Abstract
Myoclonus dystonia syndrome (MDS) is a young-onset movement disorder. A proportion of cases are due to mutations in the maternally imprinted SGCE gene. We assembled the largest cohort of MDS patients to date, and determined the frequency and type of SGCE mutations. The aim was to establish the motor phenotype in mutation carriers and utility of current diagnostic criteria. Eighty-nine probands with clinical features compatible with MDS were recruited from the UK and Ireland. Patients were phenotypically classified as “definite”, “probable” or “possible” MDS according to previous guidelines. SGCE was analyzed using direct sequencing and copy number variant analysis. In those where no mutation was found, DYT1 (GAG deletion), GCH1, THAP1 and NKX2.1 genes were also sequenced. Nineteen (21.3 %) probands had an SGCE mutation. Three patterns of motor symptoms emerged: (1) early childhood onset upper body myoclonus and dystonia, (2) early childhood onset lower limb dystonia, progressing later to more pronounced myoclonus and upper body involvement, and (3) later childhood onset upper body myoclonus and dystonia with evident cervical involvement. Five probands had large contiguous gene deletions ranging from 0.7 to 2.3 Mb in size with distinctive clinical features, including short stature, joint laxity and microcephaly. Our data confirms that SGCE mutations are most commonly identified in MDS patients with (1) age at onset ≤10 years and (2) predominant upper body involvement of a pure myoclonus-dystonia. Cases with whole SGCE gene deletions had additional clinical characteristics, which are not always predicted by deletion size or gene involvement.
Keywords: Myoclonus, Dystonia, Genetic and inherited disorders, SGCE
Introduction
Myoclonus dystonia syndrome (MDS) is a rare movement disorder with onset in the first two decades of life. The typical clinical pattern is of alcohol responsive truncal and upper limb myoclonus with cervical dystonia and/or writer’s cramp [1]. The disorder affects males and females equally [2] and is clinically consistent across ethnicities [3-5]. Previous work has shown evidence of prominent co-morbid psychiatric disorders, specifically compulsivity, anxiety disorders and excessive alcohol use [6-9].
Mutations in the epsilon-sarcoglycan gene (SGCE) are responsible for a proportion of these cases [10, 11]. SGCE mutations are inherited in an autosomal dominant manner with variable penetrance due to maternal imprinting [12,13]. SGCE encodes the epsilon-sarcoglycan protein, a single pass transmembrane protein forming part of the dystrophin-associated glycoprotein complex [14-16].
SGCE mutation rates have varied amongst previously reported cohorts, some reporting no mutations [17], while others report rates from 21 to 80 % [5, 18-21]. Genetic heterogeneity has been offered as an explanation with linkage in a large Canadian family to a locus on chromosome 18p, [22, 23] although the causative gene within this region is yet to be identified. Copy number variants (CNVs) provide another possible explanation, and more recently, both exonic [24, 25] and whole gene deletions having been identified [26, 27].
This study represents one of the largest cohorts to undergo systematic clinical and genetic evaluation, identifying the largest cohort of contiguous gene deletions involving SGCE to date. We also describe motor symptom pattern evolution from onset to examination and compare our results to the current MDS diagnostic criteria.
Methods
Patients with suspected MDS, some with previously confirmed SGCE mutations, were referred by adult and paediatric movement disorder specialists throughout the UK and Ireland. The study was approved by the Multi-Centre Research Ethics Committee for Wales (MREC 09/MRE09/56 and 09/MRE09/35). Written informed consent or assent from parents/guardians was obtained for all participants.
Cases were assessed systematically and a videotaped clinical examination performed. In patients for whom this was not possible, clinical information was obtained retrospectively using a systematic pro forma for data extraction from clinical records. We recorded the presence or absence of myoclonus, dystonia, tremor, chorea and tics, age at onset of the movement disorder and family history. Patients were classified as ‘definite’, ‘probable’ or ‘possible’ according to previously published clinical criteria [28]. Psychiatric symptoms were also assessed, the results of which have been published elsewhere [8]. Recruitment methods were also previously reported and are summarized in Supplementary Fig. 1. This cohort represents the same cohort as has been published elsewhere; however, in this article specific emphasis is placed on the demographic, clinical and genetic characteristics of the cohort [8].
Blood samples were collected from all cases and DNA was isolated from peripheral blood lymphocytes using standard protocols. All samples underwent direct sequencing of exons 1-12 (including alternatively spliced exons 1a and 11b) of the SGCE gene. In those cases where no SGCE mutation was found, MLPA analysis was performed using the commercially available probe set P099B (MRC Holland, Amsterdam, The Netherlands) according to manufacturer’s instructions. Cases with whole gene deletions were analyzed on a custom oligonucleotide CGH array platform (Roche Nimblegen) with 5,900 probes covering chr7:88000000-98000000 (NCBI36/hg18 genome build). Data was analyzed using the segment tool and visualized using SignalMap (Roche Nimblegen).
To exclude other potential genetic diagnoses, all samples negative for SGCE mutations were assessed for mutations in DYT1 (GAG deletion) GCH1, THAP1 and NXK2.1 genes by direct sequencing. Cases found with mutations in these latter genes were excluded from further analysis.
Statistical analysis was performed using the ‘R’ statistical software package. Fisher’s exact testing, binomial stepwise multiple logistic regression and ANOVA were used as appropriate.
