Abstract
The manner and extent to which birds associate with humans may influence the genetic attributes and antimicrobial resistance of their commensal Escherichia communities through strain transmission and altered selection pressures. In this study, we determined whether the distribution of the different Escherichia coli phylogenetic groups and cryptic clades, the occurrence of 49 virulence associated genes, and/or the prevalence of resistance to 12 antimicrobials differed between four groups of birds from Australia with contrasting types of human association. We found that birds sampled in suburban and wilderness areas had similar Escherichia communities. The Escherichia communities of backyard domestic poultry were phylogenetically distinct from the Escherichia communities sourced from all other birds, with a large proportion (46%) of poultry strains belonging to phylogenetic group A and a significant minority (17%) belonging to the cryptic clades. Wild birds sampled from veterinary and wildlife rehabilitation centers (in-care birds) carried Escherichia isolates that possessed particular virulence-associated genes more often than Escherichia isolates from birds sampled in suburban and wilderness areas. The Escherichia isolates from both the backyard poultry and in-care birds were more likely to be multidrug resistant than the Escherichia isolates from wild birds. We also detected a multidrug-resistant E. coli strain circulating in a wildlife rehabilitation center, reinforcing the importance of adequate hygiene practices when handling and caring for wildlife. We suggest that the relatively high frequency of antimicrobial resistance in the in-care birds and backyard poultry is due primarily to the use of antimicrobials in these animals, and we recommend that the treatment protocols used for these birds be reviewed.
INTRODUCTION
Escherichia coli is a generalist enteric bacterium that is able to colonize the lower intestinal tract of a range of vertebrate hosts, including most humans, other mammals, and birds. It is primarily a commensal; however, some strains are diarrheal pathogens and others can cause opportunistic extraintestinal infections in both humans and other animals (1). E. coli is less common in birds than mammals and can be isolated from approximately one-quarter of avian individuals (2). Birds that live close to human habitation are more likely to carry E. coli than birds in remote areas (2). This suggests that interactions and/or cohabitation between birds and humans as well as human activities and actions may influence avian E. coli communities. Here we collectively refer to these interactions and actions as “human association.” In this study, we examined whether the effect of human association on commensal avian E. coli strains extends to their genetic attributes and antimicrobial resistance traits. The distribution of these attributes and traits in avian hosts is of importance to both avian and human health, as they may affect veterinary treatment practices and birds could be reservoirs for virulent and/or antimicrobial-resistant E. coli strains, to which humans may be exposed.
Human association may influence the attributes of the commensal E. coli communities of domestic and wild birds through several processes, including strain transmission, horizontal gene transfer, and altered selection pressures. It has been shown that strains can be shared between host species (3), and thus, birds that cohabitat or come into direct contact with people may acquire human strains via fecal-oral transmission of E. coli through environmental contamination or host contact (4, 5). Additionally, genes found on plasmids or other mobile genetic elements can be readily transferred between bacteria of the same or different species (6). Thus, genes found in human strains could be transferred to avian strains by horizontal gene transfer (7). Alternatively, where humans directly interact with birds, as is the case for domestic animals and wild birds brought into rehabilitation centers, human actions such as hygiene practices and antimicrobial use may result in the selection and dissemination of particular strains of E. coli.
The genetic attributes of E. coli strains vary between host species and may reflect differences in host traits, transmission dynamics, and exposure. E. coli strains can be classified into different phylogenetic groups that differ in their ecological characteristics and virulence-associated genes (8–10). In particular, phylogenetic group B2 strains appear to be adapted to mammalian hosts and are often associated with extraintestinal infection in humans, companion animals, and avian species (1, 11, 12). In contrast, group B1 strains appear to be generalists and are more frequently isolated from ectotherms, birds, and the environment (2). The extent to which human association influences the distribution of these phylogenetic groups and virulence traits in birds is largely unknown.
In contrast to the genetic attributes of E. coli, the effect of human association on antimicrobial resistance has been relatively well documented. The use of antimicrobials selects for corresponding antimicrobial resistance in bacterial species, such as E. coli, with the prevalence of antimicrobial resistance being proportional to the extent of antimicrobial use (7). As such, selection for antimicrobial-resistant strains is most acute in institutions such as hospitals, aged care facilities, and veterinary clinics, where antimicrobial use is high. These strains can then be transferred between individuals admitted to such institutions (13, 14). Additionally, antimicrobial resistance has been shown to be more prevalent in wild animals in urban areas than in those in rural or remote regions (15, 16), presumably due to the transmission of antimicrobial-resistant E. coli strains or genes (through horizontal gene transfer) from humans and/or domestic animals.
In this study, we determined whether the distribution of the different E. coli phylogenetic groups and cryptic clades, the occurrence of 49 virulence-associated genes, and/or the proportion of isolates resistant to 12 antimicrobials differed between four groups of domestic and wild birds from Australia that had contrasting extents and types of human association. Through this analysis, we have gained insights into the probable mechanisms by which these traits have been established in avian E. coli.
MATERIALS AND METHODS
Sample collection.
Putative E. coli isolates were obtained from the feces of 594 Australian birds between 1994 and 2011. These birds belonged to 115 species, 81 genera, 42 families, and 17 orders (see Table S1 in the supplemental material). The birds were classified into four groups that reflected their probable association with humans. (i) Wild birds consisted of 68 birds that were sampled from wilderness areas (human population density, <500 per km2) and so were thought to rarely associate with humans and/or livestock. (ii) Suburban birds consisted of 126 nondomesticated birds that were sampled from suburban localities and thus had some degree of human association. The human population density varied greatly between these localities, ranging from 500 to greater to 5,000 per km2. (iii) In-care birds consisted of 246 isolates that were collected from nondomesticated birds admitted to veterinary hospitals or wildlife rehabilitation centers. (iv) Poultry consisted of 156 isolates that were collected from backyard domestic poultry; 137 of these were chickens, and the majority were sampled at the 2011 Royal Canberra National Poultry Show. The poultry included meat, egg, and fancy breeds and were kept primarily for showing by hobbyists. All poultry birds were kept in suburban backyards or on small rural properties. None of the poultry were from commercial poultry farms.
Birds were sampled from across five Australian states (Victoria, New South Wales, Queensland, Tasmania, and Western Australia [WA]) and the Australian Capital Territory (see Table S2 in the supplemental material). However, the majority of poultry and wild bird isolates came from animals in New South Wales. In general, when several samples were collected from the same bird species, they were obtained from diverse geographic locations. However, 62 long-beaked corellas and 15 rainbow lorikeets were sampled from single locations. As it could not be determined whether the Escherichia isolates from these birds reflected their locality, species, or extent of human association, data for all birds belonging to these species (n = 89) were excluded from all statistical analyses.
E. coli isolation and DNA extraction.
A single putative E. coli isolate was obtained from each sample by streaking the sample onto a MacConkey agar plate (17) that was then incubated at 35°C. Colonies were subsequently tested for citrate utilization by growth on minimal citrate agar plates. Putative E. coli isolates (lactose positive, citrate negative) were later confirmed by genetic analysis to be either E. coli or one of the cryptic clades (see “Phylogenetic group assignment” below). Isolates were maintained as freezer cultures (1 ml of lysogeny broth culture and 0.5 ml glycerol) and stored at −80°C.
DNA extractions were performed by plating the freezer cultures onto MacConkey agar plates (incubated at 35°C) and subsequently inoculating singles colonies into lysogeny broth that was then incubated for 19 h at 35°C with shaking (150 rpm). DNA was extracted from the cultures using the DNAzol genomic DNA isolation reagent (Molecular Research Center Inc.) according to the manufacturer's instructions. Precipitated DNA was resuspended in Tris-EDTA buffer.
Phylogenetic group assignment.
