Abstract
The quorum sensing (QS) system, as a well-functioning population-dependent gene switch, has been widely applied in many gene circuits in synthetic biology. In our work, an efficient cell density-controlled expression system (QS) was established via engineering of the Vibrio fischeri luxI-luxR quorum sensing system. In order to achieve in vivo programmed gene expression, a synthetic binary regulation circuit (araQS) was constructed by assembling multiple genetic components, including the quorum quenching protein AiiA and the arabinose promoter ParaBAD, into the QS system. In vitro expression assays verified that the araQS system was initiated only in the absence of arabinose in the medium at a high cell density. In vivo expression assays confirmed that the araQS system presented an in vivo-triggered and cell density-dependent expression pattern. Furthermore, the araQS system was demonstrated to function well in different bacteria, indicating a wide range of bacterial hosts for use. To explore its potential applications in vivo, the araQS system was used to control the production of a heterologous protective antigen in an attenuated Edwardsiella tarda strain, which successfully evoked efficient immune protection in a fish model. This work suggested that the araQS system could program bacterial expression in vivo and might have potential uses, including, but not limited to, bacterial vector vaccines.
INTRODUCTION
Bacteria release, detect, and respond to the accumulation of self-generated chemical signaling molecules called autoinducers (AIs), whose concentration correlates to the abundance of secreting microorganisms in the vicinity (1, 2). This process, termed quorum sensing (QS), allows bacteria to monitor a variety of processes, including competence, bioluminescence, virulence factor secretion, biofilm formation, and sporulation (3). A paradigm of quorum sensing regulation is the luminescence (lux) operon in Vibrio fischeri (4). The lux genes are organized in two divergently transcribed units (5). LuxI is the autoinducer synthase producing an acyl-homoserine lactone (AHL), N-(3-oxohexanoyl)-homoserine lactone (6). At low cell density, it is transcribed at a low basal level (7, 8). As the V. fischeri culture grows and the autoinducers accumulate to a specific threshold, cytoplasmic LuxR proteins form a complex with autoinducers and initiate a positive autoinduction circuit by binding to the luxICDABE promoter (9, 10). Numerous bacteria secrete enzymes that can degrade the QS signals of heterologous species. The approach of interfering with or destroying the QS signal is referred to as quorum quenching (11). The AiiA protein produced by Bacillus sp. is a typical AHL lactonase, which can quench quorum sensing systems by hydrolyzing lactone rings of AHL signal molecules (12). Therefore, introduction of the aiiA gene into quorum sensing circuits renders the QS system under fine negative control.
Bacterial QS signal synthases, receptors, and cognate promoter elements are important components of a wide variety of engineered biological devices (13). As a well-functioning and population-dependent gene switch, the QS system has been widely applied to many gene circuits in synthetic biology. Several synthetic biology studies have employed the entire lux regulatory module (14–16), which served as an oscillator (17), amplifier (18), and so on. For example, Basu et al. (15) constructed a synthetic multicellular system in which the engineered receiver cells were designed to form ringlike patterns by expressing reporter proteins of different colors according to AHL concentrations. You et al. (16) built and characterized a population control circuit that autonomously regulated the density of an Escherichia coli population by coupling QS with the synthesis of toxic proteins. Bulter et al. (19) developed an artificial QS system to establish a gene-metabolic network where bacterial cells exchanged acetate molecules that were produced in proportion to cell growth.
Although synthetic QS genetic circuits carried by prokaryotes performed fairly well in vitro with the addition of exogenous inducers to medium to switch the system on or off, how to achieve their in vivo application in eukaryote hosts is a new topic. Because the addition of inducers in vivo is infeasible, it is essential to design a gene switch to initiate the QS system by sensing a specific in vivo environmental signal. Guan et al. (20) established an in vivo inducible suicide circuit by introducing an iron-regulated promoter, which could activate the expression of a lysis gene in response to the iron-limiting signal in vivo and lyse the bacteria effectively. Besides low iron concentrations (21), the lack of arabinose is another typical in vivo signal. Kong et al. (22) introduced a plasmid carrying the arabinose-regulated genes murA and asdA into a Salmonella enterica serovar Typhimurium ΔmurA ΔasdA mutant, and the in vivo expression of murA and asdA was strictly turned off after the bacteria invaded host tissues.
The main purpose of our work was to achieve the self-initiation of the quorum sensing system only in an in vivo environment. It was essential to design a gene switch to initiate the QS system by sensing a specific signal in the in vivo environment. We constructed an in vivo-activated expression system called araQS by coupling an artificial QS expression module and an arabinose-regulated QS quenching control module. The expression module ensured the efficient production of the target protein by the quorum sensing system at high cell densities. The control module was composed of the arabinose-activated promoter ParaBAD and the quorum quenching gene aiiA. The quorum quenching protein AiiA effectively stopped the function of the quorum sensing system by degrading autoinducers in the presence of arabinose. The expression of AiiA was controlled by the ParaBAD promoter, which could sense arabinose in the environment. Thus, when the bacterial host harboring the araQS system was cultivated in an arabinose-rich in vitro environment, the expression of the AiiA protein inhibited the QS system and cut off the whole expression module. However, in an arabinose-free environment, such as in vivo, the absence of the AiiA protein derepressed the QS system, and the target protein was expressed effectively. Subsequent expression assays, both in vitro and in vivo, demonstrated that the araQS system was well regulated by the signals of arabinose and cell density and has great potential for in vivo applications, such as serving in multivalent bacterial vector vaccines.
MATERIALS AND METHODS
Strains and medium.
Strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown at 37°C in Luria-Bertani (LB) medium (1% tryptone, 0.5% yeast extract, 1% NaCl). Aeromonas hydrophila, Vibrio anguillarum, Salmonella Typhimurium, Staphylococcus aureus, and Edwardsiella tarda strains were grown at 30°C in LB medium. Strains were grown in 250-ml flasks with a 50-ml working volume in a shaker at 200 rpm. When required, ampicillin (100 μg/ml) or kanamycin sulfate (100 μg/ml) was added to the medium for plasmid selection, and arabinose (0.4 mg/ml) or the autoinducer (10 nM) was added for induction (23).
TABLE 1.
Strains and plasmids used in this study
| Bacterial strain or plasmid | Description | Source |
|---|---|---|
| Strains | ||
| E. coli BL21 | Expression host; F− ompT gal dcm lon hsdSB | Life Technologies |
| Vibrio fischeri MJ11 | Wild type; used for cloning of the luxR-luxI quorum sensing system | MCCCa |
| Vibrio anguillarum MVM425 | Wild type; fish pathogen, broad-range testing host | Our laboratory |
| E. tarda EIB202 | Wild type; fish pathogen, broad-range testing host | Our laboratory |
| E. tarda WED | Mutant disrupted in type III secretion system and chorismic acid synthesis; live attenuated vaccine | Our laboratory |
| S. aureus | Wild type; human pathogen, broad-range testing host | Our laboratory |
| S. Typhimurium | Wild type; human pathogen, broad-range testing host | Our laboratory |
| B. thuringiensis BT4Q7 | Gene source of the quorum quenching gene aiiA | Our laboratory |
| Plasmids | ||
| pET-28a | Expression vector; Kanr | Novagen |
| pUTat | Expression vector; Ampr | Our laboratory |
| QS1 | pUTat vector containing a Katushka reporter gene and an isolated PluxI promoter | This work |
| QS2 | QS1 plasmid inserted with intact V. fischeri QS gene elements | This work |
| QS3 | pUTat vector containing intact V. fischeri QS gene elements and a Katushka reporter gene cotranscribed with luxI | This work |
| araQS | QS3 plasmid with insertion of a ParaBAD-aiiA gene module | This work |
| araQS-G | araQS derivative in which the reporter gene was replaced by the gapA34 gene | This work |
MCCC, Marine Culture Collection of China.
Plasmid construction.
General DNA operations were conducted according to standard protocols. Seamless cloning operations were carried out by using a ClonExpress II one-step cloning kit (Vazyme Co. Ltd., China) according to standard procedures. DNA sequencing and primer synthesis (Table 2) were carried out by Generay Co. Ltd. (Shanghai, China).
TABLE 2.
Primers used in this study

Three cell density-dependent expression systems, QS1 to QS3, were designed on the basis of plasmid pUTat. The fundamental plasmid pUTat was constructed by replacing the lac promoter, the multiple-cloning site (MCS), and the replicon of pUC18 with a 506-bp sequence containing the MCS and the rrnBT1T2 terminator from plasmid pBV220 and a pAT153 replicon, respectively (24). Plasmid QS1 was constructed by inserting a luxI promoter fused with a Katushka reporter gene into pUTat. Plasmid QS2 was constructed by adding an intact luxI-luxR quorum sensing circuit into QS1. Plasmid QS3 was constructed by inserting a luxI-luxR quorum sensing circuit into pUTat, with a Katushka reporter gene fused in frame with luxI. Finally, a ParaBAD-aiiA control module was inserted into QS3 with a transcription terminator between the control module and quorum sensing module to form a binary QS system (araQS). Plasmid DNA was introduced into E. coli by heat shock and was introduced into other strains by electroporation.
Expression assay in vitro.
Bacterial cultures grown overnight were inoculated (1:1,000, vol/vol) into fresh LB medium with antibiotics, and inducers such as arabinose were preadded if needed. For fed-batch cultures, fresh LB medium with antibiotics (and arabinose or autoinducer if needed) was continuously added to the culture vessel to keep the optical density at 600 nm (OD600) lower than 0.3, below the cell density threshold for the activation of the established quorum sensing system. At different time intervals, each culture was sampled and centrifuged at 8,000 × g for 5 min, and the harvested cells were washed with phosphate-buffered saline (PBS) three times and resuspended in PBS. To measure the expression level per bacterial unit, all the samples were diluted to the same OD600 value (OD600 = 1.0), and 100 μl of each cell suspension was added to a 96-well flat-bottom polystyrene plate (Costar, USA) for fluorescence detection. Katushka protein was detected at excitation and emission wavelengths of 588 nm and 633 nm, respectively. The relative fluorescence value (RFV) per bacterial unit (RFV/OD600) was set as the evaluation parameter to compare the expression efficiencies of different systems.
Expression assay in zebrafish larvae.
Bacterial cultures of E. coli(araQS) grown overnight were inoculated (1:1,000, vol/vol) into fresh LB medium with antibiotics and arabinose. The bacteria were harvested and suspended in PBS. Zebrafish larvae at 6 to 8 days of age were immersed in the cell suspension at a concentration of 108 viable bacteria per ml for 2 h and then washed and transferred into freshwater. At the target time point, larvae were washed with PBS at least three times and anesthetized with 0.01% (wt/vol) MS-222 anesthetic. The fish larvae were observed by using an inverted fluorescence microscope (DMI3000 B; Leica). Meanwhile, 5 larvae at each time point were homogenized with lysis buffer and used to coat LB agar plates to count the corresponding number of bacteria.
