Abstract
Located in southwest China, the Aha watershed is continually contaminated by acid mine drainage (AMD) produced from upstream abandoned coal mines. The watershed is fed by creeks with elevated concentrations of aqueous Fe (total Fe > 1 g/liter) and SO42− (>6 g/liter). AMD contamination gradually decreases throughout downstream rivers and reservoirs, creating an AMD pollution gradient which has led to a suite of biogeochemical processes along the watershed. In this study, sediment samples were collected along the AMD pollution sites for geochemical and microbial community analyses. High-throughput sequencing found various bacteria associated with microbial Fe and S cycling within the watershed and AMD-impacted creek. A large proportion of Fe- and S-metabolizing bacteria were detected in this watershed. The dominant Fe- and S-metabolizing bacteria were identified as microorganisms belonging to the genera Metallibacterium, Aciditerrimonas, Halomonas, Shewanella, Ferrovum, Alicyclobacillus, and Syntrophobacter. Among them, Halomonas, Aciditerrimonas, Metallibacterium, and Shewanella have previously only rarely been detected in AMD-contaminated environments. In addition, the microbial community structures changed along the watershed with different magnitudes of AMD pollution. Moreover, the canonical correspondence analysis suggested that temperature, pH, total Fe, sulfate, and redox potentials (Eh) were significant factors that structured the microbial community compositions along the Aha watershed.
INTRODUCTION
Acid mine drainage (AMD) is one of the most environmentally threatening by-products of the mining industry. The chemical and/or microbial weathering of metal sulfide-rich rocks such as pyrite (FeS2), sphalerite (ZnS), and galena (PbS) produces sulfuric acid, which leads to the formation of AMD. AMD typically has low pH and elevated concentrations of sulfate and metals, which severely impair water and soil quality (1, 2).
Despite the extreme toxicity and acidity, the AMD-associated environments harbor numerous acidophilic microorganisms (3, 4). Recently, molecular techniques have been extensively applied to study AMD microbiology, and a variety of acidophilic and metal-tolerant microorganisms have been identified (5). Prokaryotic microorganisms that are metabolically active in acidic niche boundaries (pH < 3) were found to be phylogenetically diversified, affiliated within the phyla Proteobacteria, Firmicutes, Acitinobacteria, Nitrospirae, and Acidobacteria (5). Many of these acidophiles are capable of Fe and S cycling (e.g., oxidation and reduction of these elements in appropriate geochemical and microbial environments) and thus play an important role both in the formation of acid mine waters and in natural attenuation of AMD. However, our understanding of the roles of geochemical factors in shaping microbial community structure and the potential of microorganisms in natural attenuation of AMD and AMD-impacted watersheds is still limited.
The AMD of the Aha watershed in southwest China provides an ideal opportunity to study the interactions between microorganisms and AMD-impacted environments. The Aha watershed is located in the Southwest Coal Basin (SCB) in central Guizhou Province, which is one of the largest coal producers in China. Several abandoned coal mines are located upstream of the watershed. One of the abandoned coal mines, Dapo Coal Mine, has produced a large amount of acidic runoff (>7,000 m3/year; pH < 3) with high concentrations of Fe(II) (∼1 g/liter) and sulfate (∼7 g/liter). Dapo, along with other abandoned coal mines in this area, has led to an extensive input of iron and sulfate into the Youyu River, Aha Reservoir, and Xiaoche River (Fig. 1). Compared to the heavily contaminated creeks upstream, the downstream rivers and reservoirs were overall moderately to lightly contaminated. This watershed allows the investigation of the spatial distribution of microbial community composition along an AMD gradient. Furthermore, the contamination of the Aha Reservoir is of great concern since this reservoir is a source of drinking water for more than 500,000 people. In order to remediate these acidic mine waters, an onsite passive AMD treatment facility has been constructed in Dapo Creek, which will be operated in the near future (see Fig. S1 in the supplemental material). Therefore, a better understanding of the indigenous microbial diversity and its potential roles in natural attenuation of AMD will enable the development of molecular monitoring tools to evaluate and enhance bioremediation in this AMD treatment facility and other AMD-contaminated environments.
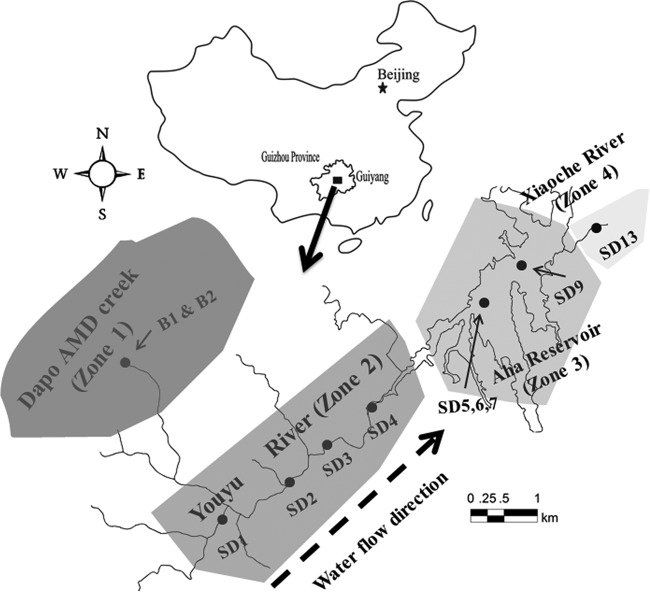
FIG 1.
Location of the Aha watershed in China and the locations of 11 sampling sites in the Aha watershed. Please note that water flows from SD1 to SD13. The map was created using CorelDraw.
In this study, we used integrated geochemical and molecular biological analyses to understand the spatial distribution of microbial communities along the pollution gradients within the Aha watershed. Our aims were (i) to characterize the microbial community structure and composition within this watershed, (ii) to explore the correlation between microbial structure and different geochemical variables, and (iii) to identify the active organisms and their metabolic potentials for natural mitigation of the AMD contamination, especially for those involved in Fe and S cycling.
MATERIALS AND METHODS
Sample collection.
The Aha watershed is located in southwest China at Guiyang, Guizhou Province (Fig. 1). Eleven samples were collected at different locations along the Aha watershed in September 2013. As illustrated by Fig. 1, samples B1 and B2 were collected from acid streamers of the mine portal (B1) and the orange sediment located 20 m downstream from mine portal (B2), respectively (see Fig. S1 in the supplemental material). Nine additional samples were collected from the downstream rivers and reservoir sediments. Specifically, samples SD1 to SD4 were collected from the Youyu River sediment, and sample SD13 was taken from Xiaoche River. The water depth ranged from 0.8 to 1.4 m for the Youyu River and was 0.6 m for the Xiaoche River. Approximately 300 g of sediment was collected per site. Samples from each site were pooled, homogenized, placed in sterile 50-ml tubes, and immediately stored at −20°C until further molecular analysis. Unlike river sediments, the Aha Reservoir has a sediment layer of more than 1 m. Thus, two sediment cores (cores 1 and 2), about 1 m in length, were taken from the reservoir using a gravity corer. The upper, middle, and bottom layers (SD5 to SD7) of core 1 and the bottom layer of core 2 (SD9) were used in all further molecular analyses.
Water chemistry analysis.
The chemistry was analyzed for all water samples from each sampling site. About 5 liters of water was immediately filtered on each sampling site through a 0.45-μm sterile membrane (Jinjing, Shanghai, China). Measurements of redox potential (Eh), pH, salinity, conductivity, total dissolved solids, and temperature were carried out using a Hach ION156 portable multiparameter meter (Loveland, CO). Remaining water was preserved with ultrapure 37% HCl for prevention of Fe(II) oxidation and then stored at 4°C in darkness for subsequent determination of metal ions.
Mineralogical and element analyses.
