Abstract
Human norovirus (HuNoV) represents a significant public health burden worldwide and can be environmentally transmitted. Copper surfaces have been shown to inactivate the cultivable surrogate murine norovirus, but no such data exist for HuNoV. The purpose of this study was to characterize the destruction of GII.4 HuNoV and virus-like particles (VLPs) during exposure to copper alloy surfaces. Fecal suspensions positive for a GII.4 HuNoV outbreak strain or GII.4 VLPs were exposed to copper alloys or stainless steel for 0 to 240 min and recovered by elution. HuNoV genome integrity was assessed by reverse transcription-quantitative PCR (RT-qPCR) (without RNase treatment), and capsid integrity was assessed by RT-qPCR (with RNase treatment), transmission electron microscopy (TEM), SDS-PAGE/Western blot analysis, and a histo-blood group antigen (HBGA) binding assay. Exposure of fecal suspensions to pure copper for 60 min reduced the GII.4 HuNoV RNA copy number by ∼3 log10 units when analyzed by RT-qPCR without RNase treatment and by 4 log10 units when a prior RNase treatment was used. The rate of reduction of the HuNoV RNA copy number was approximately proportional to the percentage of copper in each alloy. Exposure of GII.4 HuNoV VLPs to pure-copper surfaces resulted in noticeable aggregation and destruction within 240 min, an 80% reduction in the VP1 major capsid protein band intensity in 15 min, and a near-complete loss of HBGA receptor binding within 8 min. In all experiments, HuNoV remained stable on stainless steel. These results suggest that copper surfaces destroy HuNoV and may be useful in preventing environmental transmission of the virus in at-risk settings.
INTRODUCTION
Human norovirus (HuNoV) is the leading cause of viral gastroenteritis worldwide (1, 2). In the United States alone, the virus is responsible for 19 million to 21 million illnesses annually, contributing to nearly $500 million in hospital-associated costs each year (3, 4). The illness is characterized by nausea, vomiting, and diarrhea and typically lasts 24 to 48 h. While the disease is usually self-limiting, in individuals belonging to sensitive populations, symptoms can become life threatening if rehydration therapy is ignored.
HuNoV transmission occurs via the fecal-oral route, usually through ingestion of contaminated food or water, or by direct contact with an infected individual. Environmental transmission also occurs, and episodes of vomiting or diarrhea can contaminate surfaces with infectious virus particles that may persist for weeks (5). Environmental persistence of HuNoV is enhanced by its resistance to a variety of commonly used sanitizers and disinfectants, for example, alcohol-based hand sanitizers and hypochlorite at regulated concentrations (6, 7). These unique traits of HuNoV, combined with the low infectious dose and the fact that infected individuals shed a large amount of virus in both fecal material and vomitus, contribute to the high number of outbreaks observed annually in environments with close quarters such as cruise ships, restaurants, long-term-care facilities, and schools (8, 9). Clearly, there is a need for improved measures to halt the rapid spread of HuNoV in these settings.
Copper and its alloys have long been recognized for their antimicrobial properties. The first recorded medical use of copper dates to between 2600 and 2200 BC, when the Egyptians were reported to have used copper for sterilizing chest wounds and drinking water (10). The widespread use of copper as an antimicrobial has diminished with the development of commercially available antibiotics, but in recent years, interest has been renewed, due largely to the increased prevalence of antibiotic-resistant bacteria. Recently, the U.S. Environmental Protection Agency (EPA) registered several classes of copper alloys for use as antimicrobial surfaces (http://www.epa.gov/pesticides/factsheets/copper-alloy-products.htm). These alloys have been shown to have antimicrobial effects on a wide variety of bacterial and viral pathogens, including Salmonella enterica, methicillin-resistant Staphylococcus aureus (MRSA), Clostridium difficile vegetative cells and spores, and influenza A virus (H1N1), among others (11). Additionally, these copper alloys have been used to successfully reduce the microbial burden in hospital settings when integrated into high-touch surfaces such as door knobs and bed rails (12).
Given the demonstrated antiviral efficacy of copper-containing surfaces and their successful use as an antimicrobial touch surface in hospital settings, these alloys have been suggested for use in reducing the environmental transmission of HuNoV. However, little research into the efficacy of copper and its alloys against HuNoV has been performed. Recently, copper alloys were shown to be effective at inactivating murine norovirus (MNV), a cultivable surrogate for HuNoV (13, 14). Since MNV differs from HuNoV in a number of ways (15), we sought to perform similar experiments using HuNoV. Therefore, the goal of this study was to evaluate the effectiveness of copper alloy surfaces in the inactivation of HuNoV. Given the lack of a robust cell culture-based infectivity assay for HuNoV, we focused on characterizing the destruction of the HuNoV genome and capsid using combinations of reverse transcription-quantitative PCR (RT-qPCR), transmission electron microscopy (TEM), sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), Western blot analysis, and receptor binding analysis.
MATERIALS AND METHODS
HuNoV strains and virus-like particles.