Expression assay in zebrafish adults.
The bacterial suspension of E. coli(araQS) was prepared as described above. Healthy adult zebrafish with an average weight of 0.2 g were intraperitoneally injected with an E. coli(araQS) suspension at a dose of 107 CFU per tail. At 4 h, 12 h, and 24 h postinjection, the internal organs of 20 fish from each group were surgically extracted. After being homogenized and concentrated by ultrafiltration, the tissue samples were analyzed by Western blotting using an antibody specific to Katushka. Meanwhile, zebrafish internal organs taken at different time points were treated with tissue lysis buffer and used to coat LB agar plates to count the number of bacteria in zebrafish.
All in vitro and in vivo assays were repeated at least three times for accuracy.
Vaccination and challenge.
The gapA34 gene, encoding glyceraldehyde-3-phosphate dehydrogenase (GAPDH) from Aeromonas hydrophila, was inserted into araQS, and the resultant recombinant plasmid, araQS-G, was transformed into attenuated E. tarda vaccine strain WED to create WED(araQS-G). The potential multivalent vaccine candidate WED(araQS-G) was evaluated for immune protection in turbot (Scophthalmus maximus). Attenuated E. tarda strain WED was used as the control. All the vaccination and challenge experiments were done in triplicate.
The turbot, from a mariculture farm in Shandong Province, China, and weighing ∼10 g, were fed and acclimated for 30 days before the experiment. Three turbot groups were intraperitoneally injected with sterilized saline (0.1 ml per tail), a WED suspension (107 CFU per tail), and a WED(araQS-G) suspension (107 CFU per tail). The immunized fish were reared in aquaria supplied with a continuous flow of recycling water at ∼16°C to 18°C. Four weeks later, each immunized group was divided into 2 challenge subgroups, which were intramuscularly injected with wild-type A. hydrophila (8 × 107 CFU per tail) and wild-type E. tarda EIB202 (6 × 103 CFU per tail). The mortality was recorded for 4 weeks after challenge, and the observation of surviving fish was extended to 6 weeks. Significant differences and relative percent survival (RPS) values were calculated by using Fisher's exact test and according to the formula RPS = (1 − mortality rate of vaccinated fish/mortality rate of control fish) × 100, respectively.
All the animal experiments were approved by the Institutional Animal Care and Use Committee of East China University of Science and Technology.
RESULTS
Construction of cell density-dependent expression systems.
The quorum sensing elements, including luxI, luxR, and their respective promoters, were cloned from V. fischeri. Three plasmids, QS1 to QS3, were designed based on different strategies (Fig. 1A) and transformed into E. coli BL21 strains for function tests in vitro. QS1, containing only the luxI promoter, almost failed to express Katushka. This finding suggested that the luxI-luxR quorum sensing circuit from the E. coli genome could not activate the luxI promoter of V. fischeri (Fig. 1B), possibly because of species specificity (25, 26). More importantly, the single luxI promoter lost its vitality without the help of other quorum sensing components. Comparatively, both QS2 and QS3 were designed to contain intact V. fischeri QS gene elements, and the only difference between them is that the Katushka gene was transcribed from a separate PluxI promoter in QS2, while it was cotranscribed with luxI in QS3. The results (Fig. 1B) showed that QS3 displayed a dramatically increased expression level, comparable that of to highly efficient pET expression system, while QS2 displayed only a low expression level. The low efficiency of QS2 might be caused by the presence of two individual luxI promoters competitively binding with a limited number of LuxR-AI complexes, which act as transcription activators to initiate the QS system. For QS3, the expression level continued to decrease in the first 3 h and then started to increase after 3 h. These data indicated that QS3 was not activated until the cell density reached the threshold (OD of 0.47 for QS3 system) at 3 h postinoculation. To further confirm the cell density-dependent behavior of QS3, a fed-batch culture model was established to keep the cell density of the bacterial culture below the threshold by feeding fresh medium continuously. Concurrently, a contrasting experiment was conducted by adding medium with autoinducer molecules into the fed-batch culture. As shown in Fig. 1C, the RFV of the QS3 strain in the fed-batch culture dropped instantly and stayed low the entire time, showing that the quorum sensing circuit remained inactivated when the microbial population density was kept below the threshold level. This result confirmed that a small bacterial population of QS3 could not synthesize enough autoinducer molecules to activate the QS system. Simultaneously, the addition of exogenous autoinducers significantly activated QS3 in fed-batch cultivation, even with the cell density being below the threshold level. All these data confirmed that the artificial QS3 system is an efficient and cell density-regulated expression system.
FIG 1.
Construction and in vitro performance of QS. (A) Structures of the cell density-regulated expression systems QS1 to QS3. PluxI, promoter of the luxI gene; PluxR, promoter of the luxR gene; TT, transcription terminator; RBS, ribosome binding site. Katushka is a reporter gene encoding red fluorescent protein. (B) Protein expression levels of E. coli BL21 strains containing QS1 to QS3 were evaluated by determining the relative fluorescence values (RFV) of red fluorescent protein. The BL21(DE3) strain containing a traditional T7 expression system was configured as a control. (C) Cell density-regulated expression of the QS3 system. E. coli BL21 harboring the QS3 system was cultivated in a fed-batch model, and the OD600 was maintained below 0.3 by feeding fresh medium continuously. Autoinducers of the QS system were exogenously added to the medium or not.
Design of a binary QS system that responds to arabinose and cell density.