E. coli strains can be classified into seven different phylogenetic groups or subspecies (A, B1, B2, C, D, E, and F) (8). Additionally, five cryptic clades of Escherichia that are phenotypically indistinguishable from but genetically distinct from E. coli have recently been identified (18). Phylogenetic group membership was assigned by the quadruplex PCR method, as detailed in the work of Clermont et al. (19). All isolates that were identified as cryptic clade members in the quadruplex PCR were then screened to determine which cryptic clade they belonged to according to the method of Clermont et al. (20). All isolates that failed to produce a product for the quadruplex PCR were screened for the housekeeping genes gadA and uidA. Isolates that failed to yield a product for both uidA and gadA were considered not to be E. coli. All PCRs were performed as described by Blyton et al. (21).
We established whether the distribution of the different phylogenetic groups varied with the birds' extent of human association using multinomial log-linear regression models. The models were fitted using the nnet package (22) in R (v.2.15.2) (23). The response variable in the analysis was the phylogenetic group of each isolate (A, B1, B2, clade, D, or minor phylogenetic group), and the explanatory variable was the birds' human association category (wild, suburban, in-care, or poultry). As the birds in each of the human association categories differed in their characteristics, we then explored whether any additional variables could explain the distribution of the phylogenetic groups separately for each human association category. For the poultry isolates, the explanatory variables were the taxonomic order to which the birds belonged. For the isolates from wild, suburban, and in-care birds, the explanatory variables included the birds' (i) habitat (canopy, ground, or water), (ii) state or territory, (iii) diet (carnivore, granivore, herbivore, insectivore, nectarivore, omnivore), (iv) body mass (log10), and (v) taxonomic order. We then visualized the effects of the significant explanatory variables using regression trees fitted using the package Party (24) in R.
Virulence genes.
The E. coli and cryptic clade isolates were screened for the presence or absence of 49 virulence genes that belong to the variable portion of the Escherichia genome (25). While these genes were initially described for their association with intestinal or extraintestinal virulence in humans (26), they have subsequently been found to increase colonization success, persistence, and abundance within host individuals and occurrence in host populations of mammals (27–32). The genes were divided into 11 primer pools, and the isolates were screened by multiplex PCR (see Table 1 for gene names, functions, and references). The PCR mixes and amplification conditions were those described by Blyton et al. (32), except for the annealing temperatures, which were as follows: for pools 1 to 7, 63°C; for pools 8 and 9, 57°C; and for pools 10 and 11, 50°C.
TABLE 1.
Virulence genes screened by multiplex PCR
| Function | Gene(s) (primer pool no., reference) |
|---|---|
| Actin polymerization | ipaC (9, 35) |
| Bacteriocin | ColE1 (4, 55), ColK (6, 55), micV (6, 55), ColIa (6, 55), ColE6 (6, 55), ColM (7, 55), micH47 (9, 55), ColB (10, 55) |
| Capsule | K1 (10, 41), kpsMTII (2, 41) |
| Cell lysis | ehx (8, 56) |
| Iron uptake | ireA (1, 41), iroN (8, 43), sitA (3, 42), iutA (3, 57), fyuA (5, 41), eitA (1, 58) |
| Outer membrane protein | ompT (2, 43), eaeA (8, 56) |
| Toxin | hylA (1, 60), vat (2a), cdtA (3b), astA (7, 61), stx1 (8, 56), stx2 (8, 56), lta (10, 62), cdtB (11, 41), stlA (11, 62) |
| Epithelial invasion | ibeA2 (4, 41) |
| Adhesion, fimbriae, and pili | iha (1, 43), afa dra (1, 41), focH (2, 10), sfa foc (2, 63), papA (3, 41), fimH (3, 41), C1936 (4, 10), C2395 (4, 10), ppdD (4, 10), yehA (5, 10), aufA (5, 10), ygiL (5, 10), yfcV (5, 10), sfaS (5, 41), lpfA of Shigella (3, 64), lpfA of strain LF82 (7, 64), bfp (9, 35), agn43 (7, 65) |
| Plasma vapor deposition | pcvD (9, 66) |
Primers F-TCAGGACACGTTCAGGCATTCAGT and R-GGCCAGAACATTTGCTCCCTTGTT were used.
Primers F-TGCCGCTCTGACAGGTGGACTTA and R-GCCTTTAAAAACGGGGTGATACA were used.
To determine whether the E. coli and cryptic clade isolates sampled from birds in the different human association categories differed in their propensity to possess particular virulence genes, we fitted generalized linear models (family = binomial) using the lme4 package (33) in R. A separate analysis was performed for each virulence gene. The response variables were whether or not an isolate possessed a particular gene. The primary explanatory variable in the analyses was the birds' human association category. However, the occurrence of many of the virulence genes is known to vary between the different phylogenetic groups (34). Therefore, the phylogenetic group of the isolates was also included as an explanatory variable. For a particular gene, only phylogenetic groups and human association categories for which that gene occurred at a frequency of between 5% and 95% were included in the analysis (see Table 3).
TABLE 3.
Occurrence of virulence genes in Australian bird E. coli and cryptic clade isolates
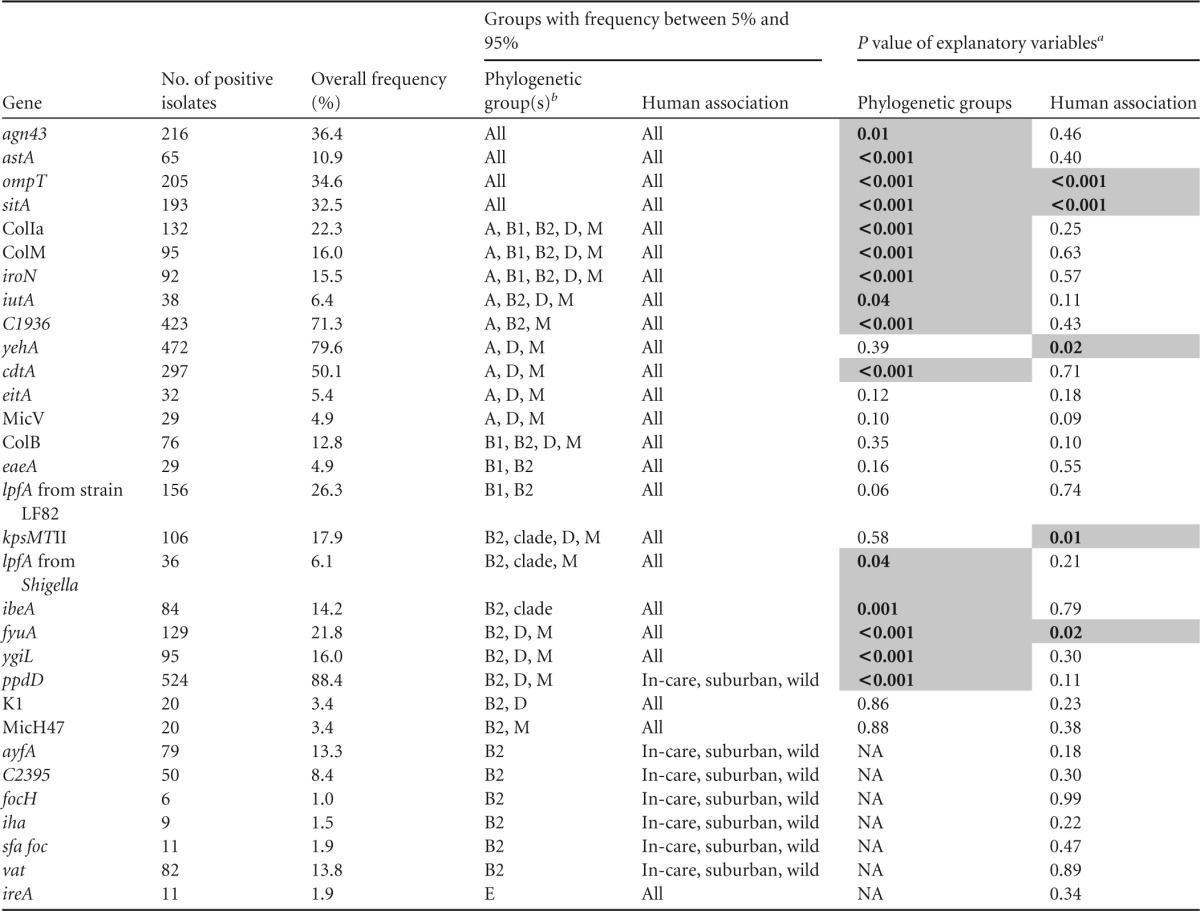
P values of <0.05 are shaded and shown in bold. NA, not applicable.