Results
Demographics
Eighty-nine probands were recruited, 50 males and 39 females, with a median age at movement disorder onset of 5 years (range 0–50 years). Nineteen (21.3 %) were found to have an SGCE mutation with a slight female predominance (8 M:11F). Median age at onset was 5 years younger in the mutation positive group compared to those without a mutation (3 vs. 8 years, respectively) with 79 % of those with an SGCE mutation developing symptoms <10 years (Table 1). With recruitment of additional affected family members the number of SGCE positive patients increased to 27 (8 additional patients) and mutation negative cases to 76 (6 additional patients) (Supplementary Fig. 1).
Table 1.
Demographic and clinic features
| Proband only cohort |
All SGCE mutation positive cases | |||
|---|---|---|---|---|
| Overall | SGCE mutation positive | SGCE mutation negative | ||
| n | 89 | 19 | 70 | 27 |
| Male:female | 50:39 | 8:11 | 45:33 | 10:27 |
| Median age at onset (range) | 5 (0–50) | 3 (1–18) | 8 (0–50) | 3 (1.5–18) |
| Age at onset | ||||
| <10 years | 56 | 15 (79 %) | 41 (59 %) | 21 (78 %) |
| 10–20 years | 20 | 4 (21 %) | 16 (23 %) | 6 (22 %) |
| >20 years | 4 | 0 | 4 (6 %) | 0 |
| Clinical likelihood of MDS | ||||
| Definite | 17 | 15 (79 %) | 2 (3 %) | 25 (93 %) |
| Probable | 8 | 4 (21 %) | 4 (6 %) | 2 (7 %) |
| Possible | 64 | 0 | 64 (91 %) | 0 |
Motor characteristics
SGCE mutation positive cohort
At onset
Myoclonus and dystonia were the only types of movement disorder observed, each being present in 19 cases. No cases were reported to have evidence of tremor, chorea or tics. Myoclonus predominantly involved the upper limbs and neck both overall and in each age-at-onset subgroup (Tables 2, 3). Dystonia was also most frequently reported in the upper limbs; however, the two age subgroups differed with lower limb involvement. Almost half of those with onset of the movement disorder <10 years had either foot or leg involvement while this was not reported in those whose symptoms developed between 10 and 20 years of age.
Table 2.
SGCE mutation positive cohort—motor characteristics at onset and examination
| Motor characteristics at onset |
Motor characteristics at examination |
|||||
|---|---|---|---|---|---|---|
| Overall | <10 years | 10–20 years | Overall | Onset <10 years | Onset 10–20 years | |
| n | 27 | 21 | 6 | 27 | 21 | 6 |
| Median age (range) | 3 (1.5–18) | 2.5 (1.5–8.5) | 10.5 (10–18) | 28 (3–74) | 22 (3–74) | 47.50 (19–63) |
| Myoclonus | ||||||
| n | 19 | 14 | 5 | 26 | 20 | 6 |
| Neck | 9 (47 %) | 6 (43 %) | 3 (60 %) | 19 (73 %) | 15 (75 %) | 4 (67 %) |
| Upper limbs | 17 (89 %) | 13 (93 %) | 4 (80 %) | 25 (96 %) | 20 (100 %) | 5 (83 %) |
| Trunk | 5 (26 %) | 5 (36 %) | 0 | 17 (65 %) | 14 (70 %) | 3 (50 %) |
| Lower limbs | 1 (5 %) | 1 (7 %) | 0 | 5 (19 %) | 4 (20 %) | 1 (17 %) |
| Dystonia | ||||||
| n | 19 | 17 | 2 | 27 | 21 | 6 |
| Neck | 5 (26 %) | 4 (24 %) | 1 (50 %) | 20 (74 %) | 14 (67 %) | 6 (100 %) |
| Voice | 1 (5 %) | 1 (6 %) | 0 | 5 (19 %) | 4 (19 %) | 1 (17 %) |
| Upper limbs | 13 (68 %) | 9 (53 %) | 2 (100 %) | 22 (81 %) | 19 (90 %) | 3 (50 %) |
| Trunk | 1 (5 %) | 1 (6 %) | 0 | 3 (11 %) | 2 (10 %) | 1 (17 %) |
| Lower limbs | 8 (42 %) | 8 (47 %) | 0 | 12 (44 %) | 11 (52 %) | 1 (17 %) |
Table 3.
Motor characteristics of SGCE mutation negative cohort
| n (%) | |
|---|---|
| Myoclonus | |
| n | 16 (23) |
| Median age at onset (range) | 4.75 (0–15) |
| Neck | 8 (50) |
| Upper limbs | 12 (75) |
| Trunk | 8 (50) |
| Lower limbs | 5 (31) |
| Dystonia | |
| n | 29 (41) |
| Median age at onset (range) | 4.5 (0–48) |
| Neck | 9 (31) |
| Voice | 4 (14) |
| Upper limbs | 15 (52) |
| Trunk | 0 |
| Lower limbs | 12 (41) |
| Tremor | |
| n | 16 (23) |
| Median age at onset (range) | 10 (0.25–48) |
| Neck | 4 (25) |
| Upper limbs | 13 (81) |
| Trunk | 0 |
| Lower limbs | 1 (6) |
| Tics | |
| n | 16 (23) |
| Median age at onset (range) | 7 (0.5–14) |
| Neck | 13 (81) |
| Voice | 8 (50) |
| Upper limbs | 11 (69) |
| Trunk | 1 (6) |
| Lower limbs | 2 (13) |
| Chorea | |
| n | 7 (10) |
| Median age at onset (range) | 4.5 (0–21) |
| Neck | 3 (43) |
| Upper limbs | 6 (86) |
| Trunk | 0 |
| Lower limbs | 3 (43) |
At examination
Median age at examination was 28 years (range 3–74 years). Myoclonus was observed in all but one case, predominantly involving the upper limbs and neck across both age subgroups. Truncal involvement was greater than had been reported at onset (65 % compared to 26 %), consistent in those with onset above and below 10 years of age. Dystonia was observed in all cases with the upper limbs most frequently affected overall and for those with onset <10 years. Cervical involvement was most common in the older age sub-group and more pronounced in those with onset <10 years than had been reported at onset (67 vs. 24 %). Lower limb dystonia demonstrated the largest difference, present in over half of the younger sub-group but only in a single case of onset between 10 and 20 years.
