Abstract
Two glucoamylase-like genes, TVN1315 and Ta0286, from the archaea Thermoplasma volcanium and T. acidophilum, respectively, were expressed in Escherichia coli. The gene products, TVN1315 and Ta0286, were identified as archaeal trehalases. These trehalases belong to the CAZy database family GH15, although they have putative (α/α)6 barrel catalytic domain structures similar to those of GH37 and GH65 family trehalases from other organisms. These newly identified trehalases function within a narrow range of acidic pH values (pH 3.2 to 4.0) and at high temperatures (50 to 60°C), and these enzymes display Km values for trehalose higher than those observed for typical trehalases. These enzymes were inhibited by validamycin A; however, the inhibition constants (Ki) were higher than those of other trehalases. Three TVN1315 mutants, corresponding to E408Q, E571Q, and E408Q/E571Q mutations, showed reduced activity, suggesting that these two glutamic acid residues are involved in trehalase catalysis in a manner similar to that of glucoamylase. To date, TVN1315 and Ta0286 are the first archaeal trehalases to be identified, and this is the first report of the heterologous expression of GH15 family trehalases. The identification of these trehalases could extend our understanding of the relationships between the structure and function of GH15 family enzymes as well as glycoside hydrolase family enzymes; additionally, these enzymes provide insight into archaeal trehalose metabolism.
INTRODUCTION
Trehalose is a stable, colorless, and odor-free disaccharide comprised of two glucose units connected by an α,α-(1,1)-, α,β-(1,1)-, or β,β-(1,1)-linkage. Only the α,α-(1,1)-trehalose form is widespread in nature and biologically active. Trehalose has been found in a wide variety of organisms, including archaea, bacteria, yeast, fungi, insects, crustaceans, invertebrates, and plants, in which trehalose plays a protective role during stress (e.g., osmolarity, heat, oxidation, desiccation, and freezing), is a source of carbon energy, and acts as a signaling molecule in select metabolic pathways (1–4). Additionally, trehalose induces autophagy, facilitating the clearance of aggregation-prone proteins, and has antiapoptotic properties that arise from the enhanced removal of proapoptotic proteins via autophagy (5, 6).
Trehalase (trehalose-glucohydrolase; EC 3.2.1.28) hydrolyzes trehalose to produce two glucose molecules and is present in nearly all tissues in various forms. Until recently, trehalase has been classified as a member of the glycoside hydrolase 37 (GH37) and 65 (GH65) families in the Carbohydrate-Active enZYmes database (CAZy; http://www.cazy.org/) that describes families of structurally related catalytic and carbohydrate-binding modules of enzymes that degrade, modify, or create glycosidic bonds. Escherichia coli periplasmic and cytoplasmic trehalases were the first GH37 family of trehalases to be studied (7–9). Saccharomyces cerevisiae has two types of trehalases, acid (extracellular) and neutral (cytosolic) (according to the pH required for their optimal activity), belonging to the GH65 and GH37 families, respectively (10, 11). Recently, Mycobacterium trehalases from M. smegmatis MSMEG4528 and M. tuberculosis MT2474 and Rv2402 have been identified and assigned to the GH15 family (12), which also contains glucoamylases (GAs) and glucodextranases (GDases). Enzymes in these families contain a common α-helix-rich catalytic domain designated the (α/α)6 barrel.
Several reports describe the presence of trehalose and the existence of multiple trehalose synthetic pathways in archaea; however, homologs of known trehalases (e.g., from bacteria and yeast) could not be identified in archaeal genomes, and the role of trehalose in archaea remains unknown (13). Recently, we identified two GA-homologous genes, TVN1315 (TVG1381191) and Ta0286, within the genomes of the thermophilic archaebacteria Thermoplasma volcanium (14, 15) and T. acidophilum (16), respectively. The deduced amino acid sequences encoded by these genes show 84% identity and 94% similarity to one another. The amino acid sequences of the putative catalytic domains exhibit 25% identity with that of Clostridium sp. strain G0005 GA (17, 18) and 38% identity with that of M. smegmatis MSMEG4528, a GH15 family trehalase (12).
With these characteristics in mind, we attempted to express the TVN1315 and Ta0286 genes in E. coli and characterize the recombinant enzymes. The purified enzymes hydrolyzed trehalose but not maltotriose. These two Thermoplasma enzymes are the first archaeal trehalases to be identified and are the first recombinant GH15 family trehalases to be expressed in E. coli in active forms.
MATERIALS AND METHODS
Bacterial strains and growth media.
Thermoplasma volcanium GSS1 (ATCC no. 51530D-5) and T. acidophilum 122-1B2 (ATCC no. 25905D-5) genomic DNA were purchased from the American Type Culture Collection (ATCC).
The E. coli strains used for expression and genetic engineering were BL21(DE3) (Novagen, San Diego, CA, USA) and DH5α (Toyobo, Osaka, Japan), respectively.
LB medium (1% Bacto tryptone, 0.5% Bacto yeast extract, 1% NaCl, pH 7.0) was used for culturing E. coli for genetic engineering and for protein expression preculturing. The solid medium contained 1.5% agar. For protein expression by E. coli, 2× YT liquid medium (1.6% Bacto tryptone, 1% Bacto yeast extract, 0.5% NaCl, pH 7.0) was used. Ampicillin (50 μg/ml) and kanamycin (20 to 50 μg/ml) were added to the medium as needed.
Genetic engineering and chemical reagents.
The genetic engineering experiments were performed as described by Sambrook and Russell (19). The enzymes used in the genetic engineering were purchased from TaKaRa Bio Inc. (Shiga, Japan) and used according to the manufacturer's instructions. Bacto tryptone and Bacto yeast extract were purchased from Becton Dickinson (Franklin Lakes, NJ, USA). Other reagents and oligosaccharide substrates were of the highest quality available and were obtained from Wako Pure Chemicals (Osaka, Japan) and Sigma-Aldrich (St. Louis, MO, USA) unless otherwise specified.
Cloning of glucoamylase-like genes from T. volcanium and T. acidophilum and expression plasmid construction.
To amplify the T. volcanium TVN1315 gene and T. acidophilum Ta0286 gene, PCR was performed with the following steps: 1 cycle at 94°C for 2 min, followed by 30 cycles of 94°C for 10 s, 60°C for 30 s for TVN1315 or 55°C for 30 s for Ta0286, respectively, and 68°C for 3 min. Reaction mixtures contained 50 ng of each genomic DNA as a template, 15 pmol each of the forward and the reverse primers described below, and 1 U of KOD DNA polymerase (Toyobo, Osaka, Japan). The primer sequences for T. volcanium were the following: forward, 5′-ACGCGTCGACGATGGCCTTTCATGGTCTTGTTAAA-3′; reverse, 5′-ACGCGTCGACTTATGAATTTATTTTTGAAGTTCTT-3′ (open reading frame [ORF] regions are underlined and SalI sites are boldfaced). The primer sequences for T. acidophilum were the following: forward, 5′-CGGGATCCCGATGGCCTTTCATGGTCTTG-3′; reverse, 5′-CGGGATCCCGTCACTCATTTATCTTTGAAACTCTC-3′ (ORF regions are underlined and BamHI sites are boldfaced). The primers were purchased from Sigma-Aldrich Life Science (Hokkaido, Japan). After trimming the amplified PCR products with SalI or BamHI, the products were inserted into the pET28c vector (TaKaRa Bio) to yield the pTvn1315 and pTa0286 expression vectors. The nucleotide sequences were confirmed using an Applied Biosystems 3130 genetic analyzer (Applied Biosystems, Foster City, CA, USA).
Trehalase expression and purification.