Clade, clades I, II, III, IV, and V; M, minor phylogenetic groups C, E, and F.
Different types of E. coli pathogens can be genetically identified by their virulence gene profiles. The set of genes screened for in this study included genes that allowed us to identify if any of the isolates were enteroaggregative, typical and atypical enteropathogenic, enteroinvasive, enterotoxigenic, enterohemorrhagic, or Shiga toxin-producing E. coli isolates, as defined by Robins-Browne et al. (35).
Antimicrobial resistance.
The E. coli and cryptic clade isolates were screened for their resistance to 12 antimicrobials using the European Committee on Antimicrobial Susceptibility Testing (EUCAST) disk diffusion method (v.3.0) (36). These antimicrobials included three cephalosporins (cefazolin, cefotaxime, and ceftazidime), two penicillins (ampicillin and amoxicillin-clavulanic acid), and two quinolones (ciprofloxacin and nalidixic acid) (Table 2). Isolates were scored as resistant, intermediate, or susceptible to an antimicrobial on the basis of BD's clinical inhibition zone diameter breakpoints. The inhibition zone diameters were measured using the automated colony counting and zone measuring instrument ProtoCol (v.3; Synbiosis). Isolates that were classified as intermediate according to their inhibition zone diameters were generally grouped with the resistant isolates for analysis. However, isolates classified as intermediate to nitrofurantoin, ampicillin, or cefazolin were grouped with the susceptible isolates, as there was no discernible break between the zone diameter distribution of those isolates and that of the susceptible isolates.
TABLE 2.
Antimicrobial resistance of Australian bird E. coli and cryptic clade isolates
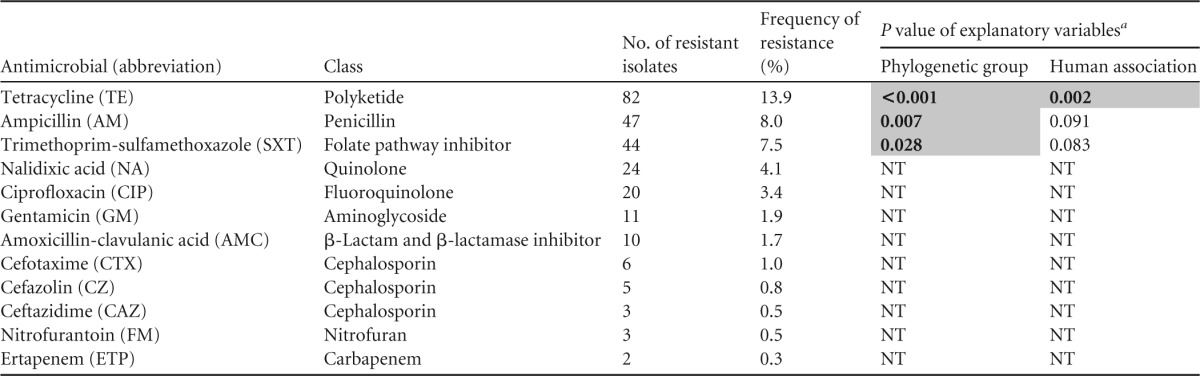
Statistical analyses did not include data for isolates from long-billed corellas, rainbow lorikeets, or the Western Australian wildlife rehabilitation clinic. P values of <0.05 are shaded and shown in bold. NT, not statistically tested.
To determine whether the E. coli and cryptic clade isolates sampled from the different bird-human association categories varied in their resistance to the antimicrobials, we performed two sets of analyses using generalized linear models (family = binomial) fitted in the lme4 package in R. In the first set of analyses, we fitted separate models for each antimicrobial to which resistance was detected in more than 5% of isolates. The response variables in these analyses were whether an isolate was resistant or susceptible to a particular antimicrobial. In the second analysis, we assessed whether or not an isolate was multidrug resistant. Isolates were classified as multidrug resistant if they were resistant to two or more classes of antimicrobials. In both sets of analyses, the primary explanatory variable was the birds' human association category. However, because antimicrobial resistance has been found to be less prevalent among phylogenetic group B2 strains (37, 38), phylogenetic group was also included as an explanatory variable.
RESULTS
Phylogenetic group.
The putative E. coli isolates were assigned to the different phylogenetic groups and cryptic clades in the following proportions: B1, 40.0% (n = 237 isolates); B2, 17.9% (n = 106); A, 17.5% (n = 104); D, 6.9% (n = 41); clade V, 4.3% (n = 26); E, 4.2% (n = 25); F, 3.7% (n = 22); clade III, 2.5% (n = 15); clade IV, 1.5% (n = 9); C <1% (n = 4); clade I, <1% (n = 3); and clade II, <1% (n = 1).
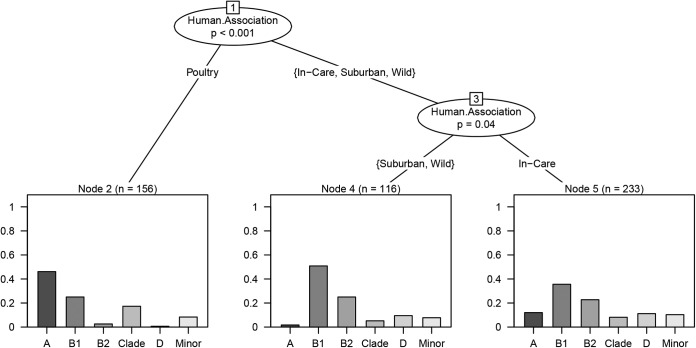
According to the results of multinomial regression analysis, the human association category of the birds had a significant effect on the phylogenetic group membership of the sampled Escherichia isolates (likelihood ratio statistic = 206.7, P < 0.001). The phylogenetic group distribution of the isolates differed between all human association categories except between wild and suburban birds (for wild versus suburban birds, P = 0.49; Fig. 1). The majority of wild and suburban bird isolates (n = 116) belonged to phylogenetic group B1 (57.5%), with B2 being the second most abundant phylogenetic group (22.8%). The major fraction of in-care bird isolates (n = 233) was also B1 (35.7%); however, group A, clade, group D, and the minor phylogenetic groups accounted for a larger proportion of the in-care bird isolates than they did of those from the suburban or wild birds. The poultry isolates (n = 156) were the most phylogenetically distinct group, being dominated by phylogenetic group A (46.1%) and including a comparative abundance of clade strains (17.3%; Fig. 1).
FIG 1.
Regression tree of how the distribution of the different phylogenetic groups differed between the different human association categories. The y axes represent proportion of isolates.
Among the poultry isolates, the various taxonomic orders of birds had significantly different phylogenetic group distributions (likelihood ratio statistic = 23.5, P < 0.001). The major fraction of Escherichia spp. from both the Anseriformes (ducks and geese) and Galliformes (chickens, quail, and guinea fowl) were phylogenetic group A. However, the Anseriformes (n = 24) possessed a very high proportion of clade strains (33.3%), whereas in the Galliformes (n = 132), the second most abundant phylogenetic group was B1 (Fig. 2).
FIG 2.
