Abstract
Inorganic phosphorus (Pi) is one of the main growth-limiting factors of diazotrophic cyanobacteria. Due to human activity, the availability of Pi has increased in water bodies, resulting in eutrophication and the formation of massive cyanobacterial blooms. In this study, we examined the molecular responses of the cyanobacterium Anabaena sp. strain 90 to phosphorus deprivation, aiming at the identification of candidate genes to monitor the Pi status in cyanobacteria. Furthermore, this study increased the basic understanding of how phosphorus affects diazotrophic and bloom-forming cyanobacteria as a major growth-limiting factor. Based on RNA sequencing data, we identified 246 differentially expressed genes after phosphorus starvation and 823 differentially expressed genes after prolonged Pi limitation, most of them related to central metabolism and cellular growth. The transcripts of the genes related to phosphorus transport and assimilation (pho regulon) were most upregulated during phosphorus depletion. One of the most increased transcripts encodes a giant protein of 1,869 amino acid residues, which contains, among others, a phytase-like domain. Our findings predict its crucial role in phosphorus starvation, but future studies are still needed. Using two-dimensional difference in gel electrophoresis (2D-DIGE) and liquid chromatography-tandem mass spectrometry (LC-MS/MS), we found 43 proteins that were differentially expressed after prolonged phosphorus stress. However, correlation analysis unraveled an association only to some extent between the transcriptomic and proteomic abundances. Based on the present results, we suggest that the method used for monitoring the Pi status in cyanobacterial bloom should contain wider combinations of pho regulon genes (e.g., PstABCS transport systems) in addition to the commonly used alkaline phosphatase gene alone.
INTRODUCTION
Nitrogen and phosphorus loading into aquatic ecosystems is a global problem causing enhanced primary production of photoautotrophic organisms (1–3). Among them, cyanobacteria are major beneficiaries of turbid and eutrophic water. They employ numerous ecophysiological strategies such as the capability for using various nutrient sources, gas vacuoles, photoprotective pigments, and UV-absorbing compounds, which enable them to survive and grow under eutrophic conditions and may lead to the formation of harmful and toxic blooms (1, 4). The theory that nutrient loading is a crucial factor causing harmful cyanobacterial blooms is supported by numerous field and laboratory studies showing that toxin production and growth rate both increase under nutrient-replete conditions (2, 5–8).
The diazotrophic Anabaena is one of the most common bloom-forming cyanobacterial genera in freshwater and brackish-water ecosystems, alongside Microcystis and Nodularia, respectively (4, 9), and some strains are capable of producing the following toxins: microcystins, anatoxin-a, anatoxin-a(S), and saxitoxins (4, 10). The ecology of these cyanobacteria thus deserves more attention. The ability to fix nitrogen gives diazotrophic cyanobacteria an advantage in surviving in nitrogen-depleted environments, and thus inorganic phosphorus (Pi), the major phosphorus source exploited by cyanobacteria, plays a key role in limiting the growth of diazotrophic cyanobacteria, alongside macronutrients and trace elements (1, 11). In eutrophic water systems, cyanobacteria are limited by Pi, especially during dense summer blooms (11). High-affinity Pi transporter systems PstABCS, similar to those in Escherichia coli, were found in numerous cyanobacteria and enable phosphorus uptake over a wide range of concentrations (12). In addition, several cyanobacterial genomes encode phosphorus acquisition and assimilation mechanisms for alternative phosphorus sources, such as organic phosphate, phosphonate, and phosphites (13–18), but the complete functioning of these mechanisms is still unclear (19).
Anabaena sp. strain 90 (here Anabaena 90) was isolated in 1987 from a toxic cyanobacterial bloom in Lake Vesijärvi, Finland (20). The strain produces many nonribosomally and ribosomally synthesized bioactive compounds, including hepatotoxic microcystin (20–27). The genome of Anabaena 90 contains two pstABCS operons and genes for several enzymes recognized as phosphonate transporters and phosphatases, leading us to the hypothesis that the strain may be capable of using various phosphorus sources (27). Furthermore, two-component regulatory systems, including the genes phoR, phoS and phoU (28), which are responsible for the regulation of phosphorus transport and acquisition, have also been recognized in the genome of Anabaena 90 (27).
The genomic variability of phosphorus acquisition genes is wide among cyanobacteria, and it was hypothesized that the responses to phosphorus starvation differ among strains (29). The effects of phosphorus stress on single-cell cyanobacteria, such as Prochlorococcus strains MIT9312, NATL2A, and SS120, Synechococystis sp. strains PCC 6803 and PCC 6714, Microcystis aeruginosa, and Synechococcus sp. strains WH 8102 and PCC 7002, have been surveyed only to some extent (5, 6, 30–35). Transcriptomic or proteomic studies with toxic, heterocystous freshwater or brackish-water cyanobacteria related to phosphorus stress have, as far as we know, not yet been performed. In this study, we investigated how short-term phosphorus starvation followed by prolonged depletion affected the transcriptome and proteome of Anabaena 90. In addition, to gain fundamental insight into the Pi stress response, we aimed at the identification of specific marker genes for various phosphorus concentrations that are suitable for development of a monitoring method to follow the Pi status in cyanobacterial blooms.
MATERIALS AND METHODS
Growth experiment and sampling.
Hepatotoxic Anabaena 90 belongs to the University of Helsinki culture collection (HAMBI, UHCC). The strain was isolated in 1987 from Lake Vesijärvi, Finland (20), purified axenic, and cultured in Z8 medium with 17.1 mg/liter of phosphate (PO43−) and nitrogen free (Z8X medium) at room temperature under continuous illumination (36). In this study, Anabaena 90 was grown in six replicates in tissue culture flasks containing 130 ml of Z8X medium at 20°C and illumination of 8.4 to 12.9 μmol photons m−2s−1. When the cultures reached an optical density at 750 nm (OD750) of 0.8, the cells were pooled, harvested, and transferred to culture flasks containing 120 ml of fresh modified Z8X without phosphate to let the cells utilize accumulated polyphosphate gathered while growing in Pi-replete medium (Fig. 1A). After 6 days in phosphate-free medium, the cells were harvested again and divided into two culture flasks containing 120 ml of Z8X medium with 0.16 mg/liter of phosphate and another containing Z8X medium with 17.1 mg/liter of phosphate, which is the original concentration of phosphate in Z8X medium. The cells were further cultured for 4 days.
FIG 1.
Experimental design. (A) Transcriptomic samples were collected at four time points (5hNP, 6dNP, 4dLP, and 4dHP) and proteomic samples at three time points (6dNP, 4dLP, and 4dHP). (B) The samples were treated and data analyzed according to the graph.
During the experiment, the chlorophyll a concentration was measured every second day by collecting cells from 1 ml of culture, followed by methanol extraction at 65°C for 10 min (Fig. 1B; see also Fig. S1 in the supplemental material). The chlorophyll a concentration was measured at 665 nm. Samples for RNA sequencing (RNA-Seq) analysis were collected from three biological replicates (bottles 1 to 3) on day 0 (control) and after growing the cells for 5 h in nonphosphate medium (5hNP), 6 days in nonphosphate medium (6dNP), and 4 days in high-phosphate medium (4dHP) or 4 days in low-phosphate medium (4dLP) (Fig. 1). The cells were harvested by centrifugation at 4°C for 8 min at 7,000 × g. Immediately after being harvested, the cells were frozen in liquid nitrogen and stored at −80°C. For the proteomic analysis, cells from three biological replicates (bottles 1 to 3) were collected by centrifugation on day 0 and 6dNP, 4dLP, and 4dHP. The pellet was resuspended in washing buffer (50 mM HEPES, 30 mM CaCl2·NaOH [pH 7.5]). After washing twice, the cells were stored at −80°C.
