Abstract
Coliphage N4 is a lytic bacteriophage discovered nearly half a century ago, and it was considered to be a “genetic orphan” until very recently, when several additional N4-like phages were discovered to infect nonenteric bacterial hosts. Interest in this genus of phages is stimulated by their unique genetic features and propagation strategies. To better understand the ecology of N4-like phages, we investigated the diversity and geographic patterns of N4-like phages by examining 56 Chesapeake Bay viral communities, using a PCR-clone library approach targeting a diagnostic N4-like DNA polymerase gene. Many new lineages of N4-like phages were found in the bay, and their genotypes shift from the lower to the upper bay. Interestingly, signature sequences of N4-like phages were recovered only from winter month samples, when water temperatures were below 4°C. An analysis of existing metagenomic libraries from various aquatic environments supports the hypothesis that N4-like phages are most prolific in colder waters. In particular, a high number of N4-like phages were detected in Organic Lake, Antarctica, a cold and hypersaline system. The prevalence of N4-like phages in the cold biosphere suggests these viruses possess yet-to-be-determined mechanisms that facilitate lytic infections under cold conditions.
INTRODUCTION
As the most abundant microbial form, viruses play important roles in shaping host population structures, mediating genetic exchange between hosts, and modulating trophic transfer in marine food webs (1–3). Marine viral metagenomic studies suggest that viruses encompass the largest genetic repertoire in the ocean (4, 5), yet the functions of ∼70% of the putative genes identified in viral metagenomic studies remain unknown (6). Recently, several novel phages infecting dominant marine bacteria have been isolated from the ocean (7, 8). Both cultivation techniques and molecular approaches suggest that a great many marine viruses await discovery.
Bacteriophage N4 was first isolated from sewer waters using Escherichia coli as a host nearly half a century ago (9) and remained a “genetic orphan” for decades, as no other genetically similar phages were characterized. Phage N4 has several unique features with regard to its morphology, physiology, and genome that have made it a focus of study. It has a 70-nm isocahedral capsid, and its genome size is 70 kb. More remarkably, N4 is the only known phage that does not rely on host RNA polymerase for early transcription (10, 11). Instead, it contains a DNA-dependent virion-encapsidated RNA polymerase (vRNAP), which is coinjected with viral DNA into its host and initiates transcription (12). The phage also exhibits a lysis-inhibited infection cycle and an extremely large burst size (ca. 3,000 phages per cell), suggestive of a novel mechanism of cell lysis regulation (13, 14).
The recent renewed interest in the ecology of N4 was prompted by the isolation from a coastal estuary of two new N4-like phages, which infect bacteria of the marine Roseobacter lineage (15). Roseobacters are a widely distributed and abundant group of marine bacteria (16). Thus, the finding of Roseobacter N4-like phages demonstrated that the phage genus is not restricted to E. coli or other enteric bacteria and suggested that the marine environment might be an important reservoir for this group of viruses. Subsequent studies have described the isolation and characterization of additional N4-like phages that infect a variety of bacterial species, including Vibrio, Pseudomonas, Salmonella, and Achromobacter spp. (17–23). With the exception of Enterobacter phage EcP1 (NC_019485), which has a relatively small capsid and genome, all N4-like phages characterized to date share morphologies, genome sizes, and genomic architectures similar to those of coliphage N4 (24). Given the recent successes in isolating N4-like phages from phylogenetically diverse bacterial taxa, we were motivated to better understand the distribution of this phage group in natural systems. The conservative nature of N4-like genomes allowed the design of molecular tools diagnostic for group members, which facilitated the isolation-independent assessments of the distribution and diversity of N4-like phages over space and time presented here.
MATERIALS AND METHODS
Sample collection and preparation.
Chesapeake Bay viral community (VC) samples were collected in 2004 and 2005 from multiple stations in the bay during research cruises for the Microbial Observatories Viral Ecology project. Viral DNA was prepared following methods described previously (25). Briefly, 50 liters of water was filtered through glass fiber filters (type A/E) and then 0.45-μm-pore-size polycarbonate filters. The viral fraction was then concentrated to ∼500 ml by ultrafiltration through a 30-kDa-cutoff filter cartridge. These concentrated VCs were further precipitated with polyethylene glycol 8000 powder at a final concentration of 100 g liter−1 and then resuspended in SM buffer (100 mM sodium chloride, 10 mM magnesium sulfate [heptahydrate], 50 mM Tris-HCl, pH 7.5). The concentrated VCs were boiled to release viral DNA, which was used as a DNA template for PCR.
A total of 56 archived viral DNA samples were analyzed for the presence of N4 phages. These samples were collected from 9 different stations across the whole bay during 2005 and from a subset of 5 of the stations during 2004 (Fig. 1). Samples were collected in four seasons (winter [February], spring [May], summer [August], and fall [October]).
FIG 1.