Clinical stool specimens obtained from GII.4 HuNoV outbreaks and confirmed to contain GII.4 HuNoV by RT-qPCR were kindly provided by S. R. Greene (North Carolina Division of Public Health, Raleigh, NC). Stool samples were prepared as 20% suspensions in phosphate-buffered saline (PBS) (pH 7.2), clarified by low-speed centrifugation (3,100 × g) for 2 min, and stored at −80°C until use. HuNoV GII.4 Grimsby and GII.4 Houston virus-like particles (VLPs) were kindly provided by R. Atmar (Baylor College of Medicine, Houston, TX). Stock VLP concentrations were 1,700 ng/μl and 5,900 ng/μl for GII.4 Grimsby and GII.4 Houston, respectively. The average size of VLPs was observed to be 30 to 38 nm.
Preparation of sample surfaces.
Copper alloys and stainless steel sheets were fabricated into coupons (2.54 by 2.54 by 0.5 mm). The composition of each alloy is listed in Table 1. Prior to exposure experiments, surfaces were degreased in acetone, washed in distilled water, air dried, and then sterilized by autoclaving. All experiments were performed under ambient conditions on a laboratory benchtop. Coupons were autoclaved and discarded after each use.
TABLE 1.
Composition of metal surfaces used in this study
Inoculation of surfaces with HuNoV fecal suspensions and VLPs.
For all experiments using HuNoV-positive 20% fecal suspensions, the inoculum consisted of 25 μl per coupon. Coupons were maintained under ambient conditions of temperature and relative humidity. Elution was performed after 0, 15, 30, 60, 120, and 240 min of exposure by using 200 μl PBS-EDTA (PBS [pH 7.2] with 20 mM EDTA). Elution was done as previously reported (16), by pipetting up and down ∼25 times, followed by collection of the total eluate volume. Eluted samples were stored at −80°C until analysis by RT-qPCR. A similar inoculation-and-elution protocol was used for the VLP experiments, with minor modifications. Specifically, the VLP inoculum volume was 1 μl, the elution volume was 20 μl, and studies were performed only on stainless steel and pure copper coupons.
Analysis of eluates by RT-qPCR.
Fecal suspension eluates both with and without prior RNase treatment were tested by RT-qPCR. The RNase treatment degrades free HuNoV genomes, whose amplification by RT-qPCR may lead to an overestimation of the amount of infectious virus. Thus, the RNase treatment followed by RT-qPCR serves as a means to estimate genome copy numbers from fully intact capsids. Omission of the RNase treatment yields an RT-qPCR assay that amplifies all intact viral RNA and RNA associated with undamaged, damaged, or destroyed capsids. This method can provide a proxy for genome integrity, since any loss of the RT-qPCR signal is attributed to an alloy effect and not RNase treatment. For RT-qPCR with RNase treatment, a 100-μl volume of eluate was mixed with 1 μl RNase One (Promega, Madison, WI) and 11 μl 10× reaction buffer, followed by incubation at 37°C for 15 min. Samples were then placed on ice, and 25 μl of ice-cold PBS was added to stop the reaction, followed by RNA extraction. For RT-qPCR without prior RNase treatment, the remaining 100 μl of the eluate was used as the input for nucleic acid extraction. Nucleic acid extraction was performed by using the automated NucliSENS easyMag system (bioMérieux, St. Louis, MO) according to the manufacturer's instructions, with reconstitution of the final pellet in 20 μl of proprietary buffer. RNA extracts were stored at −80°C until analysis by RT-qPCR.
RNase-treated samples were subjected to RT-qPCR amplification targeting the conserved region of the open reading frame 1 (ORF1)-ORF2 junction of the HuNoV genome. Primers JJV2F (5′-CAAGAGTCAATGTTTAGGTGGATGAG-3′) and COG2R (5′-TCGACGCCATCTTCATTCACA-3′) and probe RING2-P (5′-FAM [6-carboxyfluorescein]-TGGGAGGGCGATCGCAATCT-BHQ [black hole quencher]-3′) were used (17). The 25-μl RT-qPCR mixture consisted of 2.5 μl of HuNoV RNA, 200 nM forward and reverse primers, 200 nM fluorescently labeled TaqMan probe, 1× Bio-Rad PCR buffer (Bio-Rad, Hercules, CA), and 0.5 μl Bio-Rad iScript RT mix. The reaction mixture was subjected to a one-step thermal cycling profile using a CFX96 Touch real-time PCR detection system (Bio-Rad) under the following amplification conditions: (i) reverse transcription for 10 min at 50°C, (ii) an initial denaturation step for 5 min at 95°C, and (iii) 45 cycles of 15 s at 95°C and 30 s at 55°C.
Estimation of genome copy numbers was performed by comparison with a standard curve generated using RNA transcripts of amplified HuNoV genome fragments (16). These transcripts corresponded to nucleotides 5003 to 5473 (HuNoV GII.4 [NCBI accession number JX126913]). Transcript concentrations ranged from 5.33 × 1015 to 6.37 × 1015 RNA copies/μl, and these transcripts were stored at −80°C until use. RNA transcripts were serially diluted in diethylpyrocarbonate (DEPC)-treated water prior to use for the production of standard curves by using RT-qPCR. The log10-transformed RNA copy number was plotted against the threshold cycle (CT) value and analyzed by linear regression to make the standard curve.
Transmission electron microscopy.