RNA preparation and analysis by SOLiD.
Total RNA was extracted using the Ambion RiboPure-Bacteria kit (Life Technologies, Thermo Fisher Scientific Inc., Waltham, MA) with the following modifications. Cells were broken in RNAwiz-solution in MatrixE tubes (MP Biomedicals, Santa Ana, CA) by FastPrep (FastPrep-24; MP Biomedicals) for 40 s at a speed of 5 m s−1. Total RNA was extracted into the water phase by adding 0.4 volume of chloroform to the supernatant. DNA was digested with RQ1 DNase (Promega Corp., Fitchburg, WI), and total RNA was purified by phenol-chloroform extraction and ethanol precipitation. RNA was extracted from total RNA using the MICROBExpress bacterial mRNA enrichment kit (Life Technologies) and following the manufacturer's instructions. Cyanobacterium-specific oligonucleotides (unpublished; Oligomer) replaced half the amount of oligonucleotides provided with the kit to obtain the best results for removal of rRNA. Appropriate removal of rRNA and the quality of RNA were ensured by gel electrophoresis and Bioanalyzer (Agilent Technologies, Santa Clara, CA) with the RNA 6000 Nano kit (Agilent Technologies). The cDNA libraries were synthesized by the Ovation RNA-Seq System V2 (NuGEN Technologies Inc., San Carlos, CA). Sequencing by oligonucleotide ligation and detection (SOLiD) cDNA sequencing was carried out in the DNA Sequencing and Genomics Laboratory, Institute of Biotechnology, University of Helsinki.
RNA-Seq data analysis.
The SOLiD data were mapped with the short-read mapper segemehl using default parameters (37). Reads per feature were counted, using HTseq counts in the intersection-nonempty mode (38). Regions of continuous coverage not overlapping with annotated genes were also used as features for the HTseq counts to include small RNA (sRNA) candidates in the analysis. Differentially expressed genes were detected by pairwise comparison of each condition's libraries with the log-phase libraries using DESeq2 (39). For the differential expression analyses, all technical replicates were combined and outliers were replaced by the trimmed mean of the nonoutlier libraries. The RNA-Seq data were filtered with cutoff values of ≤0.05 for P and ≥1 or ≤ −1 for base two logarithmic fold change (log2FC) (see Table S2 in the supplemental material). The differentially expressed genes were divided manually into functional categories, using the published list of functional categories of Anabaena 90 genes (27) based on the Comprehensive Microbial Resources (CMR) database (see Tables S3 to S6 in the supplemental material).
Protein extraction and quantitation.
The cells were homogenized with glass beads in lysis buffer [7 M urea, 2 M thiourea, 4% (wt/vol) 3(3-cholaminopropyl diethylammonio)-1-propane sulfonate (CHAPS), 20 mM dithiothreitol (DTT)] by shaking with FastPrep (FastPrep-24; MP Biomedicals). The cell debris and glass beads were separated from the soluble proteins by centrifuging the sample at + 4°C for 30 min at 18,000 × g. The protein sample was purified by trichloroacetic acid (TCA)-acetone precipitation (10% TCA [wt/vol] and 20 mM DTT in acetone), and the centrifuged proteins were washed twice with 20 mM DTT in acetone. The air-dried protein pellet was dissolved in lysis buffer, and the protein concentration was determined, using a 2-D Quant kit (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) according to the manufacturer's instructions.
Two-dimensional difference in gel electrophoresis (2D-DIGE).
The protein samples were labeled with an Amersham CyDye DIGE fluor minimal labeling kit (GE Healthcare) by following the manufacturer's instructions. For the internal standard, equal amounts of protein samples were pooled and treated in a manner similar to other samples. Before labeling, the pH of each protein sample was adjusted to 8 to 9 by the addition of 100 mM NaOH or 100 mM HCl. For isoelectric focusing (IEF), the pooled protein samples were cup-loaded at the anodic end of the overnight-rehydrated Immobiline dry strip gels, 24 cm, nonlinear (NL) pH gradient 3 to 7 (GE Healthcare). IEF was performed with the IPGphor system (GE Healthcare) at 20°C and a current of 75 μA per strip. The following voltages were applied: during the first 3 h, 150 V, and during the next 3 h, 300 V followed by a linear increase to 1,000 V over 6 h and from 1,000 to 10,000 V within the next hour. During the last 5 h, the voltage was held at 10,000 V, achieving a total running time of 18 h.
For the second-dimension separation, sodium dodecyl sulfate polyacrylamide gels (12% and 1.0 mm) were cast between low-fluorescent glass plates, enabling accurate imaging of fluorescent dyes. Prior to a second-dimension run, the IEF strips were incubated in equilibration buffers (6 M urea, 2% [wt/vol] SDS, 50 mM Tris [pH 8.8], 0.02% bromophenol blue, 30% [vol/vol] glycerol) for 15 min with buffer A containing 5 g of DTT/liter and for another 15 min with buffer B containing 12.5 g of iodoacetamide (IAA)/liter. Electrophoresis in the second dimension was performed overnight at 25 mA and 1.5 W per gel. Water circulation at 4°C was used for cooling the apparatus.
Imaging and image analysis.
A typhoon scanner (GE Healthcare) was used for gel imaging. The gels were scanned three times with band-pass and wavelengths of 520 nm for Cy2, 580 nm for Cy3, and 670 nm for Cy5. The photomultiplier tube (PMT) voltages were assessed individually to every label for every gel so that the maximal difference between scanning values in one gel was less than 15%. A 100 μm pixel size was used for scanning. The gel images were analyzed with DeCyder 7.0 software, and the Cy2 channel was selected as a standard. The images were cropped manually to the same size, and the edges were rejected. Spot searching and matching against a master gel were performed automatically and verified manually. The differentially expressed protein spots were detected by one-way analysis of variance (ANOVA) and the Student t test. Proteins with P values less than 0.05 were chosen for further analysis.
Peptide mass fingerprinting and peptide identification.
Selected protein spots were cut from silver-stained and dried gels, and in-gel trypsin digestion was carried out, as described earlier (40, 41). The gel pieces were rehydrated in 100 mM NH4HCO3 and rinsed twice with acetonitrile, followed by freeze-drying in a vacuum centrifuge. Trypsin (0.02 μg/μl; Promega) was added to the gel pieces until they swelled and became covered with the liquid, and in-gel digestion was carried out overnight in a shaker at 37°C. The peptides were extracted using pure 10% formic acid (HCOOH) and 50% ACN–10% HCOOH solutions and lyophilized. The dried peptides were dissolved in 6 μl of 2% HCOOH and subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses.