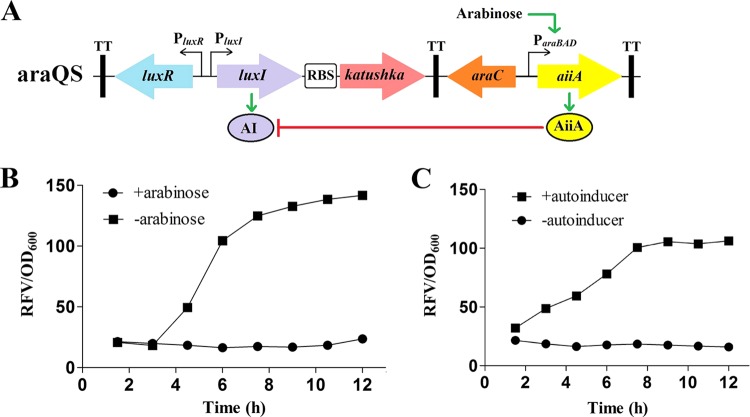
Although QS3 performed well in regulating gene expression in a cell density-dependent manner in vitro, it is rather challenging to control the switch of the QS system by a specific in vivo signal for potential applications in eukaryotic hosts. Here, a ParaBAD-aiiA gene module was inserted into QS3 to form a new binary QS system, araQS. In this circuit (Fig. 2A), arabinose, as an inducer, activates the transcription of the aiiA gene from ParaBAD (27, 28), and the expressed AiiA proteins repress the PluxI promoter by degrading autoinducers and quench the QS3 system even at high cell density. Since arabinose is often absent in eukaryotic hosts, the ParaBAD-aiiA gene module was designed to act as an arabinose-responsive molecular switch to negatively regulate the QS system in vivo.
FIG 2.
Construction and in vitro performance of araQS. (A) Linear sketch of araQS. aiiA is quorum quenching gene from B. thuringiensis; ParaBAD is the arabinose promoter. (B) Protein expression of the araQS system regulated by an arabinose signal. Arabinose was added to the medium or not. Cell cultures were harvested and adjusted to an OD600 of 1 for fluorescence detection at the indicated time points. (C) Cell density-regulated expression of araQS. E. coli BL21 harboring araQS was cultivated via fed-batch culture, and the OD600 was maintained below 0.3 by feeding fresh medium continuously. Autoinducers of QS were exogenously added to the medium or not.
The performance of araQS in the E. coli BL21 host was assayed by adding arabinose to the culture medium. The araQS system was severely inhibited in the presence of arabinose (Fig. 2B), indicating that arabinose-induced AiiA proteins remarkably repressed the QS system by degrading autoinducer molecules. On the contrary, the araQS system was significantly derepressed in the absence of arabinose because the quorum quenching protein AiiA was not synthesized. All these data confirmed that the ParaBAD-aiiA gene module could negatively control the QS system by responding to arabinose in the medium. The araQS system was further analyzed in the fed-batch culture model (Fig. 2C). As long as the cell density was below the threshold value, the araQS system was always kept switched off, even in arabinose-free medium, unless an exogenous autoinducer was added. The results confirmed that the araQS system, composed of the ParaBAD-aiiA regulation module and a quorum sensing circuit, exhibits a precise dually arabinose- and density-regulated expression feature.
In vivo performance of the araQS system.
Although araQS was demonstrated to be an efficient binary system that responds to arabinose and cell density in vitro, its performances in vivo still remained unknown. In the following work, zebrafish larvae and adults were applied as animal models to evaluate the in vivo performances of araQS. The recombinant E. coli strain carrying araQS was administered to zebrafish larvae by immersion, and both fluorescence intensity and bacterial counts in larvae were then monitored at different intervals. Until 6 h postimmersion, no obvious Katushka fluorescence signal was detected (Fig. 3A), which was in accordance with the in vivo propagation kinetics of the E. coli strains (Fig. 3B). The results indicated that the araQS system functioned well in zebrafish larvae. Responding to the arabinose-free signal in vivo, the araQS system was derepressed by switching off aiiA transcription from ParaBAD, and simultaneously, araQS was activated by locally accumulated autoinducers from an increasing microbial population in vivo. In addition, E. coli(araQS) was administered to zebrafish adults by injection, and the in vivo expression pattern of araQS was evaluated. As shown in Fig. 3C and D, the araQS system was activated significantly after 4 h postinjection upon in vivo propagation of the bacteria. All the data confirmed that the araQS system was effectively activated in the zebrafish model through responding to both arabinose-free and cell density signals, displaying a dually regulated expression feature.
FIG 3.
In vivo performance of araQS. (A and B) Protein expression of E. coli(araQS) in zebrafish larvae. Fish larvae, immersed in a bacterial suspension for 2 h, were observed by using an inverted fluorescence microscope at different time intervals, and the corresponding number of bacteria in larvae was counted by coating LB agar plates with homogenized larvae at each time point. (C and D) Protein expression of E. coli(araQS) in adult zebrafish. The internal organs of 20 fish from each group were surgically extracted at different time intervals. The tissue samples were further analyzed by Western blotting using an antibody specific to Katushka, and the corresponding number of bacteria in adult zebrafish was counted by coating LB agar plates with the tissue samples.
