Abstract
In a recent paper, we demonstrated that inactivation of the Agr system affects the patterns of survival of Listeria monocytogenes (A.-L. Vivant, D. Garmyn, L. Gal, and P. Piveteau, Front Cell Infect Microbiol 4:160, http://dx.doi.org/10.3389/fcimb.2014.00160). In this study, we investigated whether the Agr-mediated response is triggered during adaptation in soil, and we compared survival patterns in a set of 10 soils. The fate of the parental strain L. monocytogenes L9 (a rifampin-resistant mutant of L. monocytogenes EGD-e) and that of a ΔagrA deletion mutant were compared in a collection of 10 soil microcosms. The ΔagrA mutant displayed significantly reduced survival in these biotic soil microcosms, and differential transcriptome analyses showed large alterations of the transcriptome when AgrA was not functional, while the variations in the transcriptomes between the wild type and the ΔagrA deletion mutant were modest under abiotic conditions. Indeed, in biotic soil environments, 578 protein-coding genes and an extensive repertoire of noncoding RNAs (ncRNAs) were differentially transcribed. The transcription of genes coding for proteins involved in cell envelope and cellular processes, including the phosphotransferase system and ABC transporters, and proteins involved in resistance to antimicrobial peptides was affected. Under sterilized soil conditions, the differences were limited to 86 genes and 29 ncRNAs. These results suggest that the response regulator AgrA of the Agr communication system plays important roles during the saprophytic life of L. monocytogenes in soil.
INTRODUCTION
Listeria monocytogenes is the causative agent of listeriosis, a serious foodborne infection affecting essentially immunocompromised individuals, the elderly, and pregnant women (1). The pathogen is largely spread in the environment. It has been isolated from water systems (2–4), vegetation (5), soil (6–8), farms (9–12), food industries (13–15), and the feces of animals (16–18). Environmental adaptation requires that the cell have the ability to integrate environmental cues in order to adapt its physiology to the surrounding conditions through the regulation of gene expression. Genomics showed that an important part of the L. monocytogenes genome (7.3%) is dedicated to regulation and includes 209 transcriptional regulators, 15 histidine kinases, and 16 response regulators constituting two-component systems (19). Two-component systems participate in the ability of bacteria to sense and respond to fluctuating environmental conditions. AgrC/AgrA is a two-component regulatory system that is part of the Agr communication system. Initially described in Staphylococcus aureus, this communication system is organized as a four-gene operon, agrBDCA. AgrB is a membrane-bound protein that processes the propeptide AgrD into a mature autoinducing peptide (AIP). Detection of AIP by the histidine kinase AgrC induces transcriptional regulation through activation of the regulator AgrA. Detailed data concerning the role of the Agr system in the physiology of S. aureus are available (20–22). So far, its role in the adaptation of L. monocytogenes to its environment is only partially understood (23). Reports show that the Agr communication system of L. monocytogenes is involved in adhesion to abiotic surfaces (24) in the early stages of biofilm formation (24, 25) and during infection of the mammalian host (25, 26). Indeed, ΔagrA and ΔagrD in-frame deletion mutants showed defects in adherence and early biofilm development. The virulence of a ΔagrA mutant was attenuated in mice, but in vitro, adhesion and invasion in several cell lines were not altered (26), while virulence in mice and invasion of Caco-2 intestinal cells were reduced in an agrD mutant (25). This was correlated with a lower level of expression of internalin.
Recently, in a study focusing on one soil (27), we showed that the Agr system provides a benefit to the populations of L. monocytogenes, but the role of the Agr communication system in the adaptation of L. monocytogenes populations in the natural environment remains poorly understood. In order to gain a better understanding of the Agr-mediated response during the adaptation of L. monocytogenes to the telluric environment, we monitored the fate of an inoculated L. monocytogenes parental strain and an in-frame ΔagrA deletion mutant in a set of 10 soil microcosms. We further investigated the consequences of inactivation of AgrA on the transcriptome of L. monocytogenes during adaptation to the soil environment through a differential transcriptome analysis approach.
MATERIALS AND METHODS
Bacterial strains and inoculum preparation.
L. monocytogenes L9, a rifampin-resistant (Rifr) mutant of the parental strain L. monocytogenes EGD-e (28), and L. monocytogenes DG125A6, a rifampin-resistant mutant of the ΔagrA in-frame deletion mutant DG125A (24), were used. L. monocytogenes DG125A6 was isolated on polymyxin-acriflavine-lithium chloride-ceftazidime-esculin-mannitol (PALCAM) agar (AES Chemunex, Bruz, France) supplemented with 200 μg · ml−1 rifampin (Sigma-Aldrich, Saint Quentin Fallavier, France), as described by Lemunier et al. (28). This Rifr mutant was selected, as its growth rate during planktonic growth and its ability to grow as a biofilm in tryptone soy broth (TSB; AES Chemunex, Bruz, France) at 25°C without shaking were similar to those recorded with L. monocytogenes DG125A. A working stock that had been stored at −80°C was used throughout the study. Strains were grown statically at 25°C for 16 h in 5 ml of TSB. Three independent inocula were prepared by inoculating 10 ml of TSB to an optical density at 600 nm (OD600) of 0.04 and incubating statically at 25°C until the OD600 reached 0.4. The cultures were then centrifuged at 8,000 × g for 5 min at room temperature, and the pellets were suspended in NaCl (0.85%).
Soils characteristics and preparation of microcosms.
Ten soils in a country-wide soil sampling network in the Burgundy section of France were sampled on the basis of a 16- by 16-km systematic grid. Within a 20- by 20-m grid, 25 individual core samples of topsoil (depth, 0 to 30 cm) were collected with a hand auger every 5 m at the edges of the 16 inside 5- by 5-m squares. At each sampling site, the core samples were mixed into a composite sample. Soil samples were sieved to 5 mm and stored at 4°C before analysis. The attributes of the soil samples, such as the pedology, chemistry, and land use, which were extracted from the DONESOL database (29), and the bacterial abundance are listed in Table 1. Endogenous L. monocytogenes was not found in these soils. Physical and chemical analyses were performed by the Soil Analysis Laboratory of INRA (Arras, France; http://www.lille.inra.fr/las). For each soil sample, triplicate microcosms were prepared by adding 50 g of soil to sterile 180-ml capped plastic tubes.
TABLE 1.
Nomenclature, land use, soil parameters, and bacterial abundance for the soils used in this study
| Soil sample no. | Land use | Texture | pH | Organic content (g · kg−1) |
C/N ratio | CECa | WHCb (%) | Cultivable bacterial community (CFU · g−1 of soil [107]) | |
|---|---|---|---|---|---|---|---|---|---|
| C | N | ||||||||
| 748 | Agricultural field | Silt loam | 8.2 | 9.80 | 1.0 | 9.8 | 13.2 | 18.5 | 5.1 |
| 749 | Agricultural field | Clay loam | 8.0 | 19.2 | 1.8 | 10.7 | 16.6 | 24.4 | 5.2 |
| 750 | Agricultural field | Clay | 8.0 | 26.4 | 2.8 | 9.4 | 37.8 | 42.3 | 3.1 |
| 907 | Prairie | Loam | 5.9 | 25.0 | 2.4 | 10.4 | 11.6 | 20.7 | 4.1 |
| 909 | Agricultural field | Silty clay | 8.1 | 27.5 | 2.8 | 9.8 | 29.5 | 34.2 | 10.0 |
| 911 | Prairie | Silty clay | 5.9 | 38.1 | 3.7 | 10.3 | 23.6 | 40.7 | 2.6 |
| 1003 | Forest | Silty clay | 7.5 | 33.4 | 2.3 | 14.5 | 27.3 | 36.1 | 4.4 |
| 1005 | Prairie | Clay | 6.7 | 35.3 | 3.9 | 9.1 | 31.4 | 46.6 | 6.0 |
| 1051 | Prairie | Sandy loam | 5.4 | 26.2 | 2.5 | 10.5 | 8.8 | 18.8 | 4.4 |
| 1224 | Agricultural field | Clay | 7.8 | 25.6 | 2.5 | 10.2 | 27.4 | 37.6 | 3.1 |
CEC, cation-exchange capacity.
WHC, water-holding capacity.
Sterilized and biotic soil extracts were prepared for microarray experiments. Sterilized soil extracts were prepared as described by Piveteau et al. (30). Briefly, 500 g of soil was mixed with 750 ml of water for 30 min at 120 rpm and autoclaved for 1 h at 130°C. Soil suspensions were centrifuged at 10,000 × g for 20 min, and supernatants were filtered on Whatman paper (Sigma-Aldrich, Saint Quentin Fallavier, France). The particle-free soil extract obtained was used after autoclaving (20 min, 120°C). A biotic fraction was prepared by blending 100 g of soil in 300 ml of water for 1.5 min in a Waring blender. To estimate the bacterial abundance of the biotic fraction, culturable bacterial communities were enumerated by serial plating on nutrient agar (3 g · liter−1 beef extract, 5 g · liter−1 peptone, 15 g · liter−1 agar) supplemented with 100 μg · ml−1 cycloheximide (Sigma-Aldrich, Saint Quentin Fallavier, France) to suppress fungi. Biotic soil extracts were prepared by adding 1 ml of the biotic fraction to 9 ml of sterilized soil extract in order to reach a final concentration of 2 × 107 cultivable CFU/ml.
