Abstract
Inflammatory breast cancer (IBC) is a rare and highly invasive type of breast cancer, and patients diagnosed with IBC often face a very poor prognosis. IBC is characterized by the lack of primary tumor formation and the rapid accumulation of cancerous epithelial cells in the dermal lymphatic vessels. Given that normal epithelial cells require attachment to the extracellular matrix (ECM) for survival, a comprehensive examination of the molecular mechanisms underlying IBC cell survival in the lymphatic vessels is of paramount importance to our understanding of IBC pathogenesis. Here we demonstrate that, in contrast to normal mammary epithelial cells, IBC cells evade ECM-detachment-induced apoptosis (anoikis). ErbB2 and EGFR knockdown in KPL-4 and SUM149 cells, respectively, causes decreased colony growth in soft agar and increased caspase activation following ECM detachment. ERK/MAPK signaling was found to operate downstream of ErbB2 and EGFR to protect cells from anoikis by facilitating the formation of a protein complex containing Bim-EL, LC8, and Beclin-1. This complex forms as a result of Bim-EL phosphorylation on serine 59, and thus Bim-EL cannot localize to the mitochondria and cause anoikis. These results reveal a novel mechanism that could be targeted with innovative therapeutics to induce anoikis in IBC cells.
Inflammatory breast cancer (IBC) is a rare and highly invasive type of breast cancer, and patients diagnosed with IBC often face a very poor prognosis. The 5-year survival rate for patients with IBC is <40%, while the 5-year survival rate of all other breast cancers combined is approximately 90%.1, 2, 3, 4 This poor prognosis can be attributed to a number of factors, including the propensity for misdiagnosis of the disease due to its unique clinical presentation.5, 6, 7 In contrast to most breast cancers, IBC is characterized by the lack of discernible primary tumor formation and the accumulation of cancerous epithelial cells in the dermal lymphatic vessels.8 This lodging of IBC cells in the dermal lymphatics manifests as what appears to be inflammation, oftentimes causing clinicians to incorrectly diagnose the malady. Given that IBC cells are inherently aggressive, misdiagnosis is particularly problematic as a correct diagnosis or appropriate treatment is prolonged until more advanced disease is discovered. Thus it is imperative to gain a better understanding of the unique molecular mechanisms underlying IBC pathogenesis so that improved therapies can be designed to specifically eliminate IBC cells in a manner that improves patient outcome.
Unfortunately, few treatment options exist that are specifically designed to combat IBC. A review of nearly 400 IBC patients treated at The University of Texas MD Anderson Cancer Center between 1974 and 2005 demonstrated that there has been no significant improvement in prognosis for patients with IBC over the past 30 years.1 Many recent studies have focused on assessing the efficacy of chemotherapeutic regimens in IBC cells/patients where success had previously been observed only in the treatment of non-IBCs.9, 10 Some progress has been made in understanding the mechanisms underlying the invasive nature of IBC. For instance, Akt1 has been identified as a possible chemotherapeutic target that appears to be involved in the aggressive behavior of IBC cells.11 Other studies have identified RhoC, which is overexpressed in 90% of IBC tissue samples, as a potent oncogene contributing to IBC pathogenesis.11, 12, 13, 14, 15 More recently, evidence implicating the membrane protein TIG1 and the receptor tyrosine kinase Axl in the oncogenic behavior of IBC cells has been uncovered.16 However, despite these advances, knowledge of the biological mechanisms underlying IBC pathogenesis remains fairly rudimentary, and additional research dedicated to understanding the unique molecular pathways involved in IBC progression remains essential.
Given that IBC cells do not form a palpable primary tumor and instead flourish in suspension in the lymph of the dermal lymphatic vessels, we hypothesized that IBC cells must have an inherent ability to survive in the absence of attachment to the extracellular matrix (ECM). Normal mammary epithelial cells require attachment to the ECM to inhibit anoikis, which is defined as caspase-dependent cell death caused by ECM detachment.17 It has become clear that tumor progression and metastasis require cancer cells to inhibit anoikis, oftentimes through alterations in intracellular signaling pathways.18, 19, 20 Interestingly, previous studies have shown that ErbB2 and EGFR, which are hyperactivated in a substantial percentage of IBC patients,21 can effectively antagonize the anoikis program to facilitate anchorage-independent growth.22, 23, 24, 25, 26, 27, 28 However, a detailed examination of the molecular mechanisms underlying anoikis inhibition in IBC cells has yet to be completed. In this study, we demonstrate that signaling from EGFR and ErbB2 through ERK/MAPK has a major role in the ability of IBC cells to survive in the absence of ECM attachment. Surprisingly, we have discovered that ERK-mediated anoikis suppression in IBC cells is not due to targeting of the pro-apoptotic protein Bim-EL for degradation that has previously been observed in mammary epithelial cells.23, 27 Rather, ERK activation in IBC cells promotes the formation of a protein complex containing Bim-EL, Beclin-1, and LC8, which functions to sequester Bim-EL from the mitochondria and thereby block anoikis. In support of the importance of this signaling pathway in IBC patients, five of the seven IBC patient samples assayed showed discernible Bim-EL staining. Collectively, these data identify a novel mechanism utilized by IBC cells to survive during ECM detachment and reveal a potential target for the development of anoikis-inducing chemotherapeutics targeting IBC cells.
Results
ErbB2/EGFR-mediated survival of IBC cells in the absence of ECM attachment
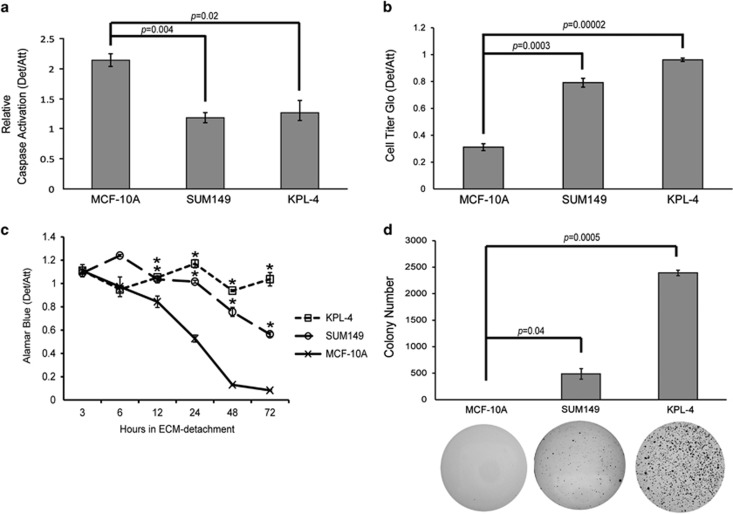
In order to determine whether IBC cells are able to block the induction of caspase activation when detached from the ECM, we obtained two IBC cell lines (SUM149 and KPL-4) and assayed caspase activation after plating the cells on non-adherent plates. As we hypothesized, both cell lines were significantly resistant to the induction of caspase activation when compared with the anoikis compliant non-transformed MCF-10A cell line (Figure 1a). To confirm that these differences in caspase activation ultimately translated to changes in cellular viability, we utilized Cell Titer Glo and alamarBlue assays to assess the viability of ECM-detached IBC cells over time. Indeed, the viability of ECM-detached KPL-4 or SUM149 cells was largely maintained, even under extended periods of ECM detachment (Figures 1b and c). To further corroborate the ability of IBC cells to survive during ECM detachment, we evaluated whether IBC cells could promote anchorage-independent growth in soft agar. In contrast to non-transformed MCF-10A cells, both SUM149 and KPL-4 cells were able to robustly grow in an anchorage-independent manner (Figure 1d).
Figure 1.