Precultures of E. coli that had been transformed with the expression vectors were grown overnight at 37°C in LB medium containing 50 μg/ml kanamycin, and then an inoculum was added to 2× YT medium. The cells were grown at 37°C to an optical density at 600 nm (OD600) of 0.8 and then induced for approximately 18 h using 0.1 mM isopropyl β-d-thiogalactopyranoside (IPTG). The cells were harvested by centrifugation and suspended in buffer containing 20 mM Tris-HCl, 0.5 M NaCl (pH 7.5), and cOmplete mini (EDTA-free) proteinase inhibitors (Roche Diagnostics, Mannheim, Germany). Following sonication, the crude extracts and supernatants (obtained by centrifugation for 20 min at 20,000 × g) were assayed for enzymatic activity, and the expression of gene products was assessed by SDS-PAGE and staining with Coomassie brilliant blue R-250 (CBB R-250) (20). Following dialysis against a buffer containing 50 mM Tris-HCl and 1 M (NH4)2SO4 (pH 7.5), the supernatants were subjected to hydrophobic chromatography (HiTrap Butyl FF; GE Healthcare, Little Chalfont, Buckinghamshire, England). The adsorbed proteins were eluted using a 1 M to 0 M linear (NH4)2SO4 gradient in 50 mM Tris-HCl (pH 7.5). The fractions containing the enzyme then were subjected to affinity chromatography using a Ni-Sepharose 6 fast flow column (GE Healthcare) that had been preequilibrated with binding buffer (20 mM Tris-HCl and 0.5 M NaCl, pH 7.5). The bound proteins were eluted using binding buffer containing 0 M, 0.05 M, and 0.5 M imidazole, pH 7.5. The enzymes were eluted at 0.5 M imidazole and were purified to homogeneity to yield a single band in SDS-PAGE after staining with CBB R-250. The fractions containing the enzyme were dialyzed against buffer containing 20 mM Tris-HCl and 0.5 M NaCl (pH 7.5), pooled at 4°C, and used for subsequent experiments.
Enzyme and protein assays.
Enzymatic activities were measured at 50°C in 50 mM Gly-HCl buffer (pH 3.7). The enzyme solution (50 μl) was added to 450 μl of an appropriate substrate solution, and the reaction mixtures were incubated for an appropriate period. To stop the reaction, 50-μl aliquots were removed from the reactions at appropriate intervals and mixed with 450 μl of 1 M Tris-HCl (pH 7.0) stop solution. The amount of glucose liberated from the substrates was measured using F-kit d-glucose (Roche Diagnostics Gmbh, Mannheim, Germany) with the glucose standard curve as a reference. One unit of enzyme was defined as the amount of the enzyme that hydrolyzed 1 μmol of trehalose in 1 min. Protein concentrations were measured using the microassay method (Bio-Rad Laboratories, Hercules, CA, USA), which is based on the Bradford method (21), using bovine serum albumin as a standard protein.
Trehalase substrate specificity, temperature and pH dependence, and temperature and pH stability.
The initial rates of hydrolysis were measured using 9 mM trehalose, nigerose (Hayashibara Biochemical Laboratories, Okayama, Japan), maltose, isomaltose, turanose, malturose, palatinose, or sucrose substrates by mixing with the enzyme solution at 50°C in 50 mM acetate buffer (pH 4.0) using F-kit d-glucose. The pH dependence of the enzymatic activities on 90 mM trehalose at 50°C was determined by measuring the initial rates at different pH values (ranging from pH 2.5 to 4.5) using 50 mM Gly-HCl buffer (pH 2.5 to 4.0) and 50 mM acetate buffer (pH 3.3 to 4.5). To study the pH stability of the trehalase, the enzyme solutions were added to various buffers (pH 3 to 11) at 4°C for 60 min, and the remaining activities on 90 mM trehalose at 50°C were determined at pH 3.7. To minimize any potential pH shift caused by the enzyme solution, the enzyme was diluted in buffer that contained 20 mM Tris-HCl and 0.5 M NaCl (pH 7.5) before activity measurements. The temperature dependence of the trehalase activity on 90 mM trehalose at pH 3.7 was examined by measuring the initial rates at 30, 40, 50, 55, 60, and 70°C. To determine the heat stability of the trehalase, the enzyme solutions were incubated at various temperatures (30 to 80°C) for 60 min and cooled rapidly, and then the remaining activities on 90 mM trehalose at 50°C were measured. The experiments were performed in duplicate.
DSF analysis.
The differential scanning fluorimetry (DSF) experiment was performed using a real-time PCR system (Mx3005p; Agilent Technologies, Santa Clara, CA, USA) with MxPro software (Agilent Technologies) based on Niesen et al.'s report (22). Tubes containing 39 μl protein solution and 1 μl SYPRO Orange (Invitrogen, Carlsbad, CA, USA) diluted 10-fold with dimethyl sulfoxide (DMSO) were loaded into the PCR instrument and subjected to temperature scanning at 1°C min−1 between 25°C and 95°C. The filter configurations were customized to accommodate the optimal excitation and emission wavelengths for SYPRO Orange (excitation, 492; emission, 610 nm). The inflection points of the transition curves, indicating the protein melting temperatures (Tms), were estimated as described in an earlier report (23). The experiments were performed in duplicate.
Kinetic analysis of trehalose hydrolysis.
The initial rates of trehalose (0 to 270 mM) hydrolysis were measured at 50°C in buffers containing 50 mM Gly-HCl (pH 3.7) or 50 mM acetate (pH 3.8) using F-kit d-glucose. The kinetic parameters, Vmax and Km, for trehalose were estimated over a range of 0 to 270 mM assuming a Michaelis-Menten kinetic model, and the DeltaGraph version 6 graphics software package (Nihon Poladigital K.K.) was used for nonlinear regression. The apparent kcat values were estimated using a molecular mass of 75 kDa deduced from the amino acid sequences of the enzymes. The experiments were performed in duplicate or triplicate.
Effects of VMA on trehalose hydrolysis.
Validamycin A (VMA), a potent trehalase inhibitor, was purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA), and dissolved in DMSO (500 mM stock solution). The effects of VMA (0 to 100 μM) on the initial rates of trehalose hydrolysis were assayed at 50°C in 50 mM Gly-HCl buffer (pH 3.7) using F-kit d-glucose. We confirmed that the addition of either VMA solution or DMSO alone had no inhibitory effects on the activities of hexokinase and glucose-6-phosphate dehydrogenase. The apparent inhibition constants (Ki) were estimated using a Dixon plot (24).
Construction of mutant expression plasmids and the expression, purification, and analysis of the mutant proteins.
Site-directed mutagenesis of the TVN1315 gene was performed using pTvn1315 as a template and by following the ODA-PCR method (Mutan-Super express Km; TaKaRa BIO). The following oligonucleotide primers were used as mutagenic primers (mismatched bases are underlined): TVN1315 E408Q, 5′-AGCATCTGGCAATTTCGTACTG-3′; TVN1315 E571Q, 5′-GGCTGTACTCACAAGAGATAGATG-3′. The nucleotide sequences surrounding the mutation site and other regions of the gene were confirmed by DNA sequencing as described above. For these mutants, the expression, purification, activity measurements, and DSF analyses were performed as described above.
Nucleotide sequence accession numbers.
The TVN1315 and Ta0286 nucleotide sequences are available from the DDBJ/EMBL/GenBank database under accession numbers LC004405 and LC004406, respectively.
RESULTS
Comparison of TVN1315 and Ta0286 sequences to those of other related enzymes and structure prediction.