Soil microcosm survival assays.
Fifty grams of each of the soil microcosms was inoculated to achieve a final concentration of 2 × 106 CFU/g of soil. The volume of the inoculum was adjusted in order to reach a final soil moisture content of 60% of the water-holding capacity (WHC). The inoculated soils were stirred with a sterile spatula, and then the soil microcosms were incubated in the dark at 25°C. The microcosms were sampled 5 times. The soils were stirred with a sterile spatula prior to sampling of 1 g. The dynamics of the L. monocytogenes populations in the soil microcosms were followed immediately after inoculation and periodically over a 14-day period by serial plating on selective PALCAM agar (AES Chemunex, Bruz, France) supplemented with 100 μg · ml−1 cycloheximide and 100 μg · ml−1 rifampin (Sigma-Aldrich, Saint Quentin Fallavier, France).
RNA extraction and cDNA synthesis.
Inocula were prepared as previously described, except that the pellets were suspended in 10 ml of sterilized or biotic soil extracts. The inoculated soil extracts were incubated statically at 25°C for 30 min. RNAs were immediately stabilized by treating the bacterial cultures with the RNAprotect bacterial reagent (Qiagen, Courtaboeuf, France) according to the manufacturer's instructions. Three independent replicates were prepared per condition. Bacteria were harvested by centrifugation at 8,000 × g for 5 min at room temperature. Pellets were suspended in 700 μl of RLT buffer (Qiagen) supplemented with β-mercaptoethanol (1%) and 0.2 g of RNase-free glass beads (diameter, 100 μm). The cells were mechanically disrupted in a Fast Prep cell disrupter (MP Bio, Illkirch Graffenstaden, France) for 3 cycles (4 m · s−1, 30 s). The cells were centrifuged at 7,000 × g for 5 min at 4°C, and the cell debris was discarded. Potassium acetate (3 M, pH 5.5) was added to the supernatants (10%), and the mixture was incubated on ice for 10 min and centrifuged at 14,000 × g for 5 min at 4°C. The aqueous phase was then precipitated with an equal volume of cold isopropanol (−20°C), washed, and dried. The pellets were suspended in 400 μl of RNase-free water and purified first on polyvinyl polypyrrolidone (PVPP; Sigma-Aldrich, Saint Quentin Fallavier, France) columns, then on Phase Lock gel (5Prime; Dominique Dutscher, Brumath, France), and finally, on a Qiagen RNeasy kit (Qiagen, Courtaboeuf, France) according to the manufacturers' protocols. Purified RNA was concentrated with a Qiagen RNeasy MinElute cleanup kit (Qiagen, Courtaboeuf, France) following the manufacturer's instructions.
cDNA strands were then synthesized using a TransPlex complete whole-transcriptome amplification kit (WTA2; Sigma, Saint Quentin Fallavier, France) according to the manufacturer's instructions. The cDNA concentration was determined spectrophotometrically by measuring the λ at 260 nm (λ260) and λ280 (λ260 = 1.0 = 50 μg DNA/ml, λ260/λ280 > 1.8). For each strain and each incubation condition, three sets of cDNAs were prepared from three sets of RNAs extracted from three independent biological repetitions and hybridized independently.
Whole-genome microarrays and data analysis.
A customized whole-genome microarray including the 2,857 annotated open reading frames (ORFs) of the genome of L. monocytogenes EGD-e was designed by NimbleGen Systems. A total of 13,456 probes were dedicated to the tiling of intergenic regions every 14 bp. Microarray hybridization, washes, raw data preprocessing, and normalization were performed by Partnerchip (Evry, France) according to NimbleGen standard protocols. DNASTAR ArrayStar software (Madison, WI) was used for analysis of the normalized results. t statistics and P values (P < 0.05) were calculated to determine differentially expressed genes. The fold change in the levels of expression between the two strains was calculated by comparing the expression levels under the same incubation conditions. Comparisons between biotic and sterilized conditions were also performed for each strain. Genes with at least a 2-fold change in expression were considered for interpretation. In order to identify the relationships between genes and biological functions, the gene ontology was subsequently searched. P values and Z-scores were calculated in order to determine the chance that a certain number of genes would be selected for any given gene ontology term. Information regarding intergenic regions (location, presence of noncoding RNA [ncRNA]) was retrieved from the web-accessible transcriptome browser (http://www.weizmann.ac.il/molgen/Sorek/listeria_browser/) provided with the publication of Wurtzel et al. (31).
qPCR.
In order to confirm the fold changes in gene expression observed in the microarray analyses, the transcript levels of several genes with higher and lower transcript levels were examined by quantitative PCR (qPCR) using the Absolute qPCR SYBR green carboxy-X-rhodamine (ROX) mix (Thermo Scientific, Dominique Dutscher, Brumath, France). Primers specific for each selected gene were designed (Table 2). Each reaction mixture contained 2× Absolute qPCR SYBR green ROX mix (7.5 μl), 0.6 μM each PCR primer, 12.5 ng of DNA template, and sterile water to 15 μl. The qPCRs were carried out in a Step One PCR system (Applied Biosystems) using the following program: an initial enzyme activation period (10 min at 95°C), followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Following PCR, absolute quantification was performed by a standard curve method. For each target, increasing concentrations of L. monocytogenes genomic DNA ranging from 10 to 80 ng/μl were amplified in order to draw a relationship between the concentration and the number of threshold cycles. Fold changes in expression were calculated as the ratio of the absolute quantity of the target gene determined in the two strains under the same incubation conditions.
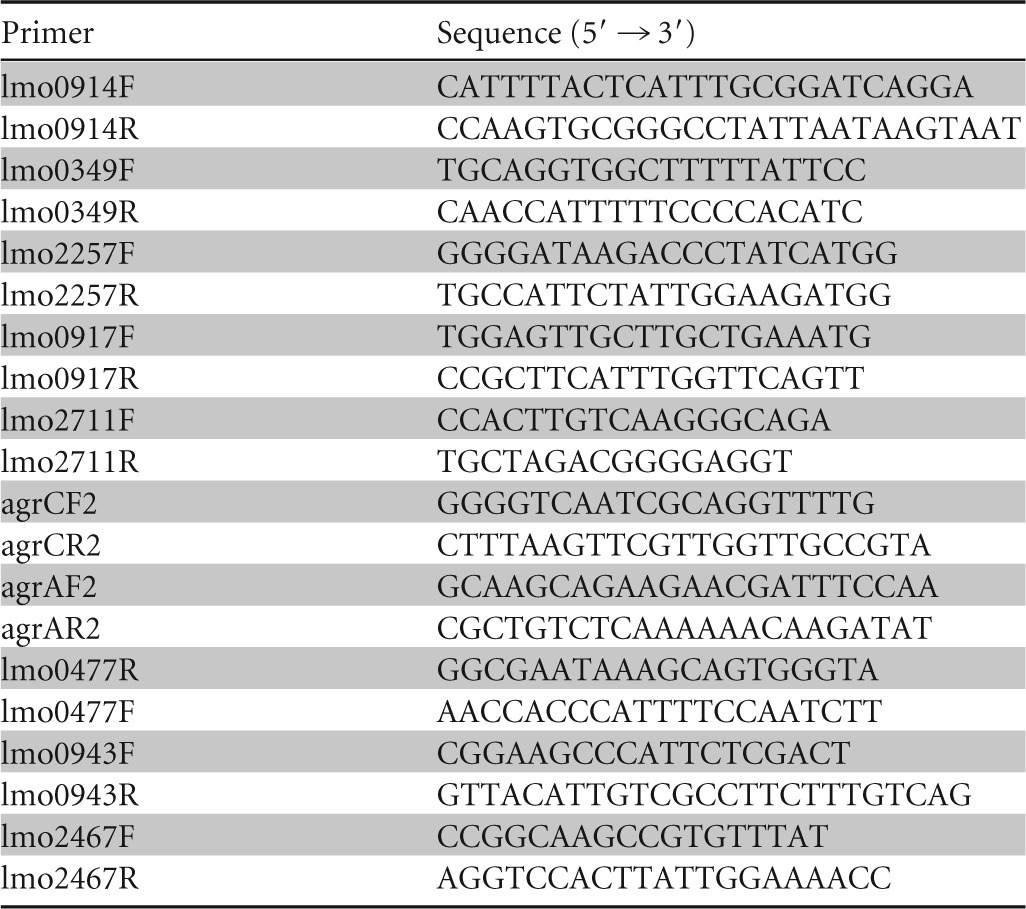
TABLE 2.