IBC cells survive in ECM-detached conditions. (a) Cells were plated on either normal or poly-HEMA coated plates, and caspase activation was measured at 48 h using the Caspase-Glo assay. (b) Cells were plated in attached or detached conditions, and cell viability was measured at 48 h post-plating via Cell Titer Glo Assay. (c) Cells were plated on either normal or poly-HEMA coated plates, and cell viability was measured at the indicated times via alamarBlue assay. (d) MCF-10A and SUM149 cells were plated at 100 000 cells/well, and KPL-4 cells were plated at 60 000 cells/well in soft agar. Colonies were stained at either 14 days (KPL-4) or 30 days (SUM149, MCF-10A) using 0.01% INT-violet and quantified with ImageJ. Error bars represent S.E.M. *P-value<0.05 compared with MCF-10A
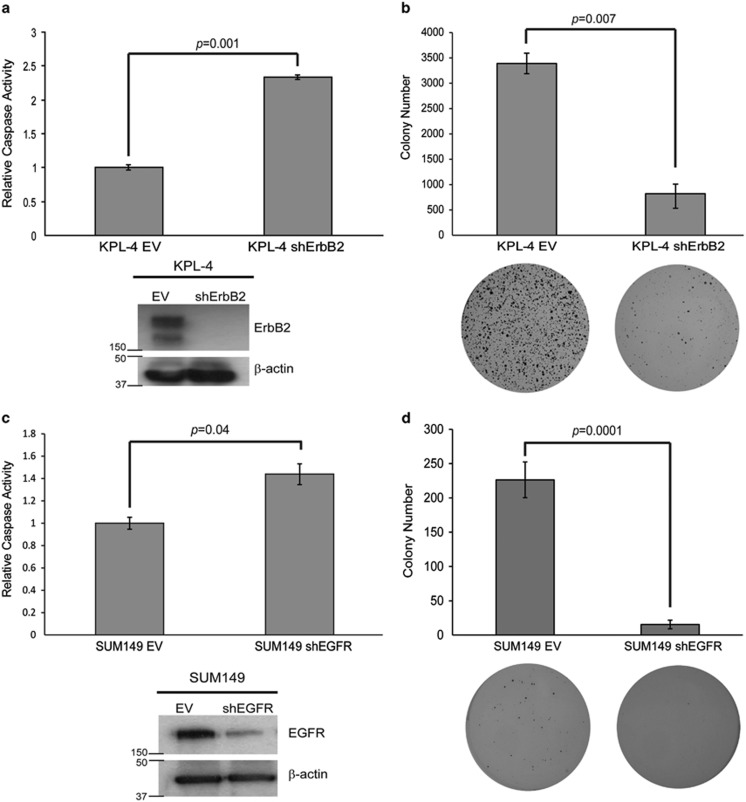
Given that IBC cells have been previously shown to have hyperactive ErbB2 or EGFR signaling,29 we hypothesized that oncogenic signaling from these receptors may be involved in the resistance to anoikis in IBC cell lines. KPL-4 cells are well known to overexpress the ErbB2 oncogene.30, 31 To determine whether this ErbB2 overexpression is significant for their ability to evade anoikis, we utilized lentiviral transduction of shRNA to generate a KPL-4 cell line that was deficient in ErbB2 (KPL-4 shErbB2) (Figure 2a). In comparison to KPL-4 cells that were transduced with an empty vector shRNA construct (KPL-4 EV), KPL-4 shErbB2 cells were significantly more susceptible to anoikis induction (Figure 2a). In addition, KPL-4 shErbB2 cells were substantially compromised in their ability to maintain anchorage-independent growth when compared with KPL-4 EV cells (Figure 2b). These data suggest that ErbB2 is necessary for the ability of KPL-4 cells to block the anoikis program.
Figure 2.
ErbB2 and EGFR are necessary for anoikis protection in KPL-4 and SUM149 cells, respectively. (a) KPL-4 cells were transduced with a lentivirus containing either an empty vector (EV) or an ErbB2 shRNA-containing vector (shErbB2), and western blotting analysis confirmed the knockdown. KPL-4 EV and shErbB2 cells were plated on poly-HEMA-coated plates, and caspase activation was quantified at 48 h as previously described. (b) KPL-4 EV and shErbB2 cells were plated at 60 000 cells/well in soft agar as described above. Colonies were stained at 14 days with 0.01% INT-violet and quantified with ImageJ. (c) SUM149 cells were transduced with a lentivirus containing either an empty vector (EV) or an EGFR shRNA-containing vector (shEGFR), and western blotting analysis confirmed the knockdown. SUM149 EV and shEGFR cells were plated on poly-HEMA-coated plates, and caspase activation was quantified at 48 h as previously described. (d) SUM149 EV and shEGFR cells were plated at 100 000 cells/well in soft agar as previously described. Colonies were stained after 30 days using 0.01% INT-violet and quantified with ImageJ. Error bars represent S.E.M.
In contrast to KPL-4 cells, SUM149 cells contain no discernible ErbB2 expression but do express high levels of EGFR.31 As we did with ErbB2 in KPL-4 cells, we utilized lentivirus to deliver shRNAs targeting EGFR to SUM149 cells and were able to achieve appreciable reduction in EGFR protein levels (Figure 2c). Much like we observed with ErbB2 knockdown in KPL-4 cells, the reduction of EGFR expression by shRNA in SUM149 cells led to a significant increase in anoikis induction (Figure 2c) and a considerable loss of anchorage-independent growth in soft agar (Figure 2d). These data suggest that EGFR is necessary for anoikis inhibition in SUM149 cells and, coupled with our data in KPL-4 cells, our results intimate that IBC cells can utilize signaling from multiple receptor tyrosine kinases to eliminate the induction of anoikis and facilitate anchorage-independent growth.
ERK/MAPK signaling is necessary for anoikis protection in IBC cells
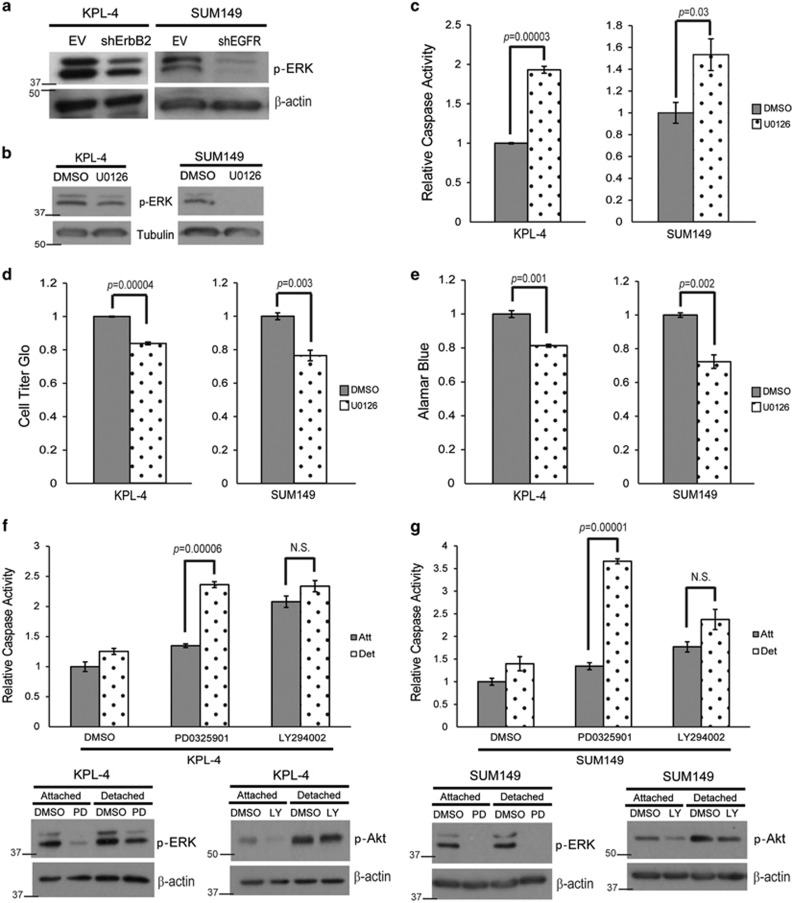
Given the respective requirement for ErbB2 or EGFR signaling in KPL-4 and SUM149 cells, we sought to determine whether there is common downstream signaling in these two cell lines that impacts the induction of anoikis. Previous studies have revealed a role for the ERK/MAPK pathway in the evasion of anoikis.21, 24, 27, 32 In order to assess the activation of ERK signaling in IBC cells, we examined phospho-ERK levels in KPL-4 cells deficient in ErbB2 or SUM149 cells deficient in EGFR. In each case, we discovered that the deficiency in receptor tyrosine kinase expression (i.e., ErbB2 in KPL-4 and EGFR in SUM149) resulted in a concomitant loss of phospho-ERK (Figure 3a). These data raise the possibility that ERK signaling is required for anoikis evasion in IBC cells downstream of aberrant receptor tyrosine kinase signaling. In order to address this possibility, we treated IBC cells with U0126, a MEK inhibitor, and found that ERK signaling was necessary for anoikis inhibition in both KPL-4 and SUM149 cells (Figures 3b and c). Cell viability assays confirmed the increase in caspase activation corresponded to decreased viability (Figures 3d and e). Interestingly, the ability of MEK inhibition to induce apoptosis is specific to ECM-detached cells as minimal caspase activation is observed when MEK is inhibited in ECM-attached cells (Figures 3f and g). These data stand in stark contrast to other survival pathways, as PI(3)K inhibition can equivalently induce caspase activation in both ECM-attached and ECM-detached IBC cells (Figures 3f and g).
