Abstract
Summary: Efforts to develop peptide-based vaccines, in particular those requiring site-specific targeting of self-proteins, rely on the ability to optimize the immunogenicity of the peptide epitopes. Currently, screening of candidate vaccines is typically performed through low-throughput, high-cost animal trials. To improve on this we present the program EpIC, which enables high-throughput prediction of peptide immunogenicity based on the endogenous occurrence of B-cell epitopes within native protein sequences. This information informs rational selection of immunogenicity-optimized epitopes for peptide vaccines.
Availability and implementation: EpIC is available as a web server at http://saphire.usask.ca/saphire/epic.
Contact: kdm449@mail.usask.ca
1 Introduction
Vaccine development efforts are increasingly based on the formulation and delivery of peptides that represent specific priority vaccine targets (epitopes). Recently, applications of immunotherapeutics have expanded beyond infectious diseases and into pathologies where the immune targets are self-molecules, such as cancer, Alzheimer’s, amyotrophic lateral sclerosis and prion diseases. Immunotherapy for such pathologies requires precise direction of immune responses against self-molecules, or even against specific regions or conformations of self-molecules. When there is the dual challenge of maintaining specificity while simultaneously overcoming immune tolerance to a self-molecule, peptide-based vaccines remain a promising option. Although advantaged in terms of cost, ease of production, safety and broad therapeutic applicability, they are often disadvantaged by poor immunogenicity (Purcell et al., 2007).
Typically, once a protein, or a region of a protein, has been identified as a vaccine target, a panel of peptides is randomly generated that collectively represent the entirety of the protein or region of interest. Often the only criterion used for creation of these peptides is length. More sophisticated investigations may incorporate in silico screening to prioritize, but not typically optimize, different regions based on predicted immunogenicity. Either approach ultimately leads to the creation of a panel of vaccines that are tested for immunogenicity in animal models. These approaches, which typically require iterative rounds of vaccine development and testing, are undesirable from efficiency, financial and ethical perspectives. A more rational approach for in silico optimization of peptide epitopes would provide a more direct route to the development of immunogenic peptide vaccines.
Several sequence-based in silico strategies have been developed to predict immunogenic regions of a protein based on a number of variables. These include predicted surface exposure, structural flexibility and occurrence of sequence elements known to correlate with immunogenicity, such as T- and B-cell epitopes (Soria-Guerra et al., 2015). These methods can facilitate peptide vaccine development by identifying a panel of candidate peptide antigens, enabling the user to effectively generate an immune response against a target protein. However, these methods fall short when antibody neutralization is restricted to a specific region of a protein. Furthermore, consideration of sequences based on surface-accessibility is complicated in the context of conformation-specific targets, where epitope selection is even more restricted. Here, a more appropriate method for generating immunogenic epitopes occurs at the level of primary structure through evaluation and selection of naturally occurring sequences containing B-cell epitopes that are predicted to possess enhanced immunogenicity.
In our previous efforts to develop peptide-based vaccines to specific regions of the PrPC protein, the inclusion or exclusion of certain residues flanking our epitope of interest profoundly influenced immunogenicity where a single residue alteration resulted in a 100-fold change in associated antibody titres (Marciniuk et al., 2014). This illustrated the challenge associated with peptide epitopes and also the opportunity to optimize epitopes through selection of appropriate expansions within the protein based on defined correlations between peptide sequence and associated immunogenicity. Through such an approach, it may be possible to utilize subtle manipulations of a peptide epitope sequence to improve immunogenicity when the epitope is limited by location constraints.
EpIC is a software program that allows the user to easily, quickly and accurately increase immunogenicity through selective inclusion of endogenous B-cell epitopes that exist within the target protein sequence near the epitope of interest. The optimized epitopes can then be fused to a carrier protein possessing T-cell epitopes required for immuno-stimulation.
2 Description of epic
Our optimization protocol involves assessing peptide epitopes that differ by a single amino acid, constructed as a panel of expansions around the core epitope of interest. These expansions are then presented in a repeat motif that was previously found to improve immunogenicity (Hedlin et al., 2010), and analyzed using the BepiPred program (Larson et al., 2006) to identify expansions of the core epitope that include B-cell epitope characteristics. BepiPred was designed to analyze an entire protein sequence to identify peptide sequences that exhibit characteristics of known immunogenic B-cell epitopes. EpIC adapts the input/output formats of the original BepiPred program specifically for manipulation of peptide epitopes, thereby increasing the potential for enhanced immunogenicity and vaccine utility.