For inorganic geochemical analyses, bulk samples were dried at 105°C and thoroughly grated using a mortar and pestle before being passed through a 200-mesh sieve. Bulk chemical analyses of major elements were performed using X-ray fluorescence spectrometry (Axios-PW4400; PANalytical, Westborough, MA) at 40 kV and 95 mA, with a detection limit below 0.01%. In this analysis, 1 g of ground sample was combusted at 900°C for 2 h, and the difference in sample weight before and after combustion was reported as loss on ignition to quantify organic carbon in the sediment. The major elements were analyzed quantitatively after the fusion of 0.1 g of combusted sample with 3.6 g of dilithium tetraborate at 1,050°C for 16 min.
Ferric and ferrous irons in water samples were measured by the spectrophotometric method (UV-9000s; METASH, Shanghai, China) with 1,10-phenanthroline at 510 nm (6, 7). Total sulfur was determined by a BaSO4-based turbidimetric method (8), and sulfate was measured by ion chromatography (ICS-90; Dionex, Sunnyvale, CA) (9). To measure pH in sediments, 10-g dry sediment samples were put into a 100-ml Erlenmeyer flask and mixed with 25 ml of distilled water (1:2.5 soil-water ratios). The mixture was left to equilibrate for 20 min after shaking for 5 min. The pH was measured using a calibrated HQ30d pH meter (Hach, Loveland, CO). Total sulfur, soluble sulfur, and total organic carbon in sediments were measured using an elemental analyzer (vario MACRO cube; Elementar, Hanau, Germany) (8, 10).
DNA extraction, PCR amplification, and sequencing analysis.
Eleven samples were selected for molecular analysis. The genomic DNA was extracted using the FastDNA spin kit (MP Biomedicals, Santa Ana, CA) by following the manufacturer's protocol. All DNA extracts were stored at −80°C until analyzed. The V4 region of the 16S rRNA gene was amplified using the 515f/806r primer set (11). 16S rRNA gene tag-encoded ultrahigh-throughput sequencing was carried out using the Illumina MiSeq platform at Novogene (Beijing, China).
Sequences were analyzed using Quantitative Insights Into Microbial Ecology (QIIME) software and the UPARSE pipeline (12). Default settings for Illumina processing in QIIME were used, and then the UPARSE pipeline was used to assign taxonomy to 97% similarity via the RDP classifier (13). Estimated species richness indicated with Chao1 and Shannon indexes for 11 libraries were calculated (14).
Statistical analyses.
The similarity of microbial communities among different sediment samples was determined using UniFrac. QIIME calculated both weighted and unweighted UniFrac. The unweighted pair group method with arithmetic mean (UPGMA) was conducted on weighted UniFrac (15). Canonical correspondence analysis (CCA) was performed to measure chemical properties that had the most significant influence on the microbial community structure. The significant correlations of the physiochemical parameters were examined by Monte Carlo permutation. The triplot was generated by CANOCO 4.5 (Biometrics Wageningen, The Netherlands). Except as otherwise noted, the figures were generated by CanoDraw 4.0 (Biometrics Wageningen).
Illumina sequencing data accession number.
The reads generated by ultrahigh-throughput sequencing were deposited into the NCBI Sequence Read Archive database under accession number SRP044396.
RESULTS
Physicochemical characterization of collected samples.
Eleven samples were collected from four geographical zones, including Dapo Creek (zone 1 for samples B1 and B2), the Youyu River (zone 2 for samples SD1 to SD4), the Aha Reservoir (zone 3 for SD5, SD6, SD7, and SD9), and the Xiaoche River (zone 4 for SD13). As shown in Tables 1 and 2, the physicochemical parameters of the water and sediments differed significantly in the samples collected from environments with different extents of AMD contamination. The pH was very low (<3) in Dapo Creek at zone 1 and increased in downstream rivers and reservoirs within zones 2 to 4, with pHs typically higher than 6.5. The total soluble Fe in the AMD-contaminated creek was much higher than that in zones 2, 3, and 4 due to the elevated solubility of iron compounds in strongly acidic environments. Most Fe(II) was rapidly oxidized to Fe(III) due to the increase in pH of the Youyu River. The increase in pH was accompanied by a significant decrease in soluble Fe in zones 2 to 4. In addition, the AMD effluents had high sulfate concentrations (∼7 g/liter). In contrast, concentrations of sulfate in downstream samples were orders of magnitude lower and ranged from 112 mg/liter to 412 mg/liter. This may be mainly due to dilution given that the volume of the Youyu River is much greater than that of AMD streams. The redox potentials were also high in the AMD-contaminated creek (375.1 to 445.2 mV) but decreased to less than 100 mV in downstream samples.
TABLE 1.
Physiochemical parameters in the water samples taken from the Aha watersheda
| Sample | pH | Temp (°C) | Eh (mV) | TDS (g/liter) | Conductivity (mS/cm) | Salinity (‰) | Fetot (mg/liter) | Fe(II) (mg/liter) | SO42− (mg/liter) |
|---|---|---|---|---|---|---|---|---|---|
| B1 | 2.81 | 20.9 | 375.1 | 3.35 | 5.8 | 3.44 | 1,304 | 1,258 | 6,646 |
| B2 | 2.81 | 25.4 | 419.2 | 3.19 | 6.19 | 3.27 | 1,273 | 971 | 6,838 |
| SD1 | 8.1 | 15.5 | 20.8 | 0.251 | 0.422 | 0.25 | 0.015 | ND | 112 |
| SD2 | 6.8 | 15.6 | 20.3 | 0.306 | 0.516 | 0.31 | 0.014 | ND | 219 |
| SD3 | 8.12 | 15.7 | 35.3 | 0.347 | 0.581 | 0.35 | 0.035 | ND | 254 |
| SD4 | 8.11 | 15.9 | 53 | 0.467 | 0.78 | 0.47 | 0.035 | ND | 412 |
| SD5 | 7.59 | 18.9 | 60 | 0.269 | 0.487 | 0.27 | 0.015 | ND | 166 |
| SD9 | 7.57 | 18.8 | 73.1 | 0.26 | 0.47 | 0.26 | 0.015 | ND | 151 |
| SD13 | 7.44 | 17.5 | −183.6 | 0.32 | 0.559 | 0.32 | 0.009 | ND | 157 |
Abbreviations: TDS, total dissolved solids; Fetot, total iron; Fe(II), ferrous; ND, not detected.
All the sediment samples taken from zones 2, 3, and 4 showed high concentrations of SiO2 and relatively high concentrations of Al2O3 (see Table S1 in the supplemental material). In comparison, Fe2O3 was not distributed evenly within the watershed and was predominant in two samples from the AMD-contaminated creek. Among samples from the other zones, however, only sample SD2, which was taken adjacent to the abandoned mines, exhibited high Fe2O3 abundance (43.72%). Fe2O3 abundance was particularly low in samples SD1 and SD9. Therefore, these sediments were probably not contaminated or only lightly contaminated by AMD. The relatively high Fe2O3 concentrations in the sediment samples from Aha Reservoir (SD6 and SD7) indicated a long-term accumulation of iron compounds from acid mine waters.
As shown in Table 2, other geochemical and physiochemical parameters (e.g., total nitrogen, total carbon [TC], total organic carbon [TOC], total hydrogen, and total and soluble sulfur) exhibited different trends across samples. The entire watershed demonstrated low concentrations of total nitrogen. The total and organic carbon was relatively high in Youyu River but was low in sample B2. Total and soluble sulfur was high in sediments of Dapo Creek and the Xiaoche River but was relatively low in sediments from the Youyu River and Aha Reservoir. One possible explanation for the high soluble sulfur in sediments from the Xiaoche River is that a pipeline was connected between the Xiaoche River and the bottom of the Aha Reservoir. Slurry at the bottom of the Aha Reservoir was discharged into the Xiaoche River. Hence, the Xiaoche River may have accumulated large amounts of sulfur-containing compounds from the Aha Reservoir.
TABLE 2.