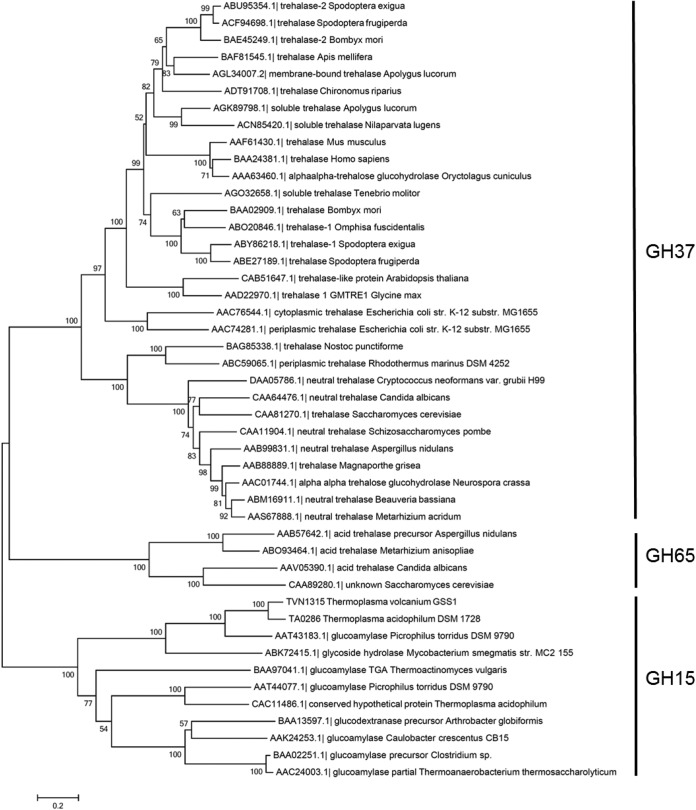
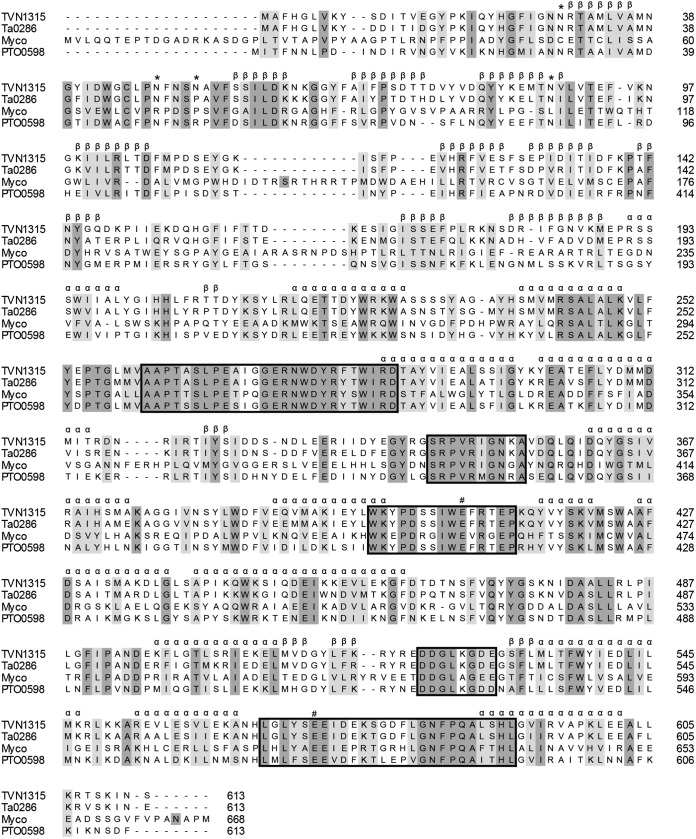
The phylogenetic relationships among GH37 and GH67 trehalases and relevant GH15 members were analyzed and are shown in Fig. 1. The analysis yielded three major branches: GH37 enzymes, which include trehalases from bacteria, yeasts, insects, plants, and animals; GH65 enzymes, which include acidic trehalases from fungi and yeast; and GH15 enzymes, which include GAs from bacteria and archaea together with two GA-like products, TVN1315 and Ta0286 (analyzed below). The two GA-like archaeal gene products appear to be more similar to acidic trehalases (GH65) than to GH37 trehalases, but the sequence homology is low even between GH65 and the two GH15 products. Similar to GH65 trehalases that lack trehalase signature consensus motifs PGGRFXEXYXWDXY (motif 1) and QWDXPXGAWPAP (motif 2) in their primary structures (8, 25), the Thermoplasma GA-like products, TVN1315 and Ta0286, do not possess those motifs or other trehalase motifs in their amino acid sequences (Fig. 2). TVN1315 and Ta0286 did not possess the carbohydrate-binding module or motif either.
FIG 1.
Phylogenetic relationship of related enzyme sequences. The phylogenetic tree was constructed by MEGA6 (52) using the neighbor-joining method. Bootstrap values based on 1,000 replicates are shown at nodes, and accession number, enzyme name, and organism name are shown.
FIG 2.
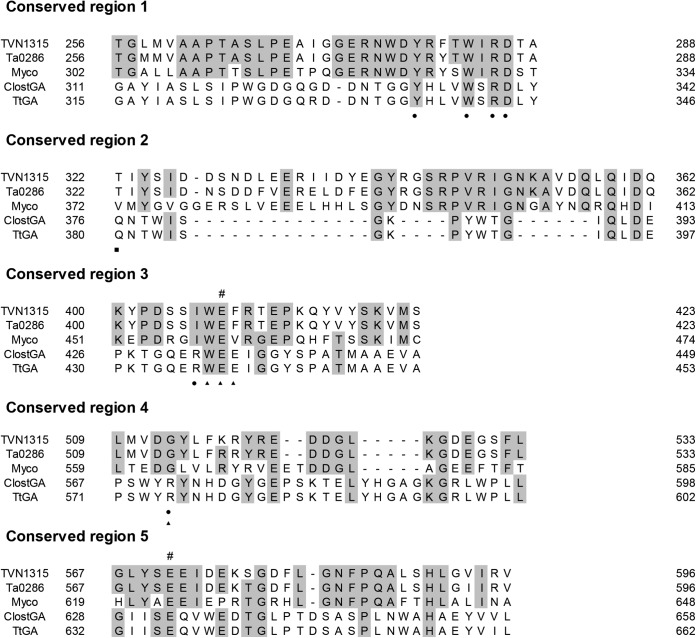
Sequence alignments of archaeal trehalases with Mycobacterium trehalases and a putative trehalase. The sequences aligned are the following: TVN1315, T. volcanium trehalase; Ta0286, T. acidophilum trehalase; Myco, Mycobacterium trehalase; PTO0598, putative Picrophilus torridus trehalase. #, putative catalytic residues. The α and β symbols denote the involvement in the putative secondary structures, α-helix and β-sheet, respectively, illustrated in the molecular models in Fig. S1 in the supplemental material. The asterisks denote potential glycosylation sites. Fully conserved amino acid residues and related amino acid residues are dark shaded and light shaded, respectively. Boxes show the five highly conserved regions.
Based on their amino acid sequences, GH37, GH65, and GH15 family enzymes are predicted to have catalytic domains with (α/α)6-barreled structures. In addition, the N-terminal domain of GH15 family enzymes can be folded into a β-sandwich structure. Therefore, the structures of the two GA-like products are assumed to be composed of an N-terminal β-sandwich and C-terminal (α/α)6-barreled structures (see Fig. S1 in the supplemental material).
E. coli expression of soluble proteins and substrate specificity assays for the GA-like genes from Thermoplasma.
TVN1315 and Ta0286 were expressed as soluble proteins in E. coli BL21(DE3) using 0.1 mM IPTG (final concentration) to induce the pET28 vectors. For Ta0286, a gene product with a 32-amino-acid N-terminal truncation also was expressed, because we did not have an a priori reason to decide that Met1 and not Met33 was the initiating Met residue; however, the truncated product was found to be insoluble (data not shown). This result suggested that the N-terminal regions of Ta0286 and likely of TVN1315 are responsible for the proper folding of the enzyme molecule, although insolubility may be caused by other reasons as well. The Met1 initiator codon was preceded by a potential promoter and purine-rich (Shine-Dalgarno) sequence (see Fig. S2 in the supplemental material).
Despite the similarity of the TVN1315 and Ta0286 gene putative catalytic sites to that of GA, cell homogenates containing the gene products did not exhibit maltotriose-hydrolyzing activity but did exhibit trehalose-hydrolyzing activity. The expressed gene products were purified using hydrophobic and affinity chromatography to yield a single band in SDS-PAGE (Fig. 3). The N-terminal sequence analysis of the TVN1315 gene product showed GSSHHHHHHSSGLV, which coincides with the expected N-terminally added sequence, including the histidine tag of the pTvn1315 construct. The N-terminal Met likely had been removed by methionine aminopeptidase. A slight difference between the expected molecular mass of the histidine-tagged TVN1315 protein (74.8 kDa) and the apparent molecular mass estimated from the mobility in the SDS-PAGE (70 kDa) was noted. A similar discrepancy was reported in the literature and was proposed to be dependent largely on the hydrophobicity and/or isoelectric point of the protein (26). The gel filtration analysis using HiLoad 16/60 Superdex 200 (GE Healthcare) provided a rough estimate of the apparent molecular mass of 97 kDa (data not shown), suggesting the monomeric nature of the expressed protein.
FIG 3.