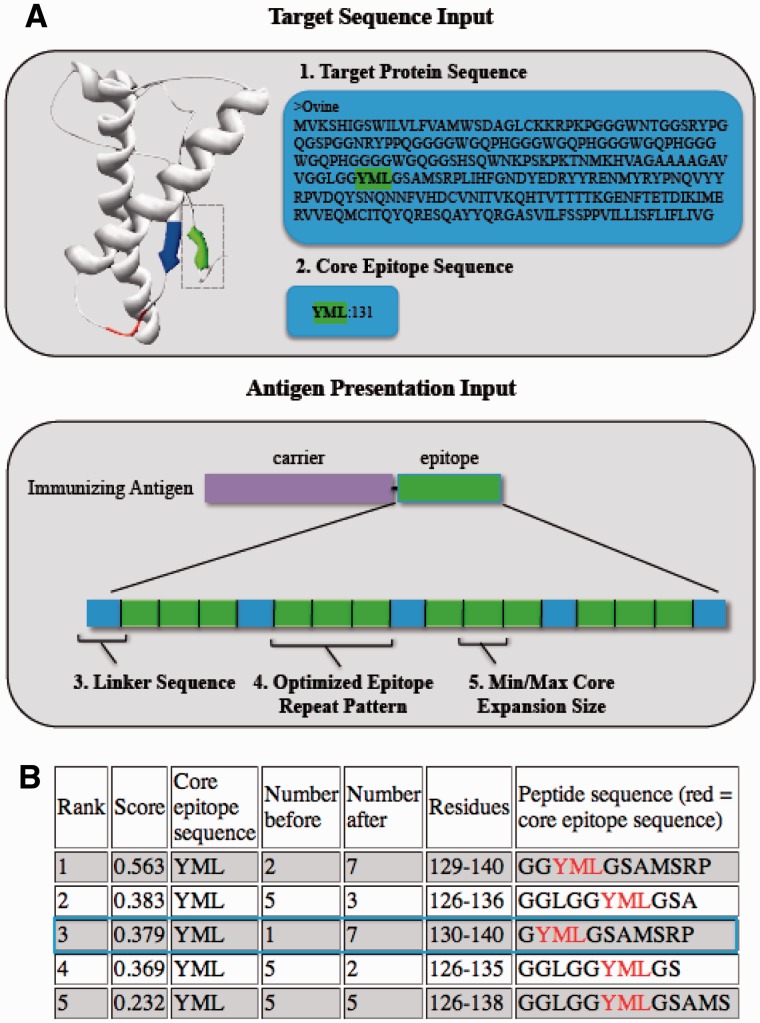
The first two input parameters for EpIC are a file containing the complete amino acid sequence of the target protein in FASTA format, and a file containing the core epitopes of interest. These core epitopes consist of the smallest variant of the desired target, the expansion analysis will be performed through the iterative addition of native residues flanking a given core epitope sequence. Additional parameters include the linker sequence, which can be inserted between expanded epitopes; the repeat pattern, which defines the repeat sequence in terms of the expanded epitope and linker sequence; and the length restrictions, which limit the size of the expanded epitope prior to presentation in the repeat motif (Fig. 1A). If analysis of epitope sequence conservation across species or isoforms is required, additional protein sequences can be added to the target protein sequence input file.
Fig. 1.