Target genes and specific primers designed for real-time PCR confirmation of the results
Statistical analyses.
Repeated-measures analysis of variance (ANOVA) was used to compare the survival patterns of the L. monocytogenes parental and ΔagrA mutant populations in soil microcosms. Principal components analysis was used to investigate the relationships between soil characteristics and the dynamics of the L. monocytogenes populations. Differences in soil texture were represented by use of the United States Department of Agriculture (USDA) pedological classification system using the statistical software R (v2.14.1). Then, Spearman's rank correlation was used to test the dependence between the following variables: the survival profiles of both the parental and mutant strains and soil attributes (texture, pH, organic C and N content, C/N ratio, and cation-exchange capacity).
RESULTS
L. monocytogenes population dynamics in the 10 soil microcosms are affected by the deletion of agrA.
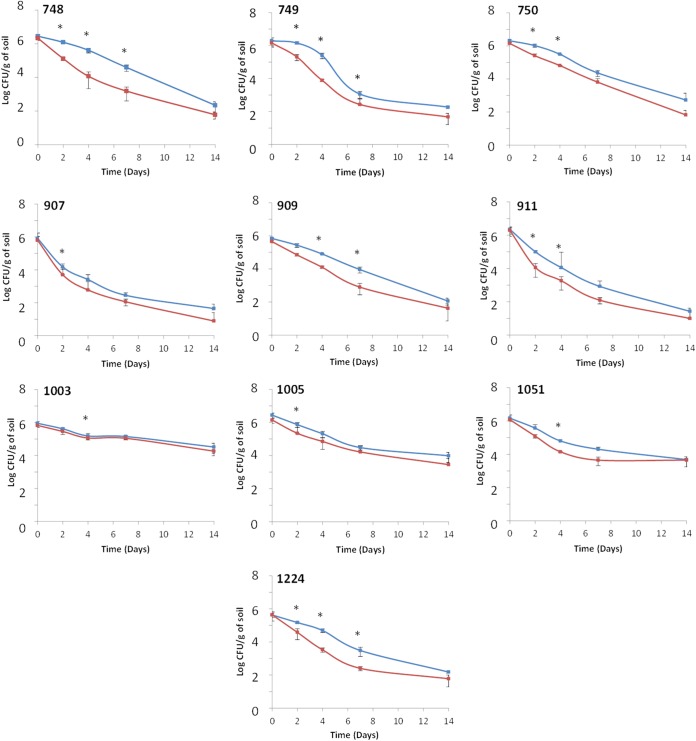
The dynamics of L. monocytogenes populations in microcosms prepared from soils for which land use and edaphic parameters were different (Table 1 and Fig. 1) were recorded over a 14-day period. During incubation in these soil microcosms, no growth of either of the two strains was detected and the populations of the parental strain and the ΔagrA mutant declined throughout the experiment (Fig. 2). However, survival patterns differed according to the strain under scrutiny. Regardless of the soil tested, the population of the mutant was reproducibly lower than the population of the parental strain in all three replicates. Repeated-measures ANOVA showed that these differences were transiently statistically significant. Indeed, in soil samples 748, 749, and 1224, the differences were significant during the first week of incubation. In soil samples 750 and 911, the differences were significant after 2 days and 4 days of incubation. In soil samples 1005 and 907, the results were statistically significant at day 2. The significance at day 4 was confirmed in soil samples 1003 and 1051. Finally, in microcosms prepared with soil sample 909, the differences were statistically significant at day 4 and day 7. The differences at day 14 were no longer statistically significant in all microcosms.
FIG 1.
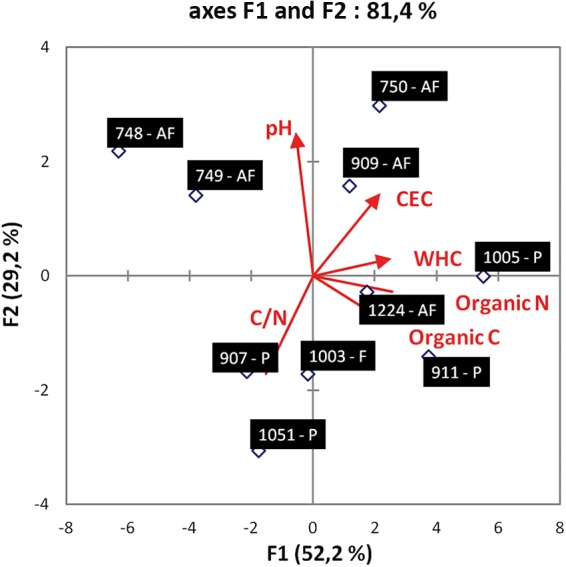
Principal component analysis (PCA) of soils. The length and direction of the lines represent the contribution of each parameter. The numbers indicate soil sample numbers, and the letters indicate whether soils were from an agricultural field (AF), a prairie (P), or a forest (F). CEC, cation-exchange capacity.
FIG 2.
L. monocytogenes parental strain (blue lines) and ΔagrA mutant (red lines) dynamics in soil microcosms. Error bars represent standard deviations from three independent replicates. *, significant differences (P < 0.05) were observed after ANOVA.
Interestingly, for each strain, the abundance at the end of the experiment was statistically significantly different according to the soil (Fig. 3). Indeed, at 14 days after incubation, it was significantly higher in soil sample 1003 (repeated-measures ANOVA, P < 0.05), where over 4 log units of the population remained detectable, than in soil samples 1005 and 1051 and in the rest of the soils (soil samples 748, 749, 750, 907, 909, 911, and 1224). Moderate survival occurred in soil samples 1005 and 1051. The weakest survival was observed in soil samples 748, 749, 750, 907, 909, 911, and 1224, where less than 3 log units were detected 14 days after inoculation. The Spearman rank correlation test was used to check correlations between the survival rate of each strain and the soils' attributes (texture, pH, organic C and N contents, C/N ratio, and cation-exchange capacity). From these six metrics, a significant negative correlation between the survival rate and pH was detected (Spearman's ρ = −0.736, P < 0.05). During the first days of the experiment, the rate of decline was steeper in the soils with pHs lower than 6 (soil samples 907, 911, and 1051). In soils with pHs higher than 6, the decline of the population either was low (soil samples 1003 and 1005) or became steeper only after 4 days of incubation (soil samples 748, 749, 750, 909, and 1224).
FIG 3.
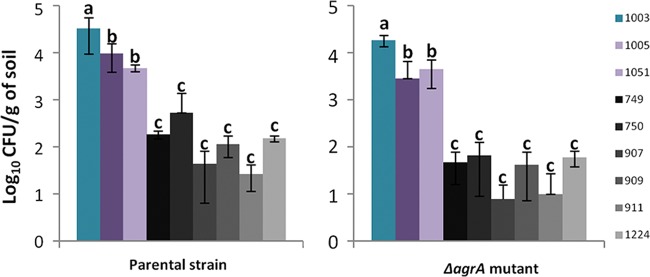
L. monocytogenes parental strain and ΔagrA mutant abundance after 14 days of incubation in soil microcosms. Error bars represent standard deviations from three independent replicates. Letters indicate that significant differences were observed after ANOVA.
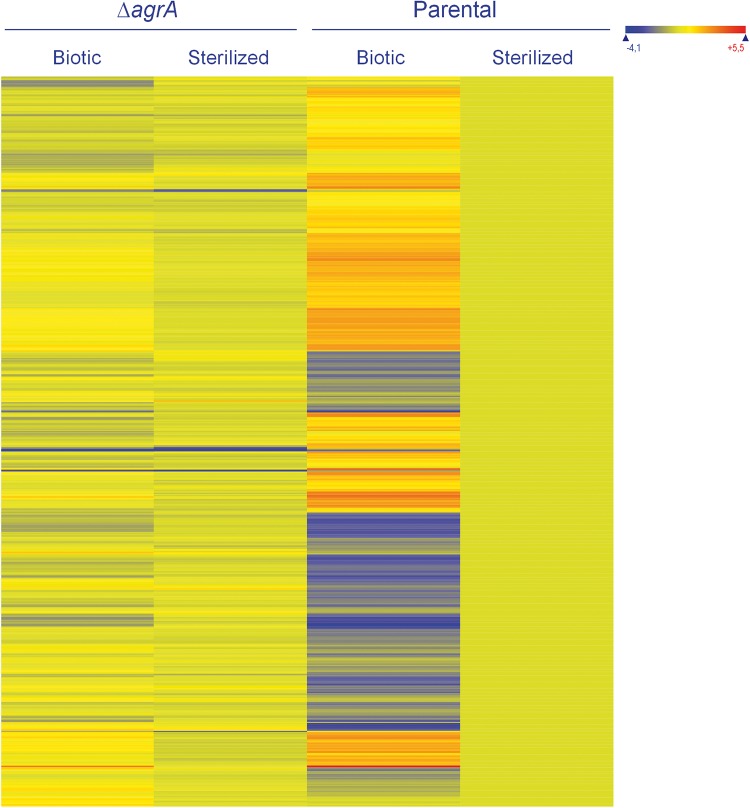
Numerous transcriptome differences between the parental strain and the ΔagrA mutant background are observed in soil environments.