Peptides originating from intensive spots were analyzed with a QSTAR Elite mass spectrometer (Applied Biosystems [Life Technologies]/MDS Sciex, Concord, Ontario, Canada) connected in-line with an Ultimate 3000 high-performance liquid chromatography (HPLC) system (Dionex Corp., Sunnyvale, CA). The peptides were separated on a C18 precolumn (5 by 0.3 mm, PepMap C18; LC Packings B.V. [Dionex], Amsterdam, The Netherlands), and a C18 nanocolumn (15 cm by 75 μm, Magic 5 μm, 200 Å, C18; Michrom BioResources Inc., Sacramento, CA), as described earlier (42), with the exception of a linear 40-min gradient from 2% to 40% solvent B that was used in this study. The peptides originating from low-intensity spots were analyzed with a QExactive mass spectrometer (Thermo Scientific, Waltham, MA) and connected in-line with an Easy-nLC HPLC system (Thermo Scientific). The peptides were separated on a C18 precolumn and a C18 nanocolumn with the characteristics described above using the separation protocol described in reference 42.
Data obtained with LC-MS/MS were analyzed using the in-house Mascot (Matrix Science, London, United Kingdom) server and the database of annotated Anabaena 90 proteins, supplemented with a list of common laboratory contaminants. During searches, carbamidomethylation of cysteine was used as a fixed modification and oxidation of methionine as a variable modification. One trypsin miscleavage was allowed. The parameters for mass tolerance were 50 ppm (peptides) and 0.01 Da (fragments) for the QSTAR Elite results and 5 ppm (peptides) and 0.02 Da (fragments) for the QExactive.
RNA sequencing data accession numbers.
All sequenced used in this study were sent to NCBI Sequence Read Archive under BioProject accession number PRJNA276792.
RESULTS AND DISCUSSION
Overview of the transcriptomic results.
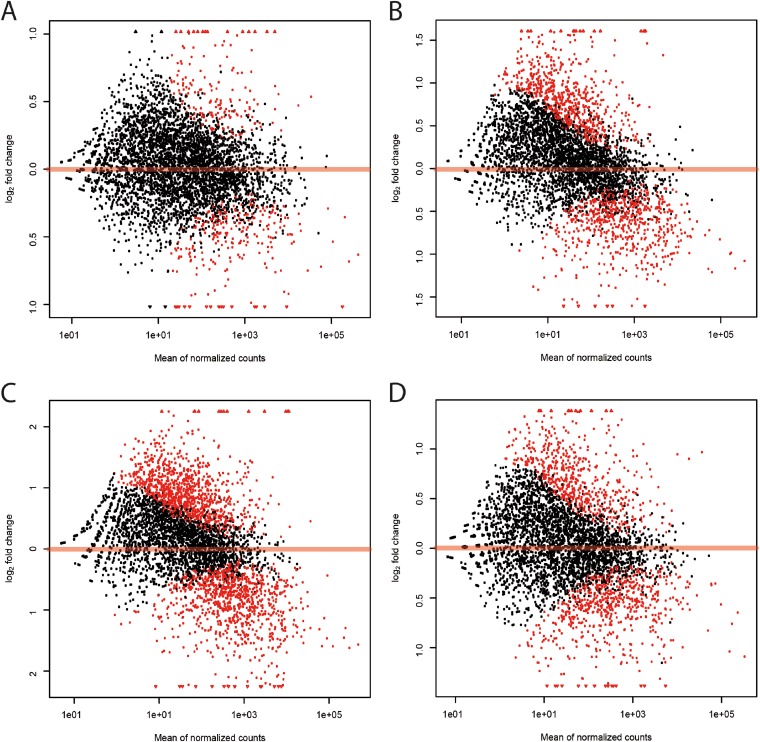
More than 155 million (71.8%) of the sequenced reads were mapped to the annotated Anabaena 90 genome, and 4,527 of the 4,739 protein-encoding genes had at least one mapped read (see Table S1 in the supplemental material). The relatively high number of unmapped reads was analyzed in more detail to investigate possible culture contaminations. With the blastn search against the NCBI nucleotide database, a more sensitive approach was chosen compared to mapping by segemehl. Over 83% of the best hits were assigned to Anabaena sp. strain 90, while no significant signs of contamination could be identified. More than half of the annotated genes without mapped reads belonged to the excision elements splitting genes nifH and nifD of the nif operon (27). These elements are lost from the developing heterocysts by homologous recombination on initiation of dinitrogen fixation, and obviously these genes were also silent in vegetative cells. The rest of the genes without sequenced reads were mainly annotated to transposable and phage elements and were not transcribed under the conditions tested. Applying the cutoff values of ≤0.05 for P and ≥1 or ≤−1 for log2FC, a total of 34 genes in 5hNP, 246 in 6dNP, 823 in 4dLP and 156 in 4dHP were differentially expressed, compared with the cells in the logarithmic growth phase (day 0) (Fig. 2; see also Tables S3 to S6 in the supplemental material). Only a minor variation in the transcriptome patterns after 5 h of starvation in Pi-free medium (5hNP) was observed, representing exploitation of the polyphosphate storages, and thus, the cells were not yet starved by Pi (Fig. 2A; see also Table S3). Furthermore, considerably less differentially expressed genes were observed after 6 days of incubation in Pi-free medium (Fig. 2B; see also Table S4) than at the next sampling point (4dLP) (Fig. 2C; see also Table S5). Based on this, we assumed that the polyphosphate storages, which were estimated to be sufficient from two to four cell divisions (43), were not completely used in 6dNP. On the other hand, the small amount of Pi added was insufficient for growth, due to the starvation period, and thus, further changes in the transcriptomic level were seen in 4dLP. In spite of the same Pi concentration in the medium as in the control and 4dHP, a surprisingly higher number of differentially expressed genes was identified in 4dHP than in the control (Fig. 2D; see also Table S6). This may have been due to the denser culture on day 0, causing self-shading and forcing cells already to prepare for the stationary growth phase.
FIG 2.
Differentially expressed transcripts during the experiments. The red dots show differentially expressed genes compared with the control condition (P < 0.05). (A) 5hNP; (B) 6dNP; (C) 4dLP; (D) 4dHP.
All genes with statistically significant differentially expressed transcripts were divided into categories according to their functional roles (27). Similar to that in previously reported studies with single-cell cyanobacteria such as Synechococcus, Microcystis, and Prochlorococcus (6, 31, 32, 44), phosphorus depletion in our study seemed to drive the culture to the stationary phase by reducing the expression of the genes related to central metabolism, photosynthetic apparatus, transcription, and translation. If culturing had lasted longer, as in the preliminary experiment (see Fig. S1 in the supplemental material), a lower chlorophyll a concentration than in the high-phosphorus culture and yellowish coloration of the starved culture could have most likely been seen. In turn, specific responses to phosphorus depletion were seen as an increased expression of the phosphorus acquisition systems.
Phosphorus transport and assimilation, nitrate/sulfonate ABC transporter.
The genes involved in the Pi transport system are well studied, not only for E. coli (12) but also for model cyanobacteria (28). Pi uptake occurs through a PstABCS transporter that is regulated by the two-component system with phosphorylation and dephosphorylation of PhoB and PhoR or their orthologues SphS and SphR (28). A third enzyme participating in the regulatory system is the putative negative regulator PhoU (or SphU), whose function is only partially resolved. The same two-component system also regulates the transport and assimilation of organic phosphorus sources and some other genes, e.g., photosynthesis and carbon fixation in cyanobacteria (16, 28). The activation of the above-described pho regulon was long believed to be a specific response in phosphorus acclimation, but a recent study showed that this particular regulon can also be activated at high cell densities of Synechococcus sp. PCC 7002 (31).