The pHs and concentrations of total nitrogen, total carbon, organic carbon, total hydrogen, and total and soluble sulfur in 11 sediment samplesa
| Geographical zone | Sample | pH | Concn (g/kg) of: |
|||||
|---|---|---|---|---|---|---|---|---|
| TN | TC | TOC | TS | SS | H | |||
| Zone 1 | B1 | 2.15 | 0.1974 | 1.2214 | 0.532 | 0.5821 | 0.555 | 0.2447 |
| B2 | 2.02 | 0.0216 | 0.1426 | 0.136 | 0.5875 | 0.563 | 0.186 | |
| Zone 2 | SD1 | 7.76 | 0.022 | 0.9281 | 0.249 | 0.0606 | 0.051 | 0.0736 |
| SD2 | 7.39 | 0.0327 | 0.465 | 0.349 | 0.0417 | 0.029 | 0.1379 | |
| SD3 | 7.84 | 0.0242 | 1.0477 | 0.855 | 0.1219 | 0.081 | 0.1188 | |
| SD4 | 7.66 | 0.0267 | 1.063 | 0.77 | 0.1121 | 0.071 | 0.1236 | |
| Zone 3 | SD5 | 7.4 | 0.0272 | 0.8025 | 0.262 | 0.0805 | 0.032 | 0.0949 |
| SD6 | 6.66 | 0.0218 | 0.8003 | 0.633 | 0.1879 | 0.151 | 0.1388 | |
| SD7 | 6.48 | 0.0238 | 0.8687 | 0.675 | 0.1701 | 0.133 | 0.1442 | |
| SD9 | 7.25 | 0.0298 | 0.4697 | 0.271 | 0.1002 | 0.083 | 0.1022 | |
| Zone 4 | SD13 | 6.86 | 0.053 | 0.7769 | 0.565 | 0.4878 | 0.423 | 0.1453 |
Abbreviations: TN, total nitrogen; TC, total carbon; TOC, total organic carbon; TS, total sulfur; SS, soluble sulfur; H, total hydrogen.
General analyses of the Illumina-derived data set.
After filtering of the low-quality reads and chimeras and trimming of the adapters, as well as bar codes and primers, a total of 691,040 valid reads (average read length, 275 bp) were identified from 11 Illumina sequencing libraries. Nonparametric indicators of community diversity (Chao1 and Shannon indexes) revealed that the diversity of communities was not uniform throughout the watershed (see Table S2 in the supplemental material). The lightly and moderately contaminated sediments, SD1 and SD3, had the richest diversity, with operational taxonomic unit (OTU) numbers of 5,402 and 5,197, respectively, followed by the sediment sample from the Youyu River (SD4; OTU number of 5,130) and the upper layer sediment sample of the Aha Reservoir (SD5; OTU number of 5,109). In comparison, the two acidic sediment samples, B1 and B2, from an AMD-polluted area displayed considerably less richness, with OTU numbers of 4,059 and 2,951, respectively.
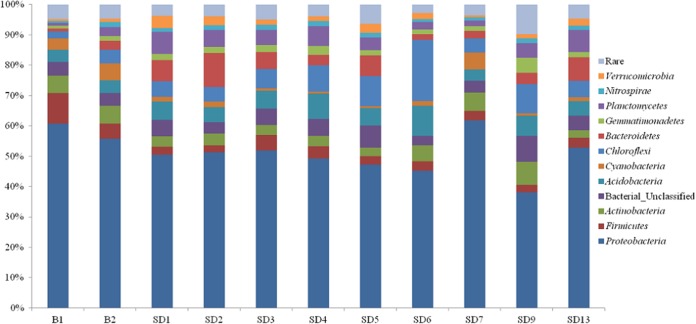
The most abundant prokaryotic organisms in all samples phylogenetically belonged to the Proteobacteria phylum, accounting for 38 to 62% of the total valid reads in all libraries (Fig. 2; see also Table S3 in the supplemental material). Within the Proteobacteria, Betaproteobacteria was the most abundant class in eight libraries, accounting for 9% to 35% of the total reads, while Gammaproteobacteria was the most abundant class in the libraries constructed from B2 (30%) and SD7 (35%). Frequently, pollution-related changes in the relative abundances of other phyla were observed. For example, Cyanobacteria and Firmicutes occurred at higher abundances in the libraries from Dapo Creek than in those from downstream samples. However, Chloroflexi showed relatively higher abundances in several libraries from the sediment samples taken from the Aha Reservoir but was less abundant in the upstream AMD-contaminated creek. Other phyla, such as Actinobacteria, Acidobacteria, Bacteroidetes, and Nitrospirae, that are most often encountered in AMD worldwide were detected in the libraries retrieved from all 11 samples. In addition to bacteria, two archaeal phyla were also detected. Crenarchaeota was detected in all samples, with relative abundances ranging from 0.02% to 2%. Euryarchaeota was detected in eight samples but not in sample B1, SD2, or SD6.
FIG 2.
Taxonomic classification of bacterial reads retrieved from different watershed samples at phylum level using the RDP classifier. “Rare” indicates phyla with relative abundances of less than 0.5%.
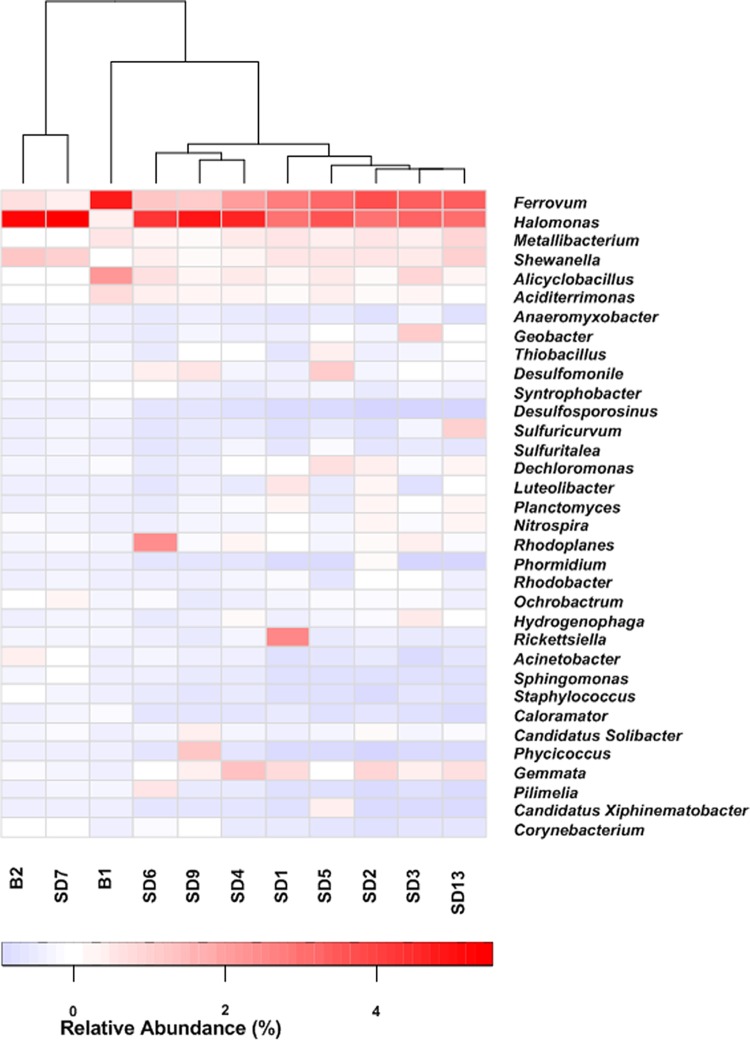
At the genus level, the microbial compositions also differed significantly across the four zones (Fig. 3; see also Table S4 in the supplemental material). Among the genera identified, Ferrovum showed relatively high abundance (>1%) in all the libraries, especially in the library retrieved from B1 (>13%). Halomonas was another genus with relative abundances over 2% in all the sequencing libraries. Other genera only demonstrated relatively high abundances in part of the samples. For instance, the sequences assigned to Shewanella were found at high abundances in libraries from B2 (4%) and SD7 (4%). The sequences related to Alicyclobacillus displayed high relative abundance in B1 (7%) but an abundance lower than 2% in all the other libraries. Detailed information about the relative abundances of dominant genera in each sample is provided in the heat map (Fig. 3) and Table S4.