Figure 3.
ERK/MAPK signaling is necessary for anoikis protection. (a) KPL-4 EV and shErbB2 and SUM149 EV and shEGFR cells were plated on poly-HEMA-coated plates for 48 h, and p-ERK levels were measured via immunoblotting. (b and c) KPL-4 and SUM149 cells were plated on poly-HEMA-coated plates in the presence of DMSO or U0126 (10 μM). Caspase activation was measured at 48 h as previously described. Inhibition of the ERK/MAPK pathway was confirmed via western blotting analysis. (d and e) KPL-4 and SUM149 cells were plated in ECM detachment with DMSO or U0126 (10 μM). Cellular viability was measured with the Cell Titer Glo Assay (d) or alamarBlue assay (e). (f and g) KPL-4 (f) and SUM149 (g) cells were plated in attached or detached conditions with DMSO, PD0325901 (1 μM), or LY294002 (25 μM). Caspase activation was measured at 24 h. Cell lysates were prepared and normalized, and protein levels were analyzed via western blotting analysis to confirm inhibitor efficacy. Error bars represent S.E.M. NS, not signifcant
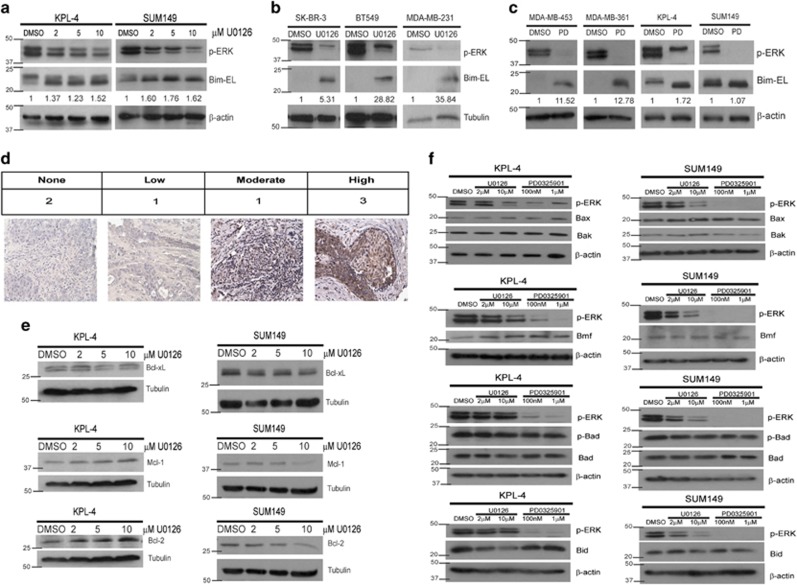
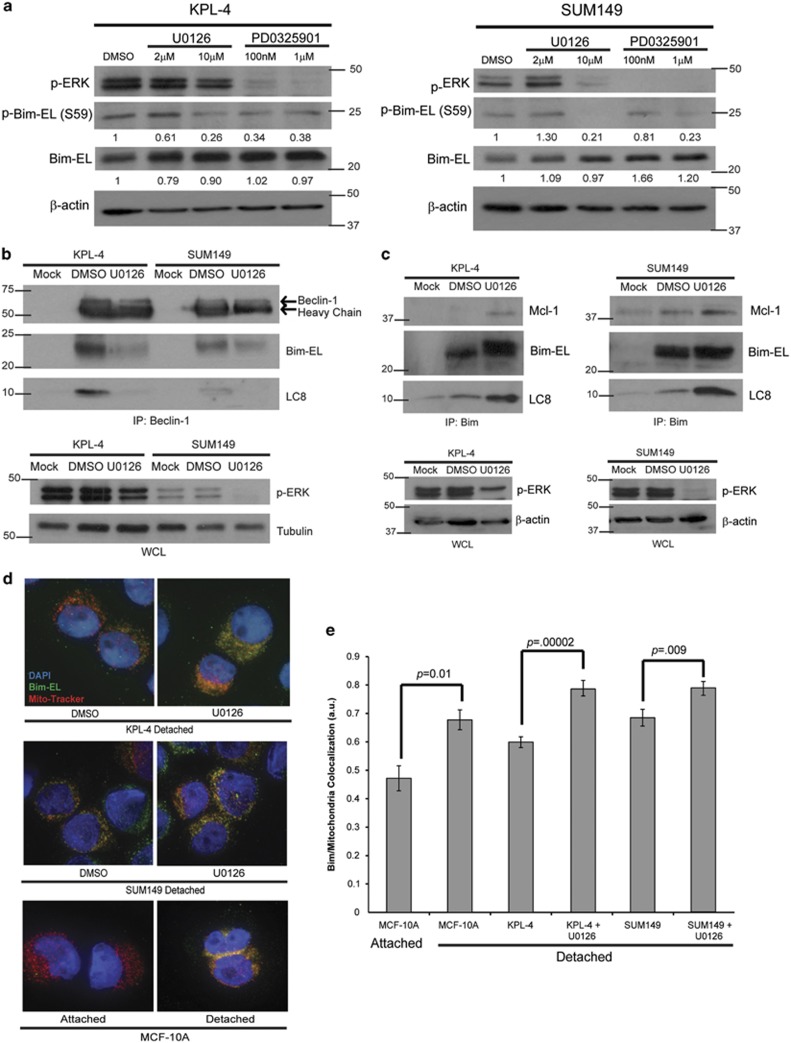
Due to the fact that ERK-mediated protection from anoikis in other cell lines has previously been shown to involve targeting of the pro-apoptotic protein Bim-EL for degradation,23 we investigated the protein levels of Bim-EL in IBC cells that have been treated with increasing doses of U0126. Much to our surprise, we found that, contrary to published reports in other cell types, inhibition of the ERK pathway did not result in dramatically increased levels of Bim-EL. Rather, in both IBC cell lines, Bim-EL levels were maintained at high levels despite effective inhibition of ERK signaling by U0126 treatment (Figure 4a). To ascertain whether this mechanism was specific to IBC cells, we conducted similar experiments with other breast cancer cell lines (non-IBC) and found that in these cell lines (SK-BR-3, BT549, and MDA-MB-231 cells), Bim-EL levels increased dramatically upon effective inhibition of ERK by U0126 (Figure 4b). To expand on these findings, we extended these studies into MDA-MB-453 (453) and MDA-MB-361 (361) cells, which have previously been published to have high basal levels of Bim-EL protein.33 However, the basal Bim-EL levels observed in the IBC cell lines are considerably elevated when compared with basal levels of Bim-EL in the 453 and 361 cells (Figure 4c). Additionally, in contrast to the IBC cell lines, ERK inhibition was sufficient to promote a significant increase in Bim-EL protein in the 453 and 361 cell lines (Figure 4c). As these data suggest that IBC cells have high basal levels of Bim-EL, we investigated whether specimens from IBC patients also had high basal Bim-EL levels. Indeed, our studies revealed that five of the seven IBC patient samples showed discernable Bim-EL staining (Figure 4d).
Figure 4.
MEK inhibition does not affect Bcl-2 family member protein levels in IBC cells. (a) Cells were plated on poly-HEMA-coated plates with DMSO or increasing amounts of U0126 for 48 h, and protein content was analyzed via immunoblotting. (b) Cells were plated in ECM-detached conditions with DMSO or U0126 (10 μM) for 48 h. Cell lysates were prepared for immunoblotting. (c) Cells were plated on poly-HEMA-coated plates with DMSO or PD0325901 (1 μM) for 48 h, and lysates were prepared for immunoblotting. (d) Seven IBC patient tissue samples were obtained, and IHC was utilized to visualize Bim protein levels in the tissue. Images were blind scored for staining intensity. (e) Cells were plated in detachment and treated with DMSO or increasing amounts of U0126 for 48 h. Lysates were prepared and normalized, and protein levels were analyzed via immunoblotting. (f) KPL-4 and SUM149 cells were plated in detachment with the indicated doses of U0126 or PD0325901 for 48 h. Protein levels were analyzed via western blotting
Given the high basal levels of Bim-EL in IBC cells, we thought that perhaps IBC cells could evade anoikis and counter high Bim-EL expression by expressing high levels of other anti-apoptotic proteins. Although we did see a minimal loss of Mcl-1 and Bcl-2 at high doses of U0126 in SUM149 cells, these proteins actually seemed to slightly increase at high doses of U0126 in KPL-4 cells (Figure 4e). No appreciable changes were observed in Bcl-xL in either cell line upon U0126 treatment. Furthermore, expression or activation of pro-apoptotic Bcl-2 family members, including Bax, Bak, Bmf, phospho-Bad (S75), total Bad, and Bid, are not consistently altered in both cell lines following treatment with MEK inhibitors (Figure 4f). Thus our data suggest that there are no alterations in the protein levels of these Bcl-2 family members that would explain the ERK-mediated inhibition of anoikis seen in both IBC cell lines. In addition, these data suggest that IBC cells utilize ERK signaling to evade anoikis through a mechanism that is distinct from other breast cancer cells.