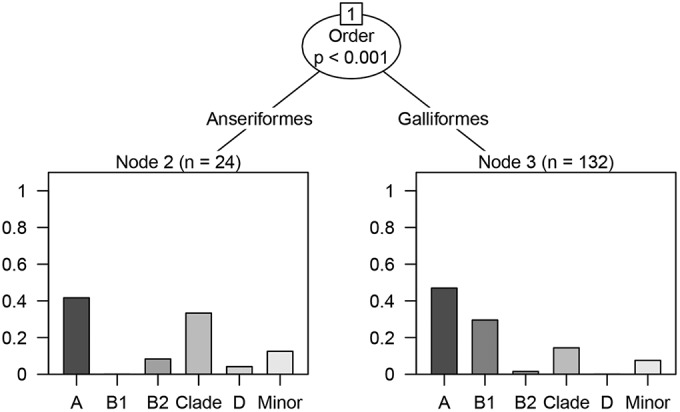
Regression tree of how the distribution of the different phylogenetic groups differed with the birds' taxonomic orders within the poultry. The y axes represent proportion of isolates.
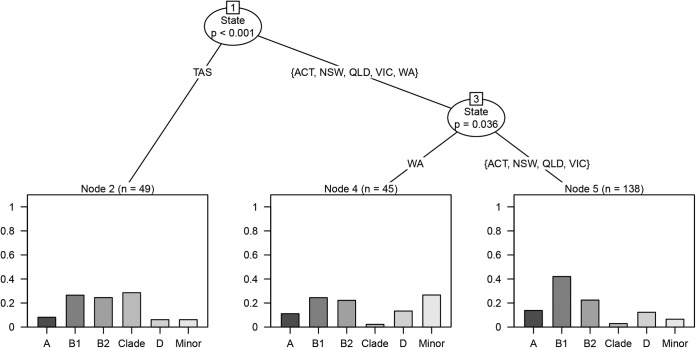
Among isolates from in-care birds, the phylogenetic group distribution varied by state or territory (likelihood ratio statistic = 63.6, P < 0.001). Isolates from Tasmania, sampled from a veterinary clinic and a wildlife rehabilitation center (n = 49), had a higher proportion of clade strains (28.6%) than isolates from the other states or territory (Fig. 3). These clade strains belonged predominantly to cryptic clade V. Additionally, the isolates from Western Australia (n = 45), sampled from a wildlife rehabilitation clinic, had a higher proportion of the minor phylogenetic groups (26.5%) than did the other in-care bird isolates. All except one of these minor phylogenetic group isolates belonged to phylogenetic group F.
FIG 3.
Regression tree of how the distribution of the different phylogenetic groups differed between the Australian states or territory within the in-care human association category. The y axes represent proportion of isolates. TAS, Tasmania; ACT, Australian Capital Territory; NSW, New South Wales; QLD, Queensland; VIC, Victoria; WA, Western Australia.
Among the suburban and wild bird isolates, none of the explanatory variables were significantly associated with the distribution of the different phylogenetic groups.
Virulence genes.
Among the 49 genes screened for, 7 were not detected in any isolate (afa dra, ColK, ColE6, ehc, stx2, ipaC, and bfp) and 17 were found in less than 5% of strains (pcvD, lta, stlA, stx1, sfaS, papA, focH, ColE1, hylA, cdtB, iha, ireA, sfa foc, MicH47, K1, MicV, and eaeA). One gene (fimH) was found at a very high frequency (96.6% of isolates).
The effect of the birds' human association category on the propensity of their E. coli and cryptic clade isolates to possess a particular trait could be assessed for 31 virulence factors that occurred at an intermediate frequency (5% to 95%) in at least one of the phylogenetic groups. There was a significant main effect of human association category on the frequency of 5 virulence genes (ompT, sitA, yehA, kpsMTII, and fyuA) after accounting for any effects of the phylogenetic groups. Among the 24 genes that were assessed across multiple phylogenetic groups, the frequency of 15 genes differed significantly between the phylogenetic groups assessed (Table 3).
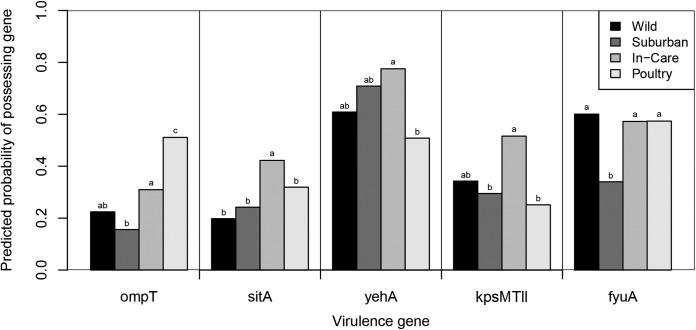
Among the five genes for which the human association category had a significant effect on their frequency, three (sitA, kpsMTII, and yehA; Fig. 4) were more prevalent among isolates from in-care birds than among isolates from birds in the other human association categories. The prevalence of sitA was significantly higher in isolates from in-care birds than those from birds in all other human association categories (Fig. 4). The prevalence of kpsMTII was significantly higher in isolates from in-care birds than those from suburban birds or poultry (Fig. 4). The prevalence of yehA was significantly higher in isolates from in-care birds than those from poultry (for in-care versus poultry isolates, P = 0.003; Fig. 4). Additionally, ompT was significantly more prevalent among poultry isolates than among isolates from birds in all other human association categories (Fig. 4) and among isolates from in-care birds than those from suburban birds (P = 0.028). Finally, fyuA was significantly less prevalent among isolates from suburban birds than those from birds in all other human association categories (Fig. 4).
FIG 4.
Predicted probability of virulence gene presence in relation to human association category. Predictions were made from the generalized linear regression models. The frequency of each phylogenetic group within each human association category was set to the overall mean for the Australian bird E. coli and cryptic clade isolates for the predictions. Probabilities for yehA were predicted for phylogenetic groups A and D and the minor phylogenetic group only. Probabilities for kpsMTII were predicted for phylogenetic group B2, cryptic clades, group D, and the minor phylogenetic groups (C, E, and F) only. Probabilities for fyuA were predicted for phylogenetic groups B2 and D and the minor phylogenetic groups (C, E, and F) only. Results for human association categories with different letters were significantly different from each other (P > 0.05).
The virulence gene profiles indicated that none of the study isolates were typical enteropathogenic, enteroinvasive, enterotoxigenic, enterohemorrhagic, or Shiga toxin-producing E. coli isolates. A single isolate from an in-care noisy minor (Manorina melanocephala) was a probable enteroaggregative pathogen, as it possessed pcvD. Twenty-nine bird isolates were potential atypical enteropathogenic E. coli isolates, as they possessed the eaeA gene. The eaeA gene occurred predominantly in isolates of phylogenetic groups B1 and B2, but it also occurred in isolates of phylogenetic group A. There was no evidence that the frequency of eaeA varied between the isolates from birds in the different human association categories (Table 3).
Antimicrobial resistance.
Antimicrobial resistance was observed for all tested antimicrobials (Table 2). Resistance to tetracycline (13.9%), ampicillin (8.0%), and trimethoprim-sulfamethoxazole (7.5%) was relatively common. Although rare, resistance to the critically important expanded-spectrum cephalosporins (cefotaxime, 1.0%; ceftazidime, 0.5%) was also observed. Antimicrobial resistance was relatively common among isolates from poultry and in-care birds (rates of resistance to one or more antimicrobials, 29.0% and 25.5%, respectively). In contrast, antimicrobial resistance was relatively uncommon among isolates from suburban and wild birds (rates of resistance to one or more antimicrobials, 4.8% and 3.0%, respectively).
We observed a high prevalence of antimicrobial resistance among isolates from a wildlife rehabilitation clinic in Western Australia (WA). In particular, 12 phylogenetic group F isolates from this clinic were multidrug resistant. All these isolates were resistant to trimethoprim-sulfamethoxazole, nalidixic acid, and a fluoroquinolone (ciprofloxacin). Ten of these isolates were also resistant to gentamicin, ampicillin, and tetracycline, and one isolate was resistant to amoxicillin-clavulanic acid. Gentamicin resistance occurred only in these phylogenetic group F isolates. Both nalidixic acid and ciprofloxacin resistance was also rare outside the WA clinic, occurring in only five and two isolates, respectively. However, among the WA clinic isolates (n = 45), 5 phylogenetic group B2 isolates and 1 phylogenetic group B1 isolate, in addition to the 12 above-mentioned group F isolates, were nalidixic acid and ciprofloxacin resistant. Due to the unusually high prevalence of antimicrobial resistance detected at the WA rehabilitation clinic, we excluded the data for that clinic's 45 isolates from our statistical analyses.