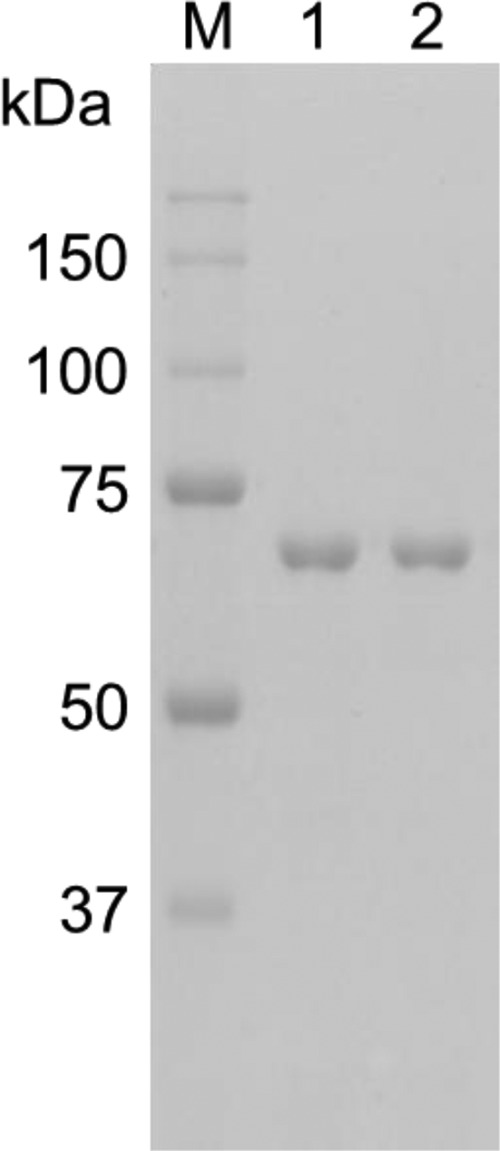
SDS-PAGE analysis of the purified proteins. Lane M, molecular size marker (Precision Plus protein standards; Bio-Rad Laboratories); lane 1, TVN1315; lane 2, Ta0286. The numbers in the margin indicate the molecular masses (kDa) of the proteins in the molecular size marker.
The purified gene products hydrolyzed trehalose to yield glucose but showed little to no activity toward the other disaccharides tested (nigerose, maltose, isomaltose, turanose, malturose, palatinose, and sucrose). In reaction mixtures containing these sugars as substrates, glucose production could not be detected even after 2 h. Consequently, we have identified the TVN1315 and Ta0286 gene products TVN1315 and Ta0286 as T. volcanium and T. acidophilum trehalases, respectively.
Effects of temperature and pH on TVN1315 and Ta0286.
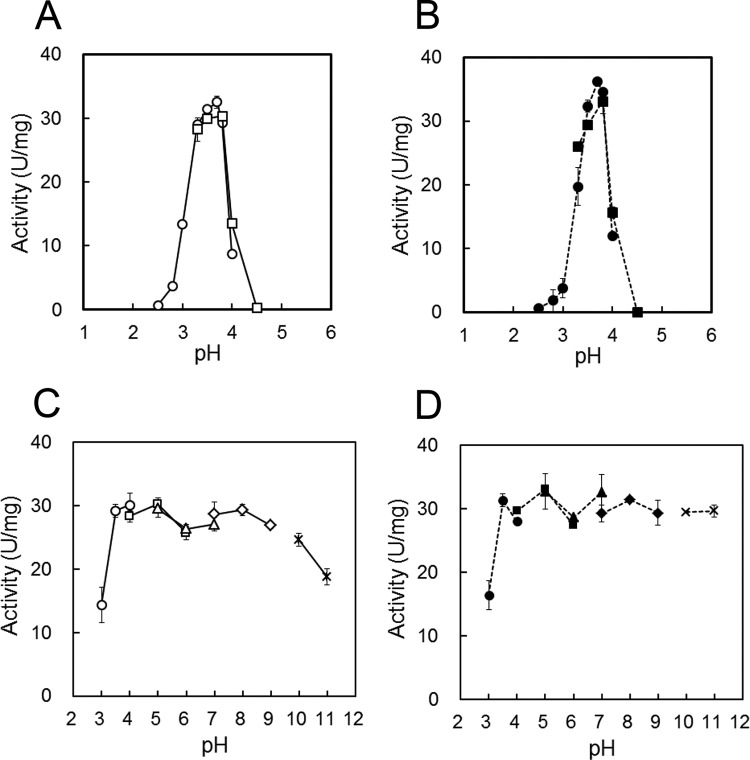
Figure 4A and B illustrates the pH dependence of TVN1315 and Ta0286 trehalase activity. Both trehalases were optimally active at approximately pH 3.7 and were active within a narrow pH range. TVN1315 and Ta0286 exhibited more than half of their maximal activities between pH 3.3 and pH 4.0 and between pH 3.2 and pH 3.9, respectively. Figure 4C and D illustrates the pH stability of the tested trehalases and shows that TVN1315 and Ta0286 retained nearly full activity after 60 min at pH 3.5 to 10 and pH 3.5 to 11, respectively.
FIG 4.
Effects of pH on the activities and stabilities of TVN1315 (A and C) and Ta0286 (B and D). (A and B) The activity was measured at 50°C in appropriate buffers at different pHs, including 50 mM Gly-HCl buffer (pH 2.5 to 4.0, circle) and 50 mM acetate buffer (pH 3.3 to 4.5, square). (C and D) Trehalases were incubated at 4°C for 60 min at various pHs, including 50 mM Gly-HCl buffer (pH 3 to 4, circle), 50 mM acetate buffer (pH 4 to 6, square), 50 mM morpholineethanesulfonic acid–NaOH buffer (pH 5 to 7, triangle), 50 mM Tris-HCl buffer (pH 7 to 9, rhombus), and 50 mM carbonate-NaOH buffer (pH 10 to 11, cross). The remaining activity was measured at 50°C and pH 3.7. The average values for TVN1315 (A and C; open symbols) and Ta0286 (B and D; closed symbols) with error bars are represented as the enzymatic activity. Experiments were performed in triplicate.
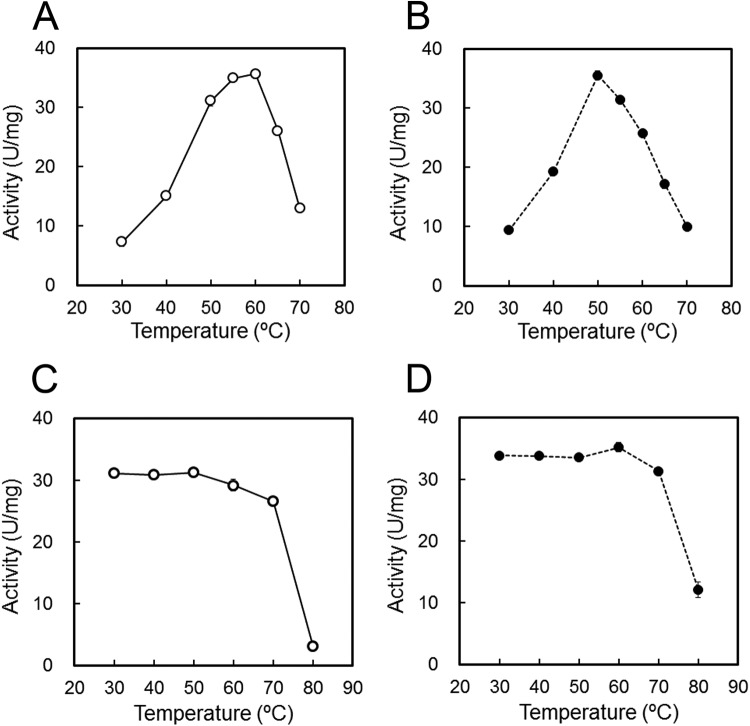
The temperature dependence of the enzymatic activities was examined using 90 mM trehalose in 50 mM Gly-HCl buffer (pH 3.7) at temperatures ranging from 30 to 80°C. Figure 5A and B shows that TVN1315 and Ta0286 activities were maximal at approximately 60°C and 50°C, respectively. The activation energies of TVN1315 and Ta0286 from 30 to 50°C were approximately 59 and 54 kJ/mol, respectively. These values are within the limits of those obtained previously for other trehalases: 46 to 81 kJ/mol (27–30).
FIG 5.
