Abstract
Traditional Chinese solid-state fermented cereal starters contain highly complex microbial communities and enzymes. Very little is known, however, about the microbial dynamics related to environmental conditions, and cellulolytic communities have never been proposed to exist during cereal starter fermentation. In this study, we performed Illumina MiSeq sequencing combined with PCR-denaturing gradient gel electrophoresis to investigate microbiota, coupled with clone library construction to trace cellulolytic communities in both fermentation stages. A succession of microbial assemblages was observed during the fermentation of starters. Lactobacillales and Saccharomycetales dominated the initial stages, with a continuous decline in relative abundance. However, thermotolerant and drought-resistant Bacillales, Eurotiales, and Mucorales were considerably accelerated during the heating stages, and these organisms dominated until the end of fermentation. Enterobacteriales were consistently ubiquitous throughout the process. For the cellulolytic communities, only the genera Sanguibacter, Beutenbergia, Agrobacterium, and Erwinia dominated the initial fermentation stages. In contrast, stages at high incubation temperature induced the appearance and dominance of Bacillus, Aspergillus, and Mucor. The enzymatic dynamics of amylase and glucoamylase also showed a similar trend, with the activities clearly increased in the first 7 days and subsequently decreased until the end of fermentation. Furthermore, β-glucosidase activity continuously and significantly increased during the fermentation process. Evidently, cellulolytic potential can adapt to environmental conditions by changes in the community structure during the fermentation of starters.
INTRODUCTION
Cereal grains are the major source of dietary nutrients worldwide. Humans have been domesticating principal “founder crops” (1) from time immemorial and fermenting their produce with the development of fermentation techniques. Generally, cereal food fermentation processes have been driven by metabolically active microorganisms and enzymes, which are oriented from raw materials, autochthonous starter cultures, or the processing surrounding environment (2) and are responsible for the degradation of polysaccharides and the release of fermentable sugars from grains. Starch is the most abundant form of “storage device” for glucose in plants. For starch utilization, microorganisms require an appropriate combination of amylolytic enzymes, including α-/β-amylase and glucoamylase to depolymerize starch polymer to oligosaccharides and smaller sugars during cereal food fermentation, α-amylase to hydrolyze linkages in the interior of starch in a random fashion and release linear and branched oligosaccharides, and β-amylase and glucoamylase to attack the substrate from the nonreducing end and produce small and well-defined oligosaccharides such as maltose and glucose (3). Traditionally, the production of fermented food is based mainly on the addition of starter cultures (4). Various types of starter cultures are widely used to accelerate and steer fermentation processes by diverse functional microbes and enzymes contained in starters (5, 6).
Similarly, traditional Chinese cereal mixtures, locally called “daqu,” which originated from the ancient Chinese culture and can be dated back to 3,000 years ago, are used as starters to produce Chinese liquor and vinegar (7). Traditional Chinese cereal grain starters are mostly fermented from barley, wheat, and pea. Preparation of cereal starter involves three stages: (i) material mixing and shaping; (ii) spontaneous solid-state fermentation with the temperature controlled within 40°C to 60°C for 20 to 25 days, which directly determines the shifts in microbial community structure and enriches the functional microbes naturally present in the raw material ingredients (barley, wheat, and pea) and its production environment (including tools, soil, air, and equipment) (8); and (iii) maturation in a fermentation room for 2 months (9). Daqu starters are primary liquefying and saccharifying agents, and the types and activities of amylolytic enzymes, which originate from microbes involved in starters, play a significant role in determining the final quality of the fermented products (6).
Cellulose is the most abundant biopolymer, constituting 35% to 50% of the grain's final dry weight (10, 11); therefore, the breakdown of this compound is a key step in the decomposition of plant material. Cellulolytic microbial communities within the phyla Acidobacteria, Actinobacteria, Proteobacteria, Bacteroidetes, Firmicutes, and Ascomycota have been recognized as potential cellulose degraders in the forest litter layer and soil ecosystems (12–14). Cellulose-degrading microbes produce cellulase enzymes that catalyze the first step of cellulose hydrolysis and release oligosaccharides, such as cellobiose; subsequently, oligosaccharides can be converted into glucose and assimilated by cellulolytic communities with the expression of β-glucosidase, which is associated with glycoside hydrolase (GH) families 1 and 3 (15). β-Glucosidase is a phylogenetically conserved enzyme that plays a rate-limiting role in cellulose degradation (16). Previous genomic analyses indicated that >80% of sequenced bacterial lineages carry β-glucosidase (17). To date, the cellulolytic diversity targeting β-glucosidase and their function have been analyzed only in regard to soil management (18, 19) and the composting process (20), but no studies have been focused on the cellulolytic diversity involved in cereal starter fermentation.
Importantly, microorganisms have both individual functions and interactions in various ecosystems. Nowadays, it is widely accepted that ecosystems are affected by environmental conditions. Recently, microbial communities in various types of cereal starters have been extensively investigated (21, 22); however, we have limited understanding on how changes in environmental conditions directly affect the microbiota dynamics related to the fermentative production of cereal starters, which leads to instability of the fermentation process and finally threatens the quality of the fermented products. In the present study, Illumina-based high-throughput sequencing and a nested PCR-denaturing gradient gel electrophoresis (DGGE) fingerprinting method were conducted to investigate the microbiota dynamics associated with environmental conditions during fermentation of traditional Chinese cereal starters. Specifically, the shift in cellulolytic structure targeting the β-glucosidase gene was characterized by clone library analysis. Abundances of the 16S/18S rRNA and β-glucosidase genes were quantified by quantitative PCR (qPCR), and the enzymatic dynamics of glucoamylase, amylase, carboxymethyl cellulase (CMCase), and β-glucosidase were also monitored. This research can provide a comprehensive microbial ecological functional map in cereal starter fermentation.
MATERIALS AND METHODS
Experiment and sampling.
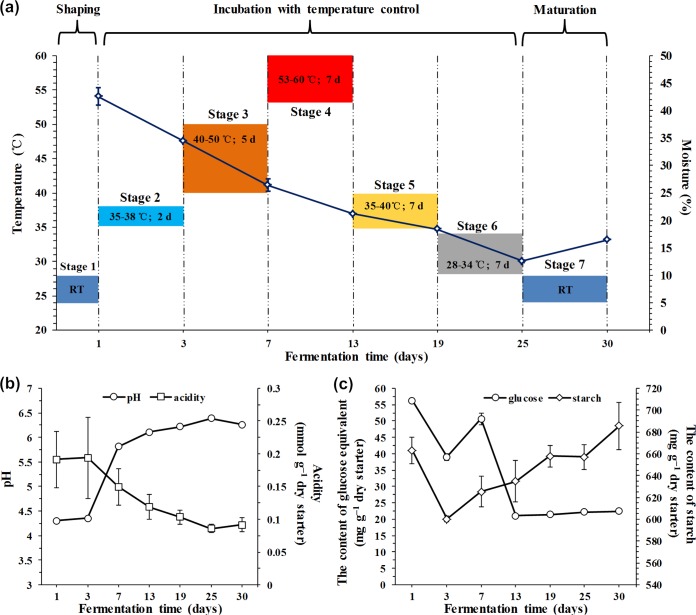
Spontaneous solid-state fermentation of cereal starters was conducted in the fermentation room of a traditional vinegar production factory in Shanxi province, China. Initially, cereal material mixtures of ground barley and pea (6:4, wt/wt) were stirred with added water (40%, wt/wt). After being shaped into bricks (32 cm by 16 cm by 6 cm) and piled in the fermentation room, mixtures were incubated for 30 days with strict temperature control (Fig. 1a). Samples were collected separately at the seven production stages based on the temperature control caused by forced ventilation during the fermentation process: 1 day (room temperature [RT]), 3 days (35°C to 38°C), 7 days (40°C to 50°C), 13 days (53°C to 60°C), 19 days (35°C to 40°C), 25 days (28°C to 34°C), and 30 days (RT) (Fig. 1a). With changes in temperature, the moisture content gradually decreased from the initial 42.53% (wt/wt) to 12.57% during 25 days of incubation and subsequently increased to 16.45% at the final stage (Fig. 1a). To obtain adequate information and representation, blocks from each stage were randomly selected from the upper, middle, and lower locations in triplicates, which were then ground, mixed, and pooled into sterile Stomacher bags (Stomacher Lab System, London, United Kingdom) to provide an experimental starter powder sample. All of the samples were stored at −20°C for further analysis.
FIG 1.
Dynamics of physicochemical characteristics during the cereal starter fermentation process. (a) Changes in incubation temperature and corresponding moisture content. Stage 3, heating stage; stage 4, high-temperature stage; stages 5 and 6, cooling stages. (b) Change in pH. (c) Changes in starch and reducing sugar (calculated as glucose) contents.