The genome of Anabaena 90 contains two closely located operons encoding two PstABCS transporter systems (ANA_C11755-ANA_C11758 and ANA_C11763-ANA_C11766) (27). One of these pstABCS operons was highly upregulated, while the other seemed to be unresponsive (Table 1). A similar difference between pstABCS operons was also observed in a previous study of Pi acquisition by Prochlorococcus (29), suggesting an unknown regulatory system and function of this unresponsive pstABCS operon. No variations in the expression of the well-known regulatory elements phoB, phoR, and phoU (ANA_C12727-ANA_C12729) were found in this study, indicating that the protein amounts were relatively stable under all conditions examined and that the regulatory function is mainly based on the phosphorylation state of these proteins (28).
TABLE 1.
Expression of the genes related to phosphorus transport and assimilation
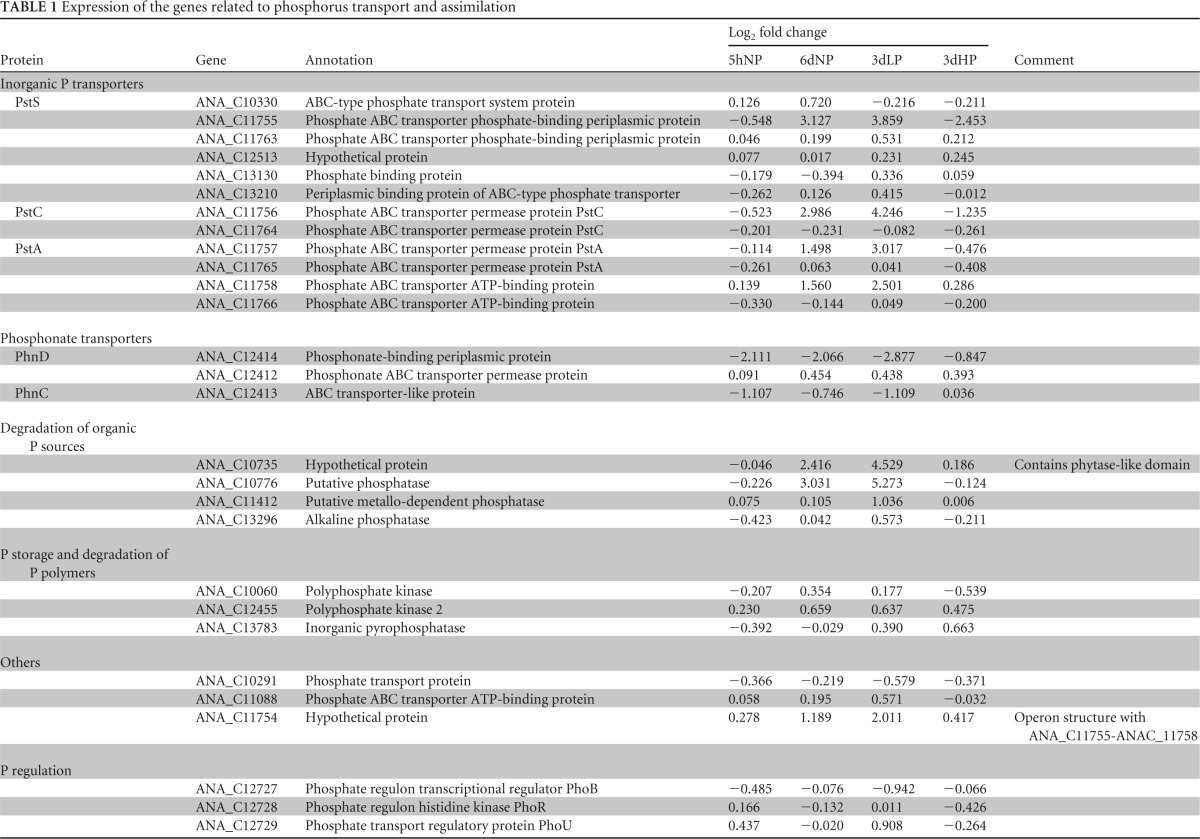
The genome sequence of Anabaena 90 indicates that the strain is also capable of transporting and assimilating organic phosphorus sources (27). The putative phosphatase (ANA_C10776) was almost 39-fold more highly transcribed in 4dLP than in the control (day 0), representing one of the most upregulated genes under low-phosphorus conditions. This particular gene was also significantly upregulated in 6dNP. Furthermore, two other phosphatase genes were annotated from the Anabaena 90 genome. The putative metallo-dependent phosphatase (ANA_C11412) showed a 2-fold increased transcription, but the alkaline phosphatase (ANA_C13296) was only slightly upregulated during phosphorus depletion. The expression levels of these genes under phosphorus depletion show that ANA_C10776 is mainly responsible for degradation of organic phosphorus sources and the other two phosphatases are less valuable for this purpose, which is in accordance with previous studies of other cyanobacteria (6, 19). Interestingly, three genes that are involved in phosphonate transport (ANA_C12412 to ANA_C12414) were downregulated under phosphorus depletion. In Escherichia coli, Pseudomonas stutzeri, and the cyanobacterium Trichodesmium, the phosphonate gene cluster consists of 10 to 14 genes (45, 46). From the genome of Anabaena 90, only three genes of the phn gene cluster were annotated (27), possibly indicating alternative functions of these particular genes. Moreover, downregulation of phn-genes gives a hint that these genes are not under the pho-regulatory system.
A strong decrease in the putative nitrate/sulfonate ABC transporter system (ANA_20529–ANA_C20532) was identified in 4dLP and may imply that this regulation releases more space on the cell membranes for the use of phosphorus transport proteins. Interestingly, this ABC transporter was more than 2-fold upregulated in 6dNP. At this time point cells were growing fast and import of nutrients was highly induced. In turn, the growth rate in 4dLP was arrested, and the cells were even more strongly starved by phosphorus, which led them to reconstruct their cell envelopes, omitting a number of nonessential molecules and replacing these with phosphorus transporter and assimilation complexes.
Expression of gene clusters of nonribosomal peptides.