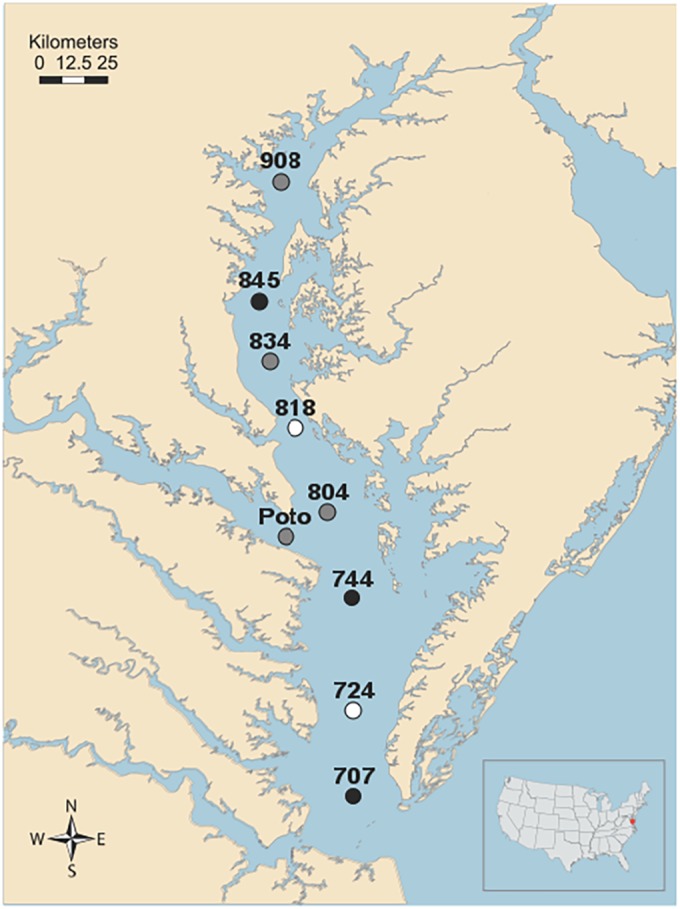
Collection locations of the winter Chesapeake Bay samples that were positive for N4-like specific DNA pol PCR products. The positive samples from the winters of both 2005 and 2004 are represented by black circles. The white circles represent the positive samples from only 1 year. The samples from stations 908, 834, 804, and Poto, which stands for Potomac River (gray circles), were available only in 2005 and not in 2004. The figure was modified from an image by Tracy Saxby and Kate Boicourt, Integration and Application Network, University of Maryland Center for Environmental Science (http://ian.umces.edu/imagelibrary/).
PCR primers and amplification.
A set of degenerate primers were designed based on available DNA polymerase (pol) genes of N4-like phages in the NCBI database through 2011, including Enterobacteria phage N4 (NC_008720), roseophage DSS3 Φ2 (NC_012697), roseophage EE36 Φ1 (NC_012696), Roseovarius Plymouth podovirus 1 (FR719956), Roseovarius sp. strain 217 phage 1 (FR682616), Enterobacter phage EcP1 (NC_019485), Pseudomonas phage LIT1 (NC_013692), Pseudomonas phage LUZ7 (NC_013691), and Sulfitobacter phage pCB2047-B (HQ317387). The forward primer was N4-DNAP-F (5′-GGI ACI ATI ACI TTY TGY TGG-3′), and the reverse primer was N4-DNAP-R (5′-RTA RTT ICC IGC RAA TYC YTG-3′). The expected PCR product size was ca. 400 bp. The PCR conditions were optimized for the template DNA and the primer concentrations, annealing temperature, magnesium chloride concentration, and cycle number.
All the PCRs were performed in 25-μl volumes containing 1× reaction buffer, 2 mM MgCl2, 200 μM deoxynucleoside triphosphate (dNTP), 2 μM each primer, 1 U of Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA), and 4 μl DNA templates. The PCR was performed with the following steps: denaturation for 10 min at 94°C and 35 cycles of denaturation (30 s at 94°C), annealing (1 min at 50°C), and extension (1 min at 72°C), with a final extension for 10 min at 72°C. The DNA of roseophage DSS3 Φ2 was used as a positive control, and a series of dilutions of the positive-control DNA were employed to determine the threshold of detection.
Clone library, sequencing, and phylogenetic analysis.
Five winter PCR amplicons, two from the upper bay (84504 and 84505), one from the middle bay (83405), and two from the lower bay (70704 and 70705), were selected to construct clone libraries (Table 1). The PCR products were purified with the QIAquick gel extraction kit (Qiagen, Venlo, The Netherlands) and cloned using the TOPO TA pcr4.0 cloning kit (Invitrogen, Carlsbad, CA, USA). Colonies were randomly picked from each clone library and sequenced using an ABI 3100 genetic analyzer (PE Applied Biosystems, Foster City, CA, USA). Homology searches (BLASTx) were performed against the NCBI database. If the top hit of the sequences was an annotated, known N4-like phage from amino acid position 206 to 317 in the N4 phage DNA polymerase (NC_008720), we identified it as an N4-like phage sequence.