Physicochemical and enzymatic analyses.
pH was determined using a Hach pH meter equipped with a pH probe. The total titratable acidity was determined by titration with 0.1 M NaOH, exhibiting a titration endpoint of pH 8.2. Changes in total starch (23) and reducing sugar (24) contents, as well as the dynamics of glucoamylase and amylase (25), CMCase (26), and β-glucosidase (27), were monitored as previously described. The details of these analyses are described in the supplemental material.
DNA isolation and qPCR of 16S/18S rRNA and β-glucosidase genes.
Direct DNA extraction from samples was performed using a soil DNA kit (Omega Bio-Tek, Norcross, GA, USA) in accordance with the manufacturer's instructions. qPCR of the 16S/18S rRNA and β-glucosidase genes was performed in quadruplicate as described in the supplemental material. Melting curve analysis and agarose gel electrophoresis confirmed the specificity of the amplification.
β-Glucosidase gene clone libraries.
Clone libraries targeting the β-glucosidase gene from all the samples were constructed to analyze the cellulolytic communities in each sample. Details of the construction of family 1 and 3 β-glucosidase gene libraries are described in the supplemental material. The representative sequences obtained and the closest matching GenBank reference sequences were used to construct the phylogenetic tree via the neighbor-joining (NJ) method with a bootstrap value of 1,000.
Illumina MiSeq sequencing of 16S/18S rRNA genes.
Direct DNA extraction from the samples was performed using a soil DNA kit (Omega Bio-Tek, Norcross, GA, USA). The V4 regions of the 16S and 18S rRNA genes were amplified as previously reported (28, 29). Illumina MiSeq sequencing of PCR amplicons is detailed in the supplemental material.
Data processing and analyses.
Paired-end reads from the original DNA fragments were merged using FLASH (30) and assigned to each sample with unique barcodes. Sequence analyses were performed using the UPARSE software package (v7.0.1001) with the UPARSE-OTU and UPARSE-OTUref algorithms (31). Sequences with >97% similarity were assigned to the same operational taxonomic units (OTUs). Representative sequences were picked for each OTU, and RDP classifier (version 2.2) was used to annotate the taxonomic information for each representative sequence. Alpha rarefaction was performed in QIIME (version 1.7.0) by using the Chao1 estimates of the species abundance, observed species estimates of the amount of unique OTUs found in each sample, and Shannon index (32). Cluster analysis was preceded by principal-component analysis (PCA). For beta diversity, QIIME calculated both the unweighted and weighted UniFrac distances, and the weighted UniFrac distance was used for principal-coordinate analysis (PCoA) and unweighted pair group method with arithmetic mean (UPGMA) clustering (33, 34). To identify the differences in microbial communities between the two groups, similarities were analyzed using the Bray-Curtis dissimilarity distance matrices (35).
DGGE fingerprints and phylogenetic analysis.
DGGE analysis of microbial diversity was performed as previously reported (9). Details of these analyses are described in the supplemental material. DGGE band patterns were scored on the basis of the band intensities, and the intensities of all distinct bands in each DGGE pattern were quantified by using the trace quantity option of Quantity One 4.6.1 (Bio-Rad). Cluster analysis was conducted using the UPGMA method, whereas PCA was performed with SPSS Statistics 19.0. The obtained sequences were compared to the GenBank database with the BLAST program. Phylogenetic analysis was conducted with MEGA 5.0 through the NJ tree with a bootstrap value of 1,000.
Nucleotide sequence accession numbers.
The sequences obtained from β-glucosidase gene clone libraries were deposited in the NCBI GenBank database under accession numbers KP663796 to KP663828. Results from the sequence read analysis are available through the GenBank Sequence Read Archive (SRA) database under BioProject number PRJNA272559. The sequences obtained from DGGE were deposited in GenBank under accession numbers KP663746 to KP663795.
RESULTS
Dynamics of physicochemical and enzymatic activities.
The pH steadily increased from 4.3 to 6.4 on days 1 to 25 and slightly declined to 6.3 on day 30 (Fig. 1b). The titratable acidity slightly increased from days 1 to 3 and reached the maximum value of about 0.19 mmol g−1 on day 3, followed by a decline (Fig. 1b). On the first 3 days, the contents of total starch and reducing sugar sharply declined from the initial 663.0 mg g−1 to 600.1 mg g−1 and from 56.2 mg g−1 to 39.0 mg g−1 dry starter, respectively. Subsequently, the concentration of the reducing sugar significantly increased and further decreased to 50.6 and 21.0 mg g−1 dry starter on process. Interestingly, the total starch content continuously increased to a final value of approximately 685.4 mg g−1 dry starter.
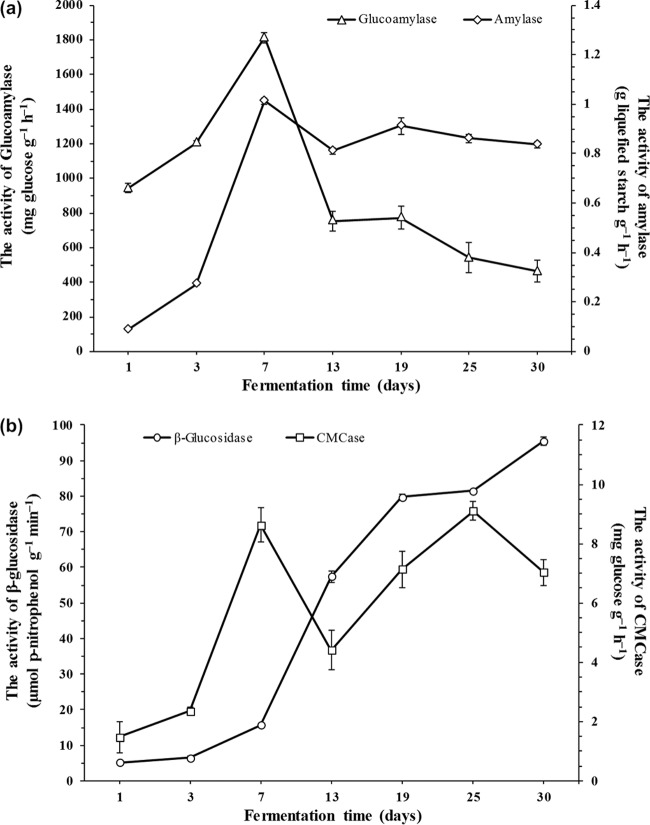
Figure 2 shows the dynamics of enzymatic activities during cereal starter fermentation. Glucoamylase and amylase activities clearly increased from 944.03 mg g−1 h−1 to 1,812.67 mg g−1 h−1 glucose and from 0.08 g g−1 h−1 to 1.01 g g−1 h−1 liquefied starch during the first 7 days, respectively. Subsequently, the glucoamylase activity sharply dropped to 463.92 mg g−1 h−1 glucose, and the amylase activity slightly decreased to 0.84 g g−1 h−1 liquefied starch at the end of fermentation (Fig. 2a). The CMCase activity also clearly increased from 1.47 mg g−1 h−1 to 8.63 mg g−1 h−1 glucose, subsequently decreased significantly, further increased to 4.40 and 9.09 mg g−1 h−1 glucose on days 13 and 25, respectively, and finally decreased to 7.03 mg g−1 h−1 glucose on day 30 (Fig. 2b). However, the β-glucosidase activity continuously and significantly increased from 5.23 μmol g−1 min−1 to 95.50 μmol g−1 min−1 p-nitrophenol during the whole fermentation process (Fig. 2b). These discrepancies in the dynamics of the physicochemical and enzymatic activities suggested that both microbial growth and metabolism became complex during the cereal starter fermentation process.
FIG 2.
Dynamics of amylase and glucoamylase (a) as well as CMCase and β-glucosidase (b) activities during the cereal starter fermentation process.
Quantification of 16S/18S rRNA and β-glucosidase genes.