SGCE mutation negative cohort
Multiple extrapyramidal features were seen, including myoclonus, dystonia, tremor, tics and chorea, with dystonia being most prevalent (41 %). The upper limbs were the most commonly affected body part for all movement disorder subtypes with the exception of tics where a more pronounced cranio-cervical involvement was observed.
Myoclonus (p < 0.0001) and dystonia (p < 0.0001) were strongly associated with the occurrence of an SGCE mutation. Tics (p = 0.0007) and tremor (p = 0.002) were more common in the SGCE negative group. Stepwise multiple logistic regression found significant associations of myoclonus (p < 0.001) and dystonia (p = 0.006) with SGCE mutations.
Genetics
SGCE mutation positive
Seventy-nine percent (15/19) had a positive family history of MDS with mutations paternally inherited in 74 % of cases (14/19). Thirteen different mutations were identified, the most prevalent of which was the nonsense mutation c.289C>T (p.97X) in exon 3, occurring in four apparently unrelated families. There were four nonsense, one missense and three splice-site mutations. The missense mutation was present in a three-generation family, co-segregating with the motor disorder and demonstrating the typical autosomal dominant pattern of inheritance with reduced penetrance due to maternal imprinting (affected proband, unaffected father and his affected mother).
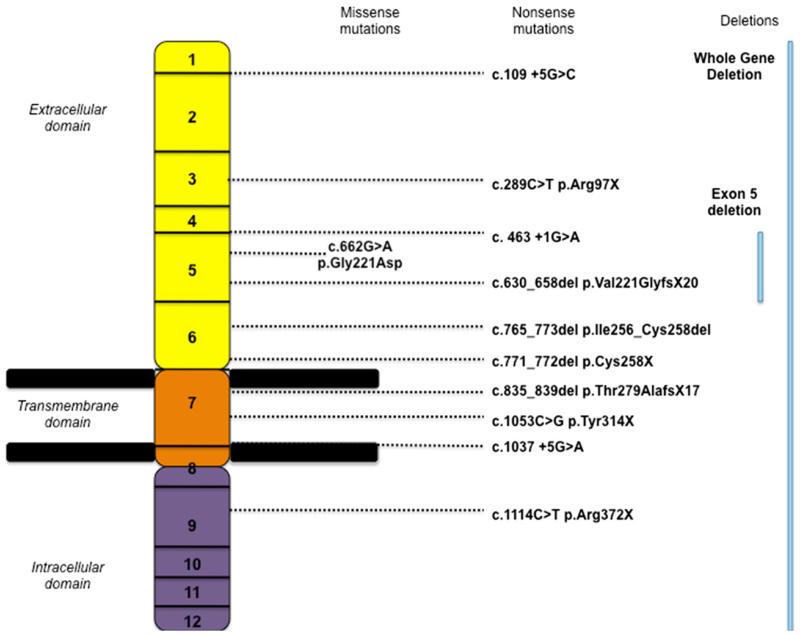
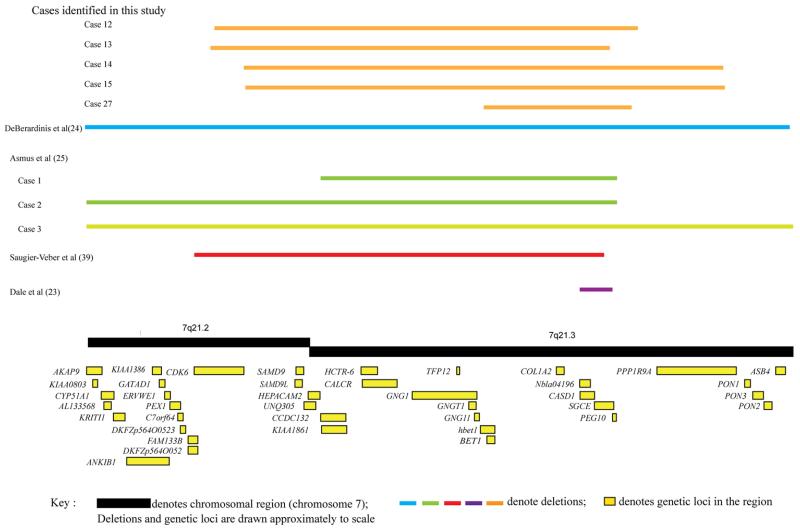
A number of deletions were also identified, including three intra-exonic deletions, one single exon deletion (of exon 5 only) and five whole gene deletions (WGD) (Fig. 1). In these patients with WGD (five cases identified in three families), the deleted region ranged in size from 0.7 to 2.3 Mb. Contiguous genes involved, in addition to SGCE, included: PEG10, PPP1R9A, CASD1, COL1A2, BET1, GNG11, GNG1, TFP12, CALCR, HCTR-6, KIAA 1861, CCDC132, HEPACAM2, SAMD9L, CDK6. Additional phenotypic features included microcephaly, intrauterine growth retardation, short stature, joint laxity, language delay, cognitive impairment and psychosis (Table 4; Fig. 2).
Fig. 1.
Diagrammatic representation of identified SGCE mutations
Table 4.