TABLE 1.
Environmental and community parameters at sampling stations
| Station | Time | Temp (°C) | Salinity (psu) | Bacterial abundance (105 cells/ml) | Viral abundance (105 VLPsa/ml) | NO2− and NO3− concn (μM) |
|---|---|---|---|---|---|---|
| 707 | February 2004 | 3.8 | 15.4 | 8.61 | 9.6 | 12.3 |
| 707 | February 2005 | 2.9 | 18 | 12.3 | 87.1 | 9.03 |
| 845 | February 2004 | 1.2 | 9.6 | 10.4 | 31.2 | 15.9 |
| 845 | February 2005 | 3.9 | 7.5 | 12.3 | 100 | 32.7 |
| 834 | February 2005 | 3.2 | 7.4 | 9.82 | 46.6 | 40.8 |
VLPs, virus-like particles.
DNA pol partial gene sequences were translated into putative protein sequences and aligned with Clustal W (26) using default parameters. A neighbor-joining (NJ) phylogenetic tree was built using MEGA 5.0 (39) and the p-distance model. Bootstrap values were determined from 1,000 resampling events.
Diversity indices and metagenomic analysis.
The operational taxonomic units (OTU), Chao index, evenness, and Shannon diversity index of each library were measured using the mothur suite of programs (27) and were based on DNA sequences at 2% divergence. Selected translated amino acid sequences from each subcluster were searched against the CAMERA portal (28) by tBLASTn with an E value threshold of ≤1E−10 and an alignment length of ≥80 amino acids. Three ocean metagenomic databases were chosen for N4-like phage recruitment, the Global Ocean Survey (GOS) microbial metagenomic database (29, 30), the Broad Phage metagenomic database (http://www.broadinstitute.org/annotation/viral/Phage/Home.html), and the Pacific Ocean Virome database (5). We also recruited N4-like phages from all CAMERA portal metagenomic libraries available by the end of 2013, including the Antarctica Aquatic Microbial Metagenome, Chesapeake Bay Virioplankton Metagenome, Chicken Cecum Microbiome, and Wastewater Metagenome from Mallard Creek (all available at http://data.imicrobe.us/project/view/104) and the Human Microbiome Project (http://hmpdacc.org/catalog/). The sequences that were related to N4-like phages and fully overlapped our clone sequences were included in a phylogenetic analysis.
Nucleotide sequence accession numbers.
All sequences have been deposited in GenBank with accession numbers KM527952 to KM528122.
RESULTS AND DISCUSSION
Detection of N4-like phages in the Chesapeake Bay.
Given that the first two marine N4-like phages were isolated from Chesapeake Bay waters, we sought to determine the prevalence and diversity of these phages over space and time in this coastal estuary. To do this, we screened a library of viral concentrates collected from the Chesapeake Bay over the course of 2 years (2004 and 2005) through the Microbial Observatory for Virioplankton Ecology Project (http://www.virusecology.org/MOVE/Home.html) using a PCR primer set targeting conserved regions of the DNA pol genes of N4-like phages. The DNA pol gene is one of the 14 conserved core genes of N4-like phages (24), making it a valuable diagnostic marker for culture-independent surveys. A total of 56 viral samples, which cover four seasons and the full transect of the Bay, were tested. Twelve of 14 winter samples yielded a PCR product of the expected size, but none of the viral samples from other seasons (spring, summer, and fall) yielded positive results. These products were sequenced and confirmed to be N4-like DNA pol gene homologs.
The 12 PCR-positive samples were collected during the winter in two consecutive years and represent samples spanning the breadth of the Chesapeake Bay watershed (Fig. 1) when water temperatures were below 4°C. It is surprising that none of the viral samples from spring, summer, and fall were PCR positive. Although no quantitative measurements have been performed to compare the abundance of N4-like phages in the winter and other seasons, the endpoint PCR results obtained here imply that N4-like phages in other seasons are either absent or below the current detection limit. It is unlikely that negative PCR results that occurred in other seasons were due to poor quality of the same viral DNA samples, because control reactions targeting DNA pol of cyanobacterial podoviruses (31) yielded positive results.
The Chesapeake Bay is a dynamic ecosystem. It has been reported that bacterial communities in the bay exhibit distinct seasonal patterns (32, 33). N4-like phages are generally thought to have a narrow host range (18). Indeed, the shift in bacterial populations and different bacterial metabolisms from cold to warm may be driving the distinct occurrence of N4-like phages observed in this study. However, an important caveat is that we cannot exclude the possibility that N4-like phages are present during other (spring, summer, and fall) seasons. It is possible that the concentration of N4-like phages in the Chesapeake Bay during the warmer seasons is below the limit of detection of the PCR assay, which was determined to be approximately 100 viruses per ml (data not shown) using DNA isolated from the positive-control roseophage DSS3 Φ2. Furthermore, not all N4-like phages have been isolated from cold environments (34).