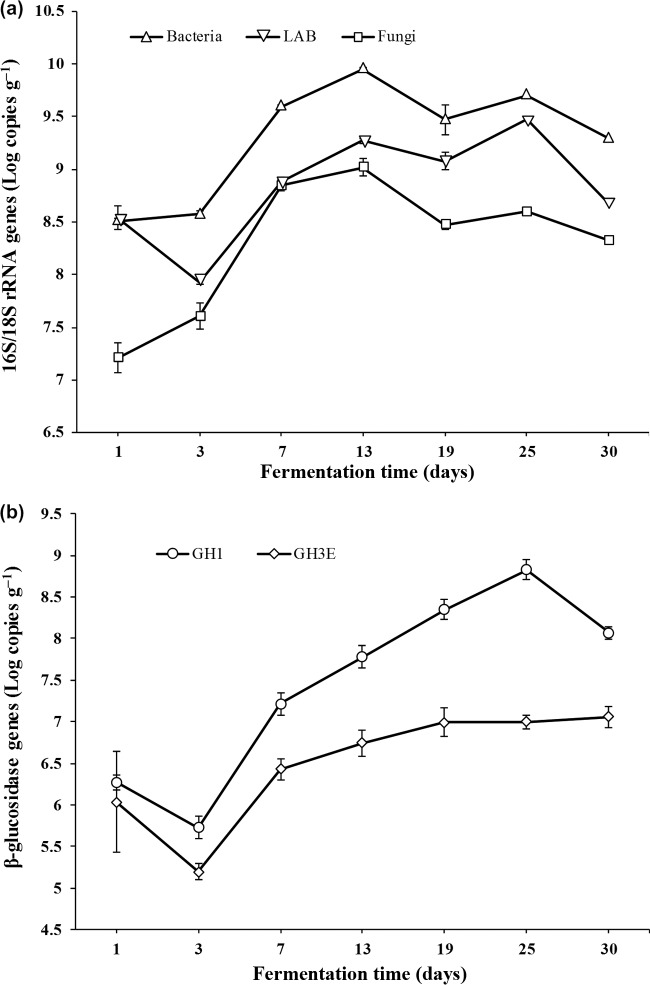
The abundances of total bacteria, lactic acid bacteria (LAB), fungi, and cellulolytic communities during cereal starter fermentation were measured by quantification of the respective 16S/18S rRNA and β-glucosidase genes (Fig. 3). The quantities of total bacteria and fungi showed a similar trend, with a rapid increase from the initial 8.51 and 7.21 log copies g−1 to 9.95 and 9.02 log copies g−1, respectively, during the first 13 days and then a decrease to 9.29 and 8.33 log copies g−1, respectively, at the end of the fermentation process (Fig. 3a). The LAB amount first declined from 8.54 log copies g−1 to 7.93 log copies g−1 on day 3, reached the highest value of 9.48 log copies g−1 on day 25, and finally decreased to 8.68 log copies g−1 on day 30 (Fig. 3a). The family 1 β-glucosidase gene abundance from bacteria and fungi and the family 3 β-glucosidase gene abundance from fungi declined from 6.27 and 6.03 log copies g−1 to 5.72 and 5.19 log copies g−1 during the first 3 days, respectively. Subsequently, the family 1 β-glucosidase gene abundance significantly increased to 8.83 log copies g−1 on day 25 and declined to 8.07 log copies g−1 on day 30, whereas the family 3 β-glucosidase gene abundance gradually increased to 7.06 log copies g−1 until the end of the fermentation process (Fig. 3b).
FIG 3.
Dynamics of 16S/18S rRNA gene (a) and β-glucosidase gene (b) abundances during the cereal starter fermentation process. Specific primers were used in qPCR: GH1, family 1 β-glucosidase genes from bacteria and fungi; GH3E, family 3 β-glucosidase genes from fungi.
Clone libraries of cellulolytic communities targeting the β-glucosidase gene.
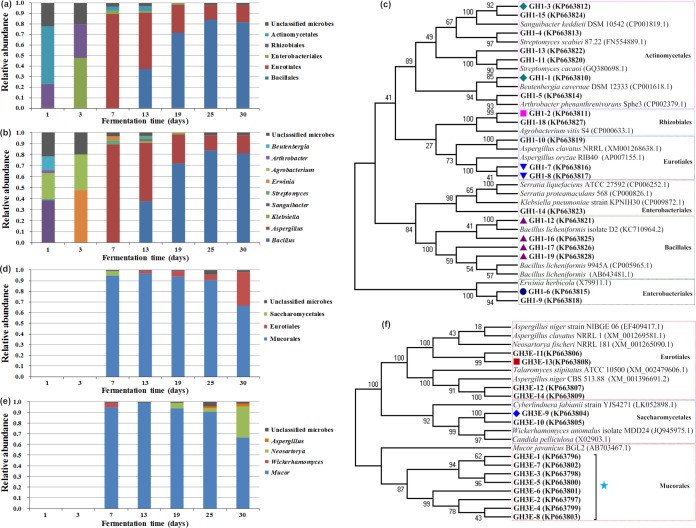
In this study, the family 1 β-glucosidase gene segments were amplified successfully from all the samples, and seven clone libraries were constructed. More than 80 clones from each library were randomly selected, and a total of 637 clones were subjected to restriction fragment length polymorphism (RFLP) analysis. A total of 132 clones were screened by RFLP and sequenced, resulting in 11 OTUs at 97% nucleotide similarity (Fig. 4b). Overall, phylogenetic analysis indicated that the family 1 β-glucosidase gene sequences were clustered within five orders (Fig. 4c) and that there were significant dynamic differences in relative structures at the order (Fig. 4a) and genus (Fig. 4b) levels during the fermentation process. On day 1, the Actinomycetales phylotypes (mainly Sanguibacter and Beutenbergia genera) were the most abundant order, which constituted 54.79% of all the microbes present. On day 3, they disappeared and were detected again only on days 7 and 13. Interestingly, Rhizobiales (mainly Agrobacterium) dominated only on days 1 and 3 and disappeared until the end of fermentation. Enterobacteriales (mainly Erwinia) dominated and comprised 47.69% on day 3, then sharply declined, and reappeared only from days 7 to 19 at low frequencies. Meanwhile, the Eurotiales phylotypes (mainly Aspergillus) first appeared on day 7 and became prominent from days 7 to 30, with relative proportions decreased from the initial 89.29% to 16.22%. Moreover, the Bacillales phylotypes (mainly Bacillus) were first found on day 13 and became another predominant order from days 13 to 30, with the relative abundance gradually increasing from 37.50% to 81.08%. Shifts in Actinomycetales and Enterobacteriales also were observed at the genus level during the fermentation (Fig. 4b).
FIG 4.
Analysis of cellulolytic communities targeting β-glucosidase genes by clone library construction. (a to c) Distribution and relative abundance of cellulolytic communities targeting family 1 β-glucosidase genes from bacteria and fungi at the order (a) and genus (b) levels and NJ phylogenetic tree based on partial family 1 β-glucosidase genes from bacteria and fungi and reference sequences from GenBank (c). ⬥, specific dominant species on day 1; ●, specific dominant species on day 3; ■, specific dominant species on days 1 and 3; ▼, specific dominant species from day 7 to 30; ▲, specific dominant species from day 13 to 30. (d to f) Distribution and relative abundance of cellulolytic communities targeting family 3 β-glucosidase genes from fungi at the order (d) and genus (e) levels (family 3 β-glucosidase genes from the fungi cannot be amplified on days 1 and 3) and NJ phylogenetic tree based on partial family 3 β-glucosidase genes from fungi and reference sequences from GenBank (f). ♦, specific dominant species on day 7; ■, specific dominant species from day 19 to 30; ⭑, specific dominant species from day 7 to 30.
For the fungal family 3 β-glucosidase gene clone library construction, except with days 1 and 3, five clone libraries were constructed, and more than 80 clones from each library were randomly selected. Thus, a total of 472 clones were analyzed by RFLP. A total of 99 clones were screened by RFLP and sequenced, resulting in only four OTUs at 97% nucleotide similarity (Fig. 4e). Overall, phylogenetic analysis indicated that the fungal family 3 β-glucosidase gene sequences were grouped into three orders (Fig. 4f). Mucorales (mainly Mucor) represented the largest fraction, ranging from 94.90% on day 7 to 90.63% on day 25 and finally declining to 66.30% on day 30 (Fig. 4d). Eurotiales was also detected from days 13 to 25 at low relative abundances and represented the other dominant phylotype on day 30, with a relative abundance of 31.52% (Fig. 4d). However, the Saccharomycetales group was retrieved only on day 7 (4.08%) (Fig. 4d).
Illumina MiSeq sequencing analysis.