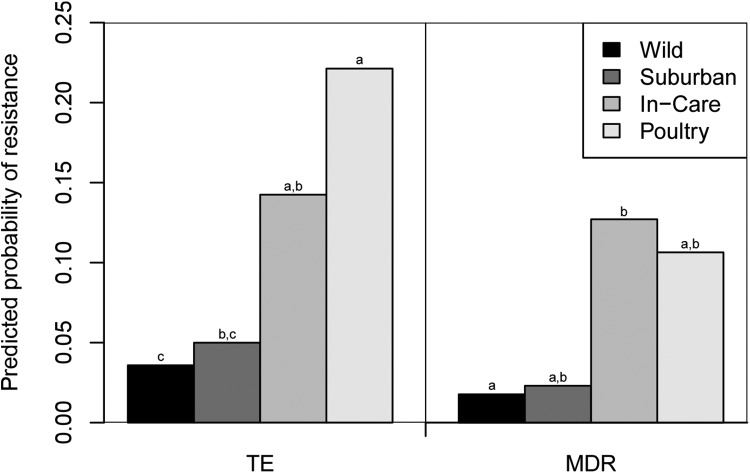
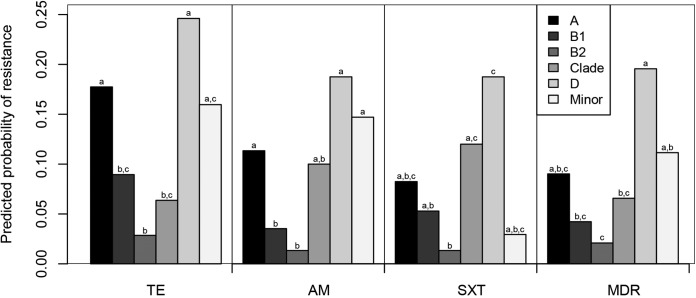
The frequency of resistance to three antimicrobials outside the WA wildlife clinic was sufficient for meaningful statistical analysis. Resistance to one of these antimicrobials (tetracycline) varied significantly by human association category, after accounting for any effect of phylogenetic group (Table 2). Tetracycline resistance was the most common in isolates from poultry (25.8%) and also was relatively common in isolates from in-care birds (15.2%) but was rare in isolates from suburban birds (2.4%) and wild birds (3.0%) (Fig. 4). Resistance to each of the three antimicrobials (tetracycline, ampicillin, and trimethoprim-sulfamethoxazole) tested in the statistical analyses was also found to vary significantly by phylogenetic group, after accounting for any effect of human association category (Table 2). Resistance to each of these antimicrobials was the highest among phylogenetic group D isolates and the lowest among phylogenetic group B2 isolates (Fig. 5).
FIG 5.
Predicted probabilities of an Escherichia isolate being resistant to tetracycline (TE) or multidrug resistant (MDR). Predictions were made from the generalized linear regression models. The frequency of each phylogenetic group within each human association category was set to the overall mean for the Australian bird E. coli and cryptic clade isolates for the predictions. The data used for the predictions did not include those from the Western Australian wildlife rehabilitation clinic. Results for human association categories with different letters were significantly different from each other (P < 0.05).
The prevalence of multidrug resistance varied significantly between the human association categories (data for WA clinic isolates were excluded; P = 0.009; Fig. 5). Multidrug resistance was relatively prevalent among isolates from the in-care birds and poultry (12.2% and 11.2%, respectively). In contrast, only a single multidrug-resistant isolate was detected in each of the suburban and wild birds (2.1% and 1.5%, respectively). The prevalence of multidrug resistance also varied significantly between the different phylogenetic groups (P = 0.004; Fig. 6). The incidence of multidrug resistance was the highest among phylogenetic group D isolates, and multidrug resistance was the least common among phylogenetic group B2 isolates.
FIG 6.
Predicted probabilities of an Escherichia isolate being resistant to an antimicrobial or multidrug resistant (MDR). TE, tetracycline; AM, ampicillin; SXT, trimethoprim-sulfamethoxazole. Predictions were made from the generalized linear regression models. Where the human association category was also a significant predictor of resistance, the human association categories were equally represented within each phylogenetic group for the predictions. The data used for the predictions did not include those from the Western Australian wildlife rehabilitation clinic. Results for phylogenetic groups with different letters were significantly different from each other (P < 0.05).
DISCUSSION
In this study, we assessed how the genetic attributes and antimicrobial resistance of commensal avian E. coli and cryptic clade isolates were influenced by the birds' type of human association, categorized into three wild bird groups, consisting of (i) birds from wilderness areas, (ii) birds from suburban locations, and (iii) birds receiving care in veterinary clinics or other wildlife rehabilitation facilities, and one domesticated bird group, consisting of backyard poultry. We found that the Escherichia spp. of backyard domestic poultry were phylogenetically distinct from the Escherichia spp. sourced from all other categories of birds. We found that Escherichia spp. from in-care birds possessed particular virulence-associated genes more often than Escherichia spp. from wild and suburban birds. Furthermore, Escherichia spp. from both the backyard poultry and in-care birds were more likely to be multidrug resistant than isolates from wild birds. In contrast, we found little difference between the Escherichia spp. isolated from wild and suburban birds. These differences and similarities between the avian E. coli and cryptic clade isolates from these different human association categories likely reflect differences in host traits, transmission dynamics, exposures, and selection pressures on the respective Escherichia communities between the groups. We discuss these potential processes for each human association category in detail below.
Wild and suburban bird E. coli and cryptic clade isolates.
The similarity between the E. coli and cryptic clade isolates from suburban and wild birds was surprising, given the findings of previous studies on the effects of human habitation on the prevalence of antimicrobial resistance. Several previous studies that compared animals in the same or different countries have found that animals in remote regions carry E. coli isolates with a lower prevalence of antimicrobial resistance than isolates carried by animals living close to human settlements (15). However, many of these studies primarily investigated mammalian hosts (for example, see reference 16), and in urban environments, the rate of transmission of strains or their genes between humans and animals may be less for birds than for mammals. Two studies that looked specifically at antimicrobial resistance in birds focused on gulls and geese that nested around waste or agricultural waters (39, 40). These waters likely had more extensive E. coli contamination from humans and domestic animals than the suburban environment of Australian cities. Thus, the low prevalence of antimicrobial resistance in both the suburban and wild bird Escherichia spp. evaluated in this study may reflect minimal exposure to human and domestic animal strains in both these groups.
E. coli and cryptic clade isolates recovered from birds in care.
It is difficult to interpret the finding that four virulence-associated genes (kpsMTII, sitA, ompT, and yehA) were overrepresented in Escherichia spp. from birds in care. These genes code for a diverse range of traits. kpsMTII encodes group 2 capsules (41), sitA encodes a protein involved in iron uptake (42), ompT encodes an outer membrane protein (43), and yehA encodes a putative fimbrial adhesion protein (10). Given the large number of statistical tests performed in this study, the results of individual analyses should be treated with caution. However, the overall finding of a greater virulence gene content in the Escherichia spp. from birds in care is convincing. The birds brought to these institutions were orphaned, had sustained an injury, or were sick. It is conceivable that the sick birds may have been carrying Escherichia spp. that more often possessed virulence-associated genes, as these genes are associated with disease-causing E. coli strains in humans. However, we do not possess individual-level information on the birds, so we were unable to determine whether those strains contributed to the birds' condition. Alternatively, the birds may have acquired Escherichia spp. possessing those virulence-associated genes from the environment at the rehabilitation centers.