N4 phages are diverse in the Chesapeake Bay.
Five of 12 positive amplicons from the Chesapeake Bay samples were selected for clone library analysis. The selected samples represent viral communities from the upper (station 845), middle (station 834), and lower (station 707) bay during the winter of 2005. Two additional samples collected from the upper (station 845) and lower (station 707) bay from the winter of 2004 were chosen to assess annual variation. All five samples were collected when the water temperature was between 1 and 4°C and represent a wide range of salinity and nutrient gradients (Table 1).
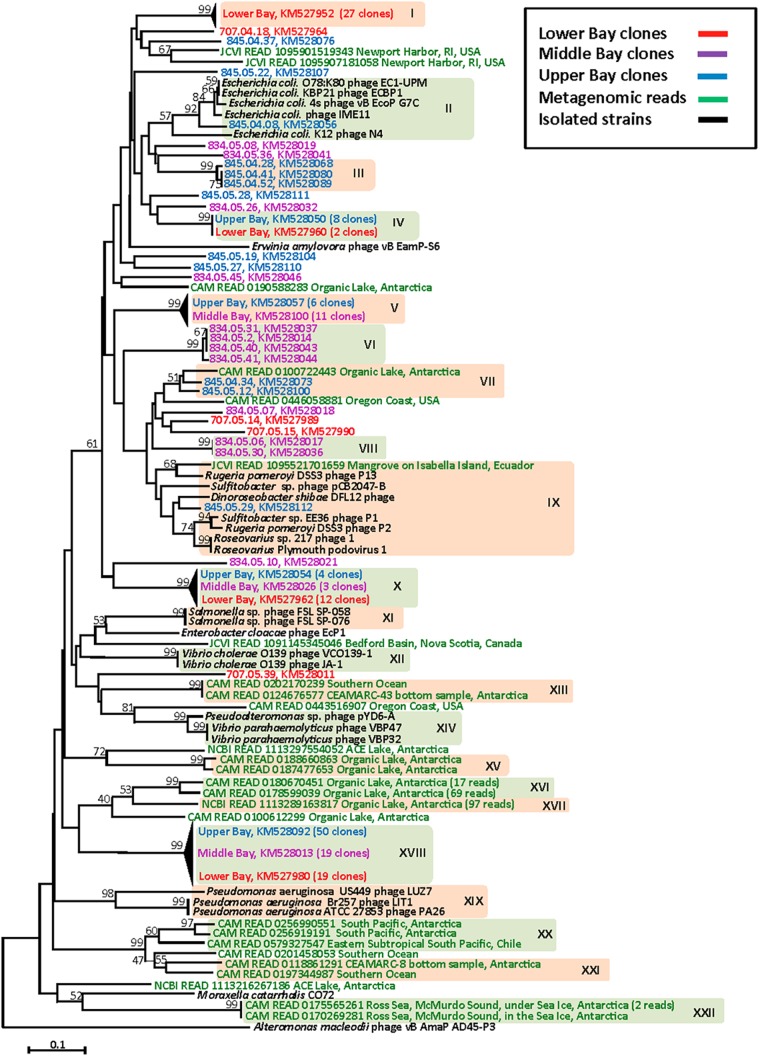
A total of 204 clones randomly selected from these five clone libraries were sequenced. Among them, 24 clones contained nontranslatable or nonspecific sequences and were not included in subsequent analyses. The DNA pol phylogeny based on the phage isolates and clonal sequences divided N4-like phages into at least 15 distinct clusters with an 80% amino acid sequence identify cutoff for each cluster (Fig. 2).
FIG 2.
Neighbor-joining phylogenetic relationships of translated DNA pol gene sequences recovered from the clone libraries, known N4-like phages, and metagenomic databases. The tree is based on the aligned amino acid sequences (ca. 130 residues). The sequences of 24 N4-like phages and 2 outgroup sequences are shown in black. The sequences recovered from metagenomic searches and upper, middle, and lower Chesapeake Bay are in green, red, purple, and blue, respectively. The clusters were defined as sequences that share greater than 80% amino acid identity. The bootstrap value was calculated with 1,000 replications. Bootstrap values of ≥50% are shown at the nodes. The scale bar represents 0.1 amino acid substitution.
For characterized N4-like phages, the N4-like DNA pol phylogeny indicates general agreement with the host phylogeny. For example, all five N4-like phages isolated on E. coli hosts fall into cluster II. Similarly, all N4-like phages isolated from Roseobacter hosts fall into cluster IX. N4-like phages that infect Vibrio spp., Salmonella spp., and Pseudomonas spp. also cluster with the host species. Overall, the phylogeny of N4-like phage genotypes based on the DNA pol gene is concordant with that previously reported based on concatenated core genes of N4-like phages (24). The congruency between the DNA pol phylogeny and the host taxonomy suggests N4 phages originated from a common ancestor and coevolved with their hosts.
