:0] and oleic [nC18:1] fatty acids) does not restore the sliding proficiency of wild-type RG4365 cells treated with a sub-MIC of cerulenin (left) but allows restart of the growth of RG4365 cells in the presence of the antibiotic under planktonic conditions (right). The graphs are the results of one representative experiment repeated five times. Symbols are as follows: red circles, 2.0 µg ⋅ ml−1 cerulenin (left graph) and 5.0 µg ⋅ ml−1 cerulenin (right graph); blue triangles, 2.0 µg ⋅ ml−1 cerulenin plus nC16:0 and nC18:1 and 5.0 µg ⋅ ml−1 cerulenin plus nC16:0 and nC18:1 (left and right graphs, respectively); green circles, no cerulenin. (B) Profile of fatty acids (FAs) synthesized de novo by wild-type B. subtilis RG4365 cells under conditions of planktonic growth in LB broth (right circle) and active sliding on soft agar LB plates (left circle). The averages from eight biological replicates are presented. Download
Transcomplementation of kinB-deficient and kinC-deficient B. subtilis strains. RG4365-derived kinB and kinC mutants were transcomplemented with wild-type kinB and kinC alleles (see Table S1), respectively, which were ectopically integrated into the nonessential locus amyE. Download
KinB harbors a potassium-sensing domain with homology to the selectivity filter of potassium channels. (A) Representative cartoon of the membrane topology of a typical prokaryotic potassium channel harboring six transmembrane domains. The region of the potassium pore (P) surrounded by two alpha helices that dips into the membrane is highlighted to indicate the components of the pore region: turret, pore helix, selectivity filter, and the loop that precedes the sixth transmembrane domain (TM6). The membrane-embedded selectivity filter for potassium is emphasized in green. (B) Representative drawing of the membrane topology of KinB. The known cytoplasmic domains (HK and ATPase) of the kinase are indicated. The position of the putative potassium-sensing domain harbored by the kinase is also highlighted in green (see text for details). (C) The 8-amino-acid sequence of KinB (S-L-K-T-N-G-T-G) with homology to the selectivity filter present in potassium channels (S/T-X-X-T-X-G-X-G) (56, 60, 78). Out of the 8 amino acids of the motif, position seven of potassium channels is generally assigned to an aromatic amino acid (mainly tyrosine [Y] but also phenylalanine can be present) (56, 57, 60, 79). In KinB, this position is occupied by a threonine (T). However, it has been shown that nonconservative substitutions of the Y residue in position 7 of the selectivity filter leave selectivity for potassium intact, and data show that an aromatic group at position 7 is not essential in determining potassium selectivity (56). More recently, it was found that other amino acids (i.e., leucine, aspartic acid, etc.) can be present in position 7 of the selectivity filter of functional potassium channels (57, 79). Download
Working model for the spatiotemporal regulation of the different social behaviors in B. subtilis. (A) When B. subtilis is attached and committed to a surface (i.e., in soft LB agar plates), its social behaviors fall under the control of the master regulator Spo0A. Initially, KinB becomes autophosphorylated under the positive input of a high intracellular concentration of potassium ions. At the same time, the high level of potassium inhibits KinC activation (autophosphorylation). Not indicated in the cartoon is the possibility that KinB might inhibit KinC in its proficiency to trigger biofilm formation (see text for details). At this early time of development, the levels of formed KinB~Pi are enough to produce low levels of active Spo0A (Spo0A~Pi) that trigger expression of the sliding machinery. Because the two phenomena, sliding and biofilm formation, share essentially similar structural components, there is the possibility that the expression levels of these components under the initial input of KinB~Pi might be different during sliding and biofilm formation in order to allow one or the other behavior and/or that other essential components of the sliding and biofilm formation machineries are differently regulated and have not yet been identified (9, 31–33). (B) At middle times of sliding development, there should be a drop in the intracellular concentration of potassium ions in cells located at the inner part of the sliding disc that leads to the downregulation of KinB and activation of KinC, respectively. KinC~Pi feeds the phosphorelay to produce higher levels of Spo0A~Pi to maintain active sliding in soft LB agar plates. Under special conditions of cultivation (i.e., in soft LBY agar plates), KinC~Pi would also trigger biofilm formation at the inner part of the sliding disc. At the border of both sliding discs (in LB and LBY agar plates), KinB and KinC remain active and inactive, respectively, due to the high intracellular concentration of potassium ions in the cells (not shown in the cartoon). (C) At later times of development, KinC remains active and KinB and KinA become activated by nutrient deficiency as previously described (53). The three activated kinases (KinA~Pi, KinB~Pi, and KinC~Pi) produce high levels of Spo0A~Pi to allow fruiting body formation (filled with spores) at the tips of the biofilm. Download
List of strains used in this study.
(A) Array analysis of spo0A and wild-type cells under sliding-permissive conditions; (B) array analysis of wild-type cells under sliding-permissive and sliding-restrictive conditions.
Oligonucleotides used in this study.