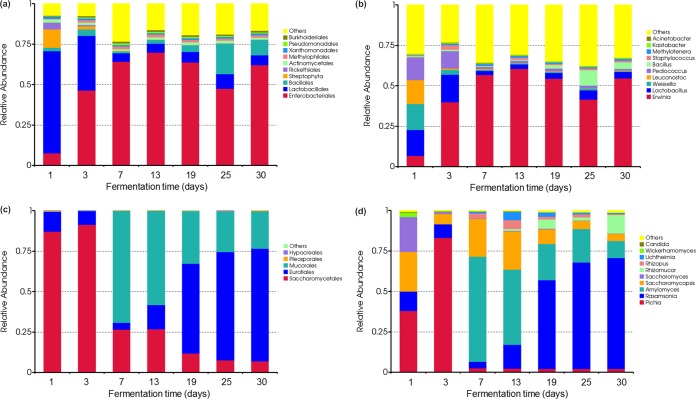
The bacterial and fungal dynamics during the fermentation processes were investigated by Illumina MiSeq sequencing analysis. For bacterial diversity and dynamics, about 24,266 to 70,457 effective tags, with different phylogenetic OTUs ranging from 617 to 721 via 97% sequence identity cutoff, were obtained from the seven samples after filtering the low-quality reads, trimming the adapters, barcodes, and primers, and detecting chimeras (Table 1). The rarefaction curves tended to approach the saturation plateau, which indicated that almost all prokaryotic communities were well represented (see Fig. S1a in the supplemental material). Taxonomic affiliations of 97% sequence similarity clusters demonstrated a distinct taxonomic shift during the fermentation process at the order (Fig. 5a) and genus (Fig. 5b) levels. The 10 most dominant orders constituted 76.63% to 92.75% of all the sequences (Fig. 5a). The phylotypes of Enterobacteriales and Lactobacillales (>5% of total sequences) dominated the whole fermentation process, with the relative abundance dramatically increased from the initial 7.71% to 62.08% and decreased from the initial 63.10% to 6.20% (Fig. 5a). Streptophyta (Cyanobacteria) was another abundant division, which accounted for 11.23% on day 1 and was retrieved at low frequencies (<2% of total sequences) until the end of fermentation (Fig. 5a), whereas Bacillales constituted only 1.05% to 4.20% from days 1 to 19 and clearly increased and dominated on days 25 and 30, with relative abundances of 18.80% and 9.34% (Fig. 5a), respectively. Variations in some small proportions in the groups Rickettsiales (0.04% to 4.20%), Actinomycetales (1.04% to 1.91%), Methylophilales (0.52% to 1.66%), Xanthomonadales (0.94% to 1.14%), Pseudomonadales (0.30% to 1.05%), and Burkholderiales (0.48% to 1.03%) were observed during the fermentation process (Fig. 5a).
TABLE 1.
Observed 16S rRNA Illumina MiSeq sequencing results and alpha diversity indices in samplesa
| Sample day | No. of: |
Q20 (%) | Q30 (%) | GC % | Effective % | Alpha diversity |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Raw tags | Clean tags | Effective tags | Nucleotides | OTU (97%) | Chao1 (97%) | Shannon (97%) | |||||
| 1 | 25,085 | 25,045 | 24,266 | 6,138,170 | 99.20 | 97.25 | 52.60 | 95.00 | 617.0 | 739.277 | 4.566 |
| 3 | 69,549 | 69,414 | 67,694 | 17,126,790 | 99.19 | 97.17 | 54.41 | 95.58 | 721.0 | 1,092.874 | 4.435 |
| 7 | 71,502 | 71,345 | 70,457 | 17,791,638 | 99.28 | 97.41 | 53.22 | 96.79 | 646.0 | 1,012.259 | 3.522 |
| 13 | 69,784 | 69,645 | 68,623 | 17,339,805 | 99.24 | 97.32 | 54.32 | 96.62 | 645.0 | 914.116 | 3.760 |
| 19 | 70,895 | 70,771 | 69,798 | 17,637,827 | 99.23 | 97.31 | 54.00 | 96.53 | 697.0 | 937.497 | 4.009 |
| 25 | 69,167 | 69,010 | 67,438 | 16,960,709 | 99.26 | 97.35 | 54.24 | 95.63 | 711.0 | 1,107.239 | 4.429 |
| 30 | 71,212 | 71,074 | 69,984 | 17,654,790 | 99.25 | 97.35 | 54.44 | 96.23 | 713.0 | 1,140.634 | 4.037 |
Q20, 99% accuracy of effective tags; Q30, 99.9% accuracy of effective tags; GC, GC content of effective tags.
FIG 5.
Dynamics of relative abundances of the major bacterial orders (a) and genera (b) belonging to Enterobacteriales, Lactobacillales, and Bacillales and fungal orders (c) and genera (d) belonging to Saccharomycetales, Eurotiales, and Mucorales in samples as obtained by Illumina MiSeq sequencing targeting the V4 regions of the 16S and 18S rRNA genes, respectively. The abundance is presented as of percentage of total effective bacterial sequences. The abundances of bacterial “other” orders and genera were <1% in all samples; a more detailed overview of bacterial “other” orders is shown in Fig. S8 in the supplemental material. Fungal “other” orders were hardly detected in samples, and the abundances of fungal “other” genera were 0.04% in all samples; a more detailed overview of bacterial “other” genera is shown in Fig. S9 in the supplemental material.
For fungal diversity and dynamics, approximately 37,617 to 105,402 effective tags, with OTUs ranging from 14 to 17 via 97% sequence identity cutoff, were obtained from the seven samples after quality control (Table 2). Rationality in fungal communities conformed to the rarefaction curves approaching the saturation plateau (see Fig. S1b in the supplemental material). Taxonomic affiliations of 97% sequence similarity clusters demonstrated a distinct taxonomic shift during the fermentation process at the order (Fig. 5c) and genus (Fig. 5d) levels. Saccharomycetales, Eurotiales, and Mucorales comprised >99% of all the sequences (Fig. 5c). Saccharomycetales and Eurotiales dominated the whole fermentation process, and Saccharomycetales represented the largest fraction, in the range of 87.12% to 91.37%, on days 1 to 3 and significantly declined to 6.90% on day 30. However, Eurotiales first decreased from 12.38% on day 1 to 4.15% on day 7 and gradually increased to 69.72% on day 30 (Fig. 5c). Mucorales accounted for only 0.16% and 0.10% on days 1 and 3, respectively, then significantly increased to 69.22% on day 7, and dominated until the end of fermentation, with the relative abundance gradually decreasing to 23.36% on day 30 (Fig. 5c). Furthermore, changes in some small proportions in the groups Pleosporales (0.01% to 0.32%) and Hypocreales (0% to 0.01%) were observed during the fermentation process (Fig. 5c).
TABLE 2.
Observed 18S rRNA Illumina MiSeq sequencing results and alpha diversity indices in samplesa
| Sample day | No. of: |
Q20 (%) | Q30 (%) | GC % | Effective % | Alpha diversity |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Raw tags | Effective tags | Nucleotides | OTU (97%) | Chao1 (97%) | Shannon (97%) | |||||
| 1 | 48,635 | 48,511 | 14,772,253 | 98.93 | 96.62 | 49.94 | 97.38 | 16.0 | 16.000 | 2.236 |
| 3 | 38,801 | 38,717 | 11,225,388 | 98.99 | 96.83 | 46.47 | 97.70 | 14.0 | 20.000 | 1.146 |
| 7 | 45,848 | 45,760 | 14,061,950 | 99.13 | 97.11 | 42.41 | 98.38 | 17.0 | 17.333 | 1.543 |
| 13 | 37,695 | 37,617 | 11,559,725 | 99.07 | 96.93 | 43.31 | 98.30 | 16.0 | 17.500 | 2.181 |
| 19 | 105,647 | 105,402 | 32,166,208 | 99.03 | 96.88 | 46.34 | 97.64 | 15.0 | 16.000 | 1.996 |
| 25 | 104,000 | 103,748 | 31,697,727 | 98.98 | 96.74 | 47.14 | 97.31 | 16.0 | 16.000 | 1.608 |
| 30 | 50,934 | 50,807 | 15,423,370 | 99.02 | 96.85 | 47.41 | 97.60 | 14.0 | 14.000 | 1.611 |
Q20, 99% accuracy of effective tags; Q30, 99.9% accuracy of effective tags; GC, GC content of effective tags.