Transmission electron microscopy (TEM) experiments were performed by using HuNoV GII.4 Grimsby VLPs eluted from copper and stainless steel surfaces after exposure times of 0, 60, 120, and 240 min. The elution buffer consisted of ultrapure water supplemented with 20 mM EDTA, since PBS is known to interfere with the staining efficacy of uranyl acetate. Eluates were fixed in carbon-coated nickel grids (Ladd Research, Williston, VT) and negatively stained with 2% uranyl acetate. The VLPs were visualized by using a JEOL 1210 transmission electron microscope (JEOL-USA, Inc., Peabody, MA) at 80 kV at the Center for Electron Microscopy (North Carolina State University, Raleigh, NC).
SDS-PAGE.
SDS-PAGE experiments were performed by using a HuNoV GII.4 Houston VLP suspension exposed to stainless steel and copper coupons from 0 to 60 min. After exposure, VLP eluates were combined 1:1 with 2× Laemmli buffer (Bio-Rad, Hercules, CA), boiled at 95°C for 5 min, and run on 12% Mini-Protean TGX precast gels (Bio-Rad) with a Spectra broad-range protein ladder (Thermo Fisher Scientific). Gels were stained with AcquaStain protein gel stain (Bulldog Bio, Portsmouth, NH) and imaged by using an AlphaImager gel documentation system (Protein Simple, Santa Clara, CA). Band intensities for each time point were determined by using ImageJ software (National Institutes of Health [http://imagej.nih.gov/ij/]).
Western blotting.
The SDS-PAGE protein gels were transferred onto 0.45-μm nitrocellulose membranes (Bio-Rad) by using a Mini Trans-Blot electrophoretic transfer cell (Bio-Rad) and blocked in 10% skim milk–PBS with 0.1% Tween 20 overnight, with gentle shaking at 4°C. The membrane was then treated with 10 ml of PBS–0.05% Tween 20 (PBST) to which 15 μg of mouse monoclonal anti-norovirus GII.4 antibody (ab80024; Abcam, Cambridge, England) had been added, followed by incubation at room temperature (RT) for 1 h with shaking. Membranes were washed three times with PBST for 10 min and then treated with goat anti-mouse IgG-alkaline phosphatase-conjugated antibody (Sigma-Aldrich, St. Louis, MO) diluted 1:5,000 in 5% skim milk–PBST for 1.5 h at room temperature. Membranes were washed three times for 10 min in PBST and developed for 10 to 15 min at room temperature with a 5-bromo-4-chloro-3-indoyl-phosphate/p-nitroblue tetrazolium chloride (BCIP/NBT) solution (MP Biomedicals, Santa Ana, CA).
HBGA receptor binding assays.
Receptor binding assays were performed by using a HuNoV GII.4 Houston VLP suspension exposed to stainless steel and copper coupons for 0 to 10 min. The histo-blood group antigen (HBGA) receptor binding assay was adapted from methods described in a previous study (18). Briefly, VLP eluates were adjusted to a final concentration of 6 ng/μl in PBS. One hundred microliters (600 ng) was then applied to individual wells of a 96-well polystyrene medium-binding flat-well plate (Costar 3591; Thermo Fisher Scientific) and incubated at 4°C overnight with gentle shaking. Excess fluid was removed, and the wells were blocked with 200 μl of 5% skim milk supplemented with 0.05% Tween containing 10 nM nonrelated DNA sequences (hlyQF, hlyQR, L23QF, and L23QR [19]) (blocking buffer) for 2 h at RT. Wells were washed twice with 200 μl PBST. They were then exposed to 100 μl of biotinylated blood type A HBGA (catalog number 01-032; GlycoTech, Gaithersburg, MD) suspended at 30 μg/ml in blocking buffer for 1 h at room temperature, followed by three more washes with PBST. Development was done by the addition of 100 μl of a streptavidin-horseradish peroxidase conjugate (1.25 mg/ml diluted 1:5,000; Invitrogen, Carlsbad, CA) and incubation for 15 min at room temperature, followed by treatment with 100 μl of 3,3′5,5′-tetramethylbenzidine (TMB) peroxidase substrate (KPL, Gaithersburg, MD). The absorbance was read at 450 nm by using a Tecan Infiniti M200 Pro plate reader (Tecan, Morrisville, NC). Every treatment time point consisted of duplicate wells per plate. Negative-control wells contained PBS instead of VLPs, while positive-control wells included VLPs without copper or stainless steel treatment. The absorbance readings of negative-control wells were subtracted from those of the sample wells to establish treatment absorbance values. Data are presented as a percentage of the absorbance of untreated VLPs (positive-control wells).
Statistical analysis.
All experiments were replicated in triplicate. Statistical analysis was performed by one-way multiple comparisons using SAS 9.2 (SAS Institute, Cary, NC), with Tukey's honestly significant difference (HSD) test being used for separation of means. A P value of <0.05 was considered statistically significant.
RESULTS
RT-qPCR assays show evidence of degradation of the HuNoV genome and capsid during exposure to copper-containing surfaces.