Effects of temperature on trehalase activity (A and B) and stability (C and D). (A and B) The activities of TVN1315 (open circle) and Ta0286 (closed circle) were measured at various temperatures (30 to 70°C) in 50 mM Gly-HCl (pH 3.7). (C and D) The trehalases were incubated at various temperatures (30 to 80°C) for 60 min, and the remaining activities of TVN1315 (open circle) and Ta0286 (closed circle) were measured at 50°C in 50 mM Gly-HCl (pH 3.7). Experiments were performed in triplicate, and the average values are shown.
To determine the temperature stability of TVN1315 and Ta0286, we determined the remaining enzymatic activities at 50°C after 60 min of heat treatment at various temperatures over a range of 30 to 80°C using 90 mM trehalose as a substrate, and the results are shown in Fig. 5C and D. The activities of both TVN1315 and Ta0286 were stable for 60 min at temperatures ranging from 30 to 70°C; however, their activities decreased significantly at 80°C after 60 min. TVN1315 was inactivated slightly more rapidly than Ta0286 at 80°C. The slightly different thermal stabilities of TVN1315 and Ta0286 activities were consistent with the DSF analysis results that determined Tm values of 80.3°C and 84.0°C, respectively, for these enzymes (see Fig. S3 in the supplemental material).
Steady-state kinetics using trehalose as the substrate.
Both TVN1315 and Ta0286 trehalases hydrolyzed the trehalose substrate containing an α,α-(1,1)-glucosidic linkage, unlike other GH15 family GAs. We determined the steady-state kinetic parameters of both trehalases for trehalose at 50°C using Delta Graph software to perform a nonlinear regression, and the results are shown in Table 1. We show the kinetic results in two different buffer solutions, because the pH values used in the present experiment, pH 3.7 and 3.8, are near the edge of the buffer capacity ranges for the two buffers. The results obtained using the two buffers at pH 3.7 and 3.8 were consistent. The hydrolysis reactions obeyed the Michaelis-Menten kinetic model. At 50°C in 50 mM Gly-HCl buffer (pH 3.7), the TVN1315 and Ta0286 kcat values were 63.0 s−1 and 66.7 s−1, respectively, and the Km values were 48.7 mM and 40.2 mM, respectively. The Km values were similar to that of a Mycobacterium trehalase (20 mM) (12) but were considerably higher than those for other GH37 and GH65 family trehalases, such as the E. coli cytoplasmic enzyme (1.9 mM) (Table 2) (8).
TABLE 1.
Steady-state kinetic parameters of trehalases using trehalose as the substrate at 50°C
| Enzyme | kcata (s−1) | Kma (mM) | kcat/Km (s−1 mM−1) | Bufferb |
|---|---|---|---|---|
| TVN1315 | 63.0 ± 0.4 | 48.7 ± 1.4 | 1.29 | Gly-HCl, pH 3.7 |
| 60.4 ± 0.8 | 49.0 ± 2.0 | 1.23 | Acetate, pH 3.8 | |
| Ta0286 | 66.7 ± 1.3 | 40.2 ± 1.5 | 1.66 | Gly-HCl, pH 3.7 |
| 59.1 ± 1.7 | 44.1 ± 1.2 | 1.34 | Acetate, pH 3.8 |
Values for kcat and Km (means ± standard deviations) are based on results using trehalose (0 to 270 mM) as the substrate. Experiments were performed in duplicate or triplicate.
Buffer solution (50 mM).
TABLE 2.
Comparison of the kinetic parameters of various trehalases using trehalose as the substratea
| Organism | GH family | kcat (s−1) | Km (mM) | kcat/Km (s−1 mM−1) | Temp (°C), pH |
|---|---|---|---|---|---|
| T. volcanium | 15 | 63.0 | 48.7 | 1.29 | 50, 3.7 |
| T. acidophilum | 15 | 66.7 | 40.2 | 1.66 | 50, 3.7 |
| M. smegmatis | 15 | NDb | 20.0 | NDb | 37, 7.1 |
| E. coli (cytoplasmic) | 37 | 57.6c | 1.9 | 30.3 | 30, 5.5 |
| E. coli (periplasmic) | 37 | 84.1c | 0.8 | 105 | 30, 5.5 |
| S. cerevisiae (acidic) | 65 | 67.7c | 4.7 | 14.4 | 37, 4.5 |
| S. cerevisiae (neutral) | 37 | 148c | 34.5 | 4.29 | 37, 7.0 |
The kinetic parameters of various trehalases and the conditions for activity measurements were obtained for T. volcanium and T. acidophilum (this study), M. smegmatis (12), E. coli (cytoplasmic) (8), E. coli (periplasmic) (49), S. cerevisiae (acidic) (50), and S. cerevisiae (neutral) (51).
ND, not determined.
The values were estimated from the Vmax for each enzyme based on its monomeric molecular mass.
VMA is known to be a potent trehalase inhibitor and has antifungal effects (31–34). To explore whether VMA affects Thermoplasma trehalase activity, we tested the effects of VMA on trehalase activity and estimated Ki values using Dixon plots of the reciprocal of the activity on the ordinate and the inhibitor concentration on the abscissa (TVN1315, 35.6 μM; Ta0286, 19.7 μM) (Fig. 6A and B). As shown in Fig. 6A and B, VMA showed competitive inhibition of Thermoplasma trehalases similar to that of other trehalases; however, the Ki values are higher than those for porcine kidney (60 nM) or soybean nodule (2 nM) trehalases (35, 36).
FIG 6.
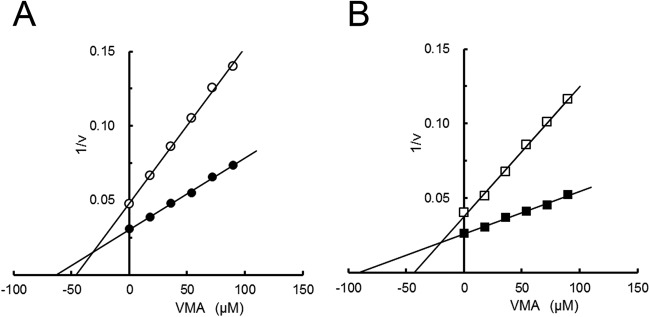
Dixon plots of TVN1315 (A) and Ta0286 (B) activities in the presence and absence of validamycin A (VMA). The changes in TVN1315 (circle) and Ta0286 (square) activities toward trehalose (36 mM [open symbols] and 90 mM [closed symbols]) in the presence of validamycin A (0 to 100 μM) were measured. Experiments were performed in duplicate, and the average values are shown.
Involvement of Glu408 and Glu571 in TVN1315 trehalase activity and thermal stability.
GH15 family enzymes, such as GAs and GDases, possess 5 conserved regions in the primary structures, and these conserved regions have been proposed to constitute the enzyme active sites. TVN1315, a newly identified GH15 family trehalase, also possesses similar regions (Fig. 7); however, the involvement of these regions in its catalysis remains unknown. For GAs and GDases, two glutamic acid residues in conserved regions 3 and 5 are thought to play important roles in the hydrolysis reactions (18, 37–39). Based on the structural similarity shown in Fig. 7, we examined the possible involvement of Glu408 and Glu571 in conserved regions 3 and 5, respectively, in the function of TVN1315. Consequently, we constructed three mutants in which Glu408, Glu571, or both Glu408 and Glu571 were replaced with glutamine residue(s). The TVN1315 E408Q, E571Q, and E408Q/E571Q mutants were expressed in E. coli and purified similarly to the wild-type TVN1315 (Fig. 8A).
FIG 7.
Sequence alignments of the conserved regions (1 to 5) of archaeal trehalases with Mycobacterium trehalases and other GAs. The sequences aligned are the following: TVN1315, T. volcanium trehalase; Ta0286, T. acidophilum trehalase; Myco, Mycobacterium trehalase; ClostGA, Clostridium sp. strain G0005 glucoamylase; TtGA, Thermoanaerobacterium thermosaccharolyticum glucoamylase. #, putative catalytic residues. Amino acid residues that are identical in three or more of the aligned enzymes are shaded. The putative amino acid residues constituting GA subsites are indicated as subsite 1 (closed circle), subsite 2 (closed triangle), and subsite 3 (closed square).
FIG 8.