ERK signaling regulates Bim-EL localization to prevent anoikis
The inherently high levels of Bim-EL (that were not modulated by ERK signaling) observed in IBC cells led us to question how IBC cells could tolerate these levels of Bim-EL without undergoing anoikis. Upon re-examining our data demonstrating stable Bim-EL levels in IBC cells, we noticed a slight shift in the molecular weight of Bim-EL upon U0126 treatment (Figure 4a). This shift is consistent with the loss of a phosphorylation event on Bim-EL, and Bim-EL has previously been shown to be a target of ERK phosphorylation.34, 35, 36 Therefore we assayed the phosphorylation of Bim-EL in IBC cells using a phospho-specific antibody. When ERK was inhibited by U0126 or PD0325901 treatment in KPL-4 or SUM149 cells, we observed a significant and dose-dependent loss of Bim-EL phosphorylation at serine 59 (S59) (Figure 5a).
Figure 5.
ERK/MAPK signaling leads to the sequestration of Bim-EL from the mitochondria to prevent anoikis. (a) Cells were plated on poly-HEMA-coated plates with DMSO or the indicated dose of U0126 or PD0325901 for 6 h. Lysates were prepared and normalized, and protein levels were analyzed via western blotting. (b) Cells were plated in detachment and treated with DMSO or U0126 (10 μM) for 3 h. Cell lysates were prepared, and immunoprecipitation was performed with a Beclin-1 antibody. Western blotting analysis was utilized to identify interacting proteins and confirm equivalent protein content across samples. (c) Cells were plated in detachment and treated with DMSO or U0126 (10 μM) for 3 h. Cell lysates were prepared and normalized, and immunoprecipitation was performed with a Bim antibody. Interacting proteins were identified via immunoblotting, and equivalent protein content across samples was confirmed. (d and e) Cells were plated on poly-HEMA-coated plates and treated with either DMSO or U0126 (10 μM) and 20 μM z-VAD-fmk for 24 h. Cells were fixed and stained with Mito-Tracker Red (200 nM), DAPI (5 μg/ml), and Bim-EL and imaged using an Applied Precision DeltaVision OMX fluorescent microscope. Co-localization was measured using the Applied Precision softWoRx software. Error bars represent S.E.M.
The role of ERK-mediated phosphorylation of Bim-EL at S59 is not well understood. Interestingly, previous studies have revealed that the S59 site on Bim-EL is in close proximity to a region on Bim-EL that has previously been shown to be critical for its interaction with LC8.37 This interaction of Bim-EL and LC8 has been more recently shown to also involve binding to Beclin-1, a critical mediator of autophagy.38 The formation of this Bim-EL–LC8–Beclin-1 complex has been proposed to block the pro-apoptotic activity of Bim-EL by sequestering it away from the mitochondria. Given these data, we hypothesized that ERK-mediated phosphorylation of Bim-EL at S59 in IBC cells could block anoikis by promoting the formation of the Bim-EL–LC8–Beclin-1 complex. Indeed, utilizing immunoprecipitation of endogenous proteins, we discovered that Bim-EL, LC8, and Beclin-1 did co-precipitate in both KPL-4 and SUM149 cells (Figure 5b). When ERK was inhibited using U0126, these proteins failed to co-precipitate suggesting that ERK signaling is necessary for the formation of the Bim-EL–LC8–Beclin-1 complex in IBC cells (Figure 5b). Furthermore, MEK inhibition resulted in enhanced binding of Bim-EL to Mcl-1 (Figure 5c), suggesting that when Bim-EL is released from this complex it can properly antagonize anti-apoptotic Bcl-2 family members. Interestingly, LC8 co-precipitates with Bim-EL in the presence and absence of U0126, suggesting that LC8 remains bound to Bim-EL following its dissociation from Beclin-1 (Figure 5c).
As mentioned above, the interaction of Bim-EL with LC8 and Beclin-1 has been proposed to abrogate apoptosis by sequestering Bim-EL from the mitochondria. We assessed the ability of Bim-EL to localize to the mitochondria in IBC cells in the presence or absence of ERK signaling. To investigate this, we examined the subcellular localization of Bim-EL relative to the mitochondria via immunofluorescence. Upon staining ECM-detached IBC cells with anti-Bim-EL and MitoTracker, we discovered that Bim-EL localization at the mitochondria was significantly enhanced when ERK was inhibited (representative pictures in Figure 5d). To quantitate this effect, we determined the Pearson's correlation coefficient for the co-localization of Bim-EL and mitochondria and found a statistically significant change in the ability of Bim-EL to localize to the mitochondria upon U0126 treatment (Figure 5e). In aggregate, these data suggest that ERK-mediated phosphorylation of Bim-EL promotes the interaction of Bim-EL with LC8 and Beclin-1 and the subsequent sequestration of Bim-EL away from Mcl-1 and the mitochondria.
Phosphorylation of Bim-EL at S59 is responsible for anoikis evasion in IBC cells
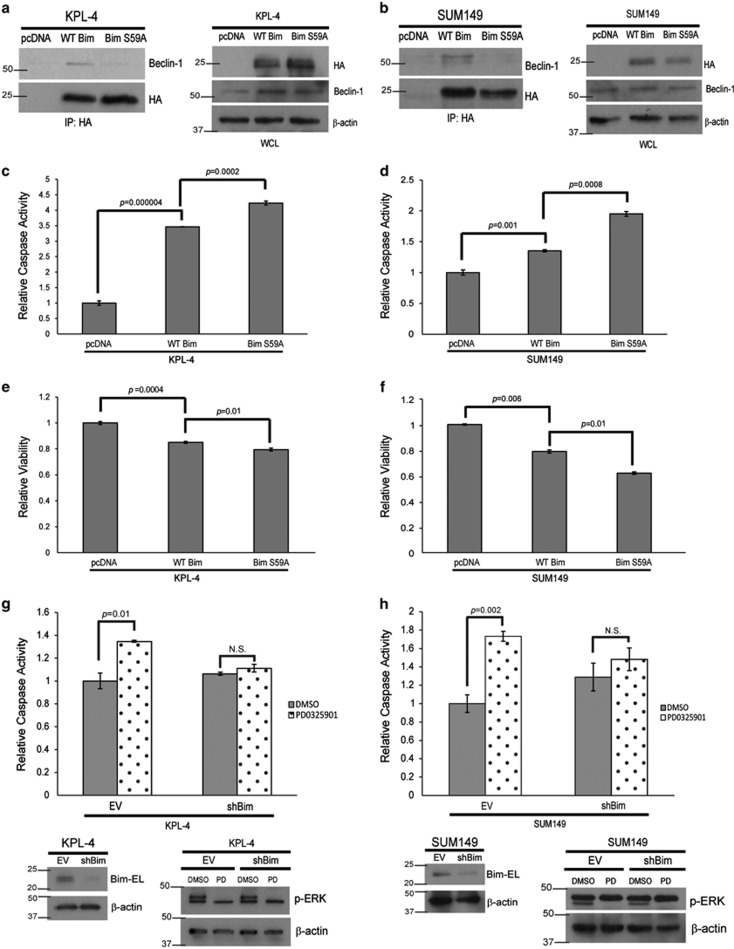
Our results above support a model in which ERK-mediated phosphorylation of Bim-EL at S59 is responsible for its interaction with LC8 and Beclin-1 and its ultimate sequestration from the mitochondria. To more directly assess the contribution of serine 59 phosphorylation, we utilized an HA-tagged Bim-EL S59A construct and expressed this construct or HA-tagged wild-type Bim-EL in IBC cells. Upon immunoprecipitation using the HA tag, we found that the interaction of Bim-EL with Beclin-1 and LC8 was markedly compromised in the presence of the Bim S59A mutation in both KPL-4 (Figure 6a) and SUM149 (Figure 6b) cells. These data suggest that ERK-mediated phosphorylation of Bim-EL at S59 is required for the formation of the Bim-EL–LC8–Beclin-1 complex in IBC cells. In addition, these data suggest that the expression of HA-Bim S59A in IBC cells may function in a dominant-negative manner to promote anoikis by failing to interact with LC8 and Beclin-1 and thus localizing to the mitochondria. Indeed, this is the case, as we see a sizeable increase in the ability of HA-tagged Bim-EL S59A to induce anoikis when compared with HA-tagged wild-type Bim-EL in KPL-4 (Figure 6c) and SUM149 (Figure 6d) cells. Furthermore, the expression of HA-tagged Bim-EL S59A led to a significant reduction in viability compared with HA-tagged wild-type Bim-EL in both KPL-4 (Figure 6e) and SUM149 (Figure 6f) cells. In order to confirm that Bim-EL is necessary for anoikis upon MEK inhibition, we engineered KPL-4 and SUM149 cells to be deficient in Bim-EL expression (shBim). When these cells were treated with a MEK inhibitor, the induction of anoikis was completely abrogated (Figures 6g and h), suggesting that the modulation of Bim-EL by ERK is necessary for anoikis induction in IBC cells.