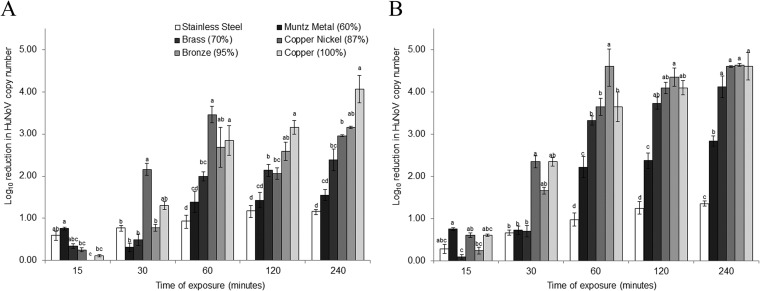
To determine the impact of copper alloy exposure on HuNoV genome integrity, GII.4-containing stool samples were exposed to each surface, and the reduction in the viral RNA copy number was measured by RT-qPCR over time. Stool samples were chosen due to their high load of HuNoV and the fact that they mimic a real-world contamination event. For the stainless steel control surfaces, there was a <1.1-log10 reduction in RNA copy number over the 240-min experimental period (Fig. 1A). On the other hand, within 60 min of exposure, a 2- to 3-log10 reduction in RNA copy number was observed for surfaces containing ≥70% copper (Fig. 1A). This reduction was statistically significant compared to that with stainless steel (P < 0.05). The reduction in the genome copy number gradually increased through the maximum exposure time of 240 min. The maximum genomic copy number reduction was ∼4 log10 units and was observed only for the 100% copper surface after 240 min of exposure (Fig. 1A).
FIG 1.
Genome and capsid degradation of GII.4 HuNoV during exposure to copper-containing surfaces. Degradation of the GII.4 HuNoV RNA genome and capsid occurs during exposure to copper-containing surfaces. Twenty-five-microliter aliquots of 20% fecal suspensions positive for GII.4 HuNoV were placed onto various alloys at room temperature and eluted at select time points. Sample eluates were analyzed by RT-qPCR without RNase treatment to determine genome integrity (A) and by RT-qPCR following RNase treatment to determine capsid integrity (B). The values in parentheses after each alloy indicate the percentage of copper in the alloy. The drying time was included in the total exposure time and was ∼20 to 30 min. The HuNoV RNA copy number was estimated by comparing CT values to a standard curve. Letters above the bars indicate statistically significant differences (P < 0.05) between alloys for each exposure time. Error bars represent standard errors of the means. All experiments were performed in triplicate.
These results are juxtaposed to those of the RT-qPCR assay with RNase pretreatment. Similar to the no-RNase samples, there was a ≤1.4-log10 reduction in RNA copy number for the stainless steel control over the entire 240-min exposure (Fig. 1B). However, for RNase-treated samples, the loss in copy number was much more rapid, with a 2- to 4-log10 reduction after 60 min of exposure for all alloys containing any copper (Fig. 1B). This reduction was statistically significant compared to that with stainless steel (P < 0.05). Furthermore, a near-maximum genome copy number reduction was achieved after 120 min of exposure for all alloys containing ≥70% copper (Fig. 1B). Again, the rate of genome copy number reduction was approximately proportional to the copper content of the alloy. Taken together, the pronounced decrease in RNA copy number over time for GII.4 HuNoV exposed to copper-containing alloys, in samples both with and without RNase treatment, suggests that copper has an impact on both genome and capsid integrity.
Further evidence that damage to the HuNoV capsid occurs during exposure to copper surfaces.
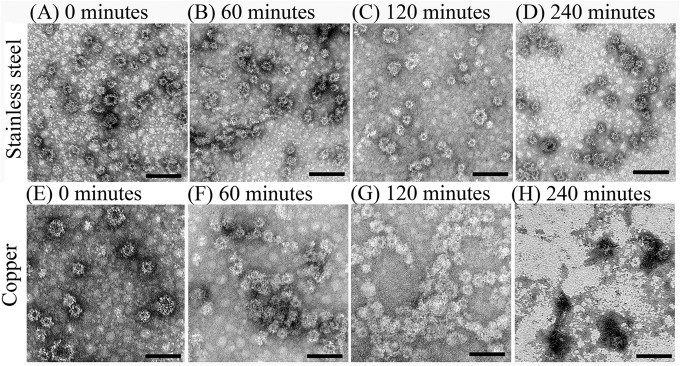
The impact of copper exposure on capsid integrity was further evaluated by a combination of TEM, SDS-PAGE, and HBGA receptor binding assays. Based on TEM applied to GII.4 Grimsby VLPs, HuNoV remained stable on stainless steel surfaces for up to 240 min, as the VLP suspensions remained evenly dispersed, and no particle clumping or protein debris fields were observed (Fig. 2A to D). Within 60 min of exposure to copper, aggregation of particles was observed (Fig. 2F), and further increasing the time to 120 min altered particle morphology, in addition to areas of clumping (Fig. 2G). By 240 min of copper exposure, widespread protein debris fields were observed, and intact particles became increasingly difficult to locate on grids (Fig. 2H).
FIG 2.
Effect of copper surfaces on the integrity of HuNoV VLPs. One microliter of purified GII.4 Grimsby HuNoV VLPs (concentration of 1,700 ng/μl) was placed onto copper or stainless steel surfaces and eluted at various time points between 0 and 240 min. Eluted samples were negatively stained with 2% uranyl acetate and visualized by transmission electron microscopy. Bar, 0.1 μm.