Clinical characteristics of cases with contiguous gene deletions involving SGCE
| Cases | Deletion size (Mb) | Genes involved | Clinical characteristics |
|---|---|---|---|
| This study | |||
| Case 1 | 2.3 | PPP1R9A, PEG10, SGCE, CASD1, COL1A2, BET1, GNG11, TFP12, GNG1, CALCR, HCTR-6, KIAA 1861, CCDC132, HEPACAM2, SAMD9, SAMD9L | Short stature, language delay |
| Case 2 | 2.3 | PPP1R9A, PEG10, SGCE, CASD1, COL1A2, BET1, GNG11, TFP12, GNG1, CALCR, HCTR-6, KIAA 1861, CCDC132, HEPACAM2, SAMD9, SAMD9L | Short stature |
| Case 3 | 2 | PEG10, SGCE, CASD1, COL1A2, BET1, GNG11, TFP12, GNG1, CALCR, HCTR-6, KIAA 1861, CCDC132, HEPACAM2, SAMD9, SAMD9L, CDK6 | Intrauterine growth retardation, microcephaly, short stature, joint laxity |
| Case 4 | 1.9 | SGCE, CASD1, COL1A2, BET1, GNG11, TFP12, GNG1, CALCR, HCTR-6, KIAA 1861, CCDC132, HEPACAM2, SAMD9, SAMD9L, CDK6 | Microcephaly, short stature, cognitive impairment |
| Case 5 | 0.7 | PEG10, SGCE, CASD1, COL1A2 | Short stature, psychosis |
| DeBerardinis et al. | |||
| Case 1 | 9–16.5 | SGCE (and contiguous genes, not further defined) | Intrauterine growth retardation, microcephaly, short stature, dysmorphic facies, language delay |
| Asmus et al. | |||
| Case 1 | 1.63 | PEG10, SGCE, COL1A2 | Short stature, joint laxity, dental caries, joint laxity, blue sclerae, cerebral cavernous malformations |
| Case 2 | 4.99 | PEG10, SGCE, COL1A2, PEX1, KRITI | |
| Case 3 | 8.78 | PEG10, SGCE, COL1A2, PEX1, KRITI, DLX5 | Dysmorphic facies, dental caries, cognitive impairment, split-hand split-foot syndrome |
| Saugier-Veber et al. | |||
| Case 1 | 1.88 | SGCE, CASD1, COL1A2, BET1, GNG11, TFP12, GNG1, CALCR, HCTR-6, KIAA 1861, CCDC132, HEPACAM2, SAMD9, SAMD9L, CDK6 | Intrauterine growth retardation, microcephaly, short stature, joint laxity, cognitive impairment |
| Dale et al. | |||
| Case 1 | 0.17 | SGCE, CASD1 | Language delay, cognitive impairment |
| Case 2 | 0.17 | SGCE, CASD1 | Nil |
| Case 3 | 0.17 | SGCE, CASD1 | Psychosis |
Fig. 2.
Diagrammatic representation of contiguous gene deletions involving SGCE identified in this cohort
SGCE mutation negative
70/89 (79 %) unrelated patients did not have an identified SGCE mutation with a male predominance (42 M: 28F). No mutations were detected in DYT1 (GAG deletion), GCH1 or THAP1. Two cases were found to have putative NKX2.1 mutations and were excluded from further analysis.
Diagnostic criteria for MDS
According to previously published MDS diagnostic criteria [28], there were 15 clinically ‘definite’, 4 clinically ‘probable’ and 0 ‘possible’ cases in the SGCE mutation positive group. In the mutation-negative group there were 2 ‘definite’, 4 ‘probable’ and 64 ‘possible’ cases (Supplementary Fig. 1). Eighty-eight percent of clinically ‘definite’ and 50 % of clinically ‘probable’ patients carried an SGCE mutation. When applied to this cohort, the ‘definite’ diagnostic criteria had 79 % sensitivity, 97 % specificity and 88 % positive predictive value (PPV) in anticipating an SGCE mutation. Applying the recently modified criteria [29], we used our previously published data of psychiatric features [8] and further refined ‘young age at onset’ to ≤10 years. Here, we found a reduced sensitivity (66.7 %), preserved specificity (97 %) and improved PPV (90 %).
Discussion
We report a large extensive cohort of MDS patients who we systematically assessed (both clinically and by molecular genetic investigation) and this is one of the few studies to include direct sequencing and CNV analysis. We have identified the largest subgroup of patients with contiguous gene deletions involving SGCE as well as allowing the assessment of motor symptom progression.
The frequency of SGCE mutations within this cohort (21.3 %) is in keeping with previously reported studies [19, 21, 30]. Significant differences in age at onset of the movement disorder were observed between those with and without SGCE mutations with the latter group manifesting a broader age range (birth–50 years). As has been observed previously [1, 2, 5, 19, 20], 95 % of those with a mutation had an age at onset of 10 years or younger and there were no cases presenting over the age of 20. This suggests that age at onset is a strong predictor of an SGCE mutation. A family history of MDS was also an important factor in determining whether cases were ‘definite’ or ‘probable’ resulting in 4 SGCE positive probands being labeled as ‘probable’. This was likely due to maternal imprinting causing ‘silencing’ of the mutated gene with several generations of maternal inheritance, however, we were unable to complete parental testing to exclude de novo mutations.