(A) Input parameters for EpIC. Two sets of parameters are required for analysis: the target sequence information and the desired presentation of the epitope in the final immunizing antigen. The above figure demonstrates the input parameters for our example prion epitope. The target protein sequence is sheep PRNP and our site-specific minimal target sequence (or core sequence) is YML, in green. EpIC enables the user to customize the presentation of the optimized epitope by inserting a linker sequence, including a repeat pattern with presentation of each repeat as forward or reverse, and the desired expansion of the core epitope can be restricted by defining a min/max. For our purposes, the most immunogenic format involved a forward, reverse, reverse presentation repeated four times with short linker sequences between the repeats. EpIC performs the immunogenicity analysis by converting the epitope expansion to the desired presentation pattern, as these features affect the overall immunogenicity of the immunizing antigen. (B) Analysis of core epitope YML using EpIC. The top five ranking epitopes based on average prediction scores are illustrated. The immunogenic YML expansion identified previously is outlined in blue
EpICs output displays the epitope expansions ranked based on average BepiPred prediction score. For each expanded epitope, the average prediction score (the sum of the scores given to each individual residue in the repeat sequence divided by the number of residues in the repeat sequence), core epitope sequence prior to manipulation, the number of N- and C-terminal residues added, the residues represented within the target protein sequence and the expanded epitope sequence are displayed (Fig. 1B). By selecting an epitope, the full repeat sequence and its conservation are indicated.
EpIC is designed mainly for optimization of linear epitopes. However, vaccine development strategies often require the use of discontinuous epitopes for effective neutralization of the target antigen. Translation of discontinuous epitopes into effective peptide-based vaccines also requires optimization of antigen presentation, formulation and delivery. In our experience, manipulations in epitope presentation patterns translate to alterations in epitope immunogenicity in vivo. Although this program cannot be used to optimize discontinuous epitopes in terms of core epitope sequence expansion, the EpIC algorithm can be employed to predict optimal presentation patterns for discontinuous epitopes and is not completely limited to linear epitope optimization.
3 Validation
Previously, to test our manual expansion-based in silico analysis, we evaluated the utility of the BepiPred B-cell epitope prediction algorithm for the analysis of expanded core epitopes specific for the misfolded prion protein PrPSc. Thermodynamic modeling and hypothesis-based predictions suggested these core epitopes are selectively exposed in the misfolded isoform. A panel of vaccines of varying degrees of predicted immunogenicity was screened in vivo. There was a strong relationship between predicted immunogenicity and magnitude of induced antibody titres (Marciniuk et al., 2014).
To validate EpIC, we analyzed PrPSc-specific epitope YML, which was previously manually optimized (Marciniuk et al., 2014). EpIC identified our epitope as a high-ranking expansion (Fig. 1B). Although 2+YML+7 ranks higher, the chosen expansion better adhered to the stringent conformation-specific restrictions required for our application. Thus, EpIC is used to identify candidate epitopes from which final selections are often determined using structural/hypothesis based approaches.
4 Conclusions
EpIC builds upon existing technologies for predicting immunogenicity of peptides based on amino acid sequence. A key modification is the ability for the user to rapidly and efficiently scan various peptide windows within the natural sequence of a protein to identify regions that are anticipated to have high immunogenicity. Although this method was developed using prion protein sequences, it is not specific to prions and is broadly applicable for peptide-based vaccine design. The development of this tool is timely, as vaccine development efforts are increasingly being applied to specific regions of self-proteins. EpIC will provide researchers a convenient mechanism for selection and optimization of peptide epitopes.
Funding
This research was made possible through funding by the Agriculture Development Fund to S.N. The Canadian Institutes of Health Research and the Saskatchewan Health Research Foundation provide funding to K.M.
Conflict of Interest: none declared.
References
- Hedlin P.D., et al. (2010) Design and delivery of a cryptic PrP(C) epitope for induction of PrP(Sc)-specific antibody responses. Vaccine, 28, 981–988. [DOI] [PubMed] [Google Scholar]
- Larson J.E.P., et al. (2006) Improved method for predicting linear B-cell epitopes. Immunome Res., 2, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniuk K., et al. (2014) Development of a multivalent, PrPSc-specific prion vaccine through rational optimization of three disease-specific epitopes. Vaccine, 32, 1988–1997. [DOI] [PubMed] [Google Scholar]
- Purcell A.W., et al. (2007) More than one reason to rethink the use of peptides in vaccine design. Nat. Rev. Drug Discov., 6, 404–414. [DOI] [PubMed] [Google Scholar]
- Soria-Guerra R.E., et al. (2015) An overview of bioinformatics tools for epitope prediction: implications on vaccine development. J. Biomed. Inform., 53, 405–414. [DOI] [PubMed] [Google Scholar]