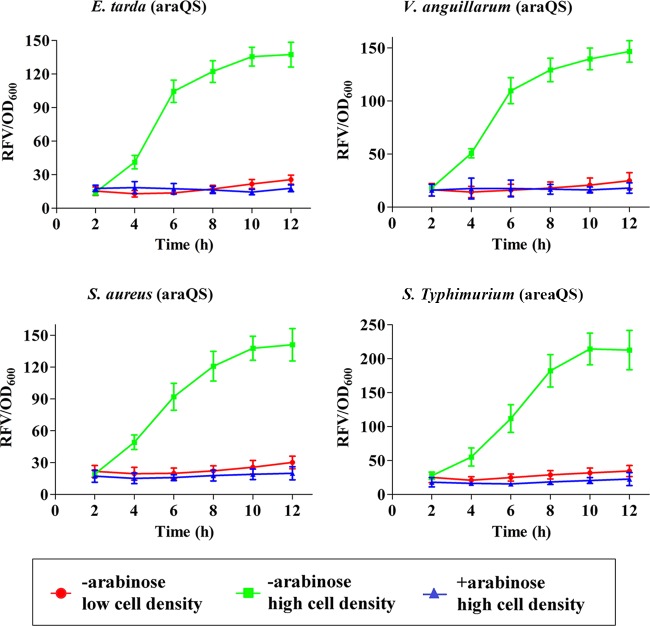
Expression of araQS in a wide range of bacterial hosts.
Adaptation to different bacterial host environments is an important indicator of a trustworthy expression system. Since araQS was composed of the quorum-related genes luxI and luxR from V. fischeri, aiiA from Bacillus thuringiensis, and the arabinose promoter, it is of interest to test whether these heterologous gene elements could be coordinated in different bacterial hosts. Here, araQS was introduced into different Gram-negative bacteria, including the fish pathogens Edwardsiella tarda and Vibrio anguillarum and the human pathogen Salmonella Typhimurium, and the Gram-positive bacterium Staphylococcus aureus. The expression performances of these recombinant strains were then assayed in vitro (Fig. 4). All four strains bearing araQS presented high protein expression levels and a binary regulation pattern similar to that in E. coli. These data suggested that the heterologous gene elements of araQS were compatible with different bacteria and might have the potential to be applied in a wide range of bacterial hosts.
FIG 4.
In vitro performance of araQS in different bacterial hosts. The araQS plasmid was transformed into Edwardsiella tarda, Vibrio anguillarum, Salmonella Typhimurium, and Staphylococcus aureus, and the resultant recombinant bacterial hosts were cultivated in LB medium with or without the addition of arabinose. Simultaneously, the recombinant bacteria were cultivated in a fed-batch model to maintain the OD600 below 0.3 by feeding fresh medium continuously. Each of the cultures was harvested and adjusted to an OD600 of 1 for fluorescence detection at the indicated time points.
Application of araQS in a multivalent bacterial vaccine.
A multivalent bacterial vaccine is constructed via the expression of a heterologous antigen in a live bacterial vector, and this is an attractive vaccine strategy to induce an immune response to the carried protective antigen. How to enhance antigen expression and delivery without reducing vaccine stability and vitality has long been challenging researchers. Constitutive expression of a heterologous antigen in attenuated bacteria often causes obvious metabolic burdens and thus results in declining in vivo colonization for the live bacterial vaccine (29). To overcome the shortcomings of constitutive expression, in vivo-inducible promoters have been used to regulate antigen expression (30). In this work, the in vivo-triggered araQS system was used to produce antigen in vivo after the live recombinant vaccine was administered to the host.
Edwardsiella tarda is an important facultative intracellular pathogen of both animals and humans (31). An attenuated E. tarda strain, called WED, was constructed by in-frame deletion of eseBCD and aroC, and this attenuated strain was proven to be an efficient live bacterial vaccine for fish (32). In order to establish a suitable expression system for application in a multivalent E. tarda vaccine, the in vivo-triggered araQS system and the in vitro-expressed QS3 system were used to express heterologous protein in WED. Compared to WED(araQS), WED(QS3) displayed significantly inhibited growth in vitro (Fig. 5A) and also an impaired colonization ability in vivo in fish (Fig. 5B), possibly due to the metabolic burden of heterologous protein expression. The results suggest that in vivo-triggered expression of a heterologous antigen facilitates bacterial colonization in vivo and thus is more suited for vector vaccine design.
FIG 5.

In vitro growth and in vivo colonization of QS3 and araQS. (A) Growth of E. tarda WED loaded with the QS3 or araQS plasmid in arabinose-rich medium. The blank plasmid pUTat was used as the control. (B) Counts of viable bacteria in zebrafish injected with plasmid-loaded WED strains (5 × 104 CFU per tail) at 12, 24, and 48 h. Ten fish were set as a pool, and three parallel experiments were done. At the indicated time points, 10 fish from each group were randomly picked and homogenized, and the corresponding number of bacteria was counted by coating LB agar plates with homogenized fish.
In order to design a multivalent E. tarda vaccine using the araQS system, the protective antigen gene gapA34 from the pathogenic organism A. hydrophila, encoding GAPDH (33, 34), was inserted into the araQS plasmid and transformed into attenuated E. tarda strain WED, forming a multivalent vaccine candidate, WED(araQS-G) (Fig. 6A). As shown in Fig. 6B, GADPH was effectively produced in WED(araQS-G) under arabinose-free and high-cell-density conditions. To further investigate the in vivo application of WED(araQS-G) as a multivalent vaccine, protection efficacy was evaluated in turbot, an important cultured fish in China. The multivalent vaccine WED(araQS-G) showed significant protection against challenge with E. tarda EIB202 (RPS = 73.3%) and A. hydrophila LSA34 (RPS = 68.6%) over the saline control (Table 3), while the monovalent vaccine WED triggered significant protection against E. tarda EIB202 (RPS = 76.7%) but only a slight response against A. hydrophila LSA34 (RPS = 13.9%). These data confirmed that araQS-mediated expression of GAPDH in WED effectively protected turbots from experimental challenge with A. hydrophila LSA34 while exerting no adverse effect on protection against E. tarda infection.