FIG 3.
Heat map analysis of the distribution of dominant phylotypes in the 11 samples. The double hierarchical dendrogram shows the microbial distribution of the 11 samples. The relative percentages for the microbial genera are depicted by the color intensity; the color key is at the bottom.
Differences between 11 microbial communities were demonstrated from the 16S rRNA amplicon data. The UPGMA tree (Fig. 4) indicated that sequencing libraries from five river sediment samples (SD1 to SD4 and SD13) and one upper layer sediment of Aha Reservoir (SD5) were clustered. However, the microbial community in sample B1 was distinct from those in the downstream rivers and reservoir. The general taxonomic patterns in Fig. 4 were mainly driven by differences in the abundances of major taxonomic groups. Some genera, such as Ferrovum, Halomonas, Alicyclobacillus, and Shewanella, were observed throughout the watershed, but the abundances of these bacteria were generally higher in the heavily AMD-contaminated area (Fig. 3). In contrast, some bacteria, including members of the genera Aciditerrimonas, Syntrophobacter, and Desulfosporosinus, accounted for relatively higher proportions in the libraries retrieved from heavily AMD-contaminated areas, suggesting that low pH may have stimulated these bacteria. In contrast, Desulfomonile, Hydrogenophaga, Geobacter, Sulfuricurvum, Dechloromonas, and Thiobacillus species appeared to be more abundant in less contaminated areas.
FIG 4.
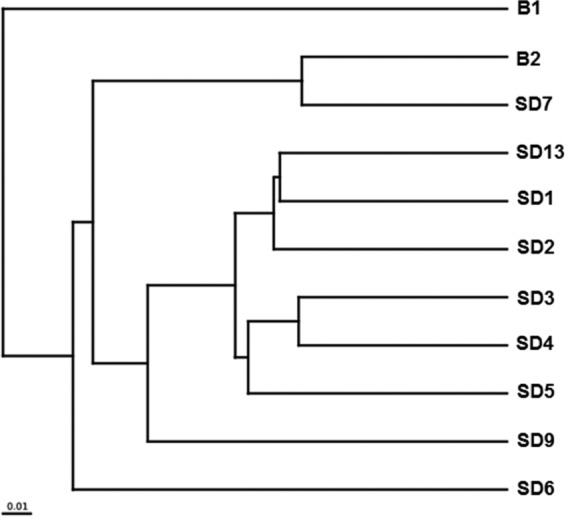
UPGMA tree showing clusters of microbial communities based on weighted UniFrac with 100% support at all nodes.
Relationship between microbial community and the environment.
The CCA that showed the relationship between environmental variables and microbial communities was conducted to examine correlations between genera with a relative abundance greater than 1% in at least one Illumina sequencing library and selected geochemical parameters (pH, temperature, Eh, SO42−, total Fe in each water sample, and concentrations of TC and TOC in each sediment sample). CCA ordination (Fig. 5) showed that specific geochemical conditions shaped the variations in microbial community compositions. CCA axis 1 and TC had a strong positive correlation. CCA axis 2 was positively correlated with pH and negatively correlated with temperature, Eh, sulfate, and total Fe. The relatively small magnitude of the TOC vector indicated that TOC was not as strongly correlated to community composition as other tested factors. Syntrophobacter, Aciditerrimonas, Ferrovum, Alicyclobacillus, and Desulfosporosinus were positively correlated with higher total aqueous Fe and sulfate concentrations and negatively correlated with pH, which may suggest that AMD environments may facilitate the growth of these bacteria. However, Geobacter and Desulfomonile were positively correlated with pH, indicating that these bacteria favor environments with higher pHs.
FIG 5.
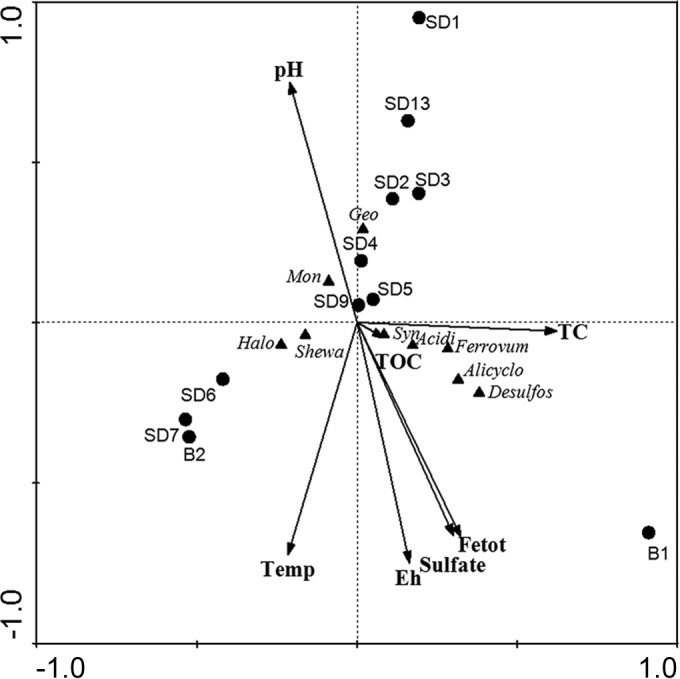
Ordination diagrams from canonical correspondence analysis (CCA) of bacterial abundances and geochemical parameters. Response variables of bacterial abundances are indicated by triangles. Arrows indicate the direction and magnitude of geochemical parameters associated with bacterial community structures. Each sample is represented by colored circles according to sampling zones. Abbreviations: Temp, temperature; Fetot, total Fe; TC, total carbon in the sediments; TOC, total organic carbon in the sediments. Bacterial abbreviations: Halo, Halomonas; Shewa, Shewanella; Mon, Desulfomonile; Geo, Geobacter; Syn, Syntrophobacter; Acidi, Aciditerrimonas; Alicyclo, Alicyclobacillus; Desulfos, Desulfosporosinus.
DISCUSSION
Spatial variations in physiochemical data.
Based on physiochemical analyses, the sites studied in the present work were grouped within three major conditions, each with a varied extent of AMD contamination. Dapo Creek exhibited the greatest extent of AMD pollution, while the downstream Youyu River was moderately polluted. The Aha Reservoir and Xiaoche River were identified as lightly AMD-contaminated areas. In previous research, environmental gradients have been observed in other AMD-contaminated watersheds, such as the Amous River region in southern France (16), the Odiel River basin in southwest Spain (17), the abandoned Coval da Mó mine in Portugal (18), and the Molonglo River in Australia (19). Similar to these AMD-contaminated environments, the Aha watershed exhibited a transition in physicochemical water quality from the upstream Dapo Creek [low in pH but dominated with Fe(II)] to the downstream rivers and reservoir (moderate-pH and low-Fe environments).
Geochemical analyses of different iron and sulfur species along the Aha watershed suggested dynamic Fe- and S-related geochemical processes. The rapid oxidation of Fe(II) and subsequent precipitation of Fe(III) at neutral pH removed a large amount of dissolved Fe from the water body. However, precipitated Fe compounds accumulated in sediments, as demonstrated by the relatively high Fe2O3 concentrations in downstream sediments. Fe2O3 may have been utilized by microorganisms as the electron acceptor and therefore contributed to biogeochemical Fe cycling in the watershed habitats. Similar to decreasing Fe(II) concentrations along the watershed, sulfate concentrations were also lower in the downstream water body. This may have been due to dilution from uncontaminated river water.
Environmental data that shape the microbial community composition.