Figure 6.
Bim is necessary and sufficient for anoikis in IBC cells (a and b) KPL-4 (a) and SUM149 (b) cells were transfected with pcDNA EV, HA-WT Bim, or HA-Bim S59A constructs using Lipofectamine LTX. Lysates were prepared and normalized. Coimmunoprecipitation experiments were conducted with an HA antibody. Immunoblotting was used to identify interacting proteins. (c and d) KPL-4 (c) and SUM149 (d) cells were transfected with pcDNA EV, HA-WT Bim, or HA-Bim S59A constructs using Lipofectamine LTX. Cells were plated in suspension and caspase activity was measured as described above. (e and f) KPL-4 (e) and SUM149 (f) cells were transfected as previously described. Cells were plated in suspension, and cellular viability was measured using Cell Titer Glo. (g and h) KPL-4 (g) and SUM149 (h) cells were transduced with shRNA-containing lentivirus to create stable Bim knockdowns (shBim). EV and shBim cells were plated in ECM detachment and treated with DMSO or PD0325901 (1 μM). Caspase activation was measured at 48 h, and western blotting analysis confirmed the Bim knockdown and inhibitor efficacy. NS, not signifcant
Discussion
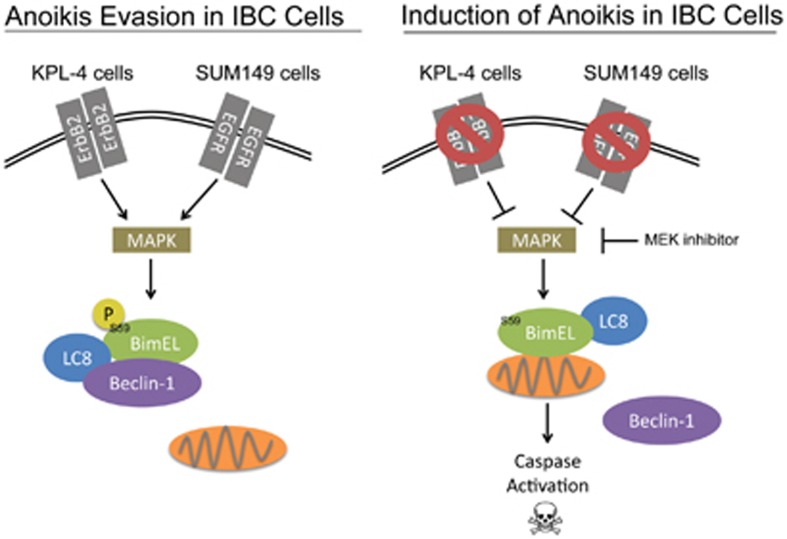
The prognosis for patients diagnosed with IBC remains poor due to the unusual presentation of the disease and the inherently aggressive nature of the cancer cells. A better understanding of the molecular mechanisms and signal transduction cascades that are utilized by IBC cells to enhance their oncogenic capabilities has the potential to unveil novel targets for the development of IBC-specific therapies. Here we uncover a molecular mechanism utilized by IBC cells to inhibit the induction of anoikis (see model in Figure 7). Anti-apoptotic signaling in IBC cells has been studied in the past, most prominently in the context of chemotherapeutic resistance.39, 40 A more recent example is the receptor tyrosine kinase Axl, which has recently been shown to have a significant role in the malignant behavior of IBC cells16 and has been previously revealed to block the induction of apoptosis through Akt and Bcl-xL.41 However, our study is the first (to our knowledge) to directly address the molecular mechanisms that allow IBC cell survival in the context of ECM detachment. In addition, our findings suggest that targeting Bim-EL in a manner that promotes mitochondrial localization may be effective in the elimination of ECM-detached IBC cells.
Figure 7.
Model for anoikis evasion in IBC cells
Targeting cancer cells for elimination through the modulation of Bcl-2 family members (like Bim-EL) has been heavily investigated for a number of years.42 However, the high basal levels of Bim-EL that we detect in IBC cells suggest an inherent vulnerability that could be exploited chemotherapeutically. Recent studies suggest that this approach may be broadly effective in both IBC and non-IBC cells that have high basal levels of Bim-EL, as the effectiveness of targeting cancer cells with kinase inhibitors has been shown to strongly correlate with Bim-EL expression.33 Our data provide a mechanistic rationale for these results and suggest that targeting ErbB2 or EGFR in IBC cells could be particularly effective given that the elevated Bim-EL expression could be rapidly targeted to the mitochondria upon kinase inhibition. Furthermore, recent studies have demonstrated that triple-negative breast cancer cells can be efficiently targeted by sequential treatment of an inhibitor of oncogenic signaling pathways followed by standard chemotherapy that causes DNA damage.43 In fact, Bim-EL is mentioned in this report as a protein that is differentially expressed using this sequential treatment pattern. Our data suggest that a similar strategy to target IBC cells (or other cancer cell lines with elevated basal Bim-EL) could be effective.
In addition, our data strongly implicate a role for ERK-mediated phosphorylation of S59 in the sequestration of Bim-EL from the mitochondria in IBC cells. These results raise some interesting questions about the regulation of Bim-EL in IBC cells. Why are basal levels of Bim-EL so high in IBC cells? Typically, Bim-EL is rapidly targeted to the proteasome in the presence of ERK signaling.44 Perhaps the phosphorylation on S59 and subsequent recruitment to LC8/Beclin-1 could impede the ability of E3 ligases to target Bim-EL for degradation. In addition, why does ERK signaling in IBC cells lead to phosphorylation at S59 as opposed to other phosphorylation sites? Previous studies have revealed that ERK-mediated phosphorylation of Bim-EL at S69 leads to proteasomal degradation.44 Interestingly, we do see evidence of S69 phosphorylation in IBC cell lines (data not shown), yet Bim-EL is clearly not targeted to the proteasome. It seems possible that an initial phosphorylation event on S69 is required for the phosphorylation on S59 to occur. Previous data in mammalian cell culture support this model, as an S69A mutation (but not an S59A mutation) has been shown to abrogate any ERK-mediated Bim-EL phosphorylation.45 These and other alternative possibilities will need to be rigorously assessed in future studies.
The fact that Beclin-1 is part of the complex sequestering Bim-EL from the mitochondria raises other interesting questions about the fate of ECM-detached IBC cells. Given that the interaction of Beclin-1 with LC8 and Bim-EL has been shown to prevent Beclin-1 from initiating autophagosome formation,38 it seems reasonable to speculate that ECM-detached IBC cells are defective in autophagy in addition to anoikis. This presents a unique problem for these cells, as autophagy has previously been shown to be critical for the survival of ECM-detached cells,46, 47 presumably to facilitate nutrient consumption. Perhaps this defect could be overcome through changes in glucose or fatty acid metabolism that have previously been reported in ECM-detached cells.24, 48 Thus it will be especially important to examine metabolic changes in IBC cells to understand how these cells deal with metabolic needs during ECM detachment. Nonetheless, the data presented here provide significant and novel insight into the molecular mechanisms utilized by IBC cells to survive in the absence of ECM attachment. We believe our data pave the way for future studies utilizing these findings aimed at the elimination of IBC cells through the modulation of Bim-EL localization.