Statistical analysis of two comparable MDS cohorts in a recent study found a disease onset in childhood and the presence of psychiatric symptoms to be the strongest factors in discriminating between those with and without SGCE mutations. When we applied these to our cohort PPV was improved (90 %), specificity preserved (97 %) but sensitivity reduced (66 %) compared to pre-existing diagnostic criteria. However, Carecchio et al. did not specify an age limit in their ‘young-onset’ definition. In keeping with the findings of this study, we imposed a ≤10 years restriction, which may in part account for the reduced sensitivity. We therefore postulate that to further aid targeted genetic testing, diagnostic guidelines could be altered to reduce the age on onset from <25 to <18 years [31].
Pure myoclonus and dystonia were identified in those with an SGCE mutation, their distribution being consistent with pre-existing definitions (predominantly upper body pattern of involvement). [1, 31] Three main patterns of motor involvement emerged with a consistent clinical phenotype: (1) the most common presentation of a young-onset predominantly upper body myoclonus and dystonia, with more pronounced truncal involvement developing during later childhood and adolescence, (2) a group presenting <10 years with isolated or prominent lower limb dystonia, later developing a larger myoclonic component with greater upper body involvement. This pattern has been reported previously in ~20 % SGCE patients with isolated dystonia, similar to the 33 % observed in this cohort [2, 30, 32, 33]. (3) A subgroup with onset >10 years with later truncal myoclonus and much more pronounced cervical dystonia. This suggests that even in the absence of myoclonus at examination, SGCE testing should still be considered in young children with focal lower limb dystonia. These patterns of clinical presentation and progression became apparent during collection and analysis of the clinical data and should be replicated in a larger cohort of SGCE mutation positive cases.
With the exception of a few cases in whom symptom progression is described [34, 35], MDS symptoms usually plateau in adulthood and are associated with a normal life span [36, 37], possibly reflecting maturation of the basal ganglia pathways [38]. There are also reports of spontaneous improvement of dystonia symptoms [39], similar to that seen with primary focal dystonia. Despite some reports of subjective improvement, no spontaneous resolution of dystonia was reported in this cohort, a feature consistent with the findings of a recent report on the impact of dystonia in children [40]. A single case had no evidence of myoclonus on examination despite this being a prominent childhood feature, which again has also been reported in other cohorts [5, 41].
Six cases had SGCE deletions detected by MLPA analysis and consistent with previous literature, no duplications were identified [26, 27, 42, 43]. One involved a single exon deletion (exon 5) with a typical MDS clinical phenotype and no additional clinical characteristics, again consistent with previous cases. [24, 28, 44] In the remaining five cases (including two sets of sibling pairs) CGH array analysis showed large contiguous gene deletions involving PEG10, PPP1R9A, CASD1, COL1A2, BET1, GNG11, GNG1, TFP12, CALCR, HCTR-6, KIAA 1861, CCDC132, HEPACAM2, SAMD9L, CDK6 genes, ranging between 0.7 and 2.3 Mb in size. All five cases had features of both myoclonus and dystonia. The size of the deletion did not dictate age of symptom onset or disease severity. For the two sibling pairs, there was evidence of intra-familial variation in motor symptom severity, the elder sibling manifesting more pronounced and disabling symptoms. This may represent a pattern of motor symptom evolution; however, it is conceivable that as with SGCE point mutations, intra-familial phenotypic variation also exists with contiguous gene deletions. There has been previous speculation that the size of the deletion may determine the presence and severity of further clinical characteristics [27].
Within this cohort, while all five cases were of short stature the number, type and nature of the additional features appeared unrelated to deletion size, or genes involved. There was also evidence of intra-familial variation despite having the same or similar size deletion, as exemplified by cases 3 and 4 (Table 4). As shown in Fig. 2 all detected deletions span a large area of chromosome 7 and involve a number of genes. COL1A2 is one of the best understood, mutations of which are associated with autosomal dominant osteogenesis imperfecta. Hence patients with CNVs involving this gene might be anticipated to develop bone fractures, hypodontia and joint laxity. Despite all five cases in this study harbouring deletions encompassing COL1A2, only a single patient (case 2) was observed to have joint laxity without a history of fractures or problems with dentition. These cases provide further evidence that patients with large contiguous gene deletions may be phenotypically distinct from those with SGCE point mutations and demonstrate significant clinical variation not predicted by deletion size or gene involved. Investigation of larger cohorts of patients with SGCE deletions are required to further delineate the clinical spectrum and elucidate the roles of other genes in the region.
Although data collection and analysis in this study was systematic and used standardized methods, a few limitations are recognized. Motor data collection combined both face-to-face interview and retrospective data collection from the clinical notes. In addition, assessment of age at onset was in most cases retrospective and dependent on patient recall. While only myoclonus and dystonia were observed in the SGCE positive group, chorea, tremor and tics were also observed in the mutation negative group. Although early clinical descriptions frequently reflected difficulty in accurately describing and classifying these movement disorders [45], improved diagnostic criteria [28, 31] suggest that presence of these additional movement disorders likely indicate that a proportion of the mutation negative group do not meet diagnostic criteria for MDS.
Conclusion
We describe a large cohort of MDS patients systematically and fully assessed SGCE mutations, with detailed delineation of motor features and additional clinical characteristics. We conclude that mutations are associated with three distinct motor patterns of presentation based on age of onset, movement phenotype and body distribution. This study also demonstrates that adherence to strict diagnostic criteria; age at onset <18 years, the presence of only myoclonus and/or dystonia as movement disorder subtypes and a positive family history of a similar movement disorder greatly increases the yield of SGCE mutations with genetic testing. Neurophysiological testing may also be used to aid diagnostic accuracy prior to genetic testing. Our cohort also reports the largest single series of whole gene deletions involving SGCE highlighting importance of microarray studies and MLPA as important diagnostic investigation for complex movement disorders [46].