The higher prevalence of multidrug resistance in the Escherichia spp. from the in-care birds than in isolates from wild and suburban birds is likely the result of selection for and acquisition of antimicrobial-resistant Escherichia spp. in those institutions. Nosocomial antimicrobial-resistant infections have been well described in humans and provide an indication of how prevalent they may be in animals where less information is available. It has been estimated that approximately 5% of people admitted to a hospital in the United States during 2002 acquired an infection while in the hospital (44). Furthermore, during 2006 and 2007, 16% of hospital-acquired infections were caused by multidrug-resistant organisms (44). Our findings show that the Escherichia isolates recovered from in-care birds were distinct from those recovered from wild or suburban birds in the same regions. In particular, we found that geographic location (state or territory) influenced the phylogenetic distribution of the strains from the in-care birds but not those from the wild or suburban birds and that a larger proportion of isolates from the in-care birds belonged to phylogenetic group A or D, the minor phylogenetic groups, and the cryptic clades. These results support the suggestion that birds at veterinary clinics and rehabilitation centers may acquire locally circulating Escherichia strains.
The circulation of several multidrug-resistant E. coli strains in veterinary teaching hospitals has been reported in Australia and the United States (13, 14). In both instances, the E. coli strains caused extraintestinal infections in dogs, were highly clonal, and were isolated from the hospital environment. Nucleotide sequencing data have shown that 11 of the phylogenetic group F isolates with similar multidrug resistance profiles from the WA clinic in our study were clonal (unpublished data). Therefore, it appears that there was a single phylogenetic group F strain circulating among the birds at the WA wildlife rehabilitation center. These phylogenetic group F isolates were resistant to at least 5 of the 12 antimicrobials tested, including the fluoroquinolone ciprofloxacin. Fluoroquinolone-resistant phylogenetic group F strains have previously been identified among fecal and urinary tract isolates from hospitalized dogs in Australia, with the vast majority belonging to sequence type 354 (ST354) (45). If the avian phylogenetic group F isolates from the WA clinic also belong to this sequence type, this would suggest that ST354 may be selected for in veterinary institutions, where antimicrobial use is common.
In addition to the circulation of the multidrug-resistant group F strain, there also appears to have been general selection for quinolone-resistant isolates at the WA wildlife rehabilitation clinic. Resistance to nalidixic acid and ciprofloxacin was rare outside this clinic, yet multiple phylogenetic group F, B2, and B1 isolates from the clinic were resistant to these antimicrobials. The fluoroquinolone enrofloxacin is commonly used to treat injured wildlife in Australia (46). Therefore, the prevalence of ciprofloxacin resistance observed in this clinic may have been the result of antimicrobial selection pressure mediated by enrofloxacin use.
Poultry E. coli and cryptic clade isolates.
The phylogenetic group distribution of the Escherichia isolates from backyard domestic poultry was broadly consistent with the findings from previous studies. In particular, the high proportion of phylogenetic group A strains fell within the range found in other studies. In four of five studies of fecal E. coli isolates from food-producing poultry, the proportion of phylogenetic group A strains ranged from 38 to 78% (47–51). Escobar-Páramo and colleagues (52) also found a higher proportion of phylogenetic group A strains in domestic birds (primarily poultry) than wild birds. It is unknown whether this high occurrence of phylogenetic group A strains in domestic poultry is a result of the avian hosts' domestic lifestyle or species-specific traits.
Consistent with the findings of Clermont and colleagues (20), in this study the isolates of the cryptic clades were more likely to be isolated from poultry than the other groups of birds. Those authors (20) found that isolates of the cryptic clades were more likely to be isolated from birds sampled in France than birds sampled in Australia, with farmyard poultry representing most of the French birds sampled. Because the cryptic Escherichia clades have only recently been described (18), relatively little is known about their patterns of occurrence and ecology. Ingle and colleagues (53) found that the cryptic clades have an enhanced ability to form biofilms compared to E. coli, can replicate at lower temperatures, and may be able to persist for longer periods in the external environment. Clade strains are generally rare in fecal samples, representing less than 3% of isolates in humans (20), although particular cryptic clade lineages may be found at higher frequencies in some species (21). Interestingly, we found that the Anseriformes (ducks and geese) had a higher proportion of cryptic clade strains than the Galliformes (chickens, quail, and guinea fowl). This could potentially reflect either a greater ability of the cryptic clades to inhabit these hosts or these hosts' association with common habitat features, such as water.
The higher prevalence of tetracycline resistance and multidrug resistance in the Escherichia spp. from backyard poultry than in isolates from suburban and wild birds is likely to be a result of antimicrobial use in the former. Tetracycline is not used to treat infections in humans in Australia; however, it is used to treat a broad spectrum of systemic infections in livestock, including poultry (54). Furthermore, backyard poultry may be medicated with a range of antimicrobials to treat bacterial diseases, particularly respiratory infections, as well as protozoan infections, including coccidiosis (54).
Conclusions.
In this study, we have shown that the extent and type of association between humans and birds influence the genetic attributes and antimicrobial resistance profiles of avian Escherichia communities. The similarity of antimicrobial resistance profiles and genetic attributes between Escherichia spp. from suburban and wild birds is reassuring, in that it suggests a low rate of transmission of Escherichia spp. between humans and birds in Australian urban environments. However, the relatively high prevalence of antimicrobial resistance and multidrug resistance detected among Escherichia spp. from in-care birds and poultry is concerning. We propose that the antimicrobial resistance observed in these groups is due primarily to the use of antimicrobials in these birds, and we advise that such use be carefully scrutinized. In particular, antimicrobials should be administered only in cases where an infection is strongly suspected or confirmed. Antimicrobial treatment should also be avoided for wildlife and poultry that have a low chance of survival. Additionally, the detection of a multidrug-resistant E. coli strain resistant to a fluoroquinolone circulating in a wildlife rehabilitation center reinforces the importance of adequate hygiene practices when handling and caring for sick as well as injured and orphaned wildlife.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00861-15.