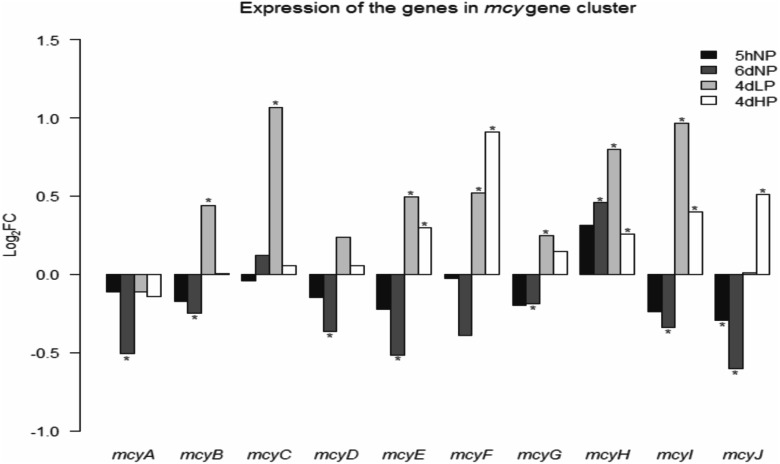
Nutrient limitation causes lower production of bioactive compounds among cyanobacteria (6–8), and thus, a closer examination of the expression of the genes encoding nonribosomal peptide synthetase (NRPS) peptides was carried out. Surprisingly, under low-phosphate conditions (4dLP), increased abundance of many transcripts belonging to the microcystin, anabaenopeptin, anabaenopeptilide, and hassallidin gene clusters were identified, but the expressions of these genes were strongly downregulated in 6dNP (see Tables S4 and S5 in the supplemental material). For instance, 7 out of 10 genes from the microcystin gene cluster were slightly or significantly upregulated in 4dLP, and only transcripts of mcyF and mcyJ were more abundant in 4dHP than in 4dLP (Fig. 3). The production pathway of NRPS is complicated, and our results gives a hint that regulation takes place at a level other than transcriptomics; thus, production of these compounds cannot be estimated by quantifying the mRNA of certain genes or gene clusters.
FIG 3.
Expression of the genes in the microcystin gene cluster. Most of the genes were upregulated under prolonged phosphorus stress, giving a hint that regulation of microcystin production happens on some level other than the transcriptomic level. The asterisks indicate significant changes in the transcription level compared with the control condition (P ≤ 0.05).
Central metabolism.
In addition to the specific acclimation processes, general stress responses can be observed in cyanobacteria under nutrient depletion (6, 31). Under nutrient stress, the cells reduce their energy demand by decreasing the activity of the central metabolism (glycolysis, pentose phosphate pathway, glycogenesis, and carbon fixation) and photosynthesis apparatus (5, 6, 47). The yellow-green coloration caused by the loss of photosynthesis pigments (bleaching or chlorosis) can clearly be seen within a few days, but changes on the molecular level are detectable even within a few hours (6, 47). Additionally, under nutrient deprivation, more prudent metabolic pathways (e.g., the pentose phosphate pathway) are favored by the induction of the key genes in a particular route (5, 48). It was also suggested that under nitrogen deprivation, the pentose phosphate pathway responds to power minimization in the cells, instead of reducing the activity of the photosynthetic apparatus (48).
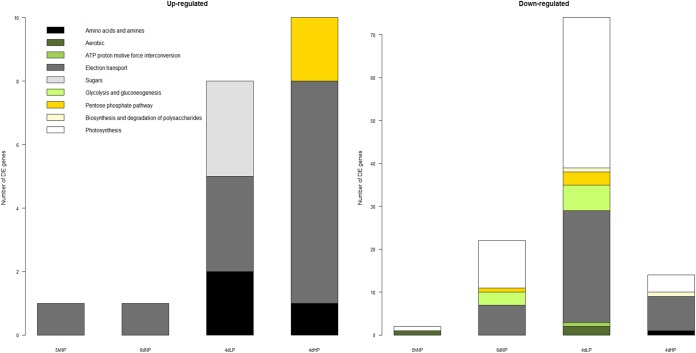
The photosynthetic apparatus of Anabaena 90 consists of photosystems I and II, cytochrome b6/f, electron transporters, F-type ATPases, and the phycobilisomes, the light harvesting complexes, containing allophycocyanins and phycocyanins (27). In the current study, the expression of genes involved in photosystems I and II as well as in the electron transport machinery and photopigment biosynthesis was reduced in 6dNP and even more strongly in 4dLP (Fig. 4). The reduced transcription and protein levels of the ribulose-bisphosphate carboxylase small and large subunits (ANA_C11377 and ANA_C11339), the key enzymes in the Calvin-Benson cycle, implement a strong reduction in carbon fixation activity. In contrast, the expression of genes participating in the pentose phosphate pathway was also arrested, which further supports lower carbohydrate production. In addition, many genes participating in glycolysis and gluconeogenesis were less transcribed, which is in line with several previously reported responses to various nutrient stressors (6). The most often described general responses in nutrient stress are a reduced expression of ribosomal proteins and a global reduction in translation and transcription (e.g., reference 6). Lower expression of the genes participating in these molecular functions was found in both 6dNP and 4dLP, and the results strongly denote arrested growth under these conditions (Fig. 4).
FIG 4.
Number of differentially expressed genes participating in central metabolism. The growth was arrested under prolonged phosphorus stress by downregulating the expression of the genes participating in central metabolism.
Expression patterns of hypothetical proteins.
Hypothetical proteins and proteins with unknown function are widely abundant among cyanobacterial genome annotations, representing the relatively incomplete knowledge of the metabolism and molecular structure of cyanobacteria (13, 27). In this study, we found 5 (5hNP), 11 (6dNP), and 19 (4dLP) proteins with unknown function with more than 4-fold transcriptional changes compared with the controls (log2FC >2 or <−2; P ≤ 0.05). The hypothetical protein ANA_C10735 was the third most upregulated gene in 4dLP, and a significant increase was already observed in 6dNP. The BLAST search revealed that this giant protein of 1896 amino-acid residues is a unique protein in GenBank, but it contains several domains recognized from other organisms, mainly filamentous cyanobacteria. The protein contains a phytase like domain that participates in Pi cleavage from organic phosphorus compounds called phytic acids and also, several Ca-Na exchangers, 6-fold beta propeller segments and one domain found in proteins participating in transport and uptake (Table 1). The other protein, significantly upregulated under phosphorus depletion, was the hypothetical protein (ANA_C11754). A protein domain search did not lead to the identification of conserved domains in ANA_C11754, but the expression pattern of this gene indicates that it constitutes most likely an operon structure with a highly induced Pi transporter (ANA_C11755–ANA_C11758) and is regulated by the pho regulon. The function and putative role of these two proteins under phosphorus stress need to be investigated further but it is difficult with the strain used since it has turned out to be recalcitrant for genetic manipulation.
The three most downregulated genes in 4dLP (ANA_C13795 to ANA_C13797) were annotated as corresponding to hypothetical proteins, while the fourth most downregulated gene, which was identified as the serralysin-like metalloprotease (ANA_C13798), is located next to ANA_C13797. The expression profile of these four genes suggests that they belong to the same operon. The putative operon was already strongly repressed in 5hNP and further in 6dNP. Among the three hypothetical proteins (corresponding to ANA_C13795 to ANA_C13797) mentioned above, no conserved domains were found and only those corresponding to ANA_C13795 and ANA_C13796 had homologues among other cyanobacteria. Due to the operon structure expected, we speculate that the three hypothetical proteins may be linked to a protease function encoded by ANA_C13798.
Differentially expressed intergenic regions under Pi-depleted conditions.
Intergenic regions may give rise to sRNAs, another type of important regulatory factors with an important role in physiological adaption and as a driver of interstrain divergence in bacteria, including cyanobacteria (33, 34, 49–51). Studies of the filamentous and nitrogen-fixing cyanobacterium Nodularia spumigena CCY9414 genome and its acclimation to stress conditions point further sRNAs crucial role in cell functioning, especially in gene expression regulation (13, 52). Due to their importance as regulatory factors, we searched intergenic regions differentially expressed under phosphorus deprivation conditions. We found 6 putative intergenic regions that were differentially expressed in the experiment, and most of them seemed not to be conserved among cyanobacteria (Table 2). In addition, we identified one sRNA that originated from an unannotated gene. This transcript actually turned out to be the RNA component of RNase P, and the gene should be called rnpB (Table 2). These examples clearly confirm that noncoding transcripts can be identified with this approach.