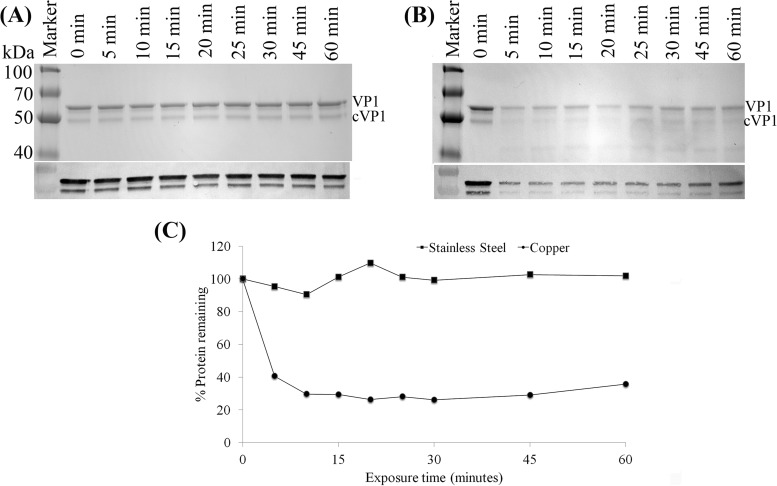
To determine whether exposure to a copper surface degraded HuNoV capsid proteins, we analyzed GII.4 Houston HuNoV VLPs after exposure to stainless steel or pure copper by SDS-PAGE and Western blotting. As shown in Fig. 3, and consistent with data described previously (20), two protein bands, with molecular masses of ∼60 and ∼50 kDa, corresponding to full-length VP1 and the cleaved form of VP1 (cVP1), respectively, were detected in VLPs. Exposure to stainless steel surfaces resulted in virtually no reduction in the HuNoV VP1 band intensity, regardless of time (Fig. 3A). However, exposure to copper for a period of 5 min resulted in degradation of VP1 observable by the reduction in the SDS-PAGE band intensity and by ImageJ density analysis (Fig. 3B and C). Extending the copper exposure to 15 min resulted in VP1 protein bands being reduced to ∼25% of the original intensity (Fig. 3C). No further reduction of VP1 was observed, even when the exposure time was increased to 120 min (data not shown). Western blotting was also performed to determine whether HuNoV VLPs exposed to copper were still able to react to a HuNoV-specific antibody. As shown in Fig. 3A and B, the amounts of HuNoV capsid proteins detected by Western blotting were reduced within 5 min of exposure to a copper surface but not during exposure to stainless steel coupons.
FIG 3.
Analysis of HuNoV capsid protein by SDS-PAGE and Western blotting. (A and B) One-microliter aliquots (representing 590 ng) of purified HuNoV GII.4 Houston VLPs were placed onto either stainless steel (A) or copper (B) surfaces and eluted at various time points in PBS-EDTA. Eluted VLPs were analyzed by 12% SDS-PAGE followed by protein staining. VP1, native full-length capsid protein; cVP1, cleaved VP1. Blots from Western blot analysis of VP1 proteins using mouse monoclonal antinorovirus GII.4 antibody are shown below each SDS-PAGE gel image. (C) Quantitative analysis of the remaining VP1 proteins detected by SDS-PAGE after surface exposure.
Receptor binding assays using synthetic HBGA type A and GII.4 Houston VLPs were used as an indirect measure of virus infectivity after exposure to copper. The VLPs rapidly lost the ability to bind to their HBGA type A receptor when exposed to copper but not a stainless steel surface. As little as 2 min of exposure to copper was sufficient to significantly (P < 0.05) reduce the receptor binding ability compared to the stainless steel control surfaces, and 8 min of copper exposure was sufficient to almost completely abolish receptor binding (Fig. 4). Extending the exposure time beyond 10 min offered no further reduction in receptor binding (data not shown). On the other hand, the receptor binding ability of the GII.4 Houston VLPs exposed to stainless steel remained largely unchanged for the duration of the exposure period (Fig. 4), even when exposure times were increased to 60 min (data not shown). Collectively, the RT-qPCR (with RNase treatment), TEM, PAGE, and HBGA binding results suggest that the capsid of HuNoV is destroyed during exposure to copper surfaces but not during exposure to stainless steel surfaces.
FIG 4.
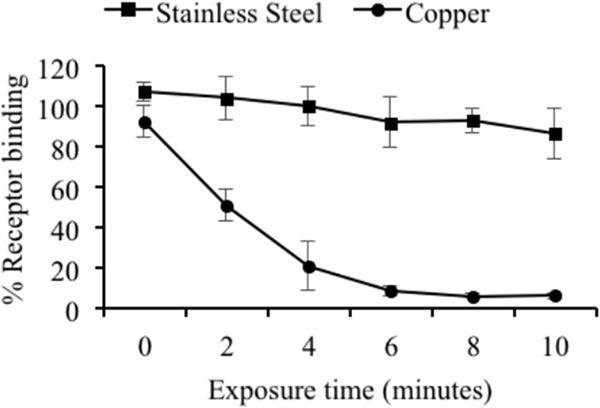
Impact of copper surface exposure on HuNoV VLP receptor binding. One-microliter aliquots (representing 590 ng) of purified HuNoV GII.4 Houston VLPs were placed onto either stainless steel (A) or copper (B) surfaces and eluted at various time points in PBS-EDTA. VLPs were then bound to a 96-well polystyrene plate and blocked, and synthetic biotinylated blood type A HBGA was added to each well. Plates were developed by using the TMB peroxidase substrate system, and receptor binding was determined by measuring the optical density at 450 nm using a microplate reader. Data are presented as percentages of the absorbance of untreated VLPs.