Our analyses revealed that microbial communities were patterned along the AMD pollution gradients. Statistical analyses suggested that microbial diversity in the heavily polluted upstream area was distinct from that in the downstream rivers and reservoir. Specifically, samples (e.g., B1 and B2) from the heavily AMD-polluted zone had a lower predicted microbial diversity than the less contaminated areas, which could be attributed to the harsh contaminated environments in zone 1. The environmental conditions, such as low pH and high Fe and sulfate concentrations, acted as environmental stressor for microbial adaptation. CCA was performed to discern how microbial communities adapt to AMD-contaminated environments. Total Fe and sulfate were the major environmental factors that affected the microbial community, as indicated by CCA (Fig. 5). With respect to AMD-contaminated environments, iron and sulfur compounds might have influenced the overall microbial communities primarily by controlling the distribution of Fe- and S-metabolizing microorganisms. This agrees with the observation that Fe-metabolizing bacteria were more frequently detected in libraries from samples in the AMD-contaminated creek. Meanwhile, sulfate-reducing bacteria (SRB) within the orders Desulfobacterales, Desulfuromonadales, Syntrophobacterales, Clostridiales, and Desulfovibrionales, which exhibited increased relative abundances in downstream rivers, may contribute to decreases in sulfate concentration in downstream rivers and reservoirs.
CCA indicated a significant relationship between overall community composition and pH. The strong selective pressure with extremely acidic conditions in zone 1 determined which microorganisms could survive. In downstream zones where pH was moderate, however, increased amounts of neutrophiles were present and therefore changed the overall microbial community. This observation is consistent with the previously published studies on AMD. For example, Kuang et al. (20) recently explored the phylogenetic differentiation among 59 microbial communities from different AMD sites across southeast China, finding that microbial communities were largely correlated with pH. In another study, pH was correlated with overall microbial communities in the Rio Tinto, which is an extremely acidic, Fe-rich location (21). Microbial communities were also significantly altered by pH in other Fe-S-rich environments, such as in black shale weathering profiles (22) and rice paddy soils which were irrigated using AMD-contaminated water (23). It is suggested that pH influences microbial survival directly and/or through control of ancillary environmental parameters that are closely related to soil pH, such as nutrient availability and cationic metal solubility (24).
There was also a significant and strong correlation between overall microbial community and temperature. Temperature has been previously reported as an important factor in shaping microbial community composition in AMD (16, 25). For example, Volant et al. used CCA and found that temperature was one of the primary environmental factors that shape the composition of bacterial communities along an acid drainage stream (16). Similarly, in the present study, temperature was strongly and significantly linked to bacterial community variance, especially to Halomonas and Shewanella populations (Fig. 5).
Biogeochemical element cycling in the watershed.
Although microorganisms have not been isolated in this study, it may be possible to predict their metabolic capabilities based on those of phylogenetically related microorganisms, since closely related microorganisms tend to have similar physiological traits (26). The majority of the most abundant genera identified in this study are known to be involved in Fe and S cycling, indicating dynamic biogeochemical Fe and S cycling in the watershed as reported previously (5, 27).
Biogeochemical Fe cycling.
Ferrovum and Alicyclobacillus were the commonly detected acidophilic Fe-oxidizing bacteria (FeOB) in AMD (5). Ferrovum spp. are aerobic, acidophilic, and autotrophic FeOB (28) and have been shown to be the predominant organisms in many AMD-impacted systems (20). Previous research reported that “Ferrovum myxofaciens” is known to produce copious amounts of extracellular polymeric substances (EPS), which are the major components of “acid streamers” (28). This agrees with our finding that Ferrovum was in highest relative abundance in the library retrieved from site B1 (acid streamers as shown in Fig. S1 in the supplemental material). Alicyclobacillus also demonstrated high abundances in many libraries. Most described Alicyclobacillus species are acidophilic, soil-dwelling Firmicutes. Isolates of these bacteria grow at a pH range of 2.0 to 6.0 (29). Prior studies that isolated these bacteria from mining areas showed that they were able to oxidize ferrous iron and reduced sulfur (30–32). In addition, many Alicyclobacillus species are able to grow using organic carbon sources (33, 34) and could grow in oxygen-free media using ferric iron as a terminal electron acceptor (35). Halomonas was abundant in all 11 libraries, with the highest relative abundances in libraries from Dapo Creek. Previous research has isolated a novel chemolithoautotrophic FeOB closely related to Halomonas from low-temperature weathering habitats in deep-sea hydrothermal areas (36, 37). Interestingly, recent studies have detected Halomonas-related bacteria in AMD environments (38, 39). However, the role of Halomonas species in AMD environments requires further investigation.
Acidithiobacillus, Thiobacillus, and Leptospirillum have frequently been detected in AMD environments (4, 40), but these FeOB showed relatively low abundances in the current study (see Table S4 in the supplemental material). Leptospirillum populations are often reported to be dominated in low-pH AMD environments (e.g., pH < 2) (41, 42) but are minor community members or absent in AMD with pHs above 2.5 (43, 44). The pH in Dapo Creek (pH > 2.5) may have been too high to facilitate the proliferation of Leptospirillum. Acidithiobacillus and Thiobacillus are reported as the dominant FeOB in AMD with pH values similar to those in the current study (3, 45). It is possible that other environmental factors may have affected the abundance of Acidithiobacillus and Thiobacillus. Gallionella is an FeOB commonly detected in neutral environments (46–48); however, it exhibited very low abundance in neutral downstream samples from this study. Other FeOB, including Aciditerrimonas, Thermomonas, and Aquabacterium, were also detected at lower abundances in the current study.
A wide variety of known Fe-reducing bacteria (FeRB) were detected in the watershed, especially in Dapo Creek. Shewanella-related sequences demonstrated relatively high abundances in libraries retrieved from most samples, especially in B2 and SD7. Shewanella species in the Gammaproteobacteria are well known for their capability of Fe(III) reduction. Shewanella species have been frequently isolated from marine environments (49–51). Our study suggested that Shewanella species were able to thrive, and may even play an important functional role, in Fe(III) reduction in AMD-impacted settings. The high abundances of Shewanella species in all sediment samples indicated that Shewanella were present in a wide range of pHs. Metallibacterium-related sequences were frequently detected in libraries derived from Dapo Creek and some downstream samples. The genus Metallibacterium is comprised of acidophilic microorganisms capable of heterotrophic or lithotrophic Fe(III) reduction (52, 53).In an increasing number of recent studies, Metallibacterium populations have been detected in high-Fe environments (53, 54). These observations may be attributed to their metabolic capability in iron reduction. Aciditerrimonas is another potential FeRB showing high abundances in the libraries retrieved from Dapo Creek, especially in sample B1. Aciditerrimonas contains thermophilic Fe(III)-reducing bacteria (55). Our current knowledge on this genus remains limited, especially concerning AMD-contaminated environments. Another important FeRB, Geobacter, was present in low abundances in libraries from the AMD creek but in higher abundances in libraries from the Youyu River, which may suggest that acidic environments may not favor this important FeRB.
Biogeochemical S cycling.
Sulfate reduction has a central role in AMD bioremediation and leads to the consumption of protons, increases the pH, and then generates sulfide as the reaction products to precipitate heavy metals (27). In contrast to the case with dissimilatory iron reduction, few acidophilic bacteria and archaea that can reduce sulfate have been described. The optimum growth conditions for SRB were found at pHs ranging from 5.0 to 9.0 (56, 57). In the present study, the low pH and high redox potentials in Dapo Creek may not have allowed for a beneficial habitat for SRB. However, many SRB were detected in acidic sediments in this study. Syntrophobacter and Desulfosporosinus, two genera containing many species involved in sulfate reduction (58–61), showed high relative abundances in libraries from Dapo Creek. Both Syntrophobacter and Desulfosporosinus have regularly been detected in AMD waters (62–65). It is noteworthy that Desulfosporosinus species are able to reduce various metals, such as Fe(III), Mn(IV), and U(VI), in acidic solutions (pH 4.2), indicating a potential role of Desulfosporosinus species for biological treatment of AMD (66). The presence of various SRB indicated that SRB may have been able to cope with extremely harsh environments and survive under acidic conditions.
Biogeochemical C cycling.