TABLE 2.
Putative differentially expressed nonencoding regions under phosphorus deprivation

Proteomic analysis.
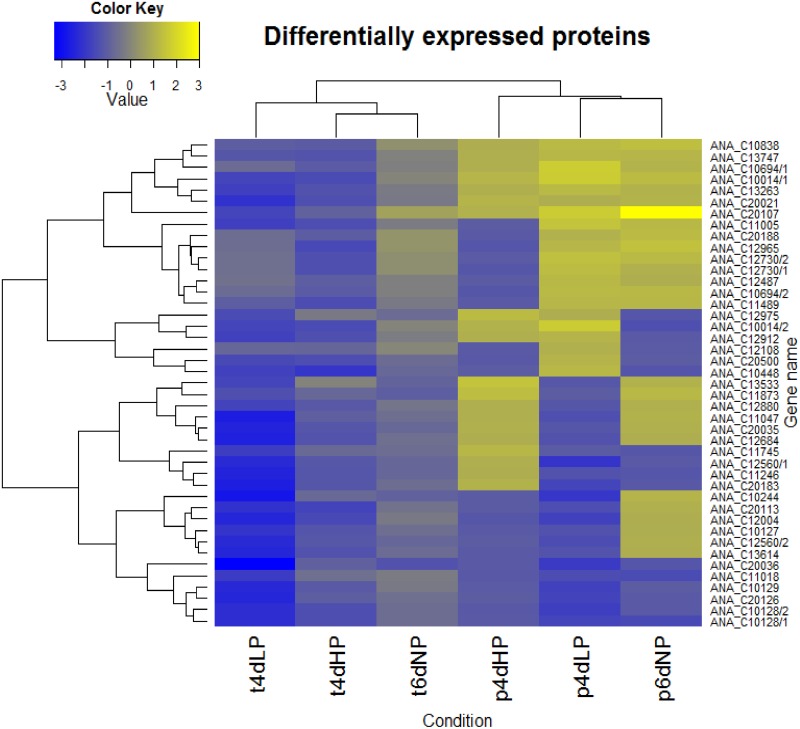
After transcriptome analysis, we performed a 2D-DIGE proteomic study to determine the response of Anabaena 90 to phosphorus limitation at the protein level. We detected differential regulation for 56 gel spots with a P value of ≤0.05 (Fig. 5) and further analyzed these with LC-MS/MS. A total of 43 proteins could be classified and analyzed further, of which five were identified in two different isoforms (see Table S7 in the supplemental material). Similar regulation of transcripts and proteins was only to some extent recognized (Pearson's correlation, 0.75), although 14 out of 38 genes were significantly downregulated at both the transcriptome and proteome levels in 4dLP. Since a weak correlation between the expression patterns of transcriptomes and proteomes is commonly known, this finding was not surprising.
FIG 5.
Significantly differentially expressed proteins (P ≤ 0.05, ANOVA) during the experiment. A heat map shows the relative abundance of 47 identified proteins and their isoforms compared to the transcriptomic results. Proteins related to transcription, translation, and photosynthesis can be seen strongly downregulated; in contrast, stress-related proteins are upregulated, representing general stress response of the bacteria. Pearson's correlation factor between transcriptomics and proteomics results was 0.75.
Eighteen identified proteins showed upregulation in 4dLP. The expressions of two different carbohydrate-selective porins (corresponding to ANA_C13263 and ANA_C12730), as well as that of the polysaccharide capsule biosynthesis protein CapD (ANA_C12487), were increased under phosphorus stress. Increased polysaccharide production was also on the basis of the slimier composition of the samples during RNA and protein extraction, as well as at the transcriptome level by increased transcription of several glycosyltransferase genes (see Tables S4 and S5). In general, many different stressors (e.g., salinity, light, and temperature) increase exopolysaccharide concentrations in cyanobacterial cells, and it has also suggested that unbalance in N/C/P ratio caused by nutrient limitation triggers this phenomenon to ensure the proper functioning of photosynthetic apparatus (53). Thus, increased amount of polysaccharides seems to be an important response to phosphorus starvation in heterocystous Anabaena sp. 90 cyanobacteria. A general response to starvation was indicated by an increased abundance of amino acid hydrolases (corresponding to ANA_C10014 and ANA_C20500), together with the outer membrane efflux protein (corresponding to ANA_C13747) and acetylornithine aminotransferase (corresponding to ANA_C11005). Moreover, we found three upregulated proteins of unknown function and one hypothetical protein whose role under phosphorus stress remains unclear. In contrast, the expression of 20 proteins was decreased under prolonged phosphorus starvation (4dLP), including 8 proteins related to protein synthesis and 7 related to central energy metabolism. The protein with the highest fold change in this experiment was ribulose-bisphosphate carboxylase small chain (relative fold change was −1.99 in 4dLP). This is well in line with the strong repression detected at the transcriptome level, giving further evidence for the strong downregulation of the Calvin-Benson cycle under phosphorus depletion. Other downregulated proteins identified in this study were related to amino acid biosynthesis (ANA_C11005 and ANA_C12965) and protochlorophyllide reductase (ANA_C12004). The findings were well consistent with transcriptome analysis and with previously published phosphorus acclimation proteomics studies of Prochlorococcus and Synechocystis sp. PCC 6803 (5, 30) and reflect the universal stress response of cyanobacteria.
Since the response to phosphorus stress is, at least to some extent, species specific (29), further studies are necessary to validate the suitability of the marker genes that can be reasonably used for environmental monitoring. The currently used method, following the activity of alkaline phosphatase or quantifying the transcript abundance of this particular gene by quantitative PCR (qPCR) alone, may not be sufficient as markers to monitor the status of Pi in cyanobacterial blooms. Based on this and on previously published studies (6, 44), a wider range of genes from the pho regulon should be used as markers to improve the adequacy of the previously used method. In this study, we also identified putative genes that are differentially expressed under phosphorus depletion, but their role in the phosphorus response among cyanobacteria is still unclear. Hence, we suggest that the role and function of these genes need to be resolved and their suitability for monitoring purposes needs to be evaluated. In addition, a time-consuming and costly proteomic approach is poorly suitable for monitoring the environmental Pi status. However, the method provides valuable clues toward a basic understanding of organism behavior under various environmental conditions, which we still have limited knowledge about concerning cyanobacteria.
Supplementary Material
ACKNOWLEDGMENTS
We thank the personnel from DNA Sequencing and Genomic Laboratory, Institute of Biotechnology, University of Helsinki, Finland, for cDNA library preparation and SOLiD RNA sequencing; Hao Wang for help in data submission; and Lyudmila Saari for maintenance of the organism.