In order to better understand the AgrA-mediated response in the adaptation of L. monocytogenes to the telluric environment, we compared the transcriptomes of the parental and ΔagrA strains during adaptation to the soil environment. We decided to focus on soil sample 748, for which high differences in survival between the parental strain and the mutant were recorded. Deletion of agrA resulted in large variations in the transcriptome (Fig. 4). A total of 578 genes were differentially transcribed. Two hundred fifty-two genes were recorded in the set of genes with higher transcript levels in the ΔagrA mutant background (see Table S1 in the supplemental material). Gene ontology did not identify any functional category to be a significant term. A focus on the genes highly expressed identified a group of 122 genes with over 3-fold higher transcript levels in the ΔagrA mutant background (see Table S2 in the supplemental material). Most of these genes code for functions related to the metabolism of carbohydrates, such as lmo2110 (mannose-6 phosphate isomerase), lmo0347 (dihydroxyacetone kinase), and lmo2824 (d-3-phosphoglycerate dehydrogenase), or functions related to the transport of proteins, such as the ABC transporter (lmo2123 to lmo2125, lmo1739) and the phosphotransferase (PTS) system (lmo2782, lmo2665, lmo2666, lmo2780). Fourteen transcriptional regulators were also in this set of genes.
FIG 4.
Heat map of the set of genes with significant differences in levels of expression between the ΔagrA mutant and the parental strain under biotic soil conditions. Expression levels were standardized so that those of the parental strain under sterilized conditions were set as the baseline. Genes are represented in rows. The expression levels of these genes under sterilized conditions are also presented.
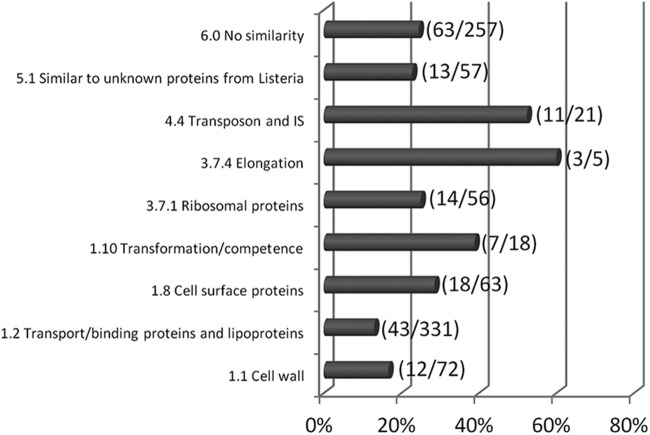
Moreover, 328 ORFs were identified in the set of genes with lower transcript levels in the ΔagrA mutant background (see Table S1 in the supplemental material). As expected, lmo0048 (agrB), lmo0049 (agrD), lmo0050 (agrC), and lmo0051 (agrA) were in this set of genes and showed, respectively, 5, 4, 4, and 9 times lower transcript levels in the ΔagrA mutant background than in the parental strain background. Gene ontology showed that functional categories 1.1, 1.2, 1.8, 1.10, 3.7.1, 3.7.4, 4.4, 5.1, and 6.0 were significant terms (Fig. 5). Eighty genes were included in category 1, cell envelope and cellular process (1.1, cell wall; 1.2, transport/binding proteins and lipoproteins; 1.8, cell surface proteins; and 1.10, transformation/competence). Among these genes, the dltABCD operon is involved in d-alanine esterification of teichoic acid, and deletion of the dlt operon affects adhesion to cell lines and antimicrobial peptide resistance (32). Several are also involved in the PTS system and ABC transporters. For example, lmo2115 (anrB), which codes for a permease component of an ABC transporter, plays a role in nisin resistance and also in bacitracin, gallidermin, and beta-lactam antibiotic resistance, and lmo0153, which codes for a probable ABC transporter, is a ZurR-regulated gene which is involved in virulence in the murine model (33). From functional category 3.7 (protein synthesis), 17 genes were identified (3.7.1, ribosomal proteins synthesis; 3.7.4, elongation). Finally, an important part of the differentially expressed genes (87 genes) was associated with other functions, such as transposons and insertion sequences (functional category 4.4), consisted of unknown proteins (functional category 5.1), or did not have any similarity to known genes.
FIG 5.
Functional categories identified to be significant by gene ontology and the percentage of genes from the set of genes with lower transcript levels in the ΔagrA mutant than in parental strain L. monocytogenes L9 under biotic conditions. The values in parentheses represent the number of genes with significant variations in transcript levels/the total number of genes in the functional category. IS, insertion sequence.
In silico analysis of intergenic regions with significant differences in transcript levels between the ΔagrA mutant and the parental strain highlighted a set of 41 ncRNAs. Their locations, the flanking genes, and the measured fold changes in expression are listed in Table 3. The levels of the transcripts of 10 genes (the rliG, Rli47, Rli80, Rli88, Rli99, Rli106, Rli120, Rli126, Rli127, and Rli140 genes) were higher in the ΔagrA background than in the parental strain, and 31 genes were identified to have lower transcript levels in the ΔagrA background than in the parental strain. To date, 271 ncRNAs have been identified and annotated in the genome of L. monocytogenes (31, 34–37). Among these ncRNAs, several have been linked to biological functions. These include the Rli28, Rli32, Rli38, and Rli50 genes, which pair to mRNAs whose products are potentially involved in bacterial adaptation and also in rodent infection (38); the Rli31 and Rli50 genes are required for full virulence in murine, larva, and macrophage infection models (37). Moreover, the S-adenosylmethionine riboswitch sreB (Rli47), which can be produced as a short transcript and act as a trans-regulator, negatively controls PrfA expression by pairing to the 5′ untranslated region of the mRNA (39). Finally, Mandin et al. predicted that the ncRNA rliI targets mRNA with biological functions related to sugar metabolism and transport (36).
TABLE 3.
Location, flanking genes, and fold change in expression of the ncRNAs found to be differentially transcribed between the ΔagrA mutant and the parental strain under biotic conditions
| ncRNA group and gene | Position |
Flanking gene |
Fold change in expression | ||
|---|---|---|---|---|---|
| Start | Stop | 5′ | 3′ | ||
| ncRNAs with significantly higher transcript levels in ΔagrA mutant | |||||
| Rli26 | 388707 | 388590 | lmo0360 | lmo0361 | 5.277 |
| Rli88 | 1320429 | 1320501 | lmo1292 | lmo1293 | 2.532 |
| Rli127 | 1473829 | 1473701 | lmo1439 | lmo1440 | 4.109 |
| Rli47 (sreB) | 2226036 | 2226481 | lmo2141 | lmo2142 | 3.457 |
| Rli120 | 226546 | 226640 | lmo0219 | lmo0220 | 2.851 |
| rliG | 2386992 | 2386715 | lmo2302 | lmo2304 | 2.676 |
| Rli99 | 2395032 | 2395236 | lmo2320 | lmo2321 | 4.911 |
| Rli140 | 2395244 | 2395032 | lmo2320 | lmo2321 | 4.911 |
| Rli106 | 2594785 | 2594561 | lmo2515 | lmo2516 | 2.309 |
| Rli52 | 552417 | 552313 | lmo0517 | lmo0518 | 3.351 |
| Rli80 | 787038 | 787254 | lmo0761 | lmo0762 | 4.516 |
| ncRNAs with significantly lower transcript levels in ΔagrA mutant | |||||