DISCUSSION
HuNoV causes a considerable disease burden and is thought to be the causative agent of at least half of all gastroenteritis outbreaks worldwide (1). The remarkable environmental stability of these viruses facilitates their efficient spread (5, 16). Touch surfaces with antimicrobial properties hold promise to control HuNoV environmental transmission. In this study, we provide evidence that both the genome and capsid of HuNoV are rapidly destroyed during exposure to copper-containing surfaces, a finding supported by data from previous studies using the genetically related and cultivable surrogate MNV (13, 14).
The results presented here are consistent with those of Warnes et al., who performed similar experiments using the cultivable surrogate MNV (13, 14). For example, both studies reported evidence of a loss of genome integrity upon exposure to copper-containing alloys. Warnes and Keevil (14) showed a >210-log reduction in genome copy number (based on RT-qPCR amplification of the Vpg gene) within 2 h of copper exposure, which was confirmed by evidence of genome degradation by using agarose gel electrophoresis. In the study reported here, the combination of RT-qPCR with and without RNase pretreatment was used to make similar conclusions. As was the case with the study by Warnes et al. (13), there was a 2- to 4-log10 reduction in genome copy number over time with exposure to copper alloys, compared to an ∼1-log10 reduction for stainless steel exposures (Fig. 1B). Furthermore, when the results from samples with and those without RNase pretreatment were compared, RNase-treated samples exposed to copper-containing surfaces showed statistically significantly higher (by ∼2 log10 units) and more rapid inactivation than did samples not pretreated with RNase (see Fig. S1 in the supplemental material). These notable differences between control and treatment groups with respect to RT-qPCR signals as a function of RNase treatment status provide further evidence that copper acts on the viral genome as well as the capsid.
Also consistent with data reported previously by Warnes et al., the efficacy (defined as both rate and total log10 inactivation) of each alloy in destroying the virus generally increased with increasing copper contents (13, 14). In both studies, the percentage of copper in the alloy at which significant inactivation of virus occurred was in the range of 60 to 80%, but the formulation of the alloy was important, and there were exceptions. Currently, the U.S. EPA recognizes copper alloys having at least a 60% copper content as being approved antimicrobial touch surfaces (a complete list of approved alloys can be found at the Copper Development Association website [http://www.copper.org/resources/properties/db/datasheets/all-alloys.pdf]). The observation that the efficacy of each alloy in destroying the viral genome increased with increasing copper content, for both MNV and HuNoV, provides strong evidence that copper is responsible for the antiviral effects. The mode of RNA degradation by copper is unknown, but it has been hypothesized that damage occurs through Fenton reactions, which result in the production of highly reactive hydroxyl radicals (21), although this has been debated by others (14).
Several additional assays (i.e., TEM, SDS-PAGE, Western blotting, and HBGA binding analysis) were employed to support the conclusion of a destructive effect of copper on HuNoV capsid integrity. Because of the need for high concentrations of virus for these experiments, it was necessary to use VLPs. Analysis by TEM showed that exposure of the VLPs to copper resulted in observable morphological changes to the particles within 240 min of exposure, specifically aggregation and degradation (Fig. 2). Copper ions have been observed to cause aggregation of hepatitis A virus, human rotavirus, human adenovirus, poliovirus, and MNV, and it has been hypothesized that this aggregation may contribute to virus inactivation (13, 22). These data are juxtaposed to data for stainless steel exposure, for which there was no observable change in VLP structure for the entire 240-min exposure period (Fig. 2). Additional evidence for capsid destruction was provided by SDS-PAGE/Western blot analysis. In this case, as little as 10 min of exposure to a copper surface was sufficient to reduce HuNoV VP1 capsid protein levels to ∼25% of the levels in the unexposed control sample (Fig. 3). Together, our TEM and SDS-PAGE/Western blot results provide direct evidence of copper surfaces both altering the HuNoV particle structure and destroying HuNoV capsid proteins. As is the case for genome destruction, it may be that Fenton reaction-generated hydroxyl radicals are the mechanism of capsid degradation (23, 24), but further experiments are needed to prove this.
Finally, we used VLPs in a HBGA binding assay to investigate the impact of copper surface exposure on the receptor binding activity of GII.4 HuNoV. As little as 2 min of exposure to copper was sufficient to significantly reduce the ability of VLPs to bind synthetic HBGA type A. When the exposure time was extended to 10 min, the receptor binding ability of the HuNoV VLPs was virtually abolished. This rapid loss of receptor biding ability was not observed for stainless steel surfaces. These results suggest that copper alloy surfaces have an impact on the HuNoV capsid in a region associated with receptor binding. Histidine and proline amino acid residues were previously identified as important sites of protein attack mediated by copper radical formation (23). Interestingly, several sites in the P2 domain of the capsid are rich in histidine and/or proline residues. This region of the capsid is known to harbor an HBGA binding pocket (25). The presence of proline or histidine residues in the binding region of the HuNoV GII.4 capsid could explain why exposure to a copper surface had a near-immediate impact on the ability of the VLPs to bind the HBGA receptor, although additional studies are necessary to confirm this hypothesis.