This work was supported by the BLUEPRINT project, funded by the European Community's Seventh Framework Programme (FP/2007-2013) under implementation agreement no. R&I/I3/2012/BONUS made with BONUS, the joint Baltic Sea Research and Development Programme, and from the Academy of Finland 1273652 to K.S., the Academy of Finland Research Center of Excellence grant 118637 to K.S. and E.-M.A., and by EU project MaCuMBA (Marine Microorganisms: Cultivation Methods for Improving their Biotechnological Applications; grant agreement number 311975) to W.R.H.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01062-15.
REFERENCES
- 1.Paerl HW, Paul VJ. 2012. Climate change: links to global expansion of harmful cyanobacteria. Water Res 46:1349–1363. doi: 10.1016/j.watres.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 2.Conley DJ, Paerl HW, Howarth RW, Boesch DF, Seitzinger SP, Havens KE, Lancelot C, Likens GE. 2009. Ecology. Controlling eutrophication: nitrogen and phosphorus. Science 323:1014–1015. [DOI] [PubMed] [Google Scholar]
- 3.Paerl HW, Huisman J. 2008. Blooms like it hot. Science 320:57–58. doi: 10.1126/science.1155398. [DOI] [PubMed] [Google Scholar]
- 4.O'Neil JM, Davis TW, Burford MA, Gobler CJ. 2012. The rise of harmful cyanobacteria blooms: the potential roles of eutrophication and climate change. Harmful Algae 14:313–334. doi: 10.1016/j.hal.2011.10.027. [DOI] [Google Scholar]
- 5.Fuszard MA, Ow SY, Gan CS, Noirel J, Ternan NG, McMullan G, Biggs CA, Reardon KF, Wright PC. 2013. The quantitative proteomic response of Synechocystis sp. PCC6803 to phosphate acclimation. Aquat Biosyst 9:5. doi: 10.1186/2046-9063-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harke MJ, Gobler CJ. 2013. Global transcriptional responses of the toxic cyanobacterium, Microcystis aeruginosa, to nitrogen stress, phosphorus stress, and growth on organic matter. PLoS One 8:e69834. doi: 10.1371/journal.pone.0069834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Repka S, Koivula M, Harjunpää V, Rouhiainen L, Sivonen K. 2004. Effects of phosphate and light on growth of and bioactive peptide production by the cyanobacterium Anabaena strain 90 and its anabaenopeptilide mutant. Appl Environ Microbiol 70:4551–4560. doi: 10.1128/AEM.70.8.4551-4560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rapala J, Sivonen K, Lyra C, Niemelä SI. 1997. Variation of microcystins, cyanobacterial hepatotoxins, in Anabaena spp. as a function of growth stimuli. Appl Environ Microbiol 63:2206–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halinen K, Jokela J, Fewer DP, Wahlsten M, Sivonen K. 2007. Direct evidence for production of microcystins by Anabaena strains from the Baltic Sea. Appl Environ Microbiol 73:6543–6550. doi: 10.1128/AEM.01377-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sivonen K, Jones G. 1999. Cyanobacterial toxins, p 41–111. In Chorus I, Bartram J (ed), Toxic cyanobacteria in water. E & FN Spon, London, United Kingdom. [Google Scholar]
- 11.Paerl HW, Fulton RS, Moisander PH, Dyble J. 2001. Harmful freshwater algal blooms, with an emphasis on cyanobacteria. ScientificWorldJournal 1:76–113. doi: 10.1100/tsw.2001.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peterson CN, Mandel MJ, Silhavy TJ. 2005. Escherichia coli starvation diets: essential nutrients weigh in distinctly. J Bacteriol 187:7549–7553. doi: 10.1128/JB.187.22.7549-7553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voss B, Bolhuis H, Fewer DP, Kopf M, Möke F, Haas F, El-Shehawy R, Hayes P, Bergman B, Sivonen K, Dittmann E, Scanlan DJ, Hagemann M, Stal LJ, Hess WR. 2013. Insights into the physiology and ecology of the brackish-water-adapted cyanobacterium Nodularia spumigena CCY9414 based on a genome-transcriptome analysis. PLoS One 8:e60224. doi: 10.1371/journal.pone.0060224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dyhrman ST, Haley ST. 2006. Phosphorus scavenging in the unicellular marine diazotroph Crocosphaera watsonii. Appl Environ Microbiol 72:1452–1458. doi: 10.1128/AEM.72.2.1452-1458.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez A, Tyson GW, Delong EF. 2010. Widespread known and novel phosphonate utilization pathways in marine bacteria revealed by functional screening and metagenomic analyses. Environ Microbiol 12:222–238. doi: 10.1111/j.1462-2920.2009.02062.x. [DOI] [PubMed] [Google Scholar]
- 16.Su Z, Olman V, Xu Y. 2007. Computational prediction of Pho regulons in cyanobacteria. BMC Genomics 8:156. doi: 10.1186/1471-2164-8-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR, Post AF, Hagemann M, Paulsen I, Partensky F. 2009. Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev 73:249–299. doi: 10.1128/MMBR.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sebastian M, Ammerman JW. 2009. The alkaline phosphatase PhoX is more widely distributed in marine bacteria than the classical PhoA. ISME J 3:563–572. doi: 10.1038/ismej.2009.10. [DOI] [PubMed] [Google Scholar]
- 19.Adams MM, Gómez-García MR, Grossman AR, Bhaya D. 2008. Phosphorus deprivation responses and phosphonate utilization in a thermophilic Synechococcus sp. from microbial mats. J Bacteriol 190:8171–8184. doi: 10.1128/JB.01011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sivonen K, Namikoshi M, Evans WR, Carmichael WW, Sun F, Rouhiainen L, Luukkainen R, Rinehart KL. 1992. Isolation and characterization of a variety of microcystins from seven strains of the cyanobacterial genus Anabaena. Appl Environ Microbiol 58:2495–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rouhiainen L, Paulin L, Suomalainen S, Hyytiäinen H, Buikema W, Haselkorn R, Sivonen K. 2000. Genes encoding synthetases of cyclic depsipeptides, anabaenopeptilides, in Anabaena strain 90. Mol Microbiol 37:156–167. [DOI] [PubMed] [Google Scholar]
- 22.Sivonen K, Leikoski N, Fewer DP, Jokela J. 2010. Cyanobactins-ribosomal cyclic peptides produced by cyanobacteria. Appl Microbiol Biotechnol 86:1213–1225. doi: 10.1007/s00253-010-2482-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujii K, Harada K-I, Suzuki M, Kondo F, Ikai Y, Oka H, Carmichael WW, Sivonen K. 1994. Occurrence of novel cyclic peptides togehter with microcystins from toxic cyanobacteria, Anabaena species, p 559–562. In Yasumoto T, Oshima Y, Fukuyo Y (ed), Harmful and toxic algal blooms. Intergovernmental Oceanographic Commission of UNESCO, Paris, France. [Google Scholar]