| Rli38 | 1152549 | 1152917 | lmo1115 | lmo1116 | 2.495 |
| Rli55 | 1198108 | 1198561 | lmo1170 | pduQ | 2.686 |
| Rli56 | 1199848 | 1199937 | pduQ | lmo1172 | 2.624 |
| Rli40 | 1275794 | 1275547 | lmo1251 | lmo1252 | 2.062 |
| Rli59 | 1702543 | 1702361 | lmo1652 | lmo1653 | 2.464 |
| Rli23 | 172171 | 172268 | lmo0172 | 2.677 | |
| Rli43 | 1861533 | 1861377 | inlC | rplS | 2.190 |
| Rli44 | 2039087 | 2039375 | lmo1964 | lmo1965 | 2.154 |
| Rli94 | 2039375 | 2039079 | lmo1964 | lmo1965 | 2.154 |
| Rli60 | 2054162 | 2054501 | lmo1982 | ilvD | 2.613 |
| Rli45 | 2154765 | 2154852 | lmo2074 | lmo2075 | 2.440 |
| Rli46 | 2155058 | 2154765 | lmo2074 | lmo2075 | 2.440 |
| Rli61 | 2275362 | 2275297 | lmo2187 | lmo2188 | 2.616 |
| Rli48 | 2361405 | 2361274 | lmo2271 | lmo2272 | 2.571 |
| Rli98 | 2361329 | 2361581 | lmo2271 | lmo2272 | 2.571 |
| Rli112 | 2782978 | 2783221 | lmo2709 | lmo2710 | 3.483 |
| Rli50 | 2783264 | 2782981 | lmo2709 | lmo2710 | 3.483 |
| Rlii | 2842199 | 2841962 | lmo2760 | lmo2761 | 2.437 |
| Rli25 | 357618 | 357516 | lmo0330 | 2.731 | |
| Rli27 | 434817 | 434929 | lmo0411 | lmo0412 | 2.812 |
| Rli28 | 507372 | 507141 | lmo0470 | lmo0471 | 3.364 |
| Rli78 | 507068 | 507473 | lmo0470 | lmo0471 | 3.364 |
| Rli29 | 507632 | 507450 | lmo0470 | lmo0471 | 2.742 |
| Rli52 | 552417 | 552313 | lmo0517 | lmo0518 | 2.423 |
| Rli31 | 597806 | 597926 | lmo0558 | lmo0559 | 2.610 |
| Rli32 | 600750 | 600604 | lmo0560 | lmo0561 | 2.334 |
| Rli33-2 | 708618 | 708860 | lmo0671 | lmo0672 | 2.501 |
| Rli35 | 855495 | 855393 | lmo0828 | 2.642 | |
| Rli36 | 859521 | 859412 | lmo0829 | lmo0830 | 2.261 |
| Rli37 | 907526 | 907832 | lmo0866 | lmo0867 | 2.395 |
| Rli53 | 955824 | 956021 | lmo0918 | lmo0919 | 2.358 |
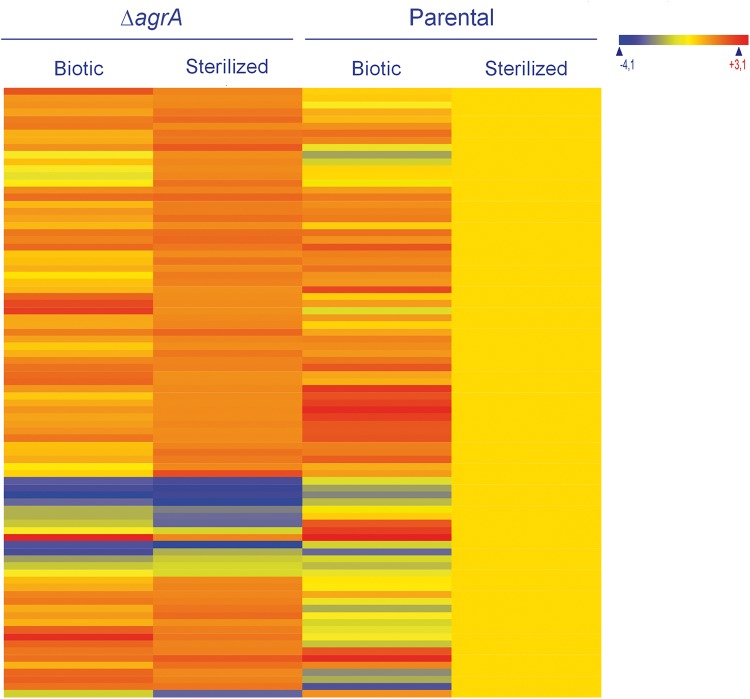
Transcriptome variations between the parental strain and the ΔagrA mutant are limited in sterilized soil environments.
In sterilized soil extracts, deletion of agrA resulted in a limited variation of the transcriptome (Fig. 6). Indeed, 86 genes and 29 intergenic regions showed significant differences in transcript levels between the ΔagrA mutant and the parental strain. Seventy-two genes had higher transcript levels in the mutant, and the fold change in expression of most of them was less than 3 (see Table S3 in the supplemental material). Gene ontology identified functional categories of intermediary metabolism 2.1.1 (specific pathways, 18 genes) and 2.5 (metabolism of coenzymes and prosthetic groups, 12 genes) to be significant terms for this set of genes. For example, genes involved in glycolytic and specific pathways (lmo0342 to lmo0351) (31) were represented, as were some genes coding for enzymes of the cobalamin synthesis pathway (lmo1190, lmo1192, lmo1193, and lmo1196).
FIG 6.
Heat map of the set of genes with significant differences in levels of expression between the ΔagrA mutant and the parental strain under sterilized soil conditions. Expression levels were standardized so that those of the parental strain under sterilized conditions were set as the baseline. Genes are represented in rows. The expression levels of these genes under biotic conditions are also presented.
In the set of genes with lower transcript levels in the ΔagrA mutant (see Table S3 in the supplemental material), most genes were also detected in the corresponding set of genes identified to have lower transcript levels under biotic conditions. As expected, the four genes of the agr operon (agrBDCA) were detected, and the greatest fold change was detected with agrD (which had a 17 times lower level of expression). Three genes coding for putative secreted proteins (lmo0477, lmo0478, and lmo0479) were also included in the top 10 genes with lower transcript levels. Their levels of expression varied by 16-, 6-, and 5-fold, respectively. Interestingly, all entries in this set of genes except lmo1798, lmo2286, and lysA were recorded in the set of genes with lower transcript levels during growth of a ΔagrA mutant of L. monocytogenes EGD-e at 25°C in rich medium (40).
The presence of the biotic fraction results in higher levels of transcription of mobility and ribosomal protein genes.
In order to extract information regarding the response of L. monocytogenes to the soil biotic fraction, we focused on the genes with high expression levels (>9 log units) and for which >3-fold changes in expression under biotic conditions in comparison to the levels of expression in a sterilized soil environment were recorded. Considering L. monocytogenes L9, a total of 369 genes met these criteria. One hundred seventeen genes had higher transcript levels (see Table S4 in the supplemental material). Of this set of genes, functional categories 3.7.1 (ribosomal proteins, 16/56 genes), 1.5 (mobility and chemotaxis, 7/30 genes), and, to a lesser extent, 3.7.4 (elongation, 3/5 genes), 4.3 (phage-related functions, 6/48 genes), 1.2 (transport/binding proteins and lipoproteins, 19/331 genes), and 2.3 (metabolism of nucleotides and nucleic acids, 7/61 genes) were identified by ontology analysis. Interestingly, 7 regulators were in this set of genes, including agrA (3.6-fold increased expression). The levels of transcripts of the genes for the global nitrogen regulator (GlnR), the ferric uptake regulator (Fur), and the pyrimidine regulatory protein (PyrR) also increased in the biotic environment. In the mutant background, only 5 genes were included in the set of genes with higher transcript levels under biotic conditions (see Table S5 in the supplemental material).
The presence of a biotic environment resulted in lower levels of transcription of 253 genes in the parental background (see Table S6 in the supplemental material). Gene ontology failed to identify functional categories in this set of genes except for category 4.3, phage-related functions (13/48 genes). Again, the set of genes with lower transcript levels under biotic conditions was more modest in the mutant background (25 genes; see Table S5 in the supplemental material).
PCR quantification of selected transcripts.
In order to confirm the results of the expression profiles, 10 target genes were selected for quantification. Four of these genes (lmo0349, lmo0914, lmo0917, and lmo2257) were in the set of genes with higher transcript levels in the mutant background, 4 (lmo0050, lmo0051, lmo0477, and lmo2711) had lower transcript levels in the ΔagrA mutant background, and 2 (lmo0943 and lmo2467) had similar trends in both the mutant and wild-type strains. qPCR analysis confirmed that all entries displayed patterns similar to those observed with the microarray results (Table 4).
TABLE 4.