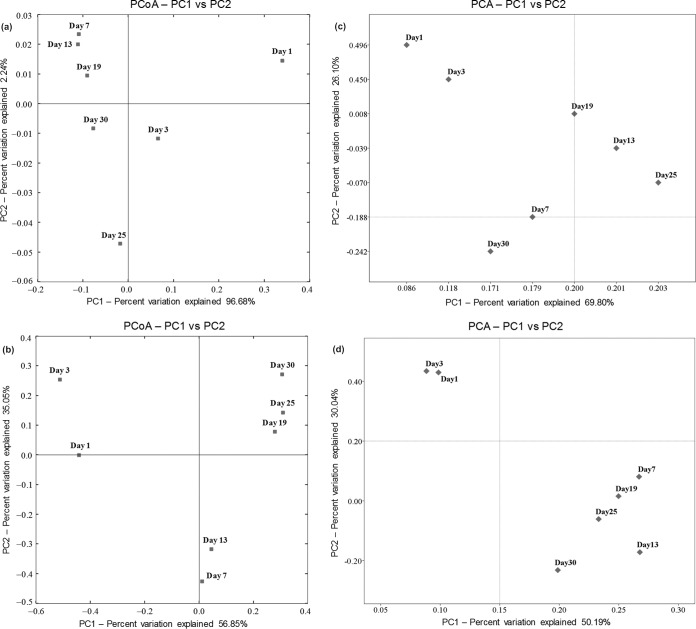
To further confirm whether our sequencing results were sufficient to analyze the food fermentation ecosystem, alpha diversity indexes (number of OTUs, Chao 1, and Shannon index) were determined, and these indexes demonstrated that the richness diversity varied during the fermentation (Tables 1 and 2). On the basis of the Shannon index, both bacterial and fungal richness diversities were significantly decreased during days 1 to 7 and days 1 to 3 and subsequently increased until days 25 and 13; these diversities then declined and became constant until the end of the fermentation process (Tables 1 and 2). PCoA and cluster analyses were conducted to evaluate the similarities in microbial communities, and similar clustering patterns were observed. Both the PCoA and cluster analyses indicated obvious differences in the bacterial communities during days 1 to 3, and the largest change occurred on days 3 to 7 and days 19 to 25. Clusters tended to form during days 3 and 25, and clusters formed on days 7 and 13 and days 19 and 30 as well (Fig. 6a; see Fig. S7a in the supplemental material). These results were consistent with the bacterial heat map of beta diversity based on weighted UniFrac distance (see Fig. S3a in the supplemental material), as expected from the unique taxonomic classifications of bacteria (Fig. 5a). However, for similarity analysis in fungal communities, the largest change occurred during days 3 to 7 and days 13 to 19; days 1 and 3, days 7 and 13, and days 19, 25, and 30 clustered with each other (Fig. 6b; see Fig. S7b in the supplemental material), which was consistent with the fungal heat map of beta diversity based on the Bray-Curtis distance (see Fig. S3b in the supplemental material), as expected from the unique taxonomic classifications of fungi (Fig. 5c).
FIG 6.
PCoA/PCA of microbial communities in samples. (a) Weighted UniFrac distance PCoA of bacterial communities in samples as obtained by Illumina MiSeq sequencing; (b) Bray-Curtis distance PCoA of fungal communities in samples as obtained by Illumina MiSeq sequencing; (c and d) PCA of DGGE profiles of partial 16S rRNA genes from bacteria (c) and 18S rRNA genes from fungi (d) in samples.
Furthermore, to clearly display the shift in microbial types and abundances during the fermentation process, including the relatively low abundance in each stage, the hierarchical heat map was based on phylogenetic classification of the top 35 most abundant bacteria (see Fig. S4 in the supplemental material) and the major fungi (see Fig. S5 in the supplemental material) at the genus level.
Comparison of Illumina MiSeq sequencing with PCR-DGGE and clone library analysis.
In this study, the microbiota dynamics during fermentation were also evaluated by DGGE analysis (see Results in the supplemental material). Overall, the predominant microbes revealed by the Illumina MiSeq sequencing and PCR-DGGE methods were generally the same at the order and genus levels (Fig. 5; see Fig. S6 in the supplemental material). However, DGGE analysis also provided some new elements compared with Illumina MiSeq sequencing; for example, Pantoea was detected as one of the predominant genera during the fermentation process by DGGE analysis (see Fig. S6 in the supplemental material) but was not detected with Illumina MiSeq sequencing (data not shown). Nevertheless, Illumina MiSeq sequencing provided an in-depth analysis of the bacterial and fungal communities compared with those determined with PCR-DGGE analysis, because some phylotypes were revealed by the Illumina MiSeq sequencing analysis but not by PCR-DGGE. For instance, relatively rare bacterial phylotypes of Streptophyta, Rickettsiales, Actinomycetales, Methylophilales, Xanthomonadales, Pseudomonadales, and Burkholderiales (Fig. 5a) and fungal phylotypes of Pleosporales and Hypocreales were shown only by Illumina MiSeq sequencing analysis (Fig. 5c).
PCA and cluster analyses were also performed to compare the similarities and differences in bacterial (Fig. 6c; see Fig. S7c in the supplemental material) and fungal (Fig. 6d; see Fig. S7d in the supplemental material) DGGE profiles at different fermentation stages. Although slight variances existed, cluster analysis indicated that the clusters formed on day 3 showed similarities with those formed on days 25 and 7 as shown by Illumina MiSeq sequencing (see Fig. S7a in the supplemental material) and PCR-DGGE (see Fig. S7c in the supplemental material), respectively; however, comparisons of PCA (PCoA) and cluster analysis presented similar trends based on the results of both methods (Fig. 6; see Fig. S7 in the supplemental material).
Moreover, the dominant cellulolytic microbial communities revealed by clone library analysis were determined by Illumina MiSeq sequencing analysis at the order level (Fig. 4 and 5; see Fig. S2 in the supplemental material). Nevertheless, some relatively rare genera, including Mucor, Neosartorya, and Agrobacterium, were more easily recognized by the clone library targeting functional genes (Fig. 4 and 5; see Fig. S2 in the supplemental material). These discrepancies were probably attributable to primer specificity issues similar to those previous reported (36, 37). Consequently, the Illumina MiSeq sequencing provided high detection of microbial communities and was superior to the PCR-DGGE in detecting the rare communities. However, the clone library targeting β-glucosidase genes was a prerequisite for integrant diversity of cellulolytic communities involved in cereal starter fermentation.
DISCUSSION
In our study, bacterial communities clearly changed in association with environmental conditions at the order level. Enterobacteriales members have been isolated from various high-temperature ecosystems, having a remarkable capacity to adapt to a wide range of temperatures and moisture levels (38, 39). Although the temperature reached a maximum of about 60°C (stage 4) and the moisture declined to 12% during fermentation (Fig. 1), Enterobacteriales also dominated the whole fermentation process effectively, with the relative abundance significantly increased (Fig. 5a; see Fig. S6a in the supplemental material). Thermophilic Bacillales, which can secrete various degradative enzymes, including amylases (40), and can better survive under low-moisture and high-temperature conditions than other bacteria (41), was another group that was present during the whole fermentation process, and it became predominant on days 25 and 30 (Fig. 5a; see Fig. S6a in the supplemental material). These results were consistent with previous studies on fen-daqu fermentation process (42). LAB play a key role in flavor formation during cereal starter fermentation (43). However, the major LAB Lactobacillales was the most abundant division on days 1 and 3, sharply decreased in relative abundance with the increase in temperature to about 40°C to 50°C from days 3 to 7, and remained stable until the end of the fermentation process (Fig. 5a). These results may be accurate for the growth of Lactobacillales members, such as the genus Leuconostoc (Fig. 5b), which were restrained when the temperature was >40°C (44, 45) and also corresponded to the pH increase and acidity decrease during days 3 to 7 (Fig. 1b). Nevertheless, despite the sharp decrease in abundance of LAB (Fig. 3a), there could have been some survivors (Fig. 5b), including the genus Lactobacillus, which was consistent with a previous report (46). Interestingly, endophytic Streptophyta was considered an order of Cyanobacteria (47, 48), which dominated only the initial fermentation process and was retrieved at low frequencies with increased temperature and decreased moisture (Fig. 5a). These results may argue that environmental conditions, including water availability and temperature, had a deleterious effect on these communities (49), and as such, endophytic Streptophyta can be considered nonimportant members during fermentation.
The fungal communities were less complex than the bacterial communities; despite these differences, fungal communities also clearly changed in relation to environmental conditions at the order level. Saccharomycetales represented the largest fraction on days 1 and 3, whereas the relative abundance dramatically declined in correlation with the temperature increase to about 40°C to 50°C during days 3 to 7, which may be due to the reduction of the nonthermotolerant genera of Pichia and Saccharomyces (50, 51) within Saccharomycetales (Fig. 5d; see Fig. S6 in the supplemental material). Nevertheless, Pichia, which was retrieved with low frequencies (Fig. 5d), is considered an ester-producing yeast and associated with flavor formation in combination with LAB (52); the thermotolerant genus Saccharomycopsis within Saccharomycetales also occurred and dominated during the entire fermentation process (Fig. 5d). This genus commonly exists in various starters (42) which demonstrate amylolytic activity to degrade and assimilate raw starch as a carbon source (53). Filamentous fungi, which are commonly involved in solid-state fermentation, are the major contributors to hydrolytic enzymatic activity (54). In the present study, the thermophilic genus Rasamsonia within Eurotiales (55) and the thermotolerant genera Rhizopus, Rhizomucor, and Amylomyces within Mucorales (46, 56), which are considered strong producers of extracellular enzymes such as amylase, glucoamylase, and cellulase in various fermentation starters, survived and were the prevailing filamentous fungi with increased temperature and decreased moisture during the fermentation process (Fig. 5d; see Fig. S6 in the supplemental material). Overall, our results show that the bacterial phylotypes of Enterobacteriales, Lactobacillales, and Bacillales and fungal phylotypes of Saccharomycetales, Eurotiales, and Mucorales were the ultimate survivors.