Of the 180 Chesapeake Bay-derived N4 DNA pol sequences, the majority fall into 11 clusters (clusters I to X and XVIII) (Fig. 2). Among these 11 clusters, 8 clusters do not contain any cultured representatives. Indeed, the vast majority of bay clones do not cluster with N4-like DNA pol sequences that were obtained from cultured isolates. One environmental clone (845.04.08) does fall into cluster II, which contains several N4-like phages infecting various E. coli strains, and another clone (845.05.29) is closely associated with N4-like phages that infect marine Roseobacter strains. N4-like phages that infect Vibrio, Salmonella, and Pseudomonas spp. belong to clusters XI, XII, XIV, and XIX; no Chesapeake Bay clones mapped to these clusters.
The compositions of N4-like phages differ between the lower and upper bay. Several clusters (I, III, VI, VII, and VIII) contain only clonal sequences from either the upper or lower bay, indicating that unique genotypes of N4-like phages occupy specific niches in the Chesapeake Bay. Meanwhile, some clusters (X and XVIII) have been detected in both upper- and lower-bay samples (Table 2 and Fig. 3). In short, Chesapeake Bay viral communities contain diverse N4-like phage sequences in winter, and different N4-like phage genotypes were found in the upper and lower bay.
TABLE 2.
Distribution of phylogenetic clusters and diversity indices among the five clone libraries
| Library | No. of clones in clustera |
No. of unclustered clones | Total no. of clones | No. of OTUb | Normalized no. of OTUc | Richness (Chao index) | Evenness | Shannon diversity index | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | VI | VII | VIII | IX | X | XVIII | ||||||||
| 70704 | 18 | 2 | 4 | 4 | 1 | 29 | 5 | 0.17 | 5.00 | 0.69 | 1.11 | |||||||
| 70705 | 9 | 8 | 15 | 3 | 35 | 8 | 0.23 | 14.00 | 0.75 | 1.56 | ||||||||
| 84504 | 1 | 3 | 6 | 4 | 1 | 3 | 25 | 1 | 44 | 11 | 0.25 | 13.50 | 0.69 | 1.66 | ||||
| 84505 | 3 | 2 | 1 | 1 | 1 | 25 | 3 | 36 | 11 | 0.31 | 56.00 | 0.52 | 1.25 | |||||
| 83405 | 1 | 11 | 4 | 2 | 3 | 10 | 5 | 36 | 19 | 0.53 | 32.20 | 0.88 | 2.60 | |||||
The clusters are defined in Fig. 2.
The number of OTU was calculated based on DNA sequences at 2% divergence.
The number of OTU was normalized to the total number of clones from a given station.
FIG 3.
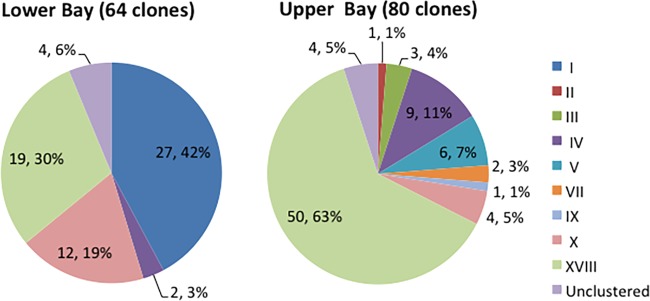
Comparison of proportions of DNA pol subclusters between the upper and lower Chesapeake Bay.
We believe that the number of environmental clones used in this study is sufficient to uncover the diversity of major populations of N4-like phages, as most clone libraries were well sampled (see Fig. S1 in the supplemental material). High-throughput sequencing could potentially yield more clusters for minor N4-like phage populations, but that is beyond the scope of this work. The presence of 11 distinct clusters of N4-like phages in the Chesapeake Bay alone suggests that N4-like phages can infect a much broader bacterial community than originally thought.
Identification of N4-like phages in cold environments.
The unexpected and apparent restriction of Chesapeake Bay N4-like phages to colder waters prompted a more global survey of their distribution and diversity. To that end, we surveyed available metagenomic libraries for N4-like DNA polymerase homologs. Representative DNA pol amino acid sequences from all subclusters were searched against the database, with an E value threshold of ≤1E−10 and an alignment length of ≥80 amino acids. The 206 homologous sequences identified were included in a DNA pol phylogenetic tree (Fig. 2). These environmental sequences form 7 unique clusters (XIII, XV, XVI, XVII, XX, XXI, and XXII), and the majority do not overlap Chesapeake Bay clones or isolated N4-like phages, suggesting that a great diversity of N4-like phages exist in other freshwater and marine environments.