Comparison of fold change in expression between the ΔagrA mutant and parental strain of selected genes in microarray and qPCR analyses
| Gene | Fold change in expression |
|
|---|---|---|
| qPCR | Array | |
| Upregulated | ||
| lmo0914 | 2.6 | 6.6 |
| lmo0349 | 4.7 | 21.4 |
| lmo2257 | 6.3 | 4.0 |
| lmo0917 | 7.2 | 3.2 |
| Downregulated | ||
| lmo2711 | −7.2 | −5.4 |
| lmo0050 (agrC) | −8.2 | −8.7 |
| lmo0051 (agrA) | −6.7 | −6.4 |
| lmo0477 | −37.9 | −16.0 |
| Unchanged | ||
| lmo0943 | 1.4 | |
| lmo2467 | 1.2 | |
DISCUSSION
One of the trademarks of bacteria of the species Listeria monocytogenes is their variety of habitats and their capacity to switch from a saprophytic mode of life to infection of animals. Soil is one of these habitats where Listeria monocytogenes can be found (41). In this study, comparison of the population dynamics in 10 soils showed that survival is dependent on the type of soil. Indeed, the final abundance of listerial populations ranged from 1.5 to 4 log units. This means that each soil sample, characterized by a combination of abiotic and biotic factors, has to be considered a unique environment. Depending on environmental factors, the chance of L. monocytogenes survival is higher in some soils than in others. These results are consistent with those of a previous study which showed that the survival of L. monocytogenes in soil depended on abiotic properties, in particular, the basic cation saturation ratio, pH, and clay content (42). In our study, pH was the predominant abiotic factor highlighted by the Spearman rank correlation. It is known that low pH is detrimental to the survival of L. monocytogenes in soil, but as pH is a structuring parameter for soil microbial communities, it is rather difficult to differentiate the actual direct effect of pH and the indirect role of the composition and structure of the microbial communities on the dynamics of the populations of L. monocytogenes (42). More generally, because the composition and structure of the microbial communities are structured by the complex characteristics of the abiotic environment (43–45), it is difficult to address the weight of a single parameter on the overall survival of L. monocytogenes in soil. Nevertheless, there is clear evidence that the biotic environment is a key player in the control of the populations of L. monocytogenes, as evidenced by comparisons of population dynamics in microcosms of sterilized and unsterilized soils (9, 46, 47). Moreover, in a study of experimental ecology, erosion of the soil microbial diversity resulted in the persistence of the population of L. monocytogenes in soil (48). The overall fate of L. monocytogenes in soil depends on a combination of abiotic characteristics, the microbial abundance, as well as the diversity and structure of the microbial communities. Other components of the soil biology could also affect the population dynamics of L. monocytogenes. For example, protozoans and/or nematodes could facilitate the survival of L. monocytogenes, and in vitro experiments suggest that they may shed the pathogen and, hence, facilitate dispersion (41). One can speculate that in soil sample 1003, collected in a forest, the abundance of protozoans could have facilitated the persistence of L. monocytogenes. The abundance of fungi could also impact survival (49–51).
In a study that focused on one silty loam soil, we previously showed that impairment of the response regulator AgrA or inactivation of the production of the propeptide AgrD reduced the fitness of L. monocytogenes in this habitat (27). The results of the present study allow the general observation that inactivation of the Agr system affects the dynamics of L. monocytogenes in other kinds of soil with textures ranging from sandy loam to clay and confirm that deletion of agrA affects the adaptation of L. monocytogenes to the soil environment. It is interesting to notice that the consequences of the inactivation of the Agr system were limited in soil sample 1003, which was permissive to L. monocytogenes. This suggests that the activity of the Agr system may be critical when L. monocytogenes has to face unfavorable conditions and when its survival is compromised. AgrA is part of the complex transcriptional regulatory circuitry of L. monocytogenes, and experimental evidence suggests that the AgrA regulon includes genes responsible for the transport and metabolism of amino acids and related molecules, genes responsible for motility and chemotaxis, and also genes that code for regulators (40). The observed interconnection between the Agr regulon and the σB regulon (40) sustains the idea that the impact of Agr activity is more pronounced under biotic and abiotic stress conditions. These observations suggest that AgrA activity participates in the overall adaptation of the cell following the integration of environmental cues.
Major variations to the transcriptome were recorded in the parental background under biotic conditions. This suggests that the sensing of the biotic environment triggers a range of physiological adaptations. This adaptation could be depicted as a scenario in which the cell surface must be rearranged as a protective action and for the development of resistance to antimicrobial compounds. Moreover, the upregulation of several transport systems and modification of metabolic activity are most likely, as evidenced by the upregulation of carbon, nitrogen, and pyrimidine metabolism regulators. Increased motility may also be a strategy developed in response to the biotic environment. The higher level of transcription of fur probably reflects the need to compete for available iron, as overexpression of this regulator was observed under iron-limiting conditions (52). The higher level of transcription of agrA under biotic conditions is another indication of the involvement of AgrA in the regulation cascade required to adapt to the biotic environment. In fact, the inactivation of AgrA impeded the appropriate rearrangement of the transcriptome in the mutant background. As a consequence, comparison of the transcriptomes of the parental and ΔagrA mutant strains showed large variations under biotic conditions. These results suggest that the extensive modification of gene expression that L. monocytogenes undergoes during adaptation to soil requires AgrA activity. In this study, inactivation of AgrA altered the transcription of genes related to cell envelope and cellular processes. It included PTS systems and ABC transporters dedicated to carbohydrate uptake. Upregulation of these genes in the parental background may illustrate a metabolic adaptation required to use resources available in the soil environment, where competition for substrates may occur (53–56). Indeed, the prevalence of nutrient acquisition mechanisms in the onset of soil adaptation has been reported previously (30). Similarly, modification of the bacterial cell surface is required for environmental adaptation and during growth and survival in soil (30, 38, 57–59). Inactivation of AgrA could have prevented appropriate adjustment of the cell surface to the conditions of the environment. Interestingly, genes known to code for proteins involved in resistance to antimicrobial peptides were transcribed to lower levels in the mutant background. This finding may indicate that the deletion of agrA impairs the onset of the appropriate mechanisms required to resist competition by interference and to resist antimicrobial weaponry.
The detection of a set of genes with higher transcript levels in the ΔagrA background is an indication of the inability of the mutant to downregulate the transcription of these targets. Similar observations were reported during incubation in the laboratory medium tryptone soy broth at 25°C and 37°C (40). At the in vivo temperature of 37°C, in the ΔagrA background, downregulation was suppressed for a set of genes coding for proteins involved in cellular processes and metabolism.
The regulatory cascades underlying these transcriptome rearrangements also probably involve ncRNAs, as the transcript levels of a repertoire of ncRNAs also varied under biotic conditions. They have been recognized to be important regulators in biological processes in eukaryotes (60) and bacteria (61, 62). So far, a large repertoire of ncRNAs has been described in L. monocytogenes, and evidence suggests that they are central to environmental adaptation (31, 35–38, 63–65). As in other bacterial models, this noncoding part of the genome is an important contributor to the regulation of virulence in L. monocytogenes (35–39). These ncRNAs may participate to the transition from saprophytic to intracellular life. For example, the L. monocytogenes-specific Rli38 gene is upregulated during invasion of the host's blood system and forms a complex with the transcripts of three genes coding for proteins involved in L. monocytogenes adaptation to blood (38). The Rli28, Rli32, Rli34, Rli38, Rli49, and Rli50 genes regulate the expression of virulence factors, such as internalins, by pairing to mRNAs (38). Beyond virulence, there is evidence that ncRNAs participate in the regulation of the transport of sugars and metabolism (36, 63), the synthesis of vitamins (35), and motility (35). The interconnection between the RNome and other regulons is evidenced by the presence of σA, σB, or PrfA boxes upstream of ncRNAs (35, 66), and deletion of lhrA affects the expression of more than 300 genes (63). In accordance with the findings of these studies, our results may suggest that the regulator AgrA also participates in the regulation of the RNome. However, further investigation will be required to decipher the actual role of these ncRNAs during the saprophytic life of L. monocytogenes in soil.
Altogether these results give more insight into the role of the AgrA-mediated response and Agr communication system during the environmental adaptation of L. monocytogenes. The activity of the Agr communication system, through AgrA-mediated regulation, is triggered in response to the soil environment. Interestingly, in soil, AgrA-mediated regulation comes in response to biotic parameters, either directly or indirectly. These results highlight that the adaptation of L. monocytogenes to soil requires reshaping of gene expression at the transcriptional level. Finally, our data suggest that, in complex environments, the regulator AgrA may have a central role in the regulation of transcription and it may participate in the modulation of the large repertoire of ncRNAs.
Supplementary Material
ACKNOWLEDGMENTS
This work received a grant from the Regional Council of Burgundy (reference PARI AGRALE 11).
The funders had no role in the study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.04134-14.