FIG 6.
Construction of the recombinant vector vaccine WED(araQS-G). (A) Plasmid construction of araQS-G. (B) Expression analysis of GAPDH in WED(araQS-G) by SDS-PAGE. Recombinant bacteria were cultivated under different conditions for 9 h. HCD, high cell density; LCD, low cell density; ara +, medium with arabinose added; ara −, arabinose-free medium; M, protein molecular marker.
TABLE 3.
Efficacy of the vaccine candidate WED(araQS-G) against E. tarda EIB202 and A. hydrophila LSA34 in turbot
| Immunogen | Vaccination dose/tail | Challenge strain | Challenge dose (CFU/tail) | No. of fishd | Mean mortality (%) ± SDa | Mean RPS ± SDb | Effectc |
|---|---|---|---|---|---|---|---|
| Saline | 0.1 ml | EIB202 | 6 × 103 | 30 | 100 ± 0 | ||
| LSA34 | 8 × 107 | 30 | 85.0 ± 5.8 | ||||
| WED | 107 CFU | EIB202 | 6 × 103 | 30 | 23.3 ± 4.7 | 76.7 ± 4.7 | + |
| LSA34 | 8 × 107 | 30 | 73.3 ± 5.8 | 13.9 ± 6.8 | − | ||
| WED(araQS-G) | 107 CFU | EIB202 | 6 × 103 | 30 | 26.7 ± 4.0 | 73.3 ± 4.0 | + |
| LSA34 | 8 × 107 | 30 | 26.7 ± 6.7 | 68.6 ± 7.8 | + |
The mortality was recorded for 4 weeks after challenge, and the observation of surviving fish was extended to 6 weeks.
RPS is the relative percent survival, which is defined relative to the saline group and is calculated as follows: RPS = (1 − mortality of vaccinated fish/mortality of control fish) × 100.
If the RPS was >60%, the vaccine candidate showed protection against challenge, indicated by a “+.” If the RPS was <30%, the vaccine candidate showed no protection against the challenge strain, indicated by a “−.”
The vaccination and challenge experiments were repeated three times.
DISCUSSION
The engineering of gene regulatory networks is a cornerstone of synthetic biology (35). Cell-cell communication represents a valuable mechanism to engineer novel signaling constructs in bacteria (36). The genetic elements of the V. fischeri quorum sensing circuit have been successfully used in several artificial genetic systems (15, 17, 37). However, all of these systems were intended to work in vitro by adding inducers to initiate the pathway of circuits. A highly efficient and cell density-dependent QS system has been designed by rewiring quorum sensing genetic elements of V. fischeri into E. coli (38). In order to engineer the QS system for use in vivo, it is essential to integrate an additional regulation module to keep the artificial network inactivated in vitro unless it senses a special in vivo signal. Generally, there are two ways to manipulate the QS circuit, either positively by an in vivo-induced activator or negatively by an in vitro-induced inhibitor. In this work, the araQS system with binary regulation was designed for in vivo applications based on the latter strategy. Because of the absence of arabinose in almost all kinds of in vivo environments, the ParaBAD-aiiA module, consisting of the arabinose-activated promoter ParaBAD and the quorum quenching gene aiiA, was constructed and coupled to the QS system. The resultant araQS system exhibited an in vivo-triggered and cell density-dependent expression pattern in in vivo environments such as zebrafish models.
The araQS system was established as a chimeric system, comprising an optimized QS circuit from V. fischeri, the arabinose promoter ParaBAD, and the aiiA gene from B. thuringiensis. The araQS system displayed a high level of compatibility with a variety of bacterial hosts, indicating araQS forms a self-contained and closed-loop expression circuit that is less dependent on hosts. Although some bacterial hosts used in this work might possess their own QS systems and produce their own AHL autoinducers, no cross talk or interference between the native bacterial host QS and the heterologous araQS systems was found. This could be attributed mainly to the fact that the AHL autoinducers differ in the shape of the acyl side chain between species and specifically activate the QS circuit of their own species (6, 26, 39). All these data suggested that the araQS system has great potential as a general expression tool in a wide variety of bacterial hosts. However, the araQS system was shown to be initiated at a very early stage, when the bacteria were still in a small population both in vitro and in vivo. This might be explained by the fact that multiple copies of the araQS plasmid make bacteria produce higher levels of autoinducers, and thus, accumulated autoinducers could reach the threshold at a lower cell density. Thus, bacteria bearing araQS exhibit a much lower threshold than V. fischeri with a single copy of the QS system on its chromosome. Therefore, if a delayed initiation of araQS in vivo is expected, a low-copy-number plasmid or single-copy integration into the chromosome is a reasonable choice.
Bacterial vector vaccines are an attractive vaccination strategy to induce multiple immune responses to both the bacterial vector and the carried protective antigen (40). The superiorities of live bacterial vaccines are essentially based on their mimicry of natural infection in hosts (41). A typical bacterial infection involves a round of sequential in vivo events, including invasion, colonization, propagation, and then dissemination. At the early stage of infection, invading bacteria are in a small population at the site of infection, and they are required to contend desperately with rigorous host stresses for successful colonization. The additional metabolic burden of heterologous expression at this stage will unwittingly attenuate the bacterium to some extent and then impair colonization. Therefore, it is rather beneficial to turn off antigen expression in bacteria until colonization is established in vivo (30, 42). In this work, the araQS system, with an in vivo-triggered and population-dependent expression pattern, was used to express a heterologous protective antigen in an attenuated bacterial strain. Compared with QS3, the araQS system kept protein expression switched off in vitro, which significantly facilitates bacterial growth in vitro and bacterial colonization in vivo. Therefore, the araQS system has great potential for the design of a bacterial vector vaccine.