REFERENCES
- 1.Johnson JR. 2011. Molecular epidemiology and population genetics of extraintestinal pathogenic Escherichia coli. In Walk ST, Feng PCH (ed), Population genetics of bacteria: a tribute to Thomas S. Whittam. ASM Press, Washington, DC. [Google Scholar]
- 2.Gordon DM, Cowling A. 2003. The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology 149:3575–3586. doi: 10.1099/mic.0.26486-0. [DOI] [PubMed] [Google Scholar]
- 3.Johnson JR, Owens K, Gajewski A, Clabots C. 2008. Escherichia coli colonization patterns among human household members and pets, with attention to acute urinary tract infection. J Infect Dis 197:218–224. doi: 10.1086/524844. [DOI] [PubMed] [Google Scholar]
- 4.Madigan MT, Martinko JM. 2006. Biology of microorganisms, 11th ed Pearson, Prentice-Hall, Upper Saddle River, NJ. [Google Scholar]
- 5.Blyton MD, Banks SC, Peakall R, Lindenmayer DB, Gordon DM. 2014. Not all types of host contacts are equal when it comes to E. coli transmission. Ecol Lett 17:970–978. doi: 10.1111/ele.12300. [DOI] [PubMed] [Google Scholar]
- 6.Syvanen M, Kado CI. 2001. Horizontal gene transfer. Academic Press, New York, NY. [Google Scholar]
- 7.van den Bogaard AE, Stobberingh EE. 2000. Epidemiology of resistance to antibiotics: links between animals and humans. Int J Antimicrob Agents 14:327–335. doi: 10.1016/S0924-8579(00)00145-X. [DOI] [PubMed] [Google Scholar]
- 8.Tenaillon O, Skurnik D, Picard B, Denamur E. 2010. The population genetics of commensal Escherichia coli. Nat Rev Microbiol 8:207–217. doi: 10.1038/nrmicro2298. [DOI] [PubMed] [Google Scholar]
- 9.Picard B, Garcia JS, Gouriou S, Duriez P, Brahimi N, Bingen E, Elion J, Denamur E. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun 67:546–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spurbeck RR, Stapleton AE, Johnson JR, Walk ST, Hooton TM, Mobley HLT. 2011. Fimbrial profiles predict virulence of uropathogenic Escherichia coli strains: contribution of Ygi and Yad fimbriae. Infect Immun 79:4753–4763. doi: 10.1128/IAI.05621-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nowrouzian FL, Adlerberth I, Wold AE. 2006. Enhanced persistence in the colonic microbiota of Escherichia coli strains belonging to phylogenetic group B2: role of virulence factors and adherence to colonic cells. Microbes Infect 8:834–840. doi: 10.1016/j.micinf.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 12.Nowrouzian FL, Wold AE, Adlerberth I. 2005. Escherichia coli strains belonging to phylogenetic group B2 have superior capacity to persist in the intestinal microflora of infants. J Infect Dis 191:1078–1083. doi: 10.1086/427996. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez S, Stevenson MM, Hudson CR, Maier M, Buffington T, Dam Q, Maurer JJ. 2002. Characterization of multidrug-resistant Escherichia coli isolates associated with nosocomial infections in dogs. J Clin Microbiol 40:3586–3595. doi: 10.1128/JCM.40.10.3586-3595.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sidjabat HE, Townsend KM, Lorentzen M, Gobius KS, Fegan N, Chin JJ-C, Bettelheim KA, Hanson ND, Bensink JC, Trott DJ. 2006. Emergence and spread of two distinct clonal groups of multidrug-resistant Escherichia coli in a veterinary teaching hospital in Australia. J Med Microbiol 55:1125–1134. doi: 10.1099/jmm.0.46598-0. [DOI] [PubMed] [Google Scholar]
- 15.Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J. 2010. Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol 8:251–259. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- 16.Skurnik D, Ruimy R, Andremont A, Amorin C, Rouquet P, Picard B, Denamur E. 2006. Effect of human vicinity on antimicrobial resistance and integrons in animal faecal Escherichia coli. J Antimicrob Chemother 57:1215–1219. doi: 10.1093/jac/dkl122. [DOI] [PubMed] [Google Scholar]
- 17.Power DA, McCuen PJ. 1988. Manual of BBL® products and laboratory procedures. Becton Dickinson Microbiology Systems, Cockeysville, MD. [Google Scholar]
- 18.Walk ST, Alm EW, Gordon DM, Ram JL, Toranzos GA, Tiedje JM, Whittam TS. 2009. Cryptic lineages of the genus Escherichia. Appl Environ Microbiol 75:6534–6544. doi: 10.1128/AEM.01262-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clermont O, Christenson JK, Denamur E, Gordon DM. 2013. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep 5:58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 20.Clermont O, Gordon DM, Brisse S, Walk ST, Denamur E. 2011. Characterization of the cryptic Escherichia lineages: rapid identification and prevalence. Environ Microbiol 13:2468–2477. doi: 10.1111/j.1462-2920.2011.02519.x. [DOI] [PubMed] [Google Scholar]
- 21.Blyton MDJ, Banks SC, Peakall R, Gordon DM. 2013. High temporal variability in commensal Escherichia coli strain communities of a herbivorous marsupial. Environ Microbiol 15:2162–2172. doi: 10.1111/1462-2920.12088. [DOI] [PubMed] [Google Scholar]
- 22.Venables WN, Ripley BD. 2002. Modern applied statistics with S, 4th ed Springer, New York, NY. [Google Scholar]
- 23.R Core Team. 2012. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- 24.Hothorn T. 2014. Party, v.1.0-17. Comprehensive R Archive Network, R Foundation for Statistical Computing, Vienna, Austria: http://cran.r-project.org/web/packages/party/index.html. [Google Scholar]
- 25.Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet 5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson JR, Delavari P, Kuskowski M, Stell AL. 2001. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J Infect Dis 183:78–88. doi: 10.1086/317656. [DOI] [PubMed] [Google Scholar]
- 27.Herias MV, Midtvedt T, Hanson LA, Wold AE. 1997. Escherichia coli K5 capsule expression enhances colonization of the large intestine in the gnotobiotic rat. Infect Immun 65:531–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nowrouzian F, Adlerberth I, Wold AE. 2001. P fimbriae, capsule and aerobactin characterize colonic resident Escherichia coli. Epidemiol Infect 126:11–18. doi: 10.1017/S0950268801005118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schierack P, Walk N, Ewers C, Wilking H, Steinrück H, Filter M, Wieler LH. 2008. ExPEC-typical virulence-associated genes correlate with successful colonization by intestinal E. coli in a small piglet group. Environ Microbiol 10:1742–1751. doi: 10.1111/j.1462-2920.2008.01595.x. [DOI] [PubMed] [Google Scholar]
- 30.Lasaro MA, Salinger N, Zhang J, Wang Y, Zhong Z, Goulian M, Zhu J. 2009. F1C fimbriae play an important role in biofilm formation and intestinal colonization by the Escherichia coli commensal strain Nissle 1917. Appl Environ Microbiol 75:246–251. doi: 10.1128/AEM.01144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diard M, Garry L, Selva M, Mosser T, Denamur E, Matic I. 2010. Pathogenicity-associated islands in extraintestinal pathogenic Escherichia coli are fitness elements involved in intestinal colonization. J Bacteriol 192:4885–4893. doi: 10.1128/JB.00804-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blyton MD, Banks SC, Peakall R, Gordon DM. 2013. Functional genotypes are associated with commensal Escherichia coli strain abundance within-host individuals and populations. Mol Ecol 22:4112–4122. doi: 10.1111/mec.12364. [DOI] [PubMed] [Google Scholar]
- 33.Bates D, Maechler M, Bolker B. 2012. lme4: linear mixed-effects models using S4 classes. R Foundation for Statistical Computing, Vienna, Austria: http://cran.r-project.org/web/packages/lme4/index.html. [Google Scholar]
- 34.Gordon DM, Stern SE, Collignon PJ. 2005. Influence of the age and sex of human hosts on the distribution of Escherichia coli ECOR groups and virulence traits. Microbiology 151:15–23. doi: 10.1099/mic.0.27425-0. [DOI] [PubMed] [Google Scholar]
- 35.Robins-Browne RM, Bordun A-M, Tauschek M, Bennett-Wood VR, Russell J, Oppedisano F, Lister NA, Bettelheim KA, Fairley CK, Sinclair MI. 2004. Escherichia coli and community-acquired gastroenteritis, Melbourne, Australia. Emerg Infect Dis 10:1797. doi: 10.3201/eid1010.031086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.EUCAST. 2013. Antimicrobial susceptibility testing EUCAST disk diffusion method. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Disk_test_documents/Manual_v_3.0_EUCAST_Disk_Test.pdf. [Google Scholar]