Consistent with our PCR-based survey of seasonally representative Chesapeake Bay samples, nearly all (97%) of the metagenome-derived DNA pol sequences were recovered from colder environments, including the Southern Ocean and sea ice (Ross Sea). Indeed, most (91%) of the metagenomic reads were retrieved from Organic Lake in Antarctica (Fig. 4). The majority (97%) of Organic Lake hits (188 in total) were classified into two narrow clusters (XVI and XVII), suggesting a relatively limited number of potential hosts in the system. It is possible that a few major bacterial populations contribute to the production of these N4-like phages. Intriguingly, these two major clusters appear to be related to cluster XVIII, which contains the largest number of the Chesapeake Bay N4-like phage sequences. Together, our results suggest that N4-like phages can thrive in cold environments. For the purposes of this discussion, we define “cold” as water masses with temperatures of ≤10°C and “warm” water masses as those with temperatures of >10°C. When considering the metagenomic library analysis, N4-like phages in cold waters are significantly more abundant than those in warm waters (t test; P < 0.01). Unfortunately, there are not presently sufficient environmental data for the metagenomic libraries to delineate the relationships between the distribution of N4-like phages and environmental parameters.
FIG 4.
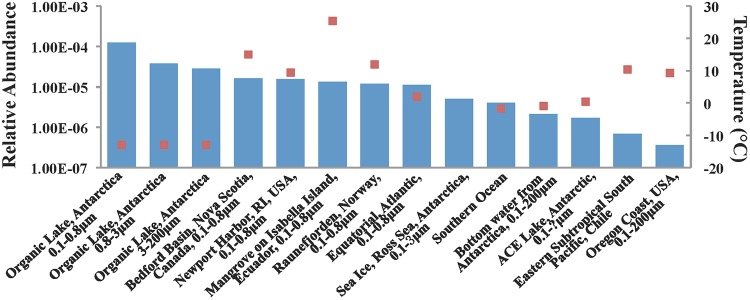
Occurrence of N4-like phage reads in metagenomic databases (blue bars) and corresponding temperatures (red squares). The temperature of the sea ice in the Ross Sea was left blank, since temperature is not provided in that metagenomic database.
Antarctica's Organic Lake is a shallow, hypersaline lake and contains the highest concentration of dimethylsulfide reported in natural water environments (35). The water temperatures are below −10°C, and the salinity exceeds 160 psu (practical salinity units) (36). Consequently, the abundance of bacteria and viruses is lower than in the oligotrophic ocean (typically, both are less than 105 ml−1), and the virus-to-bacteria ratio is approximately 1 (36). It is intriguing that a high number of N4-like phage sequences were recruited from this system despite the low viral abundance. It has been reported previously that members of the classes Alphaproteobacteria and Gammaproteobacteria dominate the bacterial composition of Organic Lake (36), and they may represent presumptive hosts for these phages.
The prevalence of N4-like phages from cold, saline waters suggests at least some of the phages may be adapted to environments with these characteristics. It could be hypothesized that the unique features of N4-like phages, especially the presence of an encapsulated RNA polymerase (37), may allow them to tolerate and replicate at cold (or even freezing) temperatures and under saline (or hypersaline) conditions. Indeed, it has been previously reported that the RNA polymerase in E. coli phage N4 can tolerate high-salt conditions (38). Further studies on cultured N4-like phages are necessary to ascertain whether cold tolerance is a defining feature of this phage class.
Conclusions.
N4-like phages are a group of bacteriophages that are unique in terms of their genomes, phylogeny, taxonomy, and ecology. This study shows that diverse N4-like phages are present in nature and that they exhibit dynamic seasonal and spatial variation. The distribution of N4-like phages in the marine environment appears to be restricted to high latitudes and/or colder seasons. The high recruitment of N4-like phages in the cold biosphere, such as Organic Lake of Antarctica, is unexpected based on prior successful cultivation efforts. The cold selection of N4-like phages could be related to their unique features, including lysis inhibition, large burst size, and viral encapsidated RNA polymerase. However, the ecological role of N4 phages and the mechanisms involved in cold adaptation remain to be elucidated.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge funding from NSF (MCB-0132070) for collecting viral communities from the Chesapeake Bay. We are grateful for the support from a Hanse-Wissenschaftskolleg Fellowship for F.C. and from the Chinese Scholar Council for Y.Z. A.B. was supported, in part, by a grant from the NSF (OCE-1061352).
We thank Kui Wang for concentrating viral particles and extracting viral DNA.
We declare no conflict of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00832-15.