It has been reported that most subsurface AMD sites receive minimum carbon input from external sources (5). However, sediment B1 demonstrated a high proportion of TC and TOC content. Consistent with high concentrations of TC in AMD-contaminated creeks, Comamonadaceae, which are metabolically versatile and can use a wide range of organic compounds as carbon sources (60, 67), were one of the most dominant families in Dapo Creek. A broad range of other heterotrophic bacteria were also detected in zone 1, indicating dynamic C cycling in acidic AMD environments.
Interaction among microorganisms with various physiological traits may be critical in maintaining the metabolic diversity in AMD environments. For example, acidophilic autotrophs (e.g., Ferrovum) are able to produce organic matter which might later be used by heterotrophic organisms. The utilization of organic matter, which is toxic to autotrophs, could conversely benefit the growth of autotrophs (5).
Conclusion.
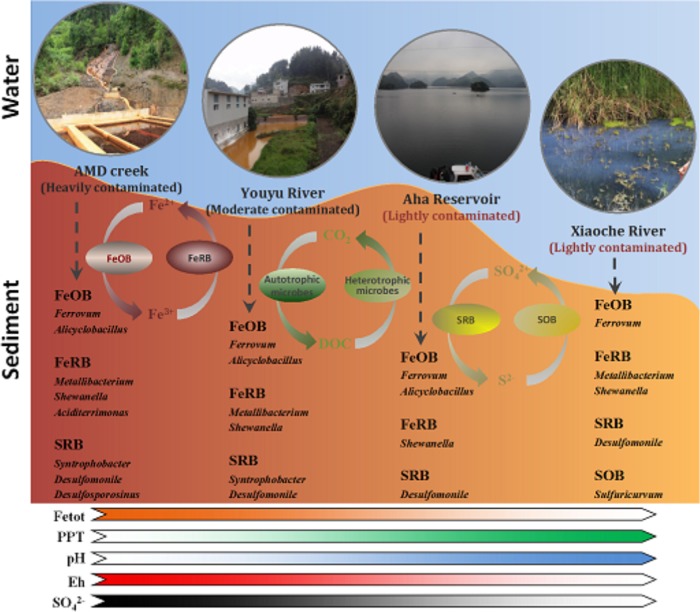
In the present study, we aimed to more accurately characterize microbial communities in an AMD-impacted watershed by high-throughput sequencing. Our results indicated that a number of Fe- and S-metabolizing bacteria exhibited high abundances in the AMD-contaminated watershed. Furthermore, dominant functional groups varied along the AMD pollution gradients (Fig. 6). The spatial distribution of microbial communities in AMD pollution gradients facilitates the understanding of the metabolic capacities of indigenous microorganisms. Such knowledge may lead to the development of approaches for remediation of AMD contamination. It has been reported that bioreactor systems using acidophilic and sulfidogenic bacteria that are indigenous to mine-impacted environments could remove iron and precipitate other metals from mine waters (68, 69). These studies suggested that iron was removed by microbial iron oxidation and stable metal sulfides were selectively precipitated by acidophilic SRB (68, 69). Therefore, the phylogenetically divergent lineages coexisting in the Aha watershed may have the potential for in situ natural attenuation of AMD pollution. Future work should explore the significance of these phylotypes in natural attenuation of AMD at this and other AMD sites.
FIG 6.
Geomicrobiological model of the Aha watershed profile. The model shows the roles of the major S- and Fe-related bacteria identified in the current study. The color of the arrows indicates the extent of the physiochemical parameters or geochemical process. A transition from saturated to transparent colors in a color ribbon indicates a decrease in the corresponding environmental factor from zone 1 to zone 4 and vice versa. Abbreviations: FeOB, Fe(II)-oxidizing bacteria; FeRB, Fe(III)-reducing bacteria; SOB, sulfur-oxidizing bacteria; SRB, sulfate-reducing bacteria; Fetot: total Fe; PPT, precipitation.
Supplementary Material
ACKNOWLEDGMENTS
This research was funded by the National Natural Science Foundation of China (41173028) and the Opening Fund of the State Key Laboratory of Environmental Geochemistry (SKLEG2013810).
We thank Ying Huang for her suggestion of and help with CCA analysis. We thank Valdis Krumins, Kaylyn Germ, Caitlin N. Ryan, and Lora M. McGuinness for thoughtful reviews of the manuscript and discussion of the topics therein.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00935-15.
REFERENCES
- 1.Dudka S, Adriano DC. 1997. Environmental impacts of metal ore mining and processing: a review. J Environ Qual 26:590–602. [Google Scholar]
- 2.Byrne P, Wood P, Reid I. 2012. The impairment of river systems by metal mine contamination: a review including remediation options. Crit Rev Environ Sci Technol 42:2017–2077. doi: 10.1080/10643389.2011.574103. [DOI] [Google Scholar]
- 3.Johnson DB. 2012. Geomicrobiology of extremely acidic subsurface environments. FEMS Microbiol Ecol 81:2–12. doi: 10.1111/j.1574-6941.2011.01293.x. [DOI] [PubMed] [Google Scholar]
- 4.Hallberg K. 2010. New perspectives in acid mine drainage microbiology. Hydrometallurgy 104:448–453. doi: 10.1016/j.hydromet.2009.12.013. [DOI] [Google Scholar]
- 5.Baker BJ, Banfield JF. 2003. Microbial communities in acid mine drainage. FEMS Microbiol Ecol 44:139–152. doi: 10.1016/S0168-6496(03)00028-X. [DOI] [PubMed] [Google Scholar]
- 6.Rice EW, Baird RB, Eaton AD, Clesceri LS (ed). 2012. Standard methods for the examination of water and wastewater, 22nd ed American Public Health Association, Washington, DC. [Google Scholar]
- 7.Tamura H, Goto K, Yotsuyanagi T, Nagayama M. 1974. Spectrophotometric determination of iron(II) with 1,10-phenanthroline in the presence of large amounts of iron(III). Talanta 21:314–318. doi: 10.1016/0039-9140(74)80012-3. [DOI] [PubMed] [Google Scholar]
- 8.Lundquist P, Mårtensson J, Sörbo B, Ohman S. 1980. Turbidimetry of inorganic sulfate, ester sulfate, and total sulfur in urine. Clin Chem 26:1178–1181. [PubMed] [Google Scholar]
- 9.Tabatabai M, Dick W. 1983. Simultaneous determination of nitrate, chloride, sulfate, and phosphate in natural waters by ion chromatography. J Environ Qual 12:209–213. [Google Scholar]
- 10.Schumacher BA. 2002. Methods for the determination of total organic carbon (TOC) in soils and sediments. Ecological Risk Assessment Support Center, US Environmental Protection Agency, Las Vegas, NV. [Google Scholar]
- 11.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuczynski J, Stombaugh J, Walters WA, González A, Caporaso JG, Knight R. 2012. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Microbiol 27:1E.5.1–1E.5.20. doi: 10.1002/9780471729259.mc01e05s27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Volant A, Bruneel O, Desoeuvre A, Héry M, Casiot C, Bru N, Delpoux S, Fahy A, Javerliat F, Bouchez O. 2014. Diversity and spatiotemporal dynamics of bacterial communities: physicochemical and other drivers along an acid mine drainage. FEMS Microbiol Ecol 90:247–263. doi: 10.1111/1574-6941.12394. [DOI] [PubMed] [Google Scholar]
- 17.Sarmiento AM, Nieto JM, Olías M, Cánovas CR. 2009. Hydrochemical characteristics and seasonal influence on the pollution by acid mine drainage in the Odiel river Basin (SW Spain). Appl Geochem 24:697–714. doi: 10.1016/j.apgeochem.2008.12.025. [DOI] [Google Scholar]
- 18.Ferreira da Silva E, Almeida SF, Nunes ML, Luís AT, Borg F, Hedlund M, de Sá CM, Patinha C, Teixeira P. 2009. Heavy metal pollution downstream the abandoned Coval da Mó mine (Portugal) and associated effects on epilithic diatom communities. Sci Total Environ 407:5620–5636. doi: 10.1016/j.scitotenv.2009.06.047. [DOI] [PubMed] [Google Scholar]
- 19.Sloane P, Norris R. 2003. Relationship of AUSRIVAS-based macroinvertebrate predictive model outputs to a metal pollution gradient. J North Am Benthol Soc 22:457–471. doi: 10.2307/1468274. [DOI] [Google Scholar]