Supplementary Material
Acknowledgments
We would like to thank the patients and their relatives for participating in this study as well as the British Neurological Surveillance Unit (BNSU) for their assistance with notification of cases. We would also like to thank Dr. Daniel Lumsden for his assistance with data collection. KJP is funded by an Ipsen Clinical Research Fellowship and supported by the Welsh Clinical Academic Track (WCAT) program. MAK is funded by GOSHCC and is a Wellcome Trust Intermediate Clinical Fellow. GK is funded by the Wellcome Trust and Medical Research Council. JPL is funded by grants from Action Medical Research, The Dystonia Society UK and Guy’s and St Thomas’ Charity. MS is funded by Parkinson’s UK, East Kent and King’s College Hospital Foundation Trusts. MDK is funded by the Children’s fund for health at The Children’s University Hospital, Temple Street, Dublin. TTW is funded by the Brain Research Trust, Cure Huntington’s Disease Initiative, The Dystonia Society, Bachmann-Strauss Dystonia Parkinson Foundation and NHS Innovations Ltd. HRM is funded by the Medical Research Council and Parkinson’s UK.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00415-014-7488-3) contains supplementary material, which is available to authorized users.
Conflicts of interest Dr JP Lin has received unrestricted educational support and Honoraria for lecturing from Medtronic Ltd. Dr Pall has received an educational grant from Medtronic to attend a movement disorders meeting. Dr B Lynch has received funding from Viropharma to attend an advisory group and epilepsy meeting. Dr Samuel has received Honoraria and sponsorship for education meetings from Medtronic, UCB, St Jude Medical Inc, Boehrinher-Ingelheim, Ipsen and Solvay. Professor Morris is on the advisory board to tev and Solvay has received speaker’s fees from UCB and Teva and a travel grant from Teva. Drs Peall, Kurian, Wardle, Waite, Hedderly, Smith, Whone, White, Lux, Jardine, O’Riordan and Professors Lynch, King, Chinnery, Warner, Blake and Owen have no conflicts of interest.
Ethical standards This study has been approved by the appropriate ethics committee and therefore has been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments. All participants in this study gave their informed consent prior to their inclusion in this study.
Contributor Information
Kathryn J. Peall, MRC Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, School of Medicine, Cardiff University, Hadyn Ellis Building, Maindy Road, Cardiff CF24 4HQ, UK
Manju A. Kurian, Neurogenetics Unit, Developmental Neurosciences, UCL-Institute of Child Health, London, UK; Department of Neurology, Great Ormond Street Hospital, London, UK
Mark Wardle, MRC Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, School of Medicine, Cardiff University, Hadyn Ellis Building, Maindy Road, Cardiff CF24 4HQ, UK.
Adrian J. Waite, MRC Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, School of Medicine, Cardiff University, Hadyn Ellis Building, Maindy Road, Cardiff CF24 4HQ, UK
Tammy Hedderly, Evelina Children’s Hospital, St Thomas’ Hospital, London, UK.
Jean-Pierre Lin, Evelina Children’s Hospital, St Thomas’ Hospital, London, UK.
Martin Smith, Birmingham Children’s Hospital, Birmingham, UK.
Alan Whone, Department of Neurology, Frenchay Hospital, Bristol, UK.
Hardev Pall, School of Clinical and Experimental Medicine, University of Birmingham, Birmingham, UK.
Cathy White, Department of Paediatrics, Singleton Hospital, Swansea, UK.
Andrew Lux, Bristol Royal Hospital for Children, Bristol, UK.
Philip E. Jardine, Bristol Royal Hospital for Children, Bristol, UK
Bryan Lynch, Children’s University Hospital, Temple Street, Dublin, Ireland.
George Kirov, MRC Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, School of Medicine, Cardiff University, Hadyn Ellis Building, Maindy Road, Cardiff CF24 4HQ, UK.
Sean O’Riordan, St. Vincent’s University Hospital, Dublin, Ireland.
Michael Samuel, Department of Neurology, East Kent Hospitals NHS Foundation Trust, Ashford, Kent; King’s College Hospital, King’s Health Partners, London, UK.
Timothy Lynch, Mater Misericordiae University Hospital, Dublin, Ireland.