REFERENCES
- 1.Fuhrman JA. 1999. Marine viruses and their biogeochemical and ecological effects. Nature 399:541–548. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- 2.Suttle CA. 2005. Viruses in the sea. Nature 437:356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- 3.Suttle CA. 2007. Marine viruses—major players in the global ecosystem. Nat Rev Microbiol 5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 4.Angly FE, Felts B, Breitbart M, Salamon P, Edward RA, Carlson C, Chan AM, Haynes M, Kelley S, Liu H, Mahaffy JM, Mueller JE, Nulton J, Olson R, Parsons R, Rayhawk S, Suttle CA, Rohwer F. 2006. The marine viromes of four oceanic regions. PLoS Biol 4:e368. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hurwitz BL, Sullivan MB. 2013. The Pacific Ocean Virome (POV): a marine viral metagenomic dataset and associated protein clusters for quantitative viral ecology. PLoS One 8:e57355. doi: 10.1371/journal.pone.0057355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosario K, Breitbart M. 2011. Exploring the viral world through metagenomics. Curr Opin Virol 1:289–297. doi: 10.1016/j.coviro.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Kang I, Oh H-M, Kang D, Cho J-C. 2013. Genome of a SAR116 bacteriophage shows the prevalence of this phage type in the oceans. Proc Natl Acad Sci U S A 110:12343–12348. doi: 10.1073/pnas.1219930110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao Y, Temperton B, Thrash JC, Schwalbach MS, Vergin KL, Landry ZC, Ellisman M, Deerinck T, Sullivan MB, Giovannoni SJ. 2013. Abundant SAR11 viruses in the ocean. Nature 494:357–360. doi: 10.1038/nature11921. [DOI] [PubMed] [Google Scholar]
- 9.Schito GC, Rialdi G, Pesce A. 1966. Biophysical properties of N4 coliphage. Biochim Biophys Acta 129:482–490. doi: 10.1016/0005-2787(66)90063-3. [DOI] [PubMed] [Google Scholar]
- 10.Davydova EK, Kaganman I, Kazmierczak KM, Rothman-Denes LB. 2009. Identification of bacteriophage N4 virion RNA polymerase-nucleic acid interactions in transcription complexes. J Biol Chem 284:1962–1970. doi: 10.1074/jbc.M807785200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falco S, Zivin R, Rothman-Denes LB. 1978. Novel template requirements of N4 virion RNA polymerase. Proc Natl Acad Sci U S A 75:3220–3224. doi: 10.1073/pnas.75.7.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kazmierczak KM, Rothman-Denes LB, Calendar R (ed). 2006. The bacteriophages, p 302–314. Oxford University Press, New York, NY. [Google Scholar]
- 13.Stojković EA, Rothman-Denes LB. 2007. Coliphage N4 N-acetylmuramidase defines a new family of murein hydrolases. J Mol Biol 366:406–419. doi: 10.1016/j.jmb.2006.11.028. [DOI] [PubMed] [Google Scholar]
- 14.Schito GC. 1974. Development of coliphage N4: ultrastructural studies. J Virol 13:186–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y, Wang K, Jiao N, Chen F. 2009. Genome sequences of two novel phages infecting marine roseobacters. Environ Microbiol 11:2055–2064. doi: 10.1111/j.1462-2920.2009.01927.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buchan A, González JM, Moran MA. 2005. Overview of the marine Roseobacter lineage. Appl Environ Microbiol 71:5665–5677. doi: 10.1128/AEM.71.10.5665-5677.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Born Y, Fieseler L, Marazzi J, Lurz R, Duffy B, Loessner MJ. 2011. Novel virulent and broad-host-range Erwinia amylovora bacteriophages reveal a high degree of mosaicism and a relationship to Enterobacteriaceae phages. Appl Environ Microbiol 77:5945–5954. doi: 10.1128/AEM.03022-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kulikov E, Kropinski AM, Golomidova A, Lingohr E, Govorun V, Serebryakova M, Prokhorov N, Letarova M, Manykin A, Strotskaya A, Letarov A. 2012. Isolation and characterization of a novel indigenous intestinal N4-related coliphage vB_EcoP_G7C. Virology 426:93–99. doi: 10.1016/j.virol.2012.01.027. [DOI] [PubMed] [Google Scholar]