REFERENCES
- 1.Cossart P, Toledo-Arana A. 2008. Listeria monocytogenes, a unique model in infection biology: an overview. Microbes Infect 10:1041–1050. doi: 10.1016/j.micinf.2008.07.043. [DOI] [PubMed] [Google Scholar]
- 2.De Luca G, Zanetti F, Fateh-Moghadm P, Stampi S. 1998. Occurrence of Listeria monocytogenes in sewage sludge. Zentralbl Hyg Umweltmed 201:269–277. [PubMed] [Google Scholar]
- 3.Garrec N, Picard-Bonnaud F, Pourcher AM. 2003. Occurrence of Listeria sp. and L. monocytogenes in sewage sludge used for land application: effect of dewatering, liming and storage in tank on survival of Listeria species. FEMS Immunol Med Microbiol 35:275–283. doi: 10.1016/S0928-8244(02)00443-1. [DOI] [PubMed] [Google Scholar]
- 4.Lyautey E, Lapen DR, Wilkes G, McCleary K, Pagotto F, Tyler K, Hartmann A, Piveteau P, Rieu A, Robertson WJ, Medeiros DT, Edge TA, Gannon V, Topp E. 2007. Distribution and characteristics of Listeria monocytogenes isolates from surface waters of the South Nation River watershed, Ontario, Canada. Appl Environ Microbiol 73:5401–5410. doi: 10.1128/AEM.00354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beuchat LR. 1996. Listeria monocytogenes: incidence on vegetables. Food Control 7:223–228. doi: 10.1016/S0956-7135(96)00039-4. [DOI] [Google Scholar]
- 6.Weis J, Seeliger HPR. 1975. Incidence of Listeria monocytogenes in nature. Appl Microbiol 30:29–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Welshimer HJ. 1960. Survival of Listeria monocytogenes in soil. J Bacteriol 80:316–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Locatelli A, Depret G, Jolivet C, Henry S, Dequiedt S, Piveteau P, Hartmann A. 2013. Nation-wide study of the occurrence of Listeria monocytogenes in French soils using culture-based and molecular detection methods. J Microbiol Methods 93:242–250. doi: 10.1016/j.mimet.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 9.Moshtaghi H, Garg SR, Mandokhot UV. 2009. Survivability of Listeria monocytogenes in agricultural field soil. Indian J Vet Res 18:1–7. [Google Scholar]
- 10.Latorre AA, Kessel JASV, Karns JS, Zurakowski MJ, Pradhan AK, Zadoks RN, Boor KJ, Schukken YH. 2009. Molecular ecology of Listeria monocytogenes: evidence for a reservoir in milking equipment on a dairy farm. Appl Environ Microbiol 75:1315–1323. doi: 10.1128/AEM.01826-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Latorre AA, Kessel JSV, Karns JS, Zurakowski MJ, Pradhan AK, Boor KJ, Jayarao BM, Houser BA, Daugherty CS, Schukken YH. 2010. Biofilm in milking equipment on a dairy farm as a potential source of bulk tank milk contamination with Listeria monocytogenes. J Dairy Sci 93:2792–2802. doi: 10.3168/jds.2009-2717. [DOI] [PubMed] [Google Scholar]
- 12.Nightingale KK, Schukken YH, Nightingale CR, Fortes ED, Ho AJ, Her Z, Grohn YT, McDonough PL, Wiedmann M. 2004. Ecology and transmission of Listeria monocytogenes infecting ruminants and in the farm environment. Appl Environ Microbiol 70:4458–4467. doi: 10.1128/AEM.70.8.4458-4467.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garrido V, Vitas AI, Garcia-Jalon I. 2009. Survey of Listeria monocytogenes in ready-to-eat products: prevalence by brands and retail establishments for exposure assessment of listeriosis in northern Spain. Food Control 20:986–991. doi: 10.1016/j.foodcont.2008.11.013. [DOI] [Google Scholar]
- 14.Goulet V, Rocourt J, Rebiere I, Jacquet C, Moyse C, Dehaumont P, Salvat G, Veit P. 1998. Listeriosis outbreak associated with the consumption of rillettes in France in 1993. J Infect Dis 177:155–160. doi: 10.1086/513814. [DOI] [PubMed] [Google Scholar]
- 15.van Houdt R, Michiels C. 2010. Biofilm formation and the food industry, a focus on the bacterial outer surface. J Appl Microbiol 109:1117–1131. doi: 10.1111/j.1365-2672.2010.04756.x. [DOI] [PubMed] [Google Scholar]
- 16.Fenlon D. 1985. Wild birds and silage as reservoirs of Listeria in the agricultural environment. J Bacteriol 59:537–543. [DOI] [PubMed] [Google Scholar]
- 17.Ho AJ, Ivanek R, Groehn YT, Nightingale KK, Wiedmann M. 2007. Listeria monocytogenes fecal shedding in dairy cattle shows high levels of day-to-day variation and includes outbreaks and sporadic cases of shedding of specific L. monocytogenes subtypes. Prev Vet Med 80:287–305. doi: 10.1016/j.prevetmed.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 18.Iida T, Kanzaki M, Maruyama T, Inoue S, Kaneuchi C. 1991. Prevalence of Listeria monocytogenes in intestinal contents of healthy animals in Japan. J Vet Med Sci 53:873–875. doi: 10.1292/jvms.53.873. [DOI] [PubMed] [Google Scholar]
- 19.Glaser P, Frangeul L, Buchrieser C, Rusniok C, Amend A, Baquero F, Berche P, Bloecker H, Brandt P, Chakraborty T, Charbit A, Chetouani F, Couvé E, de Daruvar A, Dehoux P, Domann E, Domínguez-Bernal G, Duchaud E, Durant L, Dussurget O, Entian KD, Fsihi H, Portillo FG-D, Garrido P, Gautier L, Goebel W, Gómez-López N, Hain T, Hauf J, Jackson D, Jones LM, Kaerst U, Kreft J, Kuhn M, Kunst F, Kurapkat G, Madueno E, Maitournam A, Vicente JM, Ng E, Nedjari H, Nordsiek G, Novella S, de Pablos B, Pérez-Diaz JC, Purcell R, Remmel B, Rose M, Schlueter T, Simoes N, et al. 2001. Comparative genomics of Listeria species. Science 294:849–852. doi: 10.1126/science.1063447. [DOI] [PubMed] [Google Scholar]
- 20.Novick RP, Geisinger E. 2008. Quorum sensing in staphylococci. Annu Rev Genet 42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 21.Queck SY, Jameson-Lee M, Villaruz AE, Bach T-HL, Khan BA, Sturdevant DE, Ricklefs SM, Li M, Otto M. 2008. RNAIII-independent target gene control by the Agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol Cell 32:150–158. doi: 10.1016/j.molcel.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yarwood JM, Bartels DJ, Volper EM, Greenberg EP. 2004. Quorum sensing in Staphylococcus aureus biofilms. J Bacteriol 186:1838–1850. doi: 10.1128/JB.186.6.1838-1850.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garmyn D, Gal L, Lemaitre J-P, Hartmann A, Piveteau P. 2009. Communication and autoinduction in the species Listeria monocytogenes. Commun Integr Biol 2:371–374. doi: 10.4161/cib.2.4.8610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rieu A, Weidmann S, Garmyn D, Piveteau P, Guzzo J. 2007. agr system of Listeria monocytogenes EGD-e: role in adherence and differential expression pattern. Appl Environ Microbiol 73:6125–6133. doi: 10.1128/AEM.00608-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riedel CU, Monk IR, Casey PG, Waidmann MS, Gahan CGM, Hill C. 2009. AgrD-dependent quorum sensing affects biofilm formation, invasion, virulence and global gene expression profiles in Listeria monocytogenes. Mol Microbiol 71:1177–1189. doi: 10.1111/j.1365-2958.2008.06589.x. [DOI] [PubMed] [Google Scholar]
- 26.Autret N, Raynaud C, Dubail I, Berche P, Charbit A. 2003. Identification of the agr locus of Listeria monocytogenes: role in bacterial virulence. Infect Immun 71:4463–4471. doi: 10.1128/IAI.71.8.4463-4471.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vivant AL, Garmyn D, Gal L, Piveteau P. 2014. The Agr communication system provides a benefit to the populations of Listeria monocytogenes in soil. Front Cell Infect Microbiol 4:160. doi: 10.3389/fcimb.2014.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemunier M, Francou C, Rousseaux S, Houot S, Dantigny P, Piveteau P, Guzzo J. 2005. Long-term survival of pathogenic and sanitation indicator bacteria in experimental biowaste composts. Appl Environ Microbiol 71:5779–5786. doi: 10.1128/AEM.71.10.5779-5786.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grolleau E, Bargeot L, Chafchafi A, Hardy R, Doux J, Beaudou A, Martret HL, Lacassin J-C, Fort J-L, Falipou P, Arrouays D. 2004. Le système d'information national sur les sols: DONESOL et les outils associés. Etude Gestion Sols 11:255–269. [Google Scholar]
- 30.Piveteau P, Depret G, Pivato B, Garmyn D, Hartmann A. 2011. Changes in gene expression during adaptation of Listeria monocytogenes to the soil environment. PLoS One 6:e24881. doi: 10.1371/journal.pone.0024881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wurtzel O, Sesto N, Mellin JR, Karunker I, Edelheit S, Becavin C, Archambaud C, Cossart P, Sorek R. 2012. Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol Syst Biol 8:1–14. doi: 10.1038/msb.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abachin E, Poyart C, Pellegrini E, Milohanic E, Fiedler F, Berche P, Trieu-Cuot P. 2002. Formation of d-alanyl-lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes. Mol Microbiol 43:1–14. doi: 10.1046/j.1365-2958.2002.02723.x. [DOI] [PubMed] [Google Scholar]