- 20.Kuang J-L, Huang L-N, Chen L-X, Hua Z-S, Li S-J, Hu M, Li J-T, Shu W-S. 2013. Contemporary environmental variation determines microbial diversity patterns in acid mine drainage. ISME J 7:1038–1050. doi: 10.1038/ismej.2012.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amaral-Zettler LA, Zettler ER, Theroux SM, Palacios C, Aguilera A, Amils R. 2011. Microbial community structure across the tree of life in the extreme Rio Tinto. ISME J 5:42–50. doi: 10.1038/ismej.2010.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, Sun W, Wang S, Sun Z, Lin S, Peng X. 2014. Bacteria diversity, distribution and insight into their role in S and Fe biogeochemical cycling during black shale weathering. Environ Microbiol 16:3533–3547. doi: 10.1111/1462-2920.12536. [DOI] [PubMed] [Google Scholar]
- 23.Sun M, Xiao T, Ning Z, Xiao E, Sun W. 2015. Microbial community analysis in rice paddy soils irrigated by acid mine drainage contaminated water. Appl Microbiol Biotechnol 99:2911–2922. doi: 10.1007/s00253-014-6194-5. [DOI] [PubMed] [Google Scholar]
- 24.Brady NC, Weil RR. 1996. The nature and properties of soils, 11th ed. Prentice-Hall, Upper Saddle River, NJ. [Google Scholar]
- 25.Edwards KJ, Gihring TM, Banfield JF. 1999. Seasonal variations in microbial populations and environmental conditions in an extreme acid mine drainage environment. Appl Environ Microbiol 65:3627–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bond PL, Smriga SP, Banfield JF. 2000. Phylogeny of microorganisms populating a thick, subaerial, predominantly lithotrophic biofilm at an extreme acid mine drainage site. Appl Environ Microbiol 66:3842–3849. doi: 10.1128/AEM.66.9.3842-3849.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson DB, Hallberg KB. 2005. Acid mine drainage remediation options: a review. Sci Total Environ 338:3–14. doi: 10.1016/j.scitotenv.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 28.Johnson DB, Hallberg KB, Hedrich S. 2014. Uncovering a microbial enigma: isolation and characterization of the streamer-generating, iron-oxidizing, acidophilic bacterium “Ferrovum myxofaciens.” Appl Environ Microbiol 80:672–680. doi: 10.1128/AEM.03230-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang S-S, Kang D-H. 2004. Alicyclobacillus spp. in the fruit juice industry: history, characteristics, and current isolation/detection procedures. Crit Rev Microbiol 30:55–74. doi: 10.1080/10408410490435089. [DOI] [PubMed] [Google Scholar]
- 30.Guo X, You X-Y, Liu L-J, Zhang J-Y, Liu S-J, Jiang C-Y. 2009. Alicyclobacillus aeris sp. nov., a novel ferrous-and sulfur-oxidizing bacterium isolated from a copper mine. Int J Syst Evol Microbiol 59:2415–2420. doi: 10.1099/ijs.0.008870-0. [DOI] [PubMed] [Google Scholar]
- 31.Kinnunen P-M, Robertson W, Plumb J, Gibson J, Nichols P, Franzmann P, Puhakka J. 2003. The isolation and use of iron-oxidizing, moderately thermophilic acidophiles from the Collie coal mine for the generation of ferric iron leaching solution. Appl Microbiol Biotechnol 60:748–753. doi: 10.1007/s00253-002-1185-3. [DOI] [PubMed] [Google Scholar]
- 32.Joe S-J, Suto K, Inoie C, Chida T. 2007. Isolation and characterization of acidophilic heterotrophic iron-oxidizing bacterium from enrichment culture obtained from acid mine drainage treatment plant. J Biosci Bioeng 104:117–123. doi: 10.1263/jbb.104.117. [DOI] [PubMed] [Google Scholar]
- 33.Goto K, Mochida K, Asahara M, Suzuki M, Kasai H, Yokota A. 2003. Alicyclobacillus pomorum sp. nov., a novel thermo-acidophilic, endospore-forming bacterium that does not possess ω-alicyclic fatty acids, and emended description of the genus Alicyclobacillus. Int J Syst Evol Microbiol 53:1537–1544. doi: 10.1099/ijs.0.02546-0. [DOI] [PubMed] [Google Scholar]
- 34.Goto K, Matsubara H, Mochida K, Matsumura T, Hara Y, Niwa M, Yamasato K. 2002. Alicyclobacillus herbarius sp. nov., a novel bacterium containing omega-cycloheptane fatty acids, isolated from herbal tea. Int J Syst Evol Microbiol 52:109–113. [DOI] [PubMed] [Google Scholar]
- 35.Yahya A, Hallberg KB, Johnson DB. 2008. Iron and carbon metabolism by a mineral-oxidizing Alicyclobacillus-like bacterium. Arch Microbiol 189:305–312. doi: 10.1007/s00203-007-0319-5. [DOI] [PubMed] [Google Scholar]
- 36.Edwards KJ, Rogers DR, Wirsen CO, McCollom TM. 2003. Isolation and characterization of novel psychrophilic, neutrophilic, Fe-oxidizing, chemolithoautotrophic α- and γ-Proteobacteria from the deep sea. Appl Environ Microbiol 69:2906–2913. doi: 10.1128/AEM.69.5.2906-2913.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edwards KJ, Bach W, McCollom TM, Rogers DR. 2004. Neutrophilic iron-oxidizing bacteria in the ocean: their habitats, diversity, and roles in mineral deposition, rock alteration, and biomass production in the deep-sea. Geomicrobiol J 21:393–404. doi: 10.1080/01490450490485863. [DOI] [Google Scholar]
- 38.Auld RR, Myre M, Mykytczuk N, Leduc LG, Merritt TJ. 2013. Characterization of the microbial acid mine drainage microbial community using culturing and direct sequencing techniques. J Microbiol Methods 93:108–115. doi: 10.1016/j.mimet.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 39.Santofimia E, González-Toril E, López-Pamo E, Gomariz M, Amils R, Aguilera Á. 2013. Microbial diversity and its relationship to physicochemical characteristics of the water in two extreme acidic pit lakes from the Iberian Pyrite Belt (SW Spain). PLoS One 8:e66746. doi: 10.1371/journal.pone.0066746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson DB, Hallberg KB. 2003. The microbiology of acidic mine waters. Res Microbiol 154:466–473. doi: 10.1016/S0923-2508(03)00114-1. [DOI] [PubMed] [Google Scholar]
- 41.Bond PL, Druschel GK, Banfield JF. 2000. Comparison of acid mine drainage microbial communities in physically and geochemically distinct ecosystems. Appl Environ Microbiol 66:4962–4971. doi: 10.1128/AEM.66.11.4962-4971.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.García-Moyano A, González-Toril E, Moreno-Paz M, Parro V, Amils R. 2008. Evaluation of Leptospirillum spp. in the Río Tinto, a model of interest to biohydrometallurgy. Hydrometallurgy 94:155–161. doi: 10.1016/j.hydromet.2008.05.046. [DOI] [Google Scholar]
- 43.Tan GL, Shu WS, Zhou WH, Li XL, Lan CY, Huang LN. 2009. Seasonal and spatial variations in microbial community structure and diversity in the acid stream draining across an ongoing surface mining site. FEMS Microbiol Ecol 70:277–285. doi: 10.1111/j.1574-6941.2009.00744.x. [DOI] [PubMed] [Google Scholar]