- 19.Fouts DE, Klumpp J, Bishop-Lilly KA, Rajavel M, Willner KM, Butani A, Henry M, Biswas B, Li M, Albert MJ, Loessner MJ, Calendar R, Sozhamannan S. 2013. Whole genome sequencing and comparative genomic analyses of two Vibrio cholerae O139 Bengal-specific Podoviruses to other N4-like phages reveal extensive genetic diversity. Virol J 10:165. doi: 10.1186/1743-422X-10-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gan HM, Sieo CC, Tang SGH, Omar AR, Ho YW. 2013. The complete genome sequence of EC1-UPM, a novel N4-like bacteriophage that infects Escherichia coli O78:K80. Virol J 10:308. doi: 10.1186/1743-422X-10-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan H, Fan H, An X, Huang Y, Zhang Z, Mi Z, Tong Y. 2012. Complete genome sequence of IME11, a new N4-like bacteriophage. J Virol 86:13861. doi: 10.1128/JVI.02684-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wittmann J, Dreiseikelmann B, Rohde M, Meier-Kolthoff JP, Bunk B, Rohde C. 2014. First genome sequences of Achromobacter phages reveal new members of the N4 family. Virol J 11:14. doi: 10.1186/1743-422X-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreno Switt AI, Orsi RH, Bakker HC, Vongkamjan K, Altier C, Wiedmann M. 2013. Genomic characterization provides new insight into Salmonella phage diversity. BMC Genomics 14:481. doi: 10.1186/1471-2164-14-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan JZ, Millard AD, Mann NH, Schäfer H. 2014. Comparative genomics defines the core genome of the growing N4-like phage genus and identifies N4-like Roseophage specific genes. Front Microbiol 5:506. doi: 10.3389/fmicb.2014.00506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kui W, Feng C. 2004. Genetic diversity and population dynamics of cyanophage communities in the Chesapeake Bay. Aquat Microb Ecol 34:105–116. doi: 10.3354/ame034105. [DOI] [Google Scholar]
- 26.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 27.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Horn DJV, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun S, Chen J, Li W, Altintas I, Lin A, Peltier S, Stocks K, Allen EE, Ellisman M, Grethe J, Wooley J. 2011. Community cyber infrastructure for advanced microbial ecology research and analysis: the CAMERA resource. Nucleic Acids Res 39:D546–D551. doi: 10.1093/nar/gkq1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williamson SJ, Rusch DB, Yooseph S, Halpern AL, Heidelberg KB, Glass JI, Andrews-Pfannkoch C, Fadrosh D, Miller CS, Sutton G, Frazier M, Venter JC. 2008. The Sorcerer II global ocean sampling expedition: metagenomic characterization of viruses within aquatic microbial samples. PLoS One 3:e1456. doi: 10.1371/journal.pone.0001456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson SJ, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, Beeson K, Tran B, Smith H, Baden-Tillson H, Stewart C, Thorpe J, Freeman J, Andrews-Pfannkoch C, Venter JE, Li K, Kravitz S, Heidelberg JF, Utterback T, Rogers Y-H, Falcón LI, Souza V, Bonilla-Rosso G, Eguiarte LE, Karl DM, Sathyendranath S, Platt T, Bermingham E, Gallardo V, Tamayo-Castillo G, Ferrari MR, Strausberg RL, Nealson K, Friedman R, Frazier M, Venter JC. 2007. The Sorcerer II global ocean sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol 5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen F, Wang K, Huang S, Cai H, Zhao M, Jiao N, Wommack KE. 2009. Diverse and dynamic populations of cyanobacterial podoviruses in the Chesapeake Bay unveiled through DNA polymerase gene sequences. Environ Microbiol 11:2884–2892. doi: 10.1111/j.1462-2920.2009.02033.x. [DOI] [PubMed] [Google Scholar]
- 32.Kan J, Crump B, Wang K, Chen F. 2006. Bacterioplankton community in Chesapeake Bay: predictable or random assemblages. Limnol Oceanogr 51:2157–2169. doi: 10.4319/lo.2006.51.5.2157. [DOI] [Google Scholar]
- 33.Kan J, Suzuki MT, Wang K, Evans SE, Chen F. 2007. High temporal but low spatial heterogeneity of bacterioplankton in the Chesapeake Bay. Appl Environ Microbiol 73:6776–6789. doi: 10.1128/AEM.00541-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji J, Zhang R, Jiao N. 2015. Complete genome sequence of Roseophage vB_DshP-R1, which infects Dinoroseobacter shibae DFL12. Stand Genomic Sci 9:31. doi: 10.1186/1944-3277-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Franzmann PD, Deprez PP, Burton HR, Hoff VJ. 1987. Limnology of Organic Lake, Antarctica, a meromictic lake that contains high concentrations of dimethyl sulfide. Mar Freshw Res 38:409–417. doi: 10.1071/MF9870409. [DOI] [Google Scholar]
- 36.Yau S, Lauro MF, Williams JT, DeMaere ZM, Brown MV, Rich J, Gibson JA, Cavicchioli R. 2013. Metagenomic insights into strategies of carbon conservation and unusual sulfur biogeochemistry in a hypersaline Antarctic lake. ISME J 7:1944–1961. doi: 10.1038/ismej.2013.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schito GC. 1973. The genetics and physiology of coliphage N4. Virology 55:254–265. doi: 10.1016/S0042-6822(73)81028-1. [DOI] [PubMed] [Google Scholar]
- 38.Murakami KS, Davydova EK, Rothman-Denes LB. 2008. X-ray crystal structure of the polymerase domain of the bacteriophage N4 virion RNA polymerase. Proc Natl Acad Sci U S A 105:5046–5051. doi: 10.1073/pnas.0712325105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.