Mary D. King, Children’s University Hospital, Temple Street, Dublin, Ireland
Patrick F. Chinnery, Institute of Genetic Medicine, Newcastle University, Newcastle, UK
Thomas T. Warner, Department of Clinical Neurosciences, UCL Institute of Neurology, London, UK
Derek J. Blake, MRC Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, School of Medicine, Cardiff University, Hadyn Ellis Building, Maindy Road, Cardiff CF24 4HQ, UK
Michael J. Owen, MRC Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, School of Medicine, Cardiff University, Hadyn Ellis Building, Maindy Road, Cardiff CF24 4HQ, UK
Huw R. Morris, Department of Clinical Neurosciences, UCL Institute of Neurology, London, UK
References
- 1.Asmus F, et al. Myoclonus-dystonia syndrome: epsilon-sarcoglycan mutations and phenotype. Ann Neurol. 2002;52(4):489–492. doi: 10.1002/ana.10325. [DOI] [PubMed] [Google Scholar]
- 2.Raymond D, et al. Phenotypic spectrum and sex effects in eleven myoclonus-dystonia families with epsilon-sarcoglycan mutations. Mov Disord. 2008;23(4):588–592. doi: 10.1002/mds.21785. [DOI] [PubMed] [Google Scholar]
- 3.Chung EJ, et al. Novel SGCE gene mutation in a Korean patient with myoclonus-dystonia with unique phenotype mimicking Moya-Moya disease. Mov Disord. 2007;22(8):1206–1207. doi: 10.1002/mds.21093. [DOI] [PubMed] [Google Scholar]
- 4.Chen XP, et al. A novel mutation of the epsilon-sarcoglycan gene in a Chinese family with myoclonus-dystonia syndrome. Mov Disord. 2008;23(10):1472–1475. doi: 10.1002/mds.22008. [DOI] [PubMed] [Google Scholar]
- 5.Nardocci N, et al. Myoclonus-dystonia syndrome: clinical presentation, disease course, and genetic features in 11 families. Mov Disord. 2008;23(1):28–34. doi: 10.1002/mds.21715. [DOI] [PubMed] [Google Scholar]
- 6.Doheny DO, et al. Phenotypic features of myoclonus-dystonia in three kindreds. Neurology. 2002;59(8):1187–1196. doi: 10.1212/wnl.59.8.1187. [DOI] [PubMed] [Google Scholar]
- 7.Hess CW, et al. Myoclonus-dystonia, obsessive-compulsive disorder, and alcohol dependence in SGCE mutation carriers. Neurology. 2007;68(7):522–524. doi: 10.1212/01.wnl.0000253188.76092.06. [DOI] [PubMed] [Google Scholar]
- 8.Peall KJ, et al. SGCE mutations cause psychiatric disorders: clinical and genetic characterization. Brain. 2013;136(Pt 1):294–303. doi: 10.1093/brain/aws308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weissbach A, et al. Prominent psychiatric comorbidity in the dominantly inherited movement disorder myoclonus-dystonia. Parkinsonism Relat Disord. 2013;19(4):422–425. doi: 10.1016/j.parkreldis.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 10.Zimprich A, et al. Mutations in the gene encoding epsilon-sarcoglycan cause myoclonus-dystonia syndrome. Nat Genet. 2001;29(1):66–69. doi: 10.1038/ng709. [DOI] [PubMed] [Google Scholar]
- 11.Schule B, et al. Genetic heterogeneity in ten families with myoclonus-dystonia. J Neurol Neurosurg Psychiatry. 2004;75(8):1181–1185. doi: 10.1136/jnnp.2003.027177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muller B, et al. Evidence that paternal expression of the epsilon-sarcoglycan gene accounts for reduced penetrance in myoclonus-dystonia. Am J Hum Genet. 2002;71(6):1303–1311. doi: 10.1086/344531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grabowski M, et al. The epsilon-sarcoglycan gene (SGCE), mutated in myoclonus-dystonia syndrome, is maternally imprinted. Eur J Hum Genet. 2003;11(2):138–144. doi: 10.1038/sj.ejhg.5200938. [DOI] [PubMed] [Google Scholar]
- 14.Blake DJ, et al. Function and genetics of dystrophin and dystrophin-related proteins in muscle. Physiol Rev. 2002;82(2):291–329. doi: 10.1152/physrev.00028.2001. [DOI] [PubMed] [Google Scholar]
- 15.Waite A, et al. The neurobiology of the dystrophin-associated glycoprotein complex. Ann Med. 2009;41(5):344–359. doi: 10.1080/07853890802668522. [DOI] [PubMed] [Google Scholar]
- 16.Esapa CT, et al. SGCE missense mutations that cause myoclonus-dystonia syndrome impair epsilon-sarcoglycan trafficking to the plasma membrane: modulation by ubiquitination and torsinA. Hum Mol Genet. 2007;16(3):327–342. doi: 10.1093/hmg/ddl472. [DOI] [PubMed] [Google Scholar]
- 17.Valente EM, et al. Analysis of the epsilon-sarcoglycan gene in familial and sporadic myoclonus-dystonia: evidence for genetic heterogeneity. Mov Disord. 2003;18(9):1047–1051. doi: 10.1002/mds.10476. [DOI] [PubMed] [Google Scholar]
- 18.Valente EM, et al. The epsilon-sarcoglycan gene in myoclonic syndromes. Neurology. 2005;64(4):737–739. doi: 10.1212/01.WNL.0000151979.68010.9B. [DOI] [PubMed] [Google Scholar]
- 19.du Tezenas MS, et al. Epsilon sarcoglycan mutations and phenotype in French patients with myoclonic syndromes. J Med Genet. 2006;43(5):394–400. doi: 10.1136/jmg.2005.036780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerrits MC, et al. Phenotype-genotype correlation in Dutch patients with myoclonus-dystonia. Neurology. 2006;66(5):759–761. doi: 10.1212/01.wnl.0000201192.66467.a3. [DOI] [PubMed] [Google Scholar]
- 21.Ritz K, et al. Myoclonus-dystonia: clinical and genetic evaluation of a large cohort. J Neurol Neurosurg Psychiatry. 2009;80(6):653–658. doi: 10.1136/jnnp.2008.162099. [DOI] [PubMed] [Google Scholar]