To our knowledge, this is the first study showing that copper surfaces are effective against HuNoV. Using techniques similar to those used in our study, Warnes et al. showed that copper surfaces were effective in destroying both the genome (14) and the capsid (13) of MNV. Unfortunately, cultivable surrogate viruses for HuNoV (i.e., MNV, feline calicivirus, Tulane virus, and bacteriophage MS2) do not necessarily behave in the same manner when exposed to various environmental stresses and inactivation strategies (16, 26, 27). As a result, one cannot use data from surrogates as definitive proof of the behavior of HuNoV. For example, Leon et al. (28) performed a human challenge study to evaluate the efficacy of high-pressure processing (HPP) to inactivate HuNoV in contaminated oysters. Those authors found that the pressure conditions required to prevent disease far exceeded those needed to achieve a 4-log10 reduction of the MNV copy number (29), indicating that MNV is a poor surrogate for predicting HuNoV behavior under HPP conditions. Given the limitations of surrogate viruses, our study provides direct evidence that copper alloys are effective against HuNoV. This study supports previous work performed with MNV (14, 21), and together, these data provide strong evidence that metal surfaces containing copper negatively impact the capsid and genome of GII.4 HuNoV.
To summarize, the results presented in this study demonstrate that exposure of GII.4 HuNoV or GII.4 HuNoV VLPs to copper alloy surfaces impacted the virus in three main ways: (i) degradation of the RNA genome, (ii) loss of HBGA receptor binding ability, and (iii) destruction of the viral capsid. Interestingly, the relatively shorter time to impact capsid integrity than genome integrity suggests that the former is the initial target for copper inactivation, although additional studies are needed to confirm this. In the absence of a culture-based HuNoV infectivity assay, these results provide convincing evidence that copper surfaces have the ability to inactivate HuNoV. Further studies are necessary to confirm that this phenomenon occurs with other HuNoV genotypes. While widespread use of copper alloys can be expensive, targeted replacement of high-touch surfaces (e.g., doorknobs, railings, and buttons) may be more affordable, as would the use of copper coatings. In fact, in a recent study, a hospital intensive care unit that replaced high-touch surfaces with copper experienced a >50% reduction in the overall infection rate (12). It could be that the use of copper in other venues such as cruise ships, elder care facilities, hotels, schools, and restaurants may be relevant to controlling HuNoV transmission.
Supplementary Material
ACKNOWLEDGMENTS
We thank V. Lapham from the Center for Electron Microscopy at North Carolina State University for helping to obtain the TEM images. We also thank W. House for helping to process samples.
This work was supported by Agriculture and Food Research Initiative competitive grant no. 2011-68003-30395 from the U.S. Department of Agriculture, National Institute of Food and Agriculture, through the NoroCORE project.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00388-15.
REFERENCES
- 1.Patel MM, Widdowson M-A, Glass RI, Akazawa K, Vinjé J, Parashar UD. 2008. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg Infect Dis 14:1224–1231. doi: 10.3201/eid1408.071114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel MM, Hall AJ, Vinjé J, Parashar UD. 2009. Noroviruses: a comprehensive review. J Clin Virol 44:1–8. doi: 10.1016/j.jcv.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Hall AJ, Lopman BA, Payne DC, Patel MM, Gastañaduy PA, Vinjé J, Parashar UD. 2013. Norovirus disease in the United States. Emerg Infect Dis 19:1198–1205. doi: 10.3201/eid1908.130465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lopman BA, Hall AJ, Curns AT, Parashar UD. 2011. Increasing rates of gastroenteritis hospital discharges in US adults and the contribution of norovirus, 1996-2007. Clin Infect Dis 52:466–474. doi: 10.1093/cid/ciq163. [DOI] [PubMed] [Google Scholar]
- 5.Lopman B, Gastañaduy P, Park GW, Hall AJ, Parashar UD, Vinjé J. 2012. Environmental transmission of norovirus gastroenteritis. Curr Opin Virol 2:96–102. doi: 10.1016/j.coviro.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 6.Liu P, Yuen Y, Hsiao H-M, Jaykus L-A, Moe C. 2010. Effectiveness of liquid soap and hand sanitizer against Norwalk virus on contaminated hands. Appl Environ Microbiol 76:394–399. doi: 10.1128/AEM.01729-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tung G, Macinga D, Arbogast J, Jaykus L-A. 2013. Efficacy of commonly used disinfectants for inactivation of human noroviruses and their surrogates. J Food Prot 76:1210–1217. doi: 10.4315/0362-028X.JFP-12-532. [DOI] [PubMed] [Google Scholar]
- 8.Teunis PFM, Moe CL, Liu P, Miller ES, Lindesmith L, Baric RS, Le Pendu J, Calderon RL. 2008. Norwalk virus: how infectious is it? J Med Virol 80:1468–1476. doi: 10.1002/jmv.21237. [DOI] [PubMed] [Google Scholar]