Materials and Methods
Cell culture
MCF-10A cells were grown in DMEM/F12 media (Life Technologies, Grand Island, NY, USA) with 5% horse serum (Life Technologies) and supplemented with 10 μg/ml insulin, 0.5 mg/ml hydrocortisone, 100ng/ml cholera toxin, 20 ng/ml epidermal growth factor (EGF), and 1% penicillin/streptomycin (Life Technologies). KPL-4, MDA-MB-231, MDA-MB-453, and MDA-MB-361 cells were grown in DMEM (Life Technologies) with 10% fetal bovine serum with 1% penicillin/streptomycin. SUM149 cells were cultured in F12 media (Life Technologies) with 5% fetal bovine serum and 1% penicillin/streptomycin. SK-BR-3 cells were grown in McCoy's media (Life Technologies) with 10% fetal bovine serum and 1% penicillin/streptomycin. BT549 cells were grown in RPMI media (Life Technologies) with 10% fetal bovine serum and 1% penicillin/streptomycin.
Reagents
The following reagents were used at the doses indicated in the figure legends: U0126 (Millipore, Billerica, MA, USA), z-VAD-fmk (Millipore), PD0325901 (Sigma-Aldrich, St. Louis, MO, USA), LY294002 (Millipore), and iodonitrotetrazolium violet-farmazan (INT-violet) (Sigma-Aldrich).
Caspase assays
Caspase activation was measured using the Caspase-Glo 3/7 Assay Kit (Promega, Madison, WI, USA). Cells were plated at 13 000 cells per well in white-bottom 96-well plates coated with 6 mg/ml poly-HEMA. Caspase activation was measured after the time indicated in the figure legend according to the kit protocol. Relative caspase activity was determined by dividing the raw value for caspase activation by the raw value for the control. Error bars represent S.E.M., and P-values were calculated using a two-tailed t-test. Figures show representative data from at least three individual replicates.
Cell viability assays
Cell viability was measured using the alamarBlue assays (Life Technologies) or Cell Titer Glo assays (Promega). Cells were plated at 13 000 cells per well in white-bottom 96-well plates with or without 6 mg/ml poly-HEMA coating. Cell viability was measured at the indicated time after plating according to the manufacturer's instructions. Error bars represent S.E.M., and P-values were calculated using a Student's two-tailed t-test. Figures show representative data from three replicates.
Soft agar assays
Cells (number indicated in figure legends) were added to 1.5 ml of growth media plus 0.4% low-melt agarose (Sigma-Aldrich) and layered onto a 2-ml bed of growth media plus 0.5% low-melt agarose. Media was changed every other day and imaged after the indicated times. Colonies were stained with INT-violet and quantified using ImageJ (National of Institutes of Health, Bethesda, MD, USA). The figures show representative experiments from at least three independent replicates. Error bars represent S.E.M., and P-values were calculated using a two-tailed t-test.
Immunoblotting
Cell lysates were prepared in 1% NP-40 with 1 μg/ml aprotinin, 5 μg/ml leupeptin, 20 μg/ml phenylmethylsulfonyl fluoride (PMSF), and HALT Protease and Phosphatase Inhibitor Cocktail (Pierce Biotechnology, Rockford, IL, USA) and cleared by centrifugation. Lysates were normalized using a BCA assay (Pierce Biotechnology). The following antibodies were used for immunoblotting: β-actin (Sigma-Aldrich), Bcl-2 (Millipore), Bcl-XL (Cell Signaling, Danvers, MA, USA), Mcl-1, (Millipore), ERK1&2 (pTpY185/187) (Life Technologies), Bim-EL (C34C5) (Cell Signaling), α-Tubulin (Sigma-Aldrich), Beclin-1 (Cell Signaling), LC8/DYNLL1 (Abcam, Cambridge, MA, USA), Bim-EL (pS55) (Millipore), HA-tag (6E2) (Cell Signaling), Akt (pS473) (Cell Signaling), Bax (Cell Signaling), Bak (Cell Signaling), Bmf (Cell Signaling), Bad (pS112) (Cell Signaling), Bad (Cell Signaling), and Bid (Cell Signaling). Figures show representative blots from at least three individual replicates. Densitometry was completed using Adobe Photoshop (Adobe Systems, San Jose, CA, USA) and ImageJ.
Immunofluorescence
Cells were plated in attached or detached conditions. Where indicated, MitoTracker Red CMXROS (Life Technologies) (200 nM) was used to stain mitochondria before collecting cells. Cells were deposited on slides with a Shandon Cytospin3 (Thermo Scientific, Waltham, MA, USA) at 800 r.p.m. for 5 min. Cells were fixed in 4% paraformaldehyde for 20 min at room temperature and permeabilized in 0.5% Triton-X in PBS for 10 min at 4 °C. Cells were washed with 100 mM glycine in PBS three times. Slides were blocked in IF buffer (130 mM NaCl, 7 mM Na2HPO4, 3.5 mM NaH2PO4, 7.7 mM NaH3, 0.1% bovine serum albumin, 1.2% Triton X-100, 0.05% Tween-20) and 10% goat serum (Life Technologies). Slides were stained with a Bim-EL (Cell Signaling) antibody diluted 1 : 150 in IF buffer with 10% goat serum and an Alexa Fluor 488 secondary antibody diluted 1 : 200 in IF buffer with 10% goat serum. Nuclei were stained with 5 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) and dihydrochloride (Life Technologies). Slides were mounted with ProLong Gold Antifade Reagent (Life Technologies). Imaging was completed using an Applied Precision DeltaVision OMX fluorescent microscope (Applied Precision, GE Healthcare, Issaquah, WA, USA). Colocalization measurements were determined using Applied Precision softWoRx software. Error bars represent S.E.M., and P-values were calculated using a two-tailed t-test. Images show representative images from three individual experiments. For the colocalization calculations, n=10–30.
shRNA lentiviral transduction
MISSION shRNA against ErbB2 (NM_004448; Clone number: TRCN0000010342), EGFR (NM_005228; Clone number: TRCN0000039634), and Bim (NM_138621; Clone number: TRCN0000355975) in the puromyocin-resistant pLKO.1 vector were purchased from Sigma-Aldrich along with a control empty vector pLKO.1 vector. HEK293T cells were transfected with 0.5 μg of pLKO.1 vector along with packaging vectors pCMV-D8.9 (0.5 μg) and pCMV-VSV-G (60ng) with PLUS reagent and Lipofectamine 2000 (Life Technologies). Virus was collected at 24 and 48 h posttransfection, filtered, and used for transduction of KPL-4 and SUM149 cells in the presence of 8 μg/ml polybrene. Transduced cells were selected with 2 μg/ml puromycin for 2 weeks. Knockdown efficiency was measured by immunoblotting.
Immunoprecipitation
Cells were plated on poly-HEMA-coated plates for 3 h with DMSO or 10 μM U0126. Cells were lysed in IP lysis buffer (1% Triton X-100, 50 mM NaCl, 1 mM EDTA, 20 mM HEPES, 1 μg/ml aprotinin, 5 μg/ml leupeptin, 20 μg/ PMSF, and HALT Protease and Phosphatase Inhibitor Cocktail (Pierce Biotechnology)) and cleared by centrifugation. Lysates were normalized with a BCA assay (Pierce Biotechnology) and precleared with Protein A Sepharose Fast Flow beads (GE Healthcare). One microgram of Beclin-1 antibody (Cell Signaling) or Bim antibody (Cell Signaling) was added to lysates and incubated overnight. Protein A Sepharose Fast Flow beads were used to capture the antibody, and the beads were washed with IP lysis buffer. Proteins were eluted from beads and subjected to immunoblotting. Figures show representative blots from at least three individual replicates. Mock immunoprecipitations were used as controls and were completed according to the protocol above without antibody addition.
Transient transfections
pcDNA3 empty vector (pcDNA), pcDNA3-wild type Bim-EL (WT Bim), and pcDNA3-Bim-EL S55A (Bim S55A) constructs were a kind gift from Hisashi Harada. The constructs encode for mouse Bim-EL, and the serine sites correspond with the analogous human serine sites (mouse serine 55 is equivalent to human serine 59, etc.). Cells were transfected with 10 μg of DNA (pcDNA3, WT Bim, and Bim S59A) using Lipofectamine LTX and PLUS reagent (Life Technologies) according to the manufacturer's manual. Four hours (KPL-4 cells) or 12 h (SUM149 cells) after transfection, cells were lysed in IP buffer (1% Triton X-100, 150 mM NaCl, 1 mM EDTA, 20 mM HEPES, 1 μg/ml aprotinin, 5 μg/ml leupeptin, 20 μg/ml PMSF, and HALT Protease and Phosphatase Inhibitor Cocktail (Pierce Biotechnology)) and cleared by centrifugation. Lysates were normalized with a BCA assay (Pierce Biotechnology) and precleared with Protein G Sepharose Fast Flow beads (GE Healthcare). HA-tag (6E2) antibody (0.6 μg; Cell Signaling) was added to each lysate and incubated overnight. Protein G Sepharose Fast Flow beads were used to capture the antibody, and the beads were washed with IP buffer. Proteins were eluted from beads and subjected to immunoblotting. Figures show representative data from at least three individual experiments.