In summary, an in vivo programmed expression circuit was established in this work, which was derepressed by the arabinose-free signal in vivo and then programmed to produce heterologous proteins in a cell density-dependent manner. This artificial synthetic circuit might have the potential for in vivo applications, including, but not limited to, bacterial vector vaccines.
ACKNOWLEDGMENTS
This work was financially supported by grants from the National Natural Science Funds of China (31272700), the National Key Technology Support Program of China (2012BAD17B02), the National High Technology Research and Development Program of China (2013AA093101), and the Shanghai Rising Star Program (13QA1401000).
REFERENCES
- 1.Garg N, Manchanda G, Kumar A. 2014. Bacterial quorum sensing: circuits and applications. Antonie Van Leeuwenhoek 105:289–305. doi: 10.1007/s10482-013-0082-3. [DOI] [PubMed] [Google Scholar]
- 2.Miller MB, Bassler BL. 2001. Quorum sensing in bacteria. Annu Rev Microbiol 55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 3.Bassler BL, Losick R. 2006. Bacterially speaking. Cell 125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Collins CH, Arnold FH, Leadbetter JR. 2005. Directed evolution of Vibrio fischeri LuxR for increased sensitivity to a broad spectrum of acyl-homoserine lactones. Mol Microbiol 55:712–723. doi: 10.1111/j.1365-2958.2004.04437.x. [DOI] [PubMed] [Google Scholar]
- 5.Schaefer AL, Hanzelka BL, Eberhard A, Greenberg E. 1996. Quorum sensing in Vibrio fischeri: probing autoinducer-LuxR interactions with autoinducer analogs. J Bacteriol 178:2897–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eberhard A, Burlingame A, Eberhard C, Kenyon G, Nealson K, Oppenheimer N. 1981. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry 20:2444–2449. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- 7.Egland KA, Greenberg E. 1999. Quorum sensing in Vibrio fischeri: elements of the luxI promoter. Mol Microbiol 31:1197–1204. doi: 10.1046/j.1365-2958.1999.01261.x. [DOI] [PubMed] [Google Scholar]
- 8.Kaplan HB, Greenberg E. 1985. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J Bacteriol 163:1210–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanzelka BL, Greenberg E. 1995. Evidence that the N-terminal region of the Vibrio fischeri LuxR protein constitutes an autoinducer-binding domain. J Bacteriol 177:815–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urbanowski M, Lostroh C, Greenberg E. 2004. Reversible acyl-homoserine lactone binding to purified Vibrio fischeri LuxR protein. J Bacteriol 186:631–637. doi: 10.1128/JB.186.3.631-637.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raina S, Vizio DD, Odell M, Clements M, Vanhulle S, Keshavarz T. 2009. Microbial quorum sensing: a tool or a target for antimicrobial therapy? Biotechnol Appl Biochem 54:65–84. doi: 10.1042/BA20090072. [DOI] [PubMed] [Google Scholar]
- 12.Dong Y-H, Xu J-L, Li X-Z, Zhang L-H. 2000. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc Natl Acad Sci U S A 97:3526–3531. doi: 10.1073/pnas.97.7.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choudhary S, Schmidt-Dannert C. 2010. Applications of quorum sensing in biotechnology. Appl Microbiol Biotechnol 86:1267–1279. doi: 10.1007/s00253-010-2521-7. [DOI] [PubMed] [Google Scholar]
- 14.Anderson JC, Clarke EJ, Arkin AP, Voigt CA. 2006. Environmentally controlled invasion of cancer cells by engineered bacteria. J Mol Biol 355:619–627. doi: 10.1016/j.jmb.2005.10.076. [DOI] [PubMed] [Google Scholar]
- 15.Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. 2005. A synthetic multicellular system for programmed pattern formation. Nature 434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- 16.You L, Cox RS, Weiss R, Arnold FH. 2004. Programmed population control by cell-cell communication and regulated killing. Nature 428:868–871. doi: 10.1038/nature02491. [DOI] [PubMed] [Google Scholar]
- 17.Danino T, Mondragón-Palomino O, Tsimring L, Hasty J. 2010. A synchronized quorum of genetic clocks. Nature 463:326–330. doi: 10.1038/nature08753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nistala GJ, Wu K, Rao CV, Bhalerao KD. 2010. A modular positive feedback-based gene amplifier. J Biol Eng 4:4. doi: 10.1186/1754-1611-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bulter T, Lee SG, Wong WW, Fung E, Connor MR, Liao JC. 2004. Design of artificial cell-cell communication using gene and metabolic networks. Proc Natl Acad Sci U S A 101:2299–2304. doi: 10.1073/pnas.0306484101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guan L, Mu W, Champeimont J, Wang Q, Wu H, Xiao J, Lubitz W, Zhang Y, Liu Q. 2011. Iron-regulated lysis of recombinant Escherichia coli in host releases protective antigen and confers biological containment. Infect Immun 79:2608–2618. doi: 10.1128/IAI.01219-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Touati D. 2000. Iron and oxidative stress in bacteria. Arch Biochem Biophys 373:1–6. doi: 10.1006/abbi.1999.1518. [DOI] [PubMed] [Google Scholar]