- 9.Hall AJ. 2012. Noroviruses: the perfect human pathogens? J Infect Dis 205:1622–1624. doi: 10.1093/infdis/jis251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dollwet H, Sorenson J. 1985. Historic uses of copper compounds in medicine. Trace Elem Med 2:80–87. [Google Scholar]
- 11.Grass G, Rensing C, Solioz M. 2011. Metallic copper as an antimicrobial surface. Appl Environ Microbiol 77:1541–1547. doi: 10.1128/AEM.02766-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salgado CD, Sepkowitz KA, John JF, Cantey JR, Attaway HH, Freeman KD, Sharpe PA, Michels HT, Schmidt MG. 2013. Copper surfaces reduce the rate of healthcare-acquired infections in the intensive care unit. Infect Control Hosp Epidemiol 34:479–486. doi: 10.1086/670207. [DOI] [PubMed] [Google Scholar]
- 13.Warnes SL, Summersgill EN, Keevil CW. 2015. Inactivation of murine norovirus on a range of copper alloy surfaces is accompanied by loss of capsid integrity. Appl Environ Microbiol 81:1085–1091. doi: 10.1128/AEM.03280-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warnes SL, Keevil CW. 2013. Inactivation of norovirus on dry copper alloy surfaces. PLoS One 8:e75017. doi: 10.1371/journal.pone.0075017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richards GP. 2012. Critical review of norovirus surrogates in food safety research: rationale for considering volunteer studies. Food Environ Virol 4:6–13. doi: 10.1007/s12560-011-9072-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Escudero BI, Rawsthorne H, Gensel C, Jaykus LA. 2012. Persistence and transferability of noroviruses on and between common surfaces and foods. J Food Prot 75:927–935. doi: 10.4315/0362-028X.JFP-11-460. [DOI] [PubMed] [Google Scholar]
- 17.Jothikumar N, Lowther JA, Henshilwood K, Lees DN, Hill VR, Vinjé J. 2005. Rapid and sensitive detection of noroviruses by using TaqMan-based one-step reverse transcription-PCR assays and application to naturally contaminated shellfish samples. Appl Environ Microbiol 71:1870–1875. doi: 10.1128/AEM.71.4.1870-1875.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rogers JD, Ajami NJ, Fryszczyn BG. 2013. Identification and characterization of a peptide affinity reagent for detection of noroviruses in clinical samples. J Clin Microbiol 51:1803–1808. doi: 10.1128/JCM.00295-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodríguez-Lázaro D, Hernández M. 2004. Quantitative detection of Listeria monocytogenes and Listeria innocua by real-time PCR: assessment of hly, iap, and lin02483 targets and AmpliFluor technology. Appl Environ Microbiol 70:1366–1377. doi: 10.1128/AEM.70.3.1366-1377.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lou F, Huang P, Neetoo H, Gurtler JB, Niemira BA, Chen H, Jiang X, Li J. 2012. High-pressure inactivation of human norovirus virus-like particles provides evidence that the capsid of human norovirus is highly pressure resistant. Appl Environ Microbiol 78:5320–5327. doi: 10.1128/AEM.00532-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carubelli R, Schneider JE, Pye QN, Floyd RA. 1995. Cytotoxic effects of autoxidative glycation. Free Radic Biol Med 18:265–269. doi: 10.1016/0891-5849(94)E0134-5. [DOI] [PubMed] [Google Scholar]
- 22.Abad FX, Pinto RM, Diez JM. 1994. Disinfection of human enteric viruses in water by copper and silver in combination with low levels of chlorine. Appl Environ Microbiol 60:2377–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dean RT, Wolff SP, McElligott MA. 1989. Histidine and proline are important sites of free radical damage to proteins. Free Radic Res Commun 7:97–103. doi: 10.3109/10715768909087929. [DOI] [PubMed] [Google Scholar]
- 24.Shionoiri N, Sato T, Fujimori Y, Nakayama T, Nemoto M, Matsunaga T, Tanaka T. 2012. Investigation of the antiviral properties of copper iodide nanoparticles against feline calicivirus. J Biosci Bioeng 113:580–586. doi: 10.1016/j.jbiosc.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 25.Siebenga JJ, Vennema H, Renckens B, de Bruin E, van der Veer B, Siezen RJ, Koopmans M. 2007. Epochal evolution of GGII.4 norovirus capsid proteins from 1995 to 2006. J Virol 81:9932–9941. doi: 10.1128/JVI.00674-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cromeans T, Park GW, Costantini V. 2014. Comprehensive comparison of cultivable norovirus surrogates in response to different inactivation and disinfection treatments. Appl Environ Microbiol 80:5743–5751. doi: 10.1128/AEM.01532-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoelzer K, Fanaselle W, Pouillot R. 2013. Virus inactivation on hard surfaces or in suspension by chemical disinfectants: systematic review and meta-analysis of norovirus surrogates. J Food Prot 76:1006–1016. doi: 10.4315/0362-028X.JFP-12-438. [DOI] [PubMed] [Google Scholar]
- 28.Leon JS, Kingsley DH, Montes JS. 2011. Randomized, double-blinded clinical trial for human norovirus inactivation in oysters by high hydrostatic pressure processing. Appl Environ Microbiol 77:5476–5482. doi: 10.1128/AEM.02801-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kingsley DH, Holliman DR, Calci KR, Chen H, Flick GJ. 2007. Inactivation of a norovirus by high-pressure processing. Appl Environ Microbiol 73:581–585. doi: 10.1128/AEM.02117-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.