Immunohistochemistry
Tissue sections were embedded in paraffin and sectioned at ~4 microns for immunohistochemical staining. The slides were treated with citrate buffer for epitope retrieval, avidin, biotin, and Peroxo-Block blocking solutions (Zymed Laboratories, Life Technologies), and normal serum prior to addition of the primary antibody. Endogenous levels of total Bim protein were identified utilizing Bim (C34C5) Rabbit mAb (Cell Signaling) as the primary antibody, followed by biotin-conjugated goat anti-rabbit IgG (DAKO, Glostrup, Denmark) as the secondary antibody. Streptavidin-conjugated horseradish peroxidase (BioGenex, Freemont, CA, USA) was then added, and Bim activity was identified with 3,3-diaminobenzidine as the substrate (Biomeda, Foster City, CA, USA).
Acknowledgments
We thank Calli Versagli, Raju Rayavarapu, Joshua Mason, Siyuan Zhang, Veronica Schafer, and the entire Schafer lab for helpful comments, experimental assistance, and valuable discussion. We also thank Amy Leliaert for technical and organizational assistance; Junichi Kurebayashi (Kawasaki Medical School, Japan), Zachary Hartman (Duke), H Kim Lyerly (Duke), and Naoto Ueno (MD Anderson) for facilitating access to the KPL-4 cell line; Stephen Ethier (Wayne State) for the SUM149 cells; Mary Ann McDowell (Notre Dame) for help with immunofluorescence; Hisashi Hirada (VCU) for the Bim-EL point mutant constructs; and the Tissue Procurement Core at Indiana University Simon Cancer Center and Naoto Ueno and Savitri Krishnamurthy at the Morgan Welch Inflammatory Breast Cancer Clinic (MD Anderson) for the IBC tissue samples; and Sarah Chapman for assistance with the immunohistochemistry experiments. This work was supported by a grant from the Elsa U. Pardee Foundation, a Research Incubator Award from the Boler-Parseghian Center for Rare and Neglected Diseases at Notre Dame, and funds from the Coleman Foundation to ZTS. ZTS is the recipient of a Lee National Denim Day Research Scholar Grant from the American Cancer Society (RSG-14-145-01) and a Career Catalyst Research Grant from Susan G Komen (CCR14302768).
Glossary
- IBC
inflammatory breast cancer
- ECM
extracellular matrix
- Bim-EL
Bim-extra long
- LC8
dynein light chain 8
The authors declare no conflict of interest.
Footnotes
Edited by E Gottlieb
References
- Gonzalez-Angulo AM, Hennessy BT, Broglio K, Meric-Bernstam F, Cristofanilli M, Giordano SH, et al. Trends for inflammatory breast cancer: is survival improving. Oncologist. 2007;12:904–912. doi: 10.1634/theoncologist.12-8-904. [DOI] [PubMed] [Google Scholar]
- Woodward WA, Cristofanilli M. Inflammatory breast cancer. Semin Radiat Oncol. 2009;19:256–265. doi: 10.1016/j.semradonc.2009.05.008. [DOI] [PubMed] [Google Scholar]
- Harris EE, Schultz D, Bertsch H, Fox K, Glick J, Solin LJ. Ten-year outcome after combined modality therapy for inflammatory breast cancer. Int J Radiat Oncol Biol Phys. 2003;55:1200–1208. doi: 10.1016/s0360-3016(02)04201-3. [DOI] [PubMed] [Google Scholar]
- Lerebours F, Bieche I, Lidereau R. Update on inflammatory breast cancer. Breast Cancer Res. 2005;7:52–58. doi: 10.1186/bcr997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi H, Woodward WA, Valero V, Alvarez RH, Lucci A, Buchholz TA, et al. Inflammatory breast cancer: what we know and what we need to learn. Oncologist. 2012;17:891–899. doi: 10.1634/theoncologist.2012-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane K. Elucidating an uncommon disease: inflammatory breast cancer. J Natl Cancer Inst. 2011;103:1358–1360. doi: 10.1093/jnci/djr364. [DOI] [PubMed] [Google Scholar]
- Robertson FM, Bondy M, Yang W, Yamauchi H, Wiggins S, Kamrudin S, et al. Inflammatory breast cancer: the disease, the biology, the treatment. CA Cancer J Clin. 2010;60:351–375. doi: 10.3322/caac.20082. [DOI] [PubMed] [Google Scholar]
- Kleer CG, van Golen KL, Merajver SD. Molecular biology of breast cancer metastasis. Inflammatory breast cancer: clinical syndrome and molecular determinants. Breast Cancer Res. 2000;2:423–429. doi: 10.1186/bcr89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismael G, Hegg R, Muehlbauer S, Heinzmann D, Lum B, Kim SB, et al. Subcutaneous versus intravenous administration of (neo)adjuvant trastuzumab in patients with HER2-positive, clinical stage I-III breast cancer (HannaH study): a phase 3, open-label, multicentre, randomised trial. Lancet Oncol. 2012;13:869–878. doi: 10.1016/S1470-2045(12)70329-7. [DOI] [PubMed] [Google Scholar]
- Ditsch N, Vodermaier A, Hinke A, Burghardt S, Lenhard M, Lohrs B, et al. Dose-dense intensified sequential versus conventionally-dosed anthracycline and taxane-containing neoadjuvant therapy in patients with inflammatory breast cancer. Anticancer Res. 2012;32:3539–3545. [PubMed] [Google Scholar]
- Lehman HL, Van Laere SJ, van Golen CM, Vermeulen PB, Dirix LY, van Golen KL. Regulation of inflammatory breast cancer cell invasion through Akt1/PKBalpha phosphorylation of RhoC GTPase. Mol Cancer Res. 2012;10:1306–1318. doi: 10.1158/1541-7786.MCR-12-0173. [DOI] [PubMed] [Google Scholar]
- Kleer CG, Zhang Y, Pan Q, Gallagher G, Wu M, Wu ZF, et al. WISP3 and RhoC guanosine triphosphatase cooperate in the development of inflammatory breast cancer. Breast Cancer Res. 2004;6:R110–R115. doi: 10.1186/bcr755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Golen KL, Bao L, DiVito MM, Wu Z, Prendergast GC, Merajver SD. Reversion of RhoC GTPase-induced inflammatory breast cancer phenotype by treatment with a farnesyl transferase inhibitor. Mol Cancer Ther. 2002;1:575–583. [PubMed] [Google Scholar]
- van Golen KL, Davies S, Wu ZF, Wang Y, Bucana CD, Root H, et al. A novel putative low-affinity insulin-like growth factor-binding protein, LIBC (lost in inflammatory breast cancer), and RhoC GTPase correlate with the inflammatory breast cancer phenotype. Clin Cancer Res. 1999;5:2511–2519. [PubMed] [Google Scholar]
- van Golen KL, Wu ZF, Qiao XT, Bao LW, Merajver SD. RhoC GTPase, a novel transforming oncogene for human mammary epithelial cells that partially recapitulates the inflammatory breast cancer phenotype. Cancer Res. 2000;60:5832–5838. [PubMed] [Google Scholar]
- Wang X, Saso H, Iwamoto T, Xia W, Gong Y, Pusztai L, et al. TIG1 promotes the development and progression of inflammatory breast cancer through activation of Axl kinase. Cancer Res. 2013;73:6516–6525. doi: 10.1158/0008-5472.CAN-13-0967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch SM, Screaton RA. Anoikis mechanisms. Curr Opin Cell Biol. 2001;13:555–562. doi: 10.1016/s0955-0674(00)00251-9. [DOI] [PubMed] [Google Scholar]
- Buchheit CL, Rayavarapu RR, Schafer ZT. The regulation of cancer cell death and metabolism by extracellular matrix attachment. Semin Cell Dev Biol. 2012;23:402–411. doi: 10.1016/j.semcdb.2012.04.007. [DOI] [PubMed] [Google Scholar]
- Simpson CD, Anyiwe K, Schimmer AD. Anoikis resistance and tumor metastasis. Cancer Lett. 2008;272:177–185. doi: 10.1016/j.canlet.2008.05.029. [DOI] [PubMed] [Google Scholar]
- Buchheit CL, Weigel KJ, Schafer ZT. Cancer cell survival during detachment from the ECM: multiple barriers to tumour progression. Nat Rev Cancer. 2014;14:632–641. doi: 10.1038/nrc3789. [DOI] [PubMed] [Google Scholar]