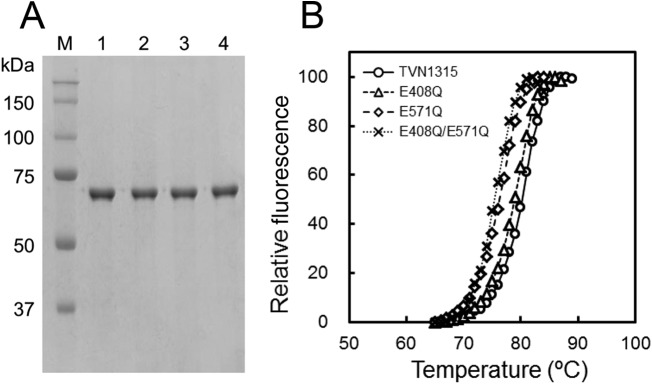
SDS-PAGE analysis (A) and heat stability analysis (B) of wild-type and mutant TVN1315 trehalase. (A) Lane M, molecular size marker (Precision Plus protein standards; Bio-Rad Laboratories); lane 1, TVN1315; lane 2, TVN1315 E408Q; lane 3, TVN1315 E571Q; and lane 4, TVN1315 E408Q/E571Q. The numbers in the margin indicate the molecular masses (in kilodaltons) of the proteins in the molecular size marker. (B) DSF analysis results for TVN1315 (circle), E408Q (triangle), E571Q (rhombus), and E408Q/E571Q (cross) are shown. The experimental procedure is detailed in Materials and Methods. Experiments were performed in triplicate, and the average values are shown.
The E408Q, E571Q, and E408Q/E571Q TVN1315 mutants exhibited reduced hydrolytic activities on 90 mM trehalose at 50°C in 50 mM Gly-HCl (pH 3.7) at 0.009, 1.3, and 0.003 U/mg, respectively, whereas that for TVN1315 was 31.4 U/mg. These results indicate the importance of these glutamic acid residues in TVN1315 catalytic activity. Notably, the E408Q, E571Q, and E408Q/E571Q mutants showed slightly lower Tm values, 79.1°C, 76.2°C, and 75.5°C, respectively, than wild-type TVN1315 (80.3°C) (Fig. 8B). The E571Q mutation had a more pronounced effect on the Tm of the enzyme than the E408Q mutation, suggesting that Glu571 contributes more to TVN1315 heat stability than Glu408.
DISCUSSION
Through our comparative studies of GAs and their mutants, we identified thermophilic GA-like genes in archaea and demonstrated that products of two of these genes hydrolyze trehalose rather than maltose or maltotriose.
TVN1315 and Ta0286 exhibited trehalase activity within a narrow acidic pH range. They showed thermophilic characteristics, and the gel filtration analysis suggested that they function as monomers. In contrast, Mycobacterium trehalase (previously identified as a GH15 family enzyme) had a pH optimum of 7.1, required both inorganic phosphate and Mg2+ for activity, and had a molecular mass of 1,500 kDa as determined by gel filtration analysis, suggesting a multimeric structure (12). The molecular basis for the considerably different properties of these GH15 family trehalases remains unclear. In the present experiment, we measured enzymatic activities in the presence of 50 mM NaCl, but we did not observe a significant increase in activity in the presence of salts such as NaCl, KCl, CaCl2, or MgCl2 over a range of 100 to 300 mM in 50 mM Gly-HCl buffer (pH 3.7). However, Thermoplasma trehalases require the presence of salt for structural stabilization, as the purified enzymes tended to become insoluble when dialyzed against buffered solution in the absence of NaCl.
For the kinetic parameters of various trehalases, the kcat values were similar among trehalases (within a 2- to 3-fold difference). In contrast, the Km values varied considerably between enzymes: a greater than 40-fold difference was noted even among trehalases belonging to the identical GH37 family. The inhibition constant (Ki) values for the competitive inhibitor VMA also varied among Thermoplasma trehalases and other trehalases (35, 36). These results suggest that all trehalase active sites are designed for the recognition of trehalose and VMA, although the actual organization of the active site may vary considerably among different enzymes. In fact, GH15 trehalases lack the trehalase signature motifs 1 and 2 that are found in GH37 trehalases. For GH15 trehalases, Thermoplasma trehalases displayed Km values on trehalose that were similar to that of a Mycobacterium trehalase, suggesting a similarity in the active site between these enzymes. The high Km values of Thermoplasma trehalases are consistent with the high cellular trehalose concentration estimated based on the trehalose content reported in the literature (40). TVN1315 and Ta0286 exhibited hydrolytic activity within a narrow acidic pH range. A number of enzymes that work within acidic pH ranges were reported in acidophilic archaea (41).
For industrial production, glucose is commonly produced from starch by α-amylase and GA. Trehalase is not useful in glucose production. In an aqueous environment, trehalose hydrolysis is overwhelmingly favored over the reverse reaction. However, if the enzyme is available in an anhydrous environment, then trehalose and other α-(1,1)-disaccharides could be obtained via the reverse reaction of trehalase. Thermophilic enzymes have a greater ability to preserve their enzymatic activity in nonaqueous media than their mesophilic counterparts. Moreover, protein engineering studies leading to enzymes with enhanced stability in organic solvents have been reported (42, 43). Investigations into the stability of archaeal trehalases in anhydrous environments are needed, and protein engineering on trehalases that remain active in nonaqueous media should be attempted in future studies.
Based on the primary structure, Thermoplasma trehalases are more similar to GAs from the identical GH15 family than to trehalases classified to the GH37 and GH65 families. We propose that comparative studies on GH15 trehalases and GAs will elucidate their structure-function relationships. GAs have five conserved regions involved in substrate binding and are important for catalytic activity. GH15 trehalases, including PTO0598 (a putative Picrophilus torridus trehalase), also possess five conserved regions at positions corresponding to, but with sequences distinct from, those of GA (Fig. 2 and 7). Two glutamic acid residues, E408 and E571 of TVN1315 in the conserved regions 3 and 5, also are conserved in GAs and are essential for the catalytic activity of both trehalases and GAs. This suggests a significant mechanistic similarity between GH15 trehalases and GAs. Differences in the amino acid sequences of the conserved regions between trehalases and GAs likely are responsible for conferring distinct substrate specificities to these enzymes. Based on the crystal structure of the complex of Thermoanaerobacter thermosaccharolyticum GA (TtGA) with acarbose (a competitive inhibitor [44]), several amino acid residues have been assigned to components of the substrate binding sites. Of these residues, Tyr337, Trp341, Arg343, and Asp344 (TtGA amino acid numbering) in subsite 1 are conserved in Thermoplasma trehalases, whereas Arg436 and Arg575 are not conserved (Fig. 7). Similarly, Trp437 and Glu438 in subsite 2 are conserved, but Glu439 and Arg575 are not. TtGA Gln380 in subsite 3 of GA is not conserved in Thermoplasma trehalases. Trehalases may not possess subsite 3. The general structure of the Thermoplasma trehalase substrate-binding site likely resembles that of the TtGA substrate-binding site, whereas distinct substrate specificity is conferred by nonconserved amino acid residues; however, the detailed structures of their substrate binding sites await future studies. The identification of GH15 family enzymes that are structurally similar to GA but display different substrate specificities would aid our future studies aimed at gaining a deeper insight into the relationship between the function and structure of GH15 family enzymes.
TVN1315 and Ta0286 are the first archaeal trehalases to be identified to date. Trehalose accumulates in archaea, such as T. acidophilum, under optimal growth conditions and is thought to be used for the stabilization of macromolecules under temperature stress and as a carbon storage molecule (40). To date, five different enzymatic pathways for trehalose synthesis have been described in various organisms. These pathways include the trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase (TPS/TPP) pathway, trehalose synthase (TreS) pathway, maltooligosyl-trehalose synthase and maltooligosyl-trehalose trehalohydrolase (TreY/TreZ) pathway, trehalose phosphorylase (TreP) pathway, and trehalose glycosyltransferring synthase (TreT) pathway (45). For trehalose degradation, trehalase (TreH), an enzyme which is directly involved in trehalose degradation, was reported in various organisms, such as E. coli and S. cerevisiae, but has not been identified in archaea, although TreT and TreP may have functions in degrading trehalose. Identifying archaeal trehalases added the third route to trehalose degradation in archaea.