Taken as a whole, although thermotolerant members may be present in phylotypes of Lactobacillales and Saccharomycetales, the abundances of Lactobacillales and Saccharomycetales continuously declined with decreased incubation temperature after the heating stage. However, thermotolerant Eurotiales and Bacillales were considerably accelerated when the incubation temperature decreased from 53 to 60°C to 35 to 40°C, thereby resulting in dry conditions (Fig. 1 and 5). Mucorales first dominated on day 7 with the incubation temperature at 40°C to 50°C despite the gradual decline in abundance, which indicated that the effect of a high incubation temperature with a corresponding low moisture level was selective in inducing the shift to thermotolerant and drought-resistant communities during the cereal starter fermentation (57, 58).
Cellulolytic species are widely distributed, and the impact of environmental changes on the cellulolytic communities has been extensively investigated in various ecosystems (59, 60). Similarly, all of the cellulolytic phylotypes targeting family 1 β-glucosidase genes from bacteria and fungi detected in this study have been previously identified in litter layer and soil ecosystems (60, 61); furthermore, significant dynamic differences in responses to environmental conditions were obtained during the cereal starter fermentation (Fig. 4). The current results showed that Actinomycetales and Rhizobiales dominated only the initial fermentation stages (days 1 and 3) and then significantly declined and nearly disappeared with the temperature increase to 40°C to 50°C on day 3 (Fig. 4), which indicated that the orders Actinomycetales and Rhizobiales may also be considered nonimportant members during fermentation. These results corroborated previous findings that the genus Agrobacterium within Rhizobiales and the genera Sanguibacter and Beutenbergia within Actinomycetales showed growth inhibition at this incubation temperature (62–64). In contrast, Bacillales and Eurotiales first prevailed at incubation temperatures of 40°C to 50°C and 53°C to 60°C, respectively, and survived (Fig. 4), which indicated that thermotolerant species, mainly the thermotolerant genus Bacillus within Bacillales and the genus Aspergillus within Eurotiales, existed through to the end of fermentation process, consistent with previous findings that thermotolerant Bacillus and Aspergillus were dominant species in high-temperature (>45°C) and low-moisture composting process (20). These findings also suggested that temperature-associated stimulation and cellulose degradation may be related to the shifts in microbial community composition. Furthermore, for family 3 β-glucosidase genes from the fungal clone library, the family 3 β-glucosidase genes cannot be amplified on days 1 to 3, which suggested that they were either below the assay detection limit or in a more restricted distribution. Mucorales, which are considered cellulose decomposers in soils (65), were another retrieved cellulolytic community that dominated even during high-temperature stages on days 7 and 13 and survived until the end of the fermentation (Fig. 4). Overall, our results show that the phylotypes of Bacillales, Eurotiales, and Mucorales were ultimate cellulolytic survivors and that the cellulolytic potential can adapt to variations in environmental conditions by changes in the thermotolerant and drought-resistant community structure during the cereal starter fermentation.
The varieties and activities of enzymes which originate from microbial communities are the main mediators of cereal starter fermentation processes. Studying the dynamics of the activities of some key enzymes in microbial metabolism can be valuable to increase our knowledge of the microbial degradation of various substrates. Amylase and glucoamylase are the major contributors to the liquefaction and saccharification processes. In this study, the activities of amylase and glucoamylase showed a similar trend, and both clearly increased during the first 7 days, after which glucoamylase activity sharply dropped, while amylase activity slightly decreased along with fermentation (Fig. 2a); these discrepancies, as previously reported, can be attributed to the thermal stability of amylase, which was much better than that of glucoamylase at high incubation temperatures of 53°C to 60°C (66). Cellulase is closely related to cellulose degradation and carbon metabolism, and β-glucosidase is involved in the hydrolysis of cellobiose to yield β-glucose (67). Earlier findings confirmed that β-glucosidase activity demonstrated a pattern of evolution similar to that of the total cellulase activity, with an increase in enzymatic activity at the initial stage of composting and a decrease during the curing phase (68, 69). However, we observed that (i) the activity of CMCase significantly increased from days 13 and 25 and (ii) the activity of β-glucosidase continuously and significantly increased during the whole fermentation process (Fig. 2b), which suggested that some thermotolerant species within Bacillales, Eurotiales, and Mucorales can synthesize the β-glucosidase requirement and are involved in the primary decomposition of cellulose (Fig. 5), regardless of whether the temperature increased to the maximum values of 53°C to 60°C. The increase in CMCase activity was also observed previously during the latter stage of municipal solid waste composting (70), and the increase in the β-glucosidase activity can be attributed to the increase of β-glucosidase gene copies, except from days 1 to 3 (Fig. 3b), despite the slight decrease in bacterial and fungal populations on days 13 to 25 (Fig. 3a).
Generally, starch is used as a renewable raw material and can be hydrolyzed by amylase and finally converted to glucose by glucoamylase. However, a continuous and clear increase in the total starch was observed the during the cereal starter fermentation on days 3 to 30 (Fig. 1). This increase may be due to (i) the decline in amylase activity on days 7 to 30 despite the increase on days 1 to 7 (Fig. 2) and (ii) the crude multienzyme, which was composed of non-starch polysaccharide-hydrolyzing enzymes, including cellulase and hemicellulose, acting synergistically to release the trapped starch granules from the fibrous cell wall structure and making starch more accessible (71, 72). This correlates with (i) thermotolerant cellulolytic survivors of Bacillales, Eurotiales, and Mucorales, which dominated from on 7 to 30 (Fig. 4), (ii) a significant increase in CMCase activity on days 13 to 25 (Fig. 2b), and (iii) a continuous and significant increase in β-glucosidase activity during the fermentation process (Fig. 2b). These results indicated that the shift in cellulolytic microbial communities played a key role by improving the availability of raw starch material during cereal starter fermentation.
In conclusion, a succession of microbial composition and cellulolytic communities was evaluated during the cereal starter fermentation process, and the results obtained support that shifts in community composition were definitely affected by environmental conditions. Our results support that the structure of the microbial community can be changed to the characteristics for thermophilic and drought-resistant communities after the high-temperature incubation stage. Elaboration of the interplay between enzymatic activities, microbial patterns, and environmental conditions can provide novel insights into the role of microbial communities in the fermentation process. These exploratory findings pose a paradigm shift in our understanding of food fermentation systems, in which cellulolytic microbial communities play key roles in improving raw material availability and productivity of the final products.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported by the National Natural Foundation of China (grant 31271924).
We declare that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01325-15.