- Masuda H, Zhang D, Bartholomeusz C, Doihara H, Hortobagyi GN, Ueno NT. Role of epidermal growth factor receptor in breast cancer. Breast Cancer Res Treat. 2012;136:331–345. doi: 10.1007/s10549-012-2289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenssen KK, Caldwell SA, Shahriari KS, Jackson SR, Whelan KA, Klein-Szanto AJ, et al. ErbB2 requires integrin alpha5 for anoikis resistance via Src regulation of receptor activity in human mammary epithelial cells. J Cell Sci. 2010;123 (Pt 8:1373–1382. doi: 10.1242/jcs.050906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reginato MJ, Mills KR, Becker EB, Lynch DK, Bonni A, Muthuswamy SK, et al. Bim regulation of lumen formation in cultured mammary epithelial acini is targeted by oncogenes. Mol Cell Biol. 2005;25:4591–4601. doi: 10.1128/MCB.25.11.4591-4601.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY, et al. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461:109–113. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan KA, Schwab LP, Karakashev SV, Franchetti L, Johannes GJ, Seagroves TN, et al. The oncogene HER2/neu (ERBB2) requires the hypoxia-inducible factor HIF-1 for mammary tumor growth and anoikis resistance. J Biol Chem. 2013;288:15865–15877. doi: 10.1074/jbc.M112.426999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Ota T, Liu P, Su C, Chien J, Shridhar V. Downregulation of HtrA1 promotes resistance to anoikis and peritoneal dissemination of ovarian cancer cells. Cancer Res. 2010;70:3109–3118. doi: 10.1158/0008-5472.CAN-09-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reginato MJ, Mills KR, Paulus JK, Lynch DK, Sgroi DC, Debnath J, et al. Integrins and EGFR coordinately regulate the pro-apoptotic protein Bim to prevent anoikis. Nat Cell Biol. 2003;5:733–740. doi: 10.1038/ncb1026. [DOI] [PubMed] [Google Scholar]
- Grassian AR, Schafer ZT, Brugge JS. ErbB2 stabilizes epidermal growth factor receptor (EGFR) expression via Erk and Sprouty2 in extracellular matrix-detached cells. J Biol Chem. 2011;286:79–90. doi: 10.1074/jbc.M110.169821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton M, Dowsett M, Ashley S, Hills M, Lowe F, Smith IE. High incidence of HER-2 positivity in inflammatory breast cancer. Breast. 2004;13:97–103. doi: 10.1016/j.breast.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Hartman ZC, Yang XY, Glass O, Lei G, Osada T, Dave SS, et al. HER2 overexpression elicits a proinflammatory IL-6 autocrine signaling loop that is critical for tumorigenesis. Cancer Res. 2011;71:4380–4391. doi: 10.1158/0008-5472.CAN-11-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, LaFortune TA, Krishnamurthy S, Esteva FJ, Cristofanilli M, Liu P, et al. Epidermal growth factor receptor tyrosine kinase inhibitor reverses mesenchymal to epithelial phenotype and inhibits metastasis in inflammatory breast cancer. Clin Cancer Res. 2009;15:6639–6648. doi: 10.1158/1078-0432.CCR-09-0951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel KJ, Jakimenko A, Conti BA, Chapman SE, Kaliney WJ, Leevy WM, et al. CAF-secreted IGFBPs regulate breast cancer cell anoikis. Mol Cancer Res. 2014;12:855–866. doi: 10.1158/1541-7786.MCR-14-0090. [DOI] [PubMed] [Google Scholar]
- Faber AC, Corcoran RB, Ebi H, Sequist LV, Waltman BA, Chung E, et al. BIM expression in treatment-naive cancers predicts responsiveness to kinase inhibitors. Cancer Discov. 2011;1:352–365. doi: 10.1158/2159-8290.CD-11-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Quearry B, Ruiz-Vela A, Korsmeyer SJ. Survival factor-induced extracellular signal-regulated kinase phosphorylates BIM, inhibiting its association with BAX and proapoptotic activity. Proc Natl Acad Sci USA. 2004;101:15313–15317. doi: 10.1073/pnas.0406837101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R, Ewings KE, Hadfield K, Cook SJ. Regulatory phosphorylation of Bim: sorting out the ERK from the JNK. Cell Death Differ. 2005;12:1008–1014. doi: 10.1038/sj.cdd.4401688. [DOI] [PubMed] [Google Scholar]
- O'Reilly LA, Kruse EA, Puthalakath H, Kelly PN, Kaufmann T, Huang DC, et al. MEK/ERK-mediated phosphorylation of Bim is required to ensure survival of T and B lymphocytes during mitogenic stimulation. J Immunol. 2009;183:261–269. doi: 10.4049/jimmunol.0803853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthalakath H, Huang DC, O'Reilly LA, King SM, Strasser A. The proapoptotic activity of the Bcl-2 family member Bim is regulated by interaction with the dynein motor complex. Mol Cell. 1999;3:287–296. doi: 10.1016/s1097-2765(00)80456-6. [DOI] [PubMed] [Google Scholar]
- Luo S, Garcia-Arencibia M, Zhao R, Puri C, Toh PP, Sadiq O, et al. Bim inhibits autophagy by recruiting Beclin 1 to microtubules. Mol Cell. 2012;47:359–370. doi: 10.1016/j.molcel.2012.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aird KM, Allensworth JL, Batinic-Haberle I, Lyerly HK, Dewhirst MW, Devi GR. ErbB1/2 tyrosine kinase inhibitor mediates oxidative stress-induced apoptosis in inflammatory breast cancer cells. Breast Cancer Res Treat. 2012;132:109–119. doi: 10.1007/s10549-011-1568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aird KM, Ding X, Baras A, Wei J, Morse MA, Clay T, et al. Trastuzumab signaling in ErbB2-overexpressing inflammatory breast cancer correlates with X-linked inhibitor of apoptosis protein expression. Mol Cancer Ther. 2008;7:38–47. doi: 10.1158/1535-7163.MCT-07-0370. [DOI] [PubMed] [Google Scholar]
- Linger RM, Cohen RA, Cummings CT, Sather S, Migdall-Wilson J, Middleton DH, et al. Mer or Axl receptor tyrosine kinase inhibition promotes apoptosis, blocks growth and enhances chemosensitivity of human non-small cell lung cancer. Oncogene. 2013;32:3420–3431. doi: 10.1038/onc.2012.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Chonghaile T, Letai A. Mimicking the BH3 domain to kill cancer cells. Oncogene. 2008;27 (Suppl 1:S149–S157. doi: 10.1038/onc.2009.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, Ye AS, Gardino AK, Heijink AM, Sorger PK, MacBeath G, et al. Sequential application of anticancer drugs enhances cell death by rewiring apoptotic signaling networks. Cell. 2012;149:780–794. doi: 10.1016/j.cell.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano F, Jacquel A, Colosetti P, Herrant M, Cagnol S, Pages G, et al. Phosphorylation of Bim-EL by Erk1/2 on serine 69 promotes its degradation via the proteasome pathway and regulates its proapoptotic function. Oncogene. 2003;22:6785–6793. doi: 10.1038/sj.onc.1206792. [DOI] [PubMed] [Google Scholar]
- Fukazawa H, Noguchi K, Masumi A, Murakami Y, Uehara Y. BimEL is an important determinant for induction of anoikis sensitivity by mitogen-activated protein/extracellular signal-regulated kinase kinase inhibitors. Mol Cancer Ther. 2004;3:1281–1288. [PubMed] [Google Scholar]
- Avivar-Valderas A, Salas E, Bobrovnikova-Marjon E, Diehl JA, Nagi C, Debnath J, et al. PERK integrates autophagy and oxidative stress responses to promote survival during extracellular matrix detachment. Mol Cell Biol. 2011;31:3616–3629. doi: 10.1128/MCB.05164-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung C, Lock R, Gao S, Salas E, Debnath J. Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol Biol Cell. 2008;19:797–806. doi: 10.1091/mbc.E07-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison CA, Durbin SM, Thau MR, Zellmer VR, Chapman SE, Diener J, et al. Antioxidant enzymes mediate survival of breast cancer cells deprived of extracellular matrix. Cancer Res. 2013;73:3704–3715. doi: 10.1158/0008-5472.CAN-12-2482. [DOI] [PubMed] [Google Scholar]