- 24.Rouhiainen L, Vakkilainen T, Siemer BL, Buikema W, Haselkorn R, Sivonen K. 2004. Genes coding for hepatotoxic heptapeptides (microcystins) in the cyanobacterium Anabaena strain 90. Appl Environ Microbiol 70:686–692. doi: 10.1128/AEM.70.2.686-692.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rouhiainen L, Jokela J, Fewer DP, Urmann M, Sivonen K. 2010. Two alternative starter modules for the non-ribosomal biosynthesis of specific anabaenopeptin variants in Anabaena (Cyanobacteria). Chem Biol 17:265–273. doi: 10.1016/j.chembiol.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 26.Vestola J, Shishido TK, Jokela J, Fewer DP, Aitio O, Permi P, Wahlsten M, Wang H, Rouhiainen L, Sivonen K. 2014. Hassallidins, antifungal glycolipopeptides, are widespread among cyanobacteria and are the end-product of a nonribosomal pathway. Proc Natl Acad Sci U S A 111:E1909–E1917. doi: 10.1073/pnas.1320913111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H, Sivonen K, Rouhiainen L, Fewer DP, Lyra C, Rantala-Ylinen, Vestola J, Jokela J, Rantasärkkä K, Li Z, Liu B. 2012. Genome-derived insights into the biology of the hepatotoxic bloom-forming cyanobacterium Anabaena sp. strain 90. BMC Genomics 13:613. doi: 10.1186/1471-2164-13-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki S, Ferjani A, Suzuki I, Murata N. 2004. The SphS-SphR two component system is the exclusive sensor for the induction of gene expression in response to phosphate limitation in Synechocystis. J Biol Chem 279:13234–13240. doi: 10.1074/jbc.M313358200. [DOI] [PubMed] [Google Scholar]
- 29.Martiny AC, Coleman ML, Chisholm SW. 2006. Phosphate acquisition genes in Prochlorococcus ecotypes: evidence for genome-wide adaptation. Proc Natl Acad Sci U S A 103:12552–12557. doi: 10.1073/pnas.0601301103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fuszard MA, Wright PC, Biggs CA. 2012. Comparative quantitative proteomics of Prochlorococcus ecotypes to a decrease in environmental phosphate concentrations. Aquat Biosyst 8:7. doi: 10.1186/2046-9063-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ludwig M, Bryant D. 2012. Acclimation of the global transcriptome of the cyanobacterium Synechococcus sp. strain PCC 7002 to nutrient limitations and different nitrogen sources. Front Microbiol 3:145. doi: 10.3389/fmicb.2012.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tetu SG, Brahamsha B, Johnson DA, Tai V, Phillippy K, Palenik B, Paulsen IT. 2009. Microarray analysis of phosphate regulation in the marine cyanobacterium Synechococcus sp. WH8102. ISME J 3:835–849. doi: 10.1038/ismej.2009.31. [DOI] [PubMed] [Google Scholar]
- 33.Kopf M, Klähn S, Scholz I, Matthiessen JKF, Hess R. 2014. Comparative analysis of the primary transcriptome of Synechocystis sp. PCC 6803. DNA Res 1–13. doi: 10.1093/dnares/dsu018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kopf M, Klähn S, Scholz I, Hess WR, Voss B. 2015. Variations in the non-coding transcriptome as a driver of inter-strain divergence and physiological adaptation in bacteria. Sci Rep 5:9560. doi: 10.1038/srep09560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reistetter EN, Krumhardt K, Callnan K, Roache-Johnson K, Saunders JK, Moore LR, Rocap G. 2013. Effects of phosphorus starvation versus limitation on the marine cyanobacterium Prochlorococcus MED4 II: gene expression. Environ Microbiol 15:2129–2143. doi: 10.1111/1462-2920.12129. [DOI] [PubMed] [Google Scholar]
- 36.Kótai J. 1972. Instructions for preparation of modified nutrient solution Z8 for algae. NIVA B–11/69. NIVA 1976. Estimation of Algal Growth Potential; Norwegian Institute for Water Research: Oslo, Norway. [Google Scholar]
- 37.Hoffmann S, Otto C, Kurtz S, Sharma CM, Khaitovich P, Vogel J, Stadler PF, Hackermüller J. 2009. Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLoS Comput Biol 5:e1000502. doi: 10.1371/journal.pcbi.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Love MI, Huber W, Anders S. 19 February 2014. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. bioRxiv. http://dx.doi.org/10.1101/002832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. 2007. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 41.Shevchenko A, Wilm M, Vorm O, Mann M. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 42.Grouneva I, Rokka A, Aro E-M. 2011. The thylakoid membrane proteome of two marine diatoms outlines both diatom-specific and species-specific features of the photosynthetic machinery. J Proteome Res 10:5338–5353. doi: 10.1021/pr200600f. [DOI] [PubMed] [Google Scholar]
- 43.Mur LR, Skulberg OM, Utkilen H. 1999. Cyanobacteria in environment, p 15–40. In Chorus I, Bartram J (ed), Toxic cyanobacteria in water. E & FN Spon, London, United Kingdom. [Google Scholar]
- 44.Orchard ED, Webb EA, Dyhrman ST. 2009. Molecular analysis of the phosphorus starvation response in Trichodesmium spp. Environ Microbiol 11:2400–2411. doi: 10.1111/j.1462-2920.2009.01968.x. [DOI] [PubMed] [Google Scholar]
- 45.Dyhrman ST, Chappell PD, Haley ST, Moffett JW, Orchard ED, Waterbury JB, Webb EA. 2006. Phosphonate utilization by the globally important marine diazotroph Trichodesmium. Nature 439:68–71. doi: 10.1038/nature04203. [DOI] [PubMed] [Google Scholar]
- 46.White AK, Metcalf WW. 2004. Two C-P lyase operons in Pseudomonas stutzeri and their roles in the oxidation of phosphonates, phosphite, and hypophosphite. J Bacteriol 186:4730–4739. doi: 10.1128/JB.186.14.4730-4739.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Collier JL, Grossman AR. 1992. Chlorosis induced by nutrient deprivation in Synechococcus sp. strain PCC 7942: not all bleaching is the same. J Bacteriol 174:4718–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Osanai T, Imamura S, Asayama M, Shirai M, Suzuki I, Murata N, Tanaka K. 2006. Nitrogen induction of sugar catabolic gene expression in Synechocystis sp. PCC 6803. DNA Res 13:185–195. doi: 10.1093/dnares/dsl010. [DOI] [PubMed] [Google Scholar]
- 49.Axmann IM, Kensche P, Vogel J, Kohl S, Herzel H, Hess WR. 2005. Identification of cyanobacterial non-coding RNAs by comparative genome analysis. Genome Biol 6:R73. doi: 10.1186/gb-2005-6-9-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steglich C, Futschik ME, Lindell D, Voss B, Chisholm SW, Hess WR. 2008. The challenge of regulation in a minimal photoautotroph: non-coding RNAs in Prochlorococcus. PLoS Genet 4:e1000173. doi: 10.1371/journal.pgen.1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kopf M, Hess WR. 2015. Regulatory RNAs in photosynthetic cyanobacteria. FEMS Microbiol Rev 39:301–315. doi: 10.1093/femsre/fuv017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kopf M, Möke F, Bauwe H, Hess WR, Hagemann M. 2015. Expression profiling of the bloom-forming cyanobacterium Nodularia CCY9414 under light and oxidative stress conditions. ISME J 9:1–14. doi: 10.1038/ismej.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pereira S, Zille A, Micheletti E, Moradas-Ferreira P, De Philippis R, Tamagnini P. 2009. Complexity of cyanobacterial exopolysaccharides: composition, structures, inducing factors and putative genes involved in their biosynthesis and assembly. FEMS Microbiol Rev 33:917–941. doi: 10.1111/j.1574-6976.2009.00183.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.