REFERENCES
- 1.Piperno DR, Weiss E, Holst I, Nadel D. 2004. Processing of wild cereal grains in the Upper Palaeolithic revealed by starch grain analysis. Nature 430:670–673. doi: 10.1038/nature02734. [DOI] [PubMed] [Google Scholar]
- 2.Bokulich NA, Bamforth CW. 2013. The microbiology of malting and brewing. Microbiol Mol Biol Rev 77:157–172. doi: 10.1128/MMBR.00060-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertoldo C, Antranikian G. 2002. Starch-hydrolyzing enzymes from thermophilic archaea and bacteria. Curr Opin Microbiol 6:151–160. [DOI] [PubMed] [Google Scholar]
- 4.Leroy F, De Vuyst L. 2004. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci Technol 15:67–78. doi: 10.1016/j.tifs.2003.09.004. [DOI] [Google Scholar]
- 5.Holzapfel W. 2002. Appropriate starter culture technologies for small-scale fermentation in developing countries. Int J Food Microbiol 75:197–212. doi: 10.1016/S0168-1605(01)00707-3. [DOI] [PubMed] [Google Scholar]
- 6.Brandt MJ. 2014. Starter cultures for cereal based foods. Food Microbiol 37:41–43. doi: 10.1016/j.fm.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Wu X-H, Zheng X-W, Han B-Z, Vervoort J, Nout MR. 2009. Characterization of Chinese liquor starter,“Daqu,” by flavor type with 1H NMR-based nontargeted analysis. J Agric Food Chem 57:11354–11359. doi: 10.1021/jf902881p. [DOI] [PubMed] [Google Scholar]
- 8.Zheng XW, Tabrizi MR, Nout M, Han BZ. 2011. Daqu—a traditional Chinese liquor fermentation starter. J Inst Brewing 117:82–90. doi: 10.1002/j.2050-0416.2011.tb00447.x. [DOI] [Google Scholar]
- 9.Li P, Li S, Cheng L, Luo L. 2014. Analyzing the relation between the microbial diversity of DaQu and the turbidity spoilage of traditional Chinese vinegar. Appl Microbiol Biotechnol 98:6073–6084. doi: 10.1007/s00253-014-5697-4. [DOI] [PubMed] [Google Scholar]
- 10.Hurkman WJ, McCue KF, Altenbach SB, Korn A, Tanaka CK, Kothari KM, Johnson EL, Bechtel DB, Wilson JD, Anderson OD. 2003. Effect of temperature on expression of genes encoding enzymes for starch biosynthesis in developing wheat endosperm. Plant Sci 164:873–881. doi: 10.1016/S0168-9452(03)00076-1. [DOI] [Google Scholar]
- 11.Yang J, Zhang J, Wang Z, Xu G, Zhu Q. 2004. Activities of key enzymes in sucrose-to-starch conversion in wheat grains subjected to water deficit during grain filling. Plant Physiol 135:1621–1629. doi: 10.1104/pp.104.041038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haichar FEZ, Achouak W, Christen R, Heulin T, Marol C, Marais MF, Mougel C, Ranjard L, Balesdent J, Berge O. 2007. Identification of cellulolytic bacteria in soil by stable isotope probing. Environ Microbiol 9:625–634. doi: 10.1111/j.1462-2920.2006.01182.x. [DOI] [PubMed] [Google Scholar]
- 13.Schellenberger S, Kolb S, Drake HL. 2010. Metabolic responses of novel cellulolytic and saccharolytic agricultural soil bacteria to oxygen. Environ Microbiol 12:845–861. doi: 10.1111/j.1462-2920.2009.02128.x. [DOI] [PubMed] [Google Scholar]
- 14.Barnard RL, Osborne CA, Firestone MK. 2013. Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J 7:2229–2241. doi: 10.1038/ismej.2013.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singhania RR, Patel AK, Sukumaran RK, Larroche C, Pandey A. 2013. Role and significance of beta-glucosidases in the hydrolysis of cellulose for bioethanol production. Bioresour Technol 127:500–507. doi: 10.1016/j.biortech.2012.09.012. [DOI] [PubMed] [Google Scholar]
- 16.Alef K, Nannipieri P. 1995. Methods in applied soil microbiology and biochemistry. Academic Press, London, United Kingdom. [Google Scholar]
- 17.Berlemont R, Martiny AC. 2013. Phylogenetic distribution of potential cellulases in bacteria. Appl Environ Microbiol 79:1545–1554. doi: 10.1128/AEM.03305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cañizares R, Benitez E, Ogunseitan OA. 2011. Molecular analyses of β-glucosidase diversity and function in soil. Eur J Soil Biol 47:1–8. doi: 10.1016/j.ejsobi.2010.11.002. [DOI] [Google Scholar]
- 19.Moreno B, Cañizares R, Nuñez R, Benitez E. 2013. Genetic diversity of bacterial β-glucosidase-encoding genes as a function of soil management. Biol Fertil Soils 49:735–745. doi: 10.1007/s00374-012-0765-3. [DOI] [Google Scholar]
- 20.Li H, Xu X, Chen H, Zhang Y, Xu J, Wang J, Lu X. 2013. Molecular analyses of the functional microbial community in composting by PCR-DGGE targeting the genes of the β-glucosidase. Bioresour Technol 134:51–58. doi: 10.1016/j.biortech.2013.01.077. [DOI] [PubMed] [Google Scholar]
- 21.Wang HY, Gao YB, Fan QW, Xu Y. 2011. Characterization and comparison of microbial community of different typical Chinese liquor Daqus by PCR-DGGE. Lett Appl Microbiol 53:134–140. doi: 10.1111/j.1472-765X.2011.03076.x. [DOI] [PubMed] [Google Scholar]
- 22.Zheng XW, Yan Z, Han BZ, Zwietering MH, Samson RA, Boekhout T, Robert Nout M. 2012. Complex microbiota of a Chinese “Fen” liquor fermentation starter (Fen Daqu), revealed by culture-dependent and culture-independent methods. Food Microbiol 31:293–300. doi: 10.1016/j.fm.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 23.Bravo L, Siddhuraju P, Saura-Calixto F. 1998. Effect of various processing methods on the in vitro starch digestibility and resistant starch content of Indian pulses. J Agric Food Chem 46:4667–4674. doi: 10.1021/jf980251f. [DOI] [Google Scholar]
- 24.Miller GL. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- 25.Bernfeld P, Colowick S, Kaplan N (ed). 1955. Methods in enzymology, 1st ed Academic Press, New York, NY. [Google Scholar]
- 26.Miller GL, Blum R, Glennon WE, Burton AL. 1960. Measurement of carboxymethylcellulase activity. Anal Biochem 1:127–132. doi: 10.1016/0003-2697(60)90004-X. [DOI] [Google Scholar]
- 27.Herr D, Baumer F, Dellweg H. 1978. Purification and properties of an extracellular β-glucosidase from Lenzites trabea. Appl Microbiol Biotechnol 5:29–36. doi: 10.1007/BF00515684. [DOI] [Google Scholar]
- 28.Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, Buckler ES, Ley RE. 2013. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci U S A 110:6548–6553. doi: 10.1073/pnas.1302837110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheung MK, Au CH, Chu KH, Kwan HS, Wong CK. 2010. Composition and genetic diversity of picoeukaryotes in subtropical coastal waters as revealed by 454 pyrosequencing. ISME J 4:1053–1059. doi: 10.1038/ismej.2010.26. [DOI] [PubMed] [Google Scholar]
- 30.Zhang J, Kobert K, Flouri T, Stamatakis A. 2014. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 32.Kemp PF, Aller JY. 2004. Bacterial diversity in aquatic and other environments: what 16S rDNA libraries can tell us. FEMS Microbiol Ecol 47:161–177. doi: 10.1016/S0168-6496(03)00257-5. [DOI] [PubMed] [Google Scholar]
- 33.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. 2011. UniFrac: an effective distance metric for microbial community comparison. ISME J 5:169. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lozupone CA, Hamady M, Kelley ST, Knight R. 2007. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCune B, Grace JB, Urban DL. 2002. Analysis of ecological communities. MjM Software Design, Gleneden Beach, OR. [Google Scholar]
- 36.Hubert CR, Oldenburg TB, Fustic M, Gray ND, Larter SR, Penn K, Rowan AK, Seshadri R, Sherry A, Swainsbury R. 2012. Massive dominance of Epsilonproteobacteria in formation waters from a Canadian oil sands reservoir containing severely biodegraded oil. Environ Microbiol 14:387–404. doi: 10.1111/j.1462-2920.2011.02521.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roh SW, Kim K-H, Nam Y-D, Chang H-W, Park E-J, Bae J-W. 2010. Investigation of archaeal and bacterial diversity in fermented seafood using barcoded pyrosequencing. ISME J 4:1–16. doi: 10.1038/ismej.2009.83. [DOI] [PubMed] [Google Scholar]
- 38.Carranza P, Grunau A, Schneider T, Hartmann I, Lehner A, Stephan R, Gehrig P, Grossmann J, Groebel K, Hoelzle LE. 2010. A gel-free quantitative proteomics approach to investigate temperature adaptation of the food-borne pathogen Cronobacter turicensis 3032. Proteomics 10:3248–3261. doi: 10.1002/pmic.200900460. [DOI] [PubMed] [Google Scholar]