- 33.Dowd GC, Casey PG, Begley M, Hill C, Gahan CGM. 2012. Investigation of the role of ZurR in the physiology and pathogenesis of Listeria monocytogenes. FEMS Microbiol Lett 327:118–125. doi: 10.1111/j.1574-6968.2011.02472.x. [DOI] [PubMed] [Google Scholar]
- 34.Behrens S, Widder S, Mannala GK, Qing X, Madhugiri R, Kefer N, Mraheil MA, Rattei T, Hain T. 2014. Ultra deep sequencing of Listeria monocytogenes sRNA transcriptome revealed new antisense RNAs. PLoS One 9:e83979. doi: 10.1371/journal.pone.0083979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mellin JR, Cossart P. 2012. The non-coding RNA world of the bacterial pathogen Listeria monocytogenes. RNA Biol 9:372–378. doi: 10.4161/rna.19235. [DOI] [PubMed] [Google Scholar]
- 36.Mandin P, Repoila F, Vergassola M, Geissmann T, Cossart P. 2007. Identification of new noncoding RNAs in Listeria monocytogenes and prediction of mRNA targets. Nucleic Acids Res 35:962–974. doi: 10.1093/nar/gkl1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mraheil MA, Billion A, Mohamed W, Mukherjee K, Kuenne C, Pischimarov J, Krawitz C, Retey J, Hartsch T, Chakraborty T, Hain T. 2011. The intracellular sRNA transcriptome of Listeria monocytogenes during growth in macrophages. Nucleic Acids Res 39:4235–4248. doi: 10.1093/nar/gkr033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, Barthelemy M, Vergassola M, Nahori MA, Soubigou G, Regnault B, Coppee JY, Lecuit M, Johansson J, Cossart P. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 39.Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J. 2009. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell 139:770–779. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]
- 40.Garmyn D, Augagneur Y, Gal L, Vivant A-L, Piveteau P. 2012. Listeria monocytogenes differential transcriptome analysis reveals temperature-dependent Agr regulation and suggests overlaps with other regulons. PLoS One 7:e43154. doi: 10.1371/journal.pone.0043154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vivant AL, Garmyn D, Piveteau P. 2013. Listeria monocytogenes, a pathogen down-to-earth. Front Cell Infect Microbiol 3:87 doi: 10.3389/fcimb.2013.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Locatelli A, Spor A, Jolivet C, Piveteau P, Hartmann A. 2013. Biotic and abiotic soil properties influence survival of Listeria monocytogenes in soil. PLoS One 8:e75969. doi: 10.1371/journal.pone.0075969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dequiedt S, Saby NPA, Lelievre M, Jolivet C, Thioulouse J, Toutain B, Arrouays D, Bispo A, Lemanceau P, Ranjard L. 2011. Biogeographical patterns of soil molecular microbial biomass as influenced by soil characteristics and management. Global Ecol Biogeogr 20:641–652. doi: 10.1111/j.1466-8238.2010.00628.x. [DOI] [Google Scholar]
- 44.Hu HW, Zhang LM, Dai Y, Di HJ, He JZ. 2013. pH-dependent distribution of soil ammonia oxidizers across a large geographical scale as revealed by high-throughput pyrosequencing. J Soils Sediments 13:1439–1449. doi: 10.1007/s11368-013-0726-y. [DOI] [Google Scholar]
- 45.Fierer N, Bradford MA, Jackson RB. 2007. Toward an ecological classification of soil bacteria. Ecology 88:1354–1364. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- 46.Dowe MJ, Jackson ED, Mori JG, Bell CR. 1997. Listeria monocytogenes survival in soil and incidence in agricultural soils. J Food Prot 60:1201–1207. [DOI] [PubMed] [Google Scholar]
- 47.McLaughlin HP, Casey PG, Cotter J, Gahan CGM, Hill C. 2011. Factors affecting survival of Listeria monocytogenes and Listeria innocua in soil samples. Arch Microbiol 193:775–785. doi: 10.1007/s00203-011-0716-7. [DOI] [PubMed] [Google Scholar]
- 48.Vivant AL, Garmyn D, Maron PA, Nowak V, Piveteau P. 2013. Microbial diversity and structure are drivers of the biological barrier effect against Listeria monocytogenes in soil. PLoS One 8:e76991. doi: 10.1371/journal.pone.0076991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alam SI, Kamran M, Sohail M, Ahmad A, Khan SA. 2011. Partial characterization of bacteriocin like inhibitory substance from Bacillus subtilis BS15, a local soil isolate. Pak J Bot 43:2195–2199. [Google Scholar]
- 50.Bigwood T, Hudson JA, Cooney J, McIntyre L, Billington C, Heinemann JA, Wall F. 2012. Inhibition of Listeria monocytogenes by Enterococcus mundtii isolated from soil. Food Microbiol 32:354–360. doi: 10.1016/j.fm.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi JA, de Castro MCM, Souza GG, Lucas EMF, Bracarense AAP, Abreu LM, Marriel IE, Oliveira MS, Floreano MB, Oliveira TS. 2008. Isolation and screening of fungal species isolated from Brazilian cerrado soil for antibacterial activity against Escherichia coli, Staphylococcus aureus, Salmonella typhimurium, Streptococcus pyogenes and Listeria monocytogenes. J Mycol Med 18:198–204. doi: 10.1016/j.mycmed.2008.08.001. [DOI] [Google Scholar]
- 52.Ledala N, Sengupta M, Muthaiyan A, Wilkinson BJ, Jayaswal RK. 2010. Transcriptomic response of Listeria monocytogenes to iron limitation and fur mutation. Appl Environ Microbiol 76:406–416. doi: 10.1128/AEM.01389-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leigh J, Fitter AH, Hodge A. 2011. Growth and symbiotic effectiveness of an arbuscular mycorrhizal fungus in organic matter in competition with soil bacteria. FEMS Microbiol Ecol 76:428–438. doi: 10.1111/j.1574-6941.2011.01066.x. [DOI] [PubMed] [Google Scholar]
- 54.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. 2010. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Elsas JD, Chiurazzi M, Mallon CA, Elhottova D, Kristufek V, Salles JF. 2012. Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc Natl Acad Sci U S A 109:1159–1164. doi: 10.1073/pnas.1109326109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marshall KC, Alexander M. 1960. Competition between soil bacteria and fusarium. Plant Soil 12:143–153. doi: 10.1007/BF01377367. [DOI] [Google Scholar]
- 57.Camejo A, Buchrieser C, Couvé E, Carvalho F, Reis O, Ferreira P, Sousa S, Cossart P, Cabanes D. 2009. In vivo transcriptional profiling of Listeria monocytogenes and mutagenesis identify new virulence factors involved in infection. PLoS Pathog 5:e1000449. doi: 10.1371/journal.ppat.1000449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hain T, Hossain H, Chatterjee SS, Machata S, Volk U, Wagner S, Brors B, Haas S, Kuenne CT, Billion A, Otten S, Pane-Farre J, Engelmann S, Chakraborty T. 2008. Temporal transcriptomic analysis of the Listeria monocytogenes EGD-e sigma B regulon. BMC Microbiol 8:20. doi: 10.1186/1471-2180-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van der Veen S, Hain T, Wouters JA, Hossain H, de Vos WM, Abee T, Chakraborty T, Wells-Bennik MHJ. 2007. The heat-shock response of Listeria monocytogenes comprises genes involved in heat shock, cell division, cell wall synthesis, and the SOS response. Microbiology 153(Pt 10):3593–3607. doi: 10.1099/mic.0.2007/006361-0. [DOI] [PubMed] [Google Scholar]
- 60.Storz G, Altuvia S, Wassarman KM. 2005. An abundance of RNA regulators. Annu Rev Biochem 74:199–217. doi: 10.1146/annurev.biochem.74.082803.133136. [DOI] [PubMed] [Google Scholar]
- 61.Repoila F, Darfeuille F. 2009. Small regulatory non-coding RNAs in bacteria: physiology and mechanistic aspects. Biol Cell 101:117–131. doi: 10.1042/BC20070137. [DOI] [PubMed] [Google Scholar]
- 62.Gottesman S. 2005. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet 21:399–404. doi: 10.1016/j.tig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 63.Nielsen JS, Larsen MH, Lillebaek EMS, Bergholz TM, Christiansen MHG, Boor KJ, Wiedmann M, Kallipolitis BH. 2011. A small RNA controls expression of the chitinase ChiA in Listeria monocytogenes. PLoS One 6:e19019. doi: 10.1371/journal.pone.0019019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nielsen JS, Olsen AS, Bonde M, Valentin-Hansen P, Kallipolitis BH. 2008. Identification of a sigma B-dependent small noncoding RNA in Listeria monocytogenes. J Bacteriol 190:6264–6270. doi: 10.1128/JB.00740-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Christiansen JK, Nielsen JS, Ebersbach T, Valentin-Hansen P, Søgaard-Andersen L, Kallipolitis BH. 2006. Identification of small Hfq-binding RNAs in Listeria monocytogenes. RNA 12:1383–1396. doi: 10.1261/rna.49706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oliver H, Orsi R, Ponnala L, Keich U, Wang W, Sun Q, Cartinhour S, Filiatrault M, Wiedmann M, Boor K. 2009. Deep RNA sequencing of Listeria monocytogenes reveals overlapping and extensive stationary phase and sigma B-dependent transcriptomes, including multiple highly transcribed noncoding RNAs. BMC Genomics 10:641. doi: 10.1186/1471-2164-10-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.