- 22.Kong W, Wanda SY, Zhang X, Bollen W, Tinge SA, Roland KL, Curtiss R III. 2008. Regulated programmed lysis of recombinant Salmonella in host tissues to release protective antigens and confer biological containment. Proc Natl Acad Sci U S A 105:9361–9366. doi: 10.1073/pnas.0803801105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hawkins AC, Arnold FH, Stuermer R, Hauer B, Leadbetter JR. 2007. Directed evolution of Vibrio fischeri LuxR for improved response to butanoyl-homoserine lactone. Appl Environ Microbiol 73:5775–5781. doi: 10.1128/AEM.00060-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao Y, Liu Q, Chen H, Zhang YX. 2011. A stable plasmid system for heterologous antigen expression in attenuated Vibrio anguillarum. Vaccine 29:6986–6993. doi: 10.1016/j.vaccine.2011.07.036. [DOI] [PubMed] [Google Scholar]
- 25.Ng WL, Bassler BL. 2009. Bacterial quorum-sensing network architectures. Annu Rev Genet 43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao Y, Martinez-Yamout MA, Dickerson TJ, Brogan AP, Wright PE, Dyson HJ. 2006. Structure of the Escherichia coli quorum sensing protein SdiA: activation of the folding switch by acyl homoserine lactones. J Mol Biol 355:262–273. doi: 10.1016/j.jmb.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 27.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reeder T, Schleif R. 1991. Mapping, sequence, and apparent lack of function of araJ, a gene of the Escherichia coli arabinose regulon. J Bacteriol 173:7765–7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loessner H, Endmann A, Leschner S, Bauer H, Zelmer A, zur Lage S, Westphal K, Weiss S. 2008. Improving live attenuated bacterial carriers for vaccination and therapy. Int J Med Microbiol 298:21–26. doi: 10.1016/j.ijmm.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 30.Medina E, Guzmán CA. 2001. Use of live bacterial vaccine vectors for antigen delivery: potential and limitations. Vaccine 19:1573–1580. doi: 10.1016/S0264-410X(00)00354-6. [DOI] [PubMed] [Google Scholar]
- 31.Leung KY, Siame BA, Tenkink BJ, Noort RJ, Mok Y-K. 2012. Edwardsiella tarda—virulence mechanisms of an emerging gastroenteritis pathogen. Microbes Infect 14:26–34. doi: 10.1016/j.micinf.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 32.Xiao J, Chen T, Liu B, Yang W, Wang Q, Qu J, Zhang Y. 2013. Edwardsiella tarda mutant disrupted in type III secretion system and chorismic acid synthesis and cured of a plasmid as a live attenuated vaccine in turbot. Fish Shellfish Immunol 35:632–641. doi: 10.1016/j.fsi.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 33.Zhou L, Wang X, Liu Q, Wang Q, Zhao Y, Zhang Y. 2010. A novel multivalent vaccine based on secretary antigen-delivery induces protective immunity against Vibrio anguillarum and Aeromonas hydrophila. J Biotechnol 146:25–30. doi: 10.1016/j.jbiotec.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 34.Mu W, Guan L, Yan Y, Liu Q, Zhang Y. 2011. A novel in vivo inducible expression system in Edwardsiella tarda for potential application in bacterial polyvalence vaccine. Fish Shellfish Immunol 31:1097–1105. doi: 10.1016/j.fsi.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 35.Benner SA. 2003. Synthetic biology: act natural. Nature 421:118. doi: 10.1038/421118a. [DOI] [PubMed] [Google Scholar]
- 36.Purnick PE, Weiss R. 2009. The second wave of synthetic biology: from modules to systems. Nat Rev Mol Cell Biol 10:410–422. doi: 10.1038/nrm2698. [DOI] [PubMed] [Google Scholar]
- 37.Neddermann P, Gargioli C, Muraglia E, Sambucini S, Bonelli F, De Francesco R, Cortese R. 2003. A novel, inducible, eukaryotic gene expression system based on the quorum-sensing transcription factor TraR. EMBO Rep 4:159–165. doi: 10.1038/sj.embor.embor734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haseltine EL, Arnold FH. 2008. Implications of rewiring bacterial quorum sensing. Appl Environ Microbiol 74:437–445. doi: 10.1128/AEM.01688-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morohoshi T, Inaba T, Kato N, Kanai K, Ikeda T. 2004. Identification of quorum-sensing signal molecules and the LuxRI homologs in fish pathogen Edwardsiella tarda. J Biosci Bioeng 98:274–281. doi: 10.1016/S1389-1723(04)00281-6. [DOI] [PubMed] [Google Scholar]
- 40.Shata MT, Stevceva L, Agwale S, Lewis GK, Hone DM. 2000. Recent advances with recombinant bacterial vaccine vectors. Mol Med Today 6:66–71. doi: 10.1016/S1357-4310(99)01633-0. [DOI] [PubMed] [Google Scholar]
- 41.Spreng S, Dietrich G, Weidinger G. 2006. Rational design of Salmonella-based vaccination strategies. Methods 38:133–143. doi: 10.1016/j.ymeth.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 42.Wang S, Li Y, Shi H, Sun W, Roland KL, Curtiss R III. 2011. Comparison of a regulated delayed antigen synthesis system with in vivo-inducible promoters for antigen delivery by live attenuated Salmonella vaccines. Infect Immun 79:937–949. doi: 10.1128/IAI.00445-10. [DOI] [PMC free article] [PubMed] [Google Scholar]