- 39.Wei C-L, Chao S-H, Tsai W-B, Lee P-S, Tsau N-H, Chen J-S, Lai W-L, Ching-Yueh Tu J, Tsai Y-C. 2013. Analysis of bacterial diversity during the fermentation of inyu, a high-temperature fermented soy sauce, using nested PCR-denaturing gradient gel electrophoresis and the plate count method. Food Microbiol 33:252–261. doi: 10.1016/j.fm.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 40.Simonen M, Palva I. 1993. Protein secretion in Bacillus species. Microbiol Mol Biol Rev 57:109–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams HE, Crump BC, Kling GW. 2010. Temperature controls on aquatic bacterial production and community dynamics in arctic lakes and streams. Environ Microbiol 12:1319–1333. doi: 10.1111/j.1462-2920.2010.02176.x. [DOI] [PubMed] [Google Scholar]
- 42.Zheng XW, Yan Z, Nout M, Smid EJ, Zwietering MH, Boekhout T, Han J-S, Han B-Z. 2014. Microbiota dynamics related to environmental conditions during the fermentative production of Fen-Daqu, a Chinese industrial fermentation starter. Int J Food Microbiol 182:57–62. doi: 10.1016/j.ijfoodmicro.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 43.Yousif NM, Huch M, Schuster T, Cho G-S, Dirar HA, Holzapfel WH, Franz CM. 2010. Diversity of lactic acid bacteria from Hussuwa, a traditional African fermented sorghum food. Food Microbiol 27:757–768. doi: 10.1016/j.fm.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 44.Murga MLf. Holgado R, Pesce A, Valdez G. 1995. Influence of the incubation temperature on the autolytic activity of Lactobacillus acidophilus. J Appl Microbiol 78:426–429. [DOI] [PubMed] [Google Scholar]
- 45.Cooper R, Collins E. 1978. Influences of temperature on growth of Leuconostoc cremoris. J Dairy Sci 61:1085–1088. doi: 10.3168/jds.S0022-0302(78)83690-X. [DOI] [Google Scholar]
- 46.Wang H, Xu Y. 2015. Effect of temperature on microbial composition of starter culture for Chinese light aroma style liquor fermentation. Lett Appl Microbiol 60:85–91. doi: 10.1111/lam.12344. [DOI] [PubMed] [Google Scholar]
- 47.Lee RE. 2008. Phycology. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 48.Reece JB, Urry LA, Cain ML, Wasserman SA, Minorsky P, Jackson RB. 2011. Campbell biology, 9th ed Benjamin Cummings, San Francisco, CA. [Google Scholar]
- 49.Botella L, Santamaría O, Diez J. 2010. Fungi associated with the decline of Pinus halepensis in Spain. Fungal Divers 40:1–11. doi: 10.1007/s13225-010-0025-5. [DOI] [Google Scholar]
- 50.Gera R, Dhamija S, Gera T, Singh D. 1997. Intergeneric ethanol producing hybrids of thermotolerant Kluyveromyces and non-thermotolerant Saccharomyces cerevisiae. Biotechnol Lett 19:189–194. doi: 10.1023/A:1018380818454. [DOI] [Google Scholar]
- 51.González SS, Gallo L, Climent MD, Barrio E, Querol A. 2007. Enological characterization of natural hybrids from Saccharomyces cerevisiae and S. kudriavzevii. Int J Food Microbiol 116:11–18. doi: 10.1016/j.ijfoodmicro.2006.10.047. [DOI] [PubMed] [Google Scholar]
- 52.Nout M. 2009. Rich nutrition from the poorest—cereal fermentations in Africa and Asia. Food Microbiol 26:685–692. doi: 10.1016/j.fm.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 53.Gonzalez C, Farina J, de Figueroa L. 2008. Optimized amylolytic enzymes production in Saccharomycopsis fibuligera DSM-70554: an approach to efficient cassava starch utilization. Enzyme Microb Technol 42:272–277. doi: 10.1016/j.enzmictec.2007.10.005. [DOI] [Google Scholar]
- 54.Hölker U, Lenz J. 2005. Solid-state fermentation—are there any biotechnological advantages? Curr Opin Microbiol 8:301–306. doi: 10.1016/j.mib.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 55.Houbraken J, Spierenburg H, Frisvad JC. 2012. Rasamsonia, a new genus comprising thermotolerant and thermophilic Talaromyces and Geosmithia species. Antononie Van Leeuwenhoek 101:403–421. doi: 10.1007/s10482-011-9647-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mouchacca J. 2007. Heat tolerant fungi and applied research: addition to the previously treated group of strictly thermotolerant species. World J Microb Biotechnol 23:1755–1770. doi: 10.1007/s11274-007-9426-3. [DOI] [PubMed] [Google Scholar]
- 57.Sheik CS, Beasley WH, Elshahed MS, Zhou X, Luo Y, Krumholz LR. 2011. Effect of warming and drought on grassland microbial communities. ISME J 5:1692–1700. doi: 10.1038/ismej.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berlemont R, Allison SD, Weihe C, Lu Y, Brodie EL, Martiny JB, Martiny AC. 2014. Cellulolytic potential under environmental changes in microbial communities from grassland litter. Front Microbiol 5:639. doi: 10.3389/fmicb.2014.00639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schneider T, Keiblinger KM, Schmid E, Sterflinger-Gleixner K, Ellersdorfer G, Roschitzki B, Richter A, Eberl L, Zechmeister-Boltenstern S, Riedel K. 2012. Who is who in litter decomposition? Metaproteomics reveals major microbial players and their biogeochemical functions. ISME J 6:1749–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Štursová M, Žifčáková L, Leigh MB, Burgess R, Baldrian P. 2012. Cellulose utilization in forest litter and soil: identification of bacterial and fungal decomposers. FEMS Microbiol Ecol 80:735–746. doi: 10.1111/j.1574-6941.2012.01343.x. [DOI] [PubMed] [Google Scholar]
- 61.Bernard L, Mougel C, Maron PA, Nowak V, Lévêque J, Henault C, Haichar F, Berge O, Marol C, Balesdent J. 2007. Dynamics and identification of soil microbial populations actively assimilating carbon from 13C-labelled wheat residue as estimated by DNA-and RNA-SIP techniques. Environ Microbiol 9:752–764. doi: 10.1111/j.1462-2920.2006.01197.x. [DOI] [PubMed] [Google Scholar]
- 62.Groth I, Schumann P, Schuetze B, Augsten K, Kramer I, Stackebrandt E. 1999. Beutenbergia cavernae gen. nov., sp. nov., an l-lysine-containing actinomycete isolated from a cave. Int J Syst Evol Microbiol 49:1733–1740. [DOI] [PubMed] [Google Scholar]
- 63.Baron C, Domke N, Beinhofer M, Hapfelmeier S. 2001. Elevated temperature differentially affects virulence, VirB protein accumulation, and T-pilus formation in different Agrobacterium tumefaciens and Agrobacterium vitis strains. J Bacteriol 183:6852–6861. doi: 10.1128/JB.183.23.6852-6861.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hong SG, Lee YK, Yim JH, Chun J, Lee HK. 2008. Sanguibacter antarcticus sp. nov., isolated from Antarctic sea sand. Int J Syst Evol Microbiol 58:50–52. doi: 10.1099/ijs.0.65031-0. [DOI] [PubMed] [Google Scholar]
- 65.Weber CF, Zak DR, Hungate BA, Jackson RB, Vilgalys R, Evans RD, Schadt CW, Megonigal JP, Kuske CR. 2011. Responses of soil cellulolytic fungal communities to elevated atmospheric CO2 are complex and variable across five ecosystems. Environ Microbiol 13:2778–2793. doi: 10.1111/j.1462-2920.2011.02548.x. [DOI] [PubMed] [Google Scholar]
- 66.Soni SK, Kaur A, Gupta JK. 2003. A solid state fermentation based bacterial α-amylase and fungal glucoamylase system and its suitability for the hydrolysis of wheat starch. Process Biochem 39:185–192. doi: 10.1016/S0032-9592(03)00058-X. [DOI] [Google Scholar]
- 67.Burns RG, Dick RP. 2002. Enzymes in the environment: activity, ecology, and applications. CRC Press, Boca Raton, FL. [Google Scholar]
- 68.Castaldi P, Garau G, Melis P. 2008. Maturity assessment of compost from municipal solid waste through the study of enzyme activities and water-soluble fractions. Waste Manag 28:534–540. doi: 10.1016/j.wasman.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 69.Cunha-Queda A, Ribeiro H, Ramos A, Cabral F. 2007. Study of biochemical and microbiological parameters during composting of pine and eucalyptus bark. Bioresour Technol 98:3213–3220. doi: 10.1016/j.biortech.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 70.Raut M, Prince William S, Bhattacharyya J, Chakrabarti T, Devotta S. 2008. Microbial dynamics and enzyme activities during rapid composting of municipal solid waste—a compost maturity analysis perspective. Bioresour Technol 99:6512–6519. doi: 10.1016/j.biortech.2007.11.030. [DOI] [PubMed] [Google Scholar]
- 71.Galante YM, De Conti A, Monteverdi R. 1998. Application of Trichoderma enzymes in the food and feed industries, p 311–326. In Harman GF, Kubicek CP (ed), Trichoderma and Gliocladium—enzymes, vol 2 Taylor & Francis, London, United Kingdom. [Google Scholar]
- 72.Divya Nair M, Padmaja G, Moorthy S. 2011. Biodegradation of cassava starch factory residue using a combination of cellulases, xylanases and hemicellulases. Biomass Bioenerg 35:1211–1218. doi: 10.1016/j.biombioe.2010.12.009. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.