A BLAST search indicated that the TPS/TPP pathway is widely distributed in archaea, including acidophilic archaea, whereas TreS, TreP, and TreY/TreZ pathways are less common in archaea (46). For instance, TreP and TreY/TreZ are not found in the acidophilic archaeon Thermoplasma, although a gene with partial similarity to TreS of Picrophilus torridus is detected in the BLAST search. The TPS/TPP pathway likely represents the major trehalose synthetic pathway in archaea.
However, the trehalose degradation pathway has not been well characterized. Qu et al. suggested that TreT participates mainly in trehalose degradation based on the fact that the treT gene of Thermococcus litoralis is found within a gene cluster encoding an ABC transporter for trehalose/maltose and that the half-maximal concentration for glucose, one of the substrates of TreT in the synthetic direction, is high compared to the physiological intracellular concentration of glucose (47). However, a BLAST search indicated that sequences similar to that of TreT are not present in P. torridus. Instead, we found a sequence similar to that of TVN1315 and Ta0286 (i.e., TreH of Thermoplasma) in P. torridus (Fig. 1 and 2). Similarly, Thermoplasma has TreH but lacks TreT. TreT also is missing from many acidophilic archaea, although a TreT-related sequence is found in Sulfolobus species. GH15 trehalase (TreH) likely serves as the major trehalose-degrading enzyme in acidophilic archaea. A TreH-related sequence is found in acidophilic archaea but not in other archaeal species. Among such organisms, select Euryarchaeota contain reversible TreT, which may participate mainly in trehalose degradation and be involved in controlling the intracellular concentration of trehalose as a compatible solute and also in providing glucose to serve as a cellular carbon source. In contrast, a unidirectional TreT that does not degrade trehalose is found in Thermoproteus tenax and in several other archaea, mainly in the Crenarchaeota (48). In these archaea, trehalose metabolic pathways for trehalose degradation have not been identified, although TreP, when present, might fulfill a catabolic role. In fact, T. tenax is not able to grow on trehalose as the only carbon source (48). The trehalose degradation pathway in archaea is poorly understood; therefore, the identification of archaeal trehalases will extend the understanding of the metabolism and biological role of trehalose in archaea.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Yukari Saisaka (High-Tech Research Center, Meiji Pharmaceutical University) for performing the N-terminal sequence analysis; to Misa Ohno, Kazuaki Okawa, and Yasuhide Nonaka for their valuable suggestions; and to Asana Takashima for technical assistance.
This study was supported in part by a grant from the Strategic Research Foundation Grant-aided Project for Private Universities of the Ministry of Education, Culture, Sport, Science and Technology, Japan (MEXT) (S1411005).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00956-15.
REFERENCES
- 1.Argüelles JC. 2000. Physiological roles of trehalose in bacteria and yeasts: a comparative analysis. Arch Microbiol 174:217–124. doi: 10.1007/s002030000192. [DOI] [PubMed] [Google Scholar]
- 2.Elbein AD, Pan YT, Pastuszak I, Carroll D. 2003. New insights on trehalose: a multifunctional molecule. Glycobiology 13:17–27. doi: 10.1093/glycob/cwg047. [DOI] [PubMed] [Google Scholar]
- 3.Paul MJ, Primavesi LF, Jhurreea D, Zhang Y. 2008. Trehalose metabolism and signaling. Annu Rev Plant Biol 59:417–441. doi: 10.1146/annurev.arplant.59.032607.092945. [DOI] [PubMed] [Google Scholar]
- 4.Friedman S. 1978. Trehalose regulation, one aspect of metabolic homeostasis. Annu Rev Entomol 23:389–407. doi: 10.1146/annurev.en.23.010178.002133. [DOI] [Google Scholar]
- 5.Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. 2007. Trehalose, a novel mTOR independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem 282:5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 6.Casarejos MJ, Solano RM, Gomez A, Perucho J, de Yebenes JG, Mena MA. 2011. The accumulation of neurotoxic proteins, induced by proteasome inhibition, is reverted by trehalose, an enhancer of autophagy, in human neuroblastoma cells. Neurochem Int 58:512–520. doi: 10.1016/j.neuint.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 7.Boos W, Ehmann U, Bremer E, Middendorf A, Postma P. 1987. Trehalase of Escherichia coli. Mapping and cloning of its structural gene and identification of the enzyme as a periplasmic protein induced under high osmolarity growth conditions J Biol Chem 262:13212–13218. [PubMed] [Google Scholar]
- 8.Horlacher R, Uhland K, Klein W, Ehrmann M, Boos W. 1996. Characterization of a cytoplasmic trehalase of Escherichia coli. J Bacteriol 178:6250–6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uhland K, Mondigler M, Spiess C, Prinz W, Ehrmann M. 2000. Determinants of translocation and folding of TreF, a trehalase of Escherichia coli. J Biol Chem 275:23439–23445. doi: 10.1074/jbc.M002793200. [DOI] [PubMed] [Google Scholar]
- 10.Destruelle M, Holzer H, Klionsky DJ. 1995. Isolation and characterization of a novel yeast gene, ATH1, that is required for vacuolar acid trehalase activity. Yeast 11:1015–1025. doi: 10.1002/yea.320111103. [DOI] [PubMed] [Google Scholar]
- 11.Nwaka S, Kopp M, Holzer H. 1995. Expression and function of the trehalase genes NTH1 and YBR0106 in Saccharomyces cerevisiae. J Biol Chem 270:10193–10198. doi: 10.1074/jbc.270.17.10193. [DOI] [PubMed] [Google Scholar]
- 12.Carroll JD, Pastuszak I, Edavana VK, Pan YT, Elbein AD. 2007. A novel trehalase from Mycobacterium smegmatis—purification, properties, requirements. FEBS J 274:1701–1714. doi: 10.1111/j.1742-4658.2007.05715.x. [DOI] [PubMed] [Google Scholar]
- 13.Zaparty M, Hagemann A, Bräsen C, Hensel R, Lupas AN, Brinkmann H, Siebers B. 2013. The first prokaryotic trehalose synthase complex identified in the hyperthermophilic crenarchaeon Thermoproteus tenax. PLoS One 8:e61354. doi: 10.1371/journal.pone.0061354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawashima T, Yamamoto Y, Aramaki H, Nunoshiba T, Kawamoto T, Watanabe K, Yamazaki M, Kanehori K, Amano N, Ohya Y, Makino K, Suzuki M. 1999. Determination of the complete genomic DNA sequence of Thermoplasma volcanium GSS1. Proc Japan Acad 75B:213–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawashima T, Amano N, Koike H, Makino S, Higuchi S, Kawashima-Ohya Y, Watanabe K, Yamazaki M, Kanehori K, Kawamoto T, Nunoshiba T, Yamamoto Y, Aramaki H, Makino K, Suzuki M. 2000. Archaeal adaptation to higher temperatures revealed by genomic sequence of Thermoplasma volcanium. Proc Natl Acad Sci U S A 97:14257–14262. doi: 10.1073/pnas.97.26.14257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruepp A, Graml W, Santos-Martinez ML, Koretke KK, Volker C, Mewes HW, Frishman D, Stocker S, Lupas AN, Baumeister W. 2000. The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature 407:508–513. doi: 10.1038/35035069. [DOI] [PubMed] [Google Scholar]
- 17.Ohnishi H, Kitamura H, Minowa T, Sakai H, Ohta T. 1992. Molecular cloning of a glucoamylase gene from a thermophilic Clostridium and kinetics of the cloned enzyme. Eur J Biochem 207:413–418. doi: 10.1111/j.1432-1033.1992.tb17064.x. [DOI] [PubMed] [Google Scholar]
- 18.Ohnishi H, Matsumoto H, Sakai H, Ohta T. 1994. Functional roles of Trp337 and Glu632 in Clostridium glucoamylase, as determined by chemical modification, mutagenesis, and the stopped-flow method. J Biol Chem 269:3503–3510. [PubMed] [Google Scholar]