- 37.Johnson JR, Kuskowski MA, Owens K, Clabots C, Singer RS. 2009. Virulence genotypes and phylogenetic background of fluoroquinolone-resistant and susceptible Escherichia coli urine isolates from dogs with urinary tract infection. Vet Microbiol 136:108–114. doi: 10.1016/j.vetmic.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 38.Skurnik D, Le Menac'h A, Zurakowski D, Mazel D, Courvalin P, Denamur E, Andremont A, Ruimy R. 2005. Integron-associated antibiotic resistance and phylogenetic grouping of Escherichia coli isolates from healthy subjects free of recent antibiotic exposure. Antimicrob Agents Chemother 49:3062–3065. doi: 10.1128/AAC.49.7.3062-3065.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cole D, Drum DJ, Stallknecht DE, White DG, Lee MD, Ayers S, Sobsey M, Maurer JJ. 2005. Free-living Canada geese and antimicrobial resistance. Emerg Infect Dis 11:935–938. doi: 10.3201/eid1106.040717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dolejska M, Cizek A, Literak I. 2007. High prevalence of antimicrobial-resistant genes and integrons in Escherichia coli isolates from black-headed gulls in the Czech Republic. J Appl Microbiol 103:11–19. doi: 10.1111/j.1365-2672.2006.03241.x. [DOI] [PubMed] [Google Scholar]
- 41.Johnson JR, Stell AL. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis 181:261–272. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- 42.Runyen-Janecky L, Reeves S, Gonzales E, Payne S. 2003. Contribution of the Shigella flexneri Sit, Iuc, and Feo iron acquisition systems to iron acquisition in vitro and in cultured cells. Infect Immun 71:1919–1928. doi: 10.1128/IAI.71.4.1919-1928.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodriguez-Siek KE, Giddings CW, Doetkott C, Johnson TJ, Nolan LK. 2005. Characterizing the APEC pathotype. Vet Res 36:241–256. doi: 10.1051/vetres:2004057. [DOI] [PubMed] [Google Scholar]
- 44.Calfee DP. 2012. Crisis in hospital-acquired, healthcare-associated infections. Annu Rev Med 63:359–371. doi: 10.1146/annurev-med-081210-144458. [DOI] [PubMed] [Google Scholar]
- 45.Guo SY, Wakeham D, Brouwers HJM, Cobbold RN, Abraham S, Mollinger JL, Johnson JR, Chapman TA, Gordon DM, Barrs VR, Trott DJ. 2015. Human-associated fluoroquinolone-resistant Escherichia coli clonal lineages, including ST354, isolated from canine feces and extraintestinal infections in Australia. Microbes Infect 17:266–274. doi: 10.1016/j.micinf.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 46.Gillett A. 2010. VetCheck: medicating wildlife. WildNews 2010(56):8–9. [Google Scholar]
- 47.Jakobsen L, Kurbasic A, Skjøt-Rasmussen L, Ejrnaes K, Porsbo LJ, Pedersen K. 2010. Escherichia coli isolates from broiler chicken meat, broiler chickens, pork, and pigs share phylo-groups and antimicrobial resistance with community dwelling humans and patients with urinary tract infection. Foodborne Pathog Dis 7:537–547. doi: 10.1089/fpd.2009.0409. [DOI] [PubMed] [Google Scholar]
- 48.Johnson TJ, Logue CM, Wannemuehler Y, Kariyawasam S, Doetkott C, DebRoy C, White DG, Nolan LK. 2009. Examination of the source and extended virulence genotypes of Escherichia coli contaminating retail poultry meat. Foodborne Pathog Dis 6:657–667. doi: 10.1089/fpd.2009.0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson TJ, Wannemuehler Y, Doetkott C, Johnson SJ, Rosenberger SC, Nolan LK. 2008. Identification of minimal predictors of avian pathogenic Escherichia coli virulence for use as a rapid diagnostic tool. J Clin Microbiol 46:3987–3996. doi: 10.1128/JCM.00816-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ewers C, Antão E-M, Diehl I, Philipp H-C, Wieler LH. 2009. Intestine and environment of the chicken as reservoirs for extraintestinal pathogenic Escherichia coli strains with zoonotic potential. Appl Environ Microbiol 75:184–192. doi: 10.1128/AEM.01324-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carlos C, Pires MM, Stoppe NC, Hachich EM, Sato MI, Gomes TA, Amaral LA, Ottoboni LM. 2010. Escherichia coli phylogenetic group determination and its application in the identification of the major animal source of fecal contamination. BMC Microbiol 10:161. doi: 10.1186/1471-2180-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Escobar-Páramo P, Menac'h L, Le Gall T, Amorin C, Gouriou S, Picard B, Skurnik D, Denamur E. 2006. Identification of forces shaping the commensal Escherichia coli genetic structure by comparing animal and human isolates. Environ Microbiol 8:1975–1984. doi: 10.1111/j.1462-2920.2006.01077.x. [DOI] [PubMed] [Google Scholar]
- 53.Ingle DJ, Clermont O, Skurnik D, Denamur E, Walk ST, Gordon DM. 2011. Biofilm formation by and thermal niche and virulence characteristics of Escherichia spp. Appl Environ Microbiol 77:2695–2700. doi: 10.1128/AEM.02401-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shaban RZ, Simon GI, Trott DJ, Turnidge J, Jordan D. 2014. Surveillance and reporting of antimicrobial resistance and antibiotic usage in animals and agriculture in Australia. Report to the Department of Agriculture, Griffith University, and University of Adelaide, Australia; Department of Agriculture, Canberra, Australia: http://www.daff.gov.au/SiteCollectionDocuments/animal-plant/animal-health/amria.pdf. [Google Scholar]
- 55.Gordon DM, O'Brien CL. 2006. Bacteriocin diversity and the frequency of multiple bacteriocin production in Escherichia coli. Microbiology 152:3239–3244. doi: 10.1099/mic.0.28690-0. [DOI] [PubMed] [Google Scholar]
- 56.Paton AW, Paton JC. 1998. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J Clin Microbiol 36:598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnson JR, Brown JJ, Carlino UB, Russo TA. 1998. Colonization with and acquisition of uropathogenic Escherichia coli as revealed by polymerase chain reaction-based detection. J Infect Dis 177:1120–1124. doi: 10.1086/517409. [DOI] [PubMed] [Google Scholar]
- 58.Johnson TJ, Siek KE, Johnson SJ, Nolan LK. 2006. DNA sequence of a ColV plasmid and prevalence of selected plasmid-encoded virulence genes among avian Escherichia coli strains. J Bacteriol 188:745–758. doi: 10.1128/JB.188.2.745-758.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maurer JJ, Brown TP, Steffens W, Thayer SG. 1998. The occurrence of ambient temperature-regulated adhesins, curli, and the temperature-sensitive hemagglutinin Tsh among avian Escherichia coli. Avian Dis 42:106–118. doi: 10.2307/1592582. [DOI] [PubMed] [Google Scholar]
- 60.Yamamoto S, Terai A, Yuri K, Kurazono H, Takeda Y, Yoshida O. 1995. Detection of urovirulence factors in Escherichia coli by multiplex polymerase chain reaction. FEMS Immunol Med Microbiol 12:85–90. doi: 10.1111/j.1574-695X.1995.tb00179.x. [DOI] [PubMed] [Google Scholar]
- 61.Ngeleka M, Pritchard J, Appleyard G, Middleton DM, Fairbrother JM. 2003. Isolation and association of Escherichia coli AIDA-I/STb, rather than EAST1 pathotype, with diarrhea in piglets and antibiotic sensitivity of isolates. J Vet Diagn Invest 15:242–252. doi: 10.1177/104063870301500305. [DOI] [PubMed] [Google Scholar]
- 62.Schultsz C, Pool GJ, Van Ketel R, De Wever B, Speelman P, Dankert J. 1994. Detection of enterotoxigenic Escherichia coli in stool samples by using nonradioactively labeled oligonucleotide DNA probes and PCR. J Clin Microbiol 32:2393–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Le Bouguenec C, Archambaud M, Labigne A. 1992. Rapid and specific detection of the pap, afa, and sfa adhesin-encoding operons in uropathogenic Escherichia coli strains by polymerase chain reaction. J Clin Microbiol 30:1189–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chassaing B, Rolhion N, de Vallée A, Salim Sa Prorok-Hamon YM, Neut C, Campbell BJ, Söderholm JD, Hugot J-P, Colombel J-F. 2011. Crohn disease-associated adherent-invasive E. coli bacteria target mouse and human Peyer's patches via long polar fimbriae. J Clin Invest 121:966. doi: 10.1172/JCI44632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang H-H, Vinopal RT, Grasso D, Smets BF. 2004. High diversity among environmental Escherichia coli isolates from a bovine feedlot. Appl Environ Microbiol 70:1528–1536. doi: 10.1128/AEM.70.3.1528-1536.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmidt H, Knop C, Franke S, Aleksic S, Heesemann J, Karch H. 1995. Development of PCR for screening of enteroaggregative Escherichia coli. J Clin Microbiol 33:701–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.