- 22.Grimes DA, et al. Inherited myoclonus-dystonia: evidence supporting genetic heterogeneity. Mov Disord. 2001;16(1):106–110. doi: 10.1002/1531-8257(200101)16:1<106::aid-mds1022>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 23.Grimes DA, et al. A novel locus for inherited myoclonus-dystonia on 18p11. Neurology. 2002;59(8):1183–1186. doi: 10.1212/wnl.59.8.1183. [DOI] [PubMed] [Google Scholar]
- 24.Asmus F, et al. Myoclonus-dystonia due to genomic deletions in the epsilon-sarcoglycan gene. Ann Neurol. 2005;58(5):792–797. doi: 10.1002/ana.20661. [DOI] [PubMed] [Google Scholar]
- 25.Dale RC, Nasti JJ, Peters GB. Familial 7q21.3 microdeletion involving epsilon-sarcoglycan causing myoclonus dystonia, cognitive impairment, and psychosis. Mov Disord. 2011;26(9):1774–1775. doi: 10.1002/mds.23639. [DOI] [PubMed] [Google Scholar]
- 26.DeBerardinis RJ, et al. Myoclonus in a patient with a deletion of the epsilon-sarcoglycan locus on chromosome 7q21. Am J Med Genet A. 2003;121A(1):31–36. doi: 10.1002/ajmg.a.20162. [DOI] [PubMed] [Google Scholar]
- 27.Asmus F, et al. Genomic deletion size at the epsilon-sarcoglycan locus determines the clinical phenotype. Brain. 2007;130(Pt 10):2736–2745. doi: 10.1093/brain/awm209. [DOI] [PubMed] [Google Scholar]
- 28.Grunewald A, et al. Myoclonus-dystonia: significance of large SGCE deletions. Hum Mutat. 2008;29(2):331–332. doi: 10.1002/humu.9521. [DOI] [PubMed] [Google Scholar]
- 29.Carecchio M, et al. Defining the epsilon-sarcoglycan (SGCE) gene phenotypic signature in myoclonus-dystonia: A reappraisal of genetic testing criteria. Mov Disord. 2013;28(6):787–794. doi: 10.1002/mds.25506. [DOI] [PubMed] [Google Scholar]
- 30.Asmus F, et al. “Jerky” dystonia in children: spectrum of phenotypes and genetic testing. Mov Disord. 2009;24(5):702–709. doi: 10.1002/mds.22426. [DOI] [PubMed] [Google Scholar]
- 31.Kinugawa K, et al. Myoclonus-dystonia: an update. Mov Disord. 2009;24(4):479–489. doi: 10.1002/mds.22425. [DOI] [PubMed] [Google Scholar]
- 32.Asmus F, Gasser T. Inherited myoclonus-dystonia. Adv Neurol. 2004;94:113–119. [PubMed] [Google Scholar]
- 33.Asmus F, et al. Clinical differentiation of genetically proven benign hereditary chorea and myoclonus-dystonia. Mov Disord. 2007;22(14):2104–2109. doi: 10.1002/mds.21692. [DOI] [PubMed] [Google Scholar]
- 34.Borges V, et al. Novel and de novo mutations of the SGCE gene in Brazilian patients with myoclonus-dystonia. Mov Disord. 2007;22(8):1208–1209. doi: 10.1002/mds.21380. [DOI] [PubMed] [Google Scholar]
- 35.Trottenberg T, et al. Neurostimulation of the ventral intermediate thalamic nucleus in inherited myoclonus-dystonia syndrome. Mov Disord. 2001;16(4):769–771. doi: 10.1002/mds.1119. [DOI] [PubMed] [Google Scholar]
- 36.Nygaard TG, et al. Localization of a gene for myoclonus-dystonia to chromosome 7q21–q31. Ann Neurol. 1999;46(5):794–798. doi: 10.1002/1531-8249(199911)46:5<794::aid-ana19>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 37.Kurlan R, et al. Myoclonus and dystonia: a family study. Adv Neurol. 1988;50:385–389. [PubMed] [Google Scholar]
- 38.Thobois S, et al. Evidence for progressive changes in clinical presentation of myoclonus-dystonia. Mov Disord. 2007;22(10):1516–1517. doi: 10.1002/mds.21483. [DOI] [PubMed] [Google Scholar]
- 39.Roze E, et al. Myoclonus-dystonia: clinical and electrophysiologic pattern related to SGCE mutations. Neurology. 2008;70(13):1010–1016. doi: 10.1212/01.wnl.0000297516.98574.c0. [DOI] [PubMed] [Google Scholar]
- 40.Lin JP, et al. The impact and prognosis for dystonia in childhood including dystonic cerebral palsy: a clinical and demographic tertiary cohort study. J Neurol Neurosurg Psychiatry. 2014 doi: 10.1136/jnnp-2013-307041. doi: 10.1136/jnnp-2013-307041 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 41.Fahn S, Sjaastad O. Hereditary essential myoclonus in a large Norwegian family. Mov Disord. 1991;6(3):237–247. doi: 10.1002/mds.870060308. [DOI] [PubMed] [Google Scholar]
- 42.Saugier-Veber P, et al. Myoclonus dystonia plus syndrome due to a novel 7q21 microdeletion. Am J Med Genet A. 2010;152A(5):1244–1249. doi: 10.1002/ajmg.a.33369. [DOI] [PubMed] [Google Scholar]
- 43.Dale RC, Nasti JJ, Peters GB. Familial 7q21.3 microdeletion involving epsilon-sarcoglycan causing myoclonus dystonia, cognitive impairment, and psychosis. Mov Disord. 2011;26(9):1774–1775. doi: 10.1002/mds.23639. [DOI] [PubMed] [Google Scholar]
- 44.Han F, et al. Large deletions account for an increasing number of mutations in SGCE. Mov Disord. 2008;23(3):456–460. doi: 10.1002/mds.21895. [DOI] [PubMed] [Google Scholar]
- 45.Quinn NP. Essential myoclonus and myoclonic dystonia. Mov Disord. 1996;11(2):119–124. doi: 10.1002/mds.870110202. [DOI] [PubMed] [Google Scholar]
- 46.Dale RC, et al. Microdeletions detected using chromosome microarray in children with suspected genetic movement disorders: a single-centre study. Dev Med Child Neurol. 2012;54(7):618–623. doi: 10.1111/j.1469-8749.2012.04287.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.