- 19.Sambrook J, Russell DW. 2012. Molecular cloning: a laboratory manual, 4th ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 20.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 7:248–254. [DOI] [PubMed] [Google Scholar]
- 22.Niesen FH, Berglund H, Vedadi M. 2007. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc 2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 23.Sakaguchi M, Matsushima Y, Nankumo T, Seino J, Miyakawa S, Honda S, Sugahara Y, Oyama F, Kawakita M. 2014. Glucoamylase of Caulobacter crescentus CB15: cloning and expression in Escherichia coli and functional identification. AMB Express 4:5. doi: 10.1186/2191-0855-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dixon M. 1953. The determination of enzyme inhibitor constants. Biochem J 55:170–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barraza A, Sánchez F. 2013. Trehalases: a neglected carbon metabolism regulator? Plant Signal Behav 8:e24778. doi: 10.4161/psb.24778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shirai A, Matsuyama A, Yashiroda Y, Hashimoto A, Kawamura Y, Arai R, Komatsu Y, Horinouchi S, Yoshida M. 2008. Global analysis of gel mobility of proteins and its use in target identification. J Biol Chem 283:10745–10752. doi: 10.1074/jbc.M709211200. [DOI] [PubMed] [Google Scholar]
- 27.Riby J, Galand G. 1985. Rat intestinal brush border membrane trehalase: some properties of the purified enzyme. Comp Biochem Physiol B 82:821–827. [DOI] [PubMed] [Google Scholar]
- 28.Killick KA. 1983. Trehalase from the cellular slime mold Dictyostelium discoideum: purification and characterization of the homogeneous enzyme from myxamoebae. Arch Biochem Biophys 222:561–573. doi: 10.1016/0003-9861(83)90554-4. [DOI] [PubMed] [Google Scholar]
- 29.Terra WR, Ferreira C, de Bianchi AG. 1978. Physical properties and Tris inhibition of an insect trehalase and a thermodynamic approach to the nature of its active site. Biochim Biophys Acta 524:131–141. doi: 10.1016/0005-2744(78)90111-0. [DOI] [PubMed] [Google Scholar]
- 30.Jorge CD, Sampaio MM, Hreggvidsson GO, Kristjánson JK, Santos H. 2007. A highly thermostable trehalase from the thermophilic bacterium Rhodothermus marinus. Extremophiles 11:115–122. doi: 10.1007/s00792-006-0021-6. [DOI] [PubMed] [Google Scholar]
- 31.Asano N, Yamaguchi T, Kameda Y, Matsui K. 1987. Effect of validamycins on glycohydrolases of Rhizoctonia solani. J Antibiot 40:526–532. doi: 10.7164/antibiotics.40.526. [DOI] [PubMed] [Google Scholar]
- 32.Li H, Su H, Kim SB, Chang YK, Hong SK, Seo YG, Kim CJ. 2012. Enhanced production of trehalose in Escherichia coli by homologous expression of otsBA in the presence of the trehalase inhibitor, validamycin A, at high osmolarity. J Biosci Bioeng 113:224–232. doi: 10.1016/j.jbiosc.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 33.Iwasa T, Yamamoto H, Shibata M. 1970. Studies on validamycins, new antibiotics. I. Streptomyces hygroscopicus var. limoneus nov. var., validamycin producing organism. Jpn J Antibiot 23:595–602. doi: 10.7164/antibiotics.23.595. [DOI] [PubMed] [Google Scholar]
- 34.Guirao-Abad JP, Sánchez-Fresneda R, Valentín E, Martínez-Esparza M, Argüelles JC. 2013. Analysis of validamycin as a potential antifungal compound against Candida albicans. Int Microbiol 16:217–225. doi: 10.2436/20.1501.01.197. [DOI] [PubMed] [Google Scholar]
- 35.Salleh HM, Honek JF. 1990. Time-dependent inhibition of porcine kidney trehalase by aminosugars. FEBS Lett 262:359–362. doi: 10.1016/0014-5793(90)80229-C. [DOI] [PubMed] [Google Scholar]
- 36.Müller J, Staehelin C, Mellor RB, Boller T, Wiemken A. 1992. Partial purification and characterization of trehalase from soybean nodules. J Plant Physiol 140:8–13. doi: 10.1016/S0176-1617(11)81048-5. [DOI] [Google Scholar]
- 37.Sierks MR, Ford C, Reilly PJ, Svensson B. 1990. Catalytic mechanism of fungal glucoamylase as defined by mutagenesis of Asp176, Glu179 and Glu180 in the enzyme from Aspergillus awamori. Protein Eng 3:193–198. doi: 10.1093/protein/3.3.193. [DOI] [PubMed] [Google Scholar]
- 38.Harris EM, Aleshin AE, Firsov LM, Honzatko RB. 1993. Refined structure for the complex of 1-deoxynojirimycin with glucoamylase from Aspergillus awamori var. X100 to 2.4-Å resolution. Biochemistry 32:1618–1626. doi: 10.1021/bi00057a028. [DOI] [PubMed] [Google Scholar]
- 39.Frandsen TP, Dupont C, Lehmbeck J, Stoffer B, Sierks MR, Honzatko RB, Svensson B. 1994. Site-directed mutagenesis of the catalytic base glutamic acid 400 in glucoamylase from Aspergillus niger and of tyrosine 48 and glutamine 401, both hydrogen-bonded to the gamma-carboxylate group of glutamic acid 400. Biochemistry 33:13808–13816. doi: 10.1021/bi00250a035. [DOI] [PubMed] [Google Scholar]
- 40.Martins LO, Huber R, Huber H, Stetter KO, Da Costa MS, Santos H. 1997. Organic solutes in hyperthermophilic archaea. Appl Environ Microbiol 63:896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma A, Kawarabayasi Y, Satyanarayana T. 2012. Acidophilic bacteria and archaea: acid stable biocatalysts and their potential applications. Extremophiles 16:1–19. doi: 10.1007/s00792-011-0402-3. [DOI] [PubMed] [Google Scholar]
- 42.Vazquez-Figueroa E, Yeh V, Broering JM, Chaparro-Riggers JF, Bommarius AS. 2008. Thermostable variants constructed via the structure-guided consensus method also show increased stability in salts solutions and homogeneous aqueous-organic media. Protein Eng Des Sel 21:673–680. doi: 10.1093/protein/gzn048. [DOI] [PubMed] [Google Scholar]
- 43.Dror A, Shemesh E, Dayan N, Fishman A. 2014. Protein engineering by random mutagenesis and structure-guided consensus of Geobacillus stearothermophilus lipase T6 for enhanced stability in methanol. Appl Environ Microbiol 80:1515–1527. doi: 10.1128/AEM.03371-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aleshin AE, Feng PH, Honzatko RB, Reilly PJ. 2003. Crystal structure and evolution of a prokaryotic glucoamylase. J Mol Biol 327:61–73. doi: 10.1016/S0022-2836(03)00084-6. [DOI] [PubMed] [Google Scholar]
- 45.Iturriaga G, Suárez R, Nova-Franco B. 2009. Trehalose metabolism: from osmoprotection to signaling. Int J Mol Sci 10:3793–3810. doi: 10.3390/ijms10093793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Avonce N, Mendoza-Vargas A, Morett E, Iturriaga G. 2006. Insights on the evolution of trehalose biosynthesis. BMC Evol Biol 6:109. doi: 10.1186/1471-2148-6-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qu Q, Lee SJ, Boos W. 2004. TreT, a novel trehalose glycosyltransferring synthase of the hyperthermophilic archaeon Thermococcus litoralis. J Biol Chem 279:47890–47897. doi: 10.1074/jbc.M404955200. [DOI] [PubMed] [Google Scholar]
- 48.Kouril T, Zaparty M, Marrero J, Brinkmann H, Siebers B. 2008. A novel trehalose synthesizing pathway in the hyperthermophilic Crenarchaeon Thermoproteus tenax: the unidirectional TreT pathway. Arch Microbiol 190:355–369. doi: 10.1007/s00203-008-0377-3. [DOI] [PubMed] [Google Scholar]
- 49.Tourinho-dos-Santos CF, Bachinski N, Paschoalin VM, Paiva CL, Silva JT, Panek AD. 1994. Periplasmic trehalase from Escherichia coli: characterization and immobilization on spherisorb. Braz J Med Biol Res 27:627–636. [PubMed] [Google Scholar]
- 50.Mittenbühler K, Holzer H. 1988. Purification and characterization of acid trehalase from the yeast suc2 mutant. J Biol Chem 263:8537–8543. [PubMed] [Google Scholar]
- 51.App H, Holzer H. 1989. Purification and characterization of neutral trehalase from the yeast ABYS1 mutant. J Biol Chem 264:17583–17588. [PubMed] [Google Scholar]
- 52.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.