- 44.Brown JF, Jones DS, Mills DB, Macalady JL, Burgos WD. 2011. Application of a depositional facies model to an acid mine drainage site. Appl Environ Microbiol 77:545–554. doi: 10.1128/AEM.01550-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bruneel O, Duran R, Casiot C, Elbaz-Poulichet F, Personné J-C. 2006. Diversity of microorganisms in Fe-As-rich acid mine drainage waters of Carnoules, France. Appl Environ Microbiol 72:551–556. doi: 10.1128/AEM.72.1.551-556.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haaijer SC, Harhangi HR, Meijerink BB, Strous M, Pol A, Smolders AJ, Verwegen K, Jetten MS, den Camp HJO. 2008. Bacteria associated with iron seeps in a sulfur-rich, neutral pH, freshwater ecosystem. ISME J 2:1231–1242. doi: 10.1038/ismej.2008.75. [DOI] [PubMed] [Google Scholar]
- 47.Emerson D, Moyer C. 1997. Isolation and characterization of novel iron-oxidizing bacteria that grow at circumneutral pH. Appl Environ Microbiol 63:4784–4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiss JV, Rentz JA, Plaia T, Neubauer SC, Merrill-Floyd M, Lilburn T, Bradburne C, Megonigal JP, Emerson D. 2007. Characterization of neutrophilic Fe(II)-oxidizing bacteria isolated from the rhizosphere of wetland plants and description of Ferritrophicum radicicola gen. nov. sp. nov., and Sideroxydans paludicola sp. nov. Geomicrobiol J 24:559–570. doi: 10.1080/01490450701670152. [DOI] [Google Scholar]
- 49.Caccavo F, Blakemore RP, Lovley DR. 1992. A hydrogen-oxidizing, Fe(III)-reducing microorganism from the Great Bay Estuary, New Hampshire. Appl Environ Microbiol 58:3211–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bozal N, Montes MJ, Tudela E, Jiménez F, Guinea J. 2002. Shewanella frigidimarina and Shewanella livingstonensis sp. nov. isolated from Antarctic coastal areas. Int J Syst Evol Microbiol 52:195–205. [DOI] [PubMed] [Google Scholar]
- 51.Brettar I, Christen R, Höfle MG. 2002. Shewanella denitrificans sp. nov., a vigorously denitrifying bacterium isolated from the oxic-anoxic interface of the Gotland Deep in the central Baltic Sea. Int J Syst Evol Microbiol 52:2211–2217. doi: 10.1099/ijs.0.02255-0. [DOI] [PubMed] [Google Scholar]
- 52.Ziegler S, Waidner B, Itoh T, Schumann P, Spring S, Gescher J. 2013. Metallibacterium scheffleri gen. nov., sp. nov., an alkalinizing gammaproteobacterium isolated from an acidic biofilm. Int J Syst Evol Microbiol 63:1499–1504. doi: 10.1099/ijs.0.042986-0. [DOI] [PubMed] [Google Scholar]
- 53.Brantner JS, Senko JM. 2014. Response of soil-associated microbial communities to intrusion of coal mine-derived acid mine drainage. Environ Sci Technol 48:8556–8563. doi: 10.1021/es502261u. [DOI] [PubMed] [Google Scholar]
- 54.Brantner JS, Haake ZJ, Burwick JE, Menge CM, Hotchkiss ST, Senko JM. 14 May 2014. Depth-dependent geochemical and microbiological gradients in Fe(III) deposits resulting from coal mine-derived acid mine drainage. Front Microbiol http://dx.doi.org/10.3389/fmicb.2014.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Itoh T, Yamanoi K, Kudo T, Ohkuma M, Takashina T. 2011. Aciditerrimonas ferrireducens gen. nov., sp. nov., an iron-reducing thermoacidophilic actinobacterium isolated from a solfataric field. Int J Syst Evol Microbiol 61:1281–1285. doi: 10.1099/ijs.0.023044-0. [DOI] [PubMed] [Google Scholar]
- 56.Grossman JP, Postgate J. 1953. The estimation of sulphate-reducing bacteria (D. desulphuricans). J Appl Microbiol 16:1–9. [Google Scholar]
- 57.Widdel F, Bak F. 1992. Gram-negative mesophilic sulfate-reducing bacteria, p 3352–3378. In Balows A, Tr̈uper HG, Dworkin M, Harder W, Schleifer K-H (ed), The prokaryotes. Springer, New York, NY. [Google Scholar]
- 58.Harmsen HJ, Van Kuijk BL, Plugge CM, Akkermans AD, De Vos WM, Stams AJ. 1998. Syntrophobacter fumaroxidans sp. nov., a syntrophic propionate-degrading sulfate-reducing bacterium. Int J Syst Bacteriol 48:1383–1387. doi: 10.1099/00207713-48-4-1383. [DOI] [PubMed] [Google Scholar]
- 59.Wallrabenstein C, Hauschild E, Schink B. 1995. Syntrophobacter pfennigii sp. nov., new syntrophically propionate-oxidizing anaerobe growing in pure culture with propionate and sulfate. Arch Microbiol 164:346–352. doi: 10.1007/BF02529981. [DOI] [Google Scholar]
- 60.Sun W, Cupples AM. 2012. Diversity of five anaerobic toluene-degrading microbial communities investigated using stable isotope probing. Appl Environ Microbiol 78:972–980. doi: 10.1128/AEM.06770-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robertson W, Bowman J, Franzmann P, Mee B. 2001. Desulfosporosinus meridiei sp. nov., a spore-forming sulfate-reducing bacterium isolated from gasolene-contaminated groundwater. Int J Syst Evol Microbiol 51:133–140. [DOI] [PubMed] [Google Scholar]
- 62.González-Toril E, Aguilera Á, Souza-Egipsy V, Pamo EL, España JS, Amils R. 2011. Geomicrobiology of La Zarza-Perrunal acid mine effluent (Iberian Pyritic Belt, Spain). Appl Environ Microbiol 77:2685–2694. doi: 10.1128/AEM.02459-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sánchez-Andrea I, Knittel K, Amann R, Amils R, Sanz JL. 2012. Quantification of Tinto River sediment microbial communities: importance of sulfate-reducing bacteria and their role in attenuating acid mine drainage. Appl Environ Microbiol 78:4638–4645. doi: 10.1128/AEM.00848-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alazard D, Joseph M, Battaglia-Brunet F, Cayol J-L, Ollivier B. 2010. Desulfosporosinus acidiphilus sp. nov.: a moderately acidophilic sulfate-reducing bacterium isolated from acid mining drainage sediments. Extremophiles 14:305–312. doi: 10.1007/s00792-010-0309-4. [DOI] [PubMed] [Google Scholar]
- 65.Sánchez-Andrea I, Stams AJ, Hedrich S, Ňancucheo I, Johnson DB. 2015. Desulfosporosinus acididurans sp. nov.: an acidophilic sulfate-reducing bacterium isolated from acidic sediments. Extremophiles 19:39–47. doi: 10.1007/s00792-014-0701-6. [DOI] [PubMed] [Google Scholar]
- 66.Senko JM, Zhang G, McDonough JT, Bruns MA, Burgos WD. 2009. Metal reduction at low pH by a Desulfosporosinus species: implications for the biological treatment of acidic mine drainage. Geomicrobiol J 26:71–82. doi: 10.1080/01490450802660193. [DOI] [Google Scholar]
- 67.Viñas M, Sabaté J, Espuny MJ, Solanas AM. 2005. Bacterial community dynamics and polycyclic aromatic hydrocarbon degradation during bioremediation of heavily creosote-contaminated soil. Appl Environ Microbiol 71:7008–7018. doi: 10.1128/AEM.71.11.7008-7018.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hedrich S, Johnson DB. 2014. Remediation and selective recovery of metals from acidic mine waters using novel modular bioreactors. Environ Sci Technol 48:12206–12212. doi: 10.1021/es5030367. [DOI] [PubMed] [Google Scholar]
- 69.Ňancucheo I, Johnson DB. 2012. Selective removal of transition metals from acidic mine waters by novel consortia of acidophilic sulfidogenic bacteria. Microb Biotechnol 5:34–44. doi: 10.1111/j.1